Technological advances in recent years have led to the development of blood-based diagnostics or “liquid biopsies” with great potential to quickly diagnose and genotype lung cancer using a minimally invasive technique. The clinical applications of liquid biopsy technologies, including circulating tumor cells, proteomics, microRNA, and cell free DNA for lung cancer, are reviewed and insight is provided into the diagnostic and therapeutic implications and challenges of these platforms.
Keywords: Non-small cell lung cancer, Liquid diagnostics, Cell-free DNA, Circulating tumor cells, MicroRNA
Abstract
A firmer understanding of the genomic landscape of lung cancer has recently led to targeted, therapeutic advances in non-small cell lung cancer. Historically, the reference standard for the diagnosis and genetic interrogation for advanced-stage patients has been tissue acquisition via computed tomography-guided core or fine needle aspiration biopsy. However, this process can frequently put the patient at risk and remains complicated by sample availability and tumor heterogeneity. In addition, the time required to complete the diagnostic assays can negatively affect clinical care. Technological advances in recent years have led to the development of blood-based diagnostics or “liquid biopsies” with great potential to quickly diagnose and genotype lung cancer using a minimally invasive technique. Recent studies have suggested that molecular alterations identified in cell-free DNA (cfDNA) or circulating tumor DNA can serve as an accurate molecular proxy of tumor biology and reliably predict the response to tyrosine kinase therapy. In addition, several trials have demonstrated the high accuracy of microRNA (miRNA) platforms in discerning cancerous versus benign nodules in high-risk, screened patients. Despite the promise of these platforms, issues remain, including varying sensitivities and specificities between competing platforms and a lack of standardization of techniques and downstream processing. In the present report, the clinical applications of liquid biopsy technologies, including circulating tumor cells, proteomics, miRNA, and cfDNA for NSCLC, are reviewed and insight is provided into the diagnostic and therapeutic implications and challenges of these platforms.
Implications for Practice:
Although tumor biopsies remain the reference standard for the diagnosis and genotyping of non-small cell lung cancer, they remain fraught with logistical complexities that can delay treatment decisions and affect clinical care. Liquid diagnostic platforms, including cell-free DNA, proteomic signatures, RNA (mRNA and microRNA), and circulating tumor cells, have the potential to overcome many of these barriers, including rapid and accurate identification of de novo and resistant genetic alterations, real-time monitoring of treatment responses, prognosis of outcomes, and identification of minimal residual disease. The present report provides insights into new liquid diagnostic platforms in non-small cell lung cancer and discusses the promise and challenges of their current and future clinical use.
Abstract
摘要
对肺癌基因组图谱的深入理解使非小细胞肺癌 (NSCLC) 的靶向治疗在近年得到了长足进展。从历史上来说, 肺癌晚期患者的诊断及基因鉴定参考标准物通常为在计算机体层摄影 (CT) 引导下粗针/细针穿刺活检获取的组织。然而, 这一步骤常使患者处于危险之中, 且结果也可能因样本可用性及肿瘤异质性的差异而较为混杂。此外, 完成这一诊断检验需要一定的时间, 这可能对临床治疗造成不利的影响。近年来临床技术的进展引领了基于血液的诊断学 (或称“液体活检”) 的飞速发展, 从而使应用微创技术对肺癌进行快速诊断和基因分型成为可能。最近有研究提示从无细胞DNA (cfDNA) 或循环肿瘤细胞中发现的分子学突变可以作为肿瘤活检的一项精准分子指标, 进而能可靠地预测患者对酪氨酸激酶抑制剂 (TKI) 治疗的反应。另外, 多项研究均证实microRNA (miRNA) 平台在高危筛查患者中鉴别恶性与良性结节时具有高准确性。尽管这些平台的前景令人瞩目, 但目前尚有许多问题, 包括各竞争平台之间敏感性和特异性的差异, 以及技术和下游操作程序缺乏规范化标准等。本文回顾总结了液体活检技术在NSCLC中的临床应用, 包括循环肿瘤细胞、蛋白质组学、miRNA及cfDNA等, 并对这些平台在诊断和治疗上的临床意义及面临的挑战提出了展望。The Oncologist 2016;21:1121–1130
对临床实践的提示: 尽管肿瘤活检是非小细胞肺癌诊断和基因分型的参考标准, 但其流程复杂, 可能延误治疗决策并影响临床治疗。液体诊断平台 [包括无细胞 DNA、蛋白质组学信号、RNA (mRNA 和 miRNA) 及循环肿瘤细胞] 有潜力克服这些障碍, 如快速并准确鉴定新发及耐药基因突变、实时监测治疗应答、预测患者转归并发现微小残留病灶等。本文对非小细胞肺癌的新型液体诊断平台提出了展望, 并讨论了当前和将来临床应用的前景和挑战。
Introduction
Lung cancer remains the leading cause of cancer-related mortality, with a 5-year survival rate of only 17% for all stages [1]. Despite advances in the development of targeted therapies and the emergence of novel immunotherapeutic approaches, significant challenges remain in the diagnosis and treatment of non-small cell lung cancer (NSCLC). First, although the adoption of low-dose helical computed tomography (LDCT) screening for high-risk patients might allow for earlier detection, roughly 65% of patients still present with advanced-stage disease, with more than half dying within 1 year [1]. Additionally, for patients receiving curative intent therapy, currently, no strategies outside of routine imaging are available to detect recurrent or minimal residual disease. Finally, standard tumor biopsies can be cumbersome, put the patient at risk, and might not accurately identify relevant molecular alterations owing to both suboptimal tissue acquisition and tumor heterogeneity. Given these challenges, additional strategies are urgently needed to help diagnose NSCLC and genotype tumor tissue from patients eligible for selection of molecularly driven therapies.
Recent technological advances have led to the development of blood-based diagnostics or “liquid biopsies” in NSCLC. This noninvasive approach allows for early detection of de novo or recurrent disease, the prognosis of outcomes, identification of genetic alterations to guide targeted therapy, and real-time monitoring of treatment response. Although liquid biopsies have historically referred to circulating tumor cells (CTCs), the definition has expanded to include proteomics, microRNA (miRNA), messenger RNA (mRNA), and, more recently, cell-free DNA (cfDNA). We review the technology of liquid biopsies in NSCLC and discuss the diagnostic and therapeutic implications of these platforms.
cfDNA
First discovered in 1948 by Mandel and Metais, fragmented DNA or cfDNA has since been associated with a number of conditions, including end-stage renal disease, myocardial infarction, stroke, and trauma [2–7]. Multiple studies have shown a correlation between the levels of cfDNA and cellular injury and necrosis, processes relevant in cancer cell survival and propagation. As tumors increase in volume, the capacity of phagocytes to eliminate and clear apoptotic and necrotic fragments can be exceeded, leading to passive release of cfDNA into the bloodstream [8]. Alternatively, in vitro studies have shown that DNA can be released by an active mechanism in which cancer cells spontaneously release DNA fragments into the circulation [9]. Depending on the tumor size and vascularity, the amount of cfDNA released in the circulation can vary from 0.01% to 90% of all DNA present in plasma [10].
