Summary
The optimal choice of cancer therapy depends upon analysis of the tumor genome for druggable molecular alterations. The spatial and temporal intratumor heterogeneity of cancers creates substantial challenges, as molecular profile depends on time and site of tumor tissue collection. To capture the entire molecular profile, multiple biopsies from primary and metastatic sites at different time points would be required, which is not feasible for ethical or economic reasons. Molecular analysis of circulating cell-free DNA offers a novel, minimally invasive method that can be performed at multiple time-points and plausibly better represents the prevailing molecular profile of the cancer. Molecular analysis of this cell-free DNA offers multiple clinically useful applications, such as identification of molecular targets for cancer therapy, monitoring of tumor molecular profile in real time, detection of emerging molecular aberrations associated with resistance to particular therapy, determination of cancer prognosis and diagnosis of cancer recurrence or progression.
Keywords: liquid biopsy, cell-free DNA, advanced cancer, targeted therapy, personalized medicine
Introduction
Despite significant progress in modern oncology, efficacy of treatment for advanced cancer remains poor; the majority of advanced tumors become resistant to available therapies and the patient ultimately succumbs to advancing metastatic disease [1]. This is mainly due to clonal evolution of the disseminated tumor and acquired resistance to cancer therapies, even if fitted to the known molecular profile [2, 3]. In the current era of personalized medicine, the optimal choice of therapy depends upon detailed analysis of the cancer genome and identification of the targetable aberrations for each individual patient [4]. This approach is substantially limited by the considerable spatial and temporal intratumor heterogeneity of advanced disease. The cancer-related aberrations in the original tumor can differ among tumor regions and distinct disease sites [5].
Molecular testing of tumor samples obtained by surgical procedures or biopsies remains the standard of care [6]. However, this approach has significant limitations because of the temporal and spatial tumor heterogeneity, which would mandate multiple biopsies from primary and metastatic sites at multiple time points. This is not feasible because of the medical condition of patients with advanced cancer, the risk of complications, and various economic and logistic considerations. To overcome these limitations, novel minimally invasive methods to detect pertinent molecular changes in tumors are being developed. Mandel and Métais in 1948 noticed the presence of cell-free nucleic acids (cfNA) in human blood [7]. However, it took several decades before reports emerged on oncogenic mutations in blood-derived cell-free DNA (cfDNA) of patients with cancer [8] or fetal cfDNA in pregnant women [9]. cfDNA also was investigated in prediction of outcome after brain trauma [10], myocardial infarction [11], and stroke [12, 13].
Fragments of cfNA such as DNA, messenger RNA, or microRNA can be detected in plasma, urine, cerebrospinal fluid (CSF), and other body fluids. In cancer patients, these cfDNA fragments can be used for detection of underlying cancer-related molecular abnormalities [8, 14]. Such approaches, which have become known as liquid biopsies, can be used to monitor a cancer molecular profile in real time with minimal invasiveness. It is assumed that fragments of cfDNA are released to blood from diverse tumor sites and perhaps better represent prevailing molecular abnormalities than single-site biopsies. In addition, molecular testing of cfDNA can be used to evaluate response to therapy, disease progression or recurrence, and emergence of molecular abnormalities that drive resistance to systemic therapy.
The Biology of cfDNA
Fragments of cfDNA can be detected in extracellular fluids, such as blood (plasma or serum), urine, CSF, or even ascites, of patients with cancer [15–21], and increased levels of cfDNA can be associated with unfavorable outcome [22, 23]. It has been demonstrated that patients with advanced cancer have higher levels of cfDNA than patients with localized cancer or individuals without cancer [24–34].
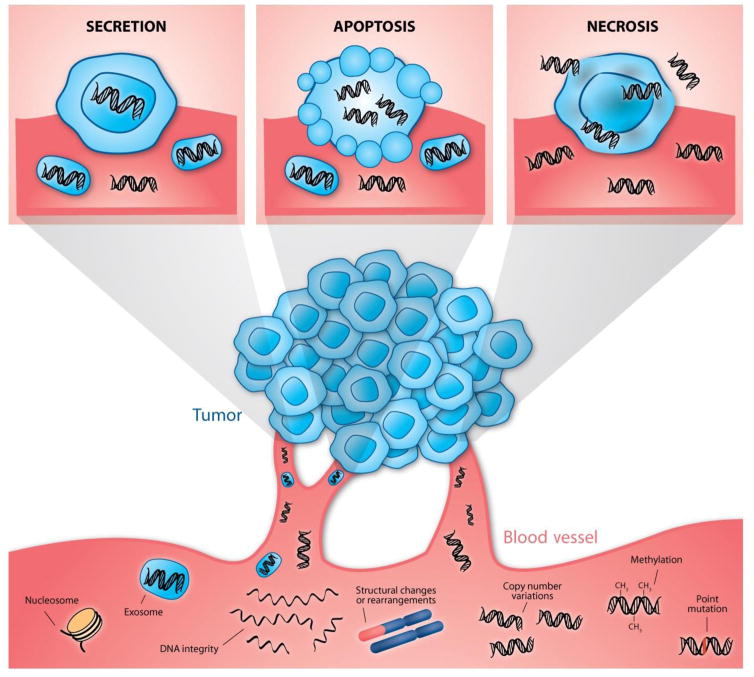
DNA can enter the circulation by several distinct mechanisms, including release of nuclear and mitochondrial DNA from dying cells during either apoptosis or necrosis (Figure 1). Other mechanisms of DNA release include autophagy and necroptosis [35, 36]. cfDNA structural characteristics differ substantially by type of release mechanism. Apoptosis is a programmed and well-controlled process of cellular destruction, and fragments of DNA released from apoptotic cells average around 160–180 bp in length [37, 38]. In contrast, necrosis is a pathological process, and the fragments of DNA are generated more randomly and usually are longer. The average lengths of cfDNA fragments from apoptotic and necrotic processes and their ratio may be assessed as an important element of the DNA integrity index, which may have prognostic implications [39]. Thierry et al. described experimental system for studying the cfDNA characteristic based on the nude mice xenografted with human HT29 or SW620 colorectal cancer cells [40]. The discrimination of cfDNA fractions from normal (murine) cells and from BRAF V600E-mutated and non-mutated tumor (human) cells was possible and the concentration of tumor (human mutated and nonmutated) but not mouse cfDNA increased significantly with tumor burden (P>0.001 and P<0.05, respectively). The higher cfDNA fragmentation was also observed in mice with bigger tumors as the integrity index decreased with tumor size. The study confirmed the predominance of mononucleosome-derived cfDNA fragments in plasma from xenografted animals and of apoptosis as a source of tumor cfDNA [40].
Figure 1.
Passive (from apoptotic and necrotic cells) and active release of DNA fragments from tumor cells into the circulation. This cell-free DNA can be used for testing of tumor-specific aberrations.
It has been proposed that plasma cfDNA can be also involve in the oncogenesis via the uptake of nucleic acids originating from tumor cells by susceptible healthy cells that consequently underwent malignant transformation, the process referred to as “genometastasis” [41, 42]. In the in-vitro study with cultures of NIH-3T3 cells treated with plasma from colorectal cancer patients, the transfer of human DNA were observed and the NIH-3T3 cells were oncogenically transformed, as shown by the development of carcinomas in nonobese diabetic–severe combined immunodeficient mice after the injection of such cells [43].
The cfDNA fragments are cleared from the circulation by the liver and kidney, with half-lives ranging from 15 minutes to a few hours [15, 44].
Technologies for cfDNA Analysis
Sample collection and processing can have significant impacts on cfDNA assessment [45]. Most often the circulating DNA is extracted from plasma; plasma is preferred to serum because of serum has higher levels of non-cancerous cfDNA due to lysis of normal leukocytes. Timely processing is paramount for success [46]. Cell-stabilizing streck tubes, which allow sample processing to be delayed for several days, have become increasingly popular for collection of blood samples intended for cfDNA analysis [47, 48]. Other materials, such urine or CSF, are less cellular and should be less prone to DNA degradation [17, 19]. At the moment, specimen collection protocols vary considerably in details such as use of ethylenediaminetetraacetic acid or even streck tubes.
