Summary
The ability to three-dimensionally interweave biological and functional materials could enable the creation of bionic devices possessing unique and compelling geometries, properties, and functionalities. Indeed, interfacing high performance active devices with biology could impact a variety of fields, including regenerative bioelectronic medicines, smart prosthetics, medical robotics, and human-machine interfaces. Biology, from the molecular scale of DNA and proteins, to the macroscopic scale of tissues and organs, is three-dimensional, often soft and stretchable, and temperature sensitive. This renders most biological platforms incompatible with the fabrication and materials processing methods that have been developed and optimized for functional electronics, which are typically planar, rigid and brittle. A number of strategies have been developed to overcome these dichotomies. One particularly novel approach is the use of extrusion-based multi-material 3D printing, which is an additive manufacturing technology that offers a freeform fabrication strategy. This approach addresses the dichotomies presented above by (1) using 3D printing and imaging for customized, hierarchical, and interwoven device architectures; (2) employing nanotechnology as an enabling route for introducing high performance materials, with the potential for exhibiting properties not found in the bulk; and (3) 3D printing a range of soft and nanoscale materials to enable the integration of a diverse palette of high quality functional nanomaterials with biology. Further, 3D printing is a multi-scale platform, allowing for the incorporation of functional nanoscale inks, the printing of microscale features, and ultimately the creation of macroscale devices. This blending of 3D printing, novel nanomaterial properties, and ‘living’ platforms may enable next-generation bionic systems. In this review, we highlight this synergistic integration of the unique properties of nanomaterials with the versatility of extrusion-based 3D printing technologies to interweave nanomaterials and fabricate novel bionic devices.
Keywords: 3D printing, 4D printing, bionic devices, nanomaterials, nanodevices, bioelectronics, cyborgs, bio-nano hybrids, electronic skins, active functional devices
Introduction
The synergistic integration of biological systems with electronic materials and devices could enable the creation of novel bionic devices. Due to the increasing miniaturization and proliferation of portable electronic devices, the field of bionics has transitioned from science fiction to an area of increasing scientific interest, with particular relevance to the fields of regenerative medicine, smart prosthetics, medical robotics and human-machine interfaces [1–4]. Most research in the field of bionics to date has focused on developing robots which behave increasingly more like humans. Similarly, an equally compelling challenge is integrating electronic and robotic components in a seamless manner onto the human body. For example, bioelectronic medicines and devices could potentially be utilized to restore or even augment the complex functionalities of naturally evolved biological systems. At the fundamental level, there are inherent materials compatibility challenges associated with integrating functional electronic materials with biology.
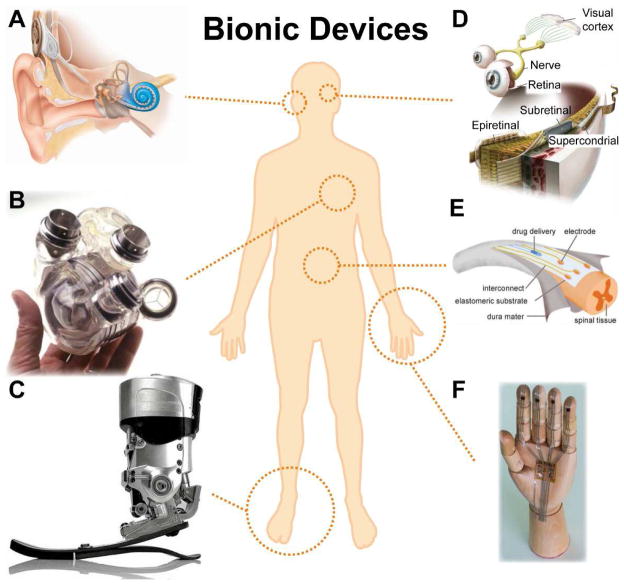
The term “bionics” is defined by Dictionary.com as, “utilizing electronic devices and mechanical parts to assist humans in performing difficult, dangerous, or intricate tasks, by supplementing or duplicating parts of the body [5].” Broadly speaking, “bionics” encompasses the functionalities of classes of systems that are formed by merging biological systems, which could be single cellular or multi-cellular systems [2,6–8], with engineered functional electronic and/or mechanical systems [2]. Our ability to develop tools which overcome the limitations of human biology has played a key role in survival and evolution [9]. Utilizing devices for regenerative medicine and as prosthetics can be traced back millennia [1]. Indeed, a very primitive bionic device from the first century AD involved the use of wrought iron for dental replacements [10]. Subsequently, bionic devices such as iron prosthetic hands (1504), contact lenses (1888), and artificial hip replacements (1905) have been used to restore or augment human function [1]. Over the past several decades, the development of active microelectronic devices has enabled the incorporation of sensing modalities [11,12], optoelectronics [13,14], actuation [15] and computational devices [16] into previously passive mechanical constructs. This has enabled an extension of the role of bionic devices toward mimicking or even augmenting the complex functionalities of biological organs. These powerful developments have been leveraged to fabricate active bionic devices such as the cochlear implant [17,18] to restore hearing (Figure 1A), pacemakers and heart replacements [1] to sustain blood flow (Figure 1B), locally powered prosthetic devices [19] to provide mobility to amputees (Figure 1C), retinal implants to provide partial restoration of vision loss due to diseases such as retinitis pigmentosa [20,21] (Figure 1D), dura mater for the spinal cord [22] (Figure 1E), and digital skin sensors and electronic skins [12,23–25] (Figure 1F). Indeed, the ability to merge a diverse palette of materials classes could enable the generation of functional devices that mimic the complex functionalities of grown biological organs [15].
Figure 1.
Bionic technologies for restorative medicine. (A) Cochlear implant [18]. (B) AbioCor self-contained replacement heart [1]. (C) Powered ankle-foot prosthetic controlled by a neuromuscular model [19]. (D) Epiretinal, subretinal, and suprachoroidal implants [21]. (E) Electronic dura mater, “e-dura,” tailored for the spinal cord [22]. (F) A skin-inspired digital mechanoreceptor, where the image shows a model hand with DiTact sensors on the fingertips connected with stretchable interconnects [12]. Reprinted with permission from Refs. [18], [1], [19], [21], [22], [12], respectively. Copyright 2009 Nature Publishing Group, 2002 American Association for the Advancement of Science, 2010 IEEE, 2013 American Association for the Advancement of Science, 2015 American Association for the Advancement of Science.
An optimized bionic device should be seamlessly merged with the human body in order to restore or augment human capabilities without causing side effects such as discomfort, infection [26] or rejection due to foreign body responses by the host [27–29]. While the continual discovery of new materials and novel properties will eventually lead to more optimized devices, ideality has been punctuated by challenges in integrating high performance materials and devices with biology. Three key challenges can be identified. First, the mechanical properties of high quality electronic materials are typically disparate from biology. For example, the typical Young’s modulus of inorganic electronics is on the order of 1–100 GPa (Si ~ 170 GPa) [30]. By contrast, the Young’s modulus of skin is on the order of 0.1–1 MPa [31]. Similarly, inorganic electronic materials typically fracture at strains (ca. 1%) [32] of up to 30× lower than human skin [33]. These significant differences in mechanical properties not only lead to obstacles in the integration of bionic devices onto the body, but can cause discomfort, agitation, rejection and injuries [34].
Second, the processing conditions inherent to high performance electronics are often incompatible with biology. Microelectronics are typically fabricated via “top down” approaches which can involve harsh chemical and temperature processing conditions. In contrast, organs and tissues have been grown from the “bottom up” under finely tuned physiological conditions [35]. Third and finally, electronic wafers are two-dimensional planar structures, whereas biology possesses intricately complex three-dimensional geometries from the molecular scale to the macroscale. These incompatibilities collectively present significant barriers in grafting independently fabricated bionic devices onto biology in a seamless manner.
A variety of novel strategies have been developed to address these issues, such as integration via intelligent device design [11,32,36,37], transfer printing processes [13,38–41] and/or assembly of prefabricated devices [42] onto three-dimensional constructs to accommodate the geometrical and material incompatibilities. This review highlights a relatively new concept in achieving a synergistic integration of bionic devices with biology: by using 3D printing. Extrusion-based 3D printing technologies may overcome the three specific challenges mentioned above. First, the use of nanoscale materials as inks in the 3D printing process and the co-printing of soft materials and functional nanoscale inks allows for a route which minimizes mechanical discrepancies. Second, while the materials may be synthesized and/or processed under harsh, high temperature conditions in order to create high quality functional nanomaterials, the printing process is typically performed under ambient conditions via a bottom-up assembly process. Finally, the 3D printing process naturally allows for the hierarchical assembly of functional materials in three dimensions, commensurate with biology. An additional benefit of 3D printing is the ability to achieve a multi-scale manufacturing approach built into the process.
This review article will first describe this multi-scale fabrication approach and highlight the unique properties of nanoscale conductive, semiconducting and plasmonic materials. Next, we will describe the challenges and strategies associated with the microscale printing and assembly of these nanoscale functional materials. We will then review progress to date in the use of 3D printing to create unique bionic architectures at various length scales.
3D Printing for Multiscale Manufacturing
3D printing is an additive manufacturing process used to build three-dimensional structures from computer-aided design (CAD) models in a layer-by-layer fashion. Developed since the 1980s [43], 3D printing has the capability to create unique architectures that cannot be made with conventional molding or subtractive manufacturing techniques. Further, the co-development of 3D imaging technologies, such as 3D scanning, allows for the acquisition of three-dimensional topological data that precisely reproduces a three-dimensional object and the incorporation of three-dimensional templates for the conformal printing of devices on non-planar substrates [44–46].
3D printing is commonly associated with either light-based or ink-based printing techniques [47]. Light-based techniques are founded on processes such as UV curing [43,48,49] and two-photon polymerization [50]. Ink-based printing can be achieved via extrusion printing, inkjet printing [51], and electro-hydrodynamic printing [52]. Extrusion-based 3D printing is a particularly interesting subset of additive manufacturing in which the materials are extruded through a nozzle [47,53–57]. Such a platform is highly versatile, affordable [58] and can be readily expanded to incorporate multiple materials [59–61]. Moreover, in contrast to other printing methods, such as inkjet printing (where the typical viscosity is limited to ca. 2–102 mPa·s [51]), extrusion-based 3D printing is capable of incorporating a wide range of materials with viscosities up to 106 mPa·s and with disparate properties [47,61]. This versatility has enabled the accommodation of different classes of materials encompassing a wide range of length-scales: including nanomaterials [61,62], fibers [63], cells [64,65], tissues [66], organs [55,67], ceramics [68,69], metals [70] and polymers such as elastomers [59,60,71], gels [72,73], and biomaterials [55,74].
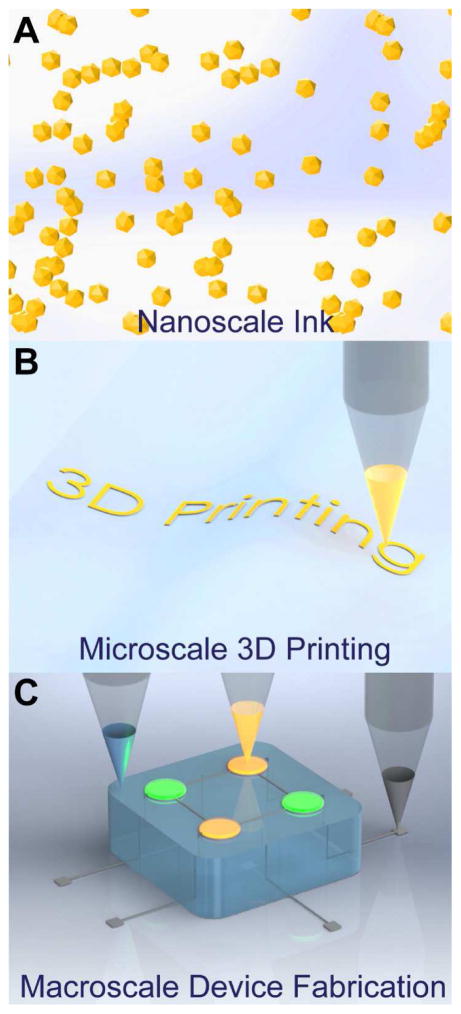
Nanomaterials represent novel building blocks in the toolset of 3D printed functionalities. These are materials which are confined such that at least one of their length scales is in the range of ca. 1–100 nm. As conceived by Richard Feynman in 1959 [75], assembling materials from the bottom-up has become an important assembly strategy for nanoscale materials [76], enabled by the ability to make such materials using scalable synthetic methods [77–81]. Functional nanomaterials can be dispersed into solvents to form solution-processable inks, as shown in Figure 2A, which can be extruded from nozzles to create microscale features (Figure 2B) commensurate with typical biological length scales. Finally, the co-printing of nanomaterials with soft, structural, and/or biological constructs enables the freeform fabrication of three-dimensional, macroscale, multi-material functional devices as illustrated in Figure 2C. Extrusion-based 3D printing thus provides a promising platform for the interweaving of different materials and functionalities.
Figure 2.
Multiscale, multi-material 3D printing. (A) Functional nanomaterials can be dispersed in solvents to form solution-processable inks. (B) The inks are then 3D printed at the microscale via extrusion from a suitable nozzle. (C) The three-dimensional co-printing of various classes of materials enables the creation of macroscale functional devices.
Functional Nanoscale Inks
Nanoscale inks are attractive for a number of reasons. Nanomaterials approach length scales such that external forces are negligible in comparison with van der Waals interactions [82]. Hence, nanomaterials can be assembled [83] or coated on arbitrarily-shaped three dimensional substrates with high adhesion. For example, a monolayer of graphene has been shown to exhibit an adhesion energy of 0.45 J·m−2 on a silicon oxide substrate [84]. Approaching the nanoscale, the surface-to-volume ratio increases as the particle size decreases [85,86]. This geometric effect has important consequences. For example, the decrease in particle size reduces the material melting temperature [87,88]. An example of this relationship between size and the melting point [89] is low melting point silver nanoparticles [90] that can be sintered to form a conductive path at lower temperatures than the bulk material. Nanomaterial properties are also highly tunable. In 1847, Michael Faraday discovered that the optical properties of gold colloids deviated from their bulk counterparts [91], introducing the prospect of tuning functional properties by tailoring the size of materials. The band gap of semiconducting nanomaterials is size-dependent below the Bohr exciton radius [85,86,92]. For example, the fluorescence of CdSe-CdS core-shell nanoparticles shifts from red to blue when the particle size decreases from 6 nm to 1.7 nm [93]. These effects offer a means of controlling the electronic and optical properties of nanomaterials by tuning their size during synthesis.
