Abstract
ProTx-II is a disulfide-rich peptide toxin from tarantula venom able to inhibit the human voltage-gated sodium channel 1.7 (hNaV1.7), a channel reported to be involved in nociception, and thus it might have potential as a pain therapeutic. ProTx-II acts by binding to the membrane-embedded voltage sensor domain of hNaV1.7, but the precise peptide channel-binding site and the importance of membrane binding on the inhibitory activity of ProTx-II remain unknown. In this study, we examined the structure and membrane-binding properties of ProTx-II and several analogues using NMR spectroscopy, surface plasmon resonance, fluorescence spectroscopy, and molecular dynamics simulations. Our results show a direct correlation between ProTx-II membrane binding affinity and its potency as an hNaV1.7 channel inhibitor. The data support a model whereby a hydrophobic patch on the ProTx-II surface anchors the molecule at the cell surface in a position that optimizes interaction of the peptide with the binding site on the voltage sensor domain. This is the first study to demonstrate that binding of ProTx-II to the lipid membrane is directly linked to its potency as an hNaV1.7 channel inhibitor.
Keywords: membrane bilayer, peptides, sodium channel, toxin, transmembrane domain, NaV1.7, analgesic, gating modifier toxin, peptide-lipid interactions, voltage-gated ion channel
Introduction
Voltage-gated ion channels (VGICs)4 are transmembrane proteins responsible for voltage-dependent movement of ions across cell membranes. They are involved in a wide range of physiological processes, including action potential generation in excitable cells, muscle and nerve relaxation, regulation of blood pressure, and sensory transduction. Many disorders are associated with VGIC abnormalities, and hence VGICs are actively pursued as drug targets for the treatment of a range of neuromuscular, neurological, or inflammatory disorders (1–3). For instance, the human voltage-gated sodium channel subtype 1.7 (hNaV1.7) is involved in pain sensation, and molecules that selectively inhibit this channel might therefore be useful as leads for the development of novel analgesics (4). However, high sequence identity and structural homology exist between the nine subtypes of voltage-gated sodium channels, and given the critical role of NaV channels in the normal electrical activity of neurons, skeletal muscles, and cardiomyocytes, subtype selectivity is crucial but often difficult to achieve with small molecule inhibitors (2).
Disulfide-rich peptide toxins isolated from animal venoms (e.g. spiders, snakes, and cone snails) have attracted much attention as potential analgesics because of their well defined three-dimensional structure and ability to inhibit voltage-gated sodium (NaV), potassium (KV), and calcium (CaV) ion channels with high potency and selectivity (5–9). Because of their stability, selectivity, and potency, these disulfide-rich peptides have been extensively characterized and are vigorously being pursued as drug leads as well as pharmacological tools to characterize VGICs (5).
In general, peptide toxins that inhibit VGICs can be divided into two main groups based on their inhibitory strategy as follows: pore blockers act by obstructing the ion-conducting pore, whereas gating modifier toxins (GMTs) bind to voltage sensor domains (VSDs; see Fig. 1 for a schematic of a sodium channel). GMTs alter the kinetics and gating behavior by changing the relative stability of the closed, open, or inactivated states of the channel (10, 11). The molecular details of the inhibitory mechanism of pore blockers are well characterized. This has been facilitated by the extracellular location of the ion-conducting pore and by the overlapping binding sites of many pore blockers (12). Because the ion-conducting pore is highly conserved among channels of the same ion selectivity (e.g. NaV or KV), pore blockers tend to show subtype promiscuity and thus low selectivity. For example, ShK, a pore blocker isolated from the sea anemone Stichodactyla helianthus, inhibits the related Kv subtypes 1.1, 1.3, 1.4, and 1.6 with similar potency (13, 14). In contrast to the ion-conducting pore, which is highly conserved, the VSDs show significant amino acid sequence variation. Thus GMTs offer the potential to inhibit VGICs with high selectivity, which makes them useful as pharmacological tools and drug leads (15). Nevertheless, there is limited understanding of the mechanism of action of GMTs as VSDs are membrane-associated and GMTs can have distinct binding sites (5, 16). This gap in knowledge is currently limiting the potential use of GMTs in therapeutic applications.
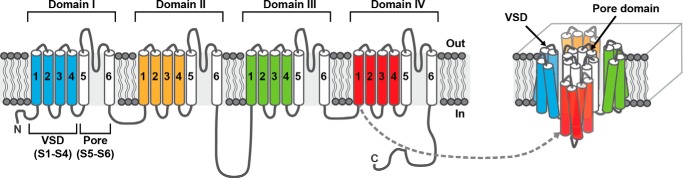
FIGURE 1.
Schematic of a NaV channel. Two-dimensional and three-dimensional representations are shown on the left and right, respectively. NaV channels consist of four homologous domains (domains I–IV) connected by intracellular linkers. Each domain is composed of six transmembrane (TM) helical segments (S1–6). S1–S4 from each domain form a voltage sensor domain (VSD; colored in blue, orange, green, and red). The S5-S6 TM regions from each of the four domains come together to form the pore domain (shown in white).
Experimental evidence suggests that VGICs are located in the cell membrane within raft domains (17), regions that are very rich in cholesterol (Chol) and sphingomyelin (SM). Because of this distinct lipid composition, raft domains display unique physical properties. Although the location of the VGICs in raft domains has been considered to be important for the pharmacological sensitivity of GMTs to VSDs (18), little is known about the importance of these lipid domains on GMT binding, potency, and selectivity. It has been suggested that for a peptide to act as a GMT, it must be able to insert into the lipid bilayer where the VSDs are located (19). This hypothesis is supported by the typical amphipathic structure of GMTs that display positively charged amino acid residues and a hydrophobic patch at the surface of the molecule, properties common to many membrane-active venom peptides (19). Studies of GMTs isolated from tarantula, such as hanatoxin (18, 20), ProTx-II (16, 21), VSTx1 (19, 22, 23), and SGTx1 (18, 24) have demonstrated their affinity for lipid bilayers, further supporting a mechanism in which the peptide gains access to its binding site via partitioning into the membrane. Nevertheless, other GMTs such as huwentoxin-IV (25) and Hd1a (26) do not show affinity for model membranes, suggesting that the ability to bind to lipid bilayers is not an obligate requirement to be a GMT (26–28).
This study investigates ProTx-II, a 30-amino acid residue peptide GMT, isolated from the venom of the Peruvian green velvet tarantula (Thrixopelma pruriens). ProTx-II has been shown to inhibit human NaV channels with ∼70–100-fold selectivity for subtype 1.7 (16, 29), making it a good pharmacological tool and potential drug lead for further optimization. ProTx-II is stabilized by three disulfide bonds and has been classified as an inhibitor cystine knot toxin (30). ProTx-II acts by inhibiting the activation of NaV channels through the reduction of peak current and the induction of a depolarizing shift in the voltage dependence of activation (21, 30). More recently, it has been shown that ProTx-II is also able to inhibit the fast inactivation of hNaV1.7 (16).
A previous report suggests that ProTx-II possesses membrane-binding properties and might insert into the membrane to interact with VSDs (21). However, the exact membrane-binding mode, the importance of the lipid composition, and how the membrane-binding properties affect the selectivity and potency of ProTx-II on NaV channels is unknown.
In this study we were specifically interested in characterizing the interaction of ProTx-II with lipid membranes and the potential role of peptide-cell membrane interactions in its inhibitory activity against human NaV channels. This is an important question as the interplay between GMTs and lipid membranes and any role in inhibiting voltage-gated ion channels is poorly understood.
We determined the three-dimensional structure of ProTx-II in solution using NMR spectroscopy. In addition, we have synthesized a set of ProTx-II analogues, conducted surface plasmon resonance (SPR) and fluorescence spectroscopy experiments, and performed molecular dynamics (MD) simulations to characterize the membrane-binding properties of ProTx-II. Our data show that ProTx-II binds to the water-lipid interface of the cell membrane and suggest that the membrane binding affinity of the peptide correlates with its inhibitory potency on hNaV1.7. In addition, the MD simulations provide mechanistic insight by characterizing the lipid-interaction surface of ProTx-II with zwitterionic and negatively charged phospholipid membranes. Information obtained in this study can be used to guide the design of more potent and selective inhibitors of hNaV1.7 with the potential for translation into analgesic drug leads.
Results
Three-dimensional Structure of ProTx-II
The solution structure of synthetic ProTx-II was calculated using 385 distance restraints derived from 1H-1H NOESY spectra (Fig. 2, PDB code 2N9T, BMRB ID 25917). Hydrogen bond restraints were derived from temperature coefficients, and hydrogen bond partners were identified from preliminary structure calculations. A total of 41 dihedral restraints (17 φ, 16 ϕ, and 8 χ1) was included in the calculations. Individual amino acid residue root mean square deviation (r.m.s.d.) and the statistical analysis were carried out on a set of 20 structures with the lowest energy and best MolProbity score (Fig. 2, A and C, and Table 1).
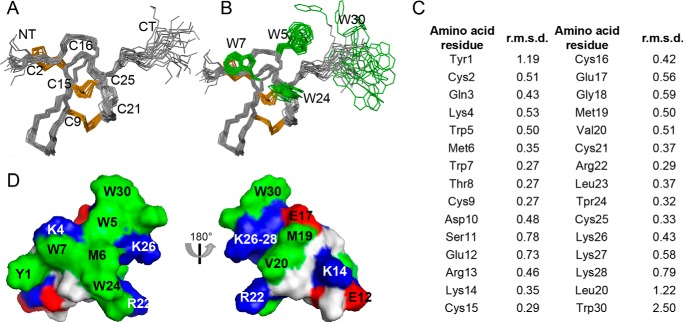
FIGURE 2.
Three-dimensional structure of ProTx-II. A, ensemble of the 20 lowest energy conformations of ProTx-II (PDB code 2N9T) superimposed over the backbone atoms of amino acid residues 2–26. Disulfide bonds are highlighted in orange and the N and C termini are labeled NT and CT, respectively. B, as for A but with the surface-exposed Trp residues shown in green. Note the disordered C-terminal region of ProTx-II, including Trp-30. C, individual amino acid residue r.m.s.d. calculated using molmol version 1.0.7. D, surface representation of ProTx-II highlighting the amphipathic nature of the peptide. Hydrophobic, cationic, and anionic amino acid residues are shown in green, blue, and red, respectively.
TABLE 1.
Structural statistics for the family of 20 lowest energy ProTx-II structures
Data are based on structures with highest overall MolProbity score (74).
| Energies (kcal/mol) | |
| Overall | −838.8 ± 40.4 |
| Bonds | 15.1 ± 1.2 |
| Angles | 50.2 ± 4.7 |
| Improper | 18.3 ± 3.2 |
| van der Waals | −130.0 ± 6.9 |
| NOE | 0.1 ± 0.02 |
| Constrained dihedrals | 1.9 ± 0.6 |
| Dihedral | 150.2 ± 1.8 |
| Electrostatic | −944.5 ± 45.4 |
| MolProbity statistics | |
| Clashes (>0.4 Å/1000 atoms) | 13.8 ± 2.6 |
| Poor rotamers | 0.15 ± 0.4 |
| Ramachandran outliers (%) | 0.0 ± 0.0 |
| Ramachandran favored (%) | 87.7 ± 2.2 |
| MolProbity score | 2.3 ± 0.1 |
| MolProbity score percentilea | 58 ± 8.4 |
| Atomic r.m.s.d. (Å) | |
| Mean global backbone (2–26) | 0.49 ± 0.1 |
| Mean global heavy (2–26) | 1.30 ± 0.2 |
| Mean global backbone (1–30) | 1.08 ± 0.3 |
| Mean global heavy (1–30) | 2.37 ± 0.5 |
| Distance restraints | |
| Intraresidue (i − j = 0) | 139 |
| Sequential (|i − j| = 1) | 133 |
| Medium range (|i − j| <5) | 51 |
| Long range (|i − j| >5) | 52 |
| Hydrogen bonds | 10 |
| Total | 385 |
| Dihedral angle restraints | |
| φ | 17 |
| Ψ | 16 |
| χ1 | 8 |
| Total | 41 |
| Violations from experimental restraints | |
| Total NOE violations exceeding 0.3 Å | 0 |
| Total dihedral violations exceeding 3.0° | 2 (highest 3.3) |
a 100th percentile is the best among structures of comparable resolution; 0 is the worst.