Multiple studies have reported elevated cfDNA in lung cancer patients compared with healthy controls and increasing cfDNA concentrations in advanced disease compared with earlier stages of disease [11, 12]. Recent genomic technologies (including digital polymerase chain reaction [D-PCR], amplification refractory mutation system [ARMS], beads, emulsion, amplification, and magnetics [BEAMing], tagged-amplicon deep sequencing, and next-generation sequencing [NGS]) can detect low levels of cfDNA in plasma or serum and identify relevant genetic alterations (Table 1). Investigators have recently used cfDNA not only to identify patients with actionable mutations, including both sensitizing (exon 19 and 21) and resistant (T790M) EGFR mutations, but also as a real-time monitor of pharmacodynamic responses to tyrosine kinase inhibitor (TKI) therapy.
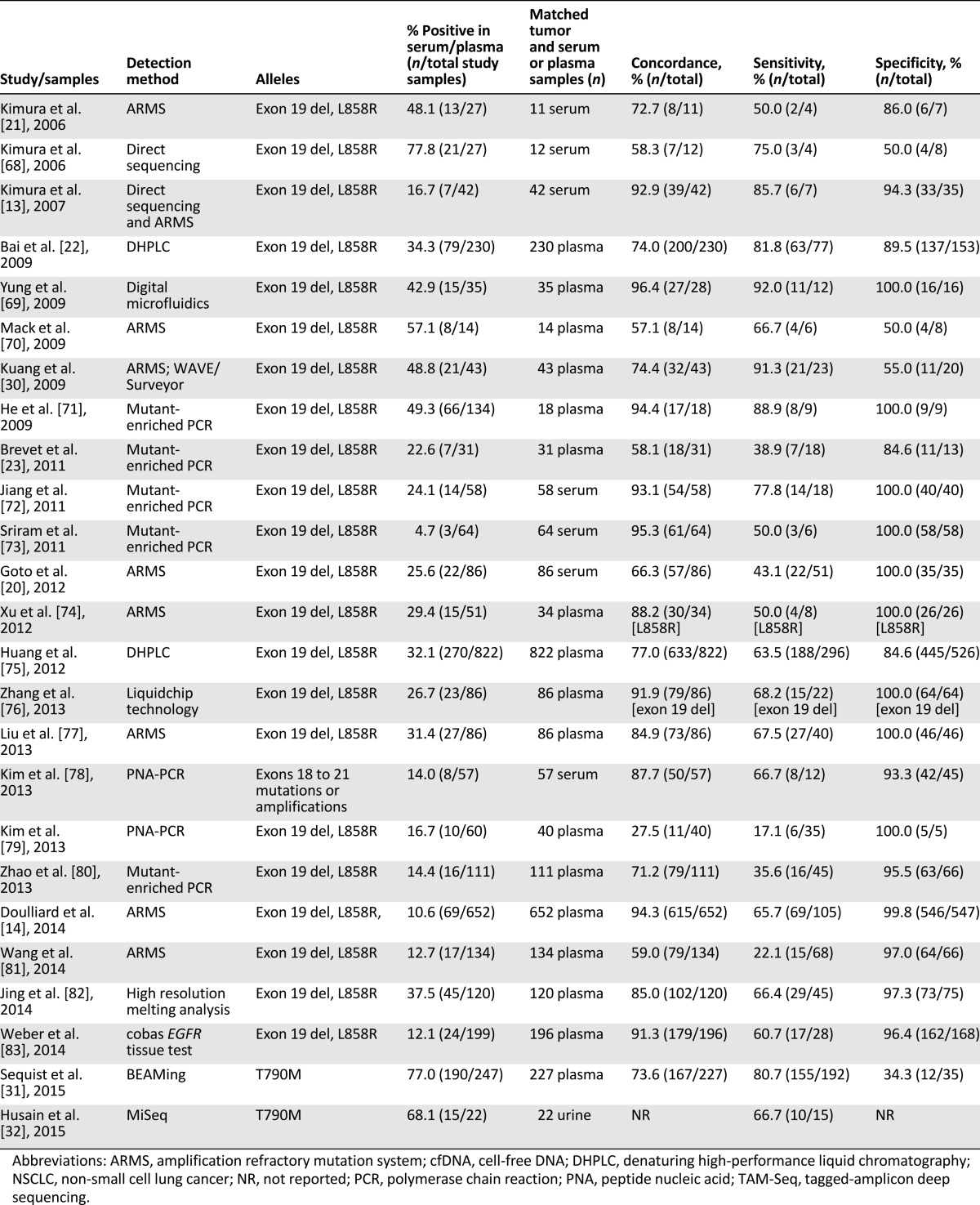
Table 1.
Diagnostic approaches for the identification of genetic alterations
Investigators have recently used cfDNA not only to identify patients with actionable mutations, including both sensitizing (exon 19 and 21) and resistant (T790M) EGFR mutations, but also as a real-time monitor of pharmacodynamic responses to tyrosine kinase inhibitor therapy.
Identifying Mutations in Treatment-Naïve Patients: Concordance With Tissue
One critical issue in blood-based genotyping is whether cfDNA can serve as an accurate molecular proxy for the corresponding tissue in the identification of EGFR mutations. Kimura et al. matched tumor and serum samples obtained from 42 advanced-stage NSCLC patients treated with gefitinib [13]. The concordance rate between EGFR mutations identified in serum and tissue was 92.9% (matching in 39 of 42 paired samples), with a sensitivity of 85.7%. High concordance rates were also demonstrated in a subset of patients from a phase IV, single-arm study evaluating first-line gefitinib in advanced-stage NSCLC patients [14]. The mutation concordance rate between 652 matched tumor and plasma samples before treatment was 94.3% (95% confidence interval [CI], 92.3–96.0), with a sensitivity of 65.7% (95% CI, 55.8–74.7) and specificity of 99.8% (95% CI, 99.0–100.0). Additional studies evaluating the concordance of plasma/serum and tissue EGFR mutations (Table 2) have reported a wide range of concordance rates (27.5%–100%) and sensitivities (17.1%–100%), with consistently high specificities (71.4%–100%). The disparities in sensitivities are heavily technology-dependent. Two recent meta-analyses assessing the diagnostic accuracy of cfDNA EGFR mutations demonstrated a pooled sensitivity of 61% and 67.4% and specificity of 90% and 93.5%, respectively [15, 16].
Table 2.
