Abstract
Dengue virus, an ∼10.7-kb positive-sense RNA virus, is the most common arthropod-communicated pathogen in the world. Despite dengue's clear epidemiological importance, mechanisms for its replication remain elusive. Here, we probed the entire dengue genome for interactions with viral RNA-dependent RNA polymerase (RdRp), and we identified the dominant interaction as a loop-forming ACAG motif in the 3′ positive-stranded terminus, complicating the prevailing model of replication. A subset of interactions coincides with known flaviviral recombination sites inside the viral protein-coding region. Specific recognition of the RNA element occurs via an arginine patch in the C-terminal thumb domain of RdRp. We also show that the highly conserved nature of the consensus RNA motif may relate to its tolerance to various mutations in the interacting region of RdRp. Disruption of the interaction resulted in loss of viral replication ability in cells. This unique RdRp-RNA interface is found throughout flaviviruses, implying possibilities for broad disease interventions.
Keywords: dengue virus (DENV), viral polymerase, viral protein, viral replication, virology, RNA-dependent RNA polymerase (RdRp), untranslated region (UTR), viral non-structural protein (NS) 5, yeast three-hybrid (Y3H)
Introduction
Positive-sense single-stranded RNA viruses initiate replication by the generation of a complementary negative (−) RNA strand via the action of viral RNA-dependent RNA polymerases (RdRps).3 In the cases of brome mosaic virus (family Bromoviridae), turnip crinkle virus (Tombusviridae), hepatitis C virus (Flaviviridae), and encephalomyocarditis virus (Picornaviridae), RNA structures in the 3′-untranslated region (UTR) of the positive strand have been characterized as RdRp promoters (1–4). In the case of dengue virus (DENV), a flavivirus with a 5′-type I cap and the absence of a polyadenylated tail, however, the current model for replication asserts that the 5′-UTR acts as a promoter for (−)-strand synthesis. This position is supported by several lines of in vitro evidence. In particular, atomic force microscopy demonstrated cyclization of the (+)-strand genomic RNA, resulting in placement of the 5′-UTR in proximity to the 3′ terminus, whereas EMSA and footprinting assays apparently documented RdRp interactions with the first hairpin element in the DENV 5′-UTR, designated stem-loop A (SLA) (5). In addition, an RNA fragment of the DENV 5′-UTR could also stimulate in vitro synthesis of a complementary product of the 3′-UTR (5, 6).
To date, investigations of RdRp actions on the flaviviral RNA genome have been limited to in vitro assays on less than 5% of the whole genome with a bias toward UTR regions; therefore, the entire interaction landscape of RdRp has not yet been revealed. It is also unclear whether flaviviral RdRps require a specific RNA promoter for de novo initiation of RNA synthesis. To gain insight into accurate mechanisms for synthesis of the viral genome in the cell, here we explored all possible interactions between RdRp and the complete RNA genome in an unbiased in vivo context using a refined yeast three-hybrid (Y3H) scan. Combining bioinformatic evidence with in vitro binding and viral replicon assays, we pinpoint amino acids and nucleotides contributing to previously undocumented RdRp-viral RNA interfaces and suggest a novel means that contributes to strong conservation of this promoter motif throughout flaviviruses, and possibly beyond.
Results
Isolation of Viral RNA Elements That Interact with RdRp via Y3H
We devised a Y3H genetic screen to explore the global interaction between viral RdRp and its own entire genomic RNA in a eukaryotic cellular environment (Fig. 1A). Y3H has been exploited to extensively identify protein-RNA interactions and has been shown to generally correlate well with other in vitro assays such as EMSA (7, 8). Because RNA inserts in the Y3H vector should generally not exceed 200 nt, as reporter signals weaken with increasing length, and at least 40 nt are required to provide sufficient length for RNA to fold into functional structures such as hairpins and pseudoknots (9), full-length cDNA from the dengue virus serotype 2 (DENV-2) was randomly digested into fragments of ∼100 bp in size via an endonuclease-V-based procedure (10), prior to non-directional ligation into a modified Y3H RNA expression vector pIIIA MS2-2 with high efficiency through a Gateway recombination protocol (see under “Experimental Procedures” for details).
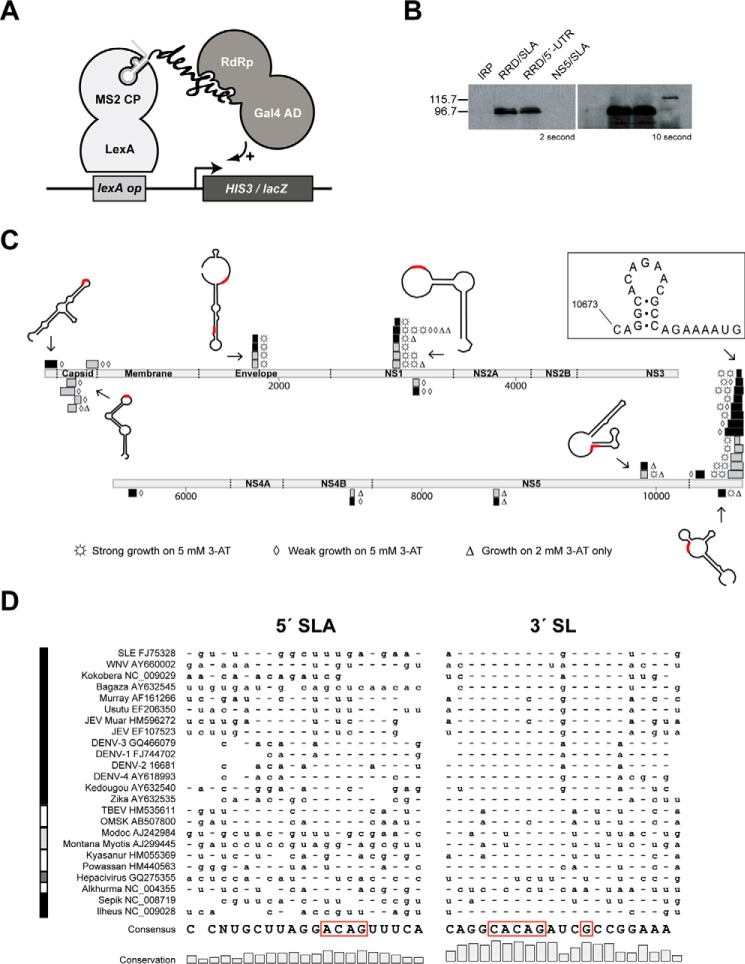
FIGURE 1.
Interaction landscape of DENV RdRp with the entire RNA genome. A, schematic diagram of Y3H in this study. B, Western blotting with polyclonal anti-NS5 antibodies (Gentex 103350) confirmed the expression of DENV-2 NS5 (120.9 kDa) and its RdRp (90.1 kDa) in Y3H with two representative RNAs, SLA and the full-length 5′-UTR. The protein IRP in pACT2 was used as a negative control. C, DENV genome is depicted as light gray rectangles and is numbered. The coding regions for viral proteins are labeled. Positive strand RNA fragments of the genome that bind the full-length NS5 and RdRp are shown above the genome as black and gray rectangles, respectively, whereas negative fragments are below. Levels of yeast growth in the presence of 3-AT are indicated by three symbols. Representative Mfold structures of RNA fragments are drawn with the CACAG motif in red. D, conservation of the ACAG loop motif is compared between the top loop regions of the 5′-SLA and the 3′-SL. Sequences are ordered according to a phylogenetic tree based on the terminal 120 nucleotides of Flaviviridae. The left vertical bar indicates the type of Flaviviridae as follows: black indicates mosquito-borne viruses; white indicates tick-borne viruses; light gray indicates viruses of no known vector; and dark gray indicates a single hepacivirus example. Consensus nucleotides are indicated once at the bottom of the figure; elsewhere, they are indicated by hyphens, with variations from the consensus indicated by the appropriate letter.
Sequencing of 100 clones verified suitable Y3H insert sizes and coverage of the entire (+)/(−) DENV genome (details under “Experimental Procedures”). Protein-RNA interactions were monitored in YBZ-1 yeast coexpressing the RNA library with either the full-length DENV-2 NS5 protein composed of N-terminal methyltransferase and C-terminal RdRp domains or the RdRp subdomain (hereafter referred to as “RdRp”) alone. Expression levels of these two proteins were confirmed via Western blotting, with the smaller RdRp being found at concentrations roughly 10-fold higher than NS5 (Fig. 1B). To cover all possible structural elements in both strands of DENV RNA, a total of ∼105 cotransformants (estimated via plating on non-selective Leu−/Ura− media) were subjected to selection with histidine deprivation. A total of 60 sequences, averaging 67 bases, mapping onto the DENV-2 genome were retrieved from yeast growing under three increasingly selective concentrations of 3-AT, a histidine metabolism inhibitor. Sampling ceased when it became apparent that four clusters of overlapping sequences dominated the mapping. Of all the sequences, the known NS5 interactor, the 5′-SLA, was identified once, with yeast growing only at the lowest levels of 3-AT (Fig. 1C). In contrast, a sequence associated with a particularly strong interaction at the last long stem-loop (SL) element in the 3′-UTR was observed on 19 occasions. In one case, a mere 23-nt fragment, consisting of the top stem-loop region of 3′-SL, was associated with robust protein-RNA interaction (Fig. 1C). Other 3′-SL-containing sequences ranged from 40 to 138 nt. Interestingly, the loop contained a highly conserved CACAG pentanucleotide sequence (Fig. 1D) that had been identified as critical for viral growth in a previous study (11). SLA also contains ACAG in a loop structure (Figs. 1D and 2), but the initial A is predicted to participate in the upper base pair of the stem, whereas that of 3′-SL is unpaired, preceded by a C that pairs with a G 8 bases downstream to clamp the loop.