ProTx-II has a well defined core structure maintained by the disulfide bonds and a flexible C-terminal region (Fig. 2, A and B) as suggested by individual amino acid residue and global r.m.s.d. values (Fig. 2C and Table 1). Amino acid residues 2–26 (i.e. excluding the flexible C terminus) have backbone and heavy atom r.m.s.d. values of 0.49 ± 0.1 and 1.30 ± 0.2 Å, respectively, whereas amino acid residues 1–30 have backbone and heavy atom r.m.s.d. of 1.08 ± 0.3 and 2.37 ± 0.5 Å, respectively. The β-sheet motif commonly present in the cystine knot of inhibitor cystine knot peptides (31) is not observed in the ProTx-II structure. ProTx-II has an amphipathic surface, mainly brought about by the high surface exposure of all Trp residues forming the central part of the hydrophobic patch surrounded by positively charged amino acid residues (Fig. 2, B and D). These features are commonly observed in other GMTs isolated from spider venoms (22, 24).
ProTx-II Interaction with Model Lipid Membranes
The ability of ProTx-II to bind to lipid bilayers was evaluated using SPR (Fig. 3, A–D) at 25 °C. We first examined the interaction of ProTx-II with model membranes composed of palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). Phospholipids containing phosphocholine (PC) headgroups are zwitterionic and are the most abundant lipids in mammalian cells (32). POPC forms bilayers in a liquid-disordered phase at 25 °C (33) that can be used to mimic the overall fluid phase and neutral outer leaflet of the lipid bilayer in cell membranes (32, 34). Binding to POPC was compared with that of POPC/Chol/SM (2.7:3.3:4 molar ratio), which forms bilayers in a liquid-ordered phase (35) and hence can be used as a model membrane to mimic lipid raft domains where VGICs are located in the cell membrane (17). In addition, the binding of ProTx-II to model membranes composed of POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE; in a 4:1 molar ratio) was examined. Phosphoethanolamine (PE) phospholipids have a zwitterionic headgroup of smaller size than PC phospholipids and are mainly located in the cytoplasmic leaflet and represent ∼20% of phospholipids in mammalian cell membranes (34).
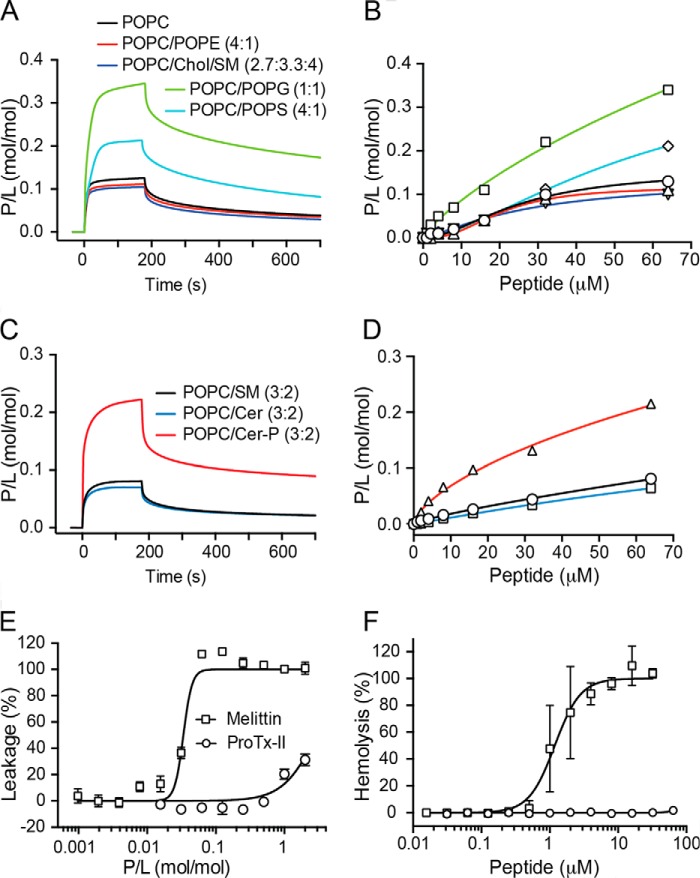
FIGURE 3.
Membrane binding and toxicity of ProTx-II. A and C, surface plasmon resonance sensorgrams obtained with 64 μm ProTx-II injected for 180 s over lipid bilayers deposited onto a L1 chip. Dissociation was monitored for 600 s. Peptide-to-lipid ratio (P/L; mol/mol) was calculated by converting RU into mass (1 RU = 1 pg·mm−2 of lipid or protein) and into moles. The amount of lipid was determined by calculating the average mass of each lipid mixture. B and D, concentration-response curves determined by calculating P/L at the end of the association phase (t = 170 s) for each peptide concentration injected over each lipid bilayer. E, leakage of POPC/POPG (1:1) vesicles induced by ProTx-II. The percentage of leakage of CF-loaded vesicles was determined by CF fluorescence dequenching (λexcitation = 485 nm/λemission = 520 nm) after 20 min of incubation of peptide (0.078–10 μm) with lipid vesicles (5 μm). The concentration-response curve is shown as a function of P/L in the sample. Data are mean ± S.D. of three replicates. F, concentration-response curve for erythrocyte hemolysis obtained by incubating 2-fold dilution of peptide with a suspension of RBCs (0.25% v/v) for 1 h at 37 °C. The percentage of hemolysis was calculated through the absorption of hemoglobin (A405) in the supernatant. Melittin, a membrane-disruptive peptide, was included as a positive control in vesicle leakage and hemolysis assays.
SPR sensorgrams obtained upon injection of ProTx-II over POPC (Fig. 3A) reveal a fast on-rate and slow off-rate. The concentration-response curve (Fig. 3B) reveals that the maximum amount of peptide bound to the membrane when 64 μm peptide is injected is reached at a peptide-to-lipid ratio (P/L) of 0.13. ProTx-II has identical membrane-binding properties when injected over POPC/POPE and POPC/Chol/SM model membranes as shown by the P/L achieved and the kinetic parameters (Fig. 3, A and B, and Table 2). Altogether, these data reveal that ProTx-II displays an affinity for both fluid phase lipid membranes and membranes with raft-like properties and does not distinguish between different zwitterionic headgroups (i.e. PC versus PE).
TABLE 2.
Affinity and kinetic parameters for the interaction of ProTx-II with model membranes as followed with SPR
The affinity of the peptide for different lipid systems is compared by P/L (mol/mol) calculated from sensorgrams obtained upon injection of 64 μm ProTx-II over deposited lipid bilayers (see sensorgrams in Fig. 3, A and C). P/L was calculated at a reporting point at the end of peptide injection (t = 170 s) at which the signal has reached equilibrium. Association and dissociation constants (ka and kd) were calculated by fitting association and dissociation phases separately with a Langmuir model (BIA-evaluation software, version 4.1; GE Healthcare). Standard errors of the fitted parameters are shown.
| Lipid | P/L | ka (103m−1 s−1) | kd (10−3 s−1) |
|---|---|---|---|
| mol/mol | |||
| POPC | 0.13 | 1.98 ± 0.04 | 5.39 ± 0.08 |
| POPC/POPE (4:1) | 0.11 | 2.02 ± 0.04 | 5.69 ± 0.10 |
| POPC/Chol/SM (2.7:3.3:4) | 0.10 | 2.01 ± 0.04 | 6.26 ± 0.12 |
| POPC/POPG (1:1) | 0.34 | 0.86 ± 0.02 | 3.46 ± 0.04 |
| POPC/POPS (4:1) | 0.21 | 0.66 ± 0.01 | 3.94 ± 0.05 |
| POPC/SM (3:2) | 0.08 | 1.00 ± 0.04 | 7.15 ± 0.09 |
| POPC/Cer (3:2) | 0.06 | 1.07 ± 0.04 | 9.87 ± 0.22 |
| POPC/Cer-P (3:2) | 0.21 | 1.13 ± 0.07 | 7.71 ± 0.14 |
The inner leaflet of mammalian cells, in addition to containing PE and PC phospholipids, also contains negatively charged phospholipids containing phosphoserine (PS) headgroups (34). Thus, the membrane binding affinity of ProTx-II, to model membranes composed of the negatively charged mixture POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS; 4:1 molar ratio), was compared with binding to POPC (Fig. 3, A and B, and Table 2). Although phospholipids containing the negatively charged phosphoglycerol (PG) headgroups are not common in mammalian cells (32), mixtures containing a high proportion of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) are commonly used in studies characterizing peptide-membrane interactions of similar peptide toxins (19, 23, 25, 36, 37). Thus, to examine the binding affinity for a highly negatively charged membrane and to see whether there is preference for a specific negatively charged phospholipid headgroup (i.e. PS versus PG), the mixture POPC/POPG (1:1 molar ratio) was also included in our study. Both POPC/POPS (4:1) and POPC/POPG (1:1) form bilayers with fluid phase properties at 25 °C.
The data reveal that ProTx-II has higher affinity, as calculated by P/L, and a slower off-rate for negatively charged membranes compared with neutral POPC membranes, and this affinity is higher with an increase in the percentage of negatively charged lipids. No selectivity for phospholipids containing PS headgroups over phospholipids containing PG headgroups was detected.
Affinity of ProTx-II for Negatively Charged Raft-like Membranes
Spider venoms are particularly rich in positively charged GMTs that modulate the activity of NaV channels (5). Interestingly, sphingomyelinase D (SMaseD), a common component of sicariid spider venoms, converts neutral SM into negatively charged ceramide 1-phosphate (Cer-P) and improves the in vitro inhibitory activity of GMTs (18). This lipid modification has profound effects on the lateral structure and morphology of raft domains by making the membrane surface more negatively charged (38).
We hypothesized that changes in the lipid charge surrounding VGICs, such as those induced by SMaseD, might increase the ability of GMTs to partition into the membrane proximal to the channel. To test this hypothesis, ProTx-II binding to a model membrane containing 40% SM (POPC/SM (3:2 molar ratio)) was compared with a membrane containing the neutral ceramide (Cer; POPC/Cer (3:2)) or the negatively charged Cer-P (POPC/Cer-P (3:2)). ProTx-II was found to have a higher affinity for Cer-P-containing membranes compared with membranes containing either SM or Cer (Fig. 3, C and D, and Table 2). This supports the proposal that SMaseD in spider venoms might increase the affinity of positively charged GMTs for the membrane surface and thus position them in closer proximity for binding to the VSDs of VGICs.
Bilayer Integrity and Toxicity of ProTx-II
The binding of peptides to lipid membranes is often associated with an increase in disorder and a loss of membrane integrity, leading to toxicity against cells. We tested the ability of ProTx-II to compromise the integrity of a lipid bilayer using a membrane leakage assay with carboxyfluorescein (CF)-loaded model membranes. No disruptive effect was detected up to P/L ratio of 1:1 against highly negatively charged vesicles (POPC/POPG (1:1), Fig. 3E). We also showed that ProTx-II does not disrupt human red blood cell (RBC) membranes at concentrations of up to 64 μm (Fig. 3F).
Based on several common features between ProTx-II and antimicrobial peptides (i.e. an overall amphipathic nature, the presence of a hydrophobic patch and a high content of positively charged amino acid residues), the antimicrobial potency of ProTx-II was tested against Gram-negative (Escherichia coli) and Gram-positive (Staphylococcus aureus) bacteria. No activity was detected up to 64 μm (data not shown).
The combined results suggest that although ProTx-II binds to lipid membranes, it does not destabilize the bilayer and is not cytotoxic. Thus, ProTx-II is a good candidate for lead optimization and development of NaV1.7-targeted analgesics, and it warrants further structure-activity relationship studies.
