This article reviews the key considerations involved in clinical decision making while reviewing a molecular genotyping report. The article presents the case of a 67-year-old postmenopausal female with metastatic estrogen receptor-positive breast cancer, whose tumor progressed on multiple endocrine therapies.
Abstract
The last decade in oncology has witnessed impressive response rates with targeted therapies, largely because of collaborative efforts at understanding tumor biology and careful patient selection based on molecular fingerprinting of the tumor. Consequently, there has been a push toward routine molecular genotyping of tumors, and large precision medicine-based clinical trials have been launched to match therapy to the molecular alteration seen in a tumor. However, selecting the “right drug” for an individual patient in clinic is a complex decision-making process, including analytical interpretation of the report, consideration of the importance of the molecular alteration in driving growth of the tumor, tumor heterogeneity, the availability of a matched targeted therapy, efficacy and toxicity considerations of the targeted therapy (compared with standard therapy), and reimbursement issues. In this article, we review the key considerations involved in clinical decision making while reviewing a molecular genotyping report. We present the case of a 67-year-old postmenopausal female with metastatic estrogen receptor-positive (ER+) breast cancer, whose tumor progressed on multiple endocrine therapies. Molecular genotyping of the metastatic lesion revealed the presence of an ESR1 mutation (encoding p.Tyr537Asn), which was absent in the primary tumor. The same ESR1 mutation was also detected in circulating tumor DNA (ctDNA) extracted from her blood. The general approach for interpretation of genotyping results, the clinical significance of the specific mutation in the particular cancer, potential strategies to target the pathway, and implications for clinical practice are reviewed in this article.
Key Points
ER+ breast tumors are known to undergo genomic evolution during treatment with the acquisition of new mutations that confer resistance to treatment.
ESR1 mutations in the ligand-binding domain of ER can lead to a ligand-independent, constitutively active form of ER and mediate resistance to aromatase inhibitors.
ESR1 mutations may be detected by genomic sequencing of tissue biopsies of the metastatic tumor or by sequencing the circulating tumor cells or tumor DNA (ctDNA).
Sequencing results may lead to a therapeutic “match” with an existing FDA-approved drug or match with an experimental agent that fits the clinical setting.
Abstract
摘要
肿瘤学在过去十年里见证了靶向疗法惊人的缓解率, 这很大程度上是缘于对肿瘤生物学的深入理解和基于肿瘤分子指纹图谱对患者进行仔细选择的通力协作。继而, 肿瘤分子基因分型的常规检测得到推动, 研究者开展了大量以精确医学为基础的临床试验以匹配治疗与肿瘤中的分子改变。但是在临床上为一名患者选择“正确的药物”是一个复杂的决策制定过程, 包括对报告进行分析性解读、考虑分子改变在驱动肿瘤生长中的重要性、肿瘤异质性、可用的匹配靶向治疗、靶向治疗的有效性和安全性考量 (与标准治疗相比) 以及费用报销问题。作者在本文中回顾了一个分子基因分型案例报告, 同时综述了临床决策制定过程中涉及的关键问题。该案例为一名罹患雌激素受体阳性 (ER+) 乳腺癌的67岁绝经后女性, 在接受多种内分泌治疗后肿瘤出现进展。转移灶的分子基因分型提示存在ESR1突变 (编码p.Tyr537Asn), 而原发肿瘤中不存在该突变。从血中提取的循环肿瘤DNA (ctDNA) 中也检测到同样的ESR1突变。本文对基因分型结果的一般解释方法、特定肿瘤中特异性突变的临床意义、针对通路进行干预的可能策略以及对临床实践的提示进行了综述。The Oncologist 2016;21:1035–1040
关键点
• 已知 ER+乳腺肿瘤在治疗过程中会发生基因组演变, 获得使肿瘤对治疗产生耐药的新突变。
• ER 配体结合域的 ESR1 突变可导致 ER 发生非配体依赖的结构活性, 介导芳香化酶抑制剂耐药的发生。
• 对转移肿瘤的组织活检标本进行基因组测序, 或者对循环肿瘤细胞或肿瘤 DNA (ctDNA) 进行测序, 可能检测到 ESR1 突变。
• 测序结果也许能够找到“匹配”的现有 FDA 批准的药物或者适用于临床环境的实验性制剂。
Patient Story
A 67-year-old female was diagnosed with metastatic breast cancer on the basis of a pleural biopsy. Her original breast cancer, diagnosed at the age of 54, was treated with neoadjuvant chemotherapy followed by surgery and adjuvant tamoxifen for ER+ breast cancer. Three years later, she noted subacute onset of rib pain. Restaging computed tomography scans revealed a pleural, extrapulmonary mass in the lower right lung. She underwent a pleural biopsy, which revealed adenocarcinoma, consistent with a breast primary. The tumor was ER+/human epidermal growth receptor 2-negative (HER−), similar to the original tumor at the time of surgery. She was started on letrozole and had stable disease for approximately 8 years, when restaging scans