Abstract
Mitotic and meiotic spindles consist primarily of microtubules, which originate from centrosomes and within the vicinity of chromatin. Indirect evidence suggested that microtubules also originate throughout the spindle, but the high microtubule density within the spindle precludes the direct observation of this phenomenon. By using meiotic Xenopus laevis egg extract and employing total internal reflection (TIRF) microscopy, microtubule nucleation from preexisting microtubules could be demonstrated and analyzed. Branching microtubule nucleation is an ideal mechanism to assemble and maintain a mitotic spindle, because microtubule numbers are amplified while preserving their polarity. Here, we describe the assays that made these findings possible and the experiments that helped identify the key molecular players involved.
Keywords: Cell division, Mitotic spindle, Meiotic spindle, Cytoskeleton, Microtubule, Microtubule nucleation, Xenopus laevis egg extract, TIRF microscopy
1 Introduction
The mitotic spindle is assembled from several microtubule (MT) organizing centers, most prominently centrosomes and chromosomes, all of which require the universal MT nucleating molecule gamma-tubulin (γ-TB). More recently, MTs were also found to originate from within the body of the spindle [1, 2], and this was shown to be important for spindle assembly [3, 4]. Furthermore, a targeting factor for γ-TB to spindle MTs was identified in a whole-genome RNAi screen and termed augmin, as it increases the MT density within the spindle [5–8]. Yet the high MT density in the spindle precludes direct observation MT nucleation events, leaving the question of how MTs are generated within the body of the spindle open.
This roadblock could be circumvented by using meiotic Xenopus egg extract, which allowed resolving individual MTs with a low background by total internal reflection (TIRF) microscopy [9]. By simultaneously imaging MTs and their growing MT plus ends via fluorescently labeled tubulin and end-binding protein 1 (EB1), respectively, it was directly demonstrated that MTs originate from the wall of preexisting MTs. Addition of recombinant proteins and immunodepletion of endogenous proteins revealed the key molecular players to be augmin, γ-TB, RanGTP and its downstream factor “targeting factor of Xklp2” (TPX2) [9]. The goal of this chapter is to describe the assay and its variations that first characterized branching MT nucleation and its molecular players in Xenopus egg extract.
2 Materials
2.1 Proteins and Antibodies
2.1.1 Unlabeled and Fluorescently Labeled Tubulin
2.1.2 Recombinant Proteins for Addition to Extract
GFP-labeled H.s. End-Binding protein 1 (EB1).
X.l. RanQ69L.
X.l. TPX2.
2.1.3 Rabbit Polyclonal Antibody Production and Purification for Immunodepletion
Polyclonal antibody serum specific against protein of interest.
Affigel-10 and -15 matrix (Bio-Rad).
Glycerol.
2.2 Branching MT Nucleation Assay in Xenopus Egg Extract
2.2.1 Preparation of Xenopus Egg Extracts
Freshly prepared Xenopus laevis CSF extract of high quality (see Note 1).
2.2.2 Flow Chamber Assembly
Frosted glass slides.
Double-sticky tape.
Razorblade.
No. 1.5 glass coverslips (22 × 22 mm).
2.2.3 Basic Branching Reaction
CSF-XB buffer (10 mM K-HEPES pH 7.7, 2 mM MgCl2, 0.1 mM CaCl2, 100 mM KCl, 5 mM EGTA, and 50 mM sucrose).
Hot candle wax or nail polish.
Objective-based TIRF microscope equipped with a 100× 1.49 NA objective, a low noise EM-CCD camera and laser power of at least 20 mW out-of-fiber.
2.2.4 Variations of the Basic Branching Reaction
Sodium orthovanadate (100 mM, NEB) or purified p150-CC1 (see Note 2).
2.2.5 Immunodepletion of CSF Extract and Add-Back of Recombinant Protein
Dynabeads Protein A (Invitrogen).
DynaMag-2 Magnet (Invitrogen) for retrieving Dynabeads.
TBS-T (50 mM Tris–HCl, 150 mM NaCl, 0.05 % Tween 20).
Antibody of desired specificity and control total IgG fraction.
CSF-XB buffer (10 mM K-HEPES pH 7.7, 2 mM MgCl2, 0.1 mM CaCl2, 100 mM KCl, 5 mM EGTA, and 50 mM sucrose).
SDS-sample buffer, SDS-PAGE and Western blot equipment.
Recombinant protein that can replace the immunodepleted endogenous protein.
2.3 Branching MT Nucleation from Stabilized MT Seeds
Passivated coverslips [12].
GpCpp/GMPCPP (Jena Bioscience).
BRB80 buffer (80 mM K-PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8 with KOH).
Anti-biotin antibody (Invitrogen).
Kappa-casein (Sigma).