Importance of Lys, Glu, and Trp Residues for ProTx-II Membrane Affinity
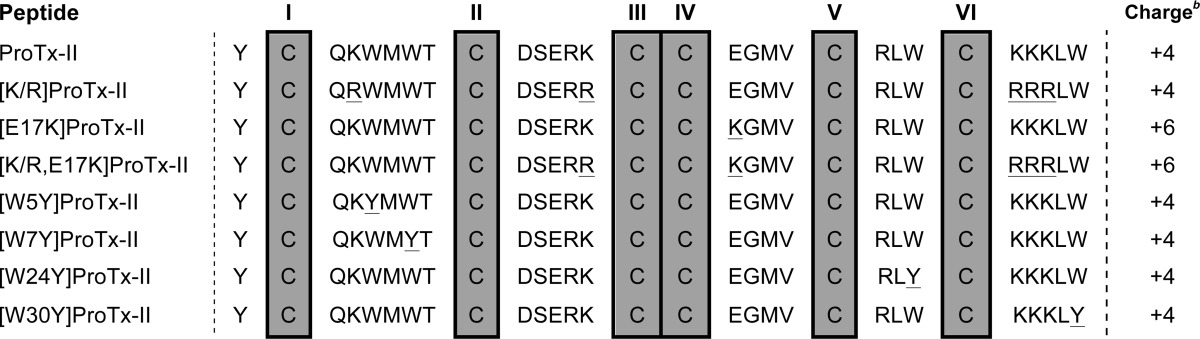
To gain further understanding of the membrane-binding properties of ProTx-II, a series of analogues was synthesized (see Table 3). We were specifically interested in examining whether the hydrophobic patch might be important for membrane binding. Previous mutagenesis studies have shown that Trp residues affect the potency (39) and/or the selectivity (40) of ProTx-II as shown with analogues with the single mutations as follows: W5A, W7A, W24L, W30A (39), W30I, or amidated Trp-30 (40). In these earlier studies, the hydrophobic aromatic Trp residues were replaced with non-aromatic amino acid residues that possess a lower tendency to bind/insert into the membrane (41). Thus, to gain more insight on the importance of the Trp residues on the membrane-binding properties of ProTx-II, we produced four analogues, each with one of the Trp residues mutated to Tyr and also a hydrophobic aromatic residue, shown to have the ability to partition into lipid membranes (41).
TABLE 3.
Amino acid sequence of ProTx-II analogues used in this studya
a Mutated amino acid residues are underlined; cysteines are labeled I–VI.
b Net charge was at pH 7.4.
Furthermore, three additional mutants were designed and synthesized to examine the effect of the overall charge of ProTx-II. Single mutations of Lys or Arg residues with Gln (i.e. K4Q, K13Q, K26Q, and K27Q) or with Ala (i.e. R22A) decreased/ablated the inhibitory potency of ProTx-II (39), suggesting that positively charged amino acid residues are important for its activity. To examine whether this effect is due to the overall charge of the peptide and/or the ability to bind to lipid bilayers, we have synthesized [K/R]ProTx-II, in which all the Lys were replaced with Arg. Arg residues are also positively charged and have been found in a number of studies to lead to tighter binding or greater disruption of cell membranes than Lys (42). We also synthesized [E17K]ProTx-II, in which Glu-17 was replaced with a Lys to increase the overall charge of the peptide. Replacement of Glu-17 with an Ala was previously shown to modestly improve the potency, suggesting that Glu-17 is not a key residue in ProTx-II activity, and it is amendable to improve potency. The third mutant, [K/R,E17K] ProTx-II is a combination mutant with all the Lys mutated with Arg and with Glu-17 replaced with Lys.
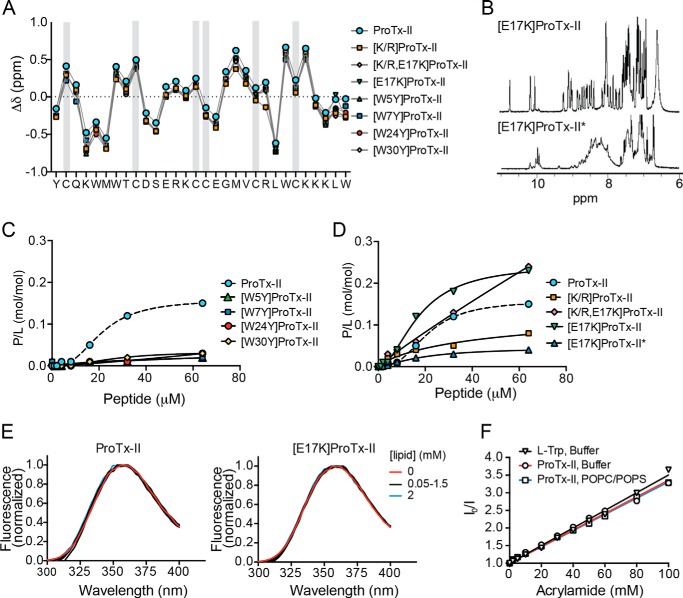
Observed masses of the synthesized peptides are identical to the calculated masses of fully oxidized peptides (i.e. with three disulfide bonds) and retention times obtained by RP-HPLC were similar for all analogues (see Table 4). Analysis of Hα proton secondary chemical shifts for each the analogues derived from 1H-1H TOCSY and NOESYs revealed that they are similar to those of ProTx-II (Fig. 4A). This suggests that all the analogues are correctly folded and have a three-dimensional structure similar to native ProTx-II. Following oxidation, most peptides folded into two isomers that were separated using RP-HPLC, and the correctly folded isomer was identified by one-dimensional NMR spectra (see [E17K]ProTx-II example in Fig. 4B).
TABLE 4.
Physicochemical characterization of synthetic peptides
| Peptide | Average calculated massa | Observed massb | RTc | ϵ280d |
|---|---|---|---|---|
| Da | Da | min | m−1·cm−1 | |
| ProTx-II | 3826.7 | 3825.2 ± 0.2 | 25.4 | 23,865 |
| [K/R]ProTx-II | 3966.7 | 3966.4 ± 0.3 | 26.6 | 23,865 |
| [E17K]ProTx-II | 3825.7 | 3825.6 ± 0.2 | 25.5 | 23,865 |
| [K/R,E17K]ProTx-II | 3965.8 | 3965.2 ± 0.1 | 26.4 | 23,865 |
| [W5Y]ProTx-II | 3803.6 | 3803.3 ± 0.1 | 24.0 | 19,855 |
| [W7Y]ProTx-II | 3803.6 | 3803.5 ± 0.3 | 25.4 | 19,855 |
| [W24Y]ProTx-II | 3803.6 | 3803.4 ± 0.2 | 24.2 | 19,855 |
| [W30Y]ProTx-II | 3803.6 | 3803.4 ± 0.2 | 24.5 | 19,855 |
a Average calculated mass from the amino acid sequence is shown.
b Observed mass was obtained from m/z from ESI-MS.
c Retention time (RT) was determined using analytical HPLC with 2%·min−1 gradient of solvent B (90% acetonitrile, 0.1% trifluoroacetic acid) against solvent A (0.1% trifluoroacetic acid) starting from 1% solvent B.
d Coefficient extinction at 280 nm (ϵ280) calculated based on contribution of disulfide bonds and Tyr and Trp residues.
FIGURE 4.
Structure and membrane-binding properties of ProTx-II analogues. A, comparison of NMR α-proton secondary chemical shifts of ProTx-II and its analogues. The amino acid sequence of native ProTx-II is shown, and the six Cys are indicated with a gray rectangle. B, one-dimensional 1H NMR spectra of amide region of two different isomers of [E17K]ProTx-II isolated using RP-HPLC following oxidation. The most abundant isomer, [E17K]ProTx-II, shows well resolved spin systems indicating native disulfide bond formation and a well structured peptide, whereas the other isomer, [E17K]ProTx-II*, shows poor separation of individual spin systems suggesting non-native disulfide bond formation. C and D, binding of ProTx-II and analogues to POPC/POPS (1:1) model membranes followed by surface plasmon resonance. Peptide samples were injected over POPC/POPS bilayers deposited onto L1 chip and concentration-response curves determined by calculating P/L (mol/mol) at the end of the association phase (t = 170 s) for each peptide concentration. E, Trp fluorescence emission spectra (λexcitation = 280 nm) of 6.25 μm ProTx-II or [E17K]ProTx-II upon titration with POPC/POPS LUVs suspension. Spectra were normalized. The absence of a blue shift upon addition of lipid indicates that Trp residues are not inserting into the hydrophobic region of the lipid bilayer. F, acrylamide quenching of ProTx-II (6.25 μm) fluorescence emission (λexcitation = 290 nm, λemission = 360 nm) in buffer, or in the presence of 1 mm POPC/POPS (4:1) vesicles, upon titration with acrylamide. l-Trp amino acid in buffer was included as a control. Data are represented as Stern-Volmer plots. Calculated Stern-Volmer constant (KSV) is 25.0 ± 0.4 μm for l-Trp in buffer, 23.6 ± 0.5 μm for ProTx-II in buffer, and 23.1 ± 0.3 μm for ProTx-II in the presence of POPC/POPS vesicles.
Hemolysis studies confirmed that none of the mutants permeabilize cell membranes or are toxic to RBCs up to 64 μm (data not shown). Thus, they are good analogues to study how the structure of ProTx-II is related to its membrane-binding and biological properties.
Membrane binding studies conducted using SPR revealed that the four analogues with the Trp/Tyr mutation lost the ability to bind both POPC and POPC/POPS membranes (see POPC/POPS in Fig. 4C). The membrane-binding properties of the other analogues follow the trend: [E17K]ProTx-II > [K/R,E17K]ProTx-II > ProTx-II > [K/R]ProTx-II (Fig. 4C). Taken together, these data show that each of the Trp residues as well as the Lys residues are important for the membrane-binding properties of ProTx-II and that an increase in the overall charge, such as with the E17K mutation, increases the affinity for lipid bilayers.
To investigate the importance of the overall structure of ProTx-II on the membrane-binding properties, we also tested an incorrectly folded isomer, as identified by one-dimensional 1H NMR (see Fig. 4B), of the most membrane-active analogue [E17K]ProTx-II. SPR results (see [E17K]ProTx-II* in Fig. 4D) reveal that this isomer has low affinity for the membrane, suggesting that the correct disulfide connectivity and overall three-dimensional structure is required for ProTx-II membrane-binding properties.
Membrane in Depth Location of ProTx-II
To evaluate whether the Trp residues insert into the lipid bilayer, the intrinsic steady-state Trp fluorescence of ProTx-II and its analogues were examined in the absence and presence of large unilamellar vesicles (LUVs). Trp fluorescence emission spectra of peptide samples were monitored upon titration with LUV suspensions. The Trp fluorescence emission spectrum typically displays a blue shift and an increase in the quantum yield when Trp residues insert into lipid bilayers.
The Trp fluorescence emission spectrum of ProTx-II, and its analogues, is identical to that of l-Trp in buffer and displays a maximum at ∼357 nm, suggesting that the Trp residues in all the analogues are solvent-exposed. The overall Trp fluorescence emission properties of ProTx-II and each of its analogues did not change upon titration with POPC/POPS vesicles (see ProTx-II and [E17K]ProTx-II examples in Fig. 4E). This suggests that when ProTx-II is bound to the membrane, Trp residues remain exposed to the aqueous solution, indicating a shallow location in the lipid bilayer. These findings were further confirmed using acrylamide, a Trp fluorescence quencher unable to enter into the hydrophobic core of lipid bilayers. Acrylamide quenches the overall fluorescence of ProTx-II in the presence of POPC/POPS membranes with the same efficiency as in the absence of lipid (see Stern-Volmer plots in Fig. 4F). Similar results were obtained for all ProTx-II analogues studied.
Insertion of peptides into membranes induces changes in the membrane dipolar potential, which can be detected by a shift in the fluorescence excitation spectrum of the membrane dye di-8-ANEPPS (43). No changes in the fluorescence excitation of di-8-ANEPPS incorporated in POPC/POPS model membranes were detected in the presence of ProTx-II or any of the analogues. These results confirm that ProTx-II as well as all its membrane-active analogues (e.g. [E17K]ProTx-II) do not partition into the hydrophobic core of the membrane and remain at the water-lipid interface when bound to the membrane.
Membrane Location and Lipid Interaction Surface of ProTx-II from MD Simulations
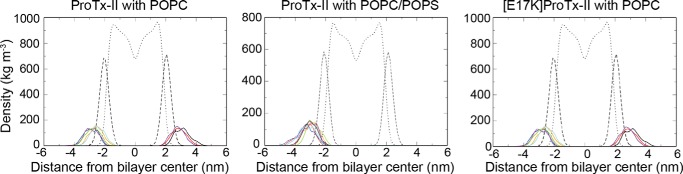
To gain further information on the extent to which ProTx-II penetrates into membranes, and to identify regions of the peptide that mediate lipid binding, MD simulations were conducted for ProTx-II in the presence of zwitterionic POPC and anionic POPC/POPS (4:1) model membranes as well as [E17K]ProTx-II in the presence of POPC. For each peptide-lipid system, eight independent 300-ns simulations were conducted. The depth of penetration in the lipid bilayer was estimated by calculating the density profiles of the lipid headgroups, lipid tails, and the peptide as a function of distance from the center of the bilayer (Fig. 5). Because of the periodic boundary conditions, the peptide can bind to the upper or lower leaflet of the lipid bilayer in the simulation box (which means the electron density of the peptide can be on either side of the lipid bilayer in Fig. 5). The leaflets are equivalent, and thus it is only the position of the peptide with respect to the headgroups and the hydrophobic core that is important. The density profiles suggest that in all three peptide-lipid systems the peptide sits at the water-lipid interface, such that approximately half of the peptide is bound to the lipid headgroups, and the other half is exposed to the extracellular aqueous environment. Further analysis of the simulations shows that the binding of peptide to the membrane surface correlates with a reduction of the solvent-accessible surface area of Trp-5, Trp-7, and Trp-24 (data not shown). However, these amino acid residues never enter the hydrophobic core of the lipid bilayer and thus remain partially solvent-accessible. These results are consistent with the data from the fluorescence experiments.