The ability to synthesize monodisperse nanowires and nanoparticles permits the development of printable inks that capture the unique properties of nanomaterials in a printable format, by suspending the nanoparticles in aqueous or organic solvents [77–81]. Stabilization in the solvent is typically achieved via the addition of polymeric materials [94] and surfactants, or via electrostatic interactions [95] to prevent aggregation and precipitation. The extrusion-based 3D printing method supports a wide range of fluid properties, and the printability can be tailored by modifying the surface tension and viscosity of the inks. Three common classes of printable inks include conducting, semiconducting and plasmonic nanomaterials. Printable conducting nanomaterials [96] can be synthesized with metallic [97] and carbon-based nanoparticles. Further, the ability to print thin conductive layers also enables the printing of transparent conductors [98,99] that can function as electrodes for optoelectronic devices [100,101]. Highly conductive metals such as silver [90,102] and gold [103,104] are suitable printable inks which, unlike other metals such as aluminum [105], are less susceptible to oxidation. Alternatively, carbon nanomaterial-based conducting inks [98] such as graphene and carbon nanotubes have been actively explored. Graphene [106–108], an atomically thick layer of carbon atoms, is attractive due its exceptionally high intrinsic mobility [109,110]. A mobility as high as 5,000 cm2 ·V−1 ·s−1 can be achieved in printed graphene films [111]. Hybrid composites can also be formed by mixing both metal- and carbon-based inks. For instance, a highly conductive composite (5.7 × 105 Ω −1·m−1) that can be strained up to 140% has been demonstrated with a silver nanotube composite [112].
Printable semiconducting nanoscale inks provide a means of introducing active electronic functionality and tuning electrical and optical properties. Quantum dots (QDs) [92] are zero-dimensional nanoscale crystals of semiconducting materials, in which quantum confinement often causes a deviation of properties from the bulk. For example, the emission wavelength of cadmium selenide QDs is tunable by changing the particle size [113]. QDs typically consist of an inorganic semiconductor core and a coating of ligands to confer solubility in solvents. A wider-bandgap inorganic material can also be coated as a shell to passivate the surface, thereby improving intrinsic properties such as photoluminescence quantum yield and photo-stability [78]. Further, the ability to synthesize highly monodisperse QDs with narrow size distributions leads to narrow emission spectra [114] that is useful for display devices with high color purity and saturation [115].
Plasmonic nanomaterials are metal nanoparticles that exhibit a localized surface plasmon resonance (LSPR), which is a collective oscillation of conduction band electrons in response to the electric field component of incident light. The unique optical properties such as large absorption and scattering cross-section, high sensitivity to the local dielectric environment, and enhanced electromagnetic field at the surface render plasmonic nanomaterials a highly attractive class of materials for a broad range of applications [116]. The LSPR wavelength is dependent on the composition, size, shape, coupling, and ambient dielectric of the nanoparticles. In biomedicine, plasmonic nanoparticles are used in applications for diagnostics and therapeutics. While tremendous progress has been made in the synthesis of size- and shape-controlled plasmonic nanostructures, their integration with other materials and application in solid-state devices is primarily either via direct fabrication (using various lithographic techniques) or immobilization on solid two-dimensional (2D) substrates such as silicon, glass, plastic, or paper.
Microscale 3D Printing of Inks
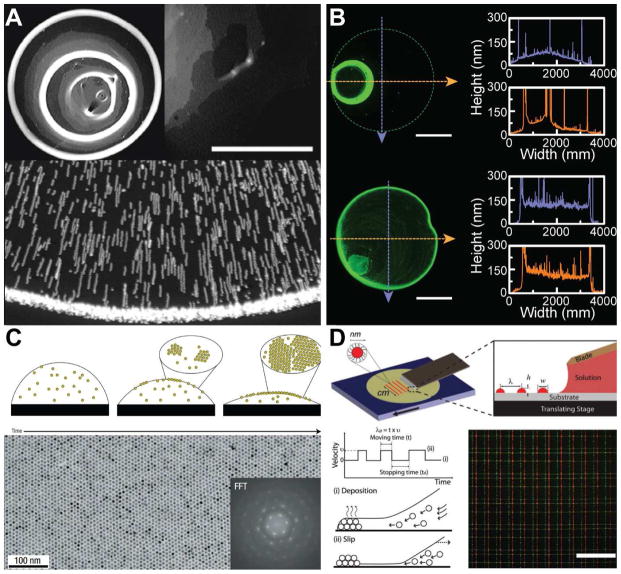
The ability to pattern solution-processed nanomaterial inks can lead to the creation of devices [117–119], where the properties of the film such as its thickness and morphology dictate the quality of the resulting device performance [120]. In microfabrication-based methods, this is typically accomplished via processes such as spin-coating [121], in which the liquid ink is spread uniformly via centrifugal forces. However, such processes require a rigid and flat substrate, as well as the use of photolithography for patterning. In addition, the spin coating of quantum dots expels 94–97% of the starting solution, which increases the material cost by up to 20-fold [122]. Yet, a colloidal ink printed onto a surface without spin-coating typically does not lead to a uniform film. When a droplet is left to evaporate on a substrate, the pinning of the contact line results in a non-uniform evaporation rate at the surface of the printed droplet. The enhanced drying rate at the edge of the droplet due to the curvature difference drives the assembly of suspended particles at the edge of the evaporating solvent, resulting in so-called “coffee rings” [123,124], as shown in Figure 3A. The formation of multiple rings can also be observed, due to the repeated pinning and unpinning of the contact lines as the printed ink evaporates [125].
Figure 3.
Microscale patterning of nanoscale inks on a surface. (A) Challenges inherent to assembling particles via convective self-assembly methods. Top left figure shows formation of so-called “coffee-rings,” typically observed when a colloidal suspension droplet dries on a surface. The photograph is of a deposit left by 100 nm microspheres with a volume fraction of 1%. Top right figure shows non-uniformity in the region of the ring, where the grey scale indicates the density of particles with the white color indicating the highest density. Scale bar is 500 μm [124]. Bottom figure show the superimposed exposures that illustrate the motion of the particles toward the edge of the droplet during the drying process [123]. (B) Non-uniformity can be reduced by introducing a co-solvent. Top figure shows the deposition of quantum dots from pure toluene, while bottom figure shows an improvement in the morphology via the introduction of 20% dichlorobenzene [61]. Scale bar is 1 mm. (C) Evaporation kinetics and particle interactions with the liquid-air interface can be tailored to achieve monolayer assembly of nanoparticles. Micrograph shows the monolayer produced by a solution of dodecanethiol-ligated 6 nm gold nanocrystals. Inset shows the fast Fourier transform (FFT) of the image [136]. (D) Arrays of quantum dots are generated via stick-slip motion of the contact line. The features are controlled by the velocity profile of the translation stage. Bottom right figure shows the fluorescent microscopy image of grid patterns of the quantum dots. Scale bar is 200 μm [142]. Reprinted with permission from Refs. [124], [123], [61], [136], [142], respectively. Copyright 2000 American Physical Society, 1997 Nature Publishing Group, 2014 American Chemical Society, 2006 Nature Publishing Group, 2010 John Wiley & Sons.
To overcome this non-homogeneous deposition, the microscale printing of nanomaterials can be achieved via directed or self-assembly based methods [83]. External forces, such as electrical forces [126], or magnetic forces [127] can be applied to drive the assembly of the particles. The assembly can also be achieved without an external field. For instance, a Marangoni effect [128–130] can be introduced to drive the particle accumulation away from the edge of the droplet. This can be achieved via the addition of a co-solvent, which introduces a surface tension gradient that generates the Marangoni flow. As shown in Figure 3B, the introduction of 20% of dicholorobenzene to a QD ink reduces the root-mean square roughness of the resulting QD film 9-fold in comparison to a single-solvent ink [61]. Similarly, a surface tension gradient can be generated via the introduction of a vapor environment. For instance, the film generated from an aqueous suspension has a higher uniformity in ethanol vapor due to the strong recirculating flow generated by the surface tension gradient [131]. Alternatively, the addition of a small ionic surfactant introduces a Marangoni eddy of particles, which improves uniformity of deposition [132]. The addition of hydrosoluble polymer additives can also mitigate pinning of the contact line via an increase in viscosity and the Marangoni effect, which suppresses the coffee ring effect [133]. Coffee rings can also be reduced by introducing temperature control [134], since temperature affects the edge evaporation rate.
Interestingly, modification of the particle shape can also affect the subsequent deposition. For instance, Yunker et al. have shown that anisotropic shaped particles, such as ellipsoids, introduce strong inter-particle interactions to form loosely packed structures that prevent the accumulation of particles at the edges of the droplet [135]. Impressively, monolayer assembly of nanoparticles has been achieved by tailoring the evaporation kinetics and particle interactions at the liquid-air interface [136], as described in Figure 3C. For example, assembly of a large nanoparticle monolayer film (3 mm × 4 mm) has been generated on a Si3N4 substrate without the application of external fields. The ligands of the nanoparticles also play a critical role in this assembly approach, and excess ligand has been found to play a key role in generating the thin film using this method [136]. The coffee-ring effect can be leveraged to create high resolution patterns and features which are otherwise challenging to create via direct-writing with a 3D printer. For instance, a transparent conductive film can be formed from silver nanoparticle rings generated with the coffee ring effect. A network of silver nanoparticle rings (10 μm width, 300 nm height, 150 μm diameter) can generate a conductive film with a resistivity of 4 × 10−7 Ω·m and a transparency of 95%. This method is not limited to silver nanoparticles, and has also been demonstrated with carbon-based particles such as carbon nanotubes [137].
Similarly, “stick-slip” motion can be used to create a uniform array of nanoparticles. Unlike the irregular ring pattern resulting from a droplet left evaporating on a surface, “bands” can be assembled by confining the evaporation rate of the inks. The exploitation of such effects have been demonstrated to yield highly regular arrays of micrometer size band structures in both planar [138,139] and confined constructs [140,141]. Indeed, the integration of such approaches could enable the microscale patterning of nanoscale inks within a 3D printing setting. For instance, as described in Figure 3D, doctor blades can provide a confinement of the evaporation rate, and the manipulation of the velocity profile of the translation stage can enable the creation of highly regular microscale quantum dot stripes on a planar surface, with control over the widths and thicknesses of the quantum dot stripes [142].
Mechanical Properties of 3D Printed Architectures
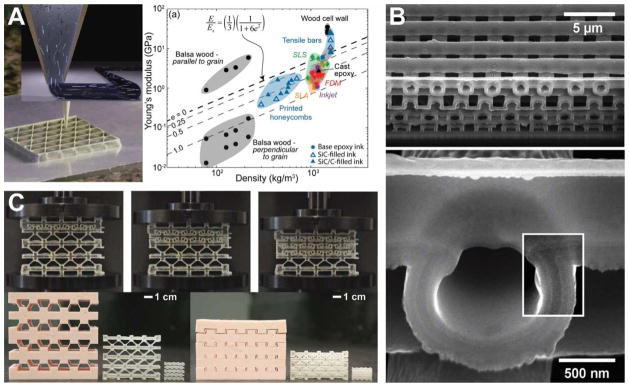
In this section, we highlight some of the unique mechanical properties enabled by extrusion-based 3D printing, which has the advantage of creating freeform architectures that can be tailored to create robust bionic devices. First, the extrusion-based 3D printing process can be combined with bioinspired inks to create lightweight composites that mimic natural materials. The co-printing of alternate layers of hard and soft materials can result in structures exhibiting outstanding strength and toughness [143] and enhanced fracture resistance [144]. Lightweight composite structures can also be achieved via shear-induced alignment of fibers embedded within epoxy [63], as illustrated in the left inset of Figure 4A. Macroscopic architectures, such as a honeycomb composite structure (left image of Figure 4A), can achieve specific and anisotropic properties to enable mechanical strength and toughness with significantly less weight. The printing of such fiber-reinforced epoxy inks could be used to create structures with exceptional toughness, impact resistance and Young’s moduli that are factors of 10–20× greater than common commercial 3D printable inks, as shown in the graph of Figure 4A [63].
Figure 4.
3D printing can create macroscale architectures exhibiting interesting mechanical properties. (A) 3D printing of lightweight cellulose composite. The inset illustrates the alignment of high aspect ratio fillers inside the nozzle (left figure). A plot of Young’s modulus vs. density of 3D printed balsa wood and 3D printed tensile bars with fillers show a factor of 10–20× higher longitudinal Young’s moduli compared to most commercially available 3D printed polymers [63]. (B) Hollow-wood pile structure, where the higher magnification SEM image (bottom image) shows a tri-layer Si/SiO2/Si tube wall [147]. (C) Multi-stable architected materials, where the top sequential images demonstrate that the structure retains a deformed shape after removal of a vertical load. Left and right bottom images show the structures before and after compression, respectively [149]. Reprinted with permission from Refs. [63], [147], [149], respectively. Copyright 2014 John Wiley & Sons, 2006 John Wiley & Sons, 2015 John Wiley & Sons.
Second, lightweight structures can be created by first printing polymer templates followed by subsequent coating of metals [145] or ceramics [146]. For example, 3D printing of polymer lattices via the removal of a sacrificial template leaves a hollow wood-pile structure coated with silicon (Figure 4B) [147]. As shown in the bottom inset of Figure 4B, a tri-layer Si/SiO2/Si tube with a wall thickness of 300 nm can be produced. However, high temperature chemical vapor deposition (CVD) processes are required, which may not be compatible with the co-printing of biological structures or low-temperature polymer substrates. Nevertheless, such designs suggest that 3D printing provides the ability to create features that are smaller than the printing resolution.
Third, the ability to create freeform architectures without the constraints of conventional planar fabrication processes also provides an opportunity for utilizing numerical analyses in the structural designs to fine-tune 3D architectures and properties. For instance, auxetic materials with enhanced shear and indentation resistances can be created via the combination of 3D printing with topology optimization. Clausen et al. have shown the 3D printing of architectures with Poisson ratios of −0.8 that were designed via numerical optimization [148]. In another example, Shan et al. demonstrated 3D printed multi-stable structures with enhanced absorption of compression energies, as shown in Figure 5C [149]. The printed beam elements are able to respond to external loading by reconfiguring, in a reversible fashion, into other stable structures without mechanical failure. Such novel geometries could potentially enable the creation of robust bionic devices with enhanced impact resistance and energy-absorbing capabilities to withstand wear and impact.