Studies evaluating cfDNA EGFR mutations in patients with NSCLC
Genetic interrogation of cfDNA via NGS has revealed additional mutations beyond EGFR. In one study, plasma-based NGS genotyped 8 of 11 patients and accurately identified two ALK rearrangements, one ROS1 rearrangement, one RET rearrangement, one EGFR G719A mutation, one KRAS G12C, and one combined KRAS G12C/PIK3CA [17]. In a second study, 42 of 54 patients evaluated using plasma NGS had at least one identifiable alteration, with 7 patients linked to a U.S. Food and Drug Administration-approved drug and 17 eligible for a targeted therapy approved for another disease type [18]. Finally, using a plasma-based digital NGS platform with the capability of capturing point mutations, insertions/deletions, fusion, and amplifications, Mack et al. retrospectively evaluated 978 patients with advanced NSCLC and identified actionable mutations in 412 patients (42%) [19]. These included activation EGFR mutations (n = 116), ALK fusions (n = 6), RET fusions (n = 9), HER2 exon 20 insertions (n = 9), BRAF mutations (n = 26), MET exon 14 skipping mutation (n = 8), met amplifications (n = 18), EGFR exon 20 insertions (n = 7), and KRAS/NF1/MEK mutations (n = 213). Although tissue was not available for concordance rates, this technology remains promising.
Predictive and Prognostic Utility of EGFR Mutations Identified by cfDNA
The clinical promise of cfDNA is rooted in its ability to serve as a predictive biomarker for targeted therapy. In the aforementioned phase IV gefitinib study, patients with EGFR mutation-positive cfDNA, regardless of mutation subtype, had a similar overall response rate (ORR) as that of patients with tissue EGFR mutation-positive tumors (76.9% and 69.8%, respectively), suggesting that the blood-based EGFR test might be as predictive to TKI treatment as tissue [14]. The ORRs were higher in patients with matched samples who harbored mutations in both plasma and tissue (76.9%; 95% CI, 65.4–85.5) compared with patients with only mutation-positive tumor tissue (59.5%; 95% CI, 43.5–73.7). Similar results were demonstrated in an exploratory analysis of 233 patients enrolled in the Iressa Pan-Asia Study (IPASS) comparing gefitinib and carboplatin-paclitaxel in 1217 treatment-naïve patients clinically enriched for EGFR mutations [20]. Of 194 patients who provided pretreatment serum samples, 46 (23.7%) had cfDNA EGFR mutations identified by Scorpion ARMS. Although the improvement in ORR in patients treated with gefitinib (n = 24) compared with chemotherapy (n = 22) was not significant (ORR, 75% vs. 64%; p = .40), statistically significant improvement was found in progression-free survival (PFS) in the cfDNA EGFR subgroup (hazard ratio [HR], 0.29; 95% CI, 0.14–0.60; p < .001). A significant interaction between cfDNA EGFR mutation status and treatment was evident for PFS (interaction test, p = .045). Multiple other studies have demonstrated EGFR mutations identified in cfDNA to be a reliable predictive biomarker of TKI treatment [13, 21–25].
Recent studies have also demonstrated the predictive value of quantitative changes in EGFR mutations in cfDNA at different time points during TKI treatment. Tseng et al. prospectively evaluated matched serum and tissue samples from 62 advanced-stage EGFR-positive patients who had received gefitinib [26]. Evaluating cfDNA using the peptide nucleic acid-zip nucleic acid PCR clamp method at 10 weeks and on progression of disease, the study demonstrated that failure to clear plasma EGFR mutations was an independent predictor of a lower disease control rate (odds ratio [OR], 5.26; 95% CI, 1.13–24.44; p = .034), shorter PFS (HR, 1.97; 95% CI, 1.33–2.91; p = .001), and decreased overall survival (OS; HR, 1.82; 95% CI, 1.04–3.18; p = .036). Mok et al. used a cobas test (Roche Molecular Diagnostics, Pleasanton, CA, http://www.molecular.roche.com) to evaluate serum/plasma EGFR mutations in patients from a phase III study randomized to receive six cycles of gemcitabine/platinum plus sequential erlotinib or placebo [25]. For patients treated in the erlotinib arm who were EGFR positive by cfDNA at baseline, the disappearance of cfDNA at cycle 3 was associated with significantly improved PFS (HR, 0.38; p = .0083) and longer OS (HR, 0.45; p = .0831) compared with patients with persistence of cfDNA EGFR. Most recently, Marchetti et al. performed PCR and ultra-deep NGS on serial plasma samples of advanced-stage patients (n = 20) with known tissue and plasma EGFR mutations before TKI treatment. Patients who had a 50% decrease in plasma EGFR copy number at 14 days (rapid responders; n = 14) had a greater mean percentage of tumor shrinkage than that of slow responders (n = 6) who had not achieved this change (70% vs. 30%; p < .0001) [27]. Finally, using a deep sequencing method (cancer personalized profiling by deep sequencing) that quantifies cancer-specific genomic alterations, Newman et al. was able to demonstrate that plasma ctDNA levels isolated from longitudinal samples in advanced-stage patients (n = 3) receiving either chemotherapy or targeted therapy highly correlated with the tumor volumes during therapy [28]. In sum, the real-time pharmacodynamic monitoring of mutations, most notably EGFR, during treatment in these studies highlights the potential of this platform to serve as an early predictor of response or resistance to therapy that could inform treatment decisions.
The prognostic utility of cfDNA has been studied by the Spanish Lung Cancer Group, which reported on a prespecified analysis from the erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small cell lung cancer (EURTAC) study of 76 patients with identifiable cfDNA EGFR mutations [29]. Evaluating subtypes of EGFR mutations in cfDNA using the TaqMan assay, the study demonstrated a shorter median OS in patients with the L858R mutation than in those with the exon 19 deletion (13.7 months; 95% CI, 7.1–17.7; vs. 30.0 months; 95% CI, 19.3–37.7; p < .001). Among the 41 patients with the L858R mutation in tissue, those in whom the L858R mutation was also detected in cfDNA had notably shorter median survival than those in whom the mutation was not detected in cfDNA (13.7 vs. 27.7 months; HR, 2.22; p = .03), suggesting a prognostic value of plasma cfDNA L858R mutations.
Identification of T790M in TKI-Resistant and -Naïve Patients
Several studies have identified resistant T790M mutations in cfDNA for both TKI-naïve and -resistant patients. Kuang et al. detected EGFR T790M in cfDNA from plasma via ARMS and/or the WAVE/Surveyor method in 15 of 28 patients with a previous clinical response to an EGFR TKI but only 4 of 14 with previous stable disease. EGFR T790M mutations detected in plasma DNA were strongly associated with a previous clinical response to TKI (p = .004) [30]. More recently, plasma BEAMing was used in a phase I/II trial evaluating rociletinib, a TKI selectively targeting both activating and T790M EGFR mutations in EGFR-positive, advanced-stage NSCLC patients who experienced disease progression during treatment with EGFR-directed therapy [31]. With tissue as the reference standard, the positive percentage of agreement for plasma T790M was 81% (155 of 192). Although the unconfirmed response rate to rociletinib in both plasma-positive and tissue-positive T790M patients was 53%, all plasma-positive patients were also tissue positive for T790M. Similar results have been demonstrated with osimertinib, another third-generation TKI, in patients with documented plasma- or tissue-positive T790M disease. Finally, using a PCR method coupled with NGS, Husain et al. identified T790M mutation in the urine of 15 of 22 EGFR-positive patients (68%) receiving TKI treatment. Of the 15 patients positive for T790M by urine, 10 patients had T790M mutation in the tissue biopsy using the Clinical Laboratory Improvement Amendments test [32].
