Abstract
Ubiquitin-specific proteases (USPs) USP15 and USP4 belong to a subset of USPs featuring an N-terminal tandem domain in USP (DUSP) and ubiquitin-like (UBL) domain. Squamous cell carcinoma antigen recognized by T-cell 3 (SART3), a spliceosome recycling factor, binds to the DUSP-UBL domain of USP15 and USP4, recruiting them to the nucleus from the cytosol to control deubiquitination of histone H2B and spliceosomal proteins, respectively. To provide structural insight, we solved crystal structures of SART3 in the apo-form and in complex with the DUSP-UBL domain of USP15 at 2.0 and 3.0 Å, respectively. Structural analysis reveals SART3 contains 12 half-a-tetratricopeptide (HAT) repeats, organized into two subdomains, HAT-N and HAT-C. SART3 dimerizes through the concave surface of HAT-C, whereas the HAT-C convex surface binds USP15 in a novel bipartite mode. Isothermal titration calorimetry measurements and mutagenesis analysis confirmed key residues of USP15 involved in the interaction and indicated USP15 binds 20-fold stronger than USP4.
Keywords: crystal structure, dimerization, protein-protein interaction, site-directed mutagenesis, ubiquitin-dependent protease, deubiquitinase, half-a-tetratricopeptide repeats
Introduction
Ubiquitination plays an important role in almost every biological process, including protein homeostasis, DNA damage response, gene transcription, protein trafficking, and RNA splicing. Deubiquitinases (DUBs)2 remove covalently conjugated ubiquitin tags from substrates and regulate ubiquitin signaling. Ubiquitin-specific proteases (USPs) comprise the largest family of DUBs. In addition to the catalytic domain, most USPs contain auxiliary domains that are involved in substrate recognition, activity regulation, or recruitment of binding partners (1, 2). USP4, USP11, and USP15 are a small set of closely related USPs, sharing a similar domain architecture as follows: a DUSP (domain in USP), followed by a ubiquitin-like (UBL) and a large catalytic domain, bifurcated by a second UBL domain and disordered region (Fig. 1A). Evolutionary analysis suggests the three USPs arose from gene duplication events (3). USP4 and USP15 are more closely related in both sequence and function compared with USP11. Both USP4 and USP15 are implicated in mRNA processing through their interaction with spliceosome components (1). USP4 is recruited by U4/U6 small nuclear RNA recycling factor SART3 (squamous cell carcinoma antigen recognized by T cells 3) to remove the Lys-63-polyubiquitin chain from pre-mRNA processing factor 3 (Prp3), and it controls the assembly of the spliceosome at distinct stages of the splicing process (4). USP15, but not USP4, is recruited by SART3 to regulate deubiquitination of free ubiquitinated histone H2B that has been evicted from the nucleosome during transcription (5).
FIGURE 1.
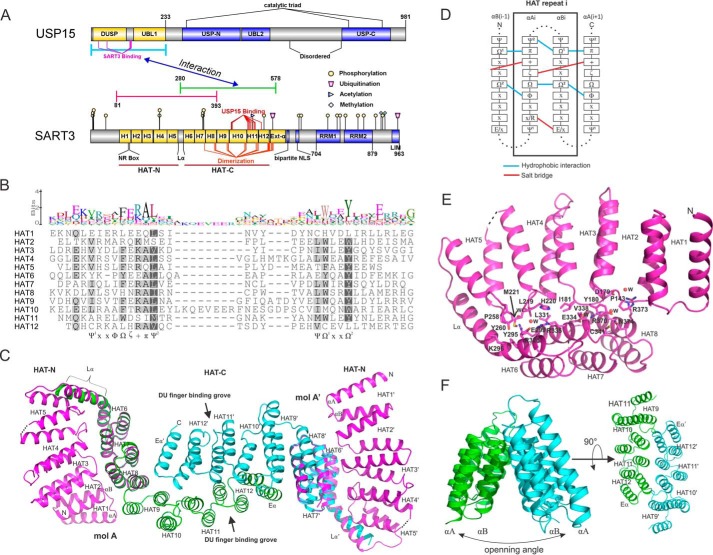
Structure and sequence analysis of SART3. A, domain architecture of USP15 and SART3, prepared using IBS software (38). The flat-ended blue, green, and magenta lines indicate the constructs used for crystallization. Post-translational modification data were retrieved from the PhosphoSite (39) database. Sites involved in binding and dimerization were obtained from structural analysis. B, sequence alignment of the 12 HAT repeats of SART3. Structure-based sequence alignment was carried out using Promals3D (40) and visualized using Geneious software (41). C, overview of SART3 HAT domain by overlapping the SART3(81–393) and SART3(280–578)-USP15 complex structures. USP15 structure was omitted for clarity. D, sequence and interaction pattern of the SART3 HAT repeats. Symbols adopt a recommended nomenclature (25). E, interaction details of the HAT-N subdomain with repeats 6–8 from HAT-C. Water molecules are shown as spheres and labeled as W. F, triangular prism shape formed by repeats 9–12. The opening angle is different in the CstF and RNA14 dimer structures.
SART3 is the mammalian homolog of yeast Prp24 protein and is essential for the formation of U4/U6 small nuclear ribonucleoprotein complex (6). SART3 plays multiple roles in mRNA splicing, viral and host gene transcription, as well as stem cell survival, proliferation, and differentiation. It is also a potential antigen for cancer immunotherapy (7–9). The biological functions of SART3 have been summarized in a recent comprehensive review (10). SART3 is a large protein of 110 kDa and contains multiple half-a-tetratricopeptide (HAT) repeats followed by a bipartite nuclear localization sequence (NLS), two RNA recognition motifs, and a C-terminal conserved LSm-interacting motif (Fig. 1A).
The HAT repeat is a repetitive sequence pattern identified in proteins involved in RNA processing (11). Sequentially and structurally similar to the tetratricopeptide repeats, it features two anti-parallel α-helices containing aromatic residues with conserved spacing. HAT repeats are involved in protein-protein (12) and protein-RNA interactions (13). The central HAT repeat region of SART3 mediates direct interactions with the DUSP-UBL domains of USP4 (4) and USP15 (5) but not of USP11. The USP15 DUSP-UBL domain has 69% sequence identity with that of USP4 and 39% with USP11. Structural details of the SART3 HAT domain and how SART3 interacts with the DUSP-UBL domain is not known. Sequence homology of SART3 to other HAT repeat-containing proteins with known structures is too low (<20%) to permit meaningful structural modeling.