FIGURE 5.
Orientation of ProTx-II in lipid bilayers calculated from MD simulations. Density profiles of ProTx-II with POPC, ProTx-II with POPC/POPS (4:1), and [E17K]ProTx-II with POPC bilayers. Densities of lipid headgroups (dashed lines) and lipid tails (dotted lines) were averaged over the eight independent simulations for the respective simulation systems. The density profiles for the peptide were calculated separately for each simulation and are shown in color.
To assess whether the peptide adopts a preferred orientation when bound to the membrane, the angle between the peptide and the membrane surface was calculated for all three peptide-membrane systems (data not shown). Although the bound orientation is not biased by the starting configuration, data from the eight replicates showed different ranges of orientations. This suggests that 300 ns was insufficient for the (average) orientation to fully converge. Nevertheless, the analysis showed that the range of orientations sampled by the peptide was narrowest in the [E17K]ProTx-II:POPC and the ProTx-II:POPC/POPS systems. This suggests that the presence of negatively charged lipids or an increase in the positive charge on the peptide helps anchor the peptide to the membrane.
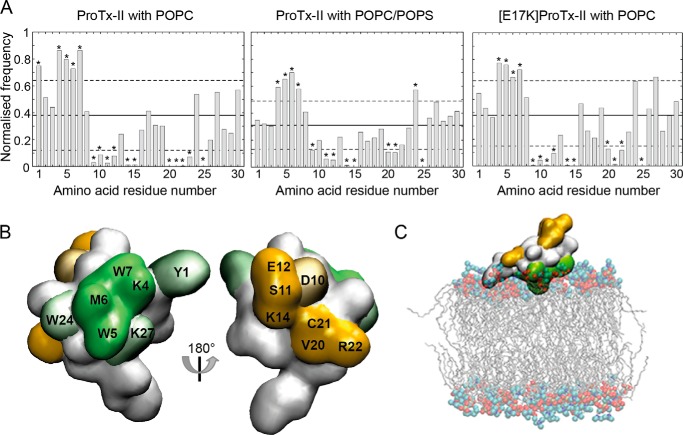
To investigate whether specific amino acid residues mediate the binding of ProTx-II to phospholipid membranes, the normalized frequency of finding a residue within 0.35 nm of the lipid bilayer was calculated. Fig. 6A shows the normalized lipid residency frequency from the simulations of ProTx-II and [E17K]ProTx-II with POPC and ProTx-II with POPC/POPS. Calculations of the normalized frequencies at increasing simulation times showed that in contrast to the relative orientation, the lipid interaction surface does converge on the time scale simulated (data not shown). The simulations suggest that, in the presence of POPC, the frequency with which amino acid residues Tyr-1, Lys-4, Trp-5, Met-6, and Trp-7 are found in close proximity to the membrane is significantly higher than that of amino acid residues Cys-9, Asp-10, Ser-11, Glu-12, Lys-14, Cys-15, Val-20, Cys-21, Arg-22, Leu-23, and Cys-25. The peptide-lipid interactions in the presence of POPC/POPS are very similar to those observed using pure POPC. The main difference is a reduced lipid interaction for Tyr-1 and an increased lipid interaction for Trp-24 and Lys-27. The [E17K]ProTx-II analogue had a very similar lipid interaction surface to ProTx-II aside from the interaction with Tyr-1 being reduced.
FIGURE 6.
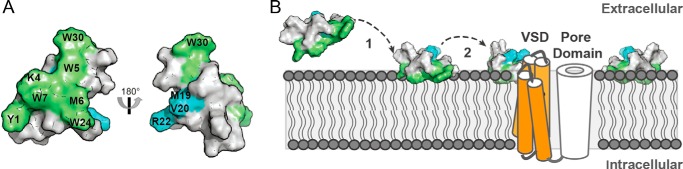
Lipid binding interface of ProTx-II calculated from MD simulations. A, normalized frequency of finding any atom of a given amino acid residue from the peptide within 0.35 nm of any atom within the lipid bilayer for ProTx-II in the presence of POPC or POPC/POPS and [E17K]ProTx-II with POPC. The distance was calculated between the center of geometry of the side chain of a given amino acid residue and the center of geometry of the headgroup of the phospholipid. The lipid residency frequency averaged over all amino acid residues is shown by the solid horizontal line, and the dashed lines indicate ±1 S.D. from the average. Amino acid residues with a normalized frequency below or above 1 S.D. from the average are marked with an asterisk. B, surface representation of ProTx-II with amino acid residues more or less likely to contact the lipid bilayer shown in green and orange, respectively. Solid colored amino acid residues are part of the interaction surface in simulations of ProTx-II in the presence of both a pure POPC bilayer and a POPC/POPS (4:1) bilayer, and shaded amino acid residues show increased or decreased frequency in either POPC bilayer or POPC/POPS. C, snapshot from the MD simulation showing a commonly adopted orientation of ProTx-II relative to the membrane; residues that are more or less likely to contact the lipid bilayer surface are colored green and orange, respectively.
In Fig. 6B these lipid residency frequencies are mapped onto the surface of ProTx-II. Amino acid residues Lys-4, Trp-5, Met-6, and Trp-7, which show higher lipid residency frequencies in all three simulation systems, form a continuous surface with amino acid residues Trp-24, Tyr-1, and Lys-27 adjacent to that surface. Amino acid residues that are less likely to interact with lipids are found on the opposite face of the peptide. Combined with the results from the analysis of the orientation, the simulations predict that, although the peptide shows certain mobility when bound to the lipid bilayer, the interaction surface formed by amino acid residues Lys-4, Trp-5, Met-6, and Trp-7 serves to position and anchor the peptide at the water-lipid interface (Fig. 6C). Trp-24 and Lys-27 also contribute to the interaction with the lipid membrane. These results are consistent with data from SPR experiments, which showed that Trp mutants of ProTx-II lacked the ability to bind to phospholipid model membranes and that the [K/R]ProTx-II analogue shows decreased binding affinity. The [E17K]ProTx-II analogue also shows enhanced membrane affinity. Analysis of the distance between peptide and bilayer as a function of time shows that in all simulations of [E17K]ProTx-II with POPC, the peptide binds to the membrane faster compared with ProTx-II, consistent with the higher on-rate in the SPR experiment. This indicates that the high affinity of the [E17K]ProTx-II analogue is more likely the result of the increase in overall charge of the peptide than the result of specific interactions with residue Lys-17.
Lipid Binding and Inhibition of hNaV1.7 in Neuronal Cells
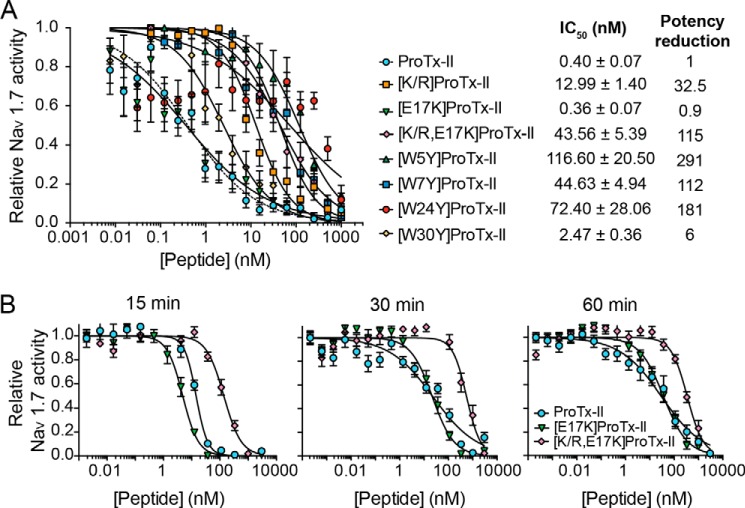
We also examined whether the ability to bind lipid bilayers was correlated with the inhibition of hNaV1.7 by ProTx-II and its analogues. FLIPR studies (Fig. 7A) revealed that synthetic ProTx-II inhibits hNaV1.7 in SH-SY5Y cells with an IC50 of 0.40 ± 0.07 nm, which is within the previously determined range (16, 29, 40). All ProTx-II analogues with mutations in Trp residues had reduced potency against hNav1.7 compared with ProTx-II. The [W30Y]ProTx-II mutant was 6-fold less potent, whereas the other Trp mutants showed a dramatic 100–300-fold reduction in potency. As all of these mutants lacked the ability to bind to model membranes (SPR data), this lower activity suggests that binding to the lipid membrane is important for ProTx-II inhibition of hNaV1.7. Differences in the potency of these analogues suggest that, in addition to affecting membrane binding, some Trp residues might also be involved in binding to the receptor-binding site, with each individual Trp residue having a different degree of involvement. For instance, a mutation in Trp-30 was previously shown to decrease the binding rate to sodium channels without affecting the off-rate, whereas Trp-24 decreased both the association and dissociation rates (39), suggesting a more direct involvement in receptor binding.
FIGURE 7.
Inhibition of hNaV1.7 in SH-SY5Y cells induced by ProTx-II and its analogues followed by fluorescence emission signal of calcium 4 dye influx using FLIPRTETRA. The relative activity of hNaV1.7 was calculated by normalizing the fluorescence emission intensity (λexcitation = 470–495 nm; λemission = 515–575 nm) to the maximum signal and by setting fluorescence emission intensity obtained with TTX (1 μm) as 0% of activity. Inhibition curves were fit as relative activity versus peptide concentration using a sigmoidal curve with Hill slope (−0.5 to −2). Data points are mean ± S.E. (n ≥3). A, comparison of the potency of ProTx-II and its analogues to inhibit hNaV1.7. Peptide samples, dye, and scorpion toxin OD1 were co-incubated with the cells for 60 min followed by stimulation of hNaV1.7 with 4 μm veratridine. IC50 and standard deviation error associated with the fitted parameter and the potency relative to native ProTx-II are shown. B, time course comparison of inhibitory activity of ProTx-II and [E17K]ProTx-II. In this modified assay, cells were pre-incubated with dye and OD1, followed by incubation with peptide samples for 15 (left), 30 (middle), or 60 (right) min. [K/R,E17K]ProTx-II was included as a control analogue with lower inhibition potency, as shown in A. The analogue [E17K]ProTx-II shows 3-fold better inactivation than native ProTx-II at the shorter incubation time of 15 min on SH-SY5Y cells, but these differences are not evident with longer incubation times.
Although the increased overall charge of the [E17K]ProTx-II mutant led to increased membrane binding in model membranes, its inhibitory potency was identical to that of ProTx-II when the peptides were pre-incubated with cells for 60 min prior to addition of the agonist (veratridine) (Fig. 7A). A modified assay performed to investigate the effect of lower peptide preincubation times (15 and 30 min) on hNav1.7 inhibitory potency revealed that [E17K]ProTx-II was consistently 2–3-fold more potent than wild-type ProTx-II at the shorter time of 15 min (Fig. 7B). This suggests a correlation between an increase in membrane binding on-rate and the more rapid inhibition of hNav1.7 for [E17K]ProTx-II with an increase in the overall charge.
The ∼30- and ∼110-fold loss of potency for the [K/R]ProTx-II and [K/R,E17K]ProTx-II analogues, respectively, suggests that one or more Lys residues are essential for the inhibitory activity of this peptide, unrelated to the charge of the residue, but likely due to one or more Lys residues directly binding to the channel and/or due to involvement in anchoring the peptide with the correct orientation in the membrane.
