Abstract
Alternatively activated macrophages (AAMs) play essential roles in the promotion of tissue remodeling, vasculogenesis, and tumor progression; however, the detailed mechanisms underlying the activation of AAMs remain largely unknown. Here, by using quantitative proteomic analysis, we identified 62 proteins that were up-regulated in IL-4-induced macrophages. Among these, Caspase-6 was increased significantly. Caspase-6 is important in the apoptotic signaling pathway; however, its role in non-apoptosis is also reported. Here, we first examined the non-apoptotic role of Caspase-6 in the alternative activation of macrophages after administration of IL-4, 4T1 tumor conditional medium, or co-culture with 4T1 cells. Both treatments promoted alternative activation of RAW264.7 cells and primary macrophages, whereas disruption of caspase-6 expression and activity could markedly suppress the biomarker levels of AAMs. Overexpression of Caspase-6 could significantly promote the activation of AAMs. Importantly, we further present evidence that caspase-6 could regulate breast cancer cell invasion by modulating MMP-2 and MMP-9 expression in 4T1 tumor-associated macrophages, as ablation of protein levels or activity of caspase-6 suppressed tumor cell invasion in vitro. In conclusion, the observed results markedly expanded our views of the dynamic changes in protein composition during alternative activation of macrophages, and they revealed a critical new role of caspase-6 in regulating this cellular biological process, which suggested that caspase-6 might be a key nod molecule to regulate immunological steady-state and be a therapeutic candidate for tumor immunotherapy.
Keywords: cell signaling, cytokine, immunology, macrophage, toll-like receptor (TLR), tumor immunology
Introduction
It is well established that macrophages play a central role in innate and adaptive immune responses (1). They contribute to the recognition, uptake, and killing of non-self agents and also the initiation and progression of the adaptive immune response (2). It has been well documented that during acute inflammation the macrophages are stimulated by toll-like receptor ligands (LPS) and IFN-γ, undergo classical M1 activation, and secrete high levels of pro-inflammatory cytokines, reactive nitrogen, and oxygen intermediates. All these factors promote Th1 response, and then enhance microbicidal and tumoricidal activity (as “killer” cells) (3, 4). Interestingly, more and more evidence shows that macrophages not only play a role in the defense mechanism but also are involved in the promotion of tissue remodeling, vasculogenesis, and tumor progression, which are defined as the alternatively activated macrophages (5–7). When stimulated by the Th2 cytokines, such as IL-4 and IL-13, macrophages express an alternative M2 activation state, which is characterized by expressing a higher level of the anti-inflammatory cytokine, such as IL-10, and a lower level of pro-inflammatory cytokines, such as IL-12. Meanwhile, the up-regulation of cell-surface mannose receptor (MRC1/CD206), hemoglobin/haptoglobin scavenger receptor (CD163), and the activity of arginase-1 are identified in M2 macrophage (3–5), which is distinct from LPS and IFN-γ-mediated M1 classical activation.
In addition to functioning in normal tissues, macrophages are also a major cellular component of the tumor microenvironment, where they are commonly termed as tumor-associated macrophages (TAMs).4 Despite the role of macrophages in ingesting pathogens, many reports showed that these TAMs act as a source of systemic and local cues to promote the proliferation, survival, migration, and invasion of cancer cells and support tumor angiogenesis and suppression of antitumor immunity (8–10). This tumor-promoting function of TAMs is consistent with clinical research that discovered a high density of macrophages in many cancer types, which are considered to be related to increased tumor vascularization, metastasis, and a poor prognosis (11, 12). Furthermore, it was proposed that tumor-promoting TAM activation is mainly toward an alternative M2 activation, which underlies their ability to support the growth and angiogenesis of tumors (4, 13, 14). Recently, much progress has been made to illustrate that TAMs are closely associated with tumor proliferation and tumor mobility, but the detailed molecular mechanism is still largely unknown.
Caspase-6, a member of the caspase family, is an effector caspase and is well known for its role in regulating apoptosis by cleaving the nuclear structural protein NuMA (nuclear mitotic apparatus protein) and the lamin A/C proteins and by inducing nuclear shrinkage and fragmentation (15–17). As such, Caspase-6 localizes in the cytoplasm and neurites of Alzheimer disease neuron-induced apoptosis (18), and Caspase-6 is also actively involved in Huntington disease by cleaving the mutated huntingtin protein (19). Caspase-6 is traditionally recognized as an important molecule in programmed cell apoptosis; however, it does not have detrimental role in this process (20). Many studies have indicated that when compared with wild-type cells, caspase-6−/− cells appear to undergo normal apoptosis following exposure to etoposide or staurosporine, and the reason for that is because the disruption of the caspase-6 gene does not affect caspase-3 and -7 expression (21, 22). Furthermore, more evidence revealed that deficiencies in caspase-6 did not result in aberrant expression of other caspases in vivo. This is due to the fact that caspase-8 can compensate developmentally for the absence of caspase-6 in constitutive knock-out animals (23, 24). All the evidence indicated that Caspase-6 is dispensable for the induction of apoptosis. Furthermore, unlike Caspase-3 and Caspase-7, recent studies have suggested an alternative non-apoptotic role for Caspase-6. For instance, by modifying cell cycle entry, Caspase-6 also regulates B cell activation and differentiation into plasma cells (25); additionally, Caspase-6 also plays critical roles in polymorphonuclear neutrophil-driven macrophage activation through the cleavage of IL-1 receptor-associated kinase M (26). All in all, the latest evidence suggests that Caspase-6 indeed has important roles in regulating non-apoptotic cellular events. Despite the important role of Caspase-6 in immunological regulation, there is still no evidence demonstrating whether Caspase-6 plays any role in the alternative activation of macrophages.
In this study, we first employed the proteomics analysis in conjunction with SILAC-based LC-MS/MS (27) to systematically investigate the profile of differentially expressed proteins in IL-4-treated macrophages. This quantitative approach provided a whole picture of the expression patterns of 3317 proteins, among which the expression level of Caspase-6 was dramatically increased in macrophages treated with IL-4. We further demonstrated that Caspase-6 could regulate alternative activation of macrophages and enhance the pro-invasiveness signature of TAMs by regulating the level of anti-inflammatory cytokines and matrix metalloproteinase, such as MMP-2 and MMP-9. Inhibition of Caspase-6 reversed the anti-inflammatory phenotype of M2 and TAMs to some extent and rescued the TAM-induced MMP-2 and MMP-9 increase and tumor cell invasion. Collectively, our findings primarily revealed a new role for Caspase-6 in regulating alternatively activated macrophages and novel molecular events, which suggested that Caspase-6 might serve as a key therapeutic target in future cancer immunotherapy.