The optimal pre-analytical handling conditions for the collected blood sample were described by Messaoudi et al. [45]. Blood samples must by drawn carefully and agitation should be avoided to prevent any hemolysis. The sample may be kept at room temperature or +4 ºC and must be processed within 4 h to prevent changes in cfDNA concentration and fragmentation. The two-steps centrifugation (1200–1600 g for 10 min and 16000 g for 10 min) is highly recommended to eliminate any cells from the plasma. The second step can be done after the storage of plasma sample at −20 ºC or −80 ºC. Plasma as well as cfDNA extracts are sensitive to freeze-thaw cycles. Plasma must be stored at −80 ºC up to maximum of nine months before the final cfDNA analysis. The extracts of cfDNA must be stored at −20 ºC for up to three months for the concentration and fragmentation analysis or up to nine months for specific mutations analysis [45].
The techniques for the quantification of total amount of cfDNA include fluorescence-based methods (such as Hoechst dye and PicoGreen staining), spectrophotometric-based methods (ultraviolet spectrometry) or quantitative real-time polymerase chain reaction (PCR) methods (such as SYBR Green and TaqMan) [35, 49, 50]. The study with plasma samples collected from 10 non-small cell lung cancer patients compared PicoGreen staining to real-time PCR methods for the quantification of cfDNA [51]. The results from PicoGreen method correlated with both the SYBR Green (R = 0.87, P < 0.0001) and TaqMan probe approach (R = 0.94, P < 0.0001). The results from another method for the cfDNA quantification using the fluorescent SYBR Gold staining without prior DNA extraction and amplification showed the high correlation with the conventional quantitative PCR assay of beta-globin (R = 0.9987, P < 0.001) [52]. Therefore fluorescence-based methods could be the rapid, accurate, and inexpensive alternatives to real-time PCR for total cfDNA quantification.
The tumor-specific fraction of the total cfDNA can be identified by the presence of cancer-specific alterations. Epigenetic modifications such as methylation patterns also are being investigated as signature markers to differentiate tumor-specific cfDNA fraction [53]. The tumor-specific fraction can vary in plasma from 0.01% to more than 90% [36, 54]. Lower-stage tumors have lower levels of cfDNA than advanced disease [28]. Therefore, highly sensitive methods are required for detection of cfDNA in early disease.
Various PCR approaches were used originally to detect tumor-related aberrations in cfDNA. These included methods such as ARMS-Scorpion PCR (amplification refractory mutation system), PCR-SSCP (single-strand conformation polymorphism), ME-PCR (mutant enriched), MASA-PCR (mutant allele–specific amplification), PAP-A amplification (pyrophosphorolysis-activated polymerization allele-specific amplification), or RFLP-PCR (restriction fragment length polymorphism) [55–60]. Even higher sensitivity is required, however, for detection of ctDNA from tumors in which specific mutations occur in very low allele fractions. For this reason, novel methods using digital PCR were introduced into cfDNA assays. Digital PCR methods include droplet-based systems [61], the use of beads, emulsions, amplification, and magnetics (BEAMing) [62], or microfluidic assays [63, 64].
Next-generation sequencing techniques (NGS), which allow detection of multiple alterations across wider regions of the cancer genome, also can be used for testing of cfDNA. The specific regions of cfDNA are analyzed by using targeted deep-sequencing techniques such as TAm-Seq (tagged amplicon deep sequencing) [63], Ion AmpliSeq [65], Safe-Seq (safe-sequencing system) [66], or CAPP-seq (cancer personalized profiling by deep sequencing) [67]. The latest and most comprehensive approaches to cfDNA analyses that do not require knowledge of preexisting mutations include whole-exome [68] as well as whole-genome sequencing of plasma samples [69, 70]. The NGS techniques and unbiased whole-exome and whole-genome assays might dominate the future of cfDNA research. The advantages of PCR-based and NGS-based approaches are summarized in Table 1.
Table 1.
Comparison of PCR-based and next genration sequencing-based approaches for cfDNA analysis and their possible applications
| Methods | PCR - based approaches | Next-generation sequencing |
|---|---|---|
| Important features | Sensitive | Less sensitive in most cases |
| Short turnaround | Longer turnaround times | |
| Tested and validated | Tested and validated | |
| Available in the CLIA-certified environment | Available in the CLIA-certified environment | |
| Limitations in terms of multiplexing | Need for bioinformatics, higher cost | |
| Possible applications | Molecular diagnosis in well-defined situation when limited mutation panel is adequate | Molecular diagnosis in clinical setting |
| Monitoring of mutation load as a surrogate for response to therapy | Discovery research | |
| Diagnosis of emergent resistance clones in malignancies with well-defined mechanisms of resistance | Malignancies with undefined patterns of resistance to targeted therapy |
Abbreviati ons: CLIA, clinical laboratory improvement amendments; PCR, polymerase chain reaction
Clinical Application of cfDNA in Cancer Management
Identification of molecular targets
The feasibility of identifying molecular targets in cfDNA as well as the level of concordance between mutations detected in tumor tissue and plasma samples are important attributes for future routine clinical use of cfDNA liquid biopsy techniques (Table 2). In a pilot study of 18 patients with metastatic colorectal cancer who were candidates for surgical resection or radiofrequency ablation, oncogenic mutations (APC, TP53, PIK3CA, and KRAS) were assessed by direct sequencing in tumor tissue [15]. At least one mutation was identified in each of the tumors. The unique molecular signature of each tumor was used for detection and quantification of tumor-derived cfDNA by the BEAMing PCR-based technology. This study demonstrated that cfDNA can be isolated from plasma samples and used to identify oncogenic mutations in cancer patients.
Table 2.
The summarization of the studies that examined the concordance between mutations detected in tumor tissue samples and cfDNA
| Cancer type | Mutated genes | Cohort no. | Results of the concordance between cfDNA and tumor tissue | Reference |
|---|---|---|---|---|
| Colorectal cancer | KRAS, BRAF | 106 | 100% specificity and sensitivity for the BRAFV600E mutation and 98% specificity and 92% sensitivity for the KRAS mutations detection | Thierry et. al. (2014) |
| Breast cancer | PIK3CA | 49 + 60 | 100% concordance if tumor and cfDNA samples collected at the same, 79% in another cohort if collected at different time points | Higgins et al. (2012) |
| PIK3CA | 29 | 93.3% sensitivity and 100% specificity for the mutations detection in pre-surgery plasma samples | Beaver et al. (2014) | |
| 50 selected genes (NGS) | 17 | 76% concordance between the analysis of primary or metastatic tumors and matched plasma samples | Rothe et al. (2014) | |
| Melanoma | BRAF | 221 | Sensitivity for BRAFV600E mutation detection in serum was 44%, in plasma 52%. Specificity was 96% in both matrices | Aung et al. (2014) |
| BRAF | 128 | 96% sensitivity and 95% specificity for BRAFV600E mutation detection in the subset of 42 stage IV patients | Panka et al. (2014) | |
| Pancreatobiliary carcinomas | 54 selected genes (NGS) | 26 | 90.3% of mutations detected in tumor biopsies were also detected in cfDNA. 92.3% sensitivity and 100% specificity across the five most frequently mutated genes | Zill et al. (2015) |
| ECD/LCH | BRAF | 30 | 100% concordance of BRAFV600E mutation between tissue and urinary cfDNA | Hyman et al. (2015) |
| Advanced cancers | BRAF, EGFR, KRAS, PIK3CA | 157 | Acceptable concordance (BRAF , 91%; EGFR , 99%; KRAS , 83%; PIK3CA , 91%) with standard-of-care mutation analysis of primary or metastatic tumor tissue | Janku et al. (2015) |
Abbreviati ons: NGS, next generati on sequencing techniques; ECD/LCH, Erdheim-Chester disease and Langerhans cell histi ocytosis
In a cohort of 49 patients with advanced breast cancer, there was 100% concordance (34 of 34 cases) between BEAMing-detected PIK3CA mutations in plasma cfDNA and in tumor tissues obtained at the same time [62]. However, the concordance decreased to 79% in an additional cohort of 60 patients when tumor samples and plasma cfDNA were obtained at different time points.