Oxygen scavenger system consisting of Trolox, protocatechuate 3,4-dioxygenase (PCD) and protocatechuic acid (PCA) according to Ref. [13] (all Sigma).
3 Methods
3.1 Proteins, Antibodies, and Xenopus Egg Extract
3.1.1 Unlabeled and Fluorescently Labeled Tubulin (See Note 3)
Tubulin is purified from bovine brain using two cycles of polymerization–depolymerization [10].
Fractions of purified tubulin are then fluorescently labeled [11] with 6((biotinoyl)amino) hexanoic acid, succinimidyl ester (Invitrogen), Alexa 568 carboxylic acid, succinimidyl ester (Invitrogen) and Cy5 NHS ester (GE). Briefly, MTs are polymerized and pelleted before being incubated with the dye at 37 °C. Labeled MTs are pelleted and depolymerized on ice. Dimeric, labeled tubulin is separated from polymerized MTs through another pelleting step at 4 °C and subsequently flash-frozen in small aliquots at 200 μM.
3.1.2 Recombinant Proteins for Extract Addition
GFP-labeled H.s. End-Binding protein 1 (EB1), X.l. RanQ69L, and X.l. TPX2 are purified as recently described [14–16].
All proteins are dialyzed into CSF-XB buffer containing 10 % sucrose, which does not disturb microtubule assembly dynamics in Xenopus laevis egg extract. During this process, the sample is further concentrated and introduced to sucrose, which serves as a cryopreservant.
Immediately after purification and dialysis, all proteins are flash frozen in small aliquots at concentrations of 20 μM (TPX2), 220 μM (RanQ69L), and 10 mg/mL (GFP-EB1).
3.1.3 Rabbit Polyclonal Antibody Production and Purification for Immunodepletion
Polyclonal antibodies for immunodepletion are generated at a commercial offsite facility, here Covance. The antigen is ideally the complete protein or protein domains, instead of peptides, to generate high affinity antibodies, as was performed for TPX2 and the augmin subunit Dgt4. Purified antibody against gamma-tubulin’s C-terminus was a gift from Christiane Weise [17].
Couple antigen covalently to a matrix, such as Affigel matrix (Bio-rad) used here.
Perform immuno-affinity purification to isolate the antibody from the serum.
Supplement antibodies with 20 % glycerol before flash-freezing them in small aliquots.
3.1.4 Xenopus Egg Extract
Preparation of Xenopus egg extracts have previously been described in great detail with complete lists of reagents required for mitotic arrested Xenopus egg extracts [18–20]. We precisely follow all steps outlined in Ref. [19]. In order to adapt the procedure to modern equipment, the packing spin during which the eggs are concentrated without lysing is no longer conducted in a clinical centrifuge, but a table centrifuge at 150 × g for 60 s followed by 598 × g for 25 s.
3.2 Branching MT Nucleation Assay in Xenopus Egg Extract
3.2.1 Preparation of Xenopus Egg Extracts
Xenopus laevis CSF extract is prepared by the standard method (Murray [18]). Freshly made extract should be stored on ice and must not subjected to shearing forces during pipetting. Extract can either be directly used (see Subheading 3.2.2) or first subjected to immunodepletion (Subheading 3.2.5) before its test in the branching assay (see Note 4).
3.2.2 Flow Chamber Assembly
Assemble a flow chamber as depicted in Fig. 1 consisting of a microscope slide, two double-sticky tape strips separated by 2–4 mm, and a glass coverslip. The chamber volume should be approximately 5 μL.
Fig. 1.
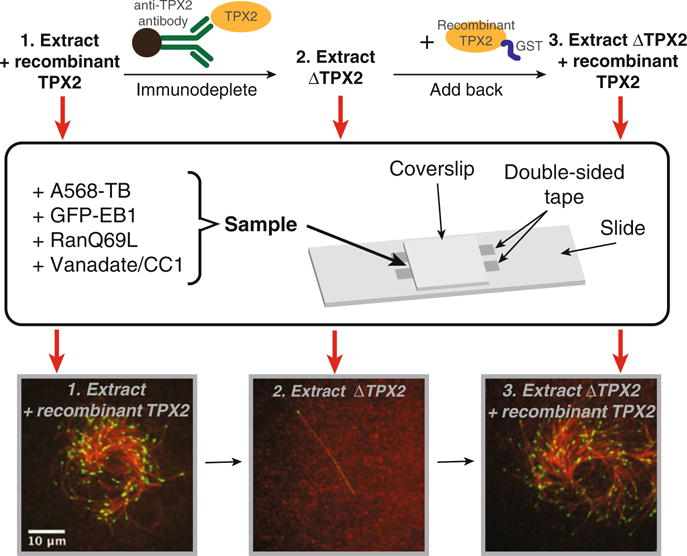
Overview of the basic branching MT nucleation assay, which is performed with Xenopus egg extract. Proteins of interest, such as TPX2, can be immunodepleted to assess their phenotype and role. Recombinantly expressed and purified proteins can then be added back to the depleted extract to test whether they complement the depleted function
3.2.3 Basic Branching Reaction (Fig. 1)
Pipette 7.5 μL CSF extract into a 1.5 mL tube on ice without shearing the extract via pipetting.