Results
Alternative Activation of RAW264.7 Macrophages
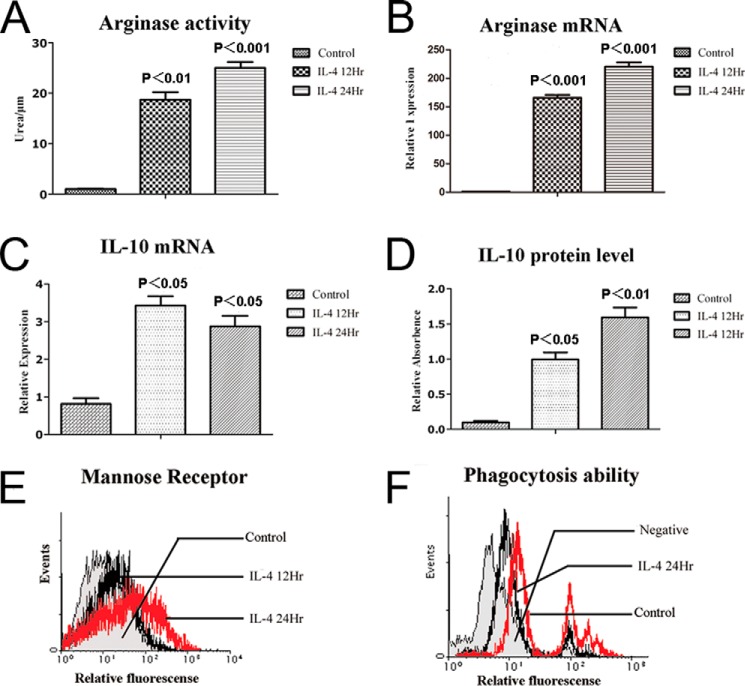
Previous studies have indicated that macrophages undergo alternative M2 activation when stimulated with Th2 cytokines, such as IL-4 and IL-13, in vitro. Importantly, these “alternatively” activated cells initiate the synthesis of arginase-1, an enzyme that suppresses NO generation and promotes them to release ornithine, which is a critical mediator in the wound-healing response (3–5). Indeed, here, after 12 or 24 h of exposure to IL-4 (10 ng/ml), RAW264.7 cells showed significantly elevated activity of arginase-1 (Fig. 1A), as measured by an arginase-1 activity assay kit, suggesting an M2-activated state of RAW264.7 cells. The activation of arginase-1 was further confirmed by quantitative real time-PCR, in which the messenger RNA level of arginase-1 was remarkably up-regulated (Fig. 1B), similar to the elevation level of arginase-1 activity measured by an arginase-1 activity assay kit. In addition, previous studies showed that IL-10 is not only able to stimulate M2c cells activation, but also an important product from M2a (5). Here, we consistently observed that in IL-4-stimulated RAW246.7 cells, both mRNA and protein levels of IL-10 were significantly enhanced (Fig. 1, C and D). Another signature molecule for the M2 state of macrophage is the mannose receptor (also known as CD206), which was also dramatically up-regulated in IL-4-stimulated RAW264.7 cells (Fig. 1E), suggesting a successful induction of M2 macrophages. Furthermore, we also tested the phagocytotic ability of macrophages by flow cytometry analysis, as its M2 macrophages generally have decreased phagocytotic capacity. Results shown in Fig. 1F, confirmed that the phagocytotic ability of RAW246.7 cells was significantly down-regulated after IL-4 stimulation. Taken together, these experimental data of molecular and functional studies indicated that stimulation of RAW264.7 cells with IL-4 was a successful method to acquire alternatively M2-activated macrophages that possess specific phenotypes.
FIGURE 1.
IL-4 induced alternative activation of macrophages. RAW264.7 cells were stimulated with IL-4 (10 ng/ml) for 12 or 24 h, respectively. A, arginase-1 enzymatic activities were measured as described. B and C, expression levels of arginase-1 and IL-10 transcript were measured by quantitative real time PCR. D, protein levels of IL-10 were measured by ELISA. E, protein levels of mannose receptor in the surface of macrophages were detected by FLC. F, phagocytosis abilities of IL-4-stimulated RAW264.7 cells were decreased compared with control, as measured by FLC. All bars were expressed as means ± S.D. (p values of plotted data of ≤0.05 were considered statistically significant).
Quantitative Proteomics Analysis of Alternatively Activated Macrophages
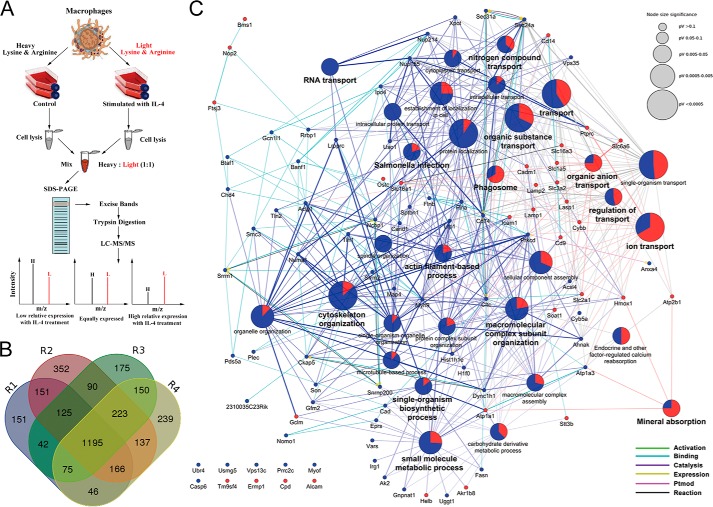
To further exploring the modulated proteins and functional phenotype of the alternatively activated macrophages, quantitative proteomic analysis was performed in conjunction with SILAC-based LC-MS/MS to acquire the relative changes in comprehensive protein expression profile of IL-4-induced alternatively activated macrophages (28, 29). SILAC analysis work flow was displayed in Fig. 2A. To start, macrophages were labeled with heavy isotopes of l-13C6-lysine and l-13C615N4-arginine (Lys-6/Arg-10) or light isotopes of l-12C6-lysine and l-12C6 14N4-arginine (Lys-0/Arg-0), respectively. The Lys-0/Arg-0 labeled (L) cells were treated with IL-4 (10 ng/ml) for 24 h, and the Lys-6/Arg-10-labeled (H) cells were used as control. Following this, identification and quantification of whole cell lysate proteins of the IL-4-induced group or the control group were performed by LC-MS/MS. All final proteomic results were generated based on four independent biological experiments R1, R2, R3, and R4. The combined 96 raw data were searched using the Andromeda search engine with 99% confidence (p < 0.01) (30), after removing pollutants, reverse assignments, and proteins without unique peptides. In total, we identified 3317 proteins in this study, and 2400 proteins (72%) were identified in at least two replicates (Fig. 2B). Only protein ratios (H/L) <0.67 were regarded as up-regulated, and these were marked in blue, and protein ratios (H/L) >1.5 was considered to be down-regulated, and there were marked in red. Finally, we obtained 62 proteins that were up-regulated and 32 proteins that were down-regulated after IL-4 challenge (supplemental Table 1).
FIGURE 2.