revealed progressive disease in the lung with an increase in pulmonary nodules and hilar lymph nodes. She underwent a bronchoscopy and transbronchial biopsy. Surgical pathology revealed metastatic carcinoma consistent with a breast primary. The tumor was ER+/HER−. The specimen was also sent for molecular genotyping. Her endocrine therapy was then switched to fulvestrant and then later to vinorelbine, both of which were discontinued because of disease progression. She was seen in medical oncology at our institution for discussion of treatment options, including consideration of clinical trials. The molecular genotyping report revealed the presence of a mutation in ESR1, the gene encoding the estrogen receptor. The molecular report and potential consideration for clinical management are reviewed in this article. The case was discussed at a Massachusetts General Hospital Molecular and Precision Medicine (MAP) tumor board meeting in 2015.
Molecular Tumor Board
With the remarkable success of imatinib targeting BCR-ABL in chronic lymphocytic leukemia, over the past decade there has been an explosion in the development of targeted therapies. A joint effort at understanding tumor biology and careful patient selection based on molecular fingerprinting of the tumor has led to impressive response rates in selected patients and has paved the unprecedented path for accelerated approval based on phase I results, such as crizotinib for ALK-positive and ROS1-rearranged lung cancer [1]. Consequently, large precision medicine-based clinical trials, such as National Cancer Institute-MATCH (Molecular Analysis for Therapy Choice) and TAPUR (Targeted Agent and Profiling Utilization Registry), have been launched to match therapy to the molecular alteration seen in the tumor [2]. Even at a federal level, there has been a tremendous push toward precision medicine: U.S. President Barack Obama launched a precision medicine initiative in 2015 and, more recently, U.S. Vice-President Joe Biden proposed a cancer moon-shot program in 2016 [3, 4]. However, selecting the “right drug” for an individual patient in clinic is a complex decision-making process, including analytical interpretation of the molecular findings, consideration of the importance of the molecular alteration in driving growth of the tumor, tumor heterogeneity, the availability of matched targeted therapy, efficacy and toxicity of targeted therapy compared with standard therapy, and reimbursement issues. The key considerations involved in clinical decision making are outlined in Table 1 and reviewed below, using the index case as an example.
Table 1.
Five considerations for clinical management while reviewing a molecular alteration report in clinic
Genotyping Results and General Interpretation
Analytical Considerations (Can I Trust the Results?)
When reviewing a genotyping report, like any test, the first question to ask is, “Can I trust the results?” because “a bad tumor biomarker test is as bad as a bad drug” [5]. Important analytical considerations include whether the test was performed in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory, the method used for sequencing, the sensitivity, the specificity, and genotyping metrics such as number of genes assayed and the depth of sequencing [6, 7]. Let us review the analytical considerations of this report.
In this case, the molecular profiling was performed by an institutional laboratory-developed test, “Snapshot-next-generation sequencing assay” (Snapshot-NGS), performed in a CLIA-certified laboratory. The Snapshot-NGS assay uses a multiplex polymerase chain reaction (PCR) technology for single nucleotide variant (SNV) and insertion/deletion (indel) detection in genomic DNA using NGS [8]. Briefly, genomic DNA is isolated from a formalin-fixed, paraffin-embedded tumor specimen (after histological review for tumor enrichment). A sequencing library targets suspected hotspot mutations in the coding region (exons) of 39 genes and is validated to detect SNV and indel variants at 5% allelic frequency or higher in target regions with sufficient read coverage.
Interpretation of the Molecular Results (What Does the Molecular Alteration Mean?)