Here, we report the crystal structure of the first eight HAT repeats of SART3 and SART3 in complex with the DUSP-UBL domain of USP15. Structure-based sequence analysis reveals SART3 contains 12 HAT repeats that can be divided into subdomains HAT-N and HAT-C. The HAT domain dimerizes through the concave surface of HAT-C, whereas the convex surface of HAT-C binds USP15. Binding affinity measurements and mutagenesis analysis confirmed the interaction between USP15 and SART3 is bivalent and revealed that USP15 binds to SART3 20-fold stronger than USP4.
Experimental Procedures
Cloning, Protein Expression, and Purification
DNAs encoding SART3(81–393), SART3(280–578), SART3(81–578), and DUSP-UBL domains (USP15(1–223), USP4(1–229), and USP11(78–286)) and DUSP domains (USP15(1–133), USP4(1–137), and USP11(80–196)) were subcloned into a modified pET28 vector encoding either a tobacco etch virus (TEV) protease (GenBankTM EF456735) or a thrombin-cleavable (GenBankTM accession number EF442785) N-terminal His6 tag. For co-expression of USP15 and SART3, a bicistronic pET28 vector was constructed by inserting a ribosomal binding site (5′-CTCGACGGAGGAATAATCAT-3′) between the genes encoding USP15(1–223) and SART3(280–578). All subclonings were completed using ligation-independent InFusionTM cloning kit (Clontech). Site-directed mutagenesis was carried out using the PrimeSTAR HS DNA polymerase (Takara) and was verified by DNA sequencing. For selenomethionine labeling, a pre-packed M9 selenomethionine growth media kit (Medicilon) was used as per the manufacturer's instructions. All proteins were overexpressed in a BL21 (DE3) Escherichia coli strain harboring a plasmid encoding tRNAs for rare codons. Expression cultures were induced using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 18 °C. All proteins were purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen), and the His6 tag was removed by TEV or thrombin proteases correspondingly. Uncleaved proteins and TEV protease were removed by another pass with nickel-nitrilotriacetic acid resin. The proteins were further purified using anion-exchange and size-exclusion chromatography (Source15Q and Superdex 200, GE Healthcare). The final concentrations of purified proteins were 10–30 mg/ml as measured by UV absorbance at 280 nm.
Crystallization
All crystals were grown at 20 °C using the vaporization method. SART3(81–393) was crystallized in 30% PEG3350, 0.2 m NaCl, 0.1 m HEPES, pH 7.5. SART3(81–578) was crystallized in 20% PEG3350, 0.2 m magnesium formate. For crystallization of the SART3(280–578)-USP15(1–223) complex, 2 μl of protein solution was mixed with 1 μl of well solution containing 19% PEG3350, 0.1 m HEPES, pH 7.5, 0.2 m MgCl2, 5% ethylene glycol, and 1 μl solution containing 0.33% (w/v) each of disodium 2,6-naphthalene-disulfonate, 2-aminobenzenesulfonic acid, disodium m-benzenedisulfonate in 20 mm HEPES, pH 6.0. Crystals were cryo-protected, when required, by paratone-N (Hampton Research).
Data Collection, Structure Determination, and Refinement
X-ray diffraction data for SART3(81–393) were collected at 100 K at the Canadian Macromolecular Crystallography Facility beamline 08ID-1 of Canadian Light Source (14). Data for SART3(81–578) and the SART3(280–578)-USP15(1–223) complex were collected at beamline 19ID at the Advanced Photon Source, Argonne National Laboratory. All datasets were collected at the selenium absorption edge and processed with the HKL-3000 suite (15). The structures were solved by single wavelength anomalous dispersion (SAD) method (16) using SOLVE/RESOLVE (17). A monomeric USP15 model (18), PDB code 3T9L, was used to build the USP15 molecule in the complex. COOT (19) was used for model building and visualization and REFMAC (20) for restrained refinement. The final models were validated by MOLPROBITY (21).
Isothermal Titration Calorimetry (ITC)
Proteins were diluted with a buffer containing 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine. All ITC measurements were performed at 25 °C either on a VP-ITC Micro Calorimeter (Malvern) or a Nano ITC (TA Instruments). For VP-ITC, a total of 26 injections, each of 10 μl of protein except for the first injection with 5 μl of protein, were delivered into a 1.4 ml sample cell containing the other protein. For Nano ITC measurement, a total of 25 injections, each of 2 μl, were delivered into a 0.167 ml sample cell at a 180 s internal. The data were analyzed using OriginTM for ITC and NanoAnalyze softwares on the instruments, respectively, and fitted to a one-site binding model.
Size Exclusion Chromatography (SEC)
For SEC analysis, 250 μl of each sample was loaded onto a Superdex 200 10/300 column (GE Healthcare) pre-equilibrated with a buffer containing 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mm dithiothreitol (DTT). The column was calibrated using gel filtration standard (Bio-Rad).
SEC-MALS
The absolute molar masses and mass distributions of the wild-type USP4(1–229) at different concentrations and the F42D mutant were determined using SEC-MALS. Samples were injected through a Superose 6 10/300 GL column (GE Healthcare) equilibrated in 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm DTT followed in line by a Dawn Heleos-II light scattering detector and a 2414 refractive index monitor (Waters). Molecular mass calculations were performed using ASTRA version 6.1.1.17 (Wyatt Technologies) assuming a refractive index increment, dn/dc, value of 0.185 ml·g−1.
Results
SART3 Is a Dimer Containing 12 HAT Repeats
We solved the crystal structures of an N-terminal fragment of SART3 (amino acids 81–393) and a central fragment of SART3 (amino acids 280–578) in complex with the USP15 DUSP-UBL domain at 2.0 and 3.0 Å, respectively, using the SAD method (Fig. 1A and Table 1). The overlapping region (amino acids 280–393) of the two SART3 constructs superimpose well with a root mean square deviation of 0.56 Å for 114 Cα atoms. A crystal for the SART3 construct containing all the HAT repeats (amino acids 81–578), in the P22121 space group diffracted to 3.4 Å. Molecular replacement using a concatenated model containing SART3(81–393) and SART3(280–578) gave a solution that fitted well into the difference density, indicating no significant change in the conformation of SART3 in the three different crystal forms. Because of the suboptimal resolution of the SART3(81–578) dataset, we hereinafter use the concatenated model for the analysis of the SART3 HAT repeats.
TABLE 1.