Quantitative proteomics analysis of the alternatively M2-activated macrophages. A, schematic outline of the SILAC-based LC-MS/MS experiment. B, in total, we identified 3317 proteins in this study by using four separate biological replicates, and 2400 proteins (72%) were identified in at least two replicates. C, results of GOBP and KEGG enrichment analysis. The up-regulated proteins are blue dots, and the down-regulated proteins are red dots, and the protein-protein interaction was shown as different colored lines. p value concerns significant enrichment of the terms, and p < 0.05 indicated significant differences.
In addition, we further manually annotated the up- and down-regulated proteins into several functional categories. These proteins were categorized using the GO biological process (GOBP) and the KEGG enrichment analysis (31). As demonstrated in Fig. 2C, the up-regulated proteins are shown by blue dots, and the down-regulated proteins are shown by red dots. GOBP and KEGG results indicated that the differentially expressed proteins were predominantly involved in RNA transport, nitrogen compound transport, ion transport, phagosome, cytoskeleton organization, macromolecular complex subunit organization, actin filament-based process, single-organism biosynthetic process, small molecule metabolic process, etc., which provided clues for future research on exploring the detailed mechanisms underlying the activation of AAMs. Importantly, among the changed proteins, the well known key players during macrophage phagocytosis such as CD14, Lamp1, and Lamp2 were significantly down-regulated (supplemental Table 1), which further supports the credibility of the proteomics analysis in this research.
Furthermore, by quantitative RT-PCR, we validated the mRNA levels of the proteins having expression profiles with more significant changes in the up- and down-regulated proteins. As shown in Fig. 3, A and B, among the up-regulated proteins Caspase-6 was remarkably increased. By closely looking into Caspase-6, we found that the change of Caspase-6 was strictly repeatable in three independent biological replicates (Table 1). Thus, we decided to focus on Caspase-6, which was often regarded as a pro-apoptosis executor; however, the role of Caspase-6 in alternatively activated macrophages has never been revealed before.
FIGURE 3.
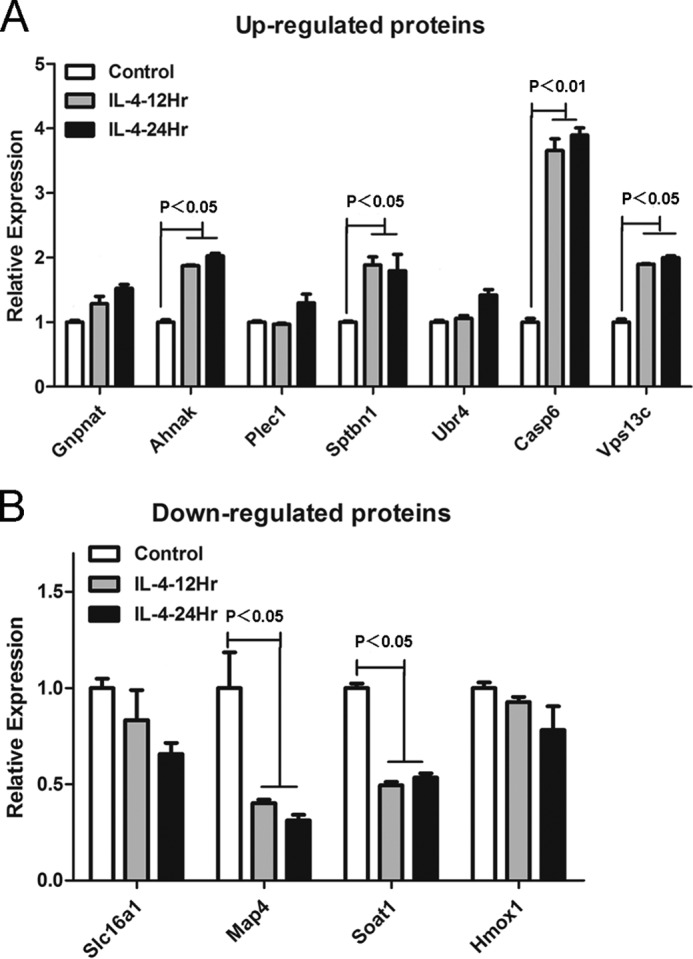
mRNA levels of the selected proteins in the quantitative proteomics results were measured by quantitative real time-PCR. A, up-regulated proteins were detected by quantitative real time-PCR; results showed that the mRNA level of caspase-6 was significantly increased in IL-4-induced macrophages. B, down-regulated proteins were tested by quantitative real time-PCR; results showed that the mRNA levels of map4 and soat1 were significantly decreased in IL-4 induced macrophages. Bars were expressed as means ± S.D. (p values of plotted data of ≤0.05 were considered statistically significant).
TABLE 1.
Quantitative proteomic data of caspase-6
| Protein ID | Protein name | Ratio H/L normalized | Ratio H/L count | |
|---|---|---|---|---|
| Group 1 | IPI00115542 | Caspase-6; apoptotic protease Mch-2; caspase-6 subunit p18; caspase-6 subunit p11 | 0.30312 | 5 |
| Group 2 | 0.34641 | 3 | ||
| Group 3 | 0.37479 | 1 |
Caspase-6 Is Not an Apoptosis Executor in Alternatively M2-activated Macrophages
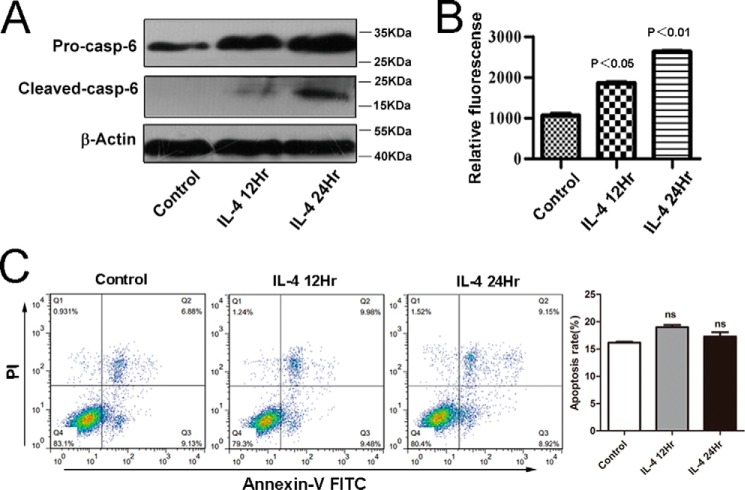
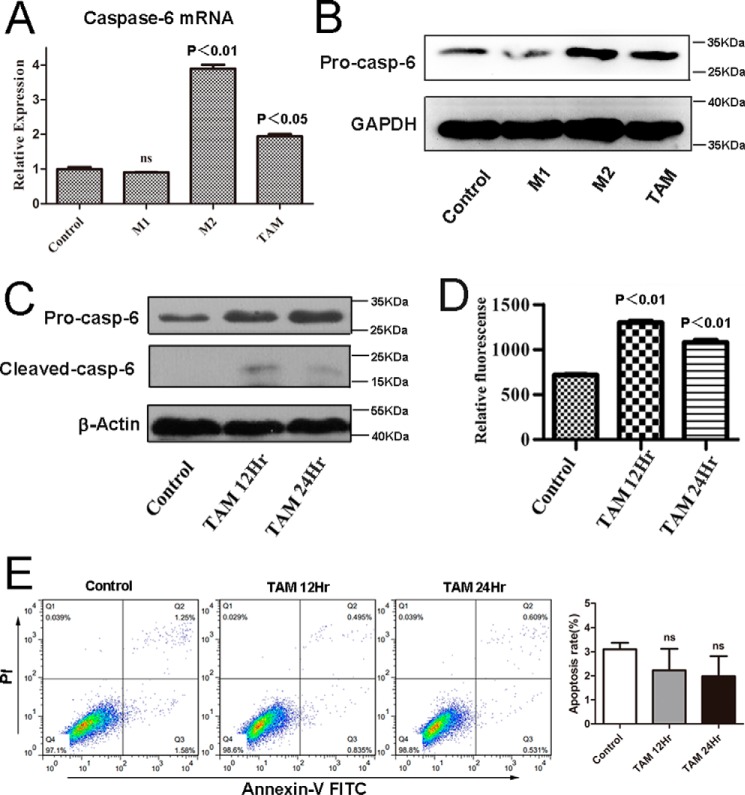
Our quantitative proteomics data had indicated that the protein levels of Caspase-6 were remarkably enriched in IL-4-induced alternatively M2-activated macrophages. To further confirm the proteomics data, we performed immunoblot assays with macrophages in the presence of IL-4. As shown in Fig. 4A, consistent with the proteomics data, the protein levels of Caspase-6 were significantly increased in IL-4-induced macrophages.