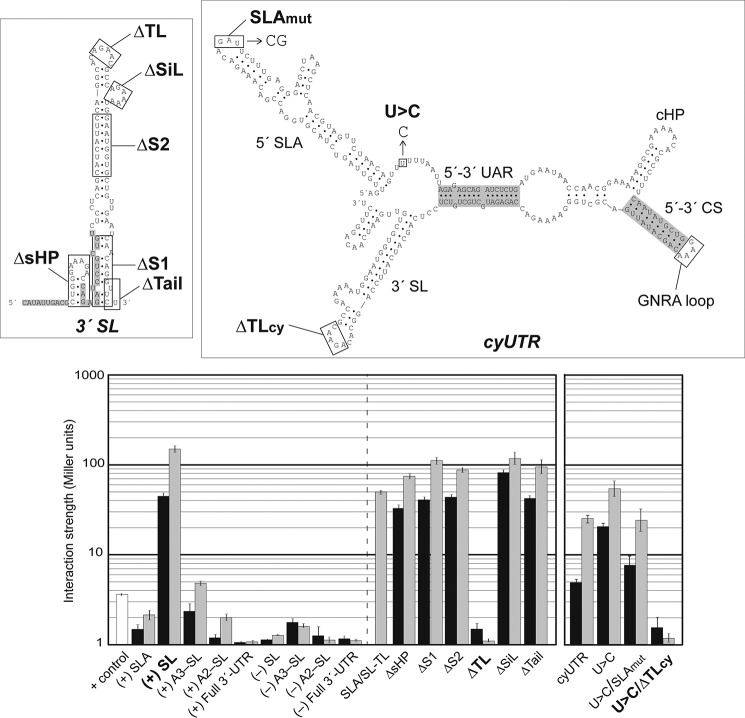
FIGURE 2.
DENV RdRp recognizes the top loop of 3′-SL. Linear 3′-SL and a series of its deletion mutants are indicated in the secondary structure with rectangles (upper left panel), whereas the model of the cyclized Y3H RNA construct containing DENV 5′- and 3′-terminal regions (cyUTR) used in this study is in the upper right panel. The long viral open reading frame was replaced by a GNRA loop that linked 5′- and 3′-UTR together in the cyUTR construct. SL mutant ΔsHP completely removed the boxed hairpin; ΔS1 removed the lower SL stem; ΔS2 removed the upper SL stem; ΔTL removed the indicated top loop nucleotides; ΔSiL removed the side loop; and ΔTail replaced the final 5′ nucleotides with CAAAA. In cyUTR, SLAmut replaced the wild-type GAU with CG, and U>C refers to the substitution of a U with a single C. Structural RNA elements are labeled. β-Galactosidase expression assays showing the interaction strengths with the full-length NS5 (black-filled bar) or RdRp (gray) are measured in Miller units. The lower left panel shows RdRp interactions with a variety of linear DENV RNAs with native sequences (left half) and mutations (right half). Results with a cyclized form of UTR are separated into the lower right panel. Error bars indicate S.E.
An RNA complementary to a dumbbell structure upstream of 3′-SL, designated A3 (the secondary structure is illustrated in Fig. 7), interacted with NS5 and RdRp at a strength comparable with that of 5′ stem-loop A ((−) A3-SL in Fig. 2), in which a highly conserved ACAG among flaviviruses is present in a predicted loop. The positive complement to this loop likely forms a loop, designated TL2 in previous work (12), which showed that mutations of TL2 resulted in significant replication impairment. Other clusters of interactors were also identified, with two clusters in coding regions being particularly obvious. Several map features are of note. First, an ACAG sequence, predicted to reside at least partially in a loop structure (13), was found in 51 of the 60 sequences in a predicted loop structure (Fig. 1C). Numerous sequences containing ACAG were found during validation of the RNA library, but these sequences were not later identified as RdRp interactors, indicating that a 4-nt sequence alone is indispensable but not sufficient for RdRp binding. Second, the majority of interactors (75%) resided on the positive strand. Because negative strand flaviviral RNA is thought to exist primarily in duplex form in infected cells, RdRp interactions with this strand might not predominate. No preference for RNA binding was observed between NS5 versus RdRp because in every case where the same interacting sequence was seen at least twice, both NS5 and RdRp were identified as interaction partners. In other words, the methylase domain likely had negligible RNA specificity. Finally, the genomic interaction sequences revealed significant correlation (p < 5E-11, see “Experimental Procedures”) with recombination regions documented in DENV-1 (14) and JEV (15) studies and also contained sites at which 5′- and 3′-DENV regions joined together to form defective interfering (DI) particles (16). It is noted that these DI-joining points are enriched in ACA and ACAG (p = 0.0029 and 0.0015, respectively, see “Experimental Procedures”). These facts would imply a possible mechanism of RNA template switching where RdRp encounters these sequences and structures.
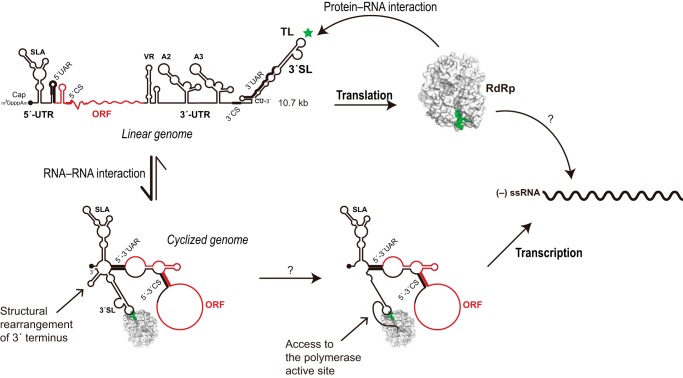
FIGURE 7.
Schematic model of DENV replication based on our results coupling with earlier studies. We propose that the structural rearrangements in both polymerase and the 3′ end region of genomic RNA upon binding of RdRp to the 3′-SL-TL and genomic cyclization play key roles in regulation of viral replication cycle. Accurate mechanisms for recruitment of the 3′ end into the active site of RdRp as well as recognition of the negative stranded genome by RdRp still remain elusive.
RdRp Specifically Recognizes the SL Top Loop in Both Genomic Forms
To validate our Y3H screen with an RNA library, various portions of the DENV 5′- and 3′-(+)/(−) UTR RNAs were constructed, and protein-RNA interactions were quantified via the ONPG-based assay for the β-galactosidase reporter gene. The results revealed that the binding affinity of RdRp to 3′-SL in Y3H exceeded that of SLA by greater than 2 orders of magnitude (Fig. 2). Previous EMSA and filter binding assays established an in vitro Kd for 5′-UTR RNA-RdRp binding at 8–16 nm (5), and surface plasmon resonance rendered a Kd of 53 nm (17), although a linear extrapolation of SL-binding strength should not be expected. It was noted that SL-binding signals diminished as the length of the RNA inserts increased from 3′-SL alone (95 nt), to SL and the A3 dumbbell structure (A3-SL, 203 nt), to SL and two dumbbell A2-A3 structures (A2-SL, 289 nt), and finally the full 3′-UTR (458 nt) (Fig. 2). This is typical Y3H behavior, one of the variables precluding usage of Y3H as a direct Kd measurement system (18). Deletions of six features of the 3′-SL showed that this robust interaction is defined entirely by the ACAG top loop (TL) motif (ΔTL in Fig. 2). In another experiment, merely replacing the SLA-TL by that of 3′-SL resulted in a 47-fold increase in binding strength in the Y3H context (SLA/SL-TL in Fig. 2). Given that the ACAG sequence in SLA is variable among flaviviruses (Fig. 1D), these results confound the current replication model.