Data collection and refinement statistics
| SART3(81–393) | SART3(280–578) + USP15(1–223) | |
|---|---|---|
| PDB code | 5JJX | 5JJW |
| Data collection | ||
| Space group | P63 | P6422 |
| Cell dimensions | ||
| a, b, c (Å) | 135.1, 135.1, 43.2 | 117.9, 117.9, 205.3 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.0–2.0 | 50.0–3.0 |
| Rsym or Rmerge | 0.067 (0.968)a | 0.197 (2.59) |
| I/σI | 43.5 (2.3) | 24.6 (1.3) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 8.3 (7.8) | 30.9 (31.6) |
| Refinement | ||
| Resolution (Å) | 50.0–2.0 | 44.7–3.0 |
| No. of reflections | 29,831 | 17,386 |
| Rwork/Rfree | 0.207/0.239 | 0.234/0.288 |
| No. of atoms | 2537 | 3325 |
| Protein | 2415 | 3310 |
| Ligand/ion | 1 | 4 |
| Water | 121 | 11 |
| B-factors | ||
| Protein | 47.7 | 91.9 |
| Ligand/ion | 51.1 | 62.3 |
| Water | 45.3 | 63.2 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.009 | 0.010 |
| Bond angles (°) | 1.213 | 0.900 |
| Ramachandran plot | ||
| Favored regions (%) | 98.3 | 95.4 |
| Allowed regions (%) | 1.7 | 4.6 |
| Disallowed regions (%) | 0.0 | 0.0 |
a Values in parentheses are for the highest resolution shell.
Previous structure and sequence analysis (11, 13, 22) suggest each HAT motif contains two anti-parallel helices (αA and αB). HAT repeats form a contiguous structure so the assignment of pairing helices in each repeat is somewhat arbitrary (22). The composite model reveals SART3(81–578) contains 12 HAT repeats (Fig. 1, A–D). The entire HAT repeat domain can be divided into two subdomains as follows: HAT-N-containing repeats 1–5 and HAT-C-containing repeats 6–12. A linker helix, Lα, connects HAT-N and HAT-C. An additional C-terminal helix, Eα, links the HAT domain to a bipartite NLS peptide (10, 23, 24). We assigned the helices in the convex surface of the subdomains as αAs, and those in the concave surface as αBs, as this gives the best alignment of the sequence motifs (Fig. 1B). The alignment indicates that HAT1 and HAT12 are less conserved than other repeats. The αA and αB of repeats HAT2 to HAT11 contain conserved motifs (Fig. 1B) of aliphatic (Ψ), hydrophobic (Φ), aromatic (Ω), hydrophilic (ζ), positively charged (+), and small aliphatic residues (π) (25). The Ω1 residue (usually Trp) of the αBi helix interacts with the aliphatic residues Ψ2 and π of the αAi and αAi + 1 helices, respectively, whereas the Ω2 residue (usually Tyr) of the αBi helix forms a hydrophobic core with the Ω and Φ residues of the αAi and αAi + 1 helices (Fig. 1D). A salt bridge is formed between the ζ and + residues of the αAi and αAi + 1 helices. Additionally, a salt bridge between an Arg and a Glu within the same repeat is seen for 5 out of the 10 conforming repeats.
Repeats 1–8 of the SART3 HAT domain form a compact moiety that folds independently and exists as a monomer in the solution, as confirmed by SEC analysis (data not shown). The pleated arc shape composed of repeats 2–5 from the HAT-N domain forms a tight interaction with the convex surface of repeats 6–8 from the HAT-C domain through polar and hydrophobic interactions and has a buried surface area of 674 Å2 excluding the Lα helix (Fig. 1, C and E).
The HAT-C subdomain exists as a dimer in solution (Fig. 2A). In the crystal structure, the two SART3 molecules dimerize through a C2 axis centered along the HAT11 repeat. Extensive interactions, mainly salt bridges between charged residues, were observed between the αB helices of repeats 7–12 and helix Eα of the two monomers. Repeats 9–12 of the two molecules form a triangular prism-like architecture (Fig. 1F), and the ridge is maintained by interaction of residues from the loops linking the αA and αB of each repeat from the two monomers. The total buried surface area of each monomer is 1155 Å2.
FIGURE 2.
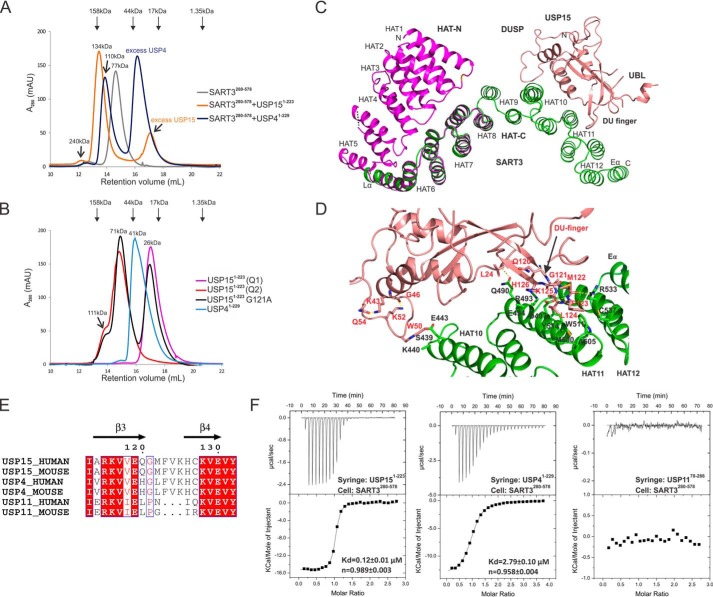
Interaction of DUSP-UBL domains with SART3. A, SEC analysis of SART3 HAT-C domain in complex with USP4 or USP15. SART3 HAT-C forms a dimer in the solution, and the apparent size of the SART3-USP15 complex matches a heterotetramer. The size of the SART3-USP4 complex is smaller due to weaker binding affinity. B, SEC analysis of USP4 and USP15. The two species of USP15 from anion-exchange Q column, Q1 and Q2, elute as monomer (pink) and dimer (red) on SEC, respectively. Only a single peak is observed for USP4 (blue). Mutation of G121A in the DU finger of USP15 leads to an equilibrium of monomer and oligomer in solution. A shoulder peak at the size of 111 kDa, corresponding to a tetramer, can be observed for the USP15 Q2 species and USP15 G121A mutant. The size of the USP15 dimers (71 kDa) is slightly larger than the calculated value (52 kDa). This could probably be caused by the elongated conformation of a domain-swapped dimer. C, interaction mode of USP15 with SART3, only one copy of SART3 and USP15 molecules are shown. D, atomic details of the interaction between USP15 and SART3. Residues Lys-43, Gly-46, Lys-52, and Gln-54 of USP15 are not involved in the direct interaction with SART3, but the hydrogen bonds they form fix the conformation of the loop and Trp-50 to a favorable position for SART3 interaction. E, sequence alignment of the DU finger of human and mouse USP4, USP11, and USP15. F, thermodiagram of ITC measurement of wild-type DUSP-UBL domains with SART3 and data fitting.