FIGURE 4.
Caspase-6 is not an apoptosis executor in alternatively M2-activated macrophages. RAW264.7 cells were stimulated with IL-4 (10 ng/ml) for 12 or 24 h, respectively. A, protein level of pro-Caspase-6 and cleaved Caspase-6 all were up-regulated in IL-4-induced macrophages; the results were detected by Western blotting with the specific antibodies. B, activity of Caspase-6 was measured using an enzymatic activity assay kit. C, apoptosis of M2 was detected by FLC. The percentages of early or late apoptosis are presented in the lower right and upper right quadrants, respectively. Columns represent the average proportions of apoptotic cells. All bars were expressed as means ±S.D. (p values of plotted data of ≤0.05 were considered statistically significant). ns indicates not significantly different.
Previous studies have indicated that Caspase-6 is a cysteine protease, and the functions of Caspase-6 are tightly related to its enzymatic activity (15–17). Hence, we explored whether the enzymatic activity of Caspase-6 is increased in alternatively activated macrophages. Because Caspase-6 was cleaved during its activation process (17), we detected the 18-kDa cleaved form of Caspase-6 by using the appropriate antibody. As revealed in Fig. 4A, we indeed detected the 18-kDa cleaved form of Caspase-6 in alternatively activated macrophages, but not in the control, suggesting Caspase-6 was activated during IL-4-induced alternative macrophage activation. Moreover, to confirm Western blotting results, we further tested the Caspase-6 activity in IL-4-induced alternatively activated macrophages by using a fluorometric assay kit. As expected, in macrophages under the challenge of IL-4, the activity of Caspase-6 increased strikingly (Fig. 4B).
As we showed above that Caspase-6 is cleaved and activated during M2 macrophage activation, we further investigated whether this event leads to apoptosis, the major role of Caspase-6. Interestingly, flow cytometry results indicated that these macrophages did not undergo apoptosis (Fig. 4C), suggesting that caspase-6 may play its non-apoptosis physiological roles in alternatively activated macrophages.
Identification of Caspase-6 as a New Regulator of Alternatively M2-activated Macrophages
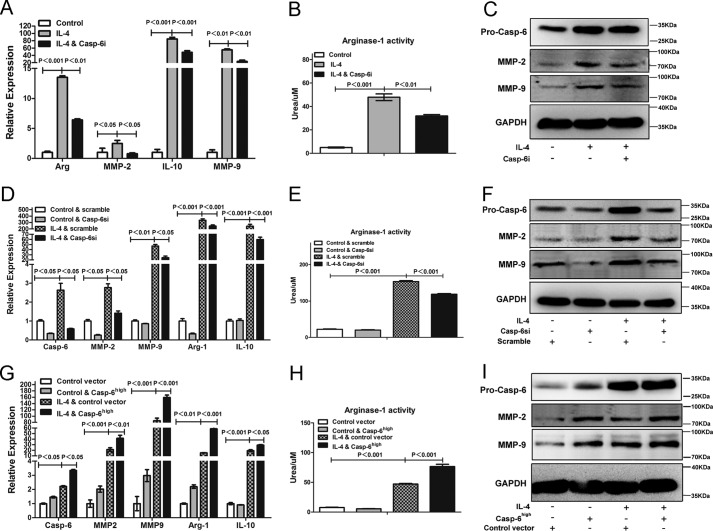
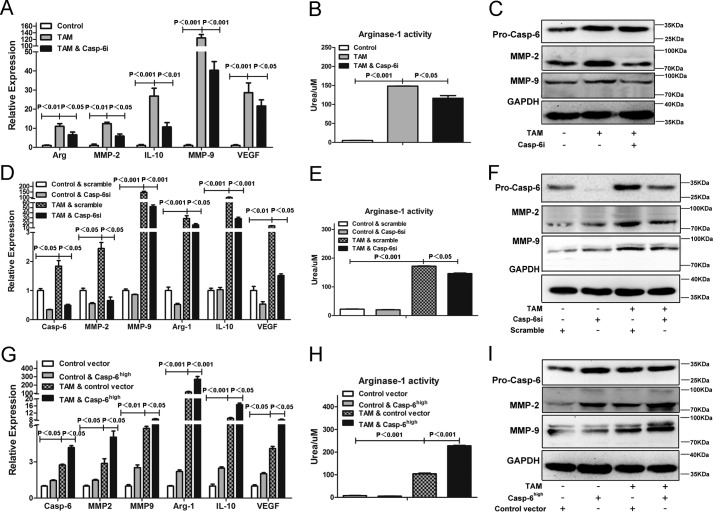
According to the results above, the up-regulated enzymatic activity and expression of caspase-6 induced by IL-4 did not lead to apoptosis of macrophages. The non-apoptosis role of caspase-6 was also reported previously in B lymphocyte cells and macrophages (20, 26). However, it remains largely unknown how caspase-6 plays an important role in regulating the alternative activation of macrophages. To explore the potential function(s) of caspase-6 in mediating alternatively activated macrophages, we utilized Z-VEID-fmk, a caspase-6 enzymatic activity inhibitor (Casp-6i), in combination with IL-4. After co-incubation with IL-4 and Casp-6i, the macrophages displayed a significant reduction in mRNA levels of arginase-1, IL-10, MMP-2, MMP-9 (Fig. 5A), and arginase-1 activity (Fig. 5B) as compared with IL-4 alone treatment, which were anti-inflammatory cytokines expressed by M2 macrophages (3–5). In addition, the protein levels of MMP-2 and MMP-9 were also reduced by Casp-6i (Fig. 5C). Similar results were acquired in the experiments where macrophages were transfected with scramble siRNA or siRNA specifically against caspase-6 (Casp-6si) prior to stimulus of IL-4. Results show that Casp-6si largely blocked the increased mRNA levels of arginase-1, IL-10, MMP-2, MMP-9 (Fig. 5, D and F), and arginase-1 activities (Fig. 5E), which were induced by IL-4, compared with scramble siRNA. In conclusion, our data convincingly supported that suppressing caspase-6, by either Z-VEID-fmk or Casp-6si, distinctly interfered with the activation of M2 macrophages by inhibiting the level of signature cytokines during this process.