In a study of 157 patients with advanced cancer that progressed on systemic therapy who were referred for treatment with experimental targeted therapies, a panel of 21 oncogenic mutations in the BRAF, EGFR, KRAS, and PIK3CA genes was assessed in plasma cfDNA by BEAMing technology. The results demonstrated acceptable concordance (BRAF, 91%; EGFR, 99%; KRAS, 83%; PIK3CA, 91%) with results of standard-of-care mutation analysis of primary or metastatic tumor tissue obtained during clinical care [71].
Thierry et al. [72] assessed the mutation status of KRAS and BRAF by using allele-specific quantitative PCR of cfDNA in 106 plasma samples from patients with metastatic colorectal cancer and compared it to the mutations detected in tissue (primary or metastatic) tested by standard-of-care methods. The cfDNA analysis showed 100% specificity and sensitivity for the BRAF V600E mutation and 98% specificity and 92% sensitivity for the KRAS mutations, with a concordance value of 96%.
The BRAF V600E mutation was recently detected in plasma and urine cfDNA samples obtained from individuals with Erdheim-Chester disease and Langerhans cell histiocytosis [18]. These patients have a high prevalence of BRAF V600E mutations and a good response to BRAF inhibitors. There was 100% concordance between tissue and urinary cfDNA genotypes assessed by droplet-digital PCR assay (ddPCR) in samples from 30 treatment-naive patients. The targetable mutation BRAF V600E was also analyzed in plasma- and serum-derived cfDNA samples from 221 patients with advanced melanoma [73]. Assay sensitivity for mutation detection was 44% in serum and 52% in plasma. Test specificity was 96% in both matrices.
Panka et al. [74] developed blood-based assay for the detection of BRAF V600E mutation and used it in the study with 128 patients with stage II-IV melanoma. The high 96% sensitivity and 95% specificity of the assay were observed for the subset of 42 stage IV patients. The area under the receiver operator curve (ROC) was 0.9929 demonstrating an excellent ability to discriminate BRAF-mutant melanoma patients. Pupilli et al. [75] investigated the role of BRAF V600E-mutated allele in plasma cfDNA from 103 patients with papillary thyroid carcinoma (PTC) as a marker for the diagnosis and follow-up. Patients with PTC showed a higher percentage of circulating BRAF V600E mutation (P = 0.035) compared to those with benign histology (n=16) and healthy controls (n=49). The assay diagnostic sensitivity and specificity were 80% and 65%, respectively.
Zill et al. [76] assessed the mutation status of 54 genes by NGS in the tumor tissue and corresponding cfDNA in plasma samples from 26 patients with pancreatobiliary carcinomas (18 pancreatic ductal adenocarcinoma cases and 8 biliary cancer cases). 90.3% of mutations detected in tumor biopsies were also detected in cfDNA. Across the five most frequently mutated genes in tumor tissue biopsies (KRAS, TP53, APC, FBXW7 and SMAD4), the assay sensitivity for the detection of such mutations in cfDNA was 92.3%, specificity was 100% and the diagnostic accuracy was 97.7%.
Forshew et al. [77] reported on using the TAm-Seq method for identification and monitoring of oncogenic mutations in plasma cfDNA. They screened 5995 genomic bases in coding regions of TP53 and PTEN and selected regions in EGFR, BRAF, KRAS, and PIK3CA for low-frequency mutations. The assay was able to detect mutations in cfDNA with sensitivity and specificity of >97%. In one patient with synchronous primary cancers of the bowel and ovary, moreover, disease relapse was identified as being derived from the original ovarian tumor. At relapse, analysis of the plasma cfDNA detected the TP53 mutation (p.R273H) originally found in the ovarian primary tumor, whereas the bowel-associated mutations were not detected.
Beaver et al. [61] showed the possibility of identifying PIK3CA mutations in plasma samples from 29 patients with early-stage breast cancer. The same mutations identified in primary tumors were detected in pre-surgery plasma samples by ddPCR with high sensitivity and specificity (93.3% and 100%, respectively). Residual disease was successfully identified by detection of the mutations in cfDNA from postoperative plasma samples. In another study of 17 patients with metastatic breast cancer, analysis of primary or metastatic tumors together with matched plasma samples for mutations in 50 selected genes by NGS yielded a concordance of 76% [78].
Bettegowda et al. [66] evaluated the possibility to detect the cfDNA point mutations and genetic rearrangements that were originally found in tumor tissue biopsies from 640 patients with various cancer types. Tumor-derived cfDNA was detected in > 75% of patients with advanced pancreatic, ovarian, colorectal, bladder, gastroesophageal, breast, melanoma, hepatocellular, and head and neck cancers. In patients with localized tumors, cfDNA was detected in 73, 57, 48, and 50% of patients with colorectal cancer, gastroesophageal cancer, pancreatic cancer, and breast adenocarcinoma, respectively [66].
Newman et al. [67] developed CAPP-Seq, an ultrasensitive method for quantifying tumor-derived plasma cfDNA by targeting recurrently mutated regions in the cancer of interest. In patients with non-small cell lung cancer, the CAPP-Seq method was able to detect cfDNA in 100% of patients with stage II–IV disease and 50% of patients with stage I disease. The method specificity was 96% for mutant allele fractions as low as 0.02%.
Assessment of prognosis
The quantification of total and/or mutant cfDNA has been studied for prognosis assessment in various tumor types. Some studies demonstrated that, in cancer patients, higher levels of cfDNA are associated with higher risk of disease recurrence and progression [15, 28, 30, 66, 79–81]. In a study by Diehl et al. [15] of 18 colorectal cancer patients, the absence of cfDNA in plasma during the first follow-up visit after surgical resection was associated with 100% recurrence-free survival.
Early limited data suggested that persistence of TP53 mutation in plasma cfDNA of patients with stage II or III breast cancer that was in remission was associated with higher likelihood of disease recurrence; however, the small sample size precluded any definitive conclusion [56].
The amount of mutant cfDNA has been found to be of prognostic significance. Spindler et al. [79] demonstrated the prognostic value of the amount of total cfDNA and KRAS mutant cfDNA in a study of 108 patients with metastatic colorectal cancer treated with third-line cetuximab and irinotecan. Patients with higher cfDNA levels had shorter progression-free survival (PFS; 2.1 vs 4.4 months; P=0.0015) and overall survival (OS; 3.6 vs 10.4 months; P<0.0001) than patients with lower cfDNA levels. Similarly, patients with higher levels of KRAS-mutant cfDNA had shorter PFS (1.8 vs 2.3 months; P=0.008) and OS (2.1 vs 5 months; P=0.0005) than patients with lower levels of KRAS-mutant cfDNA.
The mutated fraction of plasma cfDNA (mutation in codon 12 or 13 of KRAS) was assessed in another study of 206 patients with metastatic colorectal cancer [66]. Concentration of mutated cfDNA was found to provide added value in survival prediction (likelihood ratio test, P = 0.00253, df = 3) to the model of well-known prognostic factors (age, Eastern Cooperative Oncology Group performance status, and level of carcinoembryonic antigen). Also, holding other predictors constant, the 2-year survival rate steadily decreased as the plasma concentration of mutated cfDNA increased.
The study already mentioned [71] of the panel of 21 mutations in BRAF, EGFR, KRAS, and PIK3CA assessed by BEAMing technology in plasma cfDNA of 157 patients with advanced cancer also examined the prognostic impact of the amount of mutated plasma cfDNA. A higher percentage of mutant cfDNA (>1% [n = 67 patients] vs. ≤1% [n = 33 patients]), irrespective of type of mutation, was associated with a shorter OS (5.5 vs. 9.8 months; P = 0.001), which was confirmed in a multivariable analysis. Similarly, 41 patients with >1% of KRAS mutant (codon 12 or 13) cfDNA had a shorter median OS than 20 patients with ≤1% of KRAS mutant cfDNA (4.8 vs. 7.3 months; P = 0.008). The significant differences in OS were not observed for mutations in other examined genes, probably because of the smaller sample size.