Structure of the USP15-SART3 Complex
When purifying USP15 alone, monomeric and oligomeric species eluted distinctly during anion exchange, suggesting a propensity to self-associate. SEC analysis suggests the monomeric and oligomeric forms do not convert easily in the solution (Fig. 2B). The SART3-USP15 complex used for crystallization was co-expressed using a bicistronic vector and eluted as a heterotetramer on SEC, whereas no oligomeric forms of USP15 were observed during purification (Fig. 2A). The asymmetric unit of the SART3-USP15 complex contains one molecule of each protein. The second pair of the complex can be generated by a symmetry operation. Although most of the USP15 DUSP structure is well defined in the electron density map, only 21 of 98 amino acids can be modeled for the UBL domain (Fig. 2C).
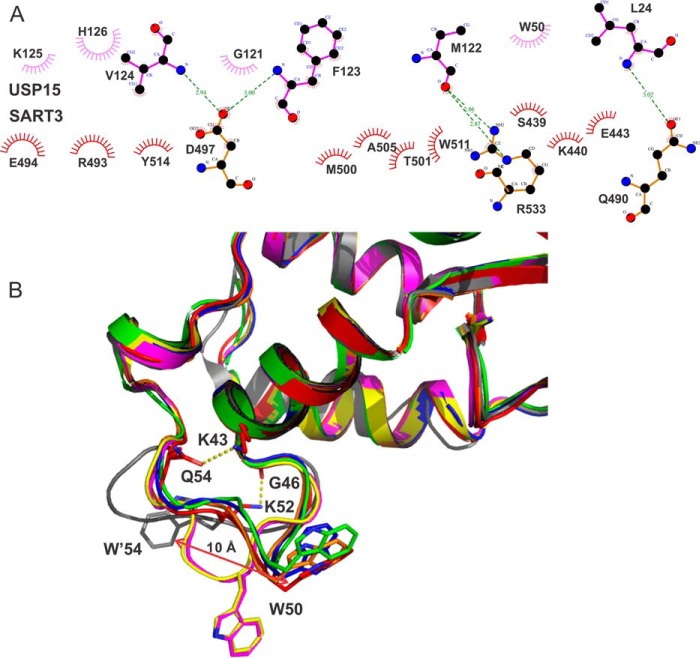
USP15 binds to the convex surface of the SART3 HAT-C domain in a bivalent mode. The DUSP-UBL domain linker forms a DU finger (18) that occupies a hydrophobic patch formed by hydrophobic and aromatic residues from helices αA11, αB11, and αA12, while the indole ring of Trp-50 forms hydrophobic interactions with the aliphatic atoms from the side chains of Ser-439, Glu-443, and Lys-440 on the linker between αB9 and αA10 of SART3 (Figs. 2D and 3A). The DU finger and the SART3 interaction also contain strong hydrogen bonds between the main chain amide group of USP15 residues Phe-123 and Val-124 with the side chain of SART3 residue Asp-497 (Fig. 2D). Overall, USP15 has a buried surface area of 568 Å2, relatively small compared with the average 800 Å2 for “standard size” protein complexes (26).
FIGURE 3.
Interaction details of USP15 with SART3. A, schematic representation of the interactions between SART3 and USP15 using LigPlot+ (42). B, comparison of the conformation of the loop containing SART3-interacting residue Trp-50 from all known structures of USP15: red (PDB code 5JJW, USP15/SART3); green (PDB code 3PV1); blue (PDB code 4A3P); yellow (PDB code 4A3O); orange (PDB code 3PPA); and magenta (PDB code 3T9L). The loop of USP4 (PDB code 3JYU) is shown in gray.
Contributing Factors to USP15/SART3 Interaction
USP15, USP4, and USP11 all contain an N-terminal DUSP-UBL domain. We measured the binding affinity of different DUSP-UBL domains with SART3 using ITC. USP11 does not bind to SART3 at all, whereas the affinity of USP4 is 20-fold weaker than that of USP15 (Table 2 and Fig. 2, E and F). Sequence alignment revealed USP11 is lacking three key residues in the DU finger critical for the interaction with SART3 (Fig. 2E), although these are conserved between USP4 and USP15.
TABLE 2.