Another important issue to consider is the specific type of molecular alteration detected (see Glossary of Genomic Terms and Nomenclature). Is it a pathogenic point mutation (single base alteration) known to affect the function of the protein encoded by the gene, an insertion/deletion that alters the coding sequence, a gene amplification, or a variant of unknown significance? Other issues to consider include allelic fraction and clonality. Because the tumor content in a pathology specimen can vary, the comparison of tumor content with allelic fraction can provide general information about whether the specific molecular alteration is clonal (present in all the tumor cells) versus subclonal (present only in a subset of tumor cells).
For example, if tumor cells are 60% of the cells in a specimen and a heterozygous mutation (occurring in one strand of the DNA duplex) is present in all tumor cells, the expected allelic frequency for the mutation would be 30% (one of the two alleles is mutated in each tumor cell). It is important to keep in mind that allelic frequency is affected by copy number changes (chromosome or gene gains and losses) and that it can be skewed by preferential amplification of certain molecules during the PCRs for target enrichment before sequencing, by sequencing errors that may result in reads being filtered out/discarded, by errors in mapping sequencing reads to the correct genomic location, and by specimen/nucleic acid quality. With these caveats, it is critical for an oncologist to know whether a mutation is subclonal in that it is detected at low allelic fraction despite a high tumor cellularity because targeted agents may not be as effective.
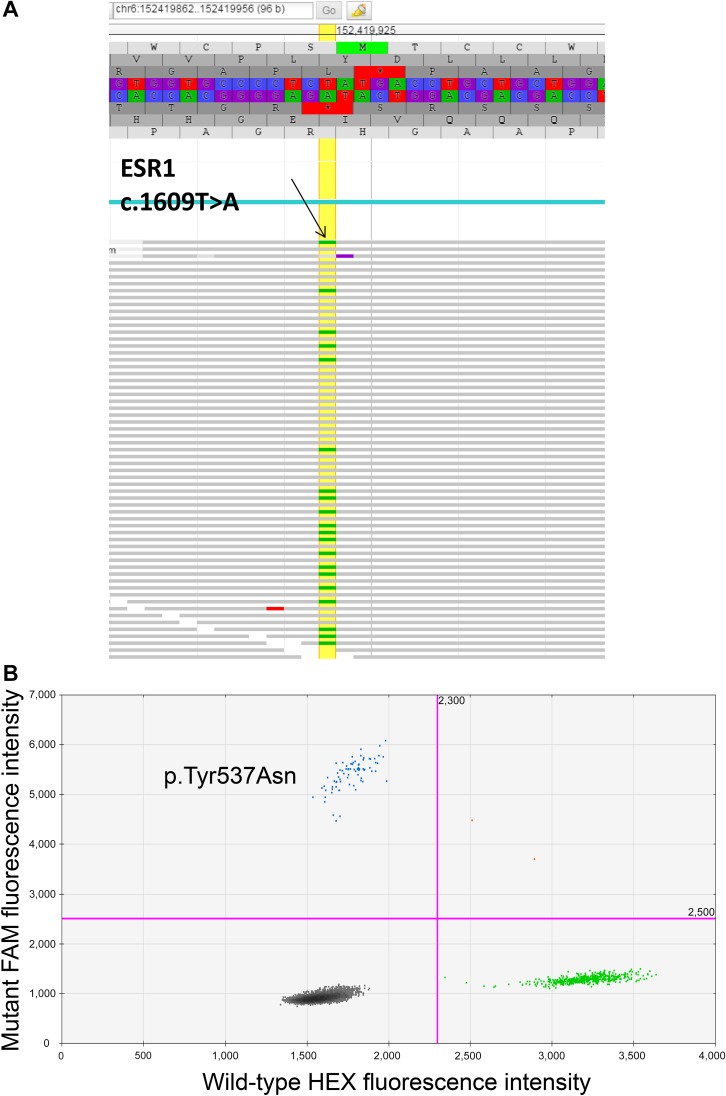
For this patient, the genotyping report revealed: ESR1: p.Tyr537Asn (ENST00000440973.1:c.1609T>A). The report, consistent with protein and coding DNA nomenclature by Human Genome Variation Society [9], indicates a molecular alteration in ESR1 (the gene encoding the estrogen receptor), with a missense mutation in which a thymine nucleotide is substituted with an adenine nucleotide at position 1,609 in the DNA sequence (c.1609T>A), resulting in a change in the encoded amino acid from a tyrosine to an asparagine at position/codon 537 (p.Tyr537Asn), as outlined in Figure 1A.