Binding of ProTx-II to Cell Membranes
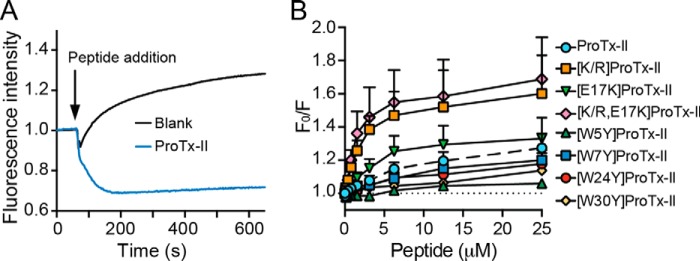
To investigate whether the affinity of ProTx-II for model lipid bilayers correlates with its ability to bind the membranes of human cells, we fluorescently labeled the outer leaflet of SH-SY5Y cells with nitrobenzoxadiazol (NBD)-labeled phospholipids and added DiSBAC2(3) to the extracellular solution. This oxonol dye has a higher affinity for depolarized membranes than polarized membranes and consequently exhibits enhanced fluorescence emission intensity in depolarized cells due to fluorescence resonance energy transfer (FRET) between NBD and DiSBAC2(3). Binding of positively charged peptides onto the membrane surface increases the membrane polarization and thus a decrease in the fluorescence emission of DiSBAC2(3)-labeled membranes is expected (44). Using this approach, we confirmed that ProTx-II binds to the membrane surface of adherent SH-SY5Y cells (Fig. 8A). An identical result was obtained upon addition of ProTx-II to SH-SY5Y cells in suspension (freshly trypsinized and washed), as measured with a cuvette-spectrofluorimeter setup. Although we did not test whether ProTx-II binds to cell membranes devoid of hNaV1.7, we anticipate that peptides with affinity for model lipid membranes also have affinity for cell lipid membranes.
FIGURE 8.
Ability of ProTx-II and analogues to bind to the membrane surface of SH-SY5Y cells. Membrane cells were labeled with NBD-labeled phospholipid, and the oxonol dye DisBAC2(3) was added extracellularly. Increase in cell membrane polarization due to peptide binding is detected by a decrease in FRET between NBD and DiSBAC2(3) as measured with excitation at 470–495 nm and emission at 565–625 nm. A, kinetics of binding showing a drop in DiSBAC2(3) fluorescence upon addition of the peptide, indicating cell membrane polarization through binding of the peptide at the cell surface. Buffer addition (blank) shows an increase in DiSBAC2(3) fluorescence indicating cell membrane depolarization due to increase in the KCl concentration. B, concentration-response curves for binding of the different peptides to the surface of SH-SY5Y cells. Fluorescence signal obtained at a fixed time point was normalized to the blank. Data are mean ± S.E. (n = 3) and are represented as ratio of fluorescence emission intensity in the absence (F0) and presence of peptide (F).
Analogues with mutation of Trp residues all showed lower affinity for cell membranes compared with native ProTx-II, whereas the remaining mutants showed increased binding in the following order [K/R,E17K]ProTx-II > [K/R]ProTx-II > [E17K]ProTx-II > ProTx-II (Fig. 8B). These data show that whereas the Lys → Arg mutation decreases lipid membrane binding and inhibitory activity on NaV1.7, it increases the amount of peptide at the cell surface. It is possible that cell surface binding of positively charged peptides such as [K/R]ProTx-II might occur due to binding to glycosaminoglycans (e.g. heparan sulfate) and other negatively charged/H-bond donor molecules rather than to the lipid membrane (45). Arg-containing peptides have been shown to possess stronger affinity to glycosaminoglycans, compared with Lys counterparts (46, 47), which can explain the higher cell surface association of [K/R]ProTx-II compared with ProTx-II or of [K/R,E17K]ProTx-II compared with [E17K]ProTx-II.
Discussion
In this study, we have investigated the membrane-binding properties and hNaV1.7 inhibitory activity of ProTx-II and a panel of ProTx-II analogues. ProTx-II binds at the water-lipid interface of the cell surface. Its ability to bind lipid bilayers was found to correlate with its inhibition of hNaV1.7, suggesting that membrane binding is the first step in the inhibitory mechanism of ProTx-II. In addition, we identified a patch of amino acid residues on the surface of ProTx-II that mediate its interaction with lipid bilayers. Overall, this study provides information on the orientation of ProTx-II within the cell membrane and on its inhibitory mechanism. Identification of the amino acid residues involved in binding to the cell membrane and to the target binding site has the potential to assist in the design of novel ProTx-II analogues with enhanced pharmacological properties.
ProTx-II Sequence and Structure Are Important for Its Membrane-binding Properties
SPR studies revealed that ProTx-II binds to neutral membranes that mimic either the overall fluidity of cell membranes (POPC) or raft domains (POPC/Chol/SM) (see Fig. 3, A and B). As no major differences were identified in binding to these two lipid systems, we hypothesize that, when added extracellularly, ProTx-II is likely to be evenly distributed across the cellular lipid membrane, instead of having a preference for raft domains where VGICs are located. Despite a high affinity for lipid bilayers, ProTx-II does not disrupt lipid bilayers nor is it toxic to mammalian cells (see Fig. 3, E and F), Gram-positive or Gram-negative bacteria.
Comparison of the membrane-binding properties of ProTx-II and analogues (see Fig. 4, C and D) suggests that the overall three-dimensional structure of the peptide, its amphipathicity, the correct orientation of the hydrophobic patch, and specific amino acid residues (Trp and Lys) all contribute to the ability of the peptide to bind lipid membranes. In addition, an increase in overall positive charge enhances the membrane binding affinity of ProTx-II, even for neutral membranes, probably due to increased electrostatic attractions for the phosphate moieties in the phospholipid headgroups.
ProTx-II Binds at the Water-Lipid Interface and Is Anchored onto the Membrane through a Specific Set of Surface-exposed Amino Acid Residues
Fluorescence studies and MD simulations indicate that ProTx-II is predominantly found at the lipid membrane surface and does not partition into the hydrophobic interior of the membrane (see Figs. 4–6). MD simulations further suggest that a continuous patch composed of amino acid residues Lys-4, Trp-5, Met-6, and Trp-7 form the main lipid interaction surface of ProTx-II (see Fig. 6). Amino acid residues Tyr-1, Trp-24, and Lys-27, which are adjacent to this surface, also contribute to lipid binding. Through this surface the peptide is “anchored” to the lipid headgroups. This binding model is consistent with our mutagenesis studies showing that mutation of Trp and Lys residues to Tyr and Arg, respectively, reduces the membrane-binding properties of ProTx-II. Neither the interacting surface nor the orientation of ProTx-II was significantly altered by the presence of negatively charged lipids or an increased overall positive charge of the peptide.
In combination, the results suggest that binding of ProTx-II to lipid bilayers is dominated by a combination of electrostatic and hydrophobic interactions. The initial attraction between the peptide and the membrane is actioned via longer range electrostatic interactions between positively charged Lys residues and negatively charged phosphate moieties in the phospholipid headgroups. Once positioned on the membrane surface, the hydrophobic face of ProTx-II orients the peptide at the water-lipid interface. This hypothesis is supported by the preference of ProTx-II for anionic lipids (e.g. POPS, Cer-P) and by the observation that the E17K mutation increases the lipid binding affinity, whereas the amino acid residue itself is not part of the interaction surface. In addition, the side chain amine group of Lys residues, particularly Lys-4 and Lys-27, may be involved in dipole-dipole interactions and H-bonds with phosphate moieties of the phospholipid headgroups (2, 48). Similar lipid-binding properties have been observed with ProTx-I, a similarly structured peptide that also inhibits VGICs as a GMT (26).
Substitution of Lys with Arg residues decreased the membrane binding affinity of ProTx-II. This observation suggests that aside from charge, the specific chemical properties of the Lys side chain are important for membrane binding. Experimental evidence suggests that Arg residues possess a higher ability to bind to lipid bilayers (42, 49, 50) and to promote attachment to cell surfaces (46) when compared with Lys residues. The Arg guanidinium group has been shown to facilitate deeper insertion of peptides into membranes, compared with that of Lys side chain amine (51), and it has been claimed by some that this is due to the potential of Arg to interact with two phosphate groups via bidentate hydrogen bonds (48, 52). In the case of ProTx-II, the presence of Arg decreased the affinity for lipid membranes, suggesting that the Lys residue side chain is indeed relevant for the binding of ProTx-II to lipid bilayers and its location at the lipid membrane surface.
Trp residues, in particular Trp-5, Trp-7, and Trp-24, appear to be responsible for anchoring ProTx-II at the water-lipid interface. Trp residues partition preferentially at membrane interfaces due to their amphipathic nature. The Trp indole side chain has a large nonpolar surface that is able to establish hydrophobic interactions with acyl groups, and a NH group able to form H-bonds with phosphate moieties. Replacement of any Trp with a Tyr residue reduced the membrane-binding properties of ProTx-II, possibly because Tyr is less suited to bind at the phospholipid interface as it is more polar than Trp (41).
Affinity of ProTx-II for Raft-like Membranes Can Be Improved by Lipid Conversion
ProTx-II does not show a preference for the membranes containing SM and Chol used to mimic raft-like domains where NaV channels are located but favors negatively charged over neutral membranes (see Fig. 3, A–D), probably due to increased electrostatic attractions between its positively charged amino acid residues and the anionic phospholipids. All reported GMTs are positively charged, and some of these toxins only bind to anionic lipid membranes (27, 28). However, negatively charged lipids are exclusively located in the cytoplasmic leaflet in healthy mammalian cells (34) and are not exposed at the cell surface where ProTx-II binds. Despite this, we do believe the preference for negatively charged membranes is functionally important. Spider venom contains many components; for instance the enzyme SMaseD has been found in sicariid spider venoms and converts the neutral SM into the anionic Cer-P. Our SPR studies with model membranes composed of POPC/Cer-P versus POPC/SM (Fig. 3, C and D) indicate that the peptide concentration in the vicinity of the channel is likely increased upon treatment with SMaseD. However Milescu et al. (18) found that the overall affinity of GMTs did not change significantly following pre-treatment of cell membranes with SMaseD. As the percentage of SM is much higher in raft domains (∼40%) (33) than the overall percentage in cell (∼5–15% (53)), the work of Milescu et al. (18) would not preclude the action of SMaseD leading to higher concentrations of the peptide in the vicinity of the channel.
Biological Activity of ProTx-II Correlates with Ability to Bind to Lipid Bilayers
Studies with SPR in model membranes and fluorescence assays with neuroblastoma cells confirmed the ability of ProTx-II to bind at the cell membrane surface, further supporting the hypothesis that inhibition of hNaV1.7 is correlated with the ability of the peptide to bind to the lipid membrane. All mutants with lower lipid membrane-binding properties compared with native ProTx-II also had lower inhibitory activity (see Fig. 4 and Table 4). Specifically, conservative mutations of ProTx-II Trp residues decreased its affinity for lipid bilayers as well as its potency against hNaV1.7. In previous mutagenesis studies, it was shown that mutations at amino acid residues shown here to be important for membrane binding by ProTx-II, particularly Trp-5, Met-6, Trp-7, Trp-24, Lys-14, and Lys-27, induced at least a 10-fold decrease in inhibitory potency against hNaV channels (39, 54). Altogether, this study suggests that binding to cell membranes is most likely required for the inhibition of NaV channels by ProTx-II.
Membrane Role in ProTx-II Binding to Human NaV Channels
In summary, our results suggest that inhibition of hNaV channels by ProTx-II is closely associated with its ability to bind to the membrane surface and that binding to the membrane may be required for the interaction with VSDs. This hypothesis is supported by our results and previous mutagenesis studies (16, 39, 40) in which amino acid residues that affect the ProTx-II on-rate/off-rate and/or its inhibitory potency and selectivity, span across a large surface area of the molecule (see Fig. 9A). The involvement of all of these residues for the binding and recognition of sodium channels is unlikely to occur unless toxin binding involves membrane-restricted receptor-binding sites.
FIGURE 9.
Model of ProTx-II binding to the membrane and sodium channels. A, ProTx-II surface representation showing the patch of amino acid residues important for binding to the membrane (green) and amino acid residues that are assumed to be important for binding to the channel (blue). B, putative location of ProTx-II within the lipidic membrane and in the presence of the channel: 1, membrane-binding patch anchors the peptide at the cell surface through binding at the water-lipid region; 2, increased peptide concentration in vicinity of channel and the peptide orientation facilitates the interaction with the receptor-binding site, likely to be located at the membrane interface (e.g. S3-S4 loop in VSD II). A model of the surface three-dimensional representation of ProTx-II (PDB code 2N9T) and a schematic of a VSD (orange) and the pore domain (white) within a phospholipid bilayer are shown.