Add Alexa 568-labeled tubulin to a final concentration of 0.86 μM to visualize MTs (see Note 5).
Add EB1-GFP to a final concentration of 0.8 μM to visualize growing MT plus ends.
Add CSF-XB to obtain a final sample volume of 10 μL.
Mix the reaction mixture by gently pipetting it up and down once.
Pipette 5 μL of the reaction mixture into the flow cell to fill the channel without introducing bubbles.
Quickly seal the sample chamber with hot candle wax or nail polish.
Start imaging with an objective-based TIRF microscope equipped with a 100× 1. 49 NA objective, a low noise EM-CCD camera and laser power of at least 20 mW out-of-fiber.
Optimize imaging parameters (laser power and exposure time) to clearly observe MTs but prevent overexposure and thus photobleaching (see Note 6).
Acquire frames every 2 s for 30 min.
Collect a stitched slide overview of fields of view using the slide explorer function in uManager [21] at equal time points (for example after 15 or 30 min; see Note 7).
3.2.4 Variations of the Basic Branching Reaction (Fig. 1)
Replace 0.5 μL CSF-XB buffer with 0.5 μL vanadate (10 mM) for a final concentration of 0.5 mM or p150-CC1 to a final concentration of 0.125 mg/mL to inhibit dynein and prevent MT gliding on glass.
Replace 0.5 μL CSF-XB buffer with RanQ69L to for a final concentration of 11 μM to release endogenous spindle assembly factors from importins, such as TPX2, and induce branching MT nucleation.
Replace 0.5 μL CSF-XB buffer with TPX2 to a final concentration of 1 μM to induce branching MT nucleation.
3.2.5 Immunodepletion of CSF Extract and Add-Back of Recombinant Protein (Fig. 1)
Wash 150 μL slurry of magnetic Dynabeads 3× in 300 μL TBS-T.
Couple 36 μg total antibody to washed Dynabeads overnight at 4 °C. Use total IgG fraction as a control.
Wash off unbound antibody with 2× 150 μL TBS-T and 2× 150 μL CSF-XB.
Split beads into three 50 μL fractions. Keep two on ice, remove CSF-XB from one and add 50 μL of CSF extract after setting aside an extract gel sample.
Incubate extract with beads for 45 min on ice and gently mix every 10 min by flicking the tube or pipetting up and down with a wide bore pipette.
Retrieve beads completely on a magnet for 5 min and retrieve a gel sample of extract after this first round of immunodepletion.
Remove CSF-XB from the second bead batch and transfer extract from first to second tube for a second round of immunodepletion.
Repeat one more time for a third round of immunodepletion (see Note 8).
Perform imaging and observe phenotype.
Add-back recombinant protein to immunodepleted extract at similar concentrations as the endogenous version and take a gel sample of the final reaction mixture to verify this. Perform imaging and observe phenotype.
Prepare SDS-PAGE gel samples by mixing 2 μL of extract with 48 μL of 1× loading buffer. Perform Western blot for immunodepleted proteins and controls, and determine extent of depletion.
3.2.6 Determining Branch Angle and Measuring MT Nucleation Kinetics (See Note 9)
Use ImageJ [22] for quantitating branch angle between the daughter and mother microtubules. A branch angle of 0° corresponds to parallel growth.
Use ImageJ [22] to duplicate the EB1 channel and apply a Gaussian Blur function with a low [1] and a high [5] radius of decay for each one.
Subtract the latter image sequence from the former.
Set a threshold to eliminate background noise before counting particles for each time frame with the Analyze Particles function in ImageJ (see Note 10).
3.3 Branching MT Nucleation from Stabilized MT Seeds
Coverslips are cleaned and passivated with dichlorodimethylsilane as described [12].
Prepare GMPCPP-stabilized MT seeds, which are biotinylated and fluorescently labeled by mixing porcine brain tubulin (1.7 mg/mL final concentration), Cy5-labeled tubulin (0.19 mg/mL final concentration, ~30 % labeled), biotin-tubulin (0.19 mg/mL final concentration), and GMPCPP (1 mM final concentration) in BRB80 buffer and incubate at 37 °C for 1 h. Store at room temperature and in the dark until use.
Prepare a flow channel with passivated coverslips.
Flow in 50 μL of 0.1 mg/mL anti-biotin antibody in BRB80 and incubate for 5 min.
Flow in 50 μL of BRB80 to wash the chamber.