Our Y3H assays were further corroborated by in vitro gel shift assay (EMSA) with highly purified components, including non-tagged DENV RdRp (17) and three RNA transcripts derived from DENV-2 3′-UTRs. We stained both protein and RNA to avoid misinterpretation of RNA EMSA because RNA could form multiple configurations, including multimers in solution, and RNA-binding proteins could have more or less non-sequence-specific binding potential due to their extensive positively charged surfaces. The results showed that RdRp comigrated, i.e. formed stable complexes, with both linSL and cirSL, which corresponded to the 3′-SL configuration as it is found in the linear (lin) and cyclized forms, respectively, of the complete genome (Fig. 3). Deletion of the 3′-SL top loop markedly impaired RdRp complex formation (ΔTL in Fig. 3). RdRp thus specifically recognizes the 3′-SL-TL because the control tRNA showed only a background level of interaction (Fig. 3). Our EMSA results strongly corroborated the Y3H assay and were in clear contrast to previous reports that failed to detect 3′-SL-binding ability (5). Next, to quantify the binding affinity of RdRp with 3′-SL, we exploited an Alpha assay with 5′-biotinylated RNA substrates and N-terminally His-tagged RdRp. The Alpha technology has recently been used to determine interactions of RNA-binding proteins in solution (19). We employed truncated forms of 5′-SLA and 3′-SL RNAs in the binding assay to narrow down the interaction site (Fig. 4A). SL binds to RdRp with a Kd of ∼3.2 nm, whereas the deletion of the top loop completely abolished the binding signal. We also performed competition assays between biotinylated SL and unlabeled RNAs (Fig. 4B). SLA RNA failed to compete with SL binding in our assay (data not shown), whereas simple substitution of the top loop of SLA by that of SL (SLA/SL-TL) obviously showed binding inhibition. Altogether, our Y3H, EMSA, and Alpha assays provided indisputable proof of specific RdRp/SL-TL interaction.
FIGURE 3.
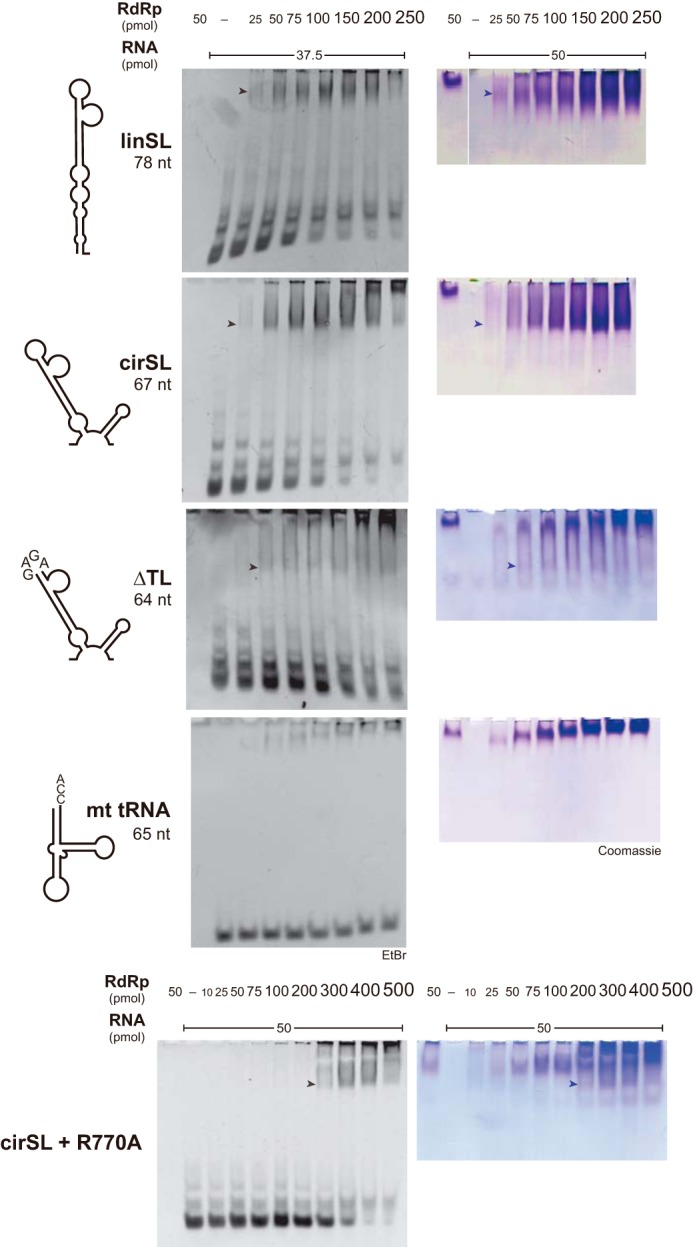
In vitro EMSA. Mfold secondary structures of RNAs are depicted in the left column. Arrows highlight protein-RNA complexes that were evident via both EtBr (middle column) and Coomassie (right column) staining. The constant amount of RNA in the reaction was increased from 37.5 to 50 pmol in the case of protein staining. The linSL and cirSL experiments were run on a single gel with one “RdRp alone” lane.
FIGURE 4.
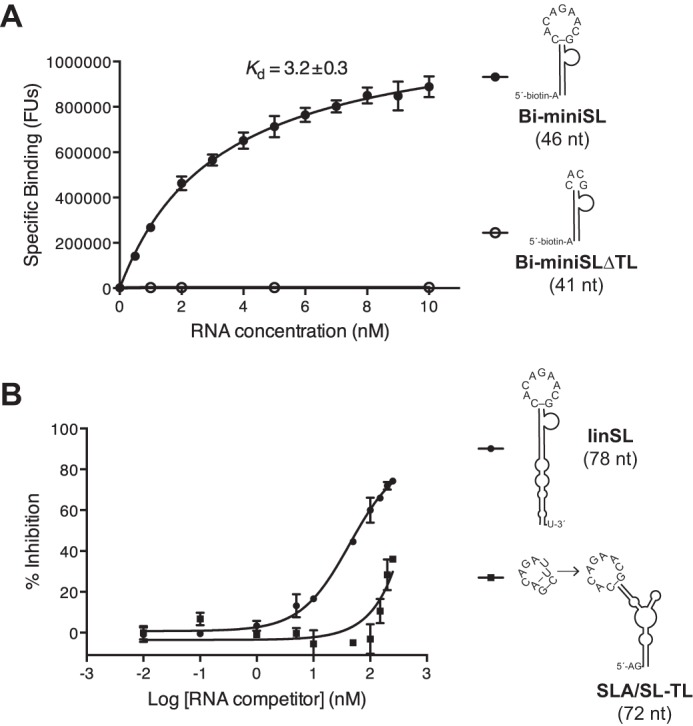
In vitro Alpha binding assay. A, saturation binding experiment of RdRp with truncated 3′-SL. The Alpha-binding signal is shown in fluorescence units (FU). B, competition assay with unlabeled RNA. RdRp and Bi-miniSL concentrations were kept constant at 200 and 10 nm, respectively. Mean and standard deviation values are derived from three independent experiments. We note that SLA did not show a binding signal in both saturation binding and competition assay.
To faithfully model NS5 interactions with genomic RNA in its cyclized form, a 254-nt RNA construct (cyUTR) containing, in order, the 5′-UTR, 5′-CS, a GNRA loop-forming sequence, the 3′-CS that binds with the 5′-CS, and the 3′-terminal SL structure was engineered (Fig. 2, right panels). In this form, the initial 144 5′-nucleotides are predicted to be placed in the vicinity of the last 106 nt of the 3′-UTR via complementary interactions mimicking the cyclized form of DENV UTRs (20). As expected, a deletion mutant disrupting the ACAG motif in 3′-SL (ΔTLcy) caused a 50-fold loss of binding affinity, which paralleled the differing affinities between the 5′-SLA and 3′-SL linear constructs. However, it could not be ruled out that SLA may be involved in a structural rearrangement of UTRs in the cyclized form because mutation of the SLA-TL decreased the potent wild-type signal by half. Nonetheless, these results emphasized the 3′-SL-TL as the predominant RdRp-binding site in both genomic forms. Moreover, a deletion mutant of the 3′-SL-TL (ΔTL) in our DENV-2 replicon (see “Experimental Procedures” for details) completely abolished viral RNA replication in BHK-21 cells, similar to the GAA mutation that inactivated the polymerase activity of NS5 (Fig. 5D). Taken together, our results suggested the ACAG motif in the 3′-SL-TL serving as a cis-element for (−) strand viral RNA synthesis in cells.
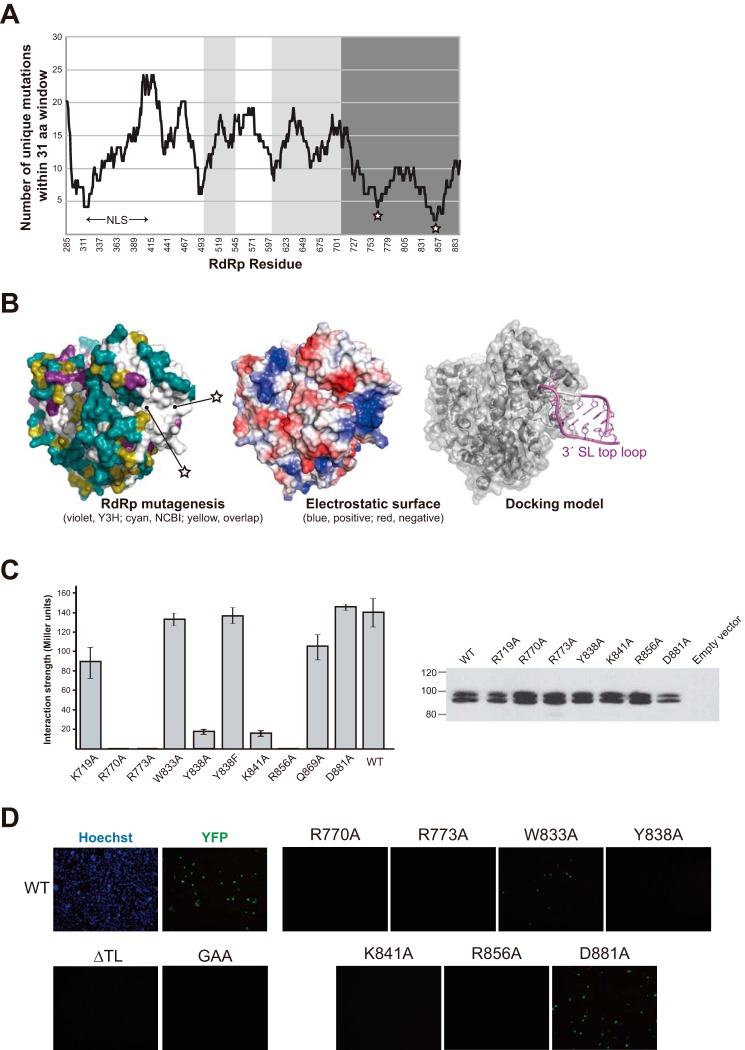
FIGURE 5.