T790M identification in cfDNA has also been clinically detected in treatment-naïve patients and for monitoring the real-time response. Wang et al. evaluated 135 patients with advanced-stage NSCLC who had clinical benefit of 6 months or more (PFS >6 months) with first- or second-line TKI treatment and retrospectively analyzed EGFR-sensitizing mutations and T790M mutation in matched pre- and post-TKI plasma via multiple assays, including D-PCR [33]. In the cohort of 83 patients with known EGFR mutations by tissue, the patients were subdivided into three groups according to the quantity of T790M in pre-TKI plasma samples by D-PCR (high, >5%, n = 7; low, 0%–5%, n = 20; and nil, 0%, n = 56). The median PFS was 7.1, 9.5, and 12.8 months (p = .001) and the median OS was 18.2, 21.2, and 32.5 months (p = .005) for the high, low, and nil groups, respectively, suggesting that a high pretreatment T790M mutational load might define patients less likely to benefit from TKI therapy. Sorensen et al. used allele-specific PCR to identify both sensitizing and resistant EGFR mutations from plasma in 23 patients with advanced adenocarcinoma receiving second-line TKI therapy as part of larger clinical trial [34]. cfDNA was evaluated at multiple time points during treatment and revealed T790M mutations in 9 patients up to 344 days before disease progression using the Response Evaluation Criteria In Solid Tumors, illustrating the potential to identify resistant disease before routine imaging. In the two aforementioned studies by Wang et al. and Sorenson et al., T790M was not interrogated in tissue samples; thus, concordance rates could not be established. It remains unclear whether identification of plasma or tissue T790M, either de novo or before clinical progression with TKI therapy, should guide treatment decisions with T790M-directed therapies.
Proteins
A number of proteins originating from the tumor and tumor microenvironment have been studied as biomarkers for NSCLC. Historically, protein biomarkers have included carcinoembryonic antigen, cytokeratin-19 fragment, and squamous cell carcinoma antigen. Although several studies have demonstrated the prognostic and predictive value of these markers, they have generally had limited clinical utility in lung cancer detection or as surrogates for response, given their relatively low sensitivity and specificity. More recently, a mass spectrometry (e.g., matrix-assisted laser desorption ionization) classifier has been used to identify proteomic signatures. These signatures are the basis of an algorithm that has resulted in a commercially available test that classifies advanced-stage patients into “poor” or “good” groups compared with a reference set. This test has served in both a predictive and a prognostic capacity for advanced-stage NSCLC patients receiving second-line therapy with either docetaxel or erlotinib [35]. Several studies have also highlighted the prognostic utility of this platform in advanced NSCLC patients receiving first-line therapy, independent of other common prognostic factors [35–38].
mRNA and miRNA
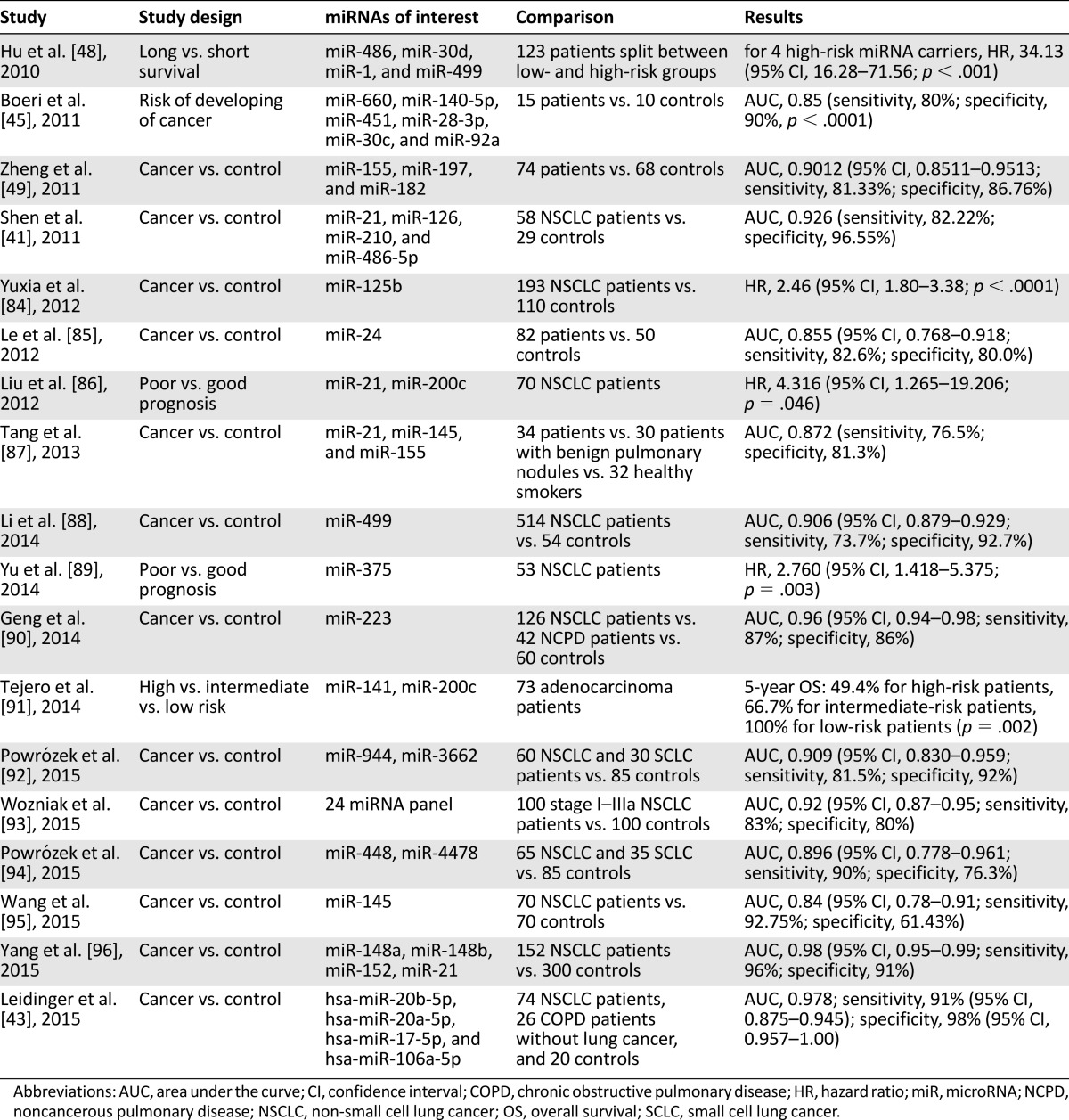
miRNAs are small noncoding RNAs predicted to regulate 10%–30% of the protein-coding genes in the human genome. They have been shown to play important roles in tumor initiation, invasion, and metastasis [39]. Circulating miRNAs are considered attractive cancer biomarkers owing to their stability and quantifiability using quantitative reverse transcription PCR (qRT-PCR) [40] (Table 3). For NSCLC, circulating miRNAs have been investigated for their potential in diagnosis, prognostic value, and predictive response to treatment. A number of miRNA signature profiles have been identified for NSCLC.