Figure 1.
Tumor genotyping results from tissue and blood, respectively. (A): Screenshot of the ESR1 mutation visualized in JBrowse. Upper: The hg19 reference genome sequence (and corresponding amino acid translation) for ESR1 codons 533–541. The horizontal gray rows indicate individual reads (i.e., individual molecules) sequenced in the forward direction. The small green bars indicate a nucleotide change (single nucleotide variant [SNV]) from the reference nucleotide T to A. The yellow vertical line marks the ESR1 nucleotides at position 1,609 in the coding sequence where an SNV was detected. Labeled arrow is pointing to an example of the presence of thymidine (T) as compared with adenine (A). (B): Droplet digital polymerase chain reaction analysis demonstrating an ESR1 p.Tyr537Asn mutation in circulating tumor DNA isolated from the patient described in this article. Blue dots represent droplets containing FAM-labeled mutant probes hybridized to mutant DNA. Green dots represent HEX-labeled wild-type (WT) probes hybridized to WT DNA. Black dots represent droplets containing both mutant and WT DNA.
Abbreviations: FAM, 5′-fluorescein amidite; HEX, hexachloro-fluorescein.
Significance of the Specific Mutation in the Particular Cancer
Functional Significance of the Molecular Alteration (How Does the Molecular Alteration Impact Tumor Biology?)
The next issue to consider is the functional and potential clinical significance of the molecular alteration in the context of breast cancer [10]. Is it a driver versus passenger mutation? Is it a known hot-spot mutation? What is the frequency of the mutation in that specific cancer? Is it a primary or an acquired mutation?
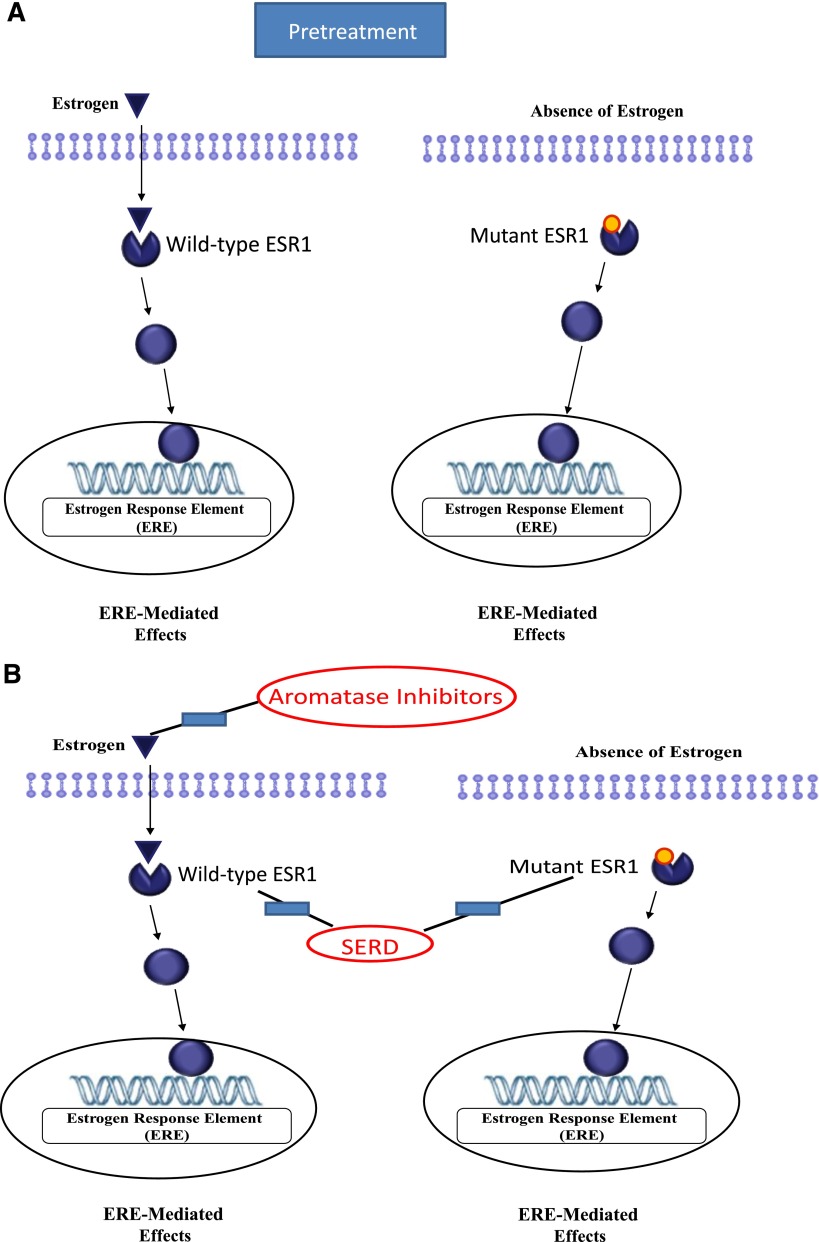
In this case, the ESR1 mutation appears to be a driver mutation because the molecular alteration occurs in the ligand-binding domain of the estrogen receptor and leads to a constitutively active form of ER that becomes ligand (or estrogen)-independent (Fig. 2A) [11]. Aromatase inhibitors (AIs) inhibit breast cancer growth by lowering estrogens, and therefore ESR1 mutations confer resistance to AIs because they allow tumors to proliferate independent of estrogen. Indeed, despite having five molecular platforms including whole-exome sequencing, mRNA expression microarrays, DNA methylation chips, Affymetrix (SNP) arrays (n5773), and miRNA sequencing, ESR1 mutations were not even mentioned in the breast cancer sequencing by TCGA [12], possibly because the molecular analysis was done on primary breast specimens. More recently, publications have reported the presence of ESR1 mutations in 17.5%–54.4% of post-AI metastatic specimens [13–16]. Because ESR1 mutations appear to be acquired alterations, the frequency of the mutation varies based on a number of factors, including prior endocrine therapies, specific specimen used for molecular analysis, and tumor burden [16].