FIGURE 5.
Caspase-6 is a regulator during alternative activation of macrophages. A–C, RAW264.7 cells were treated with or without Caspase-6-specific inhibitor (Casp-6i) in the presence of stimulus of IL-4 (10 ng/ml) for 24 h. A, Casp-6i could strikingly reduce the expression of Arginase-1, IL-10, MMP-2, and MMP-9 induced by IL-4. B, Casp-6i inhibited IL-4-induced arginase-1 activity of RAW264.7 cells. C, Western blotting analysis showed that the protein levels of MMP-2 and MMP-9 were significantly inhibited by Casp-6i. D–F, RAW264.7 cells were transfected with scramble siRNA or siRNA against caspase-6 (Casp-6si) prior to stimulus of IL-4 (10 ng/ml) for 24 h. D, Casp-6si could significantly reduce the expression of arginase-1, IL-10, MMP-2, and MMP-9 induced by IL-4, compared with scramble siRNA. E, Casp-6si inhibited IL-4-induced Arginase-1 activity of RAW264.7 cells, compared with scramble siRNA. F, Western blotting analysis showed that the protein levels of MMP-2 and MMP-9 were also inhibited by Casp-6si. G–I, RAW264.7 cells were transfected with control vector or caspase-6 overexpressed vector (Casp-6high) prior to stimulus of IL-4 (10 ng/ml) for 24 h. G, Casp-6high macrophages greatly increased the expression of arginase-1, IL-10, MMP-2, and MMP-9 induced by IL-4, compared with macrophages that transfected with control vector. H, Casp-6high macrophages promoted IL-4-induced Arginase-1 activity of RAW264.7 cells compared with macrophages that transfected with control vector. I, Western blotting analysis showed that the protein levels of MMP-2 and MMP-9 were markedly increased in Casp-6high macrophages. Values were expressed as means ± S.D. (p values of plotted data of ≤0.05 were considered statistically significant).
To further confirming the role of caspase-6 in modulating alternatively activated macrophages, we generated caspase-6 overexpressed macrophages (Casp-6high macrophages) and control vector macrophages by transfecting lentiviral vector particles of casp-6-fugw3 or fugw3-GFP into RAW264.7 cells, respectively. When stimulated with IL-4, Casp-6high macrophages provoked an even higher increase of arginase-1, IL-10, MMP-2, and MMP-9 messenger RNA levels (Fig. 5G) and arginase-1 activity (Fig. 5H), as compared with control vector macrophages that were stimulated with IL-4, suggesting a synergistic effect of IL-4 stimulation and caspase-6 expression. In addition, the protein levels of MMP-2 and MMP-9 were also significantly increased in Casp-6high macrophages (Fig. 5I). Taken together, these data indicated that Caspase-6 was indeed a regulator of alternatively activated macrophages and incapable of inducing apoptosis of M2.
Identification of Caspase-6 as an Essential Regulator of TAMs
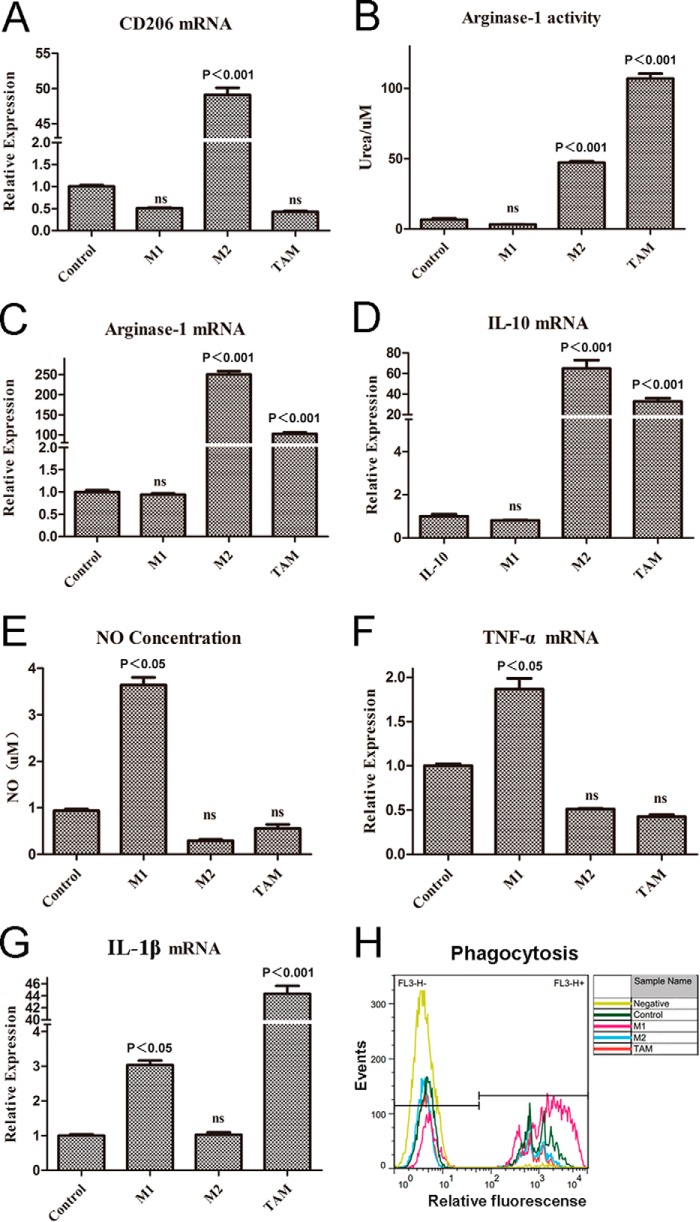
As the evidence emerges, inflammation in the immune system has recently been listed as one of the hallmarks of cancer in Weinberg's seminar cancer review (32). Importantly, TAMs are predominantly polarized toward an M2-like phenotype (4, 13, 14, 33), serving as the key regulators of the link between inflammation and cancer. To investigate this proposition, we established a TAM model in vitro by introducing the tumor-conditioned medium-treated macrophages, a model commonly used to mimic the phenotypes and functions of TAMs in vitro (34, 35). In our study, RAW264.7 cells were challenged with 4T1 tumor-conditioned medium for 24 h. Following this, we compared the phenotype of TAMs with classically activated M1 macrophages in which RAW264.7 cells were stimulated with LPS (100 ng/ml) and IFN-γ (20 ng/ml) for 24 h, and alternatively activated M2 macrophages in which RAW264.7 cells were incubated with IL-4 (10 ng/ml) for 24 h, respectively. Results showed that in TAMs, as compared with control cells, except for CD206 (Fig. 6A), the arginase-1 activity (Fig. 6B) and mRNA levels (Fig. 6C) were up-regulated; the expression of IL-10 was also up-regulated (Fig. 6D). In addition, the amount of nitric oxide release (Fig. 6E) and the level of TNF-α (Fig. 6F) were not altered as compared with control. Another exception is that the expression of IL-1β was significantly increased in TAMs (Fig. 6G), compared with control, which are the important features of M1 macrophages (3, 4, 36). Altogether, the profiles of TAMs were very similar to activated M2 macrophages (IL-4 treated), rather than M1 macrophages (LPS + IFN-γ-treated). Additionally, we also investigated the phagocytosis of macrophages by flow cytometry analysis. As shown in Fig. 6H, the phagocytic ability of M1 was significantly enhanced; however, the phagocytic ability of TAMs was significantly reduced compared with control, but there was no difference with IL-4-treated cells. Taken together, these results indicated that the phenotype of TAMs was extremely similar with IL-4-induced alternatively M2-activated macrophages. Thus, in conclusion, the TAMs model we established here was mainly toward an alternative activation in vitro.
