Abstract
All bacteria that live in oxygenated environments have to deal with oxidative stress caused by some form of exogenous or endogenous reactive oxygen species (ROS) (Imlay, 2013). Large quantities of ROS damage DNA, lipids and proteins which can eventually lead to bacterial cell death (Imlay, 2013). In contrast, smaller quantities of ROS can play more sophisticated roles in cellular signalling pathways affecting almost every process in the bacterial cell e.g. metabolism, stress responses, transcription, protein synthesis, etc. Previously, inadequate analytical methods prevented appropriate analysis of the intra-bacterial redox potential. Herein, we describe a method for the measurement of real-time changes to the intra-bacterial redox potential using redox-sensitive GFP (roGFP2) (van der Heijden et al., 2015). The roGFP2 protein is engineered to contain specific cysteine residues that form an internal disulfide bridge upon oxidation which results in a slight shift in protein conformation (Hanson et al., 2004). This shift results in two distinct protein isoforms with different fluorescence excitation spectra after excitation at 405 nm and 480 nm respectively. Consequently, the corresponding 405/480 nm ratio can be used as a measure for the intra-bacterial redox potential. The ratio-metric analysis excludes variations due to differences in roGFP2 concentrations and since the conformational shift is reversible the system allows for measurement of oxidizing as well as reducing conditions. In this protocol we describe the system by measuring the intra-bacterial redox potential inside Salmonella typhimurium (S. typhimurium) however this system can be adjusted for use in other Gram-negative bacteria.
Materials and Reagents
LB broth (Miller) (Sigma-Aldrich, catalog number: MKBS8229V)
Salmonella typhimurium (SL1344)
Carbenicillin (Goldbio, catalog number: 4800-94-6) [stock solution 100 mg/ml in 50% water/methanol (v/v) kept at −20 °C]
Saline (sodium chloride) (Sigma-Aldrich, catalog number: S9888) [0.9% sodium chloride in sterile water (w/w)]
Hydrogen peroxide [30% (w/w)] (Sigma-Aldrich, catalog number: 026K3770)
Dithiothreitol (AMRESCO, catalog number: 0363C45) (stock solution 1 M in sterile water kept at −20 °C)
pfpv25-roGFP2 vector (available by contacting bfinlay@interchange.ubc.ca) (van der Heijden et al., 2015)
10% glycerol in sterile water (v/v) (glycerol) (Sigma-Aldrich, catalog number: G5516)
Agar (Sigma-Aldrich, catalog number: A5306)
125 ml flasks (VWR International, catalog number: 89090-316)
1.5 ml sterile Eppendorf tubes (VWR International, catalog number: 89000-028)
10% glycerol in sterile water (v/v)
Luria Broth (see Recipes)
Equipment
Black, 96-well assay plate with flat clear bottom (Corning, Costar®, catalog number: 3631)
37 °C incubator/shaker (New Brunswick Scientific, model: M1025-0000)
Electroporator (Bio-Rad Laboratories, catalog number: 165-2100)
Fluorescent plate reader (Tecan Trading AG, model: Infinite M200) or comparable device able to measure 96-well plates at 37 °C, excitation at 405 nm and 480 nm, emission at 510 nm
Spectrophotometer (Thermo Fisher Scientific, model: spectronic 200)
20–200 μl Multichannel pipet (Rainin, model: L12 200R)
Electroporation cuvettes (gap size 1 mm) (VWR International, catalog number: 89047-206)
Microcentrifuge (Thermo Fisher Scientific, catalog number: 75003424)
10 ml glass culture tube (VWR International, catalog number: 47729-574)
Software
-
For fluorescent plate reader (Magellan 7)
Note: This software comes with the Tecan fluorescent plate reader. If a different plate reader is used, software comparable to Magellan 7 can be used for data acquisition of fluorescence measurements.
Procedure
A. Create roGFP2-expressing strain
Day 0
In the afternoon, streak bacterial target strain from −80 °C glycerol stock [25% (v/v) glycerol in LB] on 1.5% LB agar plates and incubate overnight at 37 °C (appropriate antibiotics added).
Day 1
In the afternoon, pick one single colony using a sterile pipette tip from the LB plate. Use the pipette tip to inoculate 5 ml of LB medium in a glass culture tube and grow overnight at 37 °C while shaking at 225 rpm (appropriate antibiotics added).