Table 3.
Studies reporting circulating miRNAs as biomarkers for NSCLC
Diagnosis and Screening of Lung Cancer
Multiple studies have reported on the potential utility of miRNAs to diagnosis NSCLC. Shen et al. determined that a panel of four miRNAs in plasma (miRNA-21, miRNA-126, miRNA-210, and miRNA-486-5p) were significantly elevated in NSCLC patients (n = 58) compared with healthy individuals (n = 29), with 86% sensitivity and 97% specificity [41]. In an exploratory phase I/II biomarker study, Hennessey et al. identified a miRNA pair (miRNA-15b and miRNA-27b) able to distinguish between NSCLC (n = 55) and cancer-free patients (n = 75), with a specificity of 84% (95% CI, 0.73–0.91) and sensitivity of 100% (95% CI,0.93–1.0) [42]. Leidinger et al. used qRT-PCR analysis to identify a set of five markers able to separate lung cancer patients (n = 74) from healthy controls (n = 20), with a specificity of 98% (95% CI, 0.95–1.0) and sensitivity of 91% (95% CI, 0.87–0.94) [43]. In addition, they were able to differentiate NSCLC patients (n = 74) from patients with chronic obstructive pulmonary disease (n = 26), with a specificity of 81% and sensitivity of 86% using 10 miRNA markers.
miRNAs have also been used to screen for individuals at risk of developing NSCLC. After identifying a set of 10 miRNAs that were differentially expressed in the serum of NSCLC patients compared with cancer-free controls, Chen et al. used the miRNA set to retrospectively analyze the serum samples of 7 NSCLC patients before and after their diagnosis [44]. They found that their miRNA profile could identify cancer up to 33 months ahead of the clinical diagnosis. Boeri et al. identified specific plasma miRNA signatures associated with lung cancer among patients enrolled in two different spiral-CT screening trials [45]. The miRNA signature classifier (MSC) algorithm was then validated in a study retrospectively evaluating 939 plasma samples (69 with lung cancer and 870 without) from the Multicenter Italian Lung Detection (MILD) clinical trial comparing LDCT (n = 652) and observation (n = 287) [46]. The MSC had a sensitivity of 87% and specificity of 81% across both arms; LDCT had a sensitivity of 79% and specificity of 81%. Using MSC and LDCT together resulted in a fivefold reduction of the LDCT false-positive rate (from 19.4% to 3.7%). The investigators calculated that the screening sensitivity could be increased to 98% using both tests [46]. Bianchi et al. compared serum samples from 93 patients enrolled in a spiral CT screening study with a diagnosis of NSCLC with serum samples from 60 individuals in the same study without cancer [47]. They found a 34-miRNA signature that could differentiate asymptomatic high-risk individuals with early-stage lung cancer (n = 34) from individuals with no cancer (n = 30), with a sensitivity of 71% and specificity of 90%. Evaluating 13 patients before and after they developed lung cancer, the investigators also applied a risk predictor algorithm based on the miRNA signature that showed a significantly increased risk index in the serum after the onset of the disease (p < .001, paired t test).
Prognosis and Predictive Response to Treatment
Using genome-wide sequencing, Hu et al. found 11 miRNAs were altered more than fivefold in the serum of lung cancer patients who survived for more than 30 months compared with those who had survived for less than 25 months [48]. Of the 11 miRNAs, 4 (miR-486, miR-30d, miR-1, and miR-499) were significantly associated with overall survival. Patients carrying two or more high-risk miRNAs had a significantly increased risk of cancer death compared with patients with one or no high-risk miRNA (log-rank test, p < .0001; HR, 3.14; 95% CI, 1.65–5.97, for two high-risk miRNA carriers; HR, 16.52; 95% CI, 8.62–31.68, for three high-risk miRNA carriers; HR, 34.13; 95% CI, 16.28–71.56, for four high-risk miRNA carriers). miR-155 and miR-197 levels have been reported to be higher in lung cancer patients with metastasis (p < .05) and to be significantly decreased in patients with a response to chemotherapy (p < .001) [49].
In addition to miRNA, messenger RNAs (mRNAs) have also been evaluated in early- and late-stage lung cancer patients. Cancer/testis antigens (CTAs) are protein antigens normally expressed in the testis but frequently expressed in lung cancer. Gumireddy et al. used a nested PCR assay to determine whether the mRNA levels of 116 CTA genes in the peripheral blood mononuclear cells of NSCLC lung cancer patients were differentially expressed compared with individuals with smoking-related benign diseases [50]. The expression of one gene, AKAP4, was able to distinguish between NSCLC patients (n = 264) and individuals with benign disease (n = 135), as well as those with benign nodules, with an area under the curve of 0.9714 (95% CI, 0.956–0.986) and 0.9825 (95% CI, 0.969–0.995), respectively. Similar to cfDNA, the investigators found that AKAP4 increased with cancer stage, with AKAP4 mRNA expression levels in stage IV NSCLC patients 3,254-fold higher than that in patients with stage I NSCLC. AKAP4 mRNA expression did not have a significant association with tobacco use, which might allow AKAP4 to be used as a screening tool for both smokers and nonsmokers.
Given that only a subpopulation of NSCLC CTCs undergo epithelial-to-mesenchymal transition and downregulate their epithelial markers, the detection rates using EpCAM-based methods have been lower.
CTCs
CTCs are tumor cells shed into the blood by solid neoplasms. They are exceedingly rare, with as few as one in every 109 blood cells in patients with metastatic disease [51]. A number of methods have been developed to detect CTCs, including the CellSearch system (Janssen Diagnostics, Raritan, NJ, http://www.janssen.com), the Intelligent System Emulation Technology (ISET) system (Aligent Technologies, Santa Clara, CA, http://www.aligent.com), flow cytometry, laser scanning cytometry, PCR-based approaches, and CTC microchip technology [52–55]. Following CTC isolation, DNA can then be extracted and analyzed in the same manner as cfDNA, which has revealed EGFR mutations [56, 57], ALK rearrangements [58, 59], and ROS1 rearrangements [60]. Although CTCs have been identified in up to 85% of small cell lung cancer patients, the reported detection rates in NSCLC using epithelial cell adhesion molecule (EpCAM)-based methods have been significantly lower [61]. Given that only a subpopulation of NSCLC CTCs undergo epithelial-to-mesenchymal transition and downregulate their epithelial markers, the detection rates using EpCAM-based methods have been lower. Although higher CTC detection rates have been reported using non-EpCAM-based methods, these methods need to be validated further in independent and large multicenter studies.