In another study of 246 patients with advanced non-small-cell lung carcinoma treated with platinum and vinorelbine chemotherapy, the patients with detectable plasma KRAS mutant (codon 12 or 13) cfDNA had a shorter median OS (4.8 vs 9.5 months; P = 0.0002) and shorter median PFS (3.0 vs 5.6 months; P = 0.0043) than patients whose cancer expressed wild-type KRAS [80]. A multivariate analysis confirmed the independent prognostic value of KRAS mutant cfDNA in OS but not in PFS. Wang et al. [82] showed the negative prognostic effect of KRAS mutation (codon 12 or 13) in plasma cfDNA of 273 patients with advanced non–small cell lung cancer. The median PFS of patients with a plasma KRAS mutation was 2.5 months, while that of patients with wild-type KRAS was 8.8 months (P < 0.001).
In a study of 44 pancreatic cancer patients, the 1-year survival rate was 0% in those with KRAS codon-12 mutation in cfDNA and 24% in those with KRAS wild-type in cfDNA (P<0.005), and plasma KRAS mutation was the only independent prognostic factor (odds ratio, 1.51; 95% confidence interval [CI], 1.02 to 2.23) [60]. In 103 patients with melanoma receiving biochemotherapy [83], those with a BRAF mutation in serum cfDNA had significantly shorter OS than those that did not have the BRAF mutation in serum cfDNA (13 vs. 30.6 months, P = 0.039).
The negative prognostic impact of increased levels of mutant cfDNA was supported by other studies in breast cancer [84], colorectal cancer [85, 86], ovarian cancer [87], and other tumor types. Furthermore, the presence of other tumor-related genomic cfDNA aberrations was associated with poor prognosis. Detection of loss of heterozygosity and microsatellite instability in cfDNA was associated with worse prognosis for patients with breast cancer [88], ovarian cancer [89], melanoma [90], lung cancer [91], or other tumor types.
Epigenetic alterations detected in cfDNA can also help determine patient prognosis. The aberrant DNA methylations were detected in cfDNA of patients with breast, lung as well as liver cancer [92–94]. Hypermethylated promoter regions of BRCA1 in serum cfDNA from 100 primary invasive ductal breast cancer patients was associated with poor DFS (14.2 months; P≤0.0001) as well as poor OS (24.3 months; P = 0.0001) [95]. Similarly cfDNA promoter hypermethylation of GSTP1 was associated with poor DFS (24.2 months; P = 0.03). Another study with 336 primary invasive breast cancer patients showed worse OS rate at 100 months (78 vs. 95%; P = 0.002) for patients with serum cfDNA hypermethylation in promoter regions of GSTP1, RASSF1A, and RARβ2 than those with negative findings [96]. In the study with 428 primary breast cancer patients, the detection of methylated PITX2 and RASSF1A in plasma cfDNA determined shorter OS in multivariate analysis (low vs. high methylation; HR 3.4, P = 0.021 and HR 3.4, P = 0.002 respectively) [97]. For distant DFS only RASSF1A showed prognostic significance (low vs. high methylation; HR 3.4, P = 0.002). The aberrant methylation of selected genes in cfDNA were associated with poor prognosis also in colorectal cancer [98], gastric cancer [99], hepatocellular carcinoma [100] and other tumor types.
Prediction of response to therapy
The liquid biopsy could provide an easy way to assess predictive biomarkers for targeted therapy as well as a minimally invasive way to monitor therapy response in real time [8, 14] (Table 3).
Table 3.
Examples of studies that assessed cfDNA for response prediction and/or therapeutic monitoring
| Cancer type | Mutated genes | Cohort no. | Therapeutic intevention | Results | Reference |
|---|---|---|---|---|---|
| Lung cancer | EGFR | 1060 | gefitinib | ORR were 76.9% (95% CI, 65.4–85.5) for EGF-mutant tumors and mutated cfDNA, and 59.5% (95% CI, 43.5–73.7) for EGFR- mutant tumors and wt cfDNA | Douillard et al . (2014) |
| EGFR | 72 | EGFR-TKIs | Failure to clear EGFR- mutant cfDNA after EGFR-TKI was an independent predictor of shorter PFS (HR 1.97, P = 0.001) and OS (HR 1.82, P = 0.036) | Tseng et al . (2014) | |
| Breast cancer | PIK3CA, TP53, structural variations | 52 | various systemic therapeutics | The levels of cfDNA in plasma generally correlated well with the treatment response assessed by CT imaging | Dawson et al . (2013) |
| Melanoma | BRAF | 160 | BRAF and/or MEK inhibitors (n=51) | TTF of 13 patients whose baseline cfDNA samples (but not tissue samples) did not have BRAFV600 mutation was significantly longer than that of 38 patients with baseline cfDNA-mutated samples (13.1 vs. 3.0 months; P=0.001) | Janku et al . (2015) |
Abbreviations:CT, computed tomography; HR, hazard ratio; ORR, overall response rate; OS, overall survival; PFS, progression-free survival; TKIs, tyrosine kinase inhibitors; TTF, time to treatment failure; wt, wild type
In a prospective study of 52 patients with metastatic breast cancer, the plasma cfDNA was monitored to qualitatively and quantitatively assess disease progression and treatment response and compare with levels of circulating tumor cells (CTC) and tumor marker cancer antigen 15-3 (CA15-3) and computed tomography (CT) imaging [63]. The cfDNA was detected by identification of the same PIK3CA and TP53 mutations and structural variations as were found in the tumor tissues. The levels of cfDNA in plasma generally correlated well with the treatment response assessed by CT imaging (as defined by Response Evaluation Criteria in Solid Tumors). However, two patients in this study had discordant correlations. In 10 of the 19 patients who experienced disease progression, the cfDNA levels increased at one or more consecutive time points, on average 5 months before progressive disease was observed on imaging. Moreover, the cfDNA was found to be a more accurate biomarker for monitoring metastatic disease than CTCs, CA 15-3, or CT imaging.
Another study in 72 patients with advanced non-small cell lung cancer examined the dynamic changes in cfDNA EGFR mutations as a predictor of response to EGFR tyrosine-kinase inhibitor (EGFR-TKI) targeted therapy [101]. Failure to clear plasma EGFR mutations after EGFR-TKI was an independent predictor for shorter PFS (hazard ratio [HR] 1.97, P = 0.001) and OS (HR 1.82, P = 0.036). The EGFR mutations were detected by ddPCR in serial plasma samples of non-small cell lung cancer patients treated with erlotinib [102]. The study demonstrated the disappearance of EGFR mutations in exon 19 and 21 and the emergence of EGFR T790M resistance mutation several weeks before radiographic disease progression.
Similarly, EGFR mutations were detected in primary tumors and corresponding plasma samples in a study of 1060 patients with advanced lung cancer treated with gefitinib [103]. Objective response rates were 76.9% (95% CI, 65.4–85.5) for patients with detected mutations in both tumor and plasma and 59.5% (95% CI, 43.5–73.7) for patients with mutation in the tumor but not in plasma. Median PFS was 9.7 months (95% CI, 8.5–11.0) for patients with mutation in the tumor sample only and 10.2 months (95% CI, 8.5–12.5) for patients with mutation in both tumor and plasma samples. This demonstrated that EGFR mutation status could be assessed in cfDNA and serve as a positive predictive biomarker for targeted therapy.
Another study [104] assessed BRAF mutations in plasma cfDNA from 160 patients with advanced cancer and known BRAF status from archival tumor samples. Patients whose formalin-fixed paraffin-embedded (FFPE) tumor had a BRAF V600 mutation (n=51) received therapy with a BRAF and/or MEK inhibitor. The time to treatment failure (TTF) of 13 patients with a BRAF V600 mutation in the tumor but not in plasma obtained before therapy was significantly longer than that of 38 patients whose baseline plasma cfDNA had a BRAF V600 mutation (13.1 vs. 3.0 months; P=0.001). The absence of BRAF V600–mutant cfDNA also was associated with longer TTF (HR, 0.31; P=0.004) in multivariate analysis.
Detection of resistance to targeted therapy
The implementation of personalized medicine principles and targeted therapy into routine oncology practice is bringing an important shift in the treatment of advanced cancers. In metastatic disease, a chronic course is no longer unusual, and patients can survive for many years [105]. However, despite the significant initial therapeutic effect of targeted therapy, the vast majority of patients eventually develop resistance and experience tumor progression. The tumor resistance results from acquisition of mutations in the targeted genes or signaling pathways of cancer cells under therapeutic selective pressure (i.e., secondary resistance). The mutations causing resistance also can be present in the infrequent subclones of pretreatment tumor cells and can predict the further failure of targeted therapy (i.e., primary resistance) [5, 14, 106].