Summary of ITC measurements of USPs and SART3 interaction
| Syringe | Cell | n | KD | ΔH | Δ |
|---|---|---|---|---|---|
| μm | kcal·mol−1 | cal·mol−1degrees−1 | |||
| USP15(1–223) | SART3 (81–578) | 0.952 ± 0.002 | 0.14 ± 0.01 | −15.03 ± 0.05 | −19.0 |
| USP4(1–229) | SART3 (81–578) | 0.808 ± 0.002 | 2.07 ± 0.04 | −16.39 ± 0.06 | −29.0 |
| USP15(1–223) | SART3 (280–578) | 0.989 ± 0.003 | 0.12 ± 0.01 | −15.24 ± 0.08 | −19.5 |
| USP4(1–229) | SART3 (280–578) | 0.958 ± 0.004 | 2.79 ± 0.10 | −13.32 ± 0.08 | −19.2 |
| SART3(280–578) | USP4 (1–229) | 0.979 ± 0.027 | 2.48 ± 0.25 | −18.41 ± 0.68 | −36.1 |
| SART3(280–578)a | USP4 (1–229) | 0.960 ± 0.075 | 2.97 ± 0.98 | −16.14 ± 1.86 | −28.9 |
| USP11(78–286) | SART3 (280–578) | NDb | ND | ||
| USP15(1–223) (Q2)c | SART3 (280–578) | 1.800 ± 0.112 | 28.01 ± 6.18 | 1.95 ± 0.21 | 27.4 |
| USP15(1–133) | SART3 (280–578) | 0.933 ± 0.010 | 1.42 ± 0.14 | −7.87 ± 0.12 | 0.4 |
| USP4(1–137) | SART3 (280–578) | 0.881 ± 0.006 | 2.40 ± 0.11 | −15.49 ± 0.14 | −26.2 |
| USP11(80–196) | SART3 (280–578) | ND | ND | ||
| USP15(1–223) L24A | SART3 (280–578) | 1.180 ± 0.005 | 0.33 ± 0.03 | −10.10 ± 0.07 | −4.2 |
| USP15(1–223) W50A | SART3 (280–578) | 0.870 ± 0.019 | 1.68 ± 0.29 | −9.36 ± 0.28 | −5.0 |
| USP15(1–223) G121A | SART3 (280–578) | 0.887 ± 0.173 | 16.34 ± 5.53 | −3.72 ± 0.85 | 10.6 |
| USP15(1–223) M122A | SART3 (280–578) | 0.868 ± 0.003 | 0.32 ± 0.02 | −13.98 ± 0.09 | −17.1 |
| USP15(1–223) M122K | SART3 (280–578) | 0.888 ± 0.001 | 0.25 ± 0.01 | −15.85 ± 0.03 | −22.9 |
| USP15(1–223) F123A | SART3 (280–578) | ND | ND | ||
| USP15(1–223) V124A | SART3 (280–578) | 0.821 ± 0.035 | 11.49 ± 1.03 | −12.74 ± 0.73 | −20.1 |
| USP15(1–223) K125A | SART3 (280–578) | 0.962 ± 0.003 | 0.07 ± 0.01 | −14.77 ± 0.09 | −16.8 |
| USP15(1–223) H126A | SART3 (280–578) | 1.030 ± 0.087 | 19.49 ± 3.21 | −5.83 ± 0.66 | 2.0 |
| SART3(280–578)a | USP4 (1–229) F42D | 0.855 ± 0.217 | 9.35 ± 3.76 | −12.14 ± 4.31 | −17.7 |
| USP4(1–229) L126K | SART3 (280–578) | 0.908 ± 0.006 | 2.87 ± 0.10 | −13.41 ± 0.11 | −19.6 |
a Measurement was performed on Nano ITC (TA Instruments).
b ND means not detected.
c Q2 refers to the second peak from anion-exchange chromatography.
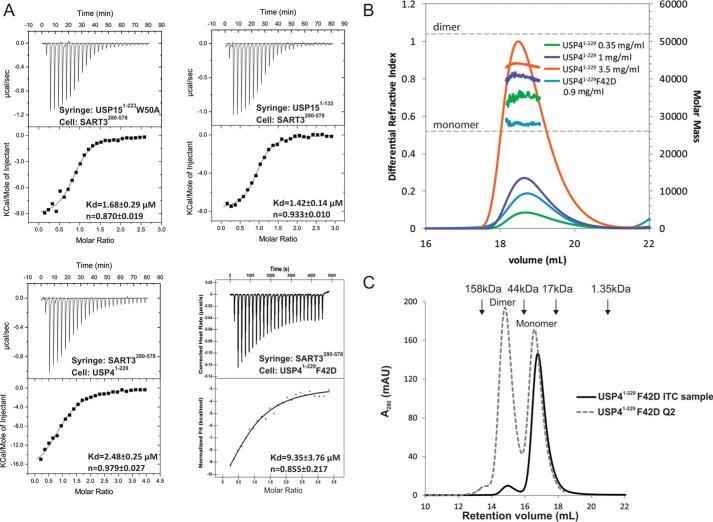
To confirm the authenticity of the interactions, we did mutagenesis based on structural analysis and measured the binding affinities. Two kinetically stable oligomeric states of USP15 that do not interconvert at room temperature or 4 °C can be separated on an ion-exchange column, corresponding to a monomeric form and an oligomeric (dimer and tetramer) form (Fig. 2B). The monomeric form of the USP15 protein was used for ITC measurement unless otherwise indicated. Wild-type USP15 binds to the HAT-C subdomain of SART3 with a dissociation constant (KD) of 0.12 μm. The HAT-N subdomain of SART3 does not interfere with the USP15 and SART3 interaction as SART3(81–578) has a similar KD value of 0.14 μm, so it is with the interaction between USP4 and SART3. Further measurement was thus carried out using the HAT-C subdomain of SART3. Mutation of residues on the USP15 DU finger, G121A, V124A, H126A, reduced the binding affinity by around 100-fold, whereas F123A abolished binding completely, suggesting the benzyl group of Phe-123 contributes most to the interaction (Table 2). Although the side chain of USP15 residue Leu-24 is within 4 Å distance from SART3 (Fig. 3A), the contribution of Leu-24 to the interaction is minimal, if any, as demonstrated by the similar binding affinity of the L24A mutant with SART3. The contribution of the USP15 Trp-50 residue to the interaction was confirmed by a 14-fold change of KD upon mutation to an alanine (Fig. 4A and Table 2).
FIGURE 4.
Identifying key contributors to tight USP15/SART3 binding. A, ITC measurement and data fitting for mutant USPs and SART3. In the lower panels, the USP4 proteins were placed in the sample cell at a lower concentration so that the monomeric form dominates. B, SEC-MALS analysis of the oligomeric states of wild-type USP4 and the F42D mutant. C, SEC shows the F42D mutant of USP4(1–229) used for ITC exists dominantly as a monomer in solution, whereas the Q2 species of the F42D mutant exists as both dimer and monomer.
Although Met-122 in USP15 is the only residue involved in the direct interaction with SART3 that is different from USP4, the side chain of Met-122 is mostly exposed, and only the main chain interacts with SART3. M122A and M122K mutations do not affect the KD value significantly (Table 2). Similarly, mutation of the corresponding Leu-126 in USP4 to a lysine also does not change its binding affinity to SART3, suggesting the small sequence difference in the DU fingers of USP4 and USP15 is not the cause of the affinity difference.
The UBL domain of USP15 does not interact with SART3 directly. However, removal of the UBL domain (USP15(1–133)) reduced its binding affinity with SART3 by almost 10-fold (Table 2 and Fig. 4A), whereas removal of the UBL domain from USP4 (USP4(1–137)) does not change the binding affinity significantly. A further inspection revealed the binding enthalpy, ΔH, upon UBL removal is increased from −15.2 kcal·mol−1 for the USP15(1–223) to −7.9 kcal·mol−1 for USP15(1–133), while the binding entropy, ΔS, is almost diminished from −19.5 to 0.35 cal·mol−1·degrees−1. On the contrary, the binding enthalpy and entropy values do not vary greatly for USP4 in the presence or absence of the UBL domain (Table 2).