Day 2
In the morning, take 150 μl from the overnight culture and inoculate 5 ml of fresh LB medium in a glass culture tube. Grow for 3 h at 37 °C and put culture on ice for 5 min.
Spin down 5 ml of the culture at 4,000 x g for 15 min and dispose of supernatant.
Wash the pellet once by adding 1 ml of sterile water and resuspension of the bacterial pellet by pipetting up and down several times. Transfer bacterial suspension to sterile Eppendorf tube and spin down at 14,000 x g for 1 min. Discard the supernatant.
Wash the pellet by adding 1 ml sterile glycerol solution [10% (v/v) glycerol in water] and spin down at 14,000 x g for 1 min. Discard the supernatant. Repeat step A4.
Resuspend the pellet in 50 μl of glycerol solution and add 2 μl of pfpv25-roGFP2 plasmid DNA (~50 ng/μl total plasmid DNA).
Transfer bacterial solution to an electro-cuvette and transform the bacteria using electroporation (V = 2.5 kV). Transfer bacterial solution to 1 ml of fresh LB medium and incubate for 1 h at 37 °C while shaking at 225 rpm prior to plating on LB plates containing 100 μg/ml carbenicillin.
Day 3
Pick one colony and grow at 37 °C overnight in 5 ml LB in a glass culture tube containing 100 μg/ml carbenicillin before making a freezer stock the next day by mixing 1 ml of bacterial culture with 1 ml of 50% glycerol/LB (v/v) for a final concentration of 25% glycerol and freeze down at −80 °C.
B. Preparing bacterial cultures for measurement
Day 0
In the afternoon, streak the bacterial strain that contains the pfpv25-roGFP2 plasmid and the original target strain (without the plasmid and therefore non-fluorescent) from −80 °C glycerol stock on LB agar plates containing appropriate antibiotics and incubate overnight at 37 °C.
Day 1
In the afternoon, pick a single colony from each plate and grow the bacteria in 5 ml of LB in a glass culture tube at containing the appropriate antibiotics overnight at 37 °C while shaking (225 rpm).
Day 2
In the morning, take 300 μl from each of the overnight cultures and inoculate 10 ml of fresh LB medium for each bacterial culture in a 125 ml Erlenmeyer flask (no antibiotic added).
Grow for 3 h (or until OD600 ~1.0) while shaking at 225 rpm. Spin down the cultures at 2,500 x g for 15 min and wash the pellets once in 5 ml saline solution (0.9% w/w in water).
Resuspend the pellet in 1 ml of saline solution and determine the OD600 of the solution in a spectrometer.
Dilute the bacterial solution to OD600=2.0 using saline solution. Transfer 100 μl of bacterial saline solution per well of a black 96-well clear bottom plate.
For each fluorescent strain, also transfer 100 μl of the identical non-fluorescent strain (at OD600=2.0) to determine background fluorescence. In Figure 1 an example of a plate layout is shown.
Figure 1. Example layout of a 96-well imaging plate.
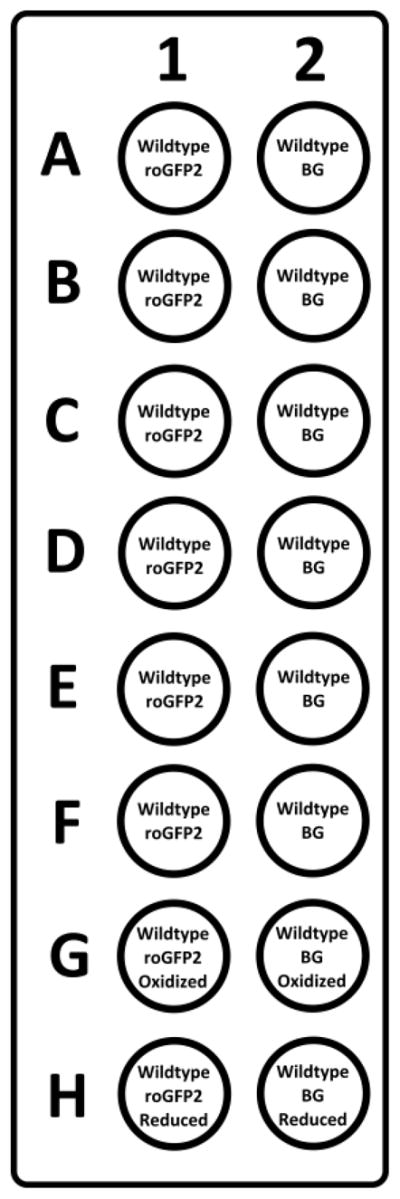
In lane 1 all bacteria express roGFP2 while in lane 2 contains well with empty-vector containing bacteria to determine the background fluorescence (BG) for each condition. The two adjacent well e.g. A1 and A2 undergo an identical challenge with oxidizing agents. In row G, 100 mM of hydrogen peroxide is added and these results are used as maximum oxidized values for normalization. In row H, 10 mM DTT is added and these results are used as maximum reduced values for normalization.