Concomitant binding of ProTx-II to the channel and the lipid membrane is a possible mechanism, but a mechanism whereby ProTx-II binds to the membrane prior to binding the sodium channel better explains our results and previous mutagenesis studies. In particular, single mutations in amino acid residues that were identified here as part of the patch that anchors the peptide to lipid membranes (e.g. Met-6, Trp-7, Lys-27, and Trp-30) were shown to decrease the association rate to sodium channels by 10–76-fold without affecting the dissociation rate (39). A plausible explanation for a decreased association rate is the involvement of these amino acid residues in bringing ProTx-II into close vicinity to the channel through binding to the lipid membrane. Once the peptide is bound to the membrane and the channel, these residues do not affect the activity. In contrast, mutation in the amino acid residues Met-19, Val-20, and Arg-22 (39) affected both the association and the dissociation rates of the peptide to the channel, suggesting a direct interaction of these amino acid residues with the receptor-binding site. Binding of ProTx-II to the lipid membrane would be, at the very least, expected to increase the peptide concentration in the channel vicinity and thus enhance interaction with its VSD-binding site.
Based on our findings and previous studies, we propose a model (Fig. 9) in which amino acid residues Tyr-1, Lys-4, Trp-5, Met-6, Trp-7, Trp-24, and Lys-27 initially anchor the peptide onto the membrane surface. An independent region, potentially formed by the amino acid residues Met-19, Val-20, and Arg-22, then directly binds to the channel. The fact that the peptide remained at the membrane surface in the experimental studies and also the MD simulations performed as part of this study suggest that ProTx-II exerts its inhibitory activity at the outer layer. Although ProTx-II possesses properties that are communally identified in cell-penetrating peptides (i.e. positively charged amino acid residues and amphipathic three-dimensional structure), its inability to deeply insert into the hydrophobic core of lipid membranes suggests that ProTx-II and its membrane-active analogues remain at the cell surface and do not penetrate cells by direct cell membrane permeation. This is in agreement with the extracellular S3-S4 loop in the VSD of channel domain II (see Fig. 1) being at least one of the binding sites for this peptide (29).
Although an interplay between GMTs and lipid membranes does not seem to be a requirement for the activity of all GMTs (26–28), those with similar structures and membrane-binding features to ProTx-II are worthy candidates for further development to improve membrane binding as a means of enhancing potency against NaV channels implicated in pain sensation.
Materials and Methods
Peptide Synthesis
ProTx-II and analogues (Table 3) were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry at 0.25-mmol scale on 2-chlorotrityl resin using an automatic synthesizer (Symphony, Protein Technologies Inc.). Side chain protecting groups used were Cys/Gln(Trt), Lys(Boc), Asp/Glu(OtBu), Tyr/Ser/Thr(tBu), and Arg(Pbf). Peptides were simultaneously released from the resin and the side chain protecting groups cleaved using a trifluoroacetic acid/triisopropylsilane/water mixture (94:3:3, v/v/v) for 3 h. ProTx-II, [K/R]ProTx-II, and [E17K]ProTx-II were oxidized in 0.1 m Tris, 2 m urea at pH 8.0 with 0.15 mm reduced (GSH) and 0.30 mm oxidized (GSSG) glutathione as before (40). For the peptides [K/R,E17K]ProTx-II, [W5Y]ProTx-II, [W7Y]ProTx-II, [W24Y]ProTx-II, and [W30Y]ProTx-II, a good folding yield was obtained with buffer containing 0.1 m Tris, 2.4 m guanidine HCl, 10 mm GSH, 1 mm GSSG at pH 8.0 (55). Peptides (0.1 mg/ml) were incubated in the oxidation buffer at room temperature for 24–48 h with stirring and purified as described previously (40). The mass of the peptides was confirmed by ESI-MS (Table 4), and correct folding was examined by two-dimensional 1H NMR spectroscopy. The overall hydrophobicity of the peptides was compared by analytical RP-HPLC retention time (see Table 4) with a 2%/min gradient of solvent B (90% v/v acetonitrile, 0.1% v/v formic acid) against solvent A (0.1% v/v formic acid).
Peptides were quantified by absorbance at 280 nm using the predicted extinction coefficient, which accounts for the contribution from disulfide bonds and aromatic amino acid residues (see Table 4). Unless otherwise stated, peptide samples were solubilized in HEPES-buffered saline (10 mm HEPES, 150 mm NaCl, pH 7.4), and measurements were conducted at 25 °C.
Solution Structure Determination by NMR Spectroscopy
Peptide samples for NMR spectroscopy were prepared at a concentration of ∼1.0 mm and pH ∼3.5 in a 90% H2O, 10% D2O mixture. All NMR spectra were recorded at 25 °C on a Bruker Avance 600 MHz spectrometer equipped with a cryoprobe. Two-dimensional 1H-1H TOCSY and NOESY spectra were recorded as described previously (56). Two-dimensional 1H-13C HSQC, 1H-15N HSQC, and ECOSY spectra were also recorded for ProTx-II to assist with structure determination. In addition, a series of 1H-1H TOCSY spectra of ProTx-II were acquired at various temperatures (7–40 °C) to calculate amide-proton temperature coefficients. Amide protons experiencing a chemical shift gradient more negative than −4.6 ppb/°C were considered shielded, and hydrogen bond restraints were added for these amide protons if the acceptor atom could be identified in preliminary structure calculations. The structure of ProTx-II was calculated as described previously (56) using the AUTO function in CYANA (57) and further refined in a water shell using CNS (58). The 20 conformers with the lowest energy and best MolProbity statistics were chosen as the final ensemble of ProTx-II structures.
Vesicle Lipid Preparation
Synthetic POPC, POPS, POPG, C18:1 oleoyl SM, C18:1 Cer-P, and C18:1 Cer were purchased from Avanti Polar Lipids. Synthetic Chol was purchased from Sigma. LUVs (diameter 100 nm) or small unilamellar vesicles (SUVs; diameter 50 nm) were prepared in HEPES-buffered saline (10 mm HEPES, 150 mm NaCl, pH 7.4) by freeze-thaw and sized by extrusion as described (59).
Peptide Membrane Binding Followed by SPR
SPR measurements were conducted using an L1 sensor chip on a BIAcore 3000 instrument (GE Healthcare) at 25 °C using protocols described previously (59, 60). Peptide and lipid samples were prepared in running buffer (10 mm HEPES, 150 mm NaCl, pH 7.4). SUV suspensions (final lipid concentration 0.5 mm) composed of POPC, POPC/POPE (4:1 molar ratio), POPC/Chol/SM (2.7:3.3:4), POPC/POPG (1:1), POPC/POPS (4:1), POPC/SM (3:2), POPC/Cer (3:2), or POPC/Cer-P (3:2) were injected onto the L1 chip surface with a flow rate of 2 μl/min for 40 min, reaching a steady-state plateau and confirming chip surface coverage of deposited lipid bilayers (60). Association of peptides to lipid bilayers was followed by injecting peptide samples with varying concentrations over deposited lipid bilayers (flow rate 5 μl/min, 180 s); peptide-lipid dissociation was followed for 600 s. Response units (RU) were normalized to compare the various peptide/lipid systems by converting RU into mol/mol (assuming 1 RU = 1 pg/mm2 of lipid and peptide) as before (60).
Vesicle Leakage Assay
Ability of ProTx-II to disrupt lipid membranes was examined by a CF fluorescence de-quenching assay using POPC/POPG (1:1) LUVs. Vesicle preparation, removal of extracellular CF, and lipid quantification were done following protocols detailed previously (61). LUVs composed of POPC/POPG (1:1) and loaded with 50 mm CF solubilized in 10 mm HEPES, 150 mm NaCl, pH 7.4, were dispensed into a 96-well plate (final lipid concentration of 5 μm) and incubated for 20 min with varying peptide concentrations (2-fold dilution starting from 10 μm). Samples with Triton X-100 (0.1% v/v) and buffer were included. Melittin, a membrane disruptive peptide, was also included as a control. CF fluorescence emission intensity (λexcitation = 485 nm, λemission = 520 nm) was measured using a fluorimeter plate reader (PHERAstar FS) and converted into percentage of leakage by considering the signal obtained with Triton X-100 and buffer as 100 and 0% of leakage, respectively.
Peptide-Membrane Binding Followed by Fluorescence Spectroscopy
The steady-state intrinsic Trp fluorescence of ProTx-II and its analogues was examined in the absence or presence of LUVs following protocols described previously (42, 62). Briefly, 6.25 μm peptide samples were titrated with LUV suspensions composed of POPC or POPC/POPS (4:1 molar ratio), and fluorescence emission spectra were recorded following excitation at 280 nm. Fluorescence emission spectra were corrected by subtracting the blank, for fluorophore dilution and light dispersion induced upon addition of LUVs suspension.
Exposure of Trp residues to the aqueous environment was examined by fluorescence emission quenching efficiency induced by acrylamide as described previously (42, 62); fluorescence emission intensity (λexcitation = 290 nm and λemission = 360 nm) of peptides (6.25 μm) in the absence or presence of 1 mm POPC/POPS (4:1) LUVs was followed upon titration with acrylamide. The quenching efficiency was quantified by calculating the Stern-Volmer constant. All fluorescence measurements were conducted using an LS50B fluorescence spectrophotometer (PerkinElmer Life Sciences).
Membrane Dipolar Potential Changes Induced in LUVs
LUVs composed of 200 μm POPC or POPC/POPS (4:1) and 4 μm di-8-ANEPPS dye were prepared and fluorescence excitation spectra (λemission = 360 nm) scanned in the absence/presence of 20 μm ProTx-II, or its analogues, using protocols previously described (43).
Modeling Studies
MD simulations were carried out as described previously (26). Briefly, all simulations were performed using GROMACS version 3.3.3 (63) in conjunction with the GROMOS 54A7 force field (64). Parameters for lipid molecules were taken from the revised GROMOS 53A6 force field for lipids (65, 66) combined with topologies from the Automatic Topology Builder (67, 68). Water was described using the simple point charge water model (69). Simulations were conducted with a 2-fs time step, and the temperature and pressure were maintained at 25 °C and 1 bar, respectively. Four simulation systems were set up with randomly oriented ProTx-II in the presence of POPC. Each system was simulated in duplicate for 300 ns giving a total of eight independent simulations. The same protocol was used for simulations of ProTx-II with POPC/POPS (4:1) and [E17K]ProTx-II with POPC. Trajectories were analyzed with GROMACS tool and custom python scripts using the MDAnalysis package (70) as outlined previously (26). Unless stated otherwise, analyses were performed using data from the last 200 ns of the simulation in which the peptide is bound to the bilayer. Convergence of properties calculated from MD trajectories such as the orientation of the bound peptide relative to the membrane, density profiles, and lipid residency frequencies were checked by comparing the property of interest averaged over blocks of increasing simulation times.
Hemolysis Studies
The hemolytic activity of ProTx-II and its analogues was examined by incubating varying concentrations of peptide solubilized in phosphate-buffered saline (PBS; 10 mm Na2HPO4, 1.8 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl) with 0.25% (v/v) of RBCs for 1 h at 37 °C as before (61). The percentage of hemolysis was quantified as described (61) by measuring hemoglobin released into the supernatant through absorption at 405 nm. All peptides were tested in triplicate.
Antibacterial Studies
ProTx-II inhibition of growth of the Gram-negative bacterium E. coli ATCC 25922 and Gram-positive S. aureus ATCC 25923 was examined using a microtiter broth dilution assay as before (42).
