Abstract
Life depends on cell proliferation and the accurate segregation of chromosomes, which are mediated by the microtubule (MT)-based mitotic spindle and ~200 essential MT-associated proteins. Yet, a mechanistic understanding of how the mitotic spindle is assembled and achieves chromosome segregation is still missing. This is mostly due to the density of MTs in the spindle, which presumably precludes their direct observation. Recent insight has been gained into the molecular building plan of the metaphase spindle using bulk and single-molecule measurements combined with computational modeling. MT nucleation was uncovered as a key principle of spindle assembly, and mechanistic details about MT nucleation pathways and their coordination are starting to be revealed. Lastly, advances in studying spindle assembly can be applied to address the molecular mechanisms of how the spindle segregates chromosomes.
Keywords: cell division, cytoskeleton, microtubule, microtubule-associated protein, microtubule nucleation, γ-tubulin ring complex
INTRODUCTION
Life depends on cell proliferation. This occurs via cell division, during which a single cell generates two daughters, each of which retains the same genetic blueprint packaged into chromosomes via reliable chromosome segregation. The importance and fidelity of cell division becomes apparent if one considers that a human body consists of ~3.72 × 1013 cells, which were generated by an equal number of successful cell divisions (1). By contrast, chromosome segregation and cell division errors can lead to aneuploidy, yielding nonviable cells, or become a first stepping stone toward cancer.
Upon breakdown of the nuclear envelope enclosing condensed chromosomes, the mitotic spindle captures and aligns chromosomes at its center, known as the metaphase plate. The iconic fusiform shape of the metaphase spindle is characterized by mirror symmetry along this equator, referred to as bipolarity, in which each chromosome’s sister chromatid faces toward one of the two spindle poles (Figure 1). Subsequently, sister chromatids are split and their segregation toward opposite poles occurs. Lastly, spindle poles are separated and cytokinesis yields two new daughter cells, each containing one full set of sister chromatids ready for replication during interphase.
Figure 1.
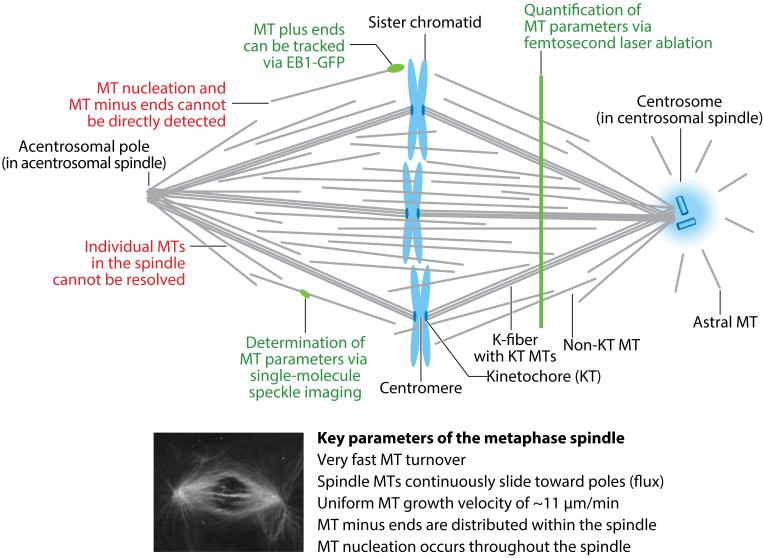
Anatomy of the metaphase spindle. To date, it has not been possible to directly determine the key parameters that describe microtubule (MT) organization within the spindle, namely the origin, dynamics, polarity, and location of each MT, presumably because the high MT density precludes their direct observation. Markers are lacking for MT minus ends and MT nucleation events, but growing MT plus ends can be tracked via the MT plus-tip tracking protein EB1-GFP (74, 75). Single-molecule speckle imaging has been applied to determine tubulin turnover and MT length distributions in the spindle (78, 79). Lastly, femtosecond laser ablation leads to MT depolymerization of newly generated plus ends, a technique used to quantify MT density and identify the locations of MT plus and minus ends (5). Altogether, these results suggest that MT nucleation combined with local MT transport govern spindle morphogenesis. Abbreviations: EB1-GFP, GFP-labeled end-binding protein 1; K-fiber, kinetochore fiber; KT, kinetochore.
Despite being central to the study of cell biology since its description in an animal cell by Flemming in 1882 (2), several aspects make the mitotic spindle one of the most challenging systems to grasp at quantitative and molecular levels. First, its size is immense, with the largest spindles measuring up to 60 μm in length (3). Second, it is complex, with a composition of up to hundreds of thousands of microtubules (MTs) as the core constituent and roughly 1,000 additional proteins in unknown copy numbers (4, 5). Third, it is one of the most dynamic cytoskeletal structures, as the mitotic spindle assembles to segregate chromosomes and again disassembles within an average of one hour using MTs that have a half-life of 60–90 s (6, 7). Fourth, it is not enclosed by a membrane, and it follows the principles of self-organization, meaning its maintenance continuously consumes and dissipates energy (8, 9).
Since Flemming’s original description, understanding the exact mechanisms of spindle assembly and chromosome segregation has been complicated by our inability to accurately determine the location of its macromolecular components in space and time. During the last decade, strides have been made in describing the metaphase spindle and its assembly at the molecular level, which is discussed in this review in the following order. Upon describing the basic spindle anatomy and defining its constituents, we first discuss bulk and single-molecule methods that have allowed inference of the dynamic behavior and organizations of MTs within the metaphase spindle and the principles on which spindle size and shape are determined. We follow with a discussion of the multiple MT nucleation pathways that play key roles in spindle assembly and how they are coordinated to generate a functional spindle. These advanced methods for probing MT organization in the metaphase spindle have yet to be applied to determine the molecular mechanisms by which chromosomes are captured, aligned, and reliably segregated during cell division.
WHAT IS THE METAPHASE SPINDLE?
Macroscopic Anatomy of the Metaphase Spindle
The overall organization of the metaphase spindle is well established (Figure 1). Kinetochore MTs connect the pole to special attachment sites of the sister chromatids called kinetochores (KTs). These connections form MT bundles (K-fibers) and directly mediate chromosome segregation. Non-KT MTs make up the rest of the spindle, and astral MTs extend from the poles outward to anchor the spindle within the cytoplasm.
In many cell types, the poles of the spindle are connected to centrosomes, which were discovered as the first cellular structures from which MTs emanate (10). Centrosomes are nearly 1 μm in diameter and consist of a centriole pair surrounded by pericentriolar material (PCM). The centrosomal proteome consists of about 100 proteins (11), and recent superresolution studies provided novel insights into the organization of the PCM (12, 13). By contrast, some cell types, such as egg meiotic cells and plant cells, do not contain centrosomes, and the two poles form the boundaries of an “acentrosomal” spindle.
Vertebrate chromosomes are characterized by the primary constriction that holds the two sister chromatids together, the centromere, which ranges from 123 base pairs in yeast to megabase pairs in metazoans (14). The centromere forms the base onto which the KT, consisting of ~60 proteins arranged into several parallel plates, of each sister chromatid is assembled (15). In addition to being the attachment site of KT MTs, KTs are responsible for detecting errors via checkpoints and for ultimately enabling the transport of sister chromatids to poles via K-fibers.
In the 1950s, the mitotic spindle was shown to consist of fibrous structures, whose submicroscopic birefringent fibrils turned out to be MTs (16). By 1968, it was known (17) that MTs are made up by the 6S GTP-binding protein tubulin, consisting of an α- and a β-subunit (55 kDa each). The subunits align in an end-on fashion to form protofilaments. On average 13 protofilaments form a hollow MT (Figure 2) (17). The ends at which the α- and β-subunits are exposed are defined as the MT minus and plus ends, respectively. MTs form above a so-called critical concentration of tubulin and in the presence of GTP with a slowly growing minus end and a fast-growing plus end. During the polymerization process, the GTP bound to β-tubulin becomes hydrolyzed to GDP, inducing a strain in the MT lattice that builds up before it is released by spontaneous depolymerization. This process results in dynamic instability, i.e., the stochastic switching between growing and shrinking phases, which is the hallmark of MTs (18).
Figure 2.