C. Setting up the plate reader
Pre-warm the Tecan fluorescent plate reader to 37 °C. The 405/480 ratio will be obtained by two subsequent measurements in which the bacteria are first excited at wavelength 405 nm while emission is measured at 510 nm. Immediately after the first measurement, the cells are excited again at 480 nm while emission is measured at 510 nm. Both fluorescent intensities are used for calculation of the normalized 405/480 nm ratio and the corresponding intra-bacterial redox potential.
These measurements can be repeated for extended periods of time to follow the intra-bacterial redox potential in real-time.
-
Program the plate reader with the following settings:
Temperature 37 °C.
Before each round of measurement shake for a duration of 2 sec.
Excitation at 405 nm and 480 nm.
Emission at 510 nm.
Reading from bottom.
Gain determined from well (65% of maximum gain for roGFP2-expressing bacteria).
Measurements for 30 min continuously.
Automatically generate excel sheet with fluorescence intensities.
Put the loaded black 96-well clear bottom plate containing bacterial solutions in the Tecan plate reader and measure for 30 min continuously. This data will be used to determine that all bacterial solutions have identical intra-bacterial redox potentials prior to challenge with oxidizing agents.
While the Tecan plate reader is measuring, prepare saline solutions with varying amounts of hydrogen peroxide (2 mM, 4 mM, 6 mM, 8 mM, 10 mM and 12 mM). Also prepare a saline solution with 100 mM hydrogen peroxide and a saline solution with 2 mM DTT. These solutions are used to determine the maximum oxidized 405/480 ratio and maximum reduced 405/480 ratio respectively. Always prepare the hydrogen peroxide solutions fresh.
After the 30 min of continuous measurements are finished, set up the machine again so that it can be immediately restarted to measure for 90 min continuously. Only when the Tecan plate reader is fully prepared and programmed, quickly add 100 μl of the pre-made hydrogen peroxide solutions from step C5 to each well. For this step use a multichannel pipet for rapid sample handling. Use the same hydrogen peroxide concentration in wells adjacent to each other (e.g. A1 and A2, B1 and B2 etc.). Add 100 μl of the 100 mM hydrogen peroxide solution to wells G1 and G2 and add 100 μl of the 2 mM DTT solution to wells H1 and H2 that were also pre-made in step C5. Immediately after the hydrogen peroxide and DTT are added start the measurement. The bacteria will detoxify hydrogen peroxide rapidly and after 90 min (or earlier depending on the experimental set up) the addition of hydrogen peroxide can be repeated for subsequent challenges.
D. Data analysis
First, we calculate the R (ratio 405/480 nm) of S. typhimurium using Equation 1 for each time point. To better explain this calculation we have used the layout from Figure 1. For each fluorescence intensity obtained from well in column 1, background fluorescence is subtracted from the corresponding well in column 2. In Equation 1 we have indicated the corresponding well from Figure 1 between brackets to better explain the calculation.
| Equation 1 |
After R is calculated using Equation 1, it will be normalized to the maximum oxidized and maximum reduced 405/480 values for better comparability using Equation 2. The maximum oxidized ratio (Rox) is calculated from fluorescence of well G1 and the maximum reduced ratio (Rred) is calculated from fluorescence of well H1 by using Equation 1 as described above. After normalization, the upper limit for oxidation given by Rnormalized is 1.0 whereas the lower limit for reduction given by Rnormalized is 0.1.