Inhibition of hNaV1.7 Assessed by Fluorimetric Imaging Plate Reader (FLIPRTETRA) Calcium Assay
Inhibition of NaV1.7 endogenously expressed in SH-SY5Y cells by ProTx-II and its analogues was quantified using a previously described assay (71) in which the fluorescence of SH-SY5Y cells loaded with calcium 4 dye (Molecular Devices) were used to indirectly measure changes in membrane depolarization when sodium channels were activated (71). Briefly, SH-SY5Y cells were plated at a density of 25,000 cells/well on 384-well black-walled imaging plates 48 h before the assay. The calcium 4 dye with 30 nm scorpion toxin OD1 (a subtype-specific allosteric modulator that delays inactivation of hNaV1.7; gift from Dr. Thomas Durek) was added to the cells. ProTx-II and its analogues, diluted in physiological salt solution (PSS; 140 mm NaCl, 11.5 mm glucose, 5.9 mm KCl, 1.4 mm MgCl2, 1.2 mm NaH2PO4, 5 mm NaHCO3, 1.8 mm CaCl2, 10 mm HEPES) with 0.1% (w/v) bovine serum albumin (BSA) were added to the cells and incubated for 60 min (37 °C, 5% CO2) in concentrations ranging from 3 μm to 0.007 nm before stimulating hNaV1.7 using 4 μm veratridine, a non-selective NaV channel activator (Abcam). Fluorescence emission intensity (excitation 470–495 nm and emission 515–575 nm) was assessed every second for 300 s after addition of veratridine. The NaV pore blocker tetrodotoxin (TTX, 1 μm; Abcam) was included as a positive control for NaV inhibition. At least three experiments each with a minimum of three replicates of ProTx-II and its analogues were performed. Fluorescence emission intensity obtained from the FLIPRTETRA was corrected against baseline and buffer-only controls using ScreenWorks 3.2.0.14 (Molecular Devices). The fluorescence emission intensity obtained for each sample was normalizing to the maximum signal (0% inhibition), and the fluorescence emission intensity obtained with TTX (1 μm) was considered as 100% inhibition. Normalized values were plotted against peptide concentration. IC50 values were determined by fitting the peptide against the normalized fluorescence emission signal (GraphPad Prism 6) with a sigmoidal curve with Hill slope. For time course experiments a modified assay was conducted, whereby pre-incubation (cells with dye and OD1) times and peptide incubation times were 45/15 min, 30/30 min, and 0/60 min, respectively, for the 15, 30, and 60 min times. Notably, the dye concentration during peptide incubation was lower (0.66×) compared with the original 60-min assay above.
Binding of Peptides to Cell Membranes
The ability of ProTx-II and its analogues to bind to SH-SY5Y cell membranes was examined using a FRET-based assay. For this assay, cell membranes were labeled with NBD-phospholipids and then DisBAC2(3), a soluble oxonol dye with higher affinity for depolarized membranes than polarized membranes (44), was added extracellularly. SH-SY5Y cells were trypsinized, washed, and seeded at 50,000 cells/well in a 96-well black-walled imaging plate 72 h before the assay. SUVs made with POPC, POPE, and NBD-dioleoylglycerophosphoethanolamine (NBD-DOPE) (POPC/POPE/NBD-DOPE (3:1:1 molar ratio)) were dispersed in PSS and prepared by extrusion as described above. NBD-labeled phospholipids were incorporated into cells by incubating POPC/POPE/NBD-DOPE SUVs (final NBD-DOPE concentration was 150 μm) for 30 min at 37 °C (72). Phospholipids with NBD label in the headgroup cannot undergo membrane flip-flop (73). Thus, upon liposome fusion with cells, labeled lipids should be restricted to the outer leaflet. Cells were washed with PSS to remove non-fused liposomes, and then 30 μl of 5 μm DiSBAC2(3) in PSS buffer was added to each well and incubated for 10 min before taking measurements. Fluorescence excitation and emission spectra, and FRET between NBD-labeled SH-SY5Y cells to DisBAC2(3) added in solution, were first confirmed using a cuvette and fluorimeter spectrophotometer setup. For this trial, SH-SY5Y cells were trypsinized after incubation with the POPC/POPE/NBD-DOPE SUVs, and the fluorescence measurements were conducted with cells in suspension. A drop in the fluorescence emission intensity of DiSBAC2(3) upon addition of ProTx-II was observed. This assay was then optimized for a high-throughput setup in which FRET of NBD/DisBAC2(3) was followed with a FLIPRTETRA using excitation at 470–495 nm and emission at 565–625 nm. KCl buffer (10 mm HEPES buffer, 150 mm KCl, pH 7.4) was added to cells at 20 μl per well (final K+ concentration was 63.5 mm) to depolarize cells and increase fluorescence emission due to higher partitioning of DisBAC2(3) at the membrane surface. Peptides solubilized in KCl buffer were added to the plate in 2-fold dilutions starting from 25 μm. A blank with buffer was also included. Fluorescence emission was recorded over time. It is worth mentioning that the peptide concentration range used in the peptide-cell membrane binding studies is higher than that used in the hNaV1.7 inhibition assays as a high amount of peptide bound to membranes is required to induce changes in cell membrane polarity measurable in the assay. Nevertheless, these higher concentrations are not expected to affect the interpretation of the results.
Author Contributions
S. T. H. and C. I. S. designed the study. S. T. H. performed and analyzed data from surface plasmon resonance, fluorescence spectroscopy, cell membrane-binding assay followed by fluorescence, hemolytic, and antibacterial studies. E. D. designed, conducted, and analyzed the molecular dynamics simulations. S. C. performed surface plasmon resonance experiments. N. L. performed and analyzed surface plasmon resonance, cell membrane binding, and hNaV1.7 inhibition assays. O. C. and P. T. synthesized and purified the peptides used in this study. I. V. and M. I. developed the hNaV1.7 inhibition assay and performed preliminary testing. C. I. S. performed and analyzed NMR experiments and calculated the three-dimensional structure of ProTx-II. S. T. H., C. I. S., N. L., and E. D. wrote the manuscript. D. J. C., A. E. M., and G. F. K. critically reviewed the manuscript and contributed to the interpretation of the data and conclusions. All the authors read the manuscript and provided specific feedback.
This work was supported in part by Australian National Health and Medical Research Center Project Grant APP1080405 (to C. I. S. and S. T. H.), Early Career Fellowship APP1071293 (to E. D.), Principal Research Fellowship APP1044414 and Program Grant APP1072113 (to G. F. K.), Australian Research Council in the form of an Australian Research Council Discovery Outstanding Researcher Grant DP130102153 (to A. E. M.), Future Fellowship FT130101215 (to I. V.), Australian Research Council Australian Laureate Fellowship FL150100146 (to D. J. C.), and an Institute for Molecular Bioscience Industry Fellowship (to C. I. S.). This research was undertaken with the assistance of resources provided at the NCI National Facility systems at the Australian National University through the National Computational Merit Allocation Scheme supported by the Australian Government. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 2N9T) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Chemical shifts have been deposited under BioMagResBank accession number 25917.
- VGIC
- voltage-gated ion channel
- LUV
- large unilamellar vesicle
- SUV
- small unilamellar vesicle
- CF
- carboxyfluorescein
- POPC
- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine
- NBD
- nitrobenzoxadiazol
- DOPE
- dioleoylglycerophosphoethanolamine
- PC
- phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
- SM
- sphingomyelin
- SMaseD
- sphingomyelinase D
- GMT
- gating modifier toxin
- RU
- response unit
- Chol
- cholesterol
- VSD
- voltage sensor domain
- SPR
- surface plasmon resonance
- MD
- molecular dynamics
- PDB
- Protein Data Bank
- PE
- phosphoethanolamine
- PS
- phosphoserine
- h
- human
- r.m.s.d.
- root mean square deviation
- Cer-1-P
- ceramide 1-phosphate
- Cer
- ceramide
- TTX
- tetrodotoxin
- P/L
- peptide-to-lipid ratio
- di-8-ANEPPS
- 4-[2-[6-(dioctylamino)-2-naphthalenyl]ethenyl]-1-(3-sulfopropyl)-pyridinium, inner salt
- DiSBAC2(3)
- bis-(1,3-diethylthiobarbituric acid)trimethine oxonol.
References
- 1. McGivern J. G. (2007) Advantages of voltage-gated ion channels as drug targets. Expert Opin. Ther. Targets. 11, 265–271 [DOI] [PubMed] [Google Scholar]
- 2. Bagal S. K., Brown A. D., Cox P. J., Omoto K., Owen R. M., Pryde D. C., Sidders B., Skerratt S. E., Stevens E. B., Storer R. I., and Swain N. A. (2013) Ion channels as therapeutic targets: a drug discovery perspective. J. Med. Chem. 56, 593–624 [DOI] [PubMed] [Google Scholar]
- 3. Wulff H., Castle N. A., and Pardo L. A. (2009) Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 8, 982–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. King G. F., and Vetter I. (2014) No gain, no pain: NaV1.7 as an analgesic target. ACS Chem. Neurosci. 5, 749–751 [DOI] [PubMed] [Google Scholar]
- 5. Klint J. K., Senff S., Rupasinghe D. B., Er S. Y., Herzig V., Nicholson G. M., and King G. F. (2012) Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon 60, 478–491 [DOI] [PubMed] [Google Scholar]
- 6. Knapp O., McArthur J. R., and Adams D. J. (2012) Conotoxins targeting neuronal voltage-gated sodium channel subtypes: potential analgesics? Toxins 4, 1236–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moran Y., Gordon D., and Gurevitz M. (2009) Sea anemone toxins affecting voltage-gated sodium channels–molecular and evolutionary features. Toxicon 54, 1089–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Norton R. S., Pennington M. W., and Wulff H. (2004) Potassium channel blockade by the sea anemone toxin ShK for the treatment of multiple sclerosis and other autoimmune diseases. Curr. Med. Chem. 11, 3041–3052 [DOI] [PubMed] [Google Scholar]
- 9. Herzig V., and King G. F. (2015) The cystine knot is responsible for the exceptional stability of the insecticidal spider toxin ω-hexatoxin-Hv1a. Toxins 7, 4366–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dilly S., Lamy C., Marrion N. V., Liégeois J.-F., and Seutin V. (2011) Ion-channel modulators: more diversity than previously thought. Chembiochem 12, 1808–1812 [DOI] [PubMed] [Google Scholar]
- 11. Bosmans F., and Swartz K. J. (2010) Targeting voltage sensors in sodium channels with spider toxins. Trends Pharmacol. Sci. 31, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tikhonov D. B., and Zhorov B. S. (2011) Possible roles of exceptionally conserved residues around the selectivity filters of sodium and calcium channels. J. Biol. Chem. 286, 2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beeton C., Pennington M. W., and Norton R. S. (2011) Analogs of the sea anemone potassium channel blocker ShK for the treatment of autoimmune diseases. Inflamm Allergy Drug Targets 10, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chi V., Pennington M. W., Norton R. S., Tarcha E. J., Londono L. M., Sims-Fahey B., Upadhyay S. K., Lakey J. T., Iadonato S., Wulff H., Beeton C., and Chandy K. G. (2012) Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon 59, 529–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Catterall W. A. (2010) Ion channel voltage sensors: structure, function, and pathophysiology. Neuron 67, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao Y., Blumenthal K., Jackson J. O. 2nd., Liang S., and Cummins T. R. (2010) The tarantula toxins ProTx-II and huwentoxin-IV differentially interact with human Nav1.7 voltage sensors to inhibit channel activation and inactivation. Mol. Pharmacol. 78, 1124–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dart C. (2010) Lipid microdomains and the regulation of ion channel function. J. Physiol. 588, 3169–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Milescu M., Bosmans F., Lee S., Alabi A. A., Kim J. I., and Swartz K. J. (2009) Interactions between lipids and voltage sensor paddles detected with tarantula toxins. Nat. Struct. Mol. Biol. 16, 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S.-Y., and MacKinnon R. (2004) A membrane-access mechanism of ion channel inhibition by voltage sensor toxins from spider venom. Nature 430, 232–235 [DOI] [PubMed] [Google Scholar]
- 20. Phillips L. R., Milescu M., Li-Smerin Y., Mindell J. A., Kim J. I., and Swartz K. J. (2005) Voltage-sensor activation with a tarantula toxin as cargo. Nature 436, 857–860 [DOI] [PubMed] [Google Scholar]