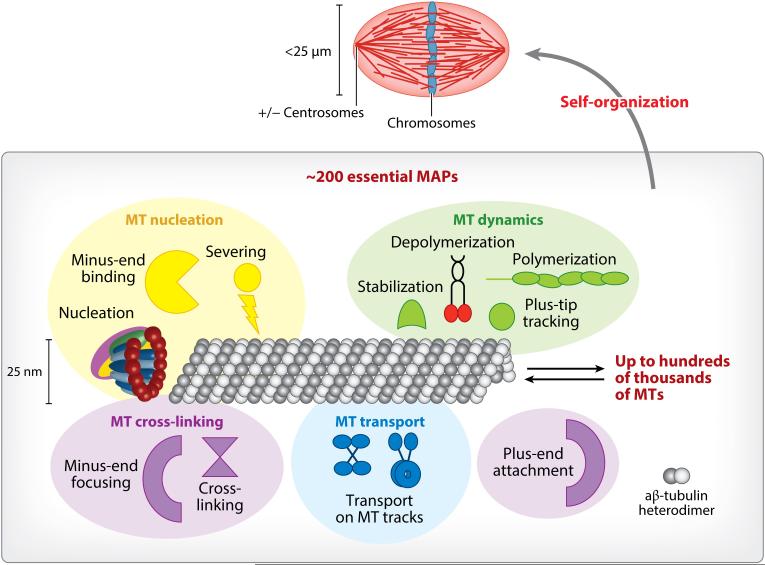
Self-organization of the metaphase spindle. The mitotic spindle is made of up to hundreds of thousands of microtubules (MTs) and roughly 1,000 MT-associated proteins (MAPs), of which ~200 are essential (22–25). Spindle MAPs can be grouped into several activity classes, namely MT nucleation, MT dynamics, MT transport, and MT cross-linking. Aside from MT nucleation by the ring-shaped γ-TuRC, the MT number can be increased by MT severing and MT minus-end-binding proteins. MT dynamics are regulated by MT polymerases, MT depolymerases, plus-end tracking proteins (+TIPs), and MT stabilization factors. MT cross-linking can occur at minus and plus ends or along the MT lattice. Finally, molecular motors are responsible for transport on MT tracks. MAPs and MTs self-organize into the functional spindle, whose maintenance continuously consumes and dissipates energy.
MTs cannot accomplish chromosome segregation by themselves. Although MTs can nucleate, grow, and shrink in solutions containing only purified tubulin and GTP, MT-associated proteins (MAPs) regulate MT nucleation, dynamics, and transport as well as MT cross-linking in cells. In the spindle, MAPs help define where and when MTs are generated, and they determine spindle MT length and dynamics as well as location and orientation. They are also involved in attaching chromosomes to MTs anchored at poles to generate tension, sense errors, and segregate chromosomes. In essence, MAPs modulate the MT parameters that ultimately define spindle shape and bipolarity, and together with MTs, they make up the microscopic organization of the spindle.
Complete List of Spindle Microtubule-Associated Proteins and Their Definition
Traditionally, individual studies identified spindle MAPs via loss-of-function experiments. This effort was first complemented by proteomic studies that defined all the components of spindle elements, such as the centrosome (11), the mitotic chromosome (19), and spindle MTs (20), as well as the complete spindle (4) and its phosphorylation sites (21). These studies identified more than 1,000 different spindle proteins, including hundreds of uncharacterized ones. The advent of high-throughput technology enabled screens at the whole-genome level that defined ~200 of these MAPs to be essential for spindle assembly (22–25), reporting their phenotype, overall location, and interaction patterns. Thereby, the complete protein inventory of the spindle is accessible. As of now, information about a particular spindle MAP still has to be looked up in each separately published database. Hence, a necessary future step to maximize the impact of these valuable studies is their integration into a unified spindle genome and proteome database.
The activity of spindle MAPs is essential for spindle formation and can be grouped into several activity classes (Figure 2). First, MT nucleation factors generate MTs from spatially discrete sites, termed MT organizing centers (MTOCs), that play a key role in constructing the mitotic spindle. This nucleation process is usually dependent on γ-tubulin, which together with γ-tubulin complex proteins GCP3, GCP4, GCP, and GCP6 exists in the form of a ring-shaped γ-tubulin ring complex (γ-TuRC) in metazoans (26–29). An outer ring of 13 γ-tubulin molecules in γ-TuRC is thought to provide a template that directly interfaces with the α-subunits of the first tubulin dimer of the MT lattice. Studies on the smaller yeast γ–tubulin complex revealed a ring radius wider than the MT lattice and suggested that a more compact ring that directly matches the MT diameter could facilitate MT nucleation (30, 31). It remains to be determined which effectors induce this conformational change, whether ring closure is the only mode of catalysis, and whether this also applies to the larger γ-TuRC. Alternative ways to possibly induce MT nucleation include the assembly and specific attachment of γ-TuRC to MTOCs. Finally, how tubulin dimers are assembled into a tubule from the γ-TuRC base is still unknown (32–34). It is possible that other MT nucleators exist (35, 36), and MT severing could generate more seeds for nucleation (37). By binding to the minus end, γ-TuRC and other MT minus-end-binding proteins cap MT minus ends in cells, thereby preventing their MT dynamics, but it is unclear whether MT minus-end proteins other than γ-TuRC also harbor MT nucleation activity (38).
Once nucleated, MAPs regulate MT dynamics by selectively stabilizing specific conformations of tubulin dimers (39). MT polymerases promote MT growth or rescue depolymerizing MTs by favoring the straight tubulin dimer conformation occurring in the MT lattice (40). By contrast, MT depolymerases promote MT catastrophe by supporting the intrinsic curvature of the tubulin dimer, which it assumes in isolation (41). MT dynamics are further regulated by MT plus-end tracking proteins (+TIPs), which recruit MAPs including MT polymerases and depolymerases (42). The +TIPs highest in the hierarchy are end-binding proteins (EBs) (43). The canonical member, EB1, acts as an MT maturation factor by transitioning GTP to GDP-tubulin in vitro (44). Recent studies have demonstrated that there is a binding hierarchy for limited binding sites at the growing MT plus ends. These guide dynamic assembly of +TIP ensembles and determine their resulting functions (45, 46). The MT lattice can also be allosterically regulated by MAPs, which do not directly contact each other, adding a new layer of complexity to the modes of how MAPs can modulate MT dynamics (47).
The precise arrangement of MTs is controlled by molecular motors and MT-bundling proteins. Kinesins walk toward MT plus ends and exist as a large superfamily with differing motor properties and cargo-binding domains (48). By contrast, only one cytoplasmic dynein fulfills the role of the minus-end-directed motor, which gets recruited by specific adapter proteins and thereby activated for transport once loaded with the right cargo (49, 50). Molecular motors can also transport MTs along MTs. If more than one motor head exists, molecular motors like dynein (51) and Eg5 (52) can slide MTs in an antiparallel fashion. MTs can also be simply tethered to each other lengthwise via MT cross-linking proteins (53). Cross-linking at MT ends can lead to aster formation (54).
Although advances at the single-molecule and structural levels are numerous for many individual MAPs, it is clear that most MAPs do not act by themselves. For example, the ~200 proteins essential for spindle assembly are estimated to arrange into 100 protein complexes (24). Thus, future studies need to take these functional assemblies into account. Regardless, the next major challenge is to understand how these MAPs work together with MTs at the nanometer scale to assemble the mitotic spindle, which is a factor of 1,000 larger than its constituents, and to orchestrate chromosome segregation, as discussed in the next section.
Molecular Building Plan of the Metaphase Spindle
How MTs achieve reliable chromosome segregation has been a mystery ever since it was clear that MTs constitute the mitotic spindle. Answering this question requires knowing the precise organization of MTs in the metaphase spindle. To date, it has not been possible to directly determine the key parameters, e.g. the origin, dynamics, polarity, and location of each MT throughout cell division. This is hampered by the fact that individual MTs in the spindle cannot be resolved by light microscopy methods, presumably because the high MT density precludes their direct observation. By contrast, electron microscopy (EM) reconstructions can resolve the location of individual MTs but lack dynamic information. Moreover, EM methods can be feasibly applied only to the smaller yeast spindle (~40 MTs and 2 μm long) (55, 56) or thin slices of larger metazoan spindles (up to hundreds of thousands of MTs and 60 μm long) (57–60).
Instead, a variety of models have been proposed that use bulk methods to infer the dynamics of MTs in the spindle (Figure 1). Two key characteristics define spindle MTs. First, early perturbation and fluorescence recovery after photobleaching studies revealed that spindle MTs exchange very quickly within tens of seconds with the soluble tubulin pool (61, 62). Second, speckle microscopy and photomarking experiments uncovered that spindle MTs continuously slide toward the poles, termed MT flux (6). How MTs can be organized into the spindle shape, display these dynamics, and effectively segregate chromosomes has remained mysterious.
Many models of spindle morphogenesis that take the high turnover rate and flux into account have been proposed. Early on, dynamic instability was suggested as a behavior responsible for the high MT turnover (6, 7). But how could a spindle be formed if its core constituents are intrinsically unstable? Different turnover rates were measured at the middle of the spindle versus near poles (63, 64), and selective MT stabilization at poles has been suggested (65). The spindle environment could also stabilize MTs (66, 67), and a mechanical matrix could support the spindle (68). Tight length regulation has been proposed as a principle to shape the spindle (69). Spindle shape could also be generated by combining the characteristics of MT flux, i.e., MT sliding toward poles, minus-end depolymerization at poles (70, 71), and a reaction-diffusion system of soluble signaling molecules that nucleate and stabilize MTs near chromosomes (67, 72). A balance of forces between different mechanical elements can maintain a spindle (73). Although combinations of these models may contribute to spindle morphogenesis and differences exist between cell types and organisms, variations in these models are also a result of the inability to directly determine MT organization in the spindle.