Flow in 50 μL kappa-casein (1 mg/mL) and incubate for 5 min.
Wash flow chamber with 50 μL of BRB80.
Flow in 50 μL MT seeds diluted 1/500 in BRB80 and incubate for 5 min.
Wash in 50 μL BRB80 with an oxygen scavenger system.
Check MT seeds for desired density with TIRF microscopy.
Flow in CSF extract with desired factors to induce branching (see Note 11).
Image as detailed above.
Acknowledgments
This work was supported by grants from the NIH/NIGMS (4R00GM100013), the Pew Scholars Program in the Biomedical Sciences, the Sidney Kimmel Foundation, and the David and Lucile Packard Foundation to S.P.
Footnotes
Only freshly prepared Xenopus laevis CSF extract of high quality remains arrested in meiosis II and displays microtubule behavior necessary for spindle assembly.
Sodium orthovanadate and p150-CC1 are both inhibitors of the minus-end directed motor protein cytoplasmic dynein.
Alternatively, unlabeled and labeled tubulin can be obtained from Cytoskeleton, Inc. if necessary.
Xenopus CSF extract only remains active for short periods of time (typically 10 h for good extract). Therefore, it is critical to plan the experiments well and minimize any delays in the procedures.
Xenopus CSF extract must not be diluted. When choosing the stock concentration of a reagent to be added, maximize the concentration and balance it with the smallest volume that can be added accurately (~0.5 μL).
TIRF angle, laser power and exposure time need to be adjusted with the first sample in order to clearly observe dynamic MTs within a bright fluorescent background. Once that is achieved, laser power and exposure time should further minimized while keeping MTs visible to prevent overexposure of the sample, which may lead to photobleaching during the course of the reaction.
uManager’s slide explorer function is useful for quantifying the number of fan structures on a slide and to assess local variation within a flow chamber.
High-affinity antibodies can deplete the protein of interest within two rounds of immunodepletion.
For this analysis, it is critical that the background signal is as low as possible. Usually this is only achieved with extract of highest quality.
Make sure that each particle detected corresponds to the EB1 signal at the growing MT ends. Each MT, even when branched, only has one EB1 comet and therefore this method directly counts MT number over time.
An additional step is to wash out the extract and flow in tubulin, which can branch from the attached seeds.
References
- 1.Lajoie-Mazenc I, Tollon Y, Detraves C, Julian M, Moisand A, Gueth-Hallonet C, Debec A, Salles-Passador I, Puget A, Mazarguil H, et al. Recruitment of antigenic gamma-tubulin during mitosis in animal cells: presence of gamma-tubulin in the mitotic spindle. J Cell Sci. 1994;107(Pt 10):2825–2837. doi: 10.1242/jcs.107.10.2825. [DOI] [PubMed] [Google Scholar]
- 2.Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubule sand mitotic spindles in cells without functional centrosomes. Curr Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 3.Brugues J, Nuzzo V, Mazur E, Needleman DJ. Nucleation and transport organize microtubules in metaphase spindles. Cell. 2012;149:554–564. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Loughlin R, Heald R, Nedelec F. A computational model predicts Xenopus meiotic spindle organization. J Cell Biol. 2010;191:1239–1249. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uehara R, Nozawa RS, Tomioka A, Petry S, Vale RD, Obuse C, Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci U S A. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, Pinchev D, Buchholz F, Peters JM, Hyman AA, Gingras AC, Pelletier L. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 11.Hyman AA. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J Cell Sci Suppl. 1991;14:125–127. doi: 10.1242/jcs.1991.supplement_14.25. [DOI] [PubMed] [Google Scholar]
- 12.Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, Schaffer E, Stear JH, Trushko A, Varga V, Widlund PO, Zanic M, Howard J. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 13.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petry S, Pugieux C, Nedelec FJ, Vale RD. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc Natl Acad Sci U S A. 2011;108:14473–14478. doi: 10.1073/pnas.1110412108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 16.Groen AC, Maresca TJ, Gatlin JC, Salmon ED, Mitchison TJ. Functional overlap of microtubule assembly factors in chromatin-promoted spindle assembly. Mol Biol Cell. 2009;20:2766–2773. doi: 10.1091/mbc.E09-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albee AJ, Wiese C. Xenopus TACC3/maskin is not required for microtubule stability but is required for anchoring microtubules at the centrosome. Mol Biol Cell. 2008;19:3347–3356. doi: 10.1091/mbc.E07-11-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 19.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 20.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 21.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using muManager software. J Biol Methods. 2014;1(2) doi: 10.14440/jbm.2014.36. pii: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasband WS. ImageJ. U.S. National Institutes of Health; Bethesda, MD, USA: 1997/2014. http://imagej.nih.gov/ij/ [Google Scholar]