Figure 2.
Targeting the estrogen receptor pathway. (A): In the presence of wild-type estrogen receptor (ESR1), estrogen binds to ESR1, leading to change to agonist conformation, which binds to the estrogen response element in the DNA strand leading to transcription of multiple genes mediating ERE-induced effects, including cellular proliferation. (B): Estrogen receptor (ER)-mediated effects can potentially be inhibited by targeting the ligand (estrogen) as done by aromatase inhibitors or targeting the receptor as done by selective ER degraders. The presence of the ESR1 mutation in the ligand-binding domain results in a constitutive active form of ER that does not require ligand (or estrogen) for activation. These cells are therefore resistant to aromatase inhibitors but could potentially be targeted by SERDs.
Abbreviations: ERE, estrogen response element; SERD, selective ER degrader.
Given the difficulties with obtaining multiple metastatic tissue biopsies, serial monitoring of mutations by blood-based assays (“liquid biopsies”) might play an important role in detection of acquired ESR1 mutations. For example, a recent study of patients with known metastatic ER+ breast cancer reported the presence of ESR1 mutations in 6 of 12 patients (50%) using a blood-based circulating tumor DNA assay, whereas no ESR1 mutations were identified in tissue biopsies of the primary specimens [17]. Similarly, another study using sequencing of ex vivo cultures from circulating tumor cells (CTCs) reported ESR1 mutations in 3/6 patient-derived CTC lines (50%), and reanalysis of the primary tumor or the pre-AI treatment biopsy of a metastatic lesion showed no evidence of ESR1 mutations [15]. All of these patients had received extensive treatment with AIs, which potentially led to the acquired ESR1 mutation.
Our patient had prolonged exposure to aromatase inhibitors (>8 years), which likely allowed for selection of the ESR1 mutation. Indeed, genotyping of the primary tumor by the institutional Snapshot-NGS assay did not identify the ESR1 mutation in the primary tumor, supporting the hypothesis that it was likely an acquired mutation. ctDNA isolated from our patient during the course of her metastatic disease management was analyzed by using a droplet digital PCR-based method. Briefly, ctDNA is extracted from patient blood and combined with probes designed to detect the presence of ESR1 mutations in thousands of water-oil emulsion droplets, permitting highly sensitive mutation detection. By using this method, the same ESR1 mutation that was detected in the patient’s metastatic tissue biopsy was also detected in ctDNA extracted from her blood (Fig. 1B).