Prognosis and Predictive Response to Treatment
CTCs have been shown to have prognostic significance in metastatic NSCLC [62, 63]. Using ISET, Hofman et al. isolated CTCs from 208 NSCLC patients and found that a cutoff value of >50 corresponded to shorter OS and PFS. In a global meta-analysis, the appearance of pretreatment CTCs in patients with different stages of lung cancer correlated with lymph node status, distant metastasis, and TNM staging [64]. Longitudinal CTC monitoring has also shown that patients with more than one time point positive for CTCs had worse survival than those with conversion from CTC positive to CTC negative or persistently negative [65].
Presence of EGFR Mutations and Disease Monitoring
Breitenbuecher et al. used combined RT-PCR and melting curve analysis to show that an increase in EGFR-mutant CTCs might be an indicator for disease relapse and EGFR-TKI resistance [56]. Additionally, just as in cfDNA, T790M identification in CTCs has been used in TKI-resistant patients and to monitor the real-time response. In NSCLC patients treated with EGFR-TKIs, Maheswaran et al. found T790M mutations in CTCs from 9 of 14 patients (64%) with clinical progression, and the presence of the mutation correlated with reduced PFS (7.7 months vs. 16.5 months; p < .001) [57].
Discussion
Despite recent therapeutic advances in NSCLC, significant diagnostic challenges remain that can bottleneck treatment decisions and negatively affect clinical care. The liquid diagnostic platforms we have reviewed have the potential to overcome many of these barriers. Perhaps the greatest immediate clinical effect will be the routine use of cfDNA to identify de novo and resistant genetic alterations in patients with advanced-stage disease (i.e., plasma genotyping). The limitations of tumor biopsies have recently been supported by a study that demonstrated up to 30% of patients at a community-based academic center did not undergo guideline-recommended molecular testing, despite an institutional reflex testing policy for tissue [66]. In addition, repeat tissue acquisition poses barriers to clinical trial enrollment, as demonstrated by a recent study retrospectively evaluating patient enrollment to 55 clinical trials at a single Canadian institution in which fewer patients received study treatment in trials mandating tissue specimens compared with trials that did not (55% vs. 83%; p < .001) [67]. Plasma cfDNA platforms offer promise in overcoming these challenges by rapidly genotyping treatment-naïve and -refractory patients and might eventually obviate the need for the repeat biopsies mandated by many clinical trials.
In addition to plasma genotyping, liquid platforms have tremendous potential to improve screening in patients at high risk of both first cancers and recurrences. The goal to identify early stage or minimal residual disease before or in combination with routine imaging remains a challenge. Although the U.S. National Lung Screening Trial (NLST) demonstrated a 20% reduction in lung cancer mortality with LDCT scans, the high false-positive rate with this modality warrants further enrichment strategies. The accuracy of mRNA and miRNA platforms in delineating benign from malignant nodules is promising, with the potential to enhance the sensitivity and specificity of screening techniques. These platforms might also contribute to surveillance strategies after curative intent therapy. A recent study of colorectal cancer patients undergoing curative-intent surgery reported cfDNA identified after resection in all patients who experienced eventual recurrence but not in the patients who remained disease free [10].
Plasma cfDNA platforms offer promise in overcoming these challenges by rapidly genotyping treatment-naïve and refractory patients and might eventually obviate the need for the repeat biopsies mandated by many clinical trials.
Despite the promise of liquid diagnostics in lung cancer, significant challenges remain. In the presence of tumor heterogeneity, it is uncertain whether cfDNA sampling provides a reliable molecular proxy of overall tumor biology and whether plasma or tissue is the true reference standard for molecular characterization of disease. Further studies evaluating the predictive utility of genetic alterations identified in cfDNA independent of those identified by tissue interrogation are urgently needed. Additionally, currently, no widely accepted standards are available for processing samples, which might contribute to the disparate test results. Performance verification and method development, including optimal preparation strategies (collecting, isolating, and storing cfDNA), and downstream analysis need to be carefully formalized. Coordination and communication between laboratories of competing platforms would help facilitate this process. Finally, given the wide range of reported sensitivities and specificities of the different platforms, it will be essential for prospective therapeutic trials to mandate the collection of plasma and/or serum to establish reproducible concordance rates with tissue for clinical validation. Special attention should be devoted to the impact of both clinical (tumor burden) and sampling variables on the accuracy and reproducibility of these platforms. It is important to note that, to date, very few, if any, published series have evaluated the accuracy of specific commercially available platforms or their concordance with paired tissue. Thus, caution should be used when interpreting the results. In addition, many of these assays’ threshold for detection have been set arbitrarily, with different internal cutoff levels, making the interpretation of some results challenging.
In addition to technical considerations, the application of liquid platforms might create new therapeutic dilemmas. For instance, whether pharmacodynamic monitoring of either EGFR or T790M in cfDNA should be performed and guide therapeutic decision making before clinical or radiographic progression for patients receiving TKI therapy remains unclear. Likewise, whether treatment of minimal residual disease identified in plasma only would improve survival for patients after curative intent therapy is also unknown. Prospective trials evaluating treatment decisions triggered by plasma tests, independent of the clinical or radiographic findings, for both early- and advanced-stage patients should be pursued. We look forward to the further development and refinement of liquid platforms and their routine use in accurately identifying recurrent and de novo disease and genotyping patients with all stages of NSCLC.