Exploration of 3′-SL-binding sites in RdRp using Y3H random mutagenesis. A, random RdRp mutants retaining SL-binding capacity were sequenced and analyzed as under “Experimental Procedures.” Results show two specific regions of interest in the thumb domain (dark gray), which were highly susceptible to mutations (stars). White areas indicate the finger domain and light gray shows the palm domain. B, mutations allowing RdRp binding in Y3H are mapped onto the crystal structure (Protein Data Bank code 2J7W) in the left panel (violet), together with naturally occurring variations found in 2,446 DENV sequences from the NCBI database (cyan). Residues overlapping between the two data sets are shown in yellow. In the middle panel, the protein solvent-accessible surfaces were generated with the program APBS handled in PyMOL and are colored according to their electrostatic potential from red to blue. Our results prompted us to manually create an RNA-docking model using YFV SL structures (Protein Data Bank code 2KPC) (right panel). RdRP and SL-TL were manually docked on PyMOL to minimize steric clashes and to reproduce mutational studies. C, Miller assays for SL interactions with specific RdRp mutants are shown (left panel), together with their expression in yeast (right panel). We have no clear explanation for the double banding pattern that was occasionally observed. D, DENV replicon assay with RdRp mutants in BHK-21 cells. Viral replication was investigated via YFP signals 4 days after transfection to monitor RNA synthesis. YFP signals are shown for each mutant. ΔTL refers to mutations that altered the 3′-SL top loop to the extent that RdRp binding was abrogated in Y3H (Fig. 2). GAA refers to an inactive RdRp by mutation of its catalytic triad, GDD.
RdRp Thumb Domain Recognizes the ACAG Motif
Because the underlying molecular basis for RNA specificity of RdRp remains obscure, we undertook a global search for the RNA-binding surface on DENV RdRp. To this end, a library of mutant RdRps in the Y3H protein expression vector was created by Mutazyme polymerase. The final optimized library contained an estimated 105 unique RdRp sequences with a 0.5% mutation rate. Following transformation into yeast expressing the 3′-SL RNA, 84 mutant RdRp sequences were retrieved from strongly growing yeast (the positive selection). Therefore, these mutations were not likely associated with RNA binding, and by deduction (see “Experimental Procedures”), RdRp regions that appeared intolerant to mutations could be discerned (Fig. 5A). Our approach minimized the false-negative binding selection that could stem from nonsense mutations and mutations that changed the local protein structure. Our analysis immediately highlighted two specific regions, including 10 residues surrounding Tyr-766 and Ile-850 residing in the C-terminal “thumb” domain of RdRp. Despite their distance in primary sequence, these two groups of residues are mapped onto the same region in the crystal structure of DENV RdRp (Fig. 5B) (21, 22). A third region encompassing Val-310 in the N terminus also appeared to be sensitive to mutations. The region is part of an interdomain linker joining fingers with thumb domains and was shown to be implicated in binding of the viral helicase NS3, as well as importin-β, a nuclear import factor (23). We further analyzed 2446 DENV RdRp sequences from the NCBI database, combined with Y3H random mutagenesis, and mapped all variable residues onto the DENV RdRp structure (Fig. 5B, left panel). Strikingly, there was only a single invariant region on the molecular surface, and its area was capable of 3′-SL-TL accommodation. The putative 3′-SL-binding region possesses strong positively charged electrostatic surfaces (Fig. 5B, middle panel), and it is located at the rim of the helical bundle in the thumb domain separated from the active site in the palm domain by ∼30 Å. There are no earlier reports of the RNA binding ability of the thumb domain nor structural similarities to any known RNA-binding modules via the DALI algorithm (24).
To pinpoint residues responsible for the RNA recognition, site-directed mutagenesis was conducted, yielding steep declines in 3′-SL-binding affinity in several cases; most strikingly, R770A, R773A, and R856A substitutions completely abrogated the interaction. Y838A and K841A mutations also had severely deleterious effects (Fig. 5C, left panel). The protein expression levels of these mutants were comparable with that of the wild type in yeast (Fig. 5C, right panel). These critical residues are strongly conserved in flaviviruses, with Arg-773 retained into Flaviviridae (Fig. 6C). Site-directed mutagenesis of RNA-binding residues in our DENV-2 replicon abolished viral replication at 4 days after transfections into BHK-21 cells, whereas mutations of irrelevant neighboring Trp-833 and Asp-881 residues showed few effects (Fig. 5D). EMSA studies with the recombinant R770A mutant corroborated these results, as binding to input RNAs was obviously weakened (Fig. 3), whereas the protein folding and polymerase activity of the mutant were retained (data not shown). These critical mutants also eliminated interactions with 5′-SLA in Y3H (data not shown), making it unlikely that more than one RNA-binding site exists in RdRp for simultaneous binding with two or more ACAG motifs. Our results emphasized an advantage of a combined Y3H random mutagenesis approach in discovering an RNA-binding motif without possible biases associated with mutagenesis strategies that are heavily reliant on conservation data or the assumption that particular amino acids (e.g. arginine and lysine) would be crucial in RNA interactions.
FIGURE 6.
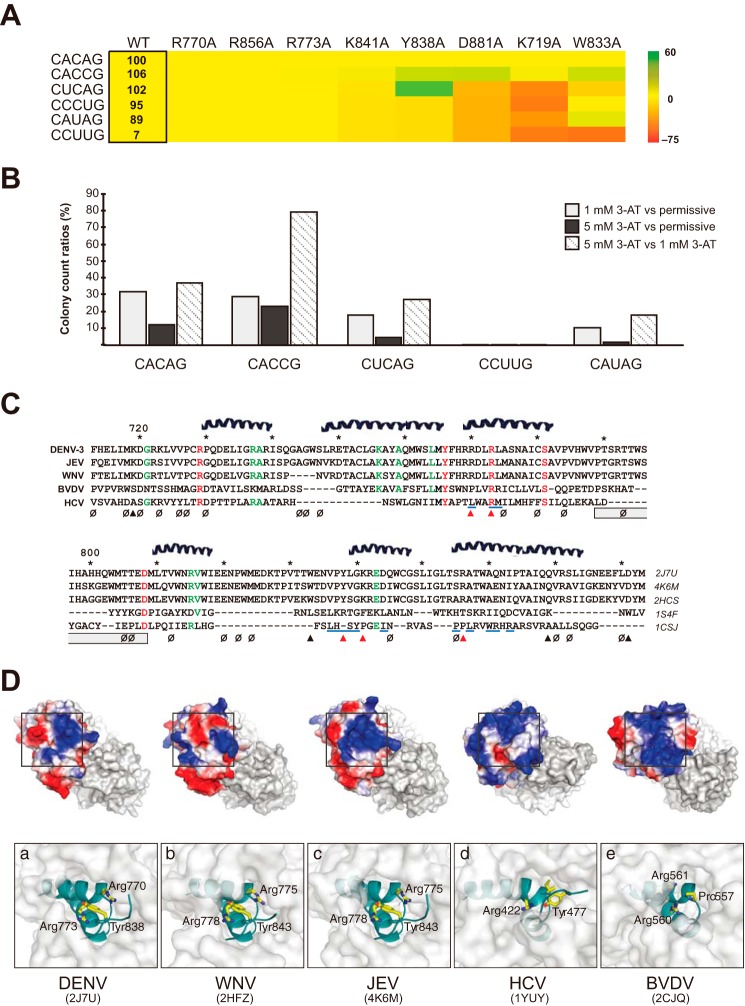
Exploring the conservation of the RdRp-RNA interface within and beyond DENV. A, heat map matrix of interaction strengths among RdRp and 3′-SL variants. Relative interaction strengths of 3′-SL variants with the DENV RdRp are shown in a box as a percentage of the wild-type (WT) RdRp-CACAG pair. Colors, indicating a 3′-SL-TL variant's sensitivity to protein alterations, were arrived at as follows: % changes in interaction strengths between mutant RdRp and 3′-SL variant combinations versus those of WT RdRp and 3′-SL variants are calculated. These percentage changes are then compared (via simple subtraction) to the relevant RdRp-CACAG interaction strengths. Numerous RNA variants, including some residing on the 5′-SLA, are not included on the table, as their interactions with RdRp were particularly weak. B, analysis of 3′-SL-TL's tolerance for mutations in Arg-773. A library of Arg-773 RdRp mutants (R773X) was examined via Y3H with five 3′-SL-TL variants under one permissive and two selective conditions. C, DALI structural alignments of the thumb domains from five available Flaviviridae RdRp structures. Residues with perfect conservation are colored in red, and ones that align in four sequences are green. The null symbols (ø) indicate mutations that had no effect in 3′-SL-binding in our Y3H random mutagenesis. Filled triangles point to residues that were chosen for site-directed mutagenesis, and red triangles indicate crucial residues in 3′-SL-TL-binding. HCV residues making contacts with thumb 2 inhibitors in previous studies are highlighted by blue lines. The gray rectangle shows the region that was deleted in an HCV structural study (45). D, superposition of five Flaviviridae RdRp structures reveals similarity in the SL-binding site of DENV RdRp comprising an Arg-770–Tyr-838–Arg-773 stacking platform. In all cases, two non-contiguous helices combine to form a positively charged pocket (blue) centering on a perfectly conserved Arg-773 in DENV.