The mechanisms of resistance are often known; however, since routine multiple sequential biopsies are not performed, we have no tools to describe these mechanisms at the level of an individual patient. Both intrinsic and adaptive resistance can occur because of pre-existing or acquired molecular abnormalities, such as gatekeeper mutations in the BCR-ABL kinase domain, which cause resistance to imatinib and other TKIs in chronic myelogenous leukemia [107]. Similarly, emergence of KRAS mutations plausibly causes resistance to EGFR monoclonal antibodies in metastatic colorectal cancer [108], and emergence of EGFR T790M mutation causes resistance to EGFR-TKIs in non-small cell lung cancer [109, 110]. Last but not least, ALK mutation L1196M or C1156Y mediates adaptive resistance to crizotinib in non-small cell lung cancer with ALK rearrangement [111], and mutations in NRAS, MEK, and BRAF amplification indicate resistance to BRAF inhibitor vemurafenib in BRAF-mutant melanoma [112]. Because liquid biopsies can be obtained at low cost at multiple time points, they offer a useful tool for monitoring molecular changes associated with resistance to certain cancer therapies (Table 4).
Table 4.
Examples of studies that assessed cfDNA as the biomarker to monitor resistance to the administrated therapy
| Cancer type | Resistance mutation | Cohort no. | Therapeutic intervention | Results | Reference |
|---|---|---|---|---|---|
| Lung cancer | EGFR T790M | 23 | EGFR-TKIs | EGFR T790M mutation was detected in 10 of 23 patients with progression on EGFR-TKI | Taniguchi et al . (2011) |
| pretreatment EGFR T790M | 135 | erlotinib, gefitinib | Pretreatment detection of mutation in plasma cfDNA predicted lower PFS (8.9 vs. 12.1 months, p =0.007) and OS (19.3 vs. 31.9 months, p = 0.001) compared to patients without mutation | Wang et a l. (2014) | |
| Colorectal cancer | KRAS mutations | 28 | panitumumab | Serum cfDNA KRAS mutations were detected in 9 of 24 originally KRAS wild-type patients | Diaz et al . (2012) |
| KRAS and/or | 62 | cetuximab, panitumumab | Acquired KRAS and/or EGFR ectodomain mutations were | Morelli et al . (2015) | |
| EGFR ectodomain mutations | detected in 44% (27/62) and 8% (5/62) plasma samples after treatment | ||||
| Breast, ovarian, and lung cancers | plasma whole- exome sequencing | 6 | various therapeutics | Activating mutations in PIK3CA after paclitaxel, RB1 after cisplatin, MED1 after tamoxifen and trastuzumab, and GAS6 after lapatinib; T790M EGFR mutation after gefitinib | Murtaza et a l. (2013) |
Abbreviations: OS, overall survival; PFS, progression-free survival; TKIs, tyrosine kinase inhibitors
For instance, PCR detection with the BEAMing approach in patients with advanced non-small cell lung cancer demonstrated EGFR T790M mutation in cfDNA from 10 of 23 patients who experienced disease progression while receiving an EGFR-TKI [113]. In a different study, digital PCR detection of the EGFR T790M resistance mutation in pretreatment cfDNA plasma samples from 135 patients with advanced non-small cell lung cancer treated with an EGFR-TKI was associated with a shorter median PFS (8.9 vs. 12.1 months; P = 0.007) and OS (19.3 vs. 31.9 months; P = 0.001) than no T790M mutation [114].
Another example of emerging resistance mutations to targeted therapy with high clinical relevance is the acquisition of tumor KRAS mutations in codon 12, 13, or 61 in patients with advanced colorectal cancer treated with anti-EGFR monoclonal antibodies cetuximab or panitumumab [16, 20]. Two landmark studies have shown the possibility of detecting and monitoring these emerging KRAS mutations in such patients in cfDNA by using BEAMing technology [16, 20]. Testing of serum cfDNA from 28 colorectal cancer patients receiving panitumumab showed that 9 of 24 patients whose tumor and cfDNA were initially KRAS wild-type had developed detectable cfDNA KRAS mutations [16]. Interestingly, multiple KRAS cfDNA mutations were detected in three individuals. The appearance of mutations generally occurred between 5 and 6 months following initiation of treatment. In the second study, emergence of KRAS aberrations was found in tumor tissue samples from metastatic sites obtained after initiation of therapy [20]. Corresponding plasma samples also showed emergence of KRAS mutation in cfDNA, which could have happened as early as 10 months before radiographic progression [20]. Furthermore, a group from MD Anderson Cancer Center, using BEAMing technology, reported acquired KRAS and/or EGFR ectodomain mutations in 44% (27/62) and 8% (5/62) of plasma samples from patients with advanced colorectal cancer treated with cetuximab or panitumumab, respectively [115]. KRAS codon 61 and 146 mutations were predominant (33% and 11%, respectively).
Even if the candidate-gene techniques to monitor emerging resistance mutations to various targeted therapeutics provide promising results, such approaches have substantial drawbacks, most notably the requirement for prior knowledge of mechanisms of resistance and corresponding mutations. Application of unbiased approaches for detection of emergence of resistant cancer cell subclones using NGS technologies directly on the plasma samples could overcome these limitations. A proof-of-principle study by Murtaza et al. [68] monitored cancer clonal evolution and the acquisition of secondary resistance mutations to various anticancer treatments in serial plasma samples from six patients with advanced breast, ovarian, or lung cancer using unbiased whole-exome sequencing. Follow-up intervals were 1–2 years, and the exome sequencing was performed on two to five plasma samples in each patient. The results revealed emergence of distinct secondary mutations, such as an activating mutation in PIK3CA after paclitaxel, a truncating mutation in RB1 after cisplatin, a truncating mutation in MED1 after tamoxifen and trastuzumab and a splicing mutation in GAS6 after subsequent treatment with lapatinib in the same patient, and a T790M EGFR mutation after treatment with gefitinib. The results of this study established that exome-wide analysis of cfDNA could complement standard biopsy to detect mutations associated with acquired resistance to therapeutic agents in advanced cancers. However, it should be noted that the detected mutant allele fractions for the aberrations were rather high (3%–45%), which can limit the applicability of such an approach to a limited subset of patients.
Overall, liquid biopsy–guided detection of clonal evolution and acquired mechanisms of resistance can be an attractive tool in cancer therapy. However, its utility needs to be tested in future prospective clinical trials that use liquid biopsies as a tool for therapeutic decision making (Table 5).
Table 5.
Overview of active clinical trials with cfDNA as their primary or secondary objective
Expert Commentary
Liquid biopsy utilizing cfDNA is an attractive tool in oncology for identification of molecular targets, determination of prognosis, assessment of response to anticancer therapy, and real-time monitoring of cancer molecular profile. However, the clinical utility of molecular profiling in cfDNA remains to be proven in prospective studies. Retrospective observations demonstrated that changes in the amount of mutant cfDNA can indicate response to anticancer therapy and that emergence of certain molecular abnormalities can predict emergent therapeutic resistance; however, it will remain unclear whether this offers any clinical advantage or alters therapeutic decisions until it is tested in prospective controlled clinical trials. Even though most cfDNA technologies have demonstrated high concordance with molecular testing of tumor tissue, there is still uncertainty whether molecular profile from cfDNA can replace tissue testing, at least in situations when the tissue is in short supply. Furthermore, cfDNA consists of both nonmalignant and tumor DNA, unlike tumor tissue, increasing the need for high sensitivity and limiting the use of technologies such as whole-genome or -exome NGS. Also, cfDNA occurs in short fragments, which can further complicate molecular analysis.