Clinical Significance of the Molecular Alteration (How Does the Molecular Alteration Impact Response to Standard Therapies?)
The next issue to consider is the clinical significance of the mutation and its impact on response to standard therapies. This is particularly an important consideration in ER+ breast cancer, where multiple therapy options are available. One has to make a choice of a standard therapy option versus experimental option. For example, on the basis of results from BOLERO-2 clinical trial [18], a standard option to consider for AI-resistant tumors would be exemestane and everolimus (mammalian target of rapamycin inhibitor). Accordingly, the question to consider is whether the presence of an ESR1 mutation would have any impact on efficacy of this regimen? This question was recently addressed by Chandralapaty et al. using plasma specimens from the BOLERO-2 trial [19]. The authors reported that the addition of everolimus to exemestane did not improve progression-free survival (PFS) in those with Y537 ESR1 mutations (4.1 versus 4.2 months), but led to improvement in those with D538 ESR1 mutations (2.7 versus 5.8 months), suggesting a differential impact dependent on the type of ESR1 mutation. In another study, the authors reported that patients with metastatic ER+ breast cancer harboring ESR1 mutations had improved PFS with fulvestrant (an ER degrader), as compared with exemestane (HR = 0.52, p = .02), but there was no difference between the endocrine therapies among patients without ESR1 mutations (HR = 1.07, p = .77), suggesting that ESR1 mutations might impact response to standard endocrine therapy [20].
Potential Strategies to Target the Pathway and Implications for Clinical Practice
Clinical Utility of the Molecular Alteration (Is the Mutation Actionable? Is There Access to Specific Targeted Therapy at the Institution? Are Clinical Trials Available?)
The final issue to consider is whether one should (and can) choose a specific therapy directly targeted against the known molecular alteration. On these lines, for tumors with ESR1 mutations, one could consider selective ER degraders (SERDs) that target the ER directly, as reviewed in Figure 2B. Although fulvestrant is a commercially available SERD, preclinical studies have suggested that fulvestrant is ineffective in suppressing the growth of ESR1-mutant cell lines at standard doses, and higher doses of fulvestrant are needed to achieve maximal inhibition. The lack of good therapeutic options for ESR1-mutant tumors has led to considerable interest in evaluation of novel SERDs, which could have greater clinical activity in this setting.
There are various clinical trials evaluating the efficacy of novel SERDs, such as GDC810 (NCT01823835), Rad1901 (NCT02338349), AZD9496 (NCT02248090), and LCZ102 (NCT02734615), in metastatic ER+ breast cancer. In a phase I/II clinical trial evaluating ARN-810 (GDC810), a novel, orally bioavailable SERD, a total loss of 18F-fluoroestradiol (FES) uptake (FES-positron emission tomography) was observed at 4 weeks of treatment, consistent with full receptor saturation and/or degradation in 95% of patients, including two patients with ESR1 mutations [21].
For the index patient, we reviewed standard therapies and potential clinical trial options, including novel SERDs. The patient enrolled in a clinical trial with a SERD. The patient has had no tumor progression after 9 months on therapy with the SERD.
Glossary of Genomic Terms and Nomenclature
Genomic DNA: coding (exons) and noncoding (introns) DNA that carries the chemical information that allows the exact transmission of genetic information from one cell to its daughter cells and from one generation to the next. The backbone included four nucleotides or bases, A, T, C, and G.
(Protein) Coding DNA Sequence (CDS): a DNA sequence (exons of a gene) that encodes for the transcription of messenger RNA, then translated into amino acids.
Codon: a unit of three consecutive bases on a DNA molecule that determines the position of a specific amino acid in a protein molecule during protein synthesis or the signal to initiate or stop synthesis.
Sequencing: determining the exact order of the bases in a strand of DNA.
Depth of Sequencing: the number of times a base is read during sequencing.
Variant Allelic Frequency: proportion of reads at a site that contain the variant.
-
Common Types of Gene Mutations:
- Missense mutation: change in one DNA base pair that results in the substitution of one amino acid for another in the protein made by a gene.