During the last decade, strides have been made by applying single-molecule methods to determine MT dynamics in the spindle more accurately (Figure 1). This has been particularly fruitful in determining MT organization in the Xenopus laevis egg extract spindle because it can be assembled in vitro and is biochemically accessible, as explained below in this section. The tracking of MT plus ends via EB1-GFP allowed determination of a plus-end polymerization velocity of 11 μm/min with no spatial variations in this speed throughout the spindle, in contrast with previous suggestions (74). Polymerizing ends near the spindle middle were enriched and could be caused by enhanced local nucleation or rescue. The tracking of MT plus ends via EB1-GFP also allowed investigators to accurately determine the distribution of growing MT ends in Drosophila S2 cells. Forming EB1-GFP punctae were observed within the spindle and from acentrosomal poles, suggesting that the spindle propagates its own architecture by promoting MT assembly throughout the spindle and not only from centrosomes or chromosomes as previously thought (75).
These findings were corroborated by the discovery that MT minus ends are not only anchored at centrosomes but are also distributed all throughout the spindle (76). By combining fluorescence speckle microscopy with cross-correlation analysis, the orientation of MTs and the localization of MT plus ends could be determined. This resulted in the slide and cluster model in which an MT travels a certain distance before it undergoes depolymerization (77). This model favored a nucleation gradient around chromosomes as the MT origin. Next, the movement of single tubulin molecules in spindles of X. laevis egg extracts were analyzed, using single-fluorophore speckle imaging. These studies revealed a mean length of MTs that was 40% of the spindle length and dependent on the antiparallel sliding activity of Eg5 (78). MTs of broadly distributed lengths displayed heterogeneous velocities and moved together when close to each other. The resulting model was described as a tiled array of individual filaments. Instead of the traditional model in which only centrosomal MTs generate tension between sister chromatids, this tiled and connected array of MTs transmits force as a system (78).
Single-molecule imaging was also used to measure tubulin turnover within a spindle in X. laevis egg extracts, showing that tubulin molecules only spend a few seconds in a polymer (79). Interestingly, tubulin residence time in MTs is short relative to transport, and MT turnover did not differ between the spindle and other MT structures. This ruled out stabilization as a mechanism to generate the high MT density within the spindle and pointed to increased MT nucleation as the major principle for spindle morphogenesis. Most recently, femtosecond laser ablation of MTs was applied to quantify the length distribution and the polarity of MTs throughout spindles, along with the density and locations of MT plus ends and minus ends (5). These elegant experiments were based on the principle that following laser ablation, newly generated MT plus ends depolymerize, whereas MT minus ends remain stable. These data suggest that, in contrast to previous views, MTs are shortest at the poles and progressively increase in length toward the center of the spindle. The findings further suggest that a nonuniform nucleation profile controls spindle length and that strictly local transport sorts MTs of a particular length to a certain space with a defined polarity, giving rise to the nucleation and transport model of spindle organization.
Although these indirect measurements at the single-molecule level have generated the most precise model of spindle morphogenesis so far, an accurate description of the molecular building plan will require new methods to directly measure parameters that are key to explaining various features of MT organization, including the point of MT nucleation, the growth dynamics, and the orientation and location of MTs in the spindle throughout the cell division process. MT plus ends can be directly tracked using fluorescently labeled EB1, but it is not possible to directly determine the locations of MT minus ends or MT nucleation events throughout the spindle (Figure 1). Although the MT nucleator γ-tubulin could be visualized in spindles (80), its location does not indicate sites where MTs are being nucleated because γ-tubulin can also cap the MT minus end. In addition, γ-tubulin data do not reveal all MT minus ends because several other MT minus-end binding proteins also cap MT minus ends in the spindle. Even though advances have been made in understanding the organization of non-KT MTs, as described above, it is necessary to characterize the MT dynamics of K-fibers and other MT populations to the same extent to ultimately achieve a quantitative understanding of how chromosomes are segregated.
Scaling of the Metaphase Spindle
The precise organization of MTs during spindle assembly remains to be determined; however, it has been possible to start addressing how spindles assume a certain size (Figure 3). Systematic, genome-wide screens for mitotic proteins that influence spindle lengths were conducted in Caenorhabditis elegans, Drosophila S2 cells, and vertebrate cultured cells (81). Biochemically accessible Xenopus extracts have proven to be an excellent system to study the effect of individual factors and have identified a particularly important role for the MT polymerase XMAP215 in regulating spindle length. By replacing endogenous XMAP215 with versions of gradually increasing enzymatic activity, MT growth velocity could be increased linearly with spindle length. This occurs without changing MT density, MT lifetime, and spindle shape, suggesting that spindle size is determined separately and by mass balance (82). Recent studies in human cells and C. elegans have shown that spindle shape anisotropically scales with spindle length and chromosome number (83, 84). The specific factors in these organisms that influence spindle shape, however, still need to be established.
Figure 3.
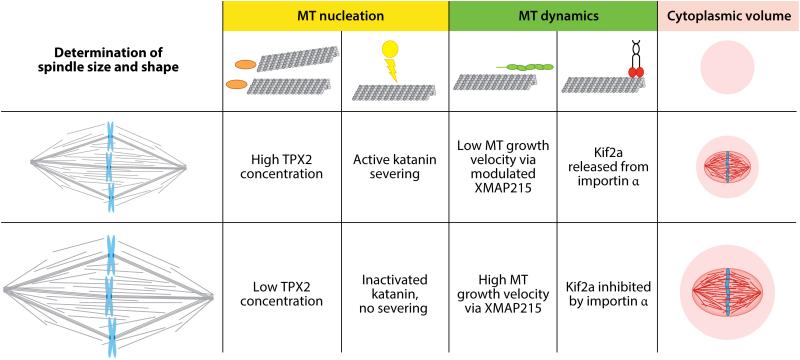
Scaling of the metaphase spindle. It is possible to start addressing how spindles assume a certain size and shape. Spindle length can be modulated by varying the concentration and activity of the microtubule (MT) polymerase XMAP215 (81, 82). During later stages of development, the MT depolymerase, Kif2a becomes inhibited and results in smaller spindles (86). Alternatively, the amount of cytoplasmic material can regulate spindle size (87, 88). Differences in the activity of katanin, as well as the activity and concentration of targeting factor for Xklp2 (TPX2), can explain spindle size and architecture changes between large Xenopus laevis and small X. tropicalis spindles (90, 91).
Although every cell has a spindle of a certain size and shape, differences exist between cell types of the same species and require spindle scaling. This becomes apparent if one considers the reduction in cell volume from a fertilized egg to a 1,000 cell embryo (85). In X. laevis, this developmental spindle scaling is driven by the kinesin 13 Kif2a, whose MT-destabilizing activity is inhibited in large, early spindles (stage 3) by the transport receptor importin α. Kif2a activity is activated in later and smaller spindles (stage 8) when importin α partitions to a membrane pool (86). Changing spindle size in developing embryos has no effect on chromosome segregation but interferes with spindle orientation. This suggests that spindle orientation is coupled to cell size through a ratiometric mechanism controlling MT destabilization (86).
This idea was further explored via an innovative system in which Xenopus egg extracts were encapsulated using microfluidic technology in two independent studies (87, 88). Both beautifully demonstrated that reductions in cytoplasmic volume, rather than developmental cues or changes in cell shape, were sufficient to replicate the spindle scaling observed in Xenopus embryos. Thus, the amount of cytoplasmic material provides a mechanism for regulating the size of intracellular structures.
Related species with size differences also display spindle scaling. Spindles in Xenopus tropicalis extract are smaller than their X. laevis counterparts (89). A major effector of this size difference is the MT-severing enzyme katanin. Katanin activity is much higher in X. tropicalis egg cytoplasm than in X. laevis, and it is more concentrated at spindle poles (90). Inhibition of katanin led to a greater increase of spindle length in X. tropicalis than X. laevis, suggesting that it acts as a scaling factor. The activity difference between katanin of the two species can be attributed to their differential regulation. The catalytic p60 subunit of X. tropicalis katanin lacks an inhibitory Aurora B kinase phosphorylation site, an absence which could explain its higher activity. Additional differences between X. laevis and X. tropicalis extract spindles exist. For example, the X. tropicalis spindle resists inhibition of two factors essential for assembly of the larger X. laevis spindles: the small GTPase Ran and the kinesin Eg5 (91). This resistance may be attributed to the level of targeting protein for Xklp2 (TPX2), a common target of RanGTP and Eg5 involved in MT generation that is threefold more abundant in X. tropicalis extracts. Interestingly, increasing the TPX2 concentration in X. laevis reduced spindle length and sensitivity to Ran and Eg5 inhibition. In addition, this concentration increase recruited more Eg5 to the spindle poles, which consequently increased local MT density. These results suggest that the balance of TPX2 and Eg5 modulates spindle architecture (91).