Five-year View
In the next 5 years, cfDNA-based liquid biopsies will be implemented in clinical studies and drug development. Such studies will provide real-time evaluation of pertinent biomarkers as well as the technology itself. Liquid biopsy approaches have been selected for testing as exploratory endpoints in national molecular matching initiatives such as the multi-arm NCI MATCH clinical trial. Furthermore, randomized studies exploring whether liquid biopsy approaches can be used for biomarker detection and subsequent treatment allocation in lieu of tumor tissue are being designed. Liquid biopsies will likely become an integral part of diagnostics in oncology; however, they are not expected to entirely replace tumor biopsies since they cannot address many important factors such as changes in and interactions with the tumor microenvironment. Furthermore, novel liquid sources of DNA will be tested and validated, including CTC and exosomes [116–120].
Key issues.
Identification of oncogenic aberrations provided key insight into cancer biology and led to discovery of new targeted therapies.
Tumor-specific aberrations are usually tested in archival tumor tissue, and limitations or absence of such tissues can preclude molecular analysis and limit the use of personalized therapy.
Small fragments of cancer cell–free DNA are released into the circulation and can be detected in blood, urine, or other biologic materials.
Testing for oncogenic mutations in cell-free DNA, which is not all from the tumor, requires highly sensitive methods capable of detecting one mutant allele in 1,000–10,000 wild-type background alleles.
PCR-based technologies are highly sensitive but do not allow testing for a broad spectrum of aberrations in cell-free DNA. Next-generation sequencing can detect multiple aberrations, but with somewhat lower sensitivity than PCR.
Detection of oncogenic aberrations in cell-free DNA demonstrated high though not absolute concordance with tumor tissue and can be used plausibly for treatment selection.
Quantity of mutant cell-free DNA seems to be of prognostic value in predicting survival.
Dynamic tracking of molecular aberrations in cell-free DNA has potential to be used for monitoring of treatment response in lieu of standard imaging.
Emergence of molecular aberrations in cell-free DNA can provide insight into mechanism of resistance at the individual patient level and can be investigated as a plausible tool for treatment guidance.
Acknowledgments
Authors would like to thank Kathryn Hale (University of Texas MD Anderson Cancer Center, Houston, TX, USA) for editorial assistance.
List of abbreviations
- ABL
abelson murine leukemia viral oncogene homolog;
- ALK
anaplastic lymphoma kinase
- APC
adenomatous polyposis coli gene
- ARMS
amplification refractory mutation system
- BCR
breakpoint cluster region
- BEAMing
beads emulsions, amplification and magnetics
- BRAF
v-raf murine sarcoma viral oncogene homologe B1
- CA 15-3
cancer antigen 15-3
- CAPP-seq
cancer personalized profiling by deep sequencing
- CEA
carcinoembryonic antigen
- cfDNA
cell-free DNA
- cfNA
cell-free nucleic acids
- CSF
cerebrospinal fluid
- CT
computed tomography
- CTC
circulating tumor cells
- ddPCR
droplet digital polymerase chain reaction
- EGFR
epidermal growth factor receptor
- FFPE
formalin-fixed paraffin-embedded
- GAS6
growth arrest-specific 6
- KRAS
kirsten rat sarcoma viral oncogene homolog
- MAPK
mitogen-activated protein kinase
- MASA
mutant allele specific amplification
- ME
mutant enriched
- MED1
mediator complex subunit 1
- MEK
mitogen-activated protein kinase kinase
- NGS
next generation sequencing techniques
- NRAS
neuroblastoma RAS viral oncogene homolog
- OS
overall survival
- PAP-A
pyrophosphorolysis-activated polymerization-allele-specific amplification
- PCR
polymerase chain reaction
- PFS
progression-free survival
- PI3K
phosphatidylinositol 3-kinase
- PIK3CA
catalytic domain p110α of the class I phosphatidylinositol 3-kinase
- PTEN
phosphatase and tensin homolog
- RB1
retinoblastoma gene
- RFLP
restriction fragment length polymorphism
- Safe-Seq
safe-sequencing system
- SSCP
single-strand conformation polymorphism
- TAm-Seq
tagged amplicon deep sequencing
- TKI
tyrosine-kinase inhibitor
- TP53
tumor protein p53
- TTF
time to treatment failure
Footnotes
Financial & competing interests disclosure
The authors were supported by MH CZ - DRO (Faculty Hospital Plzen - FNPl, 00669806) and by the National Sustainability Program I (NPU I) Nr. LO1503 provided by the Ministry of Education Youth and Sports of the Czech Republic, the Elsa U. Pardee Foundation, the Sidney Kimmel Foundation for Cancer Research, the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy, the U.S. National Center for Advancing Translational Sciences (grant no. UL1 TR000371), and the U.S. National Institutes of Health through MD Anderson’s Cancer Center Support Grant (P30 CA016672). F Janku received research support from Biocartis (Mechelen, Belgium), Trovagene (San Diego, CA, USA) and Illumina (San Diego) and is on the Scientific Advisory Board of Trovagene. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13(11):795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrell RA, Swanton C. The evolution of the unstable cancer genome. Curr Opin Genet Dev. 2014;24:61–67. doi: 10.1016/j.gde.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Garrido-Laguna I, Hidalgo M, Kurzrock R. The inverted pyramid of biomarker-driven trials. Nat Rev Clin Oncol. 2011;8(9):562–566. doi: 10.1038/nrclinonc.2011.113. [DOI] [PubMed] [Google Scholar]
- 5*.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. The paper demonstrated the tumor intrinsic heterogeneity. The cancer-related aberrations can differ among tumor regions and distinct metastatic disease sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamantis A, Magiorkinis E, Koutselini H. Fine-needle aspiration (FNA) biopsy: historical aspects. Folia Histochem Cytobiol. 2009;47(2):191–197. doi: 10.2478/v10042-009-0027-x. [DOI] [PubMed] [Google Scholar]
- 7.Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l’homme [in French] Comptes Rendus Séances Société Biol Ses Fil. 1948;142(3–4):241–243. [PubMed] [Google Scholar]
- 8.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benn P, Cuckle H, Pergament E. Non-invasive prenatal testing for aneuploidy: current status and future prospects. Ultrasound Obstet Gynecol. 2013;42(1):15–33. doi: 10.1002/uog.12513. [DOI] [PubMed] [Google Scholar]
- 10.Macher H, Egea-Guerrero JJ, Revuelto-Rey J, et al. Role of early cell-free DNA levels decrease as a predictive marker of fatal outcome after severe traumatic brain injury. Clin Chim Acta. 2012;414:12–17. doi: 10.1016/j.cca.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Jing R-R, Wang H-M, Cui M, et al. A sensitive method to quantify human cell-free circulating DNA in blood: relevance to myocardial infarction screening. Clin Biochem. 2011;44(13):1074–1079. doi: 10.1016/j.clinbiochem.2011.06.083. [DOI] [PubMed] [Google Scholar]
- 12.Tsai N-W, Lin T-K, Chen S-D, et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta. 2011;412(5–6):476–479. doi: 10.1016/j.cca.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Wang H-C, Lin Y-J, Lin W-C, et al. The value of serial plasma nuclear and mitochondrial DNA levels in acute spontaneous intra-cerebral haemorrhage. Eur J Neurol. 2012;19(12):1532–1538. doi: 10.1111/j.1468-1331.2012.03761.x. [DOI] [PubMed] [Google Scholar]
- 14.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 15**.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. This pilot study demonstrated that the tumor unique molecular signature could be found in cfDNA from patients with colorectal cancer and the patients at risk of recurrence after the surgical intervention could be detected. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Diaz LA, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. doi: 10.1038/nature11219. This landmark study demonstrated the possibility to detect KRAS mutations in serum of colorectal cancer patients receiving panitumumab whose tumor and cfDNA were initially KRAS wild-type. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janku F, Vibat CRT, Kosco K, et al. BRAF V600E mutations in urine and plasma cell-free DNA from patients with Erdheim-Chester disease. Oncotarget. 2014;5(11):3607–3610. doi: 10.18632/oncotarget.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyman DM, Diamond EL, Vibat CRT, et al. Prospective Blinded Study of BRAFV600E Mutation Detection in Cell-Free DNA of Patients with Systemic Histiocytic Disorders. Cancer Discov. 2015;5(1):64–71. doi: 10.1158/2159-8290.CD-14-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan W, Gu W, Nagpal S, et al. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem. 2015;61(3):514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. The emergence of KRAS aberrations was found in colorectal cancer tissue samples from metastatic sites obtained after initiation of anti-EGFR therapy. Corresponding plasma samples also showed emergence of KRAS mutation in cfDNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husain H, Venkatapathy S, Gomez G, et al. Cell-free DNA derived from ascites: Detection of copy number and somatic mutations using OncoScan FFPE® Assay [abstract]. Proc Annu Meet Am Assoc Cancer Res; 2015 Apr 18–22; Washington, DC. Philadelphia (PA): AACR; 2015. Abstract nr 2410. [Google Scholar]