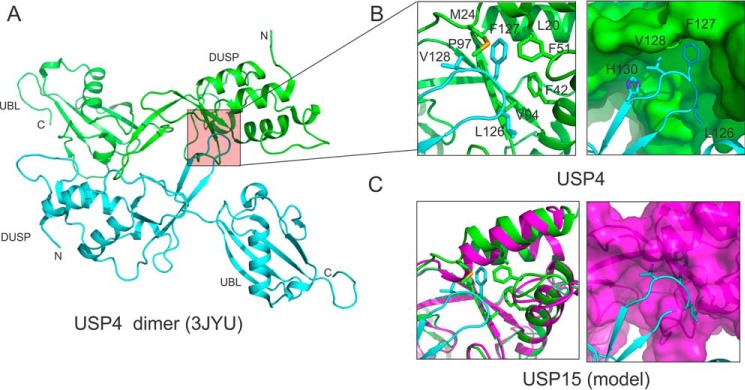
Previous studies (18, 27–29) established that the USP4 DUSP-UBL domain forms a dimer in solution and in the crystal. Detailed structural analysis revealed the DU finger of one USP4 molecule interacts with a hydrophobic patch on the DUSP domain of the other molecule (Fig. 5, A and B), forming a head-to-tail dimer. Such an interaction does not exist in USP15 due to the steric hindrance caused by an elongated nearby α-helix and protruding residues from the pocket (Fig. 5C). To understand whether the dimerization caused lower binding affinity of USP4, we first characterized the oligomeric status of USP4 DUSP-UBL domain in solution using SEC-MALS. The size of USP4 in solution is concentration-dependent, suggesting a fast monomer-dimer equilibrium (Fig. 4B). Monomeric USP4 cannot be separated from the dimeric form as in the case of USP15. We thus introduced a F42D mutation in the hydrophobic pocket of the DUSP domain that binds to the DU finger, reasoning that a negatively charged residue will disrupt the hydrophobic patch and disrupt the dimerization seen in the USP4 crystal structure. Interestingly, we observed two peaks during anion-exchange chromatography, a less charged Q1 species and a more charged Q2 species. We collected the Q1 fractions and subjected the solution to analytical SEC twice and found the final F42D mutant protein stayed dominantly as a stable monomer (Fig. 4C). The monomeric state of the F42D mutant is further confirmed by a consistent molecular mass measurement of ∼28 kDa across the elution profile as measured by SEC-MALS (Fig. 4B). This suggests the F42D mutant of USP4 behaves similarly to wild-type USP15.
FIGURE 5.
Interaction details of the DU finger with DUSP domain. A, USP4 forms a head-to-tail dimer through the interaction of the DU finger of one molecule with the DUSP domain of the other molecule. B, hydrophobic residues at the tip of the DU finger (cyan) dock into a hydrophobic pocket of the DUSP domain of a second USP4 molecule (green). C, DUSP domain of USP15 (magenta) has a very shallow pocket. When the DU finger of USP4 (cyan) is modeled onto the DUSP domain of USP15, the hydrophobic residues involved in interaction clash with the putative binding pocket of USP15.
Surprisingly, we found the KD value of the monomeric form of USP4 F42D mutant binding with SART3 to be 9.3 μm by ITC measurement, with only small changes of the binding enthalpy and entropy compared with the wild-type USP4 (Table 2). This suggests the self-association of the wild-type USP4 in the solution is not the cause of the lower binding affinity with SART3 compared with USP15. We have also ruled out the possibility of operational error by measuring the interaction of the wild-type USP4 on two different ITC instruments and by switching the samples in the syringe and the sample cell (Table 2). All measurements indicate a consistent KD of around 2.7 μm between the wild-type USP4(1–229) and SART3(280–578). The weaker binding of the wild-type USP4 with SART3 is further confirmed by the smaller apparent molecular mass of the USP4-SART3 complex at 110 kDa compared with the USP15-SART3 complex at 134 kDa on SEC (Fig. 2A).
Discussion
Comparison of SART3 with Other HAT Structures
Based on sequence pattern, eight HAT repeats were annotated for SART3 in the Uniprot database, and our structure-based analysis identified four additional repeats. It should be noted that the sequences of HAT1 and HAT12 are less conserved compared with other repeats. In our SART3 HAT-C structure, there is also an extra helix after HAT12, predicted to be part of a longer helix (about 50 amino acids) linking the HAT domain to a bipartite NLS peptide. We excluded it from the HAT repeats due to low sequence homology of this helix to other repeats in the HAT domain. Lys-568 of SART3 in this extended helix is ubiquitinated according to proteomics studies (30, 31), which suggested this helix may be involved in regulating the degradation of SART3 and is functionally distinct from other repeats.
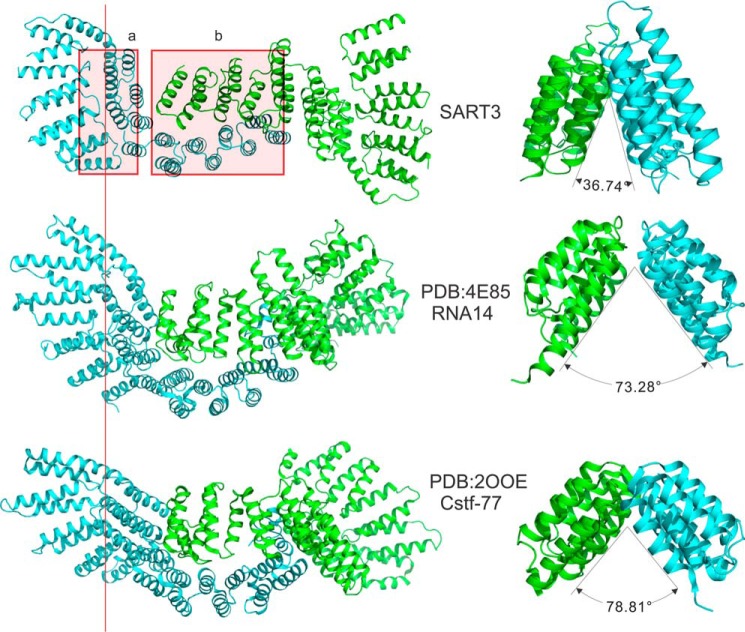
We searched the Dali server (32) for structural homologs of SART3. Top hits included mouse and fungi cleavage stimulation factor 77 (CstF-77 PDB codes 2OOE, 2OND, and 2UYL) and RNA14 (PDB codes 3EBA, 4EBH, and 4E85), which is a yeast homolog of CstF-77. Both proteins are part of the mRNA 3′-end processing machinery in different organisms. The sequence identity of these proteins to the HAT domain of SART3 is low at 16–20%. Structural analysis revealed that both CstF-77 (22) and RNA14 (33) contain 11–12 HAT repeats and also form two subdomains, HAT-N and HAT-C. CstF-77 and RNA14 both form homodimers through the concave surface of the HAT-C subdomain in a tail-to-tail mode similar to SART3, but they differ in the relative orientation of the HAT-N and HAT-C subdomains of each monomer (Figs. 1E and 6) and the opening angle between the two pleated arcs of the HAT-C subdomains of the dimer (Figs. 1F and 6).