Our results allowed manual construction of an RNA-docking model based on available DENV-3 RdRp and YFV SL structures (2J7W and 2KPC, respectively), without apparent severe clashes (Fig. 5B, right panel). Arg-773 lies inside the binding cleft and forms an in-line stack with three essential residues, Arg-770, Tyr-838, and Lys-841, likely creating a TL RNA-accommodating platform (Fig. 6D). A Y838F mutation did not affect SL binding activity, suggesting a key role of side-chain stacking in RNA recognition (Fig. 5C). The RNA-binding site is located adjacent to a bound zinc ion (∼12 Å) in the thumb domain. According to our model, the last base of the ACAG motif is ∼50 Å distant from the first base of the RNA template in the catalytic site of RdRp. Therefore, the distance formed by the 42 bases downstream of the ACAG motif to the 3′ end should be indispensable for enabling initiation of RNA synthesis in cis. Interestingly, the complex of RdRp with cirSL was more compact than that of linSL, i.e. sharp bands migrated faster in EMSA, implying that the 3′ single-stranded region might make extensive contacts near the active site of RdRp (Fig. 3).
Conservation of the ACAG Motif Relates to Tolerance for RdRp Mutants
Next, we randomly mutated the ACAG sequence in the 3′-SL-TL and challenged it against RdRp in Y3H. Although 256 variants were generated, strong yeast growth on selective media was associated only with yeast containing either ACAG or ACCG sequences. We also generated several specific SL mutants that mimic some of the more common TL variants in Flaviviridae, e.g. the AUAG that is often found in pestiviruses. A number of these mutants interacted with RdRp at wild-type levels, raising the following question: if strong interactions are retained in several TL variants, what accounts for the strong conservation of ACAG in flaviviruses? Data gathered in experiments where TL variants were cotransformed with mutant RdRps offer the possible explanation that ACAG and ACCG are especially tolerant of mutations in the RNA-binding site of RdRp (Fig. 6A). For instance, although an R719A mutation minimally reduced affinity for wild-type 3′-SL, the same mutation nearly abrogated the interaction when TL was replaced by a UCAG sequence. Our “mutation tolerance” hypothesis is further corroborated by a cotransformation of a random library of Arg-773 mutants (R773X) with specific 3′-SL variants. Transformed yeasts were then grown under one permissive and two selective conditions. Colonies were counted on the various plates, and ratios between growth on selective versus less-selective conditions were calculated (Fig. 6B). In particular, the wild-type 3′-SL (CACAG) and R773X pair gave colony counts on 1 mm 3-AT media as 32% of the counts on permissive media and on strongly selective media as 12% of those on permissive media, whereas cotransformants of 3′-SL with AUAG in TL showed only 2% of permissive media counts, suggesting that ACAG tolerates a wider spectrum of Arg-773 mutations than AUAG. It is also noted that about 80% of the CACCG colonies that grew under 1 mm 3-AT were able to grow under 5 mm 3-AT conditions.
Discussion
We used an unbiased Y3H scan to probe the entire DENV genome for interactions with its polymerase, RdRp. Despite thousands of varying sequences in our Y3H RNA library, 12% of which included double-ligated products with an average size of 122 nt representing possible long range RNA-RNA contacts, the 3′-terminal stem-loop motif emerged as the dominant interactor. Although quite compact, the 3′-SL combination of RNA sequence and structure is certainly sufficient to eliminate most of the host RNAs from candidacy for polymerization. Indeed, the vast majority of catalogued RNA-binding proteins recognize short sequences (25) and, to a lesser extent, small structures (26).
Clues for importance of the 3′-SL-TL for RdRp interaction have been reported in previous works. Observations regarding the conserved nature and predicted structure of the 3′-SL CACAG pentanucleotide date back to 1986 (27). Of particular interest, Harris and co-workers (28) were able to reduce viral replication in BHK-21 cells by 1,000-fold upon silencing 3′-SL-TL with a 20-nt morpholino oligomer. Targeting of SLA also impeded viral replication, but the oligomer was complementary to the SLA lower stem, as opposed to the TL with which RdRp has been shown to interact using RNA footprinting (6). At least two mutational studies on the 3′-SL-TL have been conducted in the case of WNV replicons, both of which revealed that various mutations severely limited viral replication, and one showed that replication, not translation, was affected (11, 29). Yet another WNV study showed complete abrogation of replicon replication when the CACAG pentanucleotide was replaced with UCUAG, implying that the CACAG bound the replicase (30). Other in vitro assays also showed RdRp interactions with the 3′-UTR in closely related viruses such as JEV (31). Contrary to earlier EMSA studies, we show that DENV RdRp formed a complex with 3′-SL in both genomic forms using in vitro EMSA and in vivo Y3H, consistent with a model in which RdRp remains bound to 3′-SL-TL in either genomic form. Although our results certainly did not exclude a role for 5′-SLA, multiple evidence from our results made it clear that 3′-SL must be included in the replication picture. Our results prompt scrutiny of the evidence supporting a role of genome cyclization in positioning the 3′ terminus in proximity of the 5′ promoter for initiation of replication (5). It should be noted that a balance between cyclized and linear forms has been suggested for numerous purposes, such as regulation of RNA synthesis versus translation to ensure generation of full-length RNA strands, and control of the ratio of positive to negative strands, and the timing of encapsidation (32–35). It was predicted that genome cyclization stimulates shortening of the 3′-SL lower stem, releasing the 3′ terminus as a short single-stranded form (20). The location of the 3′-SL-TL-binding interface with respect to both the catalytic region and the 3′ terminus thus should allow a higher level of replication after the particular viral event of cyclization by providing access of the free 3′ end of viral genome to the active site (Fig. 7). DENV and HCV studies also support our idea, showing that subtle alterations in length and strandedness (single versus double) of the 3′ templates could drastically stimulate in vitro RdRp activities (36, 37).
Sequence alignments of the SL-TL among flaviviruses show that the CACAG pentanucleotide is particularly conserved, with occasional substitutions at the 4th nucleotide (e.g. JEV: Muar HM596272, A–C, Fig. 1D). The motif is even found in tick-borne encephalitis flavivirus. Moreover, a CACAG or CAUAG sequence is often observed in the 3′-SL-TL in bovine viral diarrhea virus, a pestivirus. A perfectly conserved CACAG also resides in a stem-loop in hepacivirus; interestingly, the sequence exists in an RdRp-coding region that has been implicated in interactions with RdRp protein (38) and is fundamental for viral replication possibly via both RNA-RNA and RdRp-RNA interactions (39). The pattern may even extend beyond Flaviviridae, as an ACAG sequence has been identified in a brome mosaic virus subgenomic promoter, with point mutations to the 5′-A, -C, and -G greatly diminishing RdRp activity; however, the recognition mechanism appears to be purely sequence-dependent (40). In our work, a number of CACAG variants (e.g. CAUAG) bound to DENV RdRp with comparable affinity. We would not confine the motif's sole function to RdRp binding as, for instance, the CACAG motif in HCV is assumed to pair with a complementary sequence in the 3′-UTR to form a kissing loop (39). However, an unrecognized mechanism for RNA sequence conservation emerged when we paired SL and RdRp mutants in a Y3H context and found that multiple 3′-SL-TL variants were intolerant to RdRp mutations, although the 3′-SL-TL with ACAG appeared able to withstand some of these alterations. Consistently, ACAG is more resistant to RdRp harboring randomly mutated Arg-773 than AUAG. In other words, ACAG confers the most optimal chance for viral RdRp to specifically interact with RNA under mutational pressure. However, considerably more work involving extensive mutagenesis of both RdRp and the SL-TL would be required to firmly establish such a mutational tolerance hypothesis.