In summary, differences in the individual factors of small and large spindles provide mechanistic insight into how one MAP or condition can influence spindle size and architecture, as demonstrated by these examples. These studies also highlight the importance of quantifying the activity level and copy number of individual MAPs to understand how they contribute to the physiological size and architecture of a particular spindle. Ultimately, information about the function, activity, and location of spindle MAPs needs to be combined with the directly measured parameters of MT organization to derive a complete molecular model of the metaphase spindle.
Theory of Spindle Maintenance
Given the limited information about the precise organization of MTs and MAPs in the spindle, our understanding of spindle morphogenesis has been significantly aided by theoretical modeling (77, 92, 93). In the most recent and complete model, a bipolar structure with antiparallel fluxing MTs similar to those in a spindle was achieved in silico by incorporating dynamic MTs, an MT cross-linking force, and antiparallel sliding activity. Pole formation of this bipolar structure was mediated by the addition of a nuclear mitotic apparatus protein (NuMA)-like minus-end cross-linker, and its length was maintained by MT depolymerization activity at the minus end (71). Realistic MT lifetimes and MT length distributions required dynamic instability and minus-end depolymerization activities. Most importantly, meiotic spindle assembly could only be modeled by simulating MT nucleation throughout the spindle. This complements the experimental findings that MT nucleation and transport drive spindle assembly and that MT dynamics are constant throughout the cytoplasm (5). As the mechanistic understanding of MAPs increases, computational modeling will continue to play an important role in integrating experimental data toward a complete molecular mechanism of metaphase spindle assembly.
HOW TO ASSEMBLE A SPINDLE
In addition to a better understanding of the molecular building plan of the metaphase spindle, progress has also been made that addresses how a spindle is assembled in the first place. Upon entering mitosis, the interphase MT network is almost completely disassembled, and the MT network of the spindle is generated de novo. The first key step is the nucleation of MTs, which occurs from MTOCs. Originally, the term MTOC was synonymous with centrosome, which was believed to be the only source of MTs for the spindle. For the past two decades, new MTOCs have been added to a growing list and significantly changed the view of how the spindle is assembled.
Spindle Assembly from Centrosomes
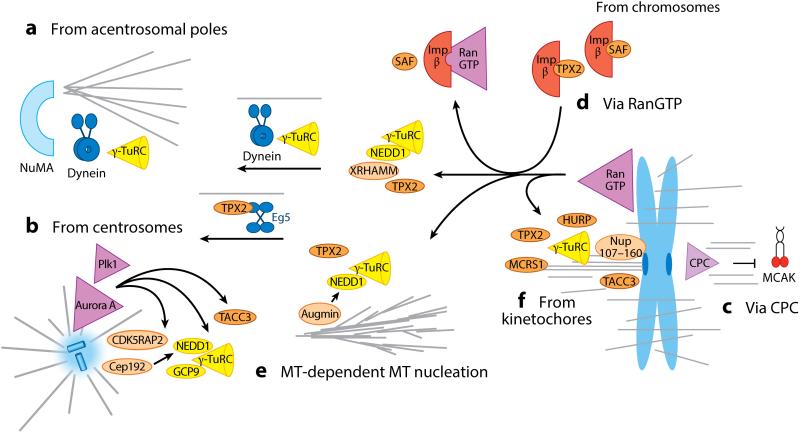
During interphase, centrosomes become duplicated at the nuclear envelope. Upon entering mitosis, centrosomes move apart from each other and mature to more active MT nucleation centers while the nuclear envelope breaks down. MT nucleation from centrosomes is mediated by γ-TuRC, and several recruiting factors have recently been identified (Figure 4b). One of these factors is the WD domain protein GCP-WD/NEDD1 (neural precursor cell expressed, developmentally down-regulated 1), which copurifies with γ-TuRC, and whose depletion impairs γ-TuRC localization and MT nucleation at the centrosome (94–96). Another protein that copurifies with γ-TuRC, and is required for centrosomal localization in mitosis, is GCP9/MOZART1 (mitotic-spindle organizing protein associated with a ring of γ-tubulin). It recruits γ-TuRC to mitotic centrosomes, and its depletion leads to spindle defects (24, 97). It is unknown whether or not GCP-WD and GCP9 act as activation factors in addition to their recruitment roles. Other proteins are implicated in the recruitment of γ-TuRC to centrosomes, such as pericentrin (98) and AKAP450 (99) with PACT domains for centrosomal targeting (100), as well as Cep192, which recruits GCP-WD (101). So far, the protein CDK5RAP2 is the only protein with defined activity in γ-TuRC attachment to centrosomes and γ-TuRC activation (102). In vitro CDK5RAP2 activates MT nucleation by the γ-TuRC 7.5 fold via a small 5.5-kDa motif, termed the γ-TuRC-mediated nucleation activator (102). The enhanced MT nucleation activity of centrosomes upon prometaphase is achieved by the recruitment of more γ-TuRC (103, 104) and components of the PCM, such as pericentrin and centrosomin/CDK5RAP2 in flies (105). It is also increased by phosphorylation of centrosomin via the kinases Polo like 1 (Plk1) (106) and Aurora A, whose complete set of targets remain to be identified. However, it is known that the Aurora A kinase phosphorylates and activates centrosomal proteins, such as TACC/Maskin, to promote MT growth (107, 108).
Figure 4.
Spindle assembly pathways. The first key step to assemble the mitotic spindle is γ-TuRC–dependent microtubule (MT) (b) nucleation from MT organizing centers. MT nucleation occurs from centrosomes to which γ-TuRC is recruited via its direct binding partners NEDD1 and GCP9 as well as via the centrosomal proteins Cep192 and CDK5RAP2 (24, 94–99, 102, 107, 174). The latter, together with the Aurora A kinase and Plk1, activates MT nucleation and polymerization (e.g., via TACC3/Maskin). (a) Less is known about MT nucleation from acentrosomal poles, which are formed by the combined action of the cross-linker NuMA and dynein (170). Dynein transports γ-TuRC poleward (59). (c) At chromosomes, the CPC inhibits MT depolymerases, such as MCAK, to induce MT formation (59, 72). (d ) The key chromosomal pathway is activated by RanGTP, which in turn releases SAFs from sequestering importins (129–131). The central SAF, TPX2, induces MT formation via its N terminus, together with XRHAMM, NEDD1, and γ-TuRC (139). (e) The C terminus of TPX2, together with augmin, induces branching MT nucleation throughout the spindle, a distribution that may be facilitated by the transport of TPX2 by the kinesin Eg5 or a poleward flux (147, 154). ( f ) The SAFs TPX2, HURP, MCRS1, and Nup107–160 play a yet-to-be-defined role in MT nucleation in the vicinity of kinetochores (133, 164, 168). Abbreviations: CPC, chromosomal passenger complex; GCP, γ-tubulin complex protein; γ-TuRC, γ-tubulin ring complex; NEDD1, neural precursor cell expressed, developmentally down-regulated 1; Plk1, Polo-like kinase 1; SAFs, spindle assembly factors; TPX2, targeting factor for Xklp2; XRHAMM, hyaluronan-mediated motility receptor.
Centrosomes were originally believed to be the only MTOCs and to generate MTs in all directions. Centrosomal MTs were shown to display dynamic instability and to become stabilized once they are captured by a KT (109, 110). This search and capture mechanism provided the first basis of spindle assembly (111). The increased nucleation rate of the centrosome and the tenfold higher MT turnover rate in mitosis allow centrosomal MTs to sample the volume of the cell faster for chromosome capture (112). However, centrosomes can be ablated (113), flies can survive without centrosomes (114), and many cell types do not contain centrosomes at all (115). These observations were the first indications that other MT nucleation pathways for spindle assembly might exist.
Spindle Assembly from Chromosomes
A centrosome-independent mechanism of MT nucleation was proposed to emanate from chromosomes, which sort MTs into an antiparallel array (116). The ultimate demonstration of chromosomal MTOCs involved the use of chromatin beads that induced the formation of acentrosomal spindles (9). When centrosomes were sufficiently separated from chromosomes, MT generation from both kinds of MTOCs was visible, demonstrating that chromosomal spindle generation is also a general mechanism (117).
The size of chromatin has a major effect on the dimensions and symmetry of spindles (118). This was examined in X. laevis egg extract using microprinting of chromatin beads. Larger circular spots loaded with many chromatin beads generated a larger number of poles. When the chromatin beads were arranged into a line, line length determined the number of poles. Thicker lines loaded with chromatin beads led to the formation of asymmetric structures in which all MTs were on the same side of a line. Thus, chromatin defines spindle shape and orientation.