Figure 5.
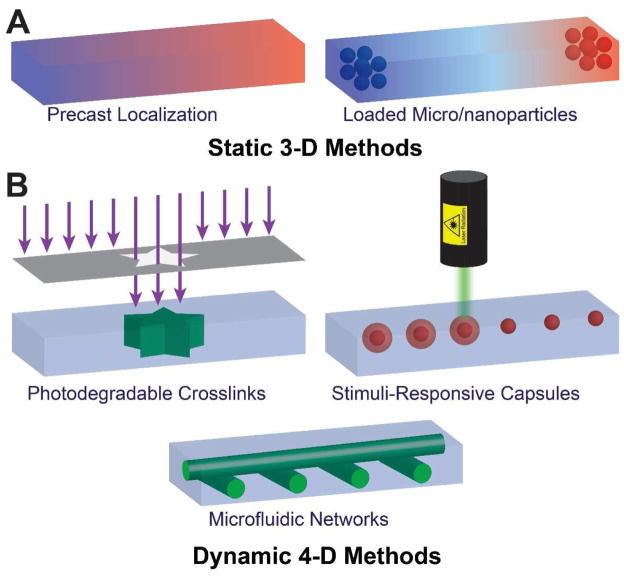
Schematic of 3D static and 4D dynamic printing methods to create chemical and biomolecular gradients. (A) Static methods allow for a preprogrammed gradient to be developed, typically based on passive diffusion from payload depots. (B) Dynamic methods allow for “on the fly” active reprogramming of gradients, by including the fourth dimension of time.
Finally, beyond serial printing using individual nozzles, a more seamless and parallel merging of multiple materials can be achieved by incorporating nozzle designs that enable the co-printing of different materials to achieve unique multi-material architectures. In addition to increasing throughput [150], multi-nozzle microfluidic printheads allow for the rapid co-printing of two different viscoelastic inks to create architectures with sharp transitions of different materials [60]. This multi-material integration has also been demonstrated via an active mixing of viscoelastic inks to achieve programmable control over local compositions [59].
3D Printing of Spatiotemporal Biomolecular Gradients
The ability to mimic the dynamic microenvironment surrounding cells in natural tissues is critical to engineering the biotic/abiotic interfaces found in bionic systems [151–153]. Indeed, cell fate is influenced by numerous molecular factors and interactions that require meticulous control for the regeneration of functional tissue [154,155]. In order to achieve such control, engineered matrices should ideally be capable of generating multiplexed spatiotemporal molecular gradients. Extensive research efforts to engineer such matrices have resulted in a number of promising methods to generate and control molecular gradients (Figure 5). These methods can generally be categorized as static 3D methods (Figure 6A) where the gradient is fixed once programmed, and dynamic 4D methods (Figure 6B) where the gradients can be reprogrammed by the user. The focus of this review is on strategies to generate biomolecular gradients that are compatible with 3D printed systems, and as such a complete description of tissue culture compatible methods to generate biomolecular gradients is beyond the scope of this review. The reader is referred to several literature sources that provide excellent in-depth discussions of such methods [151,155,156].
Figure 6.
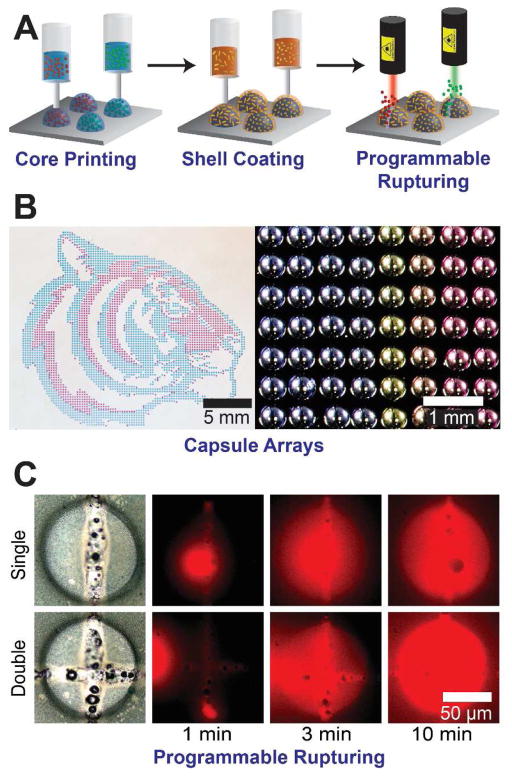
(A) Schematic showing a 3D printing strategy to creating stimuli-responsive capsules that can be selectively ruptured to release payloads in response to optical stimuli. Incorporation of gold nanorods in the shells allows the capsules to be ruptured by exposure to laser wavelengths determined by the lengths of the incorporated nanorods. (B) Optical images of complex capsule arrays including a printed ‘tiger’ and a pH gradient array with different colors from an indicator dye. (C) Programmed rupture and release of HRP from capsules by selective laser exposure [62] Reprinted with permission from Ref. [62]. Copyright 2015 American Chemical Society.
3D Static Printing of Gradients
The simplest method for creating a hydrogel scaffold with a static biomolecular gradient is through direct spatial localization of biological factors within the hydrogel. In this approach, once the spatial gradient has been generated, the temporal persistence of the gradient is based solely on the diffusivity of the biomolecule within the hydrogel, which is generally a function of the size and chemistry of the biomolecule, crosslinking density, water fraction of the hydrogel, and molecular interactions between the biomolecule and the hydrogel polymer. Scaffolds with such gradients can be made via solid freeform fabrication through sequential deposition of hydrogels containing various concentrations of differing biomolecules. This strategy is compatible with both extrusion and light-based printing methods [157]. These approaches are suitable for creating relatively simple gradients using a few biomolecules, and gradients of soluble factors typically persist for days. Applications where a gradient is desired for longer durations (weeks to months) require strategies to immobilize the biomolecule on the hydrogel polymer.
An alternative strategy to creating hydrogel gradients is to apply the gradient after the hydrogel scaffold has been formed. A common approach to doing this is inkjet printing of biomolecule solutions onto a hydrogel substrate [158–162]. As the solution droplet impacts the surface of the hydrogel, it is quickly absorbed and the biomolecules are localized to the 2D footprint of the droplet. Gradient arrays can be created by dispensing varying numbers of droplets over the same area. The precise control over droplet volumes in inkjet printing allows researchers to create gradients with precise concentrations and varying gradient profiles. Nuzzo et al. have developed a variant of this method [163]. By using soft contact printing of a microfluidic network with a permeable membrane, they were able to demonstrate transfer printing of complex gradients into the hydrogel substrate. Although these methods are most commonly utilized with flat hydrogel substrates, they can easily be integrated with methods for freeform fabrication of hydrogel scaffolds by inkjet or transfer printing of gradients at select hydrogel layers during the freeform fabrication process.
The simplicity of directly incorporating gradients in the hydrogel has made this approach widely used, particularly in fundamental studies where the effect of a particular gradient on cell behavior is being studied. However, a major consideration is the lack of control over the temporal evolution of the gradient and the cumbersome nature of incorporating multiplexed gradients. Micro/nanoparticles loaded with biomolecules represent a versatile approach to delivering multiplexed gradients with additional control over the release kinetics [164,165]. While such particles can be made via numerous methods and from a wide range of materials, they are most commonly formulated from biodegradable polymers using double emulsification or coacervation methods [166]. The particles can be efficiently loaded with a variety of biomolecular payloads, while maintaining their activities [167].
Synthesizing particles loaded with different factors and localizing them within a hydrogel matrix can lead to the generation of multiplexed spatial gradients. The payload release kinetics can be adjusted by controlling the particle properties (e.g., diameter, shell thickness, porosity, etc.). However, spatiotemporal control over the gradients is typically coarse, as most scalable methods to synthesize particles result in highly polydisperse populations. Fine control over release dose is often required, and recent research efforts have focused on achieving robust control over release kinetics. To this point, numerous groups have developed microfluidic methods to synthesize monodisperse particles with a high degree of control over particle properties [34,168,169]. While it is possible for such monodisperse particles to first be synthesized and collected, and then to be formulated into an ink for 3D printing, a more natural solution is for direct incorporation of the microfluidic devices in the printing nozzle. Currently, research efforts to develop microfluidic print nozzles have demonstrated single nozzle printing of multiple materials with varying mechanical properties, with the added ability to create discrete or graded interfaces between the materials [60].
We have developed an alternative strategy to 3D print highly monodisperse capsule arrays (Figure 7A) [62]. First, aqueous cores are printed onto a hydrophobic substrate. Next, the core is encapsulated by dispensing a biocompatible polymeric solution which rapidly evaporates, leaving a solid polymer shell. The versatility of this approach is the ability to accurately dispense multiplexed arrays over large areas with precise control over the core composition and the shell thickness. Figure 6B shows a ‘tiger’ consisting of 4,000 red and blue cores, with a spacing of 400 μm. Adjacent is an optical image of a pH gradient array generated by printing varying volumes of an acidic and basic ink. The color of each drop is a result of the pH indicator m-cresol purple. The shell thickness of the capsules was varied by adjusting the concentration of the polymer in the dispensed shell solution. In this manner, we were able to realize control over the passive release kinetics of horseradish peroxidase (HRP) from the capsule core.
Figure 7.
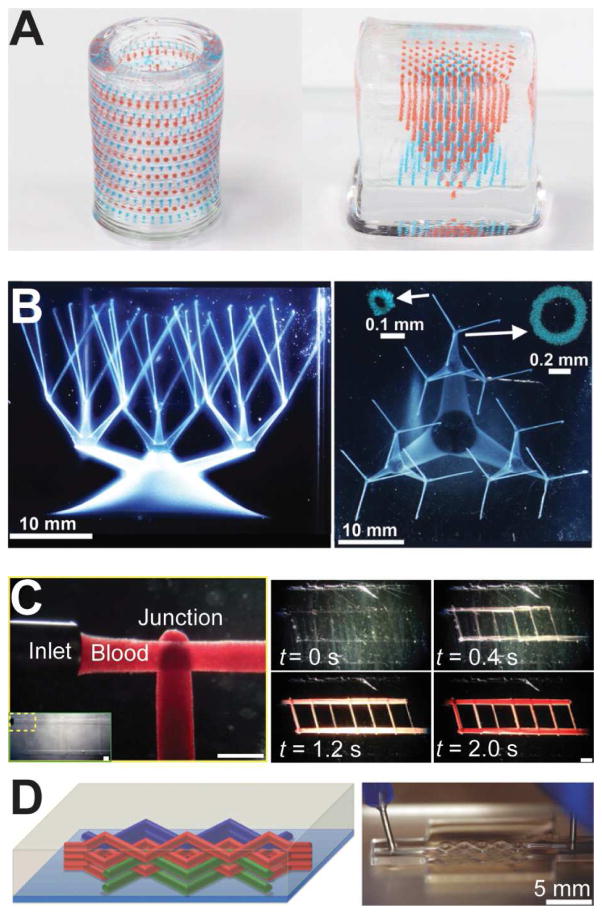
3D printing strategies to create gradients in macroscale structures. (A) An emulsion printing strategy to create stimuli-responsive multiplexed arrays of capsules within 3D hydrogel matrices (cylinder outer diameter is 8 mm; cube edge length is 10 mm) [62]. (B) Direct printing of vascular networks in granular media. Jamming of the media allows the printed network to be stabilized as it is printed [177]. (C) A carbohydrate glass is printed as a sacrificial scaffold for the vascular network. Once the gel matrix is cast, the scaffold is dissolved leaving behind open channels (scale bars are 1 mm, left; 2 mm, right) [179]. (D) In this example, the authors use a fugitive ink to create microfluidic channels in a hydrogel. After the channels are formed, the ink is removed by decreasing the temperature to fluidize the fugitive ink [178]. Reprinted with permission from Refs. [62], [177], [179], [178], respectively. Copyright 2015 American Chemical Society, 2015 American Association for the Advancement of Science, 2012 Nature Publishing Group, 2014 John Wiley & Sons.
The passive 3D methods described here to generate biomolecular gradients could represent powerful tools for studying the impact of a surrounding microenvironment on a wide range of cellular responses. The major advantage of these methods is their broad applicability to a wide range of biomolecules and hydrogel systems. For example, chemical modification of the biomolecules and hydrogel polymers is generally unnecessary. The limitation, however, is that the temporal evolution of the gradient is ultimately controlled simply by diffusion of the biomolecules through the hydrogel matrix or capsule shell. As such, there are limited opportunities to tailor the temporal response, especially in the case of multiplexed gradients. A more compelling alternative is to introduce vehicles for achieving precision control over time as well as space.
4D Dynamic Printing of Gradients
The incorporation of a selective, stimuli-responsive element in a biomolecular gradient can provide an additional level of control over its temporal evolution. The concept of stimuli-responsive controlled release has been extensively explored for applications in drug delivery. Capsules, nanoparticles, and hydrogels have been developed that release a chemical payload in response to a wide range of user-applied external stimuli such as light, heat and magnetic field, or internal biological stimuli such as pH, temperature and biomolecular signaling [170–173]. The main advantage of stimuli-responsiveness in drug delivery is the ability to specifically deliver drugs to affected cells and tissues while minimizing side effects due to interactions with healthy tissues and cells. In tissue engineering and the creation of bionic devices, the primary advantage of stimuli-responsive controlled release is the ability to reprogram gradients in response to changes in cell growth, differentiation, and/or migration.
Novel labile chemical linkers have been used to pattern and release biomolecules from polymer backbones in response to external stimuli. Most commonly, a photolabile linker is used to tether the biomolecule to the hydrogel polymer [174]. When exposed to a particular wavelength of light, the linker is degraded and the biomolecule becomes soluble. The gradient can be established by either the soluble or insoluble fraction of the biomolecule depending on the activity of the particular system. Spatial patterning can be achieved by shining light through a photomask, or by using a two-photon response where the linker is only degraded in the highly focused region of a laser beam. While this method provides excellent spatiotemporal control, selective multiplexed release requires engineering orthogonal linkers for each molecular factor to be released, which can become a major technical challenge. Additionally, the activities of many biomolecules can be decreased due to the covalent linking, and strategies to mitigate this can be challenging.