FIGURE 6.
Comparison of different HAT structures. Two regions of the HAT domain quaternary structures are dissimilar among HAT proteins with known structural models, indicated with red rectangles in the SART3 structure. The first region (a) is the interface between the HAT-N subdomain and the HAT-C subdomain. The relative orientation of the two subdomains is divergent in different structures. The second region (b) is the main dimerization interface. The opening angle of the two monomers varies significantly.
Functional Implications of the DUSP-UBL and SART3 Interaction
USP15, USP11, and USP4 compose a small set of highly networked USPs that are evolutionarily related, structurally similar, and functionally overlapping (3). The differing abilities of these USPs to bind SART3 also have variable downstream functional consequences for each of these proteins. USP11 does not bind to SART3 due to the lack of key residues in the DU finger between the DUSP and UBL domains (Fig. 2E) and is transported to the nucleus through a different mechanism (34). SART3 binds to both USP4 and USP15 and shuttles the two USPs from cytosol to nucleus (4, 5). However, USP4/SART3 controls ubiquitination of the spliceosome proteins (4), whereas USP15 is the preferred DUB for free ubiquitinated histone H2B (5). Our ITC measurements also suggest USP15 binds to SART3 at least 20-fold stronger than USP4, suggesting other factors, such as post-translational modification, may be involved in the selective recruitment of USP4 to the spliceosome.
The buried surface area of the USP15/SART3 is relatively small in reference to the low sub-micromolar binding affinity observed. Our mutagenesis data confirmed Trp-50 of the USP15 DUSP domain contributes significantly to the binding with SART3. It should be noted that in more than half of the USP15 structures deposited in the PDB, the conformation of the Trp-50 is already primed for interaction with SART3, whereas the corresponding Trp-54 in USP4 is ∼10 Å away from such a position (Fig. 3B). A further inspection of the structures revealed the hydrogen bonds formed by the side chain of Lys-52 with the main chain of Gly-46, and the side chains of Gln-54 with Lys-43, which are adjacent to Trp-50, fix the loop in a conformation preferable for interaction with SART3 (Figs. 2D and 3B). Such interactions are not observed in the USP4 structure. Although the UBL domain of USP15 does not interact with SART3 directly, our deletion mutation data suggest the UBL domain contributes to the interaction significantly, with no entropy contribution to the interaction when the UBL is removed (see ΔS of USP15(1–133) with SART3, Table 2). It is noteworthy that the conformation of the UBL domain is so flexible that most residues of the UBL domain could not be observed in the electron density map of the complex. We believe enthalpy-entropy compensation (35, 36) plays a major role in the tight binding of USP15 with SART3.
Domain Swapping and Hinge Residues
Several previous studies (18, 27–29) have analyzed the structures and dimerization of the DUSP-UBL domain of USP4, USP11, and USP15 in detail and identified residues on the DU finger and the DUSP domain to be the main cause of different oligomerization mechanisms of the USPs. USP4 exists as a dimer in the solution and in the crystal lattice. In contrast, USP15 could crystallize as a monomer (PDB codes 4A3O and 3T9L), as a domain-swapped dimer (18, 28), or a tetramer composed of a dimer of a domain-swapped dimer (PDB code 3PPA). Surprisingly, USP11 could also exist as a monomer and a domain-swapped dimer (29) even though its DU finger lacks the three residues key for SART3 binding. During purification, we observed two separate peaks during anion exchange, corresponding to different oligomeric states of USP15, which suggest the interconversion between monomer and oligomer is kinetically unfavorable. However, the kinetic barrier can be overcome by the binding of SART3, as shown by a KD of 28 μm between the oligomeric forms of USP15 with SART3 (USP15(1–233) Q2 in Table 2) or by raising the temperature of the solution (28). The DU finger of USP15 also serves as the hinge loop for domain swapping (18, 28). We observed that the G121A mutation breaks down the kinetic barrier of the conversion between the monomeric and dimeric forms of USP15 (Fig. 2B), suggesting Gly-121 is a key hinge residue for the domain swapping of USP15.
Note on the Complex Structure of SART3/USP4
When we were preparing our manuscript, Park et al. (37) reported the structures of the HAT domain of SART3 in the apo-form and in complex with the DUSP-UBL domain of USP4. The overall structures of SART3 and its complex with USP4 are similar to our results. However, we have different assignments of αA- and αB-helices for HAT repeats 6–12, and our assignment seems to give better sequence alignment and a more concise sequence pattern, whereas such a repeat-wise analysis was not done by Park et al. (37). We also confirmed that USP15 has a bivalent binding mode with SART3, where the Trp-50 (equivalent to Trp-54 in USP4) is important, the contribution of which was not analyzed in the USP4/SART3 structure. Park et al. (37) also did not explore in detail the difference of the binding affinities of USP4 or USP15 with SART3, whereas our data suggest that enthalpy-entropy compensation plays a major role.
Author Contributions
Q. Z. and Y. T. conceived and designed the experiments. Q. Z., R. H., F. H., A. D., J. R. W., and J. B. performed the experiments. Y. T., Q. Z., and R. H. analyzed the data and wrote the paper.
Acknowledgments
The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute, Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD Grant 115766), Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. X-ray diffraction data were collected at the Canadian Light Source (14) and at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, which is operated by UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5JJX and 5JJW) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DUB
- deubiquitinase
- HAT
- half-a-tetratricopeptide
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- PEG
- polyethylene glycol
- SEC
- size exclusion chromatography
- MALS
- multiangle light scattering
- USP
- ubiquitin-specific protease
- DUSP
- domain in USP
- UBL
- ubiquitin-like
- NLS
- nuclear localization sequence
- SAD
- single wavelength anomalous dispersion
- TEV
- tobacco etch virus
- DU
- DUSP-UBL.