How do chromosomes induce spindle assembly? The chromosomal proteome contains many important regulators of MT dynamics, motor proteins, and the KT, which affect spindle dynamics (19, 67). Again, the most important step in spindle assembly from chromosomes is the formation of MTs. Several pathways have been identified that lead to MT nucleation around chromosomes.
The RanGTP-Mediated Microtubule Nucleation Pathway
A key protein that mediates self-assembly of the mitotic spindle from chromosomes is the small GTPase Ran (119, 120). When added in its GTP form to Xenopus egg extract, it induces MT asters and small bipolar structures that resemble spindles. This assembly also requires γ-tubulin, Eg5, and XMAP215 (121–124). RanGTP is generated via its guanine nucleotide exchange factor RCC1, which is bound to chromatin, and its GTPase-activating protein RanGAP resides in the cytoplasm. This inherently leads to a gradient of RanGTP originating from chromatin, a distribution detected in both Xenopus (125, 126) and human cells (127). The size of the gradient can be modulated by alternating the ratio of the controlling factors. Reduction of the gradient-spanning distance results in a loss of spindle symmetry and fewer attached chromosomes (126). RCC1 inhibition or RanGTP addition impairs spindle assembly in extracts but not in cells once the spindle has formed (127). Beads coated with RCC1 were sufficient for generating whole spindles in Xenopus extract, demonstrating the central importance of RanGTP in spindle assembly (128).
How does RanGTP induce spindle self-assembly? Independent of their role in nuclear export, importins α and β sequester spindle assembly factors (SAFs) until RanGTP releases them so they can contribute to MT formation and stabilization (Table 1 and Figure 4d) (129–131). RanGTP-regulated factors involved in spindle assembly continue to be discovered. One of the most important and best-characterized SAFs is TPX2, which was already mentioned in the context of metaphase spindle scaling in X. tropicalis and X. laevis. TPX2 is sequestered in the interphase nucleus and targets the kinesin Xklp2 to spindles in mitosis (132). Its depletion causes defects in spindle and pole organization in the presence of centrosomes and blocks MT generation from chromosomes (129, 133). If the nuclear localization signal of TPX2 is mutated, TPX2 does not require Ran for MT aster formation, and the in vitro MT nucleation activity cannot be blocked by importin α (134). One of the best-characterized functions of TPX2 is that, once released, TPX2 interacts with and activates the Aurora A kinase, which phosphorylates TPX2 (135–137). This event induces the phosphorylation of other substrates, including Eg5 and TACC/Maskin (138), but how this secondary phosphorylation gradient contributes to spindle assembly remains poorly understood. Recently, TPX2 was found to form a complex with NEDD1, RHAMM (hyaluronan-mediated motility receptor), and γ-TuRC to nucleate MTs in a RanGTP-dependent manner (Figure 4) (139). TPX2 also interacts with several other SAFs and thus plays a central role in spindle assembly.
Table 1.
RanGTP-regulated spindle assembly factorsa
| Spindle assembly factors |
Activities and roles | References |
|---|---|---|
| TPX2 | Activates Aurora A kinase Organizes spindles and poles Associates with BRCA1/BARD1, NuMA, and XRHAMM to modulate spindle assembly Nucleates microtubules (MTs) Associates with XRHAMM, NEDD1, and TPX2 to nucleate MTs Is required for branching MT nucleation together with augmin and γ-TuRC |
129, 132–137, 139, 175 |
| NuMA | Provides spindle pole organization with dynein | 170 |
| Kid | Is a chromosomal kinesin Provides chromosome oscillations and alignment |
176 |
| XCTK2 | Is a kinesin Mediates spindle pole formation Provides spindle stability |
177 |
| Xnf7 | Bundles MTs Provides spindle integrity |
178 |
| Rae1 | Is an mRNA export factor Is involved in the spindle checkpoint Confers RNA-dependent MT-stabilizing activity |
179 – 181 |
| HURP | Bundles MTs Localizes to K-fibers proximal to chromosomes Is required for proper chromosome alignment Forms a complex with TPX2, Aurora A kinase, XMAP215, and Eg5 |
182, 183 |
| TACC3/Maskin | Promotes MT growth | 107, 174 |
| Lamin B | Forms a spindle matrix Associates with Eg5 and XMAP215 |
184 |
| NuSAP | Bundles MTs Contributes to interactions between MTs and chromatin |
185 |
| ISWI | Has a role in MT nucleation Bundles MTs |
186 |
| Nup103–160 | Helps form K-fibers | 164 |
| MCRS1 | Helps form K-fibers | 168 |
RanGTP induces spindle self-assembly by liberating spindle assembly factors (SAFs) from their inhibited importin-bound state. SAFs fall into all MT-associated protein activity classes. One of the best-characterized SAFs is TPX2. However, less is known about the other SAFs listed in this table.
Less is known about the other identified SAFs (Table 1). Binding of importin α or β to most SAFs alters their activities, although the exact mechanism by which this RanGTP-mediated regulation occurs remains to be determined. The RanGTP gradient is often described as the MT stabilization and nucleation gradient, but the latter may be the more critical output. It is currently unclear whether these SAFs all act in separate or in common MT nucleation pathways and how exactly they contribute to MT formation and organization toward subsequent spindle assembly.
Microtubule-Dependent Microtubule Nucleation from Within the Spindle
Evidence began to accumulate that MTs may be generated not only at centrosomes and chromosomes but also within the body of spindles (5, 75). This was particularly attractive because the nucleator γ-tubulin also localizes throughout the spindle and after all actin filaments branch with a characteristic angle of 70°. MTs had already been seen to branch off existing MTs at the cortex in plant cells in a process that was γ-tubulin-dependent and occurred with defined angles of ~40° and 0° in two independent studies (140, 141). In Schizosaccharomyces pombe, MT nucleation had been shown to produce daughter MTs that were antiparallel to the mother MT (with a 180° angle) (142). Next, whole-genome screens in Drosophila and human cells led to the identification of eight genes that specifically target γ-tubulin to spindle MTs but not to centrosomes (22, 23). The corresponding proteins form the eight-subunit augmin complex (143–145). How these subunits interact was recently identified in a recombinant augmin complex (146). In the absence of augmin, animal cells display chromosome segregation defects and a reduction in MT density within the spindle (144, 145).
Although it is clear that new MTs arise within the spindle and augmin is a γ-TuRC-targeting factor for spindle MTs, the very high MT density precludes direct observations of how MTs are made in the middle of the spindle. This limitation could be overcome by using cell-free extracts of X. laevis eggs and total internal reflection microscopy. Using fluorescent tubulin and EB1 to mark newly growing MT plus ends, individual spindle MTs could be resolved at their point of nucleation, thereby directly demonstrating that newly formed MTs grew from the wall of preexisting MTs in a process termed MT-dependent MT nucleation (147). Unexpectedly, the branch angle between the daughter and mother MT was shallow, and most often the newly nucleated MTs grew exactly parallel along the template MT. As a result, the polarity of the template MT remains preserved via this nucleation mechanism. In addition, MT-dependent MT nucleation leads to an exponential amplification of MT number over time. Thus, MT-dependent MT nucleation can quickly amplify MT numbers while preserving the original polarity of the template MT, which appears to be important for parallel, MT-rich structures such as the spindle, and which may help explain how spindles can be rapidly assembled. By propagating the architecture of existing MTs, branching MT nucleation is ideally suited to maintain MT density within the spindle and throughout the metaphase steady state. Because this mechanism is activated by RanGTP near chromosomes, MT formation is directionally biased and can help explain how boundaries can be created at the edge of the spindle.
The Xenopus extract system also allowed the identification of γ-tubulin, augmin, and TPX2 as key molecular factors of MT-dependent MT nucleation (Figure 4e). How these factors work together to induce this reaction and what the branch point looks like remain to be resolved. In parallel, augmin was identified in plants, where it plays an important role in the formation of the plant spindle, the MT-rich phragmoplast, and the plant cortex (148–151). Augmin and TPX2 provide the first clear connection between RanGTP as the upstream factor and a γ-tubulin-dependent MT nucleation pathway. Compared to models in which RanGTP activates soluble MT nucleators with randomly oriented MTs that require sorting by molecular motors (152), branching MT nucleation simplifies that task by using the information of preexisting MTs to define the polarity of new MTs. An interesting aspect of this connection is that RanGTP stimulates MT nucleation in a gradient that decays with distance from chromatin (126, 127, 153), whereas augmin and γ-TuRC are located throughout the spindle (101, 143–145) and could presumably nucleate MTs in a more uniform distribution (5, 75). Therefore the question remains of how branching MT nucleation is spatially and temporally regulated. Interestingly, TPX2 undergoes poleward transport during spindle assembly, in a process dependent on Eg5 (Figure 4e) (154). Because TPX2 is required for branching MT nucleation and is released by RanGTP in a gradient from chromatin, it is conceivable that poleward transport, possibly combined with poleward flux, is responsible for distributing the nucleation activity equally throughout the spindle. Thus, the actions of MT motors and branching nucleation can synergize to create and maintain the density and polarity of MTs in the spindle.