We have demonstrated a novel, selectively photoresponsive system using our 3D printed capsule platform [62]. In order to make the capsules photoresponsive, we loaded the shells with gold nanorods. The LSPR wavelength of the nanorod is strongly dependent on its length. In response to light exposure at the LSPR wavelength, the nanorods are rapidly heated, melting the polymeric shell, such that the capsule ruptures – quickly releasing the payload. Figure 7C shows the release of horseradish peroxidase from 2D printed capsules that have been ruptured. In this system, selective multiplexed release can be easily achieved, as no covalent modification is required to encapsulate the biomolecules, and gold nanorods with varying LSPR wavelengths can be routinely prepared in large quantities.
Microfluidic channels have been directly incorporated in hydrogels, providing a means to flow biomolecule solutions through gels as another strategy for creating dynamic gradients [175,176]. Gradients can be established in two ways. First, a gradient can be generated by the diffusion of biomolecules out of the microfluidic channel into the hydrogel matrix. Second, the flow of multiple solutions through a designed gradient generator can create precise gradients within the channel. This allows for the flexibility of generating steady-state gradients that can be maintained over long periods of time. Additionally, the concentrations of solutions can be rapidly varied for precise control of the temporal delivery of biological factors. However, multiple independent networks are required for multiplexed gradients, and an extensive pumping and fluid-handling infrastructure is required. This approach is thus more suitable for generating vasculature by consistently supplying nutrients to and removing waste from the tissue, rather than a means of generating transient gradients of biomolecules that can control cell fate at a local level.
3D Hybrid Systems with Dynamic Gradients
A longstanding challenge in tissue engineering has been the incorporation of dynamic gradients within macroscale 3D scaffolds. A significant hurdle to accomplishing this goal is the inherent challenges of multi-material 3D printing. Stimuli-responsive capsules and microfluidic networks are strategies for generating dynamic gradients that require novel materials and processes in order for them to be compatible with 3D printing. In order to 3D print complex arrays of stimuli responsive capsules, we have developed a new type of ink based on a water-in-oil emulsion [62,177–179]. The emulsion inks were prepared via high-speed dispersion of aqueous biomolecule solutions into a non-polar polymer solution. The emulsion-based ink was then directly printed into a thin layer of an aqueous hydrogel. Once printed, the solvent rapidly evaporates, leaving behind a solidified capsule sealed within the hydrogel. Thus, the hydrogel and capsules could be readily printed in a layer-by-layer fashion to create complex 3D hierarchical programmable capsule arrays. Since the 3D printing process is based on digital software control, rationally designed advanced architectures could be constructed. For instance, Figure 7A (left) shows an optical photograph of a hollow hydrogel cylinder containing alternating layers of red and blue capsules in the cylinder wall, and (right) two inverted pyramidal arrays of capsules printed within a solid hydrogel cube.
Several recent examples have demonstrated the freeform fabrication of microfluidic networks in macroscale hydrogel scaffolds [177–179] via the use of a sacrificial ink to define the channel lumen. The primary challenge in printing 3D microfluidic networks is that the sacrificial ink and hydrogel scaffold cannot be printed in a layer-by-layer manner, as doing so would not allow for continuous channels in the Z-direction of the scaffold. Bhattacharjee et al. have demonstrated the use of a granular gel medium into which highly complex continuous 3D channels could be printed (Figure 7B) [177]. Particle jamming in the media allowed for a system where the yield stress was high enough to prevent disruptions in the printed network due to differences in densities of the gel media and vascular network. Yet, the yield stress was sufficiently low to allow the print nozzle to travel through the media without leaving a void in its wake.
In contrast to the previous example, another approach to creating microfluidic networks first involves the printing of a sacrificial ink in the shape of the desired network. Then, the scaffold is deposited around the solidified network. Finally, once the scaffold is solidified the sacrificial network is removed. Miller et al. demonstrated such an approach utilizing a carbohydrate-based glass as the sacrificial ink (Figure 7C) [179]. The carbohydrate glass is printed in the molten state and allowed to cool and solidify. Once the gel scaffold is cast and solidified, the carbohydrate glass is dissolved away in water. A key advantage of this is the compatibility of the sacrificial ink with a wide range of hydrogel polymers. In another similar example, Kolesky et al. utilized inverse temperature responsive gelation of a sacrificial ink and hydrogel scaffold to create 3D microfluidic networks (Figure 7D) [178]. Both the sacrificial ink (Pluronic® F-127) and hydrogel scaffold (gelatin methacrylate) are fully gelled between 4–22 °C. The F-127 liquefies below 4 °C, whereas, the gelatin is liquid above 22 °C. Thus, to create a microfluidic network the F-127 is first printed into the shape of the channels above 4 °C. Next, above 22 °C the liquid gelatin is poured around the F-127 network and cooled to allow it to solidify. Finally, the temperature is lowered below 4 °C to liquefy the F-127, leaving channels in the gelatin scaffold. These examples show the wide diversity in materials and processes that can be utilized to generate systems with dynamic and complex biomolecular gradients.
3D Printed Anatomical Design
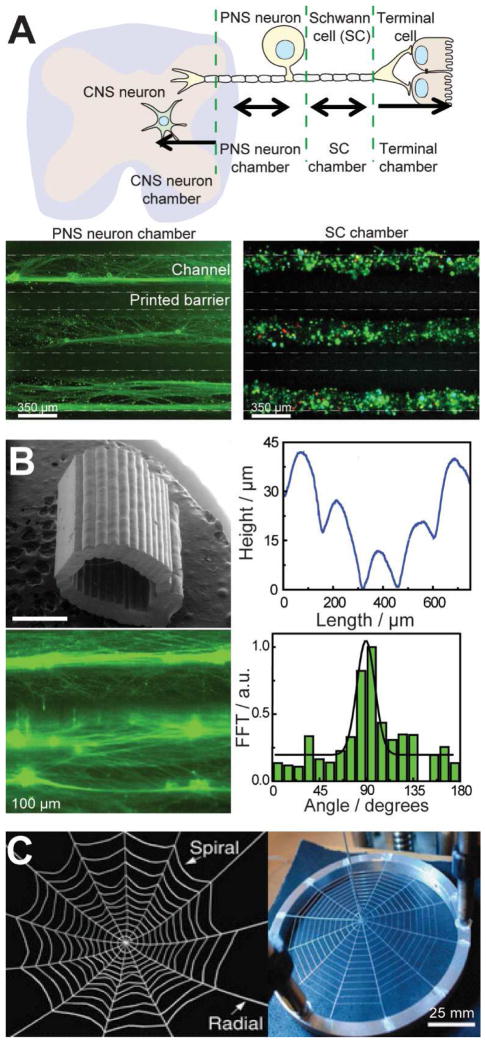
3D printed multi-scale biological systems have included artificial tissues, organs, biomedical implants, and bionic tissues. To date, 3D printed biological systems have been approached from three primary design paradigms, which include: (1) anatomical design, (2) mechanical (i.e. physical or topological) design, and (3) biochemical design. Anatomical design involves the development of 3D printed materials and devices which match the inherent anatomical structures by mimicking their 3D geometry. Some of the earliest efforts in anatomical 3D printed biological systems came in the form of engineering studies for craniofacial bone regeneration. In one such study, helical computed tomography (CT) images acquired from a dry mandible were used to generate the resultant 3D models for the printed scaffolds [180]. It was shown that the dimensional error of the 3D printed anatomical part, here the mandibular anatomy, varied from approximately 1–3% depending on the printing technique used. Recently, anatomical design of 3D printed biological systems has expanded to include: (1) heart tissues [181], (2) nerve scaffolds [64], (3) vascularized bone grafts [182], and (4) artificial skin [183].
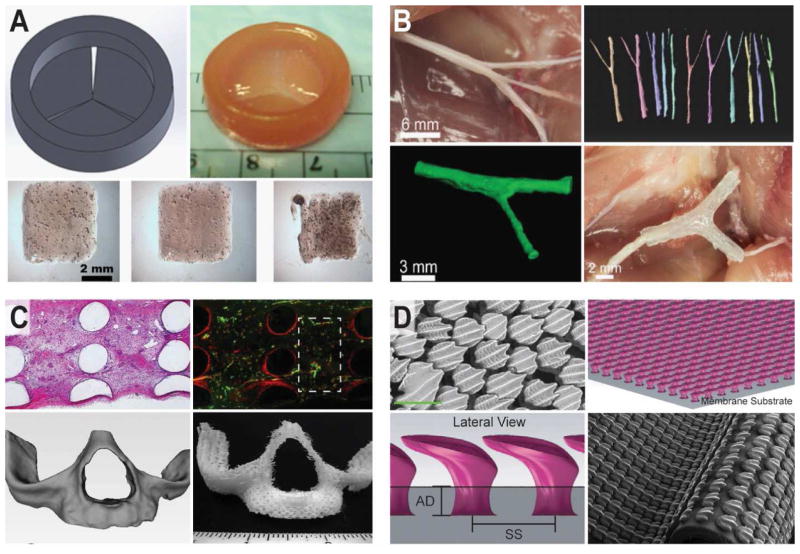
Anatomical structures include external and internal tissues and organs which range from micrometer to centimeter length scales. For example, as shown in Figure 8A, 3D printing has fabricated biomimetic tri-leaflet heart valve conduits, which use human aortic valvular interstitial cell-laden hydrogels of methacrylated hyaluronic acid and gelatin [181]. Another highlight of anatomical design was recently demonstrated in the form of 3D printed anatomical nerve regeneration pathways. As shown in Figure 8B, 3D models developed from structured light scanning of nerve tissue enabled the regeneration of complex bifurcating peripheral nerves in rats [64]. In addition to 3D printed scaffolds for heart and nerve tissue, advances in 3D printed bone and vasculature engineering have shown the ability to incorporate vasculature within anatomical bone grafts for potential connection of the tissue to the blood supply (Figure 8C) [182]. As discussed above, not only are anatomical design principles driving the manufacturing of multi-scale anatomical internal tissues and organs [55,64,67,178,181,184], but they are also guiding the exploration of external tissues which interact with the surrounding environment [183]. For example, Figure 8D shows that 3D printed artificial skins can be realized which contain micro-structured biomimetic surface topographies that achieve innovative hydrodynamic flows [183].
Figure 8.
3D printed anatomical design strategies. (A) 3D printed tri-leaflet heart valve [181]. (B) 3D printed anatomical nerve regeneration pathway [64]. (C) 3D printed vascularized bone architectures [182]. (D) 3D printed biomimetic artificial skin (green scale bar is 200 μm) [183]. Reprinted with permission from Refs. [181], [64], [182], [183] respectively. Copyright 2014 Elsevier, 2015 John Wiley & Sons, 2014 John Wiley & Sons.
3D Printed Biomechanical Design
Mechanical (or topological) design involves the mimicry of native mechanical properties and cues, such as stress-strain behavior, topographical structures, and microstructures, in 3D printed scaffold architectures. Some of the earliest efforts to examine the influence of physical cues in 3D printed biological systems included the design of porous scaffolds for bone engineering applications [185]. For example, 3D printing was used to assemble hydroxyapatite scaffolds for bone regeneration over a range of mechanical properties, such as porosity and strength. It was shown that the mechanical properties and biological consequences of 3D printed scaffolds for bone regeneration may be tuned by controlling the wall and channel thicknesses of 3D printed grid-based scaffolds. Recently, mechanical design of 3D printed biological systems has expanded to include: (1) 2D topographical cues in biomimetic in vitro models [65], (2) 3D topographical cues in 3D tissue scaffolds [64], and (3) bio-inspired mechanical systems [186].
The geometry of physical cues has included channels, grooves, and filaments. As shown in Figure 9A, we recently demonstrated that a biomimetic nervous system on a chip technology can be realized via the controlled guidance of axons within 3D printed polycaprolactone microchannels [65]. Similarly, Figure 9B shows that 3D printed physical cues in the form of microgrooves in anatomical elastomeric scaffolds for peripheral nerve regeneration resulted in the aligned growth of axonal networks and cytoskeletons of Schwann cells [64]. The parallel and orthogonal assembly of material filaments has provided flexible mechanical design opportunities in terms of controlling scaffold strength and fate of interacting cellular components. Further, the ability to control filament assembly in non-uniform geometric patterns other than layer-by-layer approaches also enables one to mimic various biomechanical systems. For example, as shown in Figure 9C, it was recently shown that radial and spiral filament strengths control the loading response in natural webbed systems, here a spider’s web [186].
Figure 9.
Mechanical design methodologies in 3D printed biological systems. (A) 3D printed microchannels control the growth of axonal networks in a 3D printed nervous system on a chip [65]. (B)3D printed microgrooves in elastomeric anatomical nerve guides control the alignment of the regenerating axonal network longitudinally toward the injury site. Scale bar is 1 mm [64]. (C) 3D printed spider web displaying interacting radial and spiral elastomeric filaments [186]. Reprinted with permission from Refs. [65], [64], [186], respectively. Copyright 2015 Centre National de la Recherche Scientifique (CNRS) and Royal Society of Chemistry, 2015 John Wiley & Sons, 2015 Nature Publishing Group.