FIGURE 6.
TAMs are mainly toward an alternative M2 activation in vitro. The established macrophage models of M1, M2, and TAMs, RAW264.7 cells were stimulated, respectively, by LPS (100 ng/ml) + IFN-γ (20 ng/ml), IL-4 (10 ng/ml), and 4T1 tumor-conditioned medium for 24 h. A, expression level of CD206 was not changed in TAMs compared with control. Result was measured by quantitative real time PCR. B, for detecting the activity of arginase-1 of the three models, like M2, the activity of arginase-1 in TAMs was up-regulated. C and D, expression levels of arginase-1, IL-10 were measured by quantitative real time-PCR. E, unlike M1, there were no nitric oxide releases in M2 and TAMs. F and G, expression levels of TNF-α and IL-1β were measured by quantitative real time-PCR. H, phagocytosis of M1 was enhanced compared with control, but the phagocytosis of M2 and TAMs was reduced, as measured by flow cytometry analysis. All bars were expressed as means ± S.D. (p values of plotted data ≤ 0.05 were considered statistically significant). ns indicates not significantly different.
As we showed with the non-apoptotic role of caspase-6 in alternatively M2-activated macrophages, we are interested to investigate whether caspase-6 showed a similar function in TAMs. Similar to alternatively M2 activated macrophages, the messenger RNA, and protein levels of pro-Caspase-6 (Fig. 7, A and B), the protein levels of cleaved Caspase-6 (Fig. 7C) and the enzymatic activity of Caspase-6 (Fig. 7D) were all increased markedly in TAMs compared with control. Likewise, as the flow cytometry analysis results showed in Fig. 7E, TAMs did not undergo apoptosis upon induction, similar to M2 macrophages. Collectively, the results we obtained here in TAMs suggested that Caspase-6 might play a similar non-apoptotic role in regulating the activation of TAMs, as it did in alternatively activated macrophages.
FIGURE 7.
Caspase-6 is not an apoptosis executor in TAMs. RAW264.7 cells were stimulated with tumor-conditioned medium for 12 or 24 h, respectively. A, relative expression of caspase-6 was up-regulated in TAMs; results were detected by quantitative real time-PCR. B, protein level of pro-Caspase-6 was up-regulated in M2 and TAMs when compared with control, and results were tested by Western blotting assay. C, protein levels of pro-Caspase-6 and cleaved Caspase-6 all were up-regulated in TAMs; results were detected by Western blotting with the specific antibodies. D, enzymatic activity of Caspase-6 was also measured using an enzymatic activity assay kit. E, apoptosis was detected by FLC. The percentages of early or late apoptosis are presented in the lower right and upper right quadrants, respectively. Columns represent the average proportions of apoptotic cells. All bars were expressed as means ± S.D. (p values of plotted data of ≤0.05 were considered statistically significant).
To further examine the role of Caspase-6 in TAMs, we suppressed the activity of Caspase-6 by Z-VEID-fmk or by inhibiting its expression by siRNA in TAMs. As expected, consistent with the results that were obtained in M2-activated macrophages, the induction of the TAM-responsive genes (arginase-1, IL-10, VEGF, MMP-2, and MMP-9) (Fig. 8, A, C, D, and F) (3, 4, 10, 36) and arginase-1 activity (Fig. 8, B and E) were drastically abolished in TAMs in response to either Z-VEID-fmk or Casp-6si treatment, respectively. Moreover, in an effort to further confirm the role of Caspase-6 in modulating TAMs, caspase-6 overexpressed macrophages were challenged with tumor-conditioned medium. The results in Fig. 8, G–I, compared with parental control, demonstrated that the Casp-6high macrophages showed a synergistic increase of arginase-1, MMP-2, MMP-9, IL-10, VEGF, and arginase-1 activity induced by tumor-conditioned medium. Taken together, these data indicate that Caspase-6 played an essential role in promoting the alternative activation of TAMs in vitro.
FIGURE 8.
Caspase-6 played an essential role in regulating the activation of TAMs. A–C, RAW264.7 cells were treated with or without Z-VEID-fmk (Casp-6i) in the presence of stimulus of 4T1 tumor-conditioned medium for 24 h. A, Casp-6i could strikingly reduce the expression of arginase-1, IL-10, MMP-2, MMP-9, and VEGF produced by TAMs. B, Casp-6i inhibited TAM-induced Arginase-1 activity of RAW264.7 cells. C, Western blotting analysis showed that the protein levels of MMP-2 and MMP-9 were inhibited by Casp-6i. D–F, RAW264.7 cells were transfected with scramble siRNA or siRNA against caspase-6 (Casp-6si) prior to stimulus of tumor-conditioned medium for 24 h. D, Casp-6si could significantly reduce the expression of arginase-1, IL-10, VEGF, MMP-2, and MMP-9 produced by TAMs, compared with scramble siRNA. E, Casp-6si inhibited TAM-induced arginase-1 activity of RAW264.7 cells compared with scramble siRNA. F, Western blotting analysis showed that the protein levels of MMP-2 and MMP-9 were inhibited by Casp-6si. G–I, RAW264.7 cells were transfected with control vector or caspase-6 overexpressed vector (Casp-6high) prior to stimulus of tumor-conditioned medium for 24 h. G, Casp-6 high macrophages greatly enhance the expression of arginase-1, IL-10, MMP-2, MMP-9, and VEGF produced by TAMs, compared with macrophages that transfected with control vector. H, Casp-6 high macrophages promoted TAM-induced arginase-1 activity of RAW264.7 cells compared with macrophages that transfected with control vector. I, Western blotting analysis showed that the protein levels of MMP-2 and MMP-9 were markedly increased in Casp-6 high macrophages. All bars were expressed as means ± S.D. (p values of plotted data of ≤0.05 were considered statistically significant).