Chromosomal Passenger Complex-Induced Microtubule Formation
A chromatin-mediated pathway that seems to function independently of RanGTP is driven by the chromosomal passenger complex (CPC). The CPC has been proposed to negatively regulate local MT-depolymerizing proteins, such as MCAK and Op18/Stathmin, thereby facilitating the stabilization of MTs and initiating spindle formation from chromosomes (Figure 4c) (72). Although inactivated by phosphatases during interphase, at mitosis the subunit Dasra/Borealin recruits the rest of the CPC consisting of Incenp, survivin, and the kinase Aurora B to chromatin, which stimulates autoactivation and spatially regulates spindle assembly (72, 155). Chromatin beads enriched with CPC can lead to local MT assembly in the absence of a RanGTP gradient (156). MT polymerization may therefore be promoted around chromatin via two separate pathways: RanGTP, originating from any chromatin, and the CPC pathway, originating from discrete centromeric sites (156). It remains to be demonstrated whether the negative regulation of MT depolymerases is the only mechanism by which the CPC operates.
Microtubule Formation from Kinetochores
From 20 to 40 MTs form the mature K-fiber in vertebrates and attach with their MT plus ends to the KT (157). Early observations suggested that MTs may also directly nucleate from or adjacent to the KT, as visualized with serial-section EM reconstructions of CHO cells after colcemid washout (158–160). MT nucleation from the KT was directly observed live via light microscopy in mammalian Ptk and S2 cells, followed by the poleward transport of distal K-fiber ends along astral MTs by dynein motors (161, 162). These MTs were more likely to form in association with KT clusters than with an individual KT, and it appears that chromosomal MTs are generated in the vicinity of the centromere rather than around chromosome arms (133). MT nucleation originating from the KT would not be consistent with the plus ends facing the KT, but if nucleation occurred near the KT, the observed polarity could be maintained, and tubulin addition could be directly linked to the poleward flux. It has been suggested that MT nucleation from the KT occurs rarely and serves as a safety mechanism operating only if a KT remains unattached for a prolonged time to assist in KT capture (163). However, it has also been proposed as a common mechanism to generate K-fibers (162), and the exact role of MT nucleation from KTs remains to be determined.
Although details about the exact organization of this MT nucleation pathway are lacking, several molecular candidates have been identified. Part of the disassembled nuclear pore complex, the Nup107–160 subcomplex, seems necessary for MT nucleation from KTs, requires RanGTP for activation, and recruits γ-TuRC to unattached KTs (Figure 4f) (164). The RanGTP-deactivating RanGAP1 is also targeted to mammalian KTs when the first MT attachments form and could possibly inactivate this pathway once KT MTs are finalized (165). Another RanGTP-regulated factor, the SAF TACC3, contributes to correct MT-KT connections and is necessary to form and sort small MT asters near KTs (166). These asters initially seem to capture KTs in a lateral fashion before forming a secure end on connection (167). A third RanGTP-activated protein that could play a role in MT nucleation around KTs is MCRS1, which binds to the minus ends of chromosomal MTs and K-fibers, protecting them from depolymerization (168). A fourth RanGTP-regulated factor that is necessary for KT-associated MT formation is TPX2, which preferentially localizes to KTs in human cells. Interestingly, the augmin subunit Dgt6/HAUS6 interacts with the KT protein Ndc80 (169), possibly suggesting that augmin and TPX2 might be involved in promoting branching MT nucleation at KTs. This would be well suited for building and maintaining parallel, high-density K-fibers, and augmin depletion indeed reduces KT fiber formation and stability (143–145, 169). Finally, the CPC subunit survivin has a role in this process because its depletion reduces the level of KT-MT formation in an MCAK-dependent manner. Thus, both RanGTP and CPC pathways may converge to generate KT MTs.
Microtubule Generation from Acentrosomal Poles
Dynein was proposed to form acentrosomal poles by carrying cross-bridged MTs toward MT minus ends (59). Once collected at the poles, the minus ends were further cross-linked by NuMA or related proteins to create a stable pole (Figure 4a) (170). MT nucleation from acentrosomal poles has also been postulated (171). Such nucleation was first detected in somatic cells without functional centrosomes by tracking EB1-GFP, which seemed to originate from acentrosomal poles and grow toward the metaphase plate (75). In addition, acentrosomal MT formation was visible in augmin-depleted acentrosomal Xenopus egg extract spindles. In such cases, the MT mass inside the spindle was drastically reduced, highlighting the origin of new MTs from acentrosomal poles (172). The poles become increasingly active in MT formation in a time-dependent manner, suggesting an increasing accumulation of γ-tubulin or an activator at the poles, perhaps mediated by dynein or other mechanisms. Thus, MT nucleation from acentrosomal poles effectively produces centrosome-like MTOCs that help to stabilize bipolar structures.
Hierarchy, Coordination, and Cell Type Differences of Spindle Assembly
Now that it has been established that multiple MT nucleation pathways lead to spindle assembly, the question arises of how these are coordinated to generate a functional spindle. This must include a temporal and spatial order in which initial MT nucleation from each MTOC becomes intertwined with the others to generate the continuous distribution of MT nucleation that characterizes the metaphase spindle.
A clue to the coordination and functional hierarchy of these pathways may be provided by the compensation observed when one of them is removed and the other is left intact. In contrast, many cells are fine without functional centrosomes. Interference of the Ran pathway inhibits spindle assembly of acentrosomal cells and significantly disrupts cell division in centrosomal cells with many chromosomes, such as human cells. An unusual exception is the acentrosomal spindle of X. tropicalis, which does not require RanGTP and only relies on TPX2 (91). Removal of the branching MT nucleation pathway can be partly compensated by the centrosomal pathway in spindles with a moderate chromosome number, such as Drosophila (143). However, larger and acentrosomal spindles, such as human cells and the X. laevis egg extract spindle, rely on this pathway (144, 145, 172). In the latter case, the augmin phenotype can be slightly reversed by increased MT nucleation from acentrosomal poles as the spindle matures (172). This confirms the notion that a cell responds to the removal of one MT nucleation pathway by increasing the use of another (173).
Regarding temporal regulation, parallel MT generation from centrosomes and chromosomes has been visualized (117). However, it is not clear if both the Ran and CPC pathways are activated at the same time, and whether there is a time difference between MT generation from chromosome arms and centromeres/KTs. Branching MT nucleation is activated with a slight delay because TPX2 and other SAFs first need to be released. In the absence of centrosomes, the major difference is that nucleation from acentrosomal poles can only begin once these are formed; thus, such nucleation represents the last pathway to be activated.
Although few studies so far have examined the coordination of MT nucleation pathways with spindle assembly, it is clear that mechanisms evolved to achieve a continuum of MT nucleation that maintains and defines the spindle. This is supported by the fact that many molecular players drive more than one MT nucleation pathway (Figure 4). The activation and transport of nucleation factors may be what connects individual pathways.
In the future, quantitative and mechanistic studies of each individual MT nucleation pathway should be performed to determine how MTs are assembled and organized to form the spindle. Although it is difficult enough to resolve individual MTs of any kind, it would be ideal if MT populations from different MTOCs could be separately detected. Independent of this challenge, it is necessary to understand how MTs of two or more MTOCs interdigitate toward creating the metaphase spindle.
CONCLUDING REMARKS
The last decade provided near-complete and overlapping lists of MAPs involved in spindle assembly, and many of those are now understood at the single-molecule and structural levels. The next challenge consists of studying the effect of more than one MAP on the generation of complexity as it occurs in the spindle. Ultimately, it will be necessary to explain how MTs and MAPs, which act at the nanometer scale, build the mitotic spindle, which is a factor of 1,000 larger, in a robust manner.
Innovative methods probing MT organization resulted in the most accurate model of the metaphase spindle to date. However, direct methods to determine exactly where MTs are nucleated, along with their MT dynamics and locations, will be necessary to precisely determine the metaphase spindle architecture. These methods also need to be applied to individual MT nucleation pathways if we want to understand how a spindle is assembled at a mechanistic level. Most importantly, such methods are needed to describe how chromosomes are captured, how chromosomes are aligned, how MTs can be securely attached to the KT, and ultimately how chromosomes are segregated. Ever since the first descriptions of cell division 130 years ago, the means by which the mitotic spindle orchestrates cell division has been a mystery. Grasping how hundreds of proteins can self-assemble into the mitotic spindle and segregate chromosomes at biochemical and structural levels is a challenge that may be finally within reach during this century.