| Equation 2 |
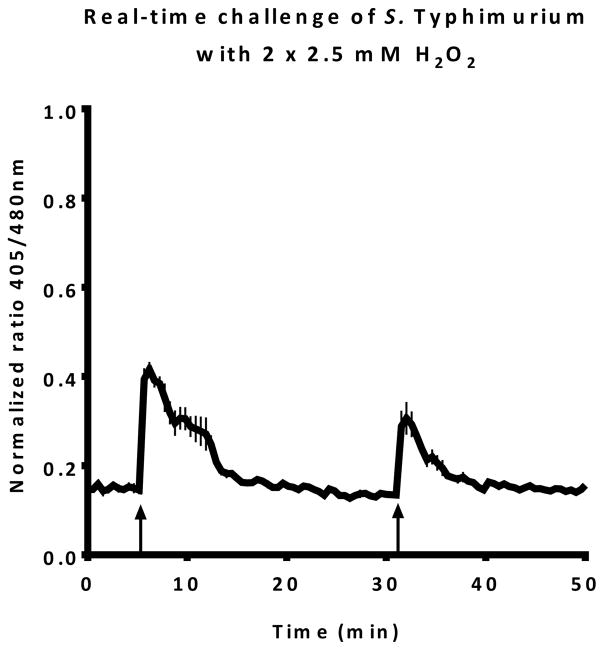
The Rnormalized for each time point can be plotted in a graph to obtain a real-time report on changes to the intra-bacterial redox potential. A typical result can be seen in Figure 2. In this figure, the upward arrows indicate a challenge with 1 mM H2O2.
Using the midpoint potential of roGFP2 (−280mV), the R, Rred and Rox can be used to calculate the actual EroGFP2 inside S. Typhimurium. This calculation has previously been described (Gutscher et al., 2008; van der Heijden et al., 2015). In addition to the R, Rred and Rox we need the fluorescence intensity at 480 nm under reduced or oxidized conditions (I480max and I480min respectively). With these values we calculated the degree of roGFP2 oxidation (OxDroGFP2) given by Equation 3.
| Equation 3 |
The degree of oxidation can then be used to calculate the intracellular sensor redox potential EroGFP2 by using Equation 4.
| Equation 4 |
In which R is the gas constant (8.315 J K−1 mol−1), T is the absolute temperature (310.15 K), z is the number of transferred electrons (Gutscher et al., 2008) and F is the Faraday constant (96,485 C mol−1). The midpoint potential of roGFP2 (E0′roGFP2) is −280 mV (Hanson et al., 2004).
Representative data
Figure 2. Real-time intra-bacterial redox potential of S. typhimurium after two subsequent challenges with 1 mM hydrogen peroxide.
Each upward arrow indicates a challenge with hydrogen peroxide.
Notes
It is important to first plate bacteria on LB instead of starting liquid cultures straight from −80 °C glycerol stocks. This can substantially alter the fitness of the bacterial population and therefore influence your final results.
In this protocol bacteria in late exponential growth phase are used however bacteria in any other growth phase can be used as well. Within one experiment, the control strains should be grown to the same growth phase.
To keep subsequent fluorescence reads frequent enough to accurately monitor the intra-bacterial redox potential, it is advised to never read more than 36 wells per experiment.
To measure the intra-bacterial redox potential in other Gram-negative bacteria, the roGFP2 gene can be cloned into an appropriate vector for the bacteria of choice under a constitutive promotor. In the pfpv25-roGFP2 vector, the roGFP2 gene is under the control of RpsM ribosomal promotor (Salmonella promotor). Promotors of highly expressed genes are recommended to allow for high expression and significant fluorescent signal. For different target strains a promotor has to be selected that allows for high level constitutive expression in the respective target strain.
Since 100 μl of the hydrogen peroxide solutions will be added to the bacterial solutions in the black 96-well clear bottom plate, the final concentration of hydrogen peroxide will be half the concentration of the hydrogen peroxide solutions.
Recipes
-
Luria Broth
25 g of LB broth (Miller) per L of sterile water
Acknowledgments
This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR). B.B.F. is the University of British Columbia Peter Wall Distinguished Professor. No competing interests exist for this work. This protocol was adapted from previous work by our laboratory (van der Heijden et al., 2015). We sincerely thank Dr. James Remington from the University of Oregon for providing the original pRSETB roGFP2 construct.
References
- 1.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5(6):553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 2.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279(13):13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 3.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11(7):443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Heijden J, Bosman ES, Reynolds LA, Finlay BB. Direct measurement of oxidative and nitrosative stress dynamics in Salmonella inside macrophages. Proc Natl Acad Sci U S A. 2015;112(2):560–565. doi: 10.1073/pnas.1414569112. [DOI] [PMC free article] [PubMed] [Google Scholar]