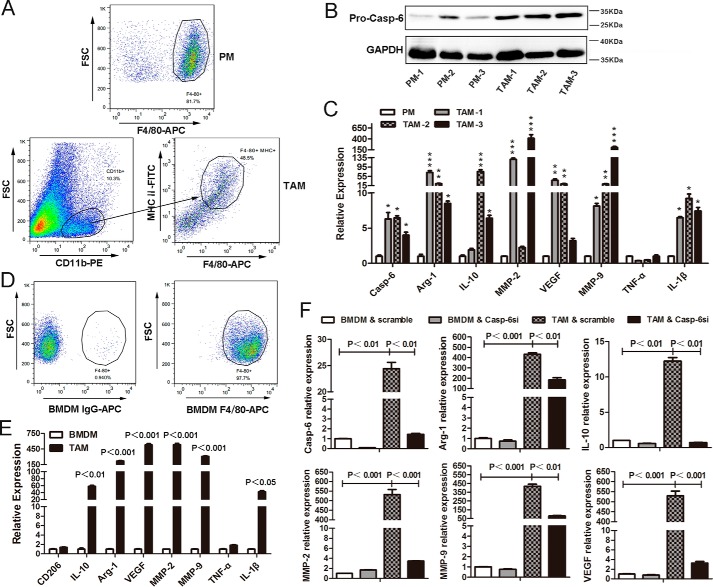
All of the results we obtained above were performed in vitro. However, we are more interested in the polarization of TAMs that infiltrated into the tumor tissues. Therefore, in this study, we isolated primary TAMs (CD11b+, F4/80+, and MHCII+) from tumor tissues of the MMTV-PyMT transgenic breast cancer mice (Fig. 9A). As compared with the peritoneal macrophages (PMs), harvested from the peritoneal cavity and the F4/80+ cells that were more than 80% (Fig. 9A), the messenger RNA and protein levels of pro-caspase-6 were significantly increased in those TAMs (Fig. 9, B and C). In addition, the real time quantitative PCR results also revealed that, in TAMs, except for TNF-α, the gene expressions of arginase-1, IL-10, VEGF, MMP-2, MMP-9, and IL-1β were dramatically up-regulated when compared with PMs (Fig. 9C). Collectively, similar to the results we obtained in vitro, we concluded that caspase-6 was highly enriched in TAMs that were isolated from tumor tissues, and these TAMs were mainly toward an M2-like phenotype.
FIGURE 9.
TAMs are mainly toward an alternative M2 activation in vivo, and caspase-6 plays an essential role in regulating the alternative activation of primary macrophages. A, PMs were harvested from the peritoneal cavity and assessed by FLC with an anti-mouse F4/80-APC antibody; TAMs were isolated from MMTV-PyMT tumor tissues and sorted by FACS with anti-mouse CD11b-PE, F4/80-APC, and MHCII-FITC antibodies. B, Western blotting analysis showed that the protein level of pro-Caspase-6 was significantly up-regulated in primary TAMs, as compared with PMs. C, real time quantitative PCR analysis revealed that the messenger levels of arginase-1, IL-10, VEGF, MMP-2, MMP-9, and IL-1β were markedly increased in TAMs when compared with PMs. D, BMDMs were derived from bone marrow cells that were isolated from FVB/Nj female mice and detected by FLC with an anti-mouse F4/80-APC antibody. E, BMDMs were co-cultured with 4T1 cells for 72 h, and the mRNA levels of arginase-1, IL-10, VEGF, MMP-2, MMP-9, and IL-1β were significantly increased. Results were measured by quantitative real time-PCR. F, BMDMs were transfected with scramble siRNA or siRNA against caspase-6 (Casp-6si) prior to co-culture with 4T1 cells for 72 h. Quantitative real time-PCR results showed that Casp-6si could significantly reduce the expression of arginase-1, IL-10, MMP-2, MMP-9, and VEGF produced by TAMs, compared with scramble siRNA. Bars were expressed as means ± S.D. (p values of plotted data of ≤ 0.05 were considered statistically significant, *, p < 0.05; **, p < 0.01; ***, p < 0.001).
To further examine the role of Caspase-6 in regulating the alternative activation of primary macrophages, we isolated bone marrow cells from FVB/Nj female mice, and we induced the cell maturation and differentiated into bone marrow-derived macrophages (BMDMs) in the presence of mouse M-CSF (20 ng/ml) in vitro. After the treatment of differentiation, flow cytometry results indicated that the F4/80+ cells were more than 90% (Fig. 9D). Based on this, we established a TAMs model in vitro, where BMDMs were co-cultured with 4T1 cells for 72 h to further investigate the polarization of TAMs. Results showed that in TAMs, as compared with untreated BMDMs, except for CD206 and TNF-α, the messenger RNA levels of arginase-1, IL-10, MMP-2, MMP-9, VEGF, and IL-1β were remarkably up-regulated (Fig. 9E). Altogether, the profile of TAMs was quite similar to alternatively activated macrophages. Furthermore, we also performed experiments where the BMDMs were transfected with scramble siRNA or siRNA specifically against caspase-6 (Casp-6si) prior to co-culture with 4T1 cells. Results showed that Casp-6si largely blocked the increased mRNA levels of arginase-1, IL-10, MMP-2, MMP-9, and VEGF (Fig. 9F), compared with scramble siRNA. In conclusion, these data convincingly support that suppressing caspase-6, by Casp-6si, distinctly interfered with the alternative activation of BMDMs by inhibiting the level of signature cytokines during this process.