ACKNOWLEDGMENTS
I thank Rebecca Heald, Tim Mitchison, Gary Brouhard, Jean Schwarzbauer, and Eric Wieschaus for helpful comments on this manuscript. Work in the Petry laboratory is supported by the National Institutes of Health (4R00GM100013), the David and Lucile Packard Foundation, the Sidney Kimmel Foundation, and the Pew Scholars Program in the Biomedical Sciences.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013;40(6):463–71. doi: 10.3109/03014460.2013.807878. [DOI] [PubMed] [Google Scholar]
- 2.Flemming W. Zellsubstanz, Kern und Zelltheilung. F.C.W. Vogel; Leipzig, Ger.: 1882. [Google Scholar]
- 3.Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, et al. Evidence for an upper limit to mitotic spindle length. Curr. Biol. 2008;18:1256–61. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH. Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics. 2005;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Brugues J, Nuzzo V, Mazur E, Needleman DJ. Nucleation and transport organize microtubules in metaphase spindles. Cell. 2012;149:554–64. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 6.Sawin KE, Mitchison TJ. Poleward microtubule flux mitotic spindles assembled in vitro. J. Cell Biol. 1991;112:941–54. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoueé S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–40. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nédélec F, Surrey T, Karsenti E. Self-organisation and forces in the microtubule cytoskeleton. Curr. Opin. Cell Biol. 2003;15:118–24. doi: 10.1016/s0955-0674(02)00014-5. [DOI] [PubMed] [Google Scholar]
- 9.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–25. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 10.Boveri T. Zellen-Studien. Gustav Fischer; Jena, Ger.: 1887–1900. [Google Scholar]
- 11.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–74. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 12.Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012;14:1159–68. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 2012;14:1148–58. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 14.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–21. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 15.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–31. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoué S. Cell division and the mitotic spindle. J. Cell Biol. 1981;91:131–47s. doi: 10.1083/jcb.91.3.131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkley W. Microtubules: a brief historical perspective. J. Struct. Biol. 1997;118:84–86. doi: 10.1006/jsbi.1997.3854. [DOI] [PubMed] [Google Scholar]
- 18.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 19.Uchiyama S, Kobayashi S, Takata H, Ishihara T, Hori N, et al. Proteome analysis of human metaphase chromosomes. J. Biol. Chem. 2005;280:16994–7004. doi: 10.1074/jbc.M412774200. [DOI] [PubMed] [Google Scholar]
- 20.Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. PNAS. 2001;98:14434–39. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. PNAS. 2006;103:5391–96. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–21. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes JR, Meireles AM, Fisher KH, Garcia A, Antrobus PR, et al. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLOS Biol. 2008;6:785–95. doi: 10.1371/journal.pbio.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchins JRA, Toyoda Y, Hegemann B, Poser I, Heriche JK, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–99. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–27. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998;142:775–86. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy SM, Preble AM, Patel UK, O’Connell KL, Dias DP, et al. GCP5 and GCP6: two new members of the human γ-tubulin complex. Mol. Biol. Cell. 2001;12:3340–52. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–83. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 29.Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, et al. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 1999;144:721–33. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, et al. Ring closure activates yeast γ TuRC for species-specific microtubule nucleation. Nat. Struct. Mol. Biol. 2015;22:132–37. doi: 10.1038/nsmb.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–82. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teixido-Travesa N, Roig J, Luders J. The where, when and how of microtubule nucleation—one ring to rule them all. J. Cell Sci. 2012;125:4445–56. doi: 10.1242/jcs.106971. [DOI] [PubMed] [Google Scholar]
- 33.Lin TC, Neuner A, Schiebel E. Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends Cell Biol. 2015;25:296–307. doi: 10.1016/j.tcb.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Petry S, Vale RD. Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 2015;17:1089–93. doi: 10.1038/ncb3220. [DOI] [PubMed] [Google Scholar]
- 35.Roostalu J, Cade NI, Surrey T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 2015;17:1422–34. doi: 10.1038/ncb3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 2015;17:907–16. doi: 10.1038/ncb3188. [DOI] [PubMed] [Google Scholar]
- 37.Roll-Mecak A, Vale RD. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 2006;175:849–51. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiese C, Zheng Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2000;2:358–64. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 39.Brouhard GJ, Rice LM. The contribution of αβ-tubulin curvature to microtubule dynamics. J. Cell Biol. 2014;207:323–34. doi: 10.1083/jcb.201407095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–29. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nogales E, Wang HW. Structural mechanisms underlying nucleotide-dependent self-assembly of tubulin and its relatives. Curr. Opin. Struct. Biol. 2006;16:221–29. doi: 10.1016/j.sbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Akhmanova A, Steinmetz MO. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–22. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 43.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–76. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 44.Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 2014;24:372–84. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts AJ, Goodman BS, Reck-Peterson SL. Reconstitution of dynein transport to the microtubule plus end by kinesin. eLife. 2014;3:e02641. doi: 10.7554/eLife.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duellberg C, Trokter M, Jha R, Sen I, Steinmetz MO, Surrey T. Reconstitution of a hierarchical+TIP interaction network controlling microtubule end tracking of dynein. Nat. Cell Biol. 2014;16:804–11. doi: 10.1038/ncb2999. [DOI] [PubMed] [Google Scholar]
- 47.Zanic M, Widlund PO, Hyman AA, Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 2013;15:688–93. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
- 48.Welburn JP. The molecular basis for kinesin functional specificity during mitosis. Cytoskeleton (Hoboken) 2013;70:476–93. doi: 10.1002/cm.21135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–68. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–41. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanenbaum ME, Vale RD, McKenney RJ. Cytoplasmic dynein crosslinks and slides anti-parallel microtubules using its two motor domains. eLife. 2013;2:e00943. doi: 10.7554/eLife.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–18. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 53.Dogterom M, Surrey T. Microtubule organization in vitro. Curr. Opin. Cell Biol. 2013;25:23–29. doi: 10.1016/j.ceb.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Subramanian R, Kapoor TM. Building complexity: insights into self-organized assembly of microtubule-based architectures. Dev. Cell. 2012;23:874–85. doi: 10.1016/j.devcel.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winey M, Morgan GP, Straight PD, Giddings TH, Jr, Mastronarde DN. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol. Biol. Cell. 2005;16:1178–88. doi: 10.1091/mbc.E04-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McIntosh JR, Landis SC. The distribution of spindle microtubules during mitosis in cultured human cells. J. Cell Biol. 1971;49:468–97. doi: 10.1083/jcb.49.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993;123:1475–89. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 1997;138:615–28. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamasaki T, O’Toole E, Kita S, Osumi M, Usukura J, et al. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol. 2013;202:25–33. doi: 10.1083/jcb.201304031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoué S, Sato H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 1967;50(Suppl.):259–92. [PMC free article] [PubMed] [Google Scholar]
- 62.Salmon ED, Leslie RJ, Saxton WM, Karow ML, McIntosh JR. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 1984;99:2165–74. doi: 10.1083/jcb.99.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pearson CG, Gardner MK, Paliulis LV, Salmon ED, Odde DJ, Bloom K. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol. Biol. Cell. 2006;17:4069–79. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J. Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Julicher F, Hyman AA. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr. Biol. 2010;20:353–58. doi: 10.1016/j.cub.2009.12.050. [DOI] [PubMed] [Google Scholar]
- 66.Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–27. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- 67.Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–47. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- 68.Schweizer N, Weiss M, Maiato H. The dynamic spindle matrix. Curr. Opin. Cell Biol. 2014;28:1–7. doi: 10.1016/j.ceb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 69.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 70.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:R797–805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 71.Loughlin R, Heald R, Nédélec F. A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol. 2010;191:1239–49. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 73.Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr. Biol. 2009;19:R749–61. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tirnauer JS, Salmon ED, Mitchison TJ. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol. Biol. Cell. 2004;15:1776–84. doi: 10.1091/mbc.E03-11-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 2006;16:564–69. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 76.Burbank KS, Groen AC, Perlman ZE, Fisher DS, Mitchison TJ. A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol. 2006;175:369–75. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr. Biol. 2007;17:1373–83. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 78.Yang G, Houghtaling BR, Gaetz J, Liu JZ, Danuser G, Kapoor TM. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat. Cell Biol. 2007;9:1233–42. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
- 79.Needleman DJ, Groen A, Ohi R, Maresca T, Mirny L, Mitchison T. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: Evidence that the spindle environment does not stabilize microtubules. Mol. Biol. Cell. 2010;21:323–33. doi: 10.1091/mbc.E09-09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lecland N, Luders J. The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 2014;16:770–78. doi: 10.1038/ncb2996. [DOI] [PubMed] [Google Scholar]
- 81.Goshima G, Scholey JM. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- 82.Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, et al. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol. 2013;15:1116–22. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- 83.Hara Y, Kimura A. An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol. Biol. Cell. 2013;24:1411–19. doi: 10.1091/mbc.E12-07-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Young S, Besson S, Welburn JP. Length-dependent anisotropic scaling of spindle shape. Biol. Open. 2014;3:1217–23. doi: 10.1242/bio.201410363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levy DL, Heald R. Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 2012;28:113–35. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- 86.Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, et al. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–56. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–60. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J. Cell Biol. 2007;176:765–70. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Loughlin R, Wilbur JD, McNally FJ, Nédélec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;147:1397–407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Helmke KJ, Heald R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 2014;206:385–93. doi: 10.1083/jcb.201401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 2005;15:828–32. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 93.Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner A. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. PNAS. 2009;106:15708–13. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu L, Wiese C. Xenopus NEDD1 is required for microtubule organization in Xenopus egg extracts. J. Cell Sci. 2008;121:578–89. doi: 10.1242/jcs.018937. [DOI] [PubMed] [Google Scholar]