Author Contributions
Conception/Design: Benjamin Levy, Zishuo I. Hu, Karen Lee, Daniel Becker
Provision of study material or patients: Benjamin Levy, Zishuo I. Hu, Karen Lee, Daniel Becker
Collection and/or assembly of data: Benjamin Levy, Zishuo I. Hu, Kristen N. Cordova, Sandra Close, Karen Lee, Daniel Becker
Data analysis and interpretation: Benjamin Levy, Zishuo I. Hu, Karen Lee, Daniel Becker
Manuscript writing: Benjamin Levy, Zishuo I. Hu, Karen Lee, Daniel Becker
Final approval of manuscript: Benjamin Levy, Zishuo I. Hu, Kristen N. Cordova, Sandra Close, Karen Lee, Daniel Becker
Disclosures
Benjamin Levy: Celgene, AstraZeneca, Pfizer, Genentech, Biodesix, Eli Lilly, Merck (C/A), Genetech, Eli Lilly (H); Kristen N. Cordova: Biodesix (Medical Affairs) (E, former); Sandra Close: Biodesix (C/A); Karen Lee: Pfizer, Bristol-Myers Squibb, Merck (H). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Beiter T, Fragasso A, Hudemann J, et al. Short-term treadmill running as a model for studying cell-free DNA kinetics in vivo. Clin Chem. 2011;57:633–636. doi: 10.1373/clinchem.2010.158030. [DOI] [PubMed] [Google Scholar]
- 3.Atamaniuk J, Kopecky C, Skoupy S, et al. Apoptotic cell-free DNA promotes inflammation in haemodialysis patients. Nephrol Dial Transplant. 2012;27:902–905. doi: 10.1093/ndt/gfr695. [DOI] [PubMed] [Google Scholar]
- 4.García Moreira V, de la Cera Martínez T, Gago González E, et al. Increase in and clearance of cell-free plasma DNA in hemodialysis quantified by real-time PCR. Clin Chem Lab Med. 2006;44:1410–1415. doi: 10.1515/CCLM.2006.252. [DOI] [PubMed] [Google Scholar]
- 5.Antonatos D, Patsilinakos S, Spanodimos S, et al. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann N Y Acad Sci. 2006;1075:278–281. doi: 10.1196/annals.1368.037. [DOI] [PubMed] [Google Scholar]
- 6.Shimony A, Zahger D, Gilutz H, et al. Cell free DNA detected by a novel method in acute ST-elevation myocardial infarction patients. Acute Card Care. 2010;12:109–111. doi: 10.3109/17482941.2010.513732. [DOI] [PubMed] [Google Scholar]
- 7.Tsai NW, Lin TK, Chen SD, et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta. 2011;412:476–479. doi: 10.1016/j.cca.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 8.Diaz LA, Jr, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilie M, Hofman V, Long E, et al. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann Transl Med. 2014;2:107. doi: 10.3978/j.issn.2305-5839.2014.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 12.Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: The new ambrosia of researchers. Biochim Biophys Acta. 2014;1846:539–546. doi: 10.1016/j.bbcan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97:778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated Caucasian NSCLC: Circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9:1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao C, Yuan JQ, Yang ZY, et al. Blood as a substitute for tumor tissue in detecting EGFR mutations for guiding EGFR TKIs treatment of nonsmall cell lung cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2015;94:e775. doi: 10.1097/MD.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: A systematic review and meta-analysis. Sci Rep. 2014;4:6269. doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher AG, Paweletz C, Alden R et al. A prospective study of rapid plasma genotyping utilizing sequential ddPCR and NGS in newly diagnosed advanced NSCLC patients. Paper presented at the World Conference Lung Cancer (WCLC); 2015, ORAL 38.01; Denver, CO. [Google Scholar]
- Villaflor V, Won B, Nagy B et al. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Paper presented at the World Conference Lung Cancer (WCLC), 2015, ORAL 38.02; Denver, CO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack PC, Gandara D, Lara P et al. Identification of actionable tumor alterations in circulating cell-free tumor DNA (cfDNA) using digital sequencing from NSCLC patients. Paper presented at the World Conference Lung Cancer (WCLC), 2015, ORAL 38.06; Denver, CO. [Google Scholar]
- 20.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: From IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7:115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 21.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 22.Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 23.Brevet M, Johnson ML, Azzoli CG, et al. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer. 2011;73:96–102. doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jian G, Songwen Z, Ling Z, et al. Prediction of epidermal growth factor receptor mutations in the plasma/pleural effusion to efficacy of gefitinib treatment in advanced non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:1341–1347. doi: 10.1007/s00432-010-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mok T, Wu YL, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21:3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 26.Tseng JS, Yang TY, Tsai CR, et al. Dynamic plasma EGFR mutation status as a predictor of EGFR-TKI efficacy in patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol. 2015;10:603–610. doi: 10.1097/JTO.0000000000000443. [DOI] [PubMed] [Google Scholar]
- 27.Marchetti A, Palma JF, Felicioni L, et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol. 2015;10:1437–1443. doi: 10.1097/JTO.0000000000000643. [DOI] [PubMed] [Google Scholar]
- 28.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol. 2015;1:149–157. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 30.Kuang Y, Rogers A, Yeap BY, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sequist L, Goldman JW, Wakelee HA, et al. Efficacy of rociletinib (CO-1686) in plasma-genotyped T790M-positive NSCLC patients. J Clin Oncol. 2015;33(suppl):8001a. [Google Scholar]
- 32.Husain H, Kosco K, Vibat CRT, et al. Kinetic monitoring of EGFR T790M in urinary circulating tumor DNA to predict radiographic progression and response in patients with metastatic lung adenocarcinoma. J Clin Oncol. 2015;33:8081a. [Google Scholar]
- 33.Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One. 2014;9:e110780. doi: 10.1371/journal.pone.0110780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120:3896–3901. doi: 10.1002/cncr.28964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregorc V, Novello S, Lazzari C, et al. Predictive value of a proteomic signature in patients with non-small-cell lung cancer treated with second-line erlotinib or chemotherapy (PROSE): A biomarker-stratified, randomised phase 3 trial. Lancet Oncol. 2014;15:713–721. doi: 10.1016/S1470-2045(14)70162-7. [DOI] [PubMed] [Google Scholar]
- 36.Carbone DP, Ding K, Roder H, et al. Prognostic and predictive role of the VeriStrat plasma test in patients with advanced non-small-cell lung cancer treated with erlotinib or placebo in the NCIC Clinical Trials Group BR.21 trial. J Thorac Oncol. 2012;7:1653–1660. doi: 10.1097/JTO.0b013e31826c1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stinchcombe TE, Roder J, Peterman AH, et al. A retrospective analysis of VeriStrat status on outcome of a randomized phase II trial of first-line therapy with gemcitabine, erlotinib, or the combination in elderly patients (age 70 years or older) with stage IIIB/IV non-small-cell lung cancer. J Thorac Oncol. 2013;8:443–451. doi: 10.1097/JTO.0b013e3182835577. [DOI] [PubMed] [Google Scholar]