3D Printed Biochemical Design
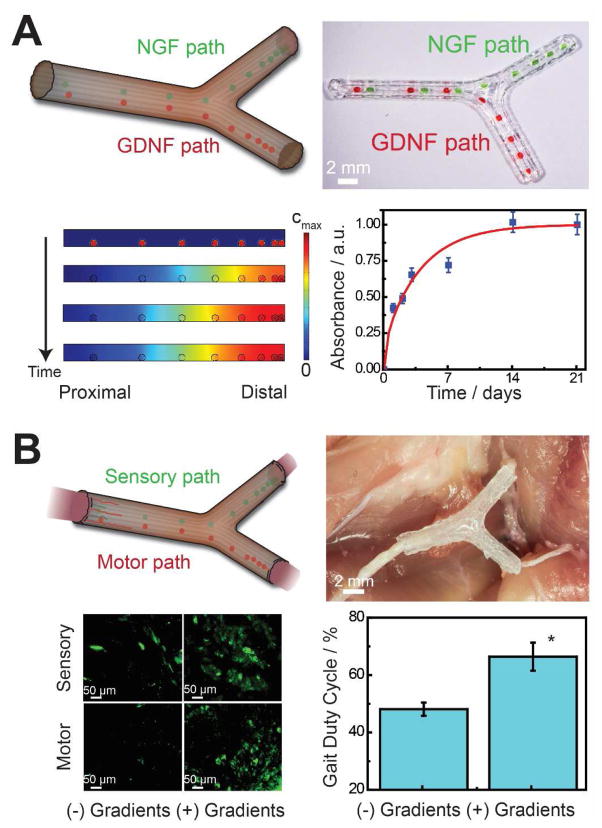
Biochemical design involves the mimicry of a native biochemical structure or profile, which may include either uniform or non-uniform distributions of biochemical factors. Some of the earliest efforts included the patterning of protein gradients on 2D substrates [158]. In this study, inkjet printing was used to print ciliary neurotrophic factor (CNTF) gradients and examine their effect on the multi-potency and differentiation of neural stem cells (NSCs) [158]. It was shown that NSCs cultured on a printed gradient of increasing levels of CNTF showed a linear increase in the numbers of cells expressing glial fibrillary associated protein (GFAP), demonstrating a functional 3D printed gradient of CNTF on a 2D substrate. As discussed, 3D printing also has the ability to apply anatomical [64,67,178,181] and mechanical [64,65,186] design principles toward the manufacturing of novel biological systems. Thus, the multiscale nature of the approach which combines anatomical, mechanical, and biochemical design paradigms also allows for the integration of biochemical gradient technologies within macroscale biological architectures in order to realize novel platform technologies and biomedical devices. In our study of nerve regeneration in bifurcating mixed nerve pathways, 3D printing was used to incorporate multi-component biochemical gradients of nerve growth factor (NGF) and glial cell-derived neurotrophic factor (GDNF) within a global scaffold architecture, to support the regeneration of both motor and sensory nerve pathways (Figure 10A) [64]. As shown in Figure 10B, scaffolds containing path-specific multi-component gradients led to regenerated tissue which possessed enhanced functional return relative to scaffolds which did not contain supplemented gradients, thus validating the potential for functional 3D printed multi-component gradients within a scaffold architecture to improve regenerative outcomes.
Figure 10.
Biochemical design strategies in 3D printed biological systems. (A) Path-specific 3D printed multi-component gradient in anatomical nerve regeneration pathways [64]. (B) Effect of the functional 3D printed path-specific regeneration on the regeneration of motor and sensory nerve pathways, and the functional return of complex regenerated peripheral nerve injuries [64]. Reprinted with permission from Ref. [64]. Copyright 2015 Royal Society of Chemistry.
3D Printed Conducting Ink Electronics
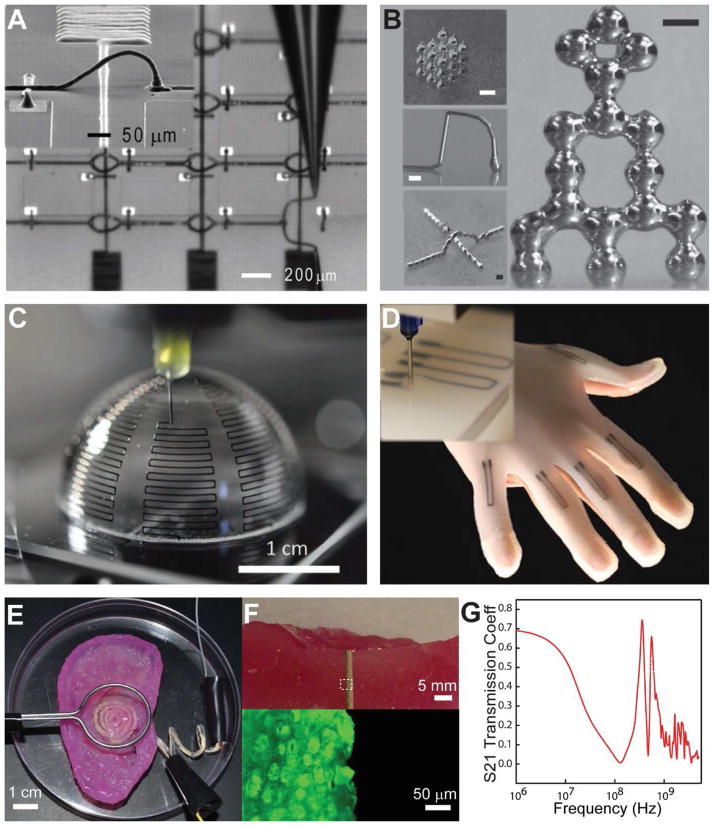
Modern prosthetics typically incorporate electronics to mimic, restore, and/or augment the complex functionalities of biological constructs [1,12,18,19,21,22]. The ability to create advanced electronics with 3D printing could lead to methods for directly printing both the mechanical prosthetics and the incorporated electronics from the same machine, replete with customization of both geometries and functionalities. This section focuses on the printing of conducting nanoscale inks such as metal nanoparticles, which can act as interconnects, or can be made into passive devices such as strain sensors and antennas. For instance, Ahn et al. described the synthesis of a highly concentrated (> 70 wt%) viscoelastic ink with silver nitrate, poly(acrylic acid) and diethanolamine [90]. Compared to previously published inkjet printed metal nanoparticle inks, this ink formulation has a key advantage in its ability to create three-dimensional interconnect arches as shown in the inset of Figure 11A. These features overcome the conventional planar constraints of traditional printing methods, allowing for interconnects that can span and stretch across circuit elements in three dimensions [90]. Further, Ahn et al. demonstrated an impressive minimum feature size of 2 μm as shown in Figure 11A. This enabled the creation of features such as a transparent conductive grid [187]. Significantly, the resistivity (5.2 × 10−7 Ω·m) approaches the resistivity of bulk silver (10−8 Ω·m), and this value can be achieved by using a relatively mild annealing process (250 °C) for a short time (30 min). The formulation of the inks and the printing of the electrodes into wavy architectures enabled the printed conductors to withstand repeated stretching and bending (maximum strain of 25%). Such attributes are important in the creation of stretchable bionic devices such as skin sensors [12].
Figure 11.
3D printing of conducting electronic inks. (A) Omni-directional printing of a concentrated silver ink to form the interconnects of an LED chip array. The inset shows an interconnect arch printed over a junction [90]. (B) 3D printing of free-standing liquid metal into a cubic array of stacked droplets (top inset), a 3D metal arch (middle inset), an arch overpassing a printed wire (bottom inset), and a tower of liquid metal droplets. Scale bars are 500 μm [70]. (C) 3D printing of a silver nanoparticle ink on a three dimensional surface to form an antenna [191]. (D) Embedded 3D printing of conducting carbon grease in an uncured elastomeric polymer (inset) enables the creation of stretchable strain sensors embedded within a glove [192]. (E) Co-printing of a conductor within a cell-laden biological scaffold to create a bionic ear. (F) Biocompatibility of the printed electronics within the biological construct. The fluorescent image (bottom) shows the viability of the neo-cartilaginous tissue in contact with the electrode (top) [67]. (G) Electromagnetic response of the 3D printed bionic ear. Plot shows the S21 transmission coefficient with frequency, demonstrating the capability of receiving signals over an expansive frequency range [67]. Reprinted with permission from Refs. [90], [70], [191], [192], [67], respectively. Copyright 2009 American Association for the Advancement of Science, 2011 John Wiley & Sons, 2014 John Wiley & Sons, 2013 American Chemical Society.
The ability of extrusion-based 3D printing to accommodate diverse materials with a wide range of viscosities allows for the incorporation of classes of electronic materials that are incompatible with other patterning processes, such as inkjet printing or dip-pen lithography [188]. For instance, eutectic gallium-indium alloy (EGaIn) [189] is a highly conductive (3.4 × 104 S cm−1) liquid metal that has recently been explored as a 3D printable conductor [70,190]. Intriguingly, despite its liquid nature at room temperature (melting point = 15.5 °C), mechanically stable three-dimensional structures can be achieved. This is due to a high surface tension (0.6 N·m−1) thin oxide film that is formed on the surface of the liquid (Figure 11B). Further, a printing resolution of 100 μm has been demonstrated, and unlike most sintered solid metal, the liquid allows for the creation of highly stretchable (up to 100% elongation) electrodes when encapsulated within an elastomeric polymer [190], enabling the creation of stretchable devices such as strain gauges. Given the work function of −4.2 eV, EGaIn has also been shown to be a suitable printable cathode. This is particularly useful for low melting point polymeric 3D printed constructs, as thermal sintering processes can be avoided [61].
The ability to pattern in three dimensions allows for the creation of electronics with unique or unusual performances in comparison with planar patterning techniques. For instance, the printing of silver nanoparticles on a three-dimensional construct allowed for the fabrication of an antenna with an order of magnitude improvement over monopole antenna designs (Figure 12C) [191]. Further, the co-printing of conductive traces with an elastomeric substrate [59,60] can result in the freeform fabrication of three dimensional structures containing electronic components. For instance, capacitive soft strain sensors can be realized via the printing of core-shell fibers with silicone and conductive fluids [71]. As shown in Figure 11D, the embedded 3D printing of conductive carbon grease within an elastomeric polymer enabled the seamless fabrication of complex arrays of strain sensors within a glove, that can be used to monitor the motion of a user’s hand. Indeed, these demonstrations highlight the many distinct advantages of 3D printing in the fabrication of electronic devices. These include the realization of devices customized to conform to the user’s morphology, and customization of functionality by incorporating different classes of materials to create functional components within soft and stretchable constructs.
Figure 12.
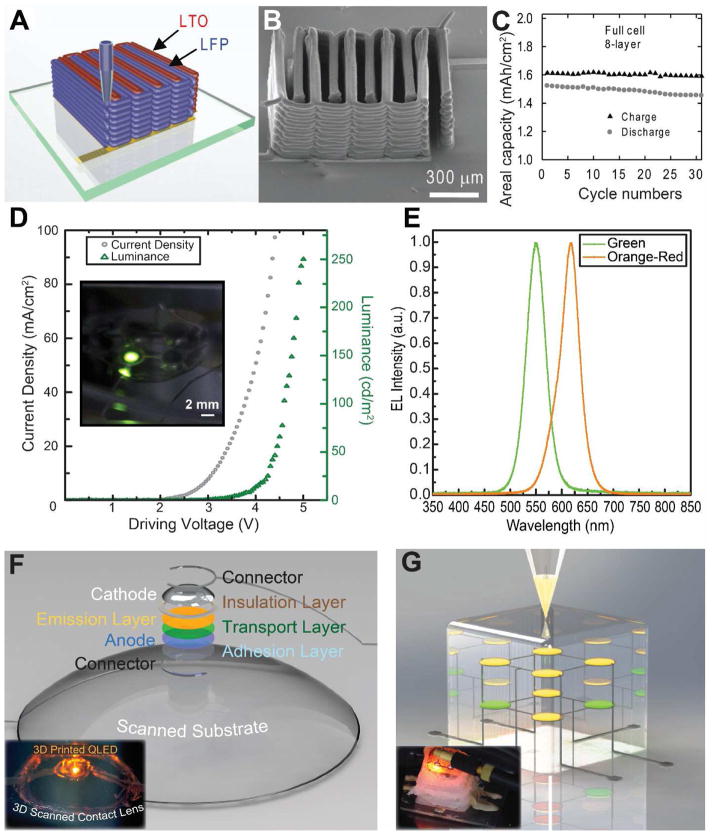
3D printing of active electronics with semiconducting inks. (A) 3D Printing of Li3Ti4O12 (LTO) and LiFePO4 (LFP) inks to create a 3D interdigitated micro-battery architecture [196]. (B) SEM images of the printed 3D interdigitated micro-battery. (C) The cycle life of the 3D printed interdigitated battery. A good cycle life is achieved due to the low-strain topotactic reactions of LFP and LTO. (D) A 3D printed quantum dot light-emitting diode (QD-LED), where the inset shows the electroluminescence output from the QD-LED and the graph shows the current density vs. voltage and forward luminance output [61]. (E) Normalized electroluminescence spectra from both green and orange-red QD-LEDS, demonstrating color tunability and high color purity of the 3D printed QD-LEDS. (F) 3D printed QD-LED on a scanned curvilinear substrate, where the figure shows the CAD model and its components. The inset shows the electroluminescence output from the printed QD-LED on a 3D scanned contact lens (lens diameter is 10 mm) [61]. (G) 3D printing of a 2 × 2 × 2 multidimensional array of embedded QD-LEDs, where the inset shows the electroluminescence from a QD-LED in the 3D matrix (cube edge length is 15 mm) [61]. Reprinted with permission from Refs. [196], [61], respectively. Copyright 2013 John Wiley & Sons, 2014 American Chemical Society.
The bottom-up nature of 3D printed electronics – and the exclusion of the harsh chemicals and temperatures found in microfabrication processes – allows for the co-printing of electronics with biological materials to yield novel constructs even including bionic organs [55,67]. This three-dimensional interweaving of electronics and biology using a multi-material 3D printing process was demonstrated by our group. Specifically, the co-printing of a cell-laden scaffold and conductive traces enabled the creation of crude yet functional ‘bionic ears’ containing an electronic device (Figure 11D) [67]. This device was fabricated by co-printing an alginate hydrogel matrix seeded with chondrocyte cells, and conducting silicone infused with silver particles, printed into a circular coil path to create an antenna. The seeded chondrocyte cells are then cultured into cartilage tissue, and the neo-cartilaginous tissue in contact with the printed electrode retains viability (Figure 11E). The interweaving of an electronic device into the organ allows the printed bionic ears to receive electromagnetic signals in a frequency range (up to 5 GHz) that is well beyond the normal perceptible range of human acoustic hearing (20 Hz to 20 kHz) as shown in Figure 11F.
3D Printed Active Semiconducting Electronics
Overcoming the planarity constraint of traditional microfabricated active electronic devices could enable the introduction of optoelectronic, sensing, and computational capabilities into non-flat, soft, flexible, and stretchable three dimensional constructs. For example, the incorporation of light emitting-diodes (LEDs) or sensors on contact lenses could provide components for on-eye wearable displays [193] or glucose sensors [194,195].
Indeed, developing the ability to 3D print active semiconducting materials is a critical and rapidly developing area which is expected to be a significant driver of 3D printing technologies going forward. For instance, Sun et al. demonstrated the 3D printing of a micro-battery via the co-printing of Li3Ti4O12` (LTO) and LiFePO4 (LFP) as cathode and anode, respectively (Figure 12A) [196]. The ability to stack the electrodes in a high aspect ratio form factor (up to 16 layers, Figure 12B) allows the 3D printed battery to achieve an exceptionally high energy density (9.7 J·cm−2 at a power density of 2.7 mW·cm−2). Further, the device exhibits a reasonable cycle life, as shown in Figure 12C, where only a small decay of areal capacity is observed after 30 charging cycles. Nevertheless, the fabrication process does include the deposition of gold on glass via electron beam evaporation. Further, a 600 °C heat treatment is required to remove organic additives and initiate the sintering process. Hence, further work is needed to 3D print such devices on three-dimensional, temperature restrictive substrates.