- 95.Luders J, Patel UK, Stearns T. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 2006;8:137–47. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 96.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 2006;172:505–15. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Teixido-Travesa N, Villen J, Lacasa C, Bertran MT, Archinti M, et al. The γTuRC revisited: A comparative analysis of interphase and mitotic human γ TuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell. 2010;21:3963–72. doi: 10.1091/mbc.E10-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 2004;15:3642–57. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-tubulin ring complex. Mol. Biol. Cell. 2002;13:3235–45. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–29. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu H, Coppinger JA, Jang CY, Yates JR, 3rd, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-γ-tubulin complex to the mitotic spindle. J. Cell Biol. 2008;183:835–48. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell Biol. 2010;191:1089–95. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khodjakov A, Rieder CL. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 1999;146:585–96. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. PNAS. 2004;101:1584–88. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–83. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLOS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Albee AJ, Tao W, Wiese C. Phosphorylation of maskin by Aurora-A is regulated by RanGTP and importin β. J. Biol. Chem. 2006;281:38293–301. doi: 10.1074/jbc.M607203200. [DOI] [PubMed] [Google Scholar]
- 108.Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–41. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kirschner MW, Mitchison T. Microtubule dynamics. Nature. 1986;324:621. doi: 10.1038/324621a0. [DOI] [PubMed] [Google Scholar]
- 110.Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. PNAS. 1994;91:5682–85. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–42. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 112.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 113.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 114.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, et al. Flies without centrioles. Cell. 2006;125:1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 115.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–98. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 116.McKim KS, Hawley RS. Chromosomal control of meiotic cell division. Science. 1995;270:1595–601. doi: 10.1126/science.270.5242.1595. [DOI] [PubMed] [Google Scholar]
- 117.Rebollo E, Llamazares S, Reina J, Gonzalez C. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLOS Biol. 2004;2:E8. doi: 10.1371/journal.pbio.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dinarina A, Pugieux C, Corral MM, Loose M, Spatz J, et al. Chromatin shapes the mitotic spindle. Cell. 2009;138:502–13. doi: 10.1016/j.cell.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 119.Dasso M. The Ran GTPase: theme and variations. Curr. Biol. 2002;12:R502–8. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- 120.Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 2002;4:E177–84. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- 121.Kalab P, Pu RT, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–84. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 122.Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–58. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 123.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–62. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 124.Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–81. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 125.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–56. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 126.Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–76. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 127.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 128.Halpin D, Kalab P, Wang J, Weis K, Heald R. Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLOS Biol. 2011;9:e1001225. doi: 10.1371/journal.pbio.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 130.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 131.Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin β in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–56. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 132.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, a novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 2000;149:1405–18. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 2006;16:536–41. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, et al. Importin α-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–70. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eyers PA, Maller JL. Regulation of Xenopus Aurora A activation by TPX2. J. Biol. Chem. 2004;279:9008–15. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- 136.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 2003;12:851–62. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 137.Tsai MY, Wiese C, Cao K, Martin O, Donovan P, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 2003;5:242–48. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 138.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, et al. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 2002;156:437–51. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Scrofani J, Sardon T, Meunier S, Vernos I. Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr. Biol. 2015;25:131–40. doi: 10.1016/j.cub.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 140.Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, et al. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol. 2005;7:961–68. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 141.Chan J, Sambade A, Calder G, Lloyd C. Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell. 2009;21:2298–306. doi: 10.1105/tpc.109.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound γ-tubulin complexes. J. Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008;181:421–29. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Uehara R, Nozawa RS, Tomioka A, Petry S, Vale RD, et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. PNAS. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, et al. HAUS, the 8-subunit human augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 2009;19:816–26. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 146.Hsia KC, Wilson-Kubalek EM, Dottore A, Hao Q, Tsai KL, et al. Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol. 2014;16:852–63. doi: 10.1038/ncb3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–77. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ho CM, Hotta T, Kong Z, Zeng CJ, Sun J, et al. Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell. 2011;23:2606–18. doi: 10.1105/tpc.111.086892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hotta T, Kong Z, Ho CM, Zeng CJ, Horio T, et al. Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell. 2012;24:1494–509. doi: 10.1105/tpc.112.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nakaoka Y, Miki T, Fujioka R, Uehara R, Tomioka A, et al. An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell. 2012;24:1478–93. doi: 10.1105/tpc.112.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu T, Tian J, Wang G, Yu Y, Wang C, et al. Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr. Biol. 2014;24:2708–13. doi: 10.1016/j.cub.2014.09.053. [DOI] [PubMed] [Google Scholar]
- 152.Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–13. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- 153.Clausen T, Ribbeck K. Self-organization of anastral spindles by synergy of dynamic instability, autocatalytic microtubule production, and a spatial signaling gradient. PLOS ONE. 2007;2:e244. doi: 10.1371/journal.pone.0000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell. 2010;21:979–88. doi: 10.1091/mbc.E09-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the Aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr. Biol. 2009;19:1210–15. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Biggins S, Walczak CE. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 2003;13:R449–60. doi: 10.1016/s0960-9822(03)00369-5. [DOI] [PubMed] [Google Scholar]
- 158.McGill M, Brinkley BR. Human chromosomes and centrioles as nucleating sites for the in vitro assembly of microtubules from bovine brain tubulin. J. Cell Biol. 1975;67:189–99. doi: 10.1083/jcb.67.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Witt PL, Ris H, Borisy GG. Origin of kinetochore microtubules in Chinese hamster ovary cells. Chromosoma. 1980;81:483–505. doi: 10.1007/BF00368158. [DOI] [PubMed] [Google Scholar]
- 160.Snyder JA, McIntosh JR. Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. J. Cell Biol. 1975;67:744–60. doi: 10.1083/jcb.67.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 2003;160:671–83. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004;167:831–40. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.LaFountain JR, Jr, Oldenbourg R. Kinetochore-driven outgrowth of microtubules is a central contributor to kinetochore fiber maturation in crane-fly spermatocytes. Mol. Biol. Cell. 2014;25:1437–45. doi: 10.1091/mbc.E14-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107–160 complex and γ-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 2010;12:164–69. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arnaoutov A, Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–65. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- 166.Bird AW, Hyman AA. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 2008;182:289–300. doi: 10.1083/jcb.200802005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fu W, Chen H, Wang G, Luo J, Deng Z, et al. Self-assembly and sorting of acentrosomal microtubules by TACC3 facilitate kinetochore capture during the mitotic spindle assembly. PNAS. 2013;110:15295–300. doi: 10.1073/pnas.1312382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Meunier S, Vernos I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 2011;13:1406–14. doi: 10.1038/ncb2372. [DOI] [PubMed] [Google Scholar]
- 169.Bucciarelli E, Pellacani C, Naim V, Palena A, Gatti M, Somma MP. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and γ-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 2009;19:1839–45. doi: 10.1016/j.cub.2009.09.043. [DOI] [PubMed] [Google Scholar]
- 170.Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–58. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 171.Verde F, Berrez JM, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J. Cell Biol. 1991;112:1177–87. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Petry S, Pugieux C, Nédélec FJ, Vale RD. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. PNAS. 2011;108:14473–78. doi: 10.1073/pnas.1110412108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hayward D, Metz J, Pellacani C, Wakefield JG. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell. 2014;28:81–93. doi: 10.1016/j.devcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW. D-TACC: A novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000;19:241–52. doi: 10.1093/emboj/19.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM functions in Ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 2004;14:1801–11. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 176.Trieselmann N, Armstrong S, Rauw J, Wilde A. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 2003;116:4791–98. doi: 10.1242/jcs.00798. [DOI] [PubMed] [Google Scholar]
- 177.Ems-McClung SC, Zheng Y, Walczak CE. Importin α/β and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Maresca TJ, Niederstrasser H, Weis K, Heald R. Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr. Biol. 2005;15:1755–61. doi: 10.1016/j.cub.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 179.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–34. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 180.Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J. Biol. Chem. 1995;270:7411–19. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- 181.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–39. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 182.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–54. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 183.Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006;173:879–91. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–93. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 185.Ribbeck K, Raemaekers T, Carmeliet G, Mattaj IW. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 2007;17:230–36. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 186.Yokoyama H, Rybina S, Santarella-Mellwig R, Mattaj IW, Karsenti E. ISWI is a RanGTP-dependent MAP required for chromosome segregation. J. Cell Biol. 2009;187:813–29. doi: 10.1083/jcb.200906020. [DOI] [PMC free article] [PubMed] [Google Scholar]