- 38.Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: A multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 39.Cortinovis D, Monica V, Pietrantonio F, et al. MicroRNAs in non-small cell lung cancer: Current status and future therapeutic promises. Curr Pharm Des. 2014;20:3982–3990. doi: 10.2174/13816128113196660755. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennessey PT, Sanford T, Choudhary A, et al. Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS One. 2012;7:e32307. doi: 10.1371/journal.pone.0032307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leidinger P, Brefort T, Backes C, et al. High-throughput qRT-PCR validation of blood microRNAs in non-small cell lung cancer. Oncotarget. 2016;7:4611–4623. doi: 10.18632/oncotarget.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Hu Z, Wang W, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 45.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: A correlative MILD trial study. J Clin Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 49.Zheng D, Haddadin S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 50.Gumireddy K, Li A, Chang DH, et al. AKAP4 is a circulating biomarker for non-small cell lung cancer. Oncotarget. 2015;6:17637–17647. doi: 10.18632/oncotarget.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allan AL, Keeney M. Circulating tumor cell analysis: Technical and statistical considerations for application to the clinic. J Oncol. 2010;2010:426218. doi: 10.1155/2010/426218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu N, Zhou J, Cui F, et al. Circulating tumor cells in lung cancer: Detection methods and clinical applications. Lung. 2015;193:157–171. doi: 10.1007/s00408-015-9697-7. [DOI] [PubMed] [Google Scholar]
- 54.Wang B, et al. Clinical test on circulating tumor cells in peripheral blood of lung cancer patients, based on novel immunomagnetic beads. Artif Cells Nanomed Biotechnol. 2016;44:892–897. doi: 10.3109/21691401.2014.998827. [DOI] [PubMed] [Google Scholar]
- 55.Vona G, Sabile A, Louha M, et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One. 2014;9:e85350. doi: 10.1371/journal.pone.0085350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: A comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol. 2012;23:2907–2913. doi: 10.1093/annonc/mds137. [DOI] [PubMed] [Google Scholar]
- 59.Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2013;31:2273–2281. doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- 60.Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol. 2015;26:1408–1415. doi: 10.1093/annonc/mdv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 62.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 63.Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hofman VJ, Long E, Ilie M, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology. 2012;23:30–38. doi: 10.1111/j.1365-2303.2010.00835.x. [DOI] [PubMed] [Google Scholar]
- 65.Li J, Shi SB, Shi WL, et al. LUNX mRNA-positive cells at different time points predict prognosis in patients with surgically resected nonsmall cell lung cancer. Transl Res. 2014;163:27–35. doi: 10.1016/j.trsl.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Inal C, Yilmaz E, Cheng H, et al. Effect of reflex testing by pathologists on molecular testing rates in lung cancer patients: Experience from a community-based academic center. J Clin Oncol. 2014;32(suppl 5):8098a. [Google Scholar]
- 67.Lim C, Sung M, Shepherd FA, et al. Patients with advanced non-small cell lung cancer: Are research biopsies a barrier to participation in clinical trials. J Thorac Oncol. 2016;11:79–84. doi: 10.1016/j.jtho.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Kimura H, Kasahara K, Shibata K, et al. EGFR mutation of tumor and serum in gefitinib-treated patients with chemotherapy-naive non-small cell lung cancer. J Thorac Oncol. 2006;1:260–267. doi: 10.1016/s1556-0864(15)31577-x. [DOI] [PubMed] [Google Scholar]
- 69.Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 70.Mack PC, Holland WS, Burich RA, et al. EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer. J Thorac Oncol. 2009;4:1466–1472. doi: 10.1097/JTO.0b013e3181bbf239. [DOI] [PubMed] [Google Scholar]
- 71.He C, Liu M, Zhou C, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer. 2009;125:2393–2399. doi: 10.1002/ijc.24653. [DOI] [PubMed] [Google Scholar]
- 72.Jiang B, Liu F, Yang L, et al. Serum detection of epidermal growth factor receptor gene mutations using mutant-enriched sequencing in Chinese patients with advanced non-small cell lung cancer. J Int Med Res. 2011;39:1392–1401. doi: 10.1177/147323001103900425. [DOI] [PubMed] [Google Scholar]
- 73.Sriram KB, Tan ME, Savarimuthu SM, et al. Screening for activating EGFR mutations in surgically resected nonsmall cell lung cancer. Eur Respir J. 2011;38:903–910. doi: 10.1183/09031936.00190110. [DOI] [PubMed] [Google Scholar]
- 74.Xu F, Wu J, Xue C, et al. Comparison of different methods for detecting epidermal growth factor receptor mutations in peripheral blood and tumor tissue of non-small cell lung cancer as a predictor of response to gefitinib. Onco Targets Ther. 2012;5:439–447. doi: 10.2147/OTT.S37289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang Z, Wang Z, Bai H, et al. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer. 2012;3:334–340. doi: 10.1111/j.1759-7714.2012.00133.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhang H, Liu D, Li S, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn. 2013;15:819–826. doi: 10.1016/j.jmoldx.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 77.Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: Comparison of methodologies. J Clin Pathol. 2013;66:1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim ST, Jung HY, Sung JS, et al. Can serum be used for analyzing the EGFR mutation status in patients with advanced non-small cell lung cancer? Am J Clin Oncol. 2013;36:57–63. doi: 10.1097/COC.0b013e31823a5217. [DOI] [PubMed] [Google Scholar]
- 79.Kim HR, Lee SY, Hyun DS, et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res. 2013;32:50. doi: 10.1186/1756-9966-32-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao X, Han RB, Zhao J, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration. 2013;85:119–125. doi: 10.1159/000338790. [DOI] [PubMed] [Google Scholar]
- 81.Wang S, Han X, Hu X, et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta. 2014;430:63–70. doi: 10.1016/j.cca.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 82.Jing CW, Wang Z, Cao HX, et al. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2014;14:6619–6623. doi: 10.7314/apjcp.2013.14.11.6619. [DOI] [PubMed] [Google Scholar]
- 83.Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer. 2014;14:294. doi: 10.1186/1471-2407-14-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuxia M, Zhennan T, Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol. 2012;138:2045–2050. doi: 10.1007/s00432-012-1285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le HB, Zhu WY, Chen DD, et al. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190–3197. doi: 10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 86.Liu XG, Zhu WY, Huang YY, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 87.Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22:540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 88.Li M, Zhang Q, Wu L, et al. Serum miR-499 as a novel diagnostic and prognostic biomarker in non-small cell lung cancer. Oncol Rep. 2014;31:1961–1967. doi: 10.3892/or.2014.3029. [DOI] [PubMed] [Google Scholar]
- 89.Yu D, Du K, Liu T, et al. Prognostic value of tumor markers, NSE, CA125 and SCC, in operable NSCLC patients. Int J Mol Sci. 2013;14:11145–11156. doi: 10.3390/ijms140611145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geng Q, Fan T, Zhang B, et al. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tejero R, Navarro A, Campayo M, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Powrózek T, Krawczyk P, Kowalski DM, et al. Plasma circulating microRNA-944 and microRNA-3662 as potential histologic type-specific early lung cancer biomarkers. Transl Res. 2015;166:315–323. doi: 10.1016/j.trsl.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 93.Wozniak MB, Scelo G, Muller DC, et al. Circulating microRNAs as non-invasive biomarkers for early detection of non-small-cell lung cancer. PLoS One. 2015;10:e0125026. doi: 10.1371/journal.pone.0125026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Powrózek T, Krawczyk P, Kowalski DM, et al. Application of plasma circulating microRNA-448, 506, 4316, and 4478 analysis for non-invasive diagnosis of lung cancer. Tumour Biol. 2016;37:2049–2055. doi: 10.1007/s13277-015-3971-4. [DOI] [PubMed] [Google Scholar]
- 95.Wang RJ, Zheng YH, Wang P, et al. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 96.Yang JS, Li BJ, Lu HW, et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]