- 22.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775(1):181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 24.Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer. 2009;64(1):92–97. doi: 10.1016/j.lungcan.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Ulivi P, Mercatali L, Casoni G-L, et al. Multiple marker detection in peripheral blood for NSCLC diagnosis. PloS One. 2013;8(2):e57401. doi: 10.1371/journal.pone.0057401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon K-A, Park S, Lee SH, et al. Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls. J Mol Diagn. 2009;11(3):182–185. doi: 10.2353/jmoldx.2009.080098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catarino R, Ferreira MM, Rodrigues H, et al. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol. 2008;27(8):415–421. doi: 10.1089/dna.2008.0744. [DOI] [PubMed] [Google Scholar]
- 28.Hashad D, Sorour A, Ghazal A, et al. Free circulating tumor DNA as a diagnostic marker for breast cancer. J Clin Lab Anal. 2012;26(6):467–472. doi: 10.1002/jcla.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong B, Xue J, Yu J, et al. Cell-free DNA in blood is a potential diagnostic biomarker of breast cancer. Oncol Lett. 2012;3(4):897–900. doi: 10.3892/ol.2012.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamat AA, Baldwin M, Urbauer D, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010;116(8):1918–1925. doi: 10.1002/cncr.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zachariah RR, Schmid S, Buerki N, et al. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 2008;112(4):843–850. doi: 10.1097/AOG.0b013e3181867bc0. [DOI] [PubMed] [Google Scholar]
- 32.Schwarzenbach H, Stoehlmacher J, Pantel K, et al. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci. 2008;1137:190–196. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 33.Boni L, Cassinotti E, Canziani M, et al. Free circulating DNA as possible tumour marker in colorectal cancer. Surg Oncol. 2007;16(Suppl 1):S29–31. doi: 10.1016/j.suronc.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Danese E, Montagnana M, Minicozzi AM, et al. Real-time polymerase chain reaction quantification of free DNA in serum of patients with polyps and colorectal cancers. Clin Chem Lab Med. 2010;48(11):1665–1668. doi: 10.1515/CCLM.2010.301. [DOI] [PubMed] [Google Scholar]
- 35.Marzese DM, Hirose H, Hoon DSB. Diagnostic and prognostic value of circulating tumor-related DNA in cancer patients. Expert Rev Mol Diagn. 2013;13(8):827–844. doi: 10.1586/14737159.2013.845088. [DOI] [PubMed] [Google Scholar]
- 36.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 37.Nagata S, Nagase H, Kawane K, et al. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 2003;10(1):108–116. doi: 10.1038/sj.cdd.4401161. [DOI] [PubMed] [Google Scholar]
- 38.Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PloS One. 2011;6(9):e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang BG, Huang H-Y, Chen Y-C, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63(14):3966–3968. [PubMed] [Google Scholar]
- 40.Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38(18):6159–6175. doi: 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Olmo DC, García-Olmo D. Biological role of cell-free nucleic acids in cancer: the theory of genometastasis. Crit Rev Oncog. 2013;18(1–2):153–161. doi: 10.1615/critrevoncog.v18.i1-2.90. [DOI] [PubMed] [Google Scholar]
- 42.García-Olmo DC, Picazo MG, García-Olmo D. Transformation of non-tumor host cells during tumor progression: theories and evidence. Expert Opin Biol Ther. 2012;12(Suppl 1):S199–207. doi: 10.1517/14712598.2012.681370. [DOI] [PubMed] [Google Scholar]
- 43.García-Olmo DC, Domínguez C, García-Arranz M, et al. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010;70(2):560–567. doi: 10.1158/0008-5472.CAN-09-3513. [DOI] [PubMed] [Google Scholar]
- 44.Emlen W, Mannik M. Effect of DNA size and strandedness on the in vivo clearance and organ localization of DNA. Clin Exp Immunol. 1984;56(1):185–192. [PMC free article] [PubMed] [Google Scholar]
- 45*.El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. The paper discussed the important preanalytical considerations that could influence cfDNA analysis.”. [DOI] [PubMed] [Google Scholar]
- 46.Ignatiadis M, Dawson S-J. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25(12):2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- 47.Wong D, Moturi S, Angkachatchai V, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem. 2013;46(12):1099–1104. doi: 10.1016/j.clinbiochem.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 48.Qin J, Williams TL, Fernando MR. A novel blood collection device stabilizes cell-free RNA in blood during sample shipping and storage. BMC Res Notes. 2013;6:380. doi: 10.1186/1756-0500-6-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuaeva NO, Abramova ZI, Sofronov VV. The origin of elevated levels of circulating DNA in blood plasma of premature neonates. Ann N Y Acad Sci. 2008;1137:27–30. doi: 10.1196/annals.1448.043. [DOI] [PubMed] [Google Scholar]
- 50.Björkman L, Reich CF, Pisetsky DS. The use of fluorometric assays to assess the immune response to DNA in murine systemic lupus erythematosus. Scand J Immunol. 2003;57(6):525–533. doi: 10.1046/j.1365-3083.2003.01261.x. [DOI] [PubMed] [Google Scholar]
- 51.Szpechcinski A, Struniawska R, Zaleska J, et al. Evaluation of fluorescence-based methods for total vs. amplifiable DNA quantification in plasma of lung cancer patients. J Physiol Pharmacol. 2008;59(Suppl 6):675–681. [PubMed] [Google Scholar]
- 52.Goldshtein H, Hausmann MJ, Douvdevani A. A rapid direct fluorescent assay for cell-free DNA quantification in biological fluids. Ann Clin Biochem. 2009;46(Pt 6):488–494. doi: 10.1258/acb.2009.009002. [DOI] [PubMed] [Google Scholar]
- 53.Legendre C, Gooden G, Johnson K, et al. Whole genome bisulfite sequencing from plasma of patients with metastatic breast cancer identifies putative biomarkers [abstract]. Proc Annu Meet Am Assoc Cancer Res; 2015 Apr 18–22; Washington, DC. Philadelphia (PA): AACR; 2015. Abstract nr 3825. [Google Scholar]
- 54.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Board RE, Wardley AM, Dixon JM, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120(2):461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 56.Chen Z, Feng J, Buzin CH, et al. Analysis of cancer mutation signatures in blood by a novel ultra-sensitive assay: monitoring of therapy or recurrence in non-metastatic breast cancer. PloS One. 2009;4(9):e7220. doi: 10.1371/journal.pone.0007220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J-Y, Hsieh J-S, Chang M-Y, et al. Molecular detection of APC, K- ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg. 2004;28(7):721–726. doi: 10.1007/s00268-004-7366-8. [DOI] [PubMed] [Google Scholar]
- 58.Frattini M, Gallino G, Signoroni S, et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008;263(2):170–181. doi: 10.1016/j.canlet.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 59.Yamada T, Nakamori S, Ohzato H, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res. 1998;4(6):1527–1532. [PubMed] [Google Scholar]
- 60.Castells A, Puig P, Móra J, et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17(2):578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- 61.Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20(10):2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higgins MJ, Jelovac D, Barnathan E, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18(12):3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dawson S-J, Tsui DWY, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 64.Yung TKF, Chan KCA, Mok TSK, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15(6):2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 65.Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6(254):254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68**.Murtaza M, Dawson S-J, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. A proof-of-principle study successfully monitored the acquisition of secondary resistance mutations to various anticancer treatments in cfDNA from six advanced cancer patients using unbiased whole-exome sequencing. [DOI] [PubMed] [Google Scholar]