References
- 1. Sowa M. E., Bennett E. J., Gygi S. P., and Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clague M. J., Barsukov I., Coulson J. M., Liu H., Rigden D. J., and Urbé S. (2013) Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 [DOI] [PubMed] [Google Scholar]
- 3. Vlasschaert C., Xia X., Coulombe J., and Gray D. A. (2015) Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol. Biol. 15, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J. W., Elledge S. J., Kirschner M. W., and Rape M. (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Long L., Thelen J. P., Furgason M., Haj-Yahya M., Brik A., Cheng D., Peng J., and Yao T. (2014) The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 289, 8916–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rader S. D., and Guthrie C. (2002) A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8, 1378–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suefuji Y., Sasatomi T., Shichijo S., Nakagawa S., Deguchi H., Koga T., Kameyama T., and Itoh K. (2001) Expression of SART3 antigen and induction of CTLs by SART3-derived peptides in breast cancer patients. Br. J. Cancer 84, 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ito M., Shichijo S., Miyagi Y., Kobayashi T., Tsuda N., Yamada A., Saito N., and Itoh K. (2000) Identification of SART3-derived peptides capable of inducing HLA-A2-restricted and tumor-specific CTLs in cancer patients with different HLA-A2 subtypes. Int. J. Cancer 88, 633–639 [DOI] [PubMed] [Google Scholar]
- 9. Furugaki K., Cui L., Kunisawa Y., Osada K., Shinkai K., Tanaka M., Kataoka K., and Nakano K. (2014) Intraperitoneal administration of a tumor-associated antigen SART3, CD40L, and GM-CSF gene-loaded polyplex micelle elicits a vaccine effect in mouse tumor models. PLoS ONE 9, e101854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whitmill A., Timani K. A., Liu Y., and He J. J. (2016) Tip110: Physical properties, primary structure, and biological functions. Life Sci. 149, 79–95 [DOI] [PubMed] [Google Scholar]
- 11. Preker P. J., and Keller W. (1998) The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 23, 15–16 [DOI] [PubMed] [Google Scholar]
- 12. Champion E. A., Lane B. H., Jackrel M. E., Regan L., and Baserga S. J. (2008) A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol. Cell. Biol. 28, 6547–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hammani K., Cook W. B., and Barkan A. (2012) RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. U.S.A. 109, 5651–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grochulski P., Fodje M. N., Gorin J., Labiuk S. L., and Berg R. (2011) Beamline 08ID-1, the prime beamline of the Canadian Macromolecular Crystallography Facility. J. Synchrotron Radiat. 18, 681–684 [DOI] [PubMed] [Google Scholar]
- 15. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 16. Rice L. M., Earnest T. N., and Brunger A. T. (2000) Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr. D Biol. Crystallogr. 56, 1413–1420 [DOI] [PubMed] [Google Scholar]
- 17. Terwilliger T. C. (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 18. Harper S., Besong T. M., Emsley J., Scott D. J., and Dreveny I. (2011) Structure of the USP15 N-terminal domains: a β-hairpin mediates close association between the DUSP and UBL domains. Biochemistry 50, 7995–8004 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bai Y., Auperin T. C., Chou C.-Y., Chang G.-G., Manley J. L., and Tong L. (2007) Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell 25, 863–875 [DOI] [PubMed] [Google Scholar]
- 23. Yang D., Nakao M., Shichijo S., Sasatomi T., Takasu H., Matsumoto H., Mori K., Hayashi A., Yamana H., Shirouzu K., and Itoh K. (1999) Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res. 59, 4056–4063 [PubMed] [Google Scholar]
- 24. Harada K., Yamada A., Yang D., Itoh K., and Shichijo S. (2001) Binding of a SART3 tumor-rejection antigen to a pre-mRNA splicing factor RNPS1: a possible regulation of splicing by a complex formation. Int. J. Cancer 93, 623–628 [DOI] [PubMed] [Google Scholar]
- 25. Aasland R., Abrams C., Ampe C., Ball L. J., Bedford M. T., Cesareni G., Gimona M., Hurley J. H., Jarchau T., Lehto V. P., Lemmon M. A., Linding R., Mayer B. J., Nagai M., Sudol M., Walter U., and Winder S. J. (2002) Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 513, 141–144 [DOI] [PubMed] [Google Scholar]
- 26. Lo Conte L., Chothia C., and Janin J. (1999) The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 27. Clerici M., Luna-Vargas M. P., Faesen A. C., and Sixma T. K. (2014) The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 5, 5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elliott P. R., Liu H., Pastok M. W., Grossmann G. J., Rigden D. J., Clague M. J., Urbé S., and Barsukov I. L. (2011) Structural variability of the ubiquitin specific protease DUSP-UBL double domains. FEBS Lett. 585, 3385–3390 [DOI] [PubMed] [Google Scholar]
- 29. Harper S., Gratton H. E., Cornaciu I., Oberer M., Scott D. J., Emsley J., and Dreveny I. (2014) Structure and catalytic regulatory function of ubiquitin specific protease 11 N-terminal and ubiquitin-like domains. Biochemistry 53, 2966–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., and Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paulson A. R., and Tong L. (2012) Crystal structure of the Rna14-Rna15 complex. RNA 18, 1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ideguchi H., Ueda A., Tanaka M., Yang J., Tsuji T., Ohno S., Hagiwara E., Aoki A., and Ishigatsubo Y. (2002) Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. Biochem. J. 367, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chodera J. D., and Mobley D. L. (2013) Entropy-enthalpy compensation: role and ramifications in biomolecular ligand recognition and design. Annu. Rev. Biophys. 42, 121–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kabiri M., and Unsworth L. D. (2014) Application of isothermal titration calorimetry for characterizing thermodynamic parameters of biomolecular interactions: peptide self-assembly and protein adsorption case studies. Biomacromolecules 15, 3463–3473 [DOI] [PubMed] [Google Scholar]
- 37. Park J. K., Das T., Song E. J., and Kim E. E. (2016) Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res. 44, 5424–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., Xue Y., and Ren J. (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pei J., Kim B.-H., and Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., and Drummond A. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laskowski R. A., and Swindells M. B. (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model 51, 2778–2786 [DOI] [PubMed] [Google Scholar]