To date, RdRp structures from five Flaviviridae, DENV, WNV, JEV, bovine viral diarrhea virus, and HCV, have been determined (21, 41–44). In all cases, a strongly positively charged RNA-binding cleft encompassing Arg-773 in DENV is obvious. Moreover, analogous stacking arrangements with adjacent arginine and tyrosine residues are also present (Fig. 6, C and D). Intriguingly, the DENV SL-TL-binding site entirely coincides with the binding sites of “thumb 2” inhibitors against HCV RdRp, and numerous direct protein-inhibitor contacts were made with residues corresponding to critical DENV residues identified in this study (Fig. 6C). This coincidence is indicative of the inhibitory action in which RNA binding is impeded, resulting in defects in viral RNA synthesis. In at least one case, an HCV inhibitor that binds to homologous SL-binding region causes a shift from a closed to open RdRp conformation (43). The recent achievement in solving an HCV RdRp complexed with double-stranded RNA revealed that the elimination of a β-hairpin loop spanning residues 442–454 (Fig. 6C), corresponding approximately to residues Pro-789–Met-804 in DENV, induces significant conformational changes to an open form with a 20° rotation of the thumb domain, allowing primer-template RNA duplex access to the catalytic core (45). Taken together, it is tempting to speculate that SL binding stimulates structural rearrangement of the thumb domain, thereby providing template RNA access to the active site and initiating de novo RNA synthesis in vivo (Fig. 7). Although RdRp has been demonstrated to promiscuously utilize numerous short templates in optimized in vitro environments, this lack of fidelity could likely prove disastrous in the complex RNA-saturated world of the cell, where the presence of random dsRNA initiates an antiviral response (46), necessitating the strong discriminatory mechanism that the promoter provides in the absence of a primer.
Our finding of strong conservation of the promoter interface in flaviviruses suggests that it could serve broadly as a drug target, although its specificity and the apparent lack of eukaryotic homologs should minimize off-target effects. In addition, interception of the interactive interfaces should reduce chances of mutational resistance because viral adaptation has to synchronously occur in both protein and RNA molecules. Conceivably, once DENV RdRp's spatial and temporal relation with its RNA targets is more fully delineated, principles of viral replication that extend well beyond Flaviviridae may be unveiled.
Experimental Procedures
Molecular Cloning
Viral RNA was isolated from DENV-2 strain 16681 and used to generate full-length cDNA according to the SuperScript III procedure (Life Technologies, Inc.). The 5′ and 3′ halves of the DENV genome were amplified using TaKaRa Ex Taq polymerase and inserted into the pSC-A vector between SacI and XbaI sites with the StrataClone PCR cloning kit (Stratagene). Clones were blue-white screened and validated by DNA sequencing.
Plasmids containing the 5′ half of the DENV-2 genome (bases 1–5428) or 3′ half (5429–10723) were used as PCR template in the presence of dUTP prior to random fragmentation with endonuclease-V to ∼100 bp according to Dyson's protocol (10). In brief, no more than 5 μg of PCR products were treated in 2× fragmentation buffer (20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 1 mm MnCl2) and 6 units of endonuclease-V (New England Biolabs) for 15 h at 37 °C. The fragmented DNA pool was excised from 1.5% agarose gel and then blunted with 2 units of T4 DNA polymerase (New England Biolabs) at 12 °C for 30 min, followed by heat inactivation at 75 °C for 20 min.
Ligation of fragments directly into a SmaI-digested and dephosphorylated pIIIA MS2 vector (9.1 kb) was found to be very inefficient. To overcome this difficulty, a Gateway-based protocol (Invitrogen) was devised to generate the desired final vectors for Y3H studies. To this end, a 3.7-kbp compact entry vector was created by inserting the MS2-2 region (925 bp) of the standard Y3H pIIIA MS2-2 plasmid, necessary for RNA expression, into the pCR8/GW/TOPO vector (Invitrogen). Subsequently, DENV-2 DNA fragments were blunt-end cloned into this entry vector at its SmaI site. A second SmaI digestion of the ligated products prior to the transformation effectively eliminated all empty vectors, giving rise to nearly 100% cloning efficiency. This step was enabled by the fact that the pCR8/MS2-2 vector's sole SmaI site is eliminated in the ligation reaction and by the fact that DENV-2 DNA contains only one SmaI site. Concurrently, a destination vector containing the remainder of the pIIIA MS2-2 backbone was constructed with the Gateway Vector Conversion System (Invitrogen). Gateway recombination finally generated Y3H pIIIAMS2-2 vectors with genomic fragment inserts.
Y3H protein expression vectors containing the full DENV-2 NS5 protein or its RdRp subunit alone (beginning from Pro272) were constructed into the pACT2 vector between BamHI and XhoI restriction sites downstream of the gal4 activation domain. Several linear forms of DENV UTR RNAs were amplified from the cDNA, and blunt-end cloned into the pIIIA MS2-2 vector at the SmaI site. The cyclized form of DENV UTR RNA was constructed via a two-step PCR procedure, by which the full 5′-UTR and the 3′-SL and CS regions were individually amplified and joined together via PCR ligation. Mutations were introduced into SL and the cyclized UTR through the QuikChange protocol (Stratagene). Earlier Y3H studies have shown that long poly-U stretches diminish Y3H signals, as RNA polymerase III is known to terminate at these repeats, eliminating key Y3H features that are required for retention of the transcript in the yeast nucleus (47). To avoid premature termination by the presence of a tandem U sequence in the SLB stem of the 5′-UTR of our cyclization-mimic, we introduced a single cytosine within the middle of the poly-U tract (U>C in Fig. 2), resulting in a 3-fold increase in NS5 and RdRp apparent affinity.
Validation of DENV RNA Library
Following DNA sequencing of 100 clones, the RNA library was found to possess an average insert length of 95 nt. 49% of the sequences were of positive orientation. None of the sequences were identical, giving a 50% probability that at least 7,200 sequences were present in the library based on the “reverse birthday problem” (i.e. if, on another planet, none of the 100 students in a room have the same birthday, what is the minimum likely length of a year?). The solution is approximated as shown in Equation 1,
where the desired probability is Pr; m corresponds to the number of non-identical sequences identified, and n is the number of sequences expected in the library at the input probability.
Coverage of the genome could be checked by two means. First, a Monte-Carlo-based algorithm was employed to simulate random placement of these sequences on the genome (21.4 kb when the minus strand is considered). Predicted random sequence coverage could then be compared with actual sequence coverage; our library showed greater coverage than 19% of computer-generated outcomes. Second, the single most strongly binding RNA motif was found in 15 non-identical sequences following probing of the library with RdRp, suggesting a high level of coverage. Therefore, the efficient generation of a quality RNA library of DENV covering both 10.7-kb (+)- and (−)-strands was demonstrated for the first time.
Random Mutagenesis of RdRp
PCR-based random mutagenesis was conducted to generate mutant RdRp products with the GeneMorph II EZCLone domain mutagenesis kit (Stratagene). In brief, specific primers upstream and downstream of the RdRp insert, 5′-AAACCTAGATATAATTGGGAAAAGAATAG-3′ and 5′-CCACAGAACTCCTGCTTCTCCC-3′, were used to introduce random mutations into the 1.9-kb PCR product via Mutazyme DNA polymerase. Varying the initial amount of DNA template from 361 to 0.95 ng was necessary to obtain the appropriate mutational rate. Products of error-prone PCR serving as megaprimers were then incorporated into the Gateway entry vector by swapping the existing wild-type with the mutants to create a random library. Initial sequencing verified an even distribution of mutations along RdRp. It was found that frameshifts caused by deletions and substitutions were common at high mutation rates, and therefore, a 0.5% nucleotide mutation rate was settled on. In vitro recombination between Gateway entry vectors carrying the mutant library and the destination vector carrying the pACT2 backbone was catalyzed through the Gateway LR Clonase II enzyme mix kit (Invitrogen), yielding a mutant RdRp library in a Y3H-suitable expression vector. Yeasts containing the Y3H DENV SL RNA expression vector were then transformed with the protein mutant library onto selective plates, and colony PCR was performed on strongly growing yeast to isolate the mutant sequences.
Y3H Screen
Yeast transformations were carried out with YBZ1 strain using the standard lithium acetate method. Single step co-transformation with 2–3 μg of each plasmid per 200 μl of yeast suspension was preferred over two single transformation steps, reducing background, and minimizing opportunities for genome or plasmid mutations. Yeast growth was initially screened on weakly selective Leu−/His− plates. Subsequently, interaction candidates were subjected to more selective conditions of Leu−/Ura−/His− with increasing concentrations of 3-AT, a histidine metabolism inhibitor, as high as 25 mm. Plasmid pAD-IRP (pACT2 with a rabbit IRP domain insert) was used as a positive control in conjunction with the RNA expression plasmid pIIIA-IRE, containing the rat ferritin light chain IRE. Numerous negative controls were used to ensure that RNA-independent or protein-independent activation of reporter expression did not occur (e.g. single transformants, pACT2-RdRp co-transformed with pIIIA-IRE). In the case of the Y3H library, colony PCR was employed to obtain sequences of interactors. Colony counts were performed with ImageJ software.