The primary challenge in developing fully 3D printable active electronic devices such as diodes and transistors [197] lies in the complications associated with the integration of diverse classes of materials exhibiting disparate properties [47]. For instance, the 3D printing of LEDs requires the integration of a printable substrate, an emission layer, charge transport layers, a cathode, an anode and interconnects, all of which could consist of metal, semiconductor and polymeric materials, with varying surface energies, viscosities, and tribological and mechanical properties [61]. In general, the selection of inks needs to fulfill three major requirements. First, the materials have to be formulated into a 3D printable ink. Second, the ink materials have to retain their performance and function following extrusion from the 3D printer. For instance, in optoelectronic devices the bands must align properly [115]. Finally, the processing conditions and ink solvents have to be compatible with the other printed layers to minimize damage or degradation to already printed materials.
Recently, we have achieved this goal of fully 3D printing active semiconducting devices in the form of quantum dot light emitting diodes [61]. First, a transparent electrode consisting of a silver nanoparticle ring was printed, where the porous and hydrophilic nature of the ink allows for the formation of good electrical contact with a subsequently printed PEDOT:PSS layer. A charge transport layer (Poly TPD) and a nanoparticle semiconductor emissive layer (CdSe/ZnS QDs) are then 3D printed. The inks are formulated to form thin and uniform layers, as described previously. Finally, EGaIn liquid metal is printed to form a conformal top electrode. The printed device achieved a maximum brightness of 250 cd/m2 at 5 V as shown in Figure 12C, and the device also exhibited pure color emission from the QD emissive layer as shown in Figure 12D [115,121]. We anticipate that further optimization of the layer thicknesses and uniformities could further improve the device performances.
3D printing allows for a liberation of the device from the constraints of conventional microfabrication processes, such that the LEDs can be printed on non-planar and polymeric substrates. For instance, 3D scanning can be used to determine the precise topography of a non-planar substrate. Then this information can be incorporated into a CAD program, such that the electronics can be conformally printed on the underlying 3D substrate. We validated this concept via the direct printing of a QD-LED on a 3D scanned contact lens (Figure 12E). This approach also allows us to build electronics up into the third dimension. Embedding these devices in three dimensions within an elastomeric structure enabled the creation of a 2 × 2 × 2 array of multicolored LEDs embedded within a silicone cube, as shown in Figure 12F. This device fabrication approach of course is generalizable to other classes of devices, such as solar cells [198,199] and transistors [197]. Further, the freeform fabrication of devices could also enable the creation of new classes of bionic devices with novel functionalities, such as soft optogenetic LED probes [14,200].
Conclusion
We have introduced a conceptually novel and comprehensive approach for the use of 3D printing as a versatile, multi-scale, multi-material tool that can address many of the fundamental challenges in the fabrication of bionic devices. 3D printing allows for the interweaving of the unique functionalities of nanomaterials with a variety of other materials, including soft and biological materials, to enable a seamless fabrication of three dimensional bionic devices. This is a multimaterial processing solution, which also achieves multiscale manufacturing – from nanoscale inks, to microscale features, to macroscale bionic devices. In this review, we have first described the unique properties of nanomaterials and their dispersion into functional inks. Second, we have reviewed the microscale printing of nanomaterials with 3D printing via convective self-assembly methods. We have also highlighted the versatility of 3D printing in creating hierarchical architectures, which can even include spatiotemporal gradients and stimuli-responsive capsules. Finally, we highlighted our ability to fabricate anatomically accurate macroscale bionic devices. The recent ability to extend these printing capabilities to active materials, such as semiconducting inks, is a critical development to impart complex functionalities into 3D printed devices that were previously simply passive constructs. Indeed, the ability to locally and directly print customized electronic devices into personalized 3D printed biomedical devices represents an exciting and extremely promising direction for future bioelectronics research. Overall, this blending of 3D printing, novel nanomaterial properties, and ‘living’ platforms may enable next-generation 3D printed bionic nanodevices.
Highlights.
Bionics involves the interfacing of functional devices directly with biology.
3D printing allows the interweaving of biological and functional materials.
We highlight the unique functionalities of nanoscale inks for 3D printing.
We highlight the 3D printing of microscale features from these nanoscale inks.
We highlight the hierarchical printing of macroscale bionic architectures.
Acknowledgments
M.C.M. thanks the following agencies for their generous support of the studies from the authors’ laboratory that contributed to this review: The Defense Advanced Research Projects Agency (No. D12AP00245), the Air Force Office of Scientific Research (No. FA9550-12-1-0368), the Intelligence Community (Award No. 2013-13070300004), and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (Award No. 1DP2EB020537).
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lavine M, Roberts L, Smith O. Science. 2002;295:995. [Google Scholar]
- 2.Craelius W. Science. 2002;295:1018–1021. doi: 10.1126/science.295.5557.1018. [DOI] [PubMed] [Google Scholar]
- 3.Clausen J. Nature. 2009;457:1080–1081. doi: 10.1038/4571080a. [DOI] [PubMed] [Google Scholar]
- 4.Vogel G. Science. 2002;295:1020. doi: 10.1126/science.295.5557.1020. [DOI] [PubMed] [Google Scholar]
- 5.Dictionary.com. Unabridged. Random House, Inc; [Google Scholar]
- 6.Service RF. Science. 2013;340:1162–1165. doi: 10.1126/science.340.6137.1162. [DOI] [PubMed] [Google Scholar]
- 7.Giraldo JP, Landry MP, Faltermeier SM, McNicholas TP, Iverson NM, Boghossian AA, Reuel NF, Hilmer AJ, Sen F, Brew JA, Strano MS. Nat Mater. 2014;13:400–408. doi: 10.1038/nmat3890. [DOI] [PubMed] [Google Scholar]
- 8.Stavrinidou E, Gabrielsson R, Gomez E, Crispin X, Nilsson O, Simon DT, Berggren M. Sci Adv. 2015;1:e1501136. doi: 10.1126/sciadv.1501136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haslam M, Hernandez-Aguilar A, Ling V, Carvalho S, de la Torre I, DeStefano A, Du A, Hardy B, Harris J, Marchant L, Matsuzawa T, McGrew W, Mercader J, Mora R, Petraglia M, Roche H, Visalberghi E, Warren R. Nature. 2009;460:339–344. doi: 10.1038/nature08188. [DOI] [PubMed] [Google Scholar]
- 10.Crubzy E, Murail P, Girard L, Bernadou JP. Nature. 1998;391:29. doi: 10.1038/34067. [DOI] [PubMed] [Google Scholar]
- 11.Kim DH, Lu N, Ma R, Kim YS, Kim RH, Wang S, Wu J, Won SM, Tao H, Islam A, Yu KJ, Kim TI, Chowdhury R, Ying M, Xu L, Li M, Chung HJ, Keum H, McCormick M, Liu P, Zhang YW, Omenetto FG, Huang Y, Coleman T, Rogers JA. Science. 2011;333:838–843. doi: 10.1126/science.1206157. [DOI] [PubMed] [Google Scholar]
- 12.Tee BC, Chortos A, Berndt A, Nguyen AK, Tom A, McGuire A, Lin ZC, Tien K, Bae WG, Wang H, Mei P, Chou HH, Cui B, Deisseroth K, Ng TN, Bao Z. Science. 2015;350:313–316. doi: 10.1126/science.aaa9306. [DOI] [PubMed] [Google Scholar]
- 13.Song YM, Xie Y, Malyarchuk V, Xiao J, Jung I, Choi KJ, Liu Z, Park H, Lu C, Kim RH, Li R, Crozier KB, Huang Y, Rogers JA. Nature. 2013;497:95–99. doi: 10.1038/nature12083. [DOI] [PubMed] [Google Scholar]
- 14.Park SI, Brenner DS, Shin G, Morgan CD, Copits BA, Chung HU, Pullen MY, Noh KN, Davidson S, Oh SJ, Yoon J, Jang KI, Samineni VK, Norman M, Grajales-Reyes JG, Vogt SK, Sundaram SS, Wilson KM, Ha JS, Xu R, Pan T, Kim TI, Huang Y, Montana MC, Golden JP, Bruchas MR, Gereau RW, Rogers JA. Nat Biotechnol. 2015;33:1280–1286. doi: 10.1038/nbt.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McEvoy MA, Correll N. Science. 2015;347:1261689. doi: 10.1126/science.1261689. [DOI] [PubMed] [Google Scholar]
- 16.Kim DH, Ahn JH, Choi WM, Kim HS, Kim TH, Song J, Huang YY, Liu Z, Lu C, Rogers JA. Science. 2008;320:507–511. doi: 10.1126/science.1154367. [DOI] [PubMed] [Google Scholar]
- 17.Wilson BS, Finley CC, Lawson DT, Wolford RD, Eddington DK, Rabinowitz WM. Nature. 1991;352:236–238. doi: 10.1038/352236a0. [DOI] [PubMed] [Google Scholar]
- 18.Moore DR, Shannon RV. Nature Neuro. 2009;12:686–691. doi: 10.1038/nn.2326. [DOI] [PubMed] [Google Scholar]
- 19.Eilenberg MF, Geyer H, Herr H. IEEE Trans Neural Syst Rehabil Eng. 2010;18:164–173. doi: 10.1109/TNSRE.2009.2039620. [DOI] [PubMed] [Google Scholar]
- 20.da Cruz L, Coley BF, Dorn J, Merlini F, Filley E, Christopher P, Chen FK, Wuyyuru V, Sahel J, Stanga P, Humayun M, Greenberg RJ, Dagnelie G. Br J Ophthalmol. 2013;97:632–636. doi: 10.1136/bjophthalmol-2012-301525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zrenner E. Sci Transl Med. 2013;5:210ps216. doi: 10.1126/scitranslmed.3007399. [DOI] [PubMed] [Google Scholar]
- 22.Minev IR, Musienko P, Hirsch A, Barraud Q, Wenger N, Moraud EM, Gandar J, Capogrosso M, Milekovic T, Asboth L, Torres RF, Vachicouras N, Liu Q, Pavlova N, Duis S, Larmagnac A, Voros J, Micera S, Suo Z, Courtine G, Lacour SP. Science. 2015;347:159–163. doi: 10.1126/science.1260318. [DOI] [PubMed] [Google Scholar]
- 23.Hammock ML, Chortos A, Tee BCK, Tok JBH, Bao Z. Adv Mater. 2013;25:5997–6038. doi: 10.1002/adma.201302240. [DOI] [PubMed] [Google Scholar]
- 24.Someya T, Sekitani T, Iba S, Kato Y, Kawaguchi H, Sakurai T. Proc Nat Acad Sci USA. 2004;101:9966–9970. doi: 10.1073/pnas.0401918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipomi DJ, Vosgueritchian M, Tee BCK, Hellstrom SL, Lee JA, Fox CH, Bao Z. Nat Nanotechnol. 2011;6:788–792. doi: 10.1038/nnano.2011.184. [DOI] [PubMed] [Google Scholar]
- 26.Farina D, Aszmann O. Sci Transl Med. 2014;6:257ps212. doi: 10.1126/scitranslmed.3010453. [DOI] [PubMed] [Google Scholar]
- 27.Anderson JM, McNally AK. Semin Immunopathol. 2011;33:221–233. doi: 10.1007/s00281-011-0244-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Cao Z, Bai T, Carr L, Ella-Menye JR, Irvin C, Ratner BD, Jiang S. Nat Biotechnol. 2013;31:553–556. doi: 10.1038/nbt.2580. [DOI] [PubMed] [Google Scholar]
- 29.Veiseh O, Doloff JC, Ma M, Vegas AJ, Tam HH, Bader AR, Li J, Langan E, Wyckoff J, Loo WS, Jhunjhunwala S, Chiu A, Siebert S, Tang K, Hollister-Lock J, Aresta-Dasilva S, Bochenek M, Mendoza-Elias J, Wang Y, Qi M, Lavin DM, Chen M, Dholakia N, Thakrar R, Lacik I, Weir GC, Oberholzer J, Greiner DL, Langer R, Anderson DG. Nat Mater. 2015;14:643–651. doi: 10.1038/nmat4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashby MF. Materials Selection in Mechanical Design. 1. Pergamon Press; Oxford; New York: 1992. [Google Scholar]