- Nonsense mutation: change in one DNA base pair that results in the substitution of one amino acid with a STOP codon resulting in premature termination of the protein.
- Deletion: mutation resulting in loss of any number of nucleotides.
- Insertion: mutations resulting in gain of any number of nucleotide.
- Frame shift mutations: addition or loss of DNA bases changing a gene’s reading frame, shifting the grouping of the bases forming codons, and thereby changing the code for amino acids. The resulting protein is usually shortened and nonfunctional.
Author Contributions
Conception/Design: Aditya Bardia, John A. Iafrate, Tilak Sundaresan, Jerry Younger, Valentina Nardi
Provision of study material or patients: Aditya Bardia, John A. Iafrate, Tilak Sundaresan, Jerry Younger, Valentina Nardi
Collection and/or assembly of data: Aditya Bardia, John A. Iafrate, Tilak Sundaresan, Jerry Younger, Valentina Nardi
Data analysis and interpretation: Aditya Bardia, John A. Iafrate, Tilak Sundaresan, Jerry Younger, Valentina Nardi
Manuscript writing: Aditya Bardia, John A. Iafrate, Tilak Sundaresan, Jerry Younger, Valentina Nardi
Final approval of manuscript: Aditya Bardia, John A. Iafrate, Tilak Sundaresan, Jerry Younger, Valentina Nardi
Disclosures
John A. Iafrate: Roche, Chugai, Constellation, Pfizer (C/A), ArcherDx (OI, IP); Tilak Sundaresan: coinventor for a patent describing a noninvasive method for ESR1 genotyping (IP). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chabner BA, Ellisen LW, Iafrate AJ. Personalized medicine: Hype or reality. The Oncologist. 2013;18:640–643. doi: 10.1634/theoncologist.2013-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy M. US president endorses “moonshot” effort to cure cancer. BMJ. 2016;352:i213. doi: 10.1136/bmj.i213. [DOI] [PubMed] [Google Scholar]
- 5.Hayes DF. Biomarker validation and testing. Mol Oncol. 2015;9:960–966. doi: 10.1016/j.molonc.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennello GA. Analytical and clinical evaluation of biomarkers assays: When are biomarkers ready for prime time? Clin Trials. 2013;10:666–676. doi: 10.1177/1740774513497541. [DOI] [PubMed] [Google Scholar]
- 7.Isler JA, Vesterqvist OE, Burczynski ME. Analytical validation of genotyping assays in the biomarker laboratory. Pharmacogenomics. 2007;8:353–368. doi: 10.2217/14622416.8.4.353. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 9.Human Genome Variation Society Sequence Variant Nomenclature. Available at http://varnomen.hgvs.org. Accessed August 15, 2016.
- 10.Ellis MJ. Mutational analysis of breast cancer: Guiding personalized treatments. Breast. 2013;22(suppl 2):S19–S21. doi: 10.1016/j.breast.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-α: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73:6856–6864. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 14.Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu D, Paoletti C, Gersch C, et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin Cancer Res. 2016;22:993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandarlapaty S, Sung P, Chen D, et al. cfDNA analysis from BOLERO-2 plasma samples identifies a high rate of ESR1 mutations: Exploratory analysis for prognostic and predictive correlation of mutations reveals different efficacy outcomes of endocrine-therapy-based regimens. Paper presented at: San Antonio Breast Cancer Symposium; December 8–12, 2015, San Antonio, TX. [Google Scholar]
- 20.O’Leary B, Fribbens C, Kilburn L, et al. ESR1 mutations in circulating tumour DNA predict outcome to endocrine treatment in patients with estrogen receptor positive advanced breast cancer; analysis of 521 patients in the SoFEA and PALOMA3 trials. Paper presented at: American Association for Cancer Research Annual Meeting; April 16–20, 2016, New Orleans, LA. [Google Scholar]
- 21.Bardia A, Dickler M, Mayer IA, et al. Phase I study of ARN-810, a novel and potent oral selective estrogen receptor degrader, in postmenopausal women with metastatic estrogen receptor positive (ER+), HER2- breast cancer. Paper presented at: San Antonio Breast Cancer Symposium. December 9–13, 2014. San Antonio, TX. [Google Scholar]