- 69.Chan KCA, Jiang P, Zheng YWL, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59(1):211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 70.Heitzer E, Ulz P, Belic J, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5(4):30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janku F, Angenendt P, Tsimberidou AM, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. 2015;6(14):12809–21. doi: 10.18632/oncotarget.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 73.Aung KL, Donald E, Ellison G, et al. Analytical validation of BRAF mutation testing from circulating free DNA using the amplification refractory mutation testing system. J Mol Diagn. 2014;16(3):343–349. doi: 10.1016/j.jmoldx.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Panka DJ, Buchbinder E, Giobbie-Hurder A, et al. Clinical utility of a blood-based BRAF(V600E) mutation assay in melanoma. Mol Cancer Ther. 2014;13(12):3210–3218. doi: 10.1158/1535-7163.MCT-14-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pupilli C, Pinzani P, Salvianti F, et al. Circulating BRAFV600E in the diagnosis and follow-up of differentiated papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98(8):3359–3365. doi: 10.1210/jc.2013-1072. [DOI] [PubMed] [Google Scholar]
- 76.Zill OA, Greene C, Sebisanovic D, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 78.Rothé F, Laes J-F, Lambrechts D, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25(10):1959–1965. doi: 10.1093/annonc/mdu288. [DOI] [PubMed] [Google Scholar]
- 79*.Spindler K-LG, Pallisgaard N, Vogelius I, et al. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18(4):1177–1185. doi: 10.1158/1078-0432.CCR-11-0564. The study demonstrated the prognostic value of the amount of total cfDNA as well as KRAS mutant cfDNA for metastatic colorectal cancer patients. [DOI] [PubMed] [Google Scholar]
- 80.Nygaard AD, Garm Spindler K-L, Pallisgaard N, et al. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79(3):312–317. doi: 10.1016/j.lungcan.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 81.Divella R, Tommasi S, Lacalamita R, et al. Circulating hTERT DNA in early breast cancer. Anticancer Res. 2009;29(7):2845–2849. [PubMed] [Google Scholar]
- 82.Wang S, An T, Wang J, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16(4):1324–1330. doi: 10.1158/1078-0432.CCR-09-2672. [DOI] [PubMed] [Google Scholar]
- 83.Shinozaki M, O’Day SJ, Kitago M, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13(7):2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Silva JM, Silva J, Sanchez A, et al. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin Cancer Res. 2002;8(12):3761–3766. [PubMed] [Google Scholar]
- 85.Lefebure B, Charbonnier F, Di Fiore F, et al. Prognostic value of circulating mutant DNA in unresectable metastatic colorectal cancer. Ann Surg. 2010;251(2):275–280. doi: 10.1097/SLA.0b013e3181c35c87. [DOI] [PubMed] [Google Scholar]
- 86.Trevisiol C, Di Fabio F, Nascimbeni R, et al. Prognostic value of circulating KRAS2 gene mutations in colorectal cancer with distant metastases. Int J Biol Markers. 2006;21(4):223–228. doi: 10.5301/jbm.2008.3336. [DOI] [PubMed] [Google Scholar]
- 87.Swisher EM, Wollan M, Mahtani SM, et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193(3 Pt 1):662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 88.Schwarzenbach H, Eichelser C, Kropidlowski J, et al. Loss of heterozygosity at tumor suppressor genes detectable on fractionated circulating cell-free tumor DNA as indicator of breast cancer progression. Clin Cancer Res. 2012;18(20):5719–5730. doi: 10.1158/1078-0432.CCR-12-0142. [DOI] [PubMed] [Google Scholar]
- 89.Kuhlmann JD, Schwarzenbach H, Wimberger P, et al. LOH at 6q and 10q in fractionated circulating DNA of ovarian cancer patients is predictive for tumor cell spread and overall survival. BMC Cancer. 2012;12:325. doi: 10.1186/1471-2407-12-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fujimoto A, O’Day SJ, Taback B, et al. Allelic imbalance on 12q22–23 in serum circulating DNA of melanoma patients predicts disease outcome. Cancer Res. 2004;64(12):4085–4088. doi: 10.1158/0008-5472.CAN-04-0957. [DOI] [PubMed] [Google Scholar]
- 91.Sozzi G, Conte D, Mariani L, et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001;61(12):4675–4678. [PubMed] [Google Scholar]
- 92.Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59(1):67–70. [PubMed] [Google Scholar]
- 93.Silva JM, Dominguez G, Villanueva MJ, et al. Aberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patients. Br J Cancer. 1999;80(8):1262–1264. doi: 10.1038/sj.bjc.6690495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong IH, Lo YM, Zhang J, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59(1):71–73. [PubMed] [Google Scholar]
- 95.Sharma G, Mirza S, Parshad R, et al. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci. 2010;87(3–4):83–91. doi: 10.1016/j.lfs.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 96.Fujita N, Nakayama T, Yamamoto N, et al. Methylated DNA and total DNA in serum detected by one-step methylation-specific PCR is predictive of poor prognosis for breast cancer patients. Oncology. 2012;83(5):273–282. doi: 10.1159/000342083. [DOI] [PubMed] [Google Scholar]
- 97.Göbel G, Auer D, Gaugg I, et al. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res Treat. 2011;130(1):109–117. doi: 10.1007/s10549-010-1335-8. [DOI] [PubMed] [Google Scholar]
- 98.Philipp AB, Stieber P, Nagel D, et al. Prognostic role of methylated free circulating DNA in colorectal cancer. Int J Cancer. 2012;131(10):2308–2319. doi: 10.1002/ijc.27505. [DOI] [PubMed] [Google Scholar]
- 99.Balgkouranidou I, Karayiannakis A, Matthaios D, et al. Assessment of SOX17 DNA methylation in cell free DNA from patients with operable gastric cancer. Association with prognostic variables and survival. Clin Chem Lab Med. 2013;51(7):1505–1510. doi: 10.1515/cclm-2012-0320. [DOI] [PubMed] [Google Scholar]
- 100.Sun F-K, Fan Y-C, Zhao J, et al. Detection of TFPI2 methylation in the serum of hepatocellular carcinoma patients. Dig Dis Sci. 2013;58(4):1010–1015. doi: 10.1007/s10620-012-2462-3. [DOI] [PubMed] [Google Scholar]
- 101.Tseng J-S, Yang T-Y, Tsai C-R, et al. Dynamic plasma EGFR mutation status as a predictor of EGFR-TKI efficacy in patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol. 2015;10(4):603–10. doi: 10.1097/JTO.0000000000000443. [DOI] [PubMed] [Google Scholar]
- 102.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103*.Douillard J-Y, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9(9):1345–1353. doi: 10.1097/JTO.0000000000000263. This paper demonstrated that EGFR mutation status could be assessed in cfDNA from patients with advanced lung cancer and served as a positive predictive biomarker for targeted therapy with gefitinib. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Janku F, Huang H, Claes B, et al. Rapid, automated BRAF mutation testing of cell-free DNA from plasma of patients with advanced cancers using the novel Idylla platform [abstract]. Proc Annu Meet Am Assoc Cancer Res; 2015 Apr 18–22; Washington, DC. Philadelphia (PA): AACR; 2015. Abstract nr 2413. [Google Scholar]
- 105.Normanno N, Rachiglio AM, Roma C, et al. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. J Cell Biochem. 2013;114(3):514–524. doi: 10.1002/jcb.24401. [DOI] [PubMed] [Google Scholar]
- 106.Ramos P, Bentires-Alj M. Mechanism-based cancer therapy: resistance to therapy, therapy for resistance. Oncogene. 2015;34(28):3617–26. doi: 10.1038/onc.2014.314. [DOI] [PubMed] [Google Scholar]
- 107.Soverini S, Branford S, Nicolini FE, et al. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leuk Res. 2014;38(1):10–20. doi: 10.1016/j.leukres.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 108.Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, et al. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol. 2014;8(6):1084–1094. doi: 10.1016/j.molonc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yun C-H, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su K-Y, Chen H-Y, Li K-C, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(4):433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 111.Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363(18):1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 112.Holderfield M, Deuker MM, McCormick F, et al. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14(7):455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17(24):7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 114.Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PloS One. 2014;9(11):e110780. doi: 10.1371/journal.pone.0110780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26(4):731–6. doi: 10.1093/annonc/mdv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 117.Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12(14 Pt 1):4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 118.De Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 119.Cohen SJ, Punt CJA, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 120.Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]