- 31.Agache PG, Monneur C, Leveque JL, De Rigal J. Arch Dermatol Res. 1980;269:221–232. doi: 10.1007/BF00406415. [DOI] [PubMed] [Google Scholar]
- 32.Rogers JA, Someya T, Huang Y. Science. 2010;327:1603–1607. doi: 10.1126/science.1182383. [DOI] [PubMed] [Google Scholar]
- 33.Nicolelis MA, Dimitrov D, Carmena JM, Crist R, Lehew G, Kralik JD, Wise SP. Proc Nat Acad Sci USA. 2003;100:11041–11046. doi: 10.1073/pnas.1934665100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai S, Debnath S, Gibson K, Schlicht B, Bayne L, Zagnoni M, Ulijn RV. Small. 2014;10:285–293. doi: 10.1002/smll.201301333. [DOI] [PubMed] [Google Scholar]
- 35.Fratzl P, Weinkamer R. Prog Mater Sci. 2007;52:1263–1334. [Google Scholar]
- 36.Kim RH, Kim DH, Xiao J, Kim BH, Park SI, Panilaitis B, Ghaffari R, Yao J, Li M, Liu Z, Malyarchuk V, Kim DG, Le AP, Nuzzo RG, Kaplan DL, Omenetto FG, Huang Y, Kang Z, Rogers JA. Nat Mater. 2010;9:929–937. doi: 10.1038/nmat2879. [DOI] [PubMed] [Google Scholar]
- 37.Rogers JA, Lagally MG, Nuzzo RG. Nature. 2011;477:45–53. doi: 10.1038/nature10381. [DOI] [PubMed] [Google Scholar]
- 38.McAlpine MC, Ahmad H, Wang D, Heath JR. Nat Mater. 2007;6:379–384. doi: 10.1038/nmat1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dagdeviren C, Yang BD, Su Y, Tran PL, Joe P, Anderson E, Xia J, Doraiswamy V, Dehdashti B, Feng X, Lu B, Poston R, Khalpey Z, Ghaffari R, Huang Y, Slepian MJ, Rogers JA. Proc Nat Acad Sci USA. 2014;111:1927–1932. doi: 10.1073/pnas.1317233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim DH, Viventi J, Amsden JJ, Xiao J, Vigeland L, Kim YS, Blanco JA, Panilaitis B, Frechette ES, Contreras D, Kaplan DL, Omenetto FG, Huang Y, Hwang KC, Zakin MR, Litt B, Rogers JA. Nat Mater. 2010;9:511–517. doi: 10.1038/nmat2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DH, Lu N, Ghaffari R, Rogers JA. NPG Asia Mater. 2012;4:e15. [Google Scholar]
- 42.Morin SA, Shevchenko Y, Lessing J, Kwok SW, Shepherd RF, Stokes AA, Whitesides GM. Adv Mater. 2014;26:5991–5999. doi: 10.1002/adma.201401642. [DOI] [PubMed] [Google Scholar]
- 43.Hull CW. UVP, Inc; US: Mar 11, 1986. [Google Scholar]
- 44.Jarvis RA. IEEE Trans Pattern Anal Mach Intell. 1983;PAMI-5:122–139. doi: 10.1109/tpami.1983.4767365. [DOI] [PubMed] [Google Scholar]
- 45.Rocchini C, Cignoni P, Montani C, Pingi P, Scopigno R. Comput Graph Forum. 2001;20:299–308. [Google Scholar]
- 46.Salvi J, Pagès J, Batlle J. Pattern Recognit. 2004;37:827–849. [Google Scholar]
- 47.Raney JR, Lewis JA. MRS Bull. 2015;40:943–950. [Google Scholar]
- 48.Zhang X, Jiang X, Sun C. Sens Actuat A - Phys. 1999;77:149–156. [Google Scholar]
- 49.Sun C, Fang N, Wu D, Zhang X. Sens Actuat A - Phys. 2005;121:113–120. [Google Scholar]
- 50.Cumpston BH, Ananthavel SP, Barlow S, Dyer DL, Ehrlich JE, Erskine LL, Heikal AA, Kuebler SM, Lee IYS, McCord-Maughon D. Nature. 1999;398:51–54. [Google Scholar]
- 51.Calvert P. Chem Mater. 2001;13:3299–3305. [Google Scholar]
- 52.Onses MS, Sutanto E, Ferreira PM, Alleyne AG, Rogers JA. Small. 2015;11:4237–4266. doi: 10.1002/smll.201500593. [DOI] [PubMed] [Google Scholar]
- 53.Lipson H, Kurman M. Fabricated : The New World of 3D Printing. John Wiley and Sons, Inc; Indianapolis, IN: 2013. [Google Scholar]
- 54.Malone E, Lipson H. Rapid Prototyping J. 2007;13:245–255. [Google Scholar]
- 55.Murphy SV, Atala A. Nat Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 56.Lewis JA, Smay JE, Stuecker J, Cesarano J. J Am Ceram Soc. 2006;89:3599–3609. [Google Scholar]
- 57.Lewis JA. Adv Funct Mater. 2006;16:2193–2204. [Google Scholar]
- 58.Pearce JM. Science. 2012;337:1303–1304. doi: 10.1126/science.1228183. [DOI] [PubMed] [Google Scholar]
- 59.Ober TJ, Foresti D, Lewis JA. Proc Nat Acad Sci USA. 2015;112:12293–12298. doi: 10.1073/pnas.1509224112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hardin JO, Ober TJ, Valentine AD, Lewis JA. Adv Mater. 2015;27:3279–3284. doi: 10.1002/adma.201500222. [DOI] [PubMed] [Google Scholar]
- 61.Kong YL, Tamargo IA, Kim H, Johnson BN, Gupta MK, Koh TW, Chin HA, Steingart DA, Rand BP, McAlpine MC. Nano Lett. 2014;14:7017–7023. doi: 10.1021/nl5033292. [DOI] [PubMed] [Google Scholar]
- 62.Gupta MK, Meng F, Johnson BN, Kong YL, Tian L, Yeh YW, Masters N, Singamaneni S, McAlpine MC. Nano Lett. 2015;15:5321–5329. doi: 10.1021/acs.nanolett.5b01688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Compton BG, Lewis JA. Adv Mater. 2014;26:5930–5935. doi: 10.1002/adma.201401804. [DOI] [PubMed] [Google Scholar]
- 64.Johnson BN, Lancaster KZ, Zhen G, He J, Gupta MK, Kong YL, Engel EA, Krick KD, Ju A, Meng F, Enquist LW, Jia X, McAlpine MC. Adv Funct Mater. 2015;25:6205–6217. doi: 10.1002/adfm.201501760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson BN, Lancaster KZ, Hogue IB, Meng F, Kong YL, Enquist LW, McAlpine MC. Lab Chip. 2015 [Google Scholar]
- 66.Lewis JA. Tissue Eng Pt A. 2015;21:S22–S22. [Google Scholar]
- 67.Mannoor MS, Jiang Z, James T, Kong YL, Malatesta KA, Soboyejo WO, Verma N, Gracias DH, McAlpine MC. Nano Lett. 2013;13:2634–2639. doi: 10.1021/nl4007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis JA. Curr Opin Solid State Mater Sci. 2002;6:245–250. [Google Scholar]
- 69.Morissette SL, Lewis JA, Clem PG, Cesarano J, Dimos DB. J Am Ceram Soc. 2001;84:2462–2468. [Google Scholar]
- 70.Ladd C, So JH, Muth J, Dickey MD. Adv Mater. 2013;25:5081–5085. doi: 10.1002/adma.201301400. [DOI] [PubMed] [Google Scholar]
- 71.Frutiger A, Muth JT, Vogt DM, Menguc Y, Campo A, Valentine AD, Walsh CJ, Lewis JA. Adv Mater. 2015;27:2440–2446. doi: 10.1002/adma.201500072. [DOI] [PubMed] [Google Scholar]
- 72.Hanson Shepherd JN, Parker ST, Shepherd RF, Gillette MU, Lewis JA, Nuzzo RG. Adv Funct Mater. 2011;21:47–54. doi: 10.1002/adfm.201001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barry RA, Shepherd RF, Hanson JN, Nuzzo RG, Wiltzius P, Lewis JA. Adv Mater. 2009;21:2407–2410. [Google Scholar]
- 74.Webber MJ, Appel EA, Meijer EW, Langer R. Nat Mater. 2015;15:13–26. doi: 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- 75.Feynman RP. Eng Sci. 1960;23:22–36. [Google Scholar]
- 76.Huang Y, Duan X, Wei Q, Lieber CM. Science. 2001;291:630–633. doi: 10.1126/science.291.5504.630. [DOI] [PubMed] [Google Scholar]
- 77.Chen X, Mao SS. Chem Rev. 2007;107:2891–2959. doi: 10.1021/cr0500535. [DOI] [PubMed] [Google Scholar]
- 78.Dabbousi B, Rodriguez-Viejo J, Mikulec FV, Heine J, Mattoussi H, Ober R, Jensen K, Bawendi M. J Phys Chem B. 1997;101:9463–9475. [Google Scholar]
- 79.Edelstein AS, Cammaratra R. Nanomaterials: synthesis, properties and applications. CRC Press; 1998. [Google Scholar]
- 80.Murray CB, Sun S, Gaschler W, Doyle H, Betley TA, Kagan CR. IBM J Res Dev. 2001;45:47–56. [Google Scholar]
- 81.Rao CNR, Müller A, Cheetham AK. The chemistry of nanomaterials: synthesis, properties and applications. John Wiley & Sons; 2006. [Google Scholar]
- 82.Israelachvili JN. Intermolecular and surface forces. 3. Academic Press; Burlington, MA: 2011. [Google Scholar]
- 83.Min Y, Akbulut M, Kristiansen K, Golan Y, Israelachvili J. Nat Mater. 2008;7:527–538. doi: 10.1038/nmat2206. [DOI] [PubMed] [Google Scholar]
- 84.Koenig SP, Boddeti NG, Dunn ML, Bunch JS. Nat Nanotechnol. 2011;6:543–546. doi: 10.1038/nnano.2011.123. [DOI] [PubMed] [Google Scholar]
- 85.Roduner E. Chem Soc Rev. 2006;35:583–592. doi: 10.1039/b502142c. [DOI] [PubMed] [Google Scholar]
- 86.Goesmann H, Feldmann C. Angew Chem Int Ed. 2010;49:1362–1395. doi: 10.1002/anie.200903053. [DOI] [PubMed] [Google Scholar]
- 87.Couchman P, Jesser W. Nature. 1977;269:481–483. [Google Scholar]
- 88.Lai SL, Guo JY, Petrova VV, Ramanath G, Allen LH. Phys Rev Lett. 1996;77:99–102. doi: 10.1103/PhysRevLett.77.99. [DOI] [PubMed] [Google Scholar]
- 89.Thomson W. Proc R Soc Edinburgh. 1872;7:63–68. [Google Scholar]
- 90.Ahn BY, Duoss EB, Motala MJ, Guo X, Park SI, Xiong Y, Yoon J, Nuzzo RG, Rogers JA, Lewis JA. Science. 2009;323:1590–1593. doi: 10.1126/science.1168375. [DOI] [PubMed] [Google Scholar]
- 91.Faraday M. Phil Trans R Soc Lond. 1857;147:145–181. [Google Scholar]
- 92.Alivisatos AP. Science. 1996;271:933–937. [Google Scholar]
- 93.Eychmüller A. J Phys Chem B. 2000;104:6514–6528. [Google Scholar]
- 94.Witten T, Pincus P. Macromolecules. 1986;19:2509–2513. [Google Scholar]
- 95.Li D, Muller MB, Gilje S, Kaner RB, Wallace GG. Nat Nanotechnol. 2008;3:101–105. doi: 10.1038/nnano.2007.451. [DOI] [PubMed] [Google Scholar]
- 96.Kamyshny A, Magdassi S. Small. 2014;10:3515–3535. doi: 10.1002/smll.201303000. [DOI] [PubMed] [Google Scholar]
- 97.Sun Y, Xia Y. Science. 2002;298:2176–2179. doi: 10.1126/science.1077229. [DOI] [PubMed] [Google Scholar]
- 98.Hecht DS, Hu L, Irvin G. Adv Mater. 2011;23:1482–1513. doi: 10.1002/adma.201003188. [DOI] [PubMed] [Google Scholar]
- 99.Chopra K, Major S, Pandya D. Thin Solid Films. 1983;102:1–46. [Google Scholar]
- 100.Bonaccorso F, Sun Z, Hasan T, Ferrari A. Nat Photon. 2010;4:611–622. [Google Scholar]
- 101.Eda G, Fanchini G, Chhowalla M. Nat Nanotechnol. 2008;3:270–274. doi: 10.1038/nnano.2008.83. [DOI] [PubMed] [Google Scholar]
- 102.Li Y, Wu Y, Ong BS. J Am Chem Soc. 2005;127:3266–3267. doi: 10.1021/ja043425k. [DOI] [PubMed] [Google Scholar]
- 103.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994:801–802. [Google Scholar]
- 104.Brust M, Fink J, Bethell D, Schiffrin D, Kiely C. J Chem Soc, Chem Commun. 1995:1655–1656. [Google Scholar]
- 105.Campbell T, Kalia RK, Nakano A, Vashishta P, Ogata S, Rodgers S. Phys Rev Lett. 1999;82:4866. [Google Scholar]
- 106.Novoselov KS, Jiang D, Schedin F, Booth TJ, Khotkevich VV, Morozov SV, Geim AK. Proc Nat Acad Sci USA. 2005;102:10451–10453. doi: 10.1073/pnas.0502848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Geim AK, Novoselov KS. Nat Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 108.Geim AK. Science. 2009;324:1530–1534. doi: 10.1126/science.1158877. [DOI] [PubMed] [Google Scholar]
- 109.Bolotin KI, Sikes K, Jiang Z, Klima M, Fudenberg G, Hone J, Kim P, Stormer H. Solid State Commun. 2008;146:351–355. [Google Scholar]
- 110.Castro Neto AH, Guinea F, Peres NMR, Novoselov KS, Geim AK. Rev Mod Phys. 2009;81:109–162. [Google Scholar]
- 111.Wang S, Ang PK, Wang Z, Tang AL, Thong JT, Loh KP. Nano Lett. 2010;10:92–98. doi: 10.1021/nl9028736. [DOI] [PubMed] [Google Scholar]
- 112.Chun KY, Oh Y, Rho J, Ahn JH, Kim YJ, Choi HR, Baik S. Nat Nanotechnol. 2010;5:853–857. doi: 10.1038/nnano.2010.232. [DOI] [PubMed] [Google Scholar]
- 113.Anikeeva PO, Halpert JE, Bawendi MG, Bulović V. Nano Lett. 2007;7:2196–2200. doi: 10.1021/nl0703424. [DOI] [PubMed] [Google Scholar]
- 114.Chen O, Zhao J, Chauhan VP, Cui J, Wong C, Harris DK, Wei H, Han HS, Fukumura D, Jain RK, Bawendi MG. Nat Mater. 2013;12:445–451. doi: 10.1038/nmat3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shirasaki Y, Supran GJ, Bawendi MG, Bulovic V. Nat Photon. 2013;7:13–23. [Google Scholar]
- 116.Austin LA, Kang B, El-Sayed MA. Nano Today. 2015;10:542–558. [Google Scholar]
- 117.Gur I, Fromer NA, Geier ML, Alivisatos AP. Science. 2005;310:462–465. doi: 10.1126/science.1117908. [DOI] [PubMed] [Google Scholar]
- 118.McDonald SA, Konstantatos G, Zhang S, Cyr PW, Klem EJ, Levina L, Sargent EH. Nat Mater. 2005;4:138–142. doi: 10.1038/nmat1299. [DOI] [PubMed] [Google Scholar]
- 119.Graetzel M, Janssen RA, Mitzi DB, Sargent EH. Nature. 2012;488:304–312. doi: 10.1038/nature11476. [DOI] [PubMed] [Google Scholar]