The 2-nitrophenyl β-d-galactopyranoside (ONPG)-based Miller assay was used to quantify interaction strengths of protein-RNA pairs and to confirm that screening results based on expression of HIS3 reporter gene were not spurious. The assays were performed at least in triplicate as earlier described (48) with slight modifications. In brief, 1-ml yeast cultures grown to 0.5–1.0 A600 were harvested and permeabilized in 220 μl of solution containing ∼23% chloroform, 0.03% 2-mercaptoethanol, and 0.01% SDS. 700 μl of 1 mg/ml ONPG was added to the lysate, and the reactions were quenched with 500 μl of 1 m Na2CO3 prior to measurement of A420. The Miller assay formula is (1000 × A420)/(A600 × V × time), where V is the reaction volume and time is in minutes.
In Vitro EMSA
Non-tagged and N-terminally hexahistidine-tagged DENV-2 RdRp (NS5 residues 277–900) was prepared as described earlier in our report (17). Five RdRp mutations that severely impaired SL binding in Y3H were also generated via the QuikChange procedure, but only R770A showed comparable protein expression and solubility levels to the wild type. To avoid misinterpretation that might be caused by a structural alteration in protein, in in vitro assays only the recombinant R770A mutant was pursued. The folding of purified R770A was further assessed by circular dichroism (CD). Purified RdRp was stored in 10% glycerol-containing buffer at −80 °C until use. DENV-2 RNAs, including 5′-UTR (nucleotides 1–159), SLA (nucleotide 1–69), cirSL (nucleotides 10,657–10,723), and linSL (nucleotides 10,646–10,723), were generated by in vitro transcription using T7 RNA polymerase. In the case of biotin labeling, the transcription reaction consisted of 1 part biotinylated UTP (Thermo Scientific catalog no. AM8450, carrying an 11-carbon linker) to 3 parts UTP, which should give a mean of 4 biotin moieties per transcript. Transcripts were purified via gel excision and refolded while in 0.3 m NaOAc, pH 6.0, 0.5 mm EDTA, 5 mm Mg(OAc)2, and 0.1% SDS overnight at 4 °C.
The RdRp-RNA interaction was determined by EMSA. The binding reaction (10 μl) was performed by mixing 37.5 or 50 pmol of RNAs with increasing concentrations of the RdRp protein in a binding buffer consisting of 50 mm Tris-HCl, pH 8.8, 10 mm Mg(OAc)2, 65 mm NH4OAc, 1 mm EDTA, 1 unit of RNase inhibitor, and 5% glycerol at room temperature for 20 min. RdRp-RNA complexes were resolved on native 5% polyacrylamide gels supplemented with 5% glycerol at 4 °C. Gels were stained by ethidium bromide and Coomassie Blue to visualize RNA and protein, respectively. It was noted that the DENV-2 RdRp protein alone scarcely migrated into the native gel due to its relatively large size (73.2 kDa) and high pI (> 8.0).
In Vitro Alpha Assay
AlphaScreen histidine (nickel chelate) detection kit (PerkinElmer Life Sciences) was exploited in this study. 5′-Biotinylated RNAs were purchased from Integrated DNA Technologies (Coralville, IA), including truncated SL (Bi-miniSL, 5′-biotin-ACAGCAUCAUUCCAGGCACAGAACGCCAGAAAAUGGAAUGGUGCUG), truncated SL with deletion in the top loop (Bi-miniSLΔTL, 5′-biotin-ACAGCAUCAUUCCAGGCACGCCAGAAAAUGGAAUGGUGCUG), and truncated SLA (Bi-miniSLA, 5′-biotin-ACUACGUGGACCGACAAAGACAGAUUCUUUGAGGGAGCUAAGCUCAACGUAG). All RNAs were pre-heated at 70 °C for 3 min, and then slowly cooled to room temperature in a folding buffer (20 mm HEPES, pH 7.4, 50 mm NaCl, 2 mm EDTA) before use. All assays were performed in 384-well white-gray Alpha Plates (PerkinElmer Life Sciences; catalog no. 6005350) in 25-μl reactions at room temperature. Initially, assay conditions were extensively optimized by cross-titration between protein and RNA concentrations to yield the maximum Alpha signal and to determine the “hooking zone,” where quenching of the signal is observed due to an excess of the binding partner. 200 nm RdRp and RNA (total 12 μl) were incubated in the reaction mixture containing 25 mm HEPES, pH 7.4, 100 mm NaCl, 2 mm MgCl2, and 0.1% BSA for 30 min. Then the nickel-chelated acceptor beads (30 μg/ml final concentration) were added and incubated in the dark for 30 min, following by addition of the streptavidin donor beads (20 μg/ml final concentration) and a 30-min incubation in the dark before the assay was conducted on an EnSpireTM 2300 Multilabel Plate Reader (PerkinElmer Life Sciences). Equilibrium dissociation constants (Kd) between SL and RdRp were quantified from the saturation binding experiments. The data were then fitted on a nonlinear regression curve by using the one-site binding mode in GraphPad Prism software (San Diego).
The Alpha competition assay was performed in a 25-μl reaction with non-biotinylated RNAs as competitor. 200 nm His-tagged RdRp was allowed to react with RNA competitors in various concentrations ranging from 0.01 to 250 nm at room temperature for 20 min. Bi-miniSL RNA was then added to 10 nm and reacted for 30 min. The signal was developed as mentioned above with 30 μg/ml acceptor and 20 μg/ml donor beads.
Replicon Construction
Full-length DENV-2 16,681 cDNA (10,723 bp) in the pUC19 vector was used for construction of the DENV-2 replicon in this study. The pre-M/E genes were replaced by the YFP gene fused with autoproteolytic FMDV 2A sequence from foot and mouth disease virus, and the natural junction cleavage sites, C/preM and E/NS1, were retained for maintaining the proteolytic processing of polyprotein (49). All DENV replicon mutants in this study were constructed by QuikChange site-directed mutagenesis and confirmed by sequencing.
RNA Transcription, Transfection, and Fluorescence Imaging
DNA templates for the in vitro run-off transcription were generated by PCR with the forward primer, 5′-GAAATTAATACGACTCACTATTAGTTGTTAGTCTACGTGGACCGAC-3′, carrying the T7 promoter sequence (underline), and the reverse primer, 5′-AGAACCTGTTGATTCAACAGCACC-3′, and purified using the QIAquick gel extraction kit (Qiagen). DENV replicon RNAs were synthesized using the RiboMAX large scale RNA production system kit (Promega) with the following optimized protocol: the 50-μl reaction mixtures containing 7.5 mm each GTP, CTP, and UTP, 2 mm ATP, 3 mm m7GpppA cap analog (New England Biolabs), and 5 μg of DNA template were incubated for 4 h at 37 °C. DNA templates were eliminated by DNase I treatment, and subsequently, transcripts were purified using RNeasy mini kit (Qiagen). Transcribed RNA quality was analyzed by gel electrophoresis. 500 ng of replicon RNAs were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions into BHK-21 cells plated in a 24-well plate with 2 × 104 cells/well. After 4 days, cells were fixed, stained with Hoechst 33258, and imaged. At this late stage, the YFP signal is indicative of the level of viral RNA synthesis inside the cell (12).
Bioinformatics
To calculate the probability that known recombination regions coincide with RdRp interaction sites, we first noted that 43 out of 60 interaction points fell within known recombination points. We asked the following question. What is the probability that 43 (or more) interaction points fall within the known recombination regions (that occupy 30.3% of the genome)? We then used the probability mass function for the calculation. If DI joining points are included in the calculation, the probability diminishes to 2.6 × 10−15. If we view clusters, instead of individual sequencing results, there are 13 clusters, 6 of which land in recombination regions, p = 0.17. If DI joining points are considered, p = 0.019.
Li et al. (16) have underlined the sequences where 5′–3′-joining occurred to form DI particles in a previous study. To account for the possibility that joining may have occurred at the 5′ or 3′ ends of an ACA sequence, we extended these joining point sequences by 6 nt (3 nt to each side) and calculated the probability that a specific 3-nt sequence could be found in the extended joining points. Under these parameters, there are 152 frames in which ACA could be found. We then applied the probability mass function to determine the probability that ACA occurs 8 (or more) times in 152 possible frames, with a probability of 0.0156 (1/64) in any particular frame (assuming a G/C ratio of 1.0). Similarly, ACAG is seen 4 times in 124 possible frames with a probability of 0.0039 of appearing in any particular frame. These are conservative estimates, as the calculations assume that, for example, two ACA sequences could be found in a 4-nt sequence (which is not possible).
To determine RdRp regions that were insensitive to mutations introduced via random mutagenesis (Fig. 5A), all mutant sequences from strongly growing yeast were aligned, and only unique mutations were counted and tallied in each alignment column. These tallies were summed over all possible 31 column windows (15 to the left + 1 + 15 to the right of the “center” amino acid) and plotted residue-by-residue. This method is actually a simplification of a protocol previously developed in our laboratory (50). Programs relating to Monte Carlo-based determination of RNA library quality and for calculating the sequence-to-sequence volatility of all N-nucleotide motifs in a database were created in-house.
Author Contributions
S. C. designed the study; K. H., C. T., and M. K. performed the experiments and analyzed the data; P. Y. and T. L. provided materials and advice; S. C., K. H., and C. T. wrote the manuscript, which was commented on by all authors.