- 21. Smith J. J., Alphy S., Seibert A. L., and Blumenthal K. M. (2005) Differential phospholipid binding by site 3 and site 4 toxins. Implications for structural variability between voltage-sensitive sodium channel domains. J. Biol. Chem. 280, 11127–11133 [DOI] [PubMed] [Google Scholar]
- 22. Jung H. J., Lee J. Y., Kim S. H., Eu Y.-J., Shin S. Y., Milescu M., Swartz K. J., and Kim J. I. (2005) Solution structure and lipid membrane partitioning of VSTx1, an inhibitor of the KvAP potassium channel. Biochemistry 44, 6015–6023 [DOI] [PubMed] [Google Scholar]
- 23. Mihailescu M., Krepkiy D., Milescu M., Gawrisch K., Swartz K. J., and White S. (2014) Structural interactions of a voltage sensor toxin with lipid membranes. Proc. Natl. Acad. Sci. U.S.A. 111, E5463–E5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jung H. H., Jung H. J., Milescu M., Lee C. W., Lee S., Lee J. Y., Eu Y.-J., Kim H. H., Swartz K. J., and Kim J. I. (2010) Structure and orientation of a voltage-sensor toxin in lipid membranes. Biophys. J. 99, 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao Y., Luo X., Kuang F., Deng M., Wang M., Zeng X., and Liang S. (2008) Synthesis and characterization of huwentoxin-IV, a neurotoxin inhibiting central neuronal sodium channels. Toxicon 51, 230–239 [DOI] [PubMed] [Google Scholar]
- 26. Deplazes E., Henriques S. T., Smith J. J., King G. F., Craik D. J., Mark A. E., and Schroeder C. I. (2016) Membrane-binding properties of gating modifier and pore-blocking toxins: membrane interaction is not a prerequisite for modification of channel gating. Biochim. Biophys. Acta 1858, 872–882 [DOI] [PubMed] [Google Scholar]
- 27. Posokhov Y. O., Gottlieb P. A., Morales M. J., Sachs F., and Ladokhin A. S. (2007) Is lipid bilayer binding a common property of inhibitor cysteine knot ion-channel blockers? Biophys. J. 93, L20–L22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cohen L., Gilles N., Karbat I., Ilan N., Gordon D., and Gurevitz M. (2006) Direct evidence that receptor site-4 of sodium channel gating modifiers is not dipped in the phospholipid bilayer of neuronal membranes. J. Biol. Chem. 281, 20673–20679 [DOI] [PubMed] [Google Scholar]
- 29. Schmalhofer W. A., Calhoun J., Burrows R., Bailey T., Kohler M. G., Weinglass A. B., Kaczorowski G. J., Garcia M. L., Koltzenburg M., and Priest B. T. (2008) ProTx-II, a selective inhibitor of NaV1.7 sodium channels, blocks action potential propagation in nociceptors. Mol. Pharmacol. 74, 1476–1484 [DOI] [PubMed] [Google Scholar]
- 30. Middleton R. E., Warren V. A., Kraus R. L., Hwang J. C., Liu C. J., Dai G., Brochu R. M., Kohler M. G., Gao Y.-D., Garsky V. M., Bogusky M. J., Mehl J. T., Cohen C. J., and Smith M. M. (2002) Two tarantula peptides inhibit activation of multiple sodium channels. Biochemistry 41, 14734–14747 [DOI] [PubMed] [Google Scholar]
- 31. Pallaghy P. K., Nielsen K. J., Craik D. J., and Norton R. S. (1994) A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 3, 1833–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Meer G., Voelker D. R., and Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Almeida R. F., Fedorov A., and Prieto M. (2003) Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys. J. 85, 2406–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Daleke D. L. (2008) Regulation of phospholipid asymmetry in the erythrocyte membrane. Curr. Opin. Hematol. 15, 191–195 [DOI] [PubMed] [Google Scholar]
- 35. Pokorny A., Yandek L. E., Elegbede A. I., Hinderliter A., and Almeida P. F. (2006) Temperature and composition dependence of the interaction of delta-lysin with ternary mixtures of sphingomyelin/cholesterol/POPC. Biophys. J. 91, 2184–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bae C., Anselmi C., Kalia J., Jara-Oseguera A., Schwieters C. D., Krepkiy D., Won Lee C., Kim E.-H., Kim J. I., Faraldo-Gómez J. D., and Swartz K. J. (2016) Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin. Elife 5, e11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta K., Zamanian M., Bae C., Milescu M., Krepkiy D., Tilley D. C., Sack J. T., Yarov-Yarovoy V., Kim J. I., and Swartz K. J. (2015) Tarantula toxins use common surfaces for interacting with Kv and ASIC ion channels. Elife 4, e06774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stock R. P., Brewer J., Wagner K., Ramos-Cerrillo B., Duelund L., Jernshøj K. D., Olsen L. F., and Bagatolli L. A. (2012) Sphingomyelinase D activity in model membranes: structural effects of in situ generation of ceramide-1-phosphate. PLoS ONE 7, e36003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith J. J., Cummins T. R., Alphy S., and Blumenthal K. M. (2007) Molecular interactions of the gating modifier toxin ProTx-II with NaV 1.5: implied existence of a novel toxin binding site coupled to activation. J. Biol. Chem. 282, 12687–12697 [DOI] [PubMed] [Google Scholar]
- 40. Park J. H., Carlin K. P., Wu G., Ilyin V. I., Musza L. L., Blake P. R., and Kyle D. J. (2014) Studies examining the relationship between the chemical structure of protoxin II and its activity on voltage gated sodium channels. J. Med. Chem. 57, 6623–6631 [DOI] [PubMed] [Google Scholar]
- 41. White S. H., and Wimley W. C. (1998) Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta 1376, 339–352 [DOI] [PubMed] [Google Scholar]
- 42. Torcato I. M., Huang Y.-H., Franquelim H. G., Gaspar D., Craik D. J., Castanho M. A., and Troeira Henriques S. (2013) Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim. Biophys. Acta 1828, 944–955 [DOI] [PubMed] [Google Scholar]
- 43. Henriques S. T., Pattenden L. K., Aguilar M.-I., and Castanho M. A. (2009) The toxicity of prion protein fragment PrP(106–126) is not mediated by membrane permeabilization as shown by a M112W substitution. Biochemistry 48, 4198–4208 [DOI] [PubMed] [Google Scholar]
- 44. Henriques S. T., Costa J., and Castanho M. A. (2005) Re-evaluating the role of strongly charged sequences in amphipathic cell-penetrating peptides: a fluorescence study using Pep-1. FEBS Lett. 579, 4498–4502 [DOI] [PubMed] [Google Scholar]
- 45. Prevette L. E., Benish N. C., Schoenecker A. R., and Braden K. J. (2015) Cell-penetrating compounds preferentially bind glycosaminoglycans over plasma membrane lipids in a charge density- and stereochemistry-dependent manner. Biophys. Chem. 207, 40–50 [DOI] [PubMed] [Google Scholar]
- 46. Amand H. L., Rydberg H. A., Fornander L. H., Lincoln P., Nordén B., and Esbjörner E. K. (2012) Cell surface binding and uptake of arginine- and lysine-rich penetratin peptides in absence and presence of proteoglycans. Biochim. Biophys. Acta 1818, 2669–2678 [DOI] [PubMed] [Google Scholar]
- 47. Rothbard J. B., Jessop T. C., Lewis R. S., Murray B. A., and Wender P. A. (2004) Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 126, 9506–9507 [DOI] [PubMed] [Google Scholar]
- 48. Mishra A., Lai G. H., Schmidt N. W., Sun V. Z., Rodriguez A. R., Tong R., Tang L., Cheng J., Deming T. J., Kamei D. T., and Wong G. C. (2011) Translocation of HIV TAT peptide and analogues induced by multiplexed membrane and cytoskeletal interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rothbard J. B., Jessop T. C., and Wender P. A. (2005) Adaptive translocation: the role of hydrogen bonding and membrane potential in the uptake of guanidinium-rich transporters into cells. Adv. Drug Deliv. Rev. 57, 495–504 [DOI] [PubMed] [Google Scholar]
- 50. Yang S.-T., Shin S. Y., Lee C. W., Kim Y.-C., Hahm K.-S., and Kim J. I. (2003) Selective cytotoxicity following Arg-to-Lys substitution in tritrpticin adopting a unique amphipathic turn structure. FEBS Lett. 540, 229–233 [DOI] [PubMed] [Google Scholar]
- 51. Gallivan J. P., and Dougherty D. A. (1999) Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. U.S.A. 96, 9459–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang M., Waring A. J., and Hong M. (2007) Phosphate-mediated arginine insertion into lipid membranes and pore formation by a cationic membrane peptide from solid-state NMR. J. Am. Chem. Soc. 129, 11438–11446 [DOI] [PubMed] [Google Scholar]
- 53. Zager R. A., Burkhart K. M., and Johnson A. (2000) Sphingomyelinase and membrane sphingomyelin content: determinants of proximal tubule cell susceptibility to injury. J. Am. Soc. Nephrol. 11, 894–902 [DOI] [PubMed] [Google Scholar]
- 54. Priest B. T., Blumenthal K. M., Smith J. J., Warren V. A., and Smith M. M. (2007) ProTx-I and ProTx-II: gating modifiers of voltage-gated sodium channels. Toxicon 49, 194–201 [DOI] [PubMed] [Google Scholar]
- 55. Pan M., He Y., Wen M., Wu F., Sun D., Li S., Zhang L., Li Y., and Tian C. (2014) One-pot hydrazide-based native chemical ligation for efficient chemical synthesis and structure determination of toxin Mambalgin-1. Chem. Commun. 50, 5837–5839 [DOI] [PubMed] [Google Scholar]
- 56. Conibear A. C., Rosengren K. J., Harvey P. J., and Craik D. J. (2012) Structural characterization of the cyclic cystine ladder motif of θ-defensins. Biochemistry 51, 9718–9726 [DOI] [PubMed] [Google Scholar]
- 57. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 58. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 59. Henriques S. T., Pattenden L. K., Aguilar M.-I., and Castanho M. A. (2008) PrP(106–126) does not interact with membranes under physiological conditions. Biophys. J. 95, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Henriques S. T., Huang Y.-H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., and Craik D. J. (2011) Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J. Biol. Chem. 286, 24231–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang Y.-H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., and Craik D. J. (2009) The biological activity of the prototypic cyclotide kalata B1 is modulated by the formation of multimeric pores. J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Henriques S. T., and Castanho M. A. (2005) Environmental factors that enhance the action of the cell penetrating peptide pep-1 A spectroscopic study using lipidic vesicles. Biochim. Biophys. Acta 1669, 75–86 [DOI] [PubMed] [Google Scholar]
- 63. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., and Berendsen H. J. (2005) GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 64. Schmid N., Eichenberger A. P., Choutko A., Riniker S., Winger M., Mark A. E., and van Gunsteren W. F. (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 40, 843–856 [DOI] [PubMed] [Google Scholar]
- 65. Poger D., and Mark A. E. (2012) Lipid bilayers: The effect of force field on ordering and dynamics. J. Chem. Theory Comput. 8, 4807–4817 [DOI] [PubMed] [Google Scholar]
- 66. Poger D., Van Gunsteren W. F., and Mark A. E. (2010) A new force field for simulating phosphatidylcholine bilayers. J. Comput. Chem. 31, 1117–1125 [DOI] [PubMed] [Google Scholar]
- 67. Koziara K. B., Stroet M., Malde A. K., and Mark A. E. (2014) Testing and validation of the Automated Topology Builder (ATB) version 2.0: prediction of hydration free enthalpies. J. Comput. Aided Mol. Des. 28, 221–233 [DOI] [PubMed] [Google Scholar]
- 68. Malde A. K., Zuo L., Breeze M., Stroet M., Poger D., Nair P. C., Oostenbrink C., and Mark A. E. (2011) An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 7, 4026–4037 [DOI] [PubMed] [Google Scholar]
- 69. Berendsen H. J., Postma J. P., van Gunsteren W. F., and Hermans J. (1981) in Intermolecular Forces (Pullman B., ed) pp. 331–342, Reidel Publishing Co., Dordrecht, Netherlands [Google Scholar]
- 70. Michaud-Agrawal N., Denning E. J., Woolf T. B., and Beckstein O. (2011) MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vetter I., Mozar C. A., Durek T., Wingerd J. S., Alewood P. F., Christie M. J., and Lewis R. J. (2012) Characterisation of Nav types endogenously expressed in human SH-SY5Y neuroblastoma cells. Biochem. Pharmacol. 83, 1562–1571 [DOI] [PubMed] [Google Scholar]
- 72. Henriques S. T., Huang Y.-H., Chaousis S., Sani M.-A., Poth A. G., Separovic F., and Craik D. J. (2015) The prototypic cyclotide Kalata B1 has a unique mechanism of entering cells. Chem. Biol. 22, 1087–1097 [DOI] [PubMed] [Google Scholar]
- 73. Maier O., Oberle V., and Hoekstra D. (2002) Fluorescent lipid probes: some properties and applications (a review). Chem. Phys. Lipids. 116, 3–18 [DOI] [PubMed] [Google Scholar]
- 74. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd., Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]