- 120.Yang J, Choi MK, Kim DH, Hyeon T. Adv Mater. 2015 doi: 10.1002/adma.201502851. [DOI] [PubMed] [Google Scholar]
- 121.Qian L, Zheng Y, Xue J, Holloway PH. Nat Photon. 2011;5:543–548. [Google Scholar]
- 122.Supran GJ, Shirasaki Y, Song KW, Caruge JM, Kazlas PT, Coe-Sullivan S, Andrew TL, Bawendi MG, Bulović V. MRS Bull. 2013;38:703–711. [Google Scholar]
- 123.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Nature. 1997;389:827–829. doi: 10.1103/physreve.62.756. [DOI] [PubMed] [Google Scholar]
- 124.Deegan RD. Phys Rev E. 2000;61:475–485. doi: 10.1103/physreve.61.475. [DOI] [PubMed] [Google Scholar]
- 125.Shmuylovich L, Shen AQ, Stone HA. Langmuir. 2002;18:3441–3445. [Google Scholar]
- 126.Ryan KM, Mastroianni A, Stancil KA, Liu H, Alivisatos AP. Nano Lett. 2006;6:1479–1482. doi: 10.1021/nl060866o. [DOI] [PubMed] [Google Scholar]
- 127.Tsai SSH, Wexler JS, Wan J, Stone HA. Appl Phys Lett. 2011;99:153509. [Google Scholar]
- 128.Park J, Moon J. Langmuir. 2006;22:3506–3513. doi: 10.1021/la053450j. [DOI] [PubMed] [Google Scholar]
- 129.Harris DJ, Lewis JA. Langmuir. 2008;24:3681–3685. doi: 10.1021/la8000637. [DOI] [PubMed] [Google Scholar]
- 130.Hu H, Larson RG. J Phys Chem B. 2006;110:7090–7094. doi: 10.1021/jp0609232. [DOI] [PubMed] [Google Scholar]
- 131.Majumder M, Rendall CS, Eukel JA, Wang JYL, Behabtu N, Pint CL, Liu TY, Orbaek AW, Mirri F, Nam J, Barron AR, Hauge RH, Schmidt HK, Pasquali M. J Phys Chem B. 2012;116:6536–6542. doi: 10.1021/jp3009628. [DOI] [PubMed] [Google Scholar]
- 132.Still T, Yunker PJ, Yodh AG. Langmuir. 2012;28:4984–4988. doi: 10.1021/la204928m. [DOI] [PubMed] [Google Scholar]
- 133.Cui L, Zhang J, Zhang X, Huang L, Wang Z, Li Y, Gao H, Zhu S, Wang T, Yang B. ACS Appl Mater Interfaces. 2012;4:2775–2780. doi: 10.1021/am300423p. [DOI] [PubMed] [Google Scholar]
- 134.Soltman D, Subramanian V. Langmuir. 2008;24:2224–2231. doi: 10.1021/la7026847. [DOI] [PubMed] [Google Scholar]
- 135.Yunker PJ, Still T, Lohr MA, Yodh AG. Nature. 2011;476:308–311. doi: 10.1038/nature10344. [DOI] [PubMed] [Google Scholar]
- 136.Bigioni TP, Lin XM, Nguyen TT, Corwin EI, Witten TA, Jaeger HM. Nat Mater. 2006;5:265–270. doi: 10.1038/nmat1611. [DOI] [PubMed] [Google Scholar]
- 137.Shimoni A, Azoubel S, Magdassi S. Nanoscale. 2014;6:11084–11089. doi: 10.1039/c4nr02133a. [DOI] [PubMed] [Google Scholar]
- 138.Xu J, Xia J, Lin Z. Angew Chem Int Ed. 2007;46:1860–1863. doi: 10.1002/anie.200604540. [DOI] [PubMed] [Google Scholar]
- 139.Watanabe S, Inukai K, Mizuta S, Miyahara MT. Langmuir. 2009;25:7287–7295. doi: 10.1021/la900315h. [DOI] [PubMed] [Google Scholar]
- 140.Abkarian M, Nunes J, Stone HA. J Am Chem Soc. 2004;126:5978–5979. doi: 10.1021/ja049775o. [DOI] [PubMed] [Google Scholar]
- 141.Kong YL, Boulogne F, Kim H, Nunes J, Feng J, Stone HA. Langmuir. 2015;31:12560–12566. doi: 10.1021/acs.langmuir.5b03443. [DOI] [PubMed] [Google Scholar]
- 142.Kim HS, Lee CH, Sudeep PK, Emrick T, Crosby AJ. Adv Mater. 2010;22:4600–4604. doi: 10.1002/adma.201001892. [DOI] [PubMed] [Google Scholar]
- 143.Wang L, Lau J, Thomas EL, Boyce MC. Adv Mater. 2011;23:1524–1529. doi: 10.1002/adma.201003956. [DOI] [PubMed] [Google Scholar]
- 144.Dimas LS, Bratzel GH, Eylon I, Buehler MJ. Adv Funct Mater. 2013;23:4629–4638. [Google Scholar]
- 145.Schaedler TA, Jacobsen AJ, Torrents A, Sorensen AE, Lian J, Greer JR, Valdevit L, Carter WB. Science. 2011;334:962–965. doi: 10.1126/science.1211649. [DOI] [PubMed] [Google Scholar]
- 146.Meza LR, Das S, Greer JR. Science. 2014;345:1322–1326. doi: 10.1126/science.1255908. [DOI] [PubMed] [Google Scholar]
- 147.Gratson GM, García-Santamaría F, Lousse V, Xu M, Fan S, Lewis JA, Braun PV. Adv Mater. 2006;18:461–465. [Google Scholar]
- 148.Clausen A, Wang FW, Jensen JS, Sigmund O, Lewis JA. Adv Mater. 2015;27:5523–5527. doi: 10.1002/adma.201502485. [DOI] [PubMed] [Google Scholar]
- 149.Shan S, Kang SH, Raney JR, Wang P, Fang L, Candido F, Lewis JA, Bertoldi K. Adv Mater. 2015;27:4296–4301. doi: 10.1002/adma.201501708. [DOI] [PubMed] [Google Scholar]
- 150.Hansen CJ, Saksena R, Kolesky DB, Vericella JJ, Kranz SJ, Muldowney GP, Christensen KT, Lewis JA. Adv Mater. 2013;25:96–102. doi: 10.1002/adma.201203321. [DOI] [PubMed] [Google Scholar]
- 151.Biondi M, Ungaro F, Quaglia F, Netti PA. Adv Drug Deliv Rev. 2008;60:229–242. doi: 10.1016/j.addr.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 152.Griffith LG, Swartz MA. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 153.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen FM, Zhang M, Wu ZF. Biomaterials. 2010;31:6279–6308. doi: 10.1016/j.biomaterials.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 155.Keenan TM, Folch A. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lutolf MP, Blau HM. Adv Mater. 2009;21:3255–3268. doi: 10.1002/adma.200802582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U. Adv Mater. 2013;25:1192–1198. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ilkhanizadeh S, Teixeira AI, Hermanson O. Biomaterials. 2007;28:3936–3943. doi: 10.1016/j.biomaterials.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 159.Ker ED, Chu B, Phillippi JA, Gharaibeh B, Huard J, Weiss LE, Campbell PG. Biomaterials. 2011;32:3413–3422. doi: 10.1016/j.biomaterials.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ker ED, Nain AS, Weiss LE, Wang J, Suhan J, Amon CH, Campbell PG. Biomaterials. 2011;32:8097–8107. doi: 10.1016/j.biomaterials.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 161.Miller ED, Li K, Kanade T, Weiss LE, Walker LM, Campbell PG. Biomaterials. 2011;32:2775–2785. doi: 10.1016/j.biomaterials.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Miller ED, Phillippi JA, Fisher GW, Campbell PG, Walker LM, Weiss LE. Comb Chem High Throughput Screen. 2009;12:604–618. doi: 10.2174/138620709788681907. [DOI] [PubMed] [Google Scholar]
- 163.Zhang H, Shepherd JNH, Nuzzo RG. Soft Matter. 2010;6:2238–2245. [Google Scholar]
- 164.Chien MP, Rush AM, Thompson MP, Gianneschi NC. Angew Chem Int Ed. 2010;49:5076–5080. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee MH, Hribar KC, Brugarolas T, Kamat NP, Burdick JA, Lee D. Adv Funct Mater. 2012;22:131–138. [Google Scholar]
- 166.Mohamed F, van der Walle CF. J Pharm Sci. 2008;97:71–87. doi: 10.1002/jps.21082. [DOI] [PubMed] [Google Scholar]
- 167.Shah RK, Shum HC, Rowat AC, Lee D, Agresti JJ, Utada AS, Chu LY, Kim JW, Fernandez-Nieves A, Martinez CJ. Mater Today. 2008;11:18–27. [Google Scholar]
- 168.Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 169.Xu S, Nie Z, Seo M, Lewis P, Kumacheva E, Stone HA, Garstecki P, Weibel DB, Gitlin I, Whitesides GM. Angew Chem Int Ed. 2005;117:734–738. doi: 10.1002/anie.200462226. [DOI] [PubMed] [Google Scholar]
- 170.Shah RK, Kim JW, Weitz DA. Langmuir. 2010;26:1561–1565. doi: 10.1021/la9041327. [DOI] [PubMed] [Google Scholar]
- 171.Tian L, Gandra N, Singamaneni S. ACS Nano. 2013;7:4252–4260. doi: 10.1021/nn400728t. [DOI] [PubMed] [Google Scholar]
- 172.Timko BP, Arruebo M, Shankarappa SA, McAlvin JB, Okonkwo OS, Mizrahi B, Stefanescu CF, Gomez L, Zhu J, Zhu A, Santamaria J, Langer R, Kohane DS. Proc Nat Acad Sci USA. 2014;111:1349–1354. doi: 10.1073/pnas.1322651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang Y, Byrne JD, Napier ME, DeSimone JM. Adv Drug Deliv Rev. 2012;64:1021–1030. doi: 10.1016/j.addr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Luo Y, Shoichet MS. Nat Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 175.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Nat Mater. 2007;6:908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 176.Kobel S, Lutolf MP. Curr Opin Biotechnol. 2011;22:690–697. doi: 10.1016/j.copbio.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 177.Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, Angelini TE. Sci Adv. 2015;1:e1500655. doi: 10.1126/sciadv.1500655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. Adv Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 179.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ibrahim D, Broilo TL, Heitz C, de Oliveira MG, de Oliveira HW, Nobre SMW, dos Santos Filho JHG, Silva DN. J Craniomaxillofac Surg. 2009;37:167–173. doi: 10.1016/j.jcms.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 181.Duan B, Kapetanovic E, Hockaday LA, Butcher JT. Acta Biomater. 2014;10:1836–1846. doi: 10.1016/j.actbio.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Temple JP, Hutton DL, Hung BP, Huri PY, Cook CA, Kondragunta R, Jia X, Grayson WL. Journal of Biomedical Materials Research Part A. 2014;102:4317–4325. doi: 10.1002/jbm.a.35107. [DOI] [PubMed] [Google Scholar]
- 183.Wen L, Weaver JC, Lauder GV. J Exp Biol. 2014;217:1656–1666. doi: 10.1242/jeb.097097. [DOI] [PubMed] [Google Scholar]
- 184.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Trends Biotechnol. 2003;21:157–161. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 185.Seitz H, Rieder W, Irsen S, Leukers B, Tille C. J Biomed Mater Res B Appl Biomater. 2005;74:782–788. doi: 10.1002/jbm.b.30291. [DOI] [PubMed] [Google Scholar]
- 186.Qin Z, Compton BG, Lewis JA, Buehler MJ. Nat Commun. 2015;6:7038. doi: 10.1038/ncomms8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ahn BY, Lorang DJ, Lewis JA. Nanoscale. 2011;3:2700–2702. doi: 10.1039/c1nr10048c. [DOI] [PubMed] [Google Scholar]
- 188.Piner RD, Zhu J, Xu F, Hong S, Mirkin CA. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 189.Chiechi RC, Weiss EA, Dickey MD, Whitesides GM. Angew Chem Int Ed. 2008;47:142–144. doi: 10.1002/anie.200703642. [DOI] [PubMed] [Google Scholar]
- 190.Boley JW, White EL, Chiu GTC, Kramer RK. Adv Funct Mater. 2014;24:3501–3507. [Google Scholar]
- 191.Adams JJ, Duoss EB, Malkowski TF, Motala MJ, Ahn BY, Nuzzo RG, Bernhard JT, Lewis JA. Adv Mater. 2011;23:1335–1340. doi: 10.1002/adma.201003734. [DOI] [PubMed] [Google Scholar]
- 192.Muth JT, Vogt DM, Truby RL, Menguc Y, Kolesky DB, Wood RJ, Lewis JA. Adv Mater. 2014;26:6307–6312. doi: 10.1002/adma.201400334. [DOI] [PubMed] [Google Scholar]
- 193.Lee MS, Lee K, Kim SY, Lee H, Park J, Choi KH, Kim HK, Kim DG, Lee DY, Nam S, Park JU. Nano Lett. 2013;13:2814–2821. doi: 10.1021/nl401070p. [DOI] [PubMed] [Google Scholar]
- 194.Yao H, Shum AJ, Cowan M, Lahdesmaki I, Parviz BA. Biosens Bioelectron. 2011;26:3290–3296. doi: 10.1016/j.bios.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bandodkar AJ, Wang J. Trends Biotechnol. 2014;32:363–371. doi: 10.1016/j.tibtech.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 196.Sun K, Wei TS, Ahn BY, Seo JY, Dillon SJ, Lewis JA. Adv Mater. 2013;25:4539–4543. doi: 10.1002/adma.201301036. [DOI] [PubMed] [Google Scholar]
- 197.Franklin AD. Science. 2015;349:aab2750. doi: 10.1126/science.aab2750. [DOI] [PubMed] [Google Scholar]
- 198.Sargent EH. Adv Mater. 2008;20:3958–3964. [Google Scholar]
- 199.Pattantyus-Abraham AG, Kramer IJ, Barkhouse AR, Wang X, Konstantatos G, Debnath R, Levina L, Raabe I, Nazeeruddin MK, Gratzel M, Sargent EH. ACS Nano. 2010;4:3374–3380. doi: 10.1021/nn100335g. [DOI] [PubMed] [Google Scholar]
- 200.Goßler C, Bierbrauer C, Moser R, Kunzer M, Holc K, Pletschen W, Köhler K, Wagner J, Schwaerzle M, Ruther P, Paul O, Neef J, Keppeler D, Hoch G, Moser T, Schwarz UT. J Phys D: Appl Phys. 2014;47:205401. [Google Scholar]