Acknowledgments
We thank Prof. Marvin Wickens for kindly providing Y3H plasmids and Sasiprapa Khunchai for sharing Y2H methods and experience. We also thank Nunghatai Sawasdee for managerial assistance.
This work was supported by grants-in-aid for scientific research from Mahidol University, Thailand (to S. C. and P. Y.), a senior research scholar grant from the Thailand Research Fund (to P. Y.), and Mid-Career Grant RSA5680054 from Thailand Research Fund (to S. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- RdRp
- RNA-dependent RNA polymerase
- DENV
- dengue virus
- SLA
- stem-loop A
- Y3H
- yeast three-hybrid
- nt
- nucleotide
- DI
- defective interfering
- HCV
- hepatitis C virus
- SL
- stem-loop
- TL
- top loop
- 3-AT
- 3-aminotriazole
- ONPG
- o-nitrophenyl β-d-galactopyranoside
- cy
- cyclized
- cir
- circular
- lin
- linear
- CS
- cyclization sequence
- WNV
- West Nile virus
- JEV
- Japanese encephalitis virus
- YFV
- yellow fever virus.
References
- 1. Dreher T. W., and Hall T. C. (1988) Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 201, 31–40 [DOI] [PubMed] [Google Scholar]
- 2. Song C., and Simon A. E. (1995) Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254, 6–14 [DOI] [PubMed] [Google Scholar]
- 3. Oh J. W., Sheu G. T., and Lai M. M. (2000) Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275, 17710–17717 [DOI] [PubMed] [Google Scholar]
- 4. Cui T., and Porter A. G. (1995) Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3′-noncoding region of the viral RNA. Nucleic Acids Res. 23, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Filomatori C. V., Lodeiro M. F., Alvarez D. E., Samsa M. M., Pietrasanta L., and Gamarnik A. V. (2006) A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 20, 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Filomatori C. V., Iglesias N. G., Villordo S. M., Alvarez D. E., and Gamarnik A. V. (2011) RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 286, 6929–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hook B., Bernstein D., Zhang B., and Wickens M. (2005) RNA-protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11, 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wurster S. E., and Maher L. J. 3rd. (2010) Selections that optimize RNA display in the yeast three-hybrid system. RNA 16, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sassanfar M., and Szostak J. W. (1993) An RNA motif that binds ATP. Nature 364, 550–553 [DOI] [PubMed] [Google Scholar]
- 10. Dyson M. R., Perera R. L., Shadbolt S. P., Biderman L., Bromek K., Murzina N. V., and McCafferty J. (2008) Identification of soluble protein fragments by gene fragmentation and genetic selection. Nucleic Acids Res. 36, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tilgner M., Deas T. S., and Shi P. Y. (2005) The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 331, 375–386 [DOI] [PubMed] [Google Scholar]
- 12. Manzano M., Reichert E. D., Polo S., Falgout B., Kasprzak W., Shapiro B. A., and Padmanabhan R. (2011) Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 286, 22521–22534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen S. P., Yu M., Jiang T., Deng Y. Q., Qin C. F., Han J. F., and Qin E. D. (2008) Identification of a recombinant dengue virus type 1 with 3 recombination regions in natural populations in Guangdong province, China. Arch. Virol. 153, 1175–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chuang C. K., and Chen W. J. (2009) Experimental evidence that RNA recombination occurs in the Japanese encephalitis virus. Virology 394, 286–297 [DOI] [PubMed] [Google Scholar]
- 16. Li D., Lott W. B., Lowry K., Jones A., Thu H. M., and Aaskov J. (2011) Defective interfering viral particles in acute dengue infections. PloS one 6, e19447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamkaew M., and Chimnaronk S. (2015) Characterization of soluble RNA-dependent RNA polymerase from dengue virus serotype 2: the polyhistidine tag compromises the polymerase activity. Protein Expr. Purif. 112, 43–49 [DOI] [PubMed] [Google Scholar]
- 18. Bernstein D. S., Buter N., Stumpf C., and Wickens M. (2002) Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26, 123–141 [DOI] [PubMed] [Google Scholar]
- 19. D'Agostino V. G., Adami V., and Provenzani A. (2013) A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation. PLoS One 8, e72426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sztuba-Solinska J., Teramoto T., Rausch J. W., Shapiro B. A., Padmanabhan R., and Le Grice S. F. (2013) Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 41, 5075–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yap T. L., Xu T., Chen Y. L., Malet H., Egloff M. P., Canard B., Vasudevan S. G., and Lescar J. (2007) Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81, 4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noble C. G., Lim S. P., Chen Y. L., Liew C. W., Yap L., Lescar J., and Shi P. Y. (2013) Conformational flexibility of the Dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 87, 5291–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brooks A. J., Johansson M., John A. V., Xu Y., Jans D. A., and Vasudevan S. G. (2002) The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin β1 and importin α/β-recognized nuclear localization signals. J. Biol. Chem. 277, 36399–36407 [DOI] [PubMed] [Google Scholar]
- 24. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cook K. B., Kazan H., Zuberi K., Morris Q., and Hughes T. R. (2011) RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 39, D301–D308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Auweter S. D., Oberstrass F. C., and Allain F. H. (2006) Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 34, 4943–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brinton M. A., Fernandez A. V., and Dispoto J. H. (1986) The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153, 113–121 [DOI] [PubMed] [Google Scholar]
- 28. Holden K. L., Stein D. A., Pierson T. C., Ahmed A. A., Clyde K., Iversen P. L., and Harris E. (2006) Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem-loop structure. Virology 344, 439–452 [DOI] [PubMed] [Google Scholar]
- 29. Elghonemy S., Davis W. G., and Brinton M. A. (2005) The majority of the nucleotides in the top loop of the genomic 3′ terminal stem loop structure are cis-acting in a West Nile virus infectious clone. Virology 331, 238–246 [DOI] [PubMed] [Google Scholar]
- 30. Khromykh A. A., Kondratieva N., Sgro J. Y., Palmenberg A., and Westaway E. G. (2003) Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 77, 10623–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen C. J., Kuo M. D., Chien L. J., Hsu S. L., Wang Y. M., and Lin J. H. (1997) RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71, 3466–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khromykh A. A., Meka H., Guyatt K. J., and Westaway E. G. (2001) Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75, 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Villordo S. M., Alvarez D. E., and Gamarnik A. V. (2010) A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 16, 2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hahn C. S., Hahn Y. S., Rice C. M., Lee E., Dalgarno L., Strauss E. G., and Strauss J. H. (1987) Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198, 33–41 [DOI] [PubMed] [Google Scholar]
- 35. Proutski V., Gould E. A., and Holmes E. C. (1997) Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25, 1194–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. You S., and Padmanabhan R. (1999) A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274, 33714–33722 [DOI] [PubMed] [Google Scholar]
- 37. Kao C. C., Yang X., Kline A., Wang Q. M., Barket D., and Heinz B. A. (2000) Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74, 11121–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng J. C., Chang M. F., and Chang S. C. (1999) Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73, 7044–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Friebe P., Boudet J., Simorre J. P., and Bartenschlager R. (2005) Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siegel R. W., Adkins S., and Kao C. C. (1997) Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 94, 11238–11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malet H., Egloff M. P., Selisko B., Butcher R. E., Wright P. J., Roberts M., Gruez A., Sulzenbacher G., Vonrhein C., Bricogne G., Mackenzie J. M., Khromykh A. A., Davidson A. D., and Canard B. (2007) Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 282, 10678–10689 [DOI] [PubMed] [Google Scholar]
- 42. Lu G., and Gong P. (2013) Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 9, e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biswal B. K., Cherney M. M., Wang M., Chan L., Yannopoulos C. G., Bilimoria D., Nicolas O., Bedard J., and James M. N. (2005) Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280, 18202–18210 [DOI] [PubMed] [Google Scholar]
- 44. Choi K. H., Gallei A., Becher P., and Rossmann M. G. (2006) The structure of bovine viral diarrhea virus RNA-dependent RNA polymerase and its amino-terminal domain. Structure 14, 1107–1113 [DOI] [PubMed] [Google Scholar]
- 45. Mosley R. T., Edwards T. E., Murakami E., Lam A. M., Grice R. L., Du J., Sofia M. J., Furman P. A., and Otto M. J. (2012) Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 86, 6503–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karpala A. J., Doran T. J., and Bean A. G. (2005) Immune responses to dsRNA: implications for gene silencing technologies. Immunol. Cell Biol. 83, 211–216 [DOI] [PubMed] [Google Scholar]
- 47. SenGupta D. J., Zhang B., Kraemer B., Pochart P., Fields S., and Wickens M. (1996) A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. U.S.A 93, 8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dolan J. W., and Fields S. (1990) Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 4, 492–502 [DOI] [PubMed] [Google Scholar]
- 49. Alvarez D. E., De Lella Ezcurra A. L., Fucito S., and Gamarnik A. V. (2005) Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339, 200–212 [DOI] [PubMed] [Google Scholar]
- 50. Choksupmanee O., Hodge K., Katzenmeier G., and Chimnaronk S. (2012) Structural platform for the autolytic activity of an intact NS2B-NS3 protease complex from dengue virus. Biochemistry 51, 2840–2851 [DOI] [PubMed] [Google Scholar]