Caspase-6 Enhances Pro-invasive Signature of TAMs by Up-regulating MMP-2 and MMP-9 Production
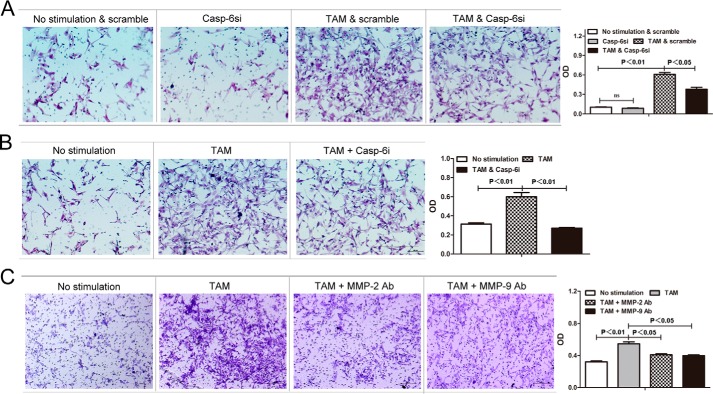
In the tumor microenvironment, the key function of tumor-associated macrophages is to promote tumor cell invasion and migration (8–10, 37–39). MMP-2 and MMP-9 are enzymes that belong to the matrix metalloproteinase family that is involved in the degradation and remodeling of the extracellular matrix (40, 41) and has been demonstrated to promote tumor invasiveness in various tumor types, such as lung cancer, gastric cancer, mammary carcinoma, esophageal cancer, thyroid cancer, etc. (42, 43). As we discovered above that caspase-6 regulates expression of MMP-2 and MMP-9 that is associated with activation of M2 macrophages and TAMs, we hypothesized here that caspase-6 may play an important role in modulating the pro-invasiveness signature of TAMs through up-regulating MMP-2 and MMP-9 production. To address this, RAW264.7 macrophages were transfected with siRNAs against caspase-6 (Casp-6si) or scramble siRNA, respectively, and then induced cells into TAMs by applying tumor conditional medium, and the tumor cell invasion assay was performed in those cells. As we expected, the decrease of caspase-6 in TAMs led to a significant suppression in the pro-invasion capacity of these cells (Fig. 10A). Additionally, incubation with Z-VEID-fmk significantly reduced the pro-invasion capacity of TAMs, too (Fig. 10B), further confirming the essential role of Caspase-6 in promoting tumor progression activity of TAMs. Finally, to further explore the significance of MMP-2 and MMP-9 in the regulation of TAMs-enhanced tumor invasiveness, we conducted a cell invasion assay where MMP-2 and MMP-9 were neutralized with special antibody (Bioworld Co.), respectively. As expected, the neutralization of MMP-2 and MMP-9 remarkably decreased the pro-tumor cell invasion ability of TAMs (Fig. 10C). Hence, these data again proved that Caspase-6 could up-regulate the pro-invasive capacity of TAMs through increasing MMP-2 and MMP-9 levels. In conclusion, these results clearly revealed functional relevance of Caspase-6 in the promotion of tumor invasion by tumor-associated macrophages.
FIGURE 10.
Caspase-6 enhances pro-invasiveness signature of TAMs by up-regulating production of MMP-2 and MMP-9. RAW264.7 macrophages stimulated with or without 4T1 tumor-conditioned medium for 24 h were subjected to invasion assay. “No stimulation” represents unstimulated macrophages, and “TAM” represents macrophages incubated with 4T1 tumor-conditioned medium. A, normal RAW264.7 cells transfected with scramble siRNA (no stimulation & scramble), normal RAW264.7 cells with small interference RNA (siRNA)-induced caspase-6 silencing (Casp-6si), RAW264.7 cells transfected with scramble siRNA prior to stimulus of tumor conditional medium (TAM & scramble), and RAW264.7 cells transfected with siRNA against caspase-6 prior to stimulus of tumor conditional medium (TAM & Casp-6si) were co-cultured with 4T1 cells and subjected to invasion assay. The results are shown as both qualitative data (microscopic images) and quantitative data (OD) values of resolubilized crystal violet measured at a wavelength of 600 nm. B, normal RAW264.7 cells (No stimulation), TAM, TAM with caspase-6 activity inhibitor (TAM & Casp-6i) were co-cultured with 4T1 cells and subjected to an in vitro invasion test. The results are shown as both microscopic pictures (qualitative data) and optical density (OD). C, to address the significance of MMP-2/MMP-9 in TAM-induced tumor invasion, TAMs were subjected to an in vitro invasion assay with or without the addition of 1:50 MMP-2/MMP-9 antibody in the co-culture medium. Both qualitative (microscopic pictures) and quantitative (OD) results are shown herein. Values are expressed as means ± S.D. (p values of plotted data of ≤0.05 were considered statistically significant).
Discussion
In the immune system, cytokines are the hormonal messengers that are responsible for most of the biological effects. Among them, Th2-type cytokines, including IL4, IL5, and IL13, tend to promote the generation of IgE and eosinophilic responses in atopy, which has more of an anti-inflammatory response and is mostly triggered by extracellular parasitic infections, asthma, and other allergic inflammation (44–46). Major Th2 cytokine, IL4, stimulates the activation of AAMs, which work toward resolution of inflammation and promotion of wound repair and angiogenic activities, and importantly, they are different from classically activated macrophages (47). In addition to its beneficial activities, the AAMs have been implicated in several pathologies, the most prominent of which are allergy and asthma. Such groups of alternatively activated macrophages are widely named as M2 macrophages. Recently, more and more evidence showed the molecular profile of M2 macrophages, including the activation of Jak-Stat6 signaling pathways, up-regulation of arginase-1, CD206, ym1, fizz, IL-10, TGF-β, VEGF, etc. (48). In addition, there are several other cytosolic proteins that are capable of promoting M2 macrophage activation, including the PPAR protein family, such as PPARγ, PPARδ, and PPARγ co-activator-1 β (PGC-1 β) (48–51). Other examples are c-Myc and Klf4, which are able to amplify Stat6 signaling and eventually promote the polarization of M2 macrophages (48, 52, 53). Another aspect involved in the activation of AAMs is the epigenetic alteration. For example, Jmjd3 (Jumonji Domain Containing 3), a histone H3K27 demethylase, regulates the expression of Arginase-1, Cbi313, and Retnla, all of which are signature proteins in M2 macrophages (48, 54, 55). However, the detailed molecular mechanisms underlying activation of AAMs are still largely unknown.
In this study, we established a comprehensive quantitative protein expression profile during M2 macrophage polarization induced by IL-4 by utilizing highly sensitive LC-MS/MS in conjunction with SILAC protein-labeling technique. Proteomic analysis showed that, among the 3317 proteins we obtained, there were 62 up-regulated proteins and 32 down-regulated proteins (supplemental Table 1). Then, we discovered Caspase-6, which was up-regulated significantly, and the increasing trend of caspase-6 expression was highly reproducible during four experiments. Thus, our following research focused on Caspase-6. Further investigation showed that the enzymatic activity of Caspase-6 was largely induced at M2 macrophages that were stimulated by IL-4 in vitro. Interestingly, such an increase does not induce apoptosis of M2 macrophages (Fig. 4). Previous existing reporters have shown such non-apoptotic roles of Caspase-6. Caspase-6 regulates the transition of B cells from G0 to G1 phases, which is essential in maturation of B cells and B cell differentiation to plasma cells (20). Caspase-6 is responsible for the cleavage of IL-1 receptor-associated kinase M, mediating polymorphonuclear neutrophil-induced activation of macrophages (26). Given the above evidence, we postulated that caspase-6 might play a non-apoptotic role in regulating the activation of AAMs. Our research showed that inhibiting caspase-6 by either Z-VEID-fmk, the specific caspase-6 inhibitor, or siRNA against caspase-6 could dramatically suppress the expression/activation of arginase-1 and IL-10, the signature molecules for IL-4-induced AAMs (Fig. 5). Furthermore, overexpression of caspase-6 significantly enhanced the levels/activation of Arginase-1 (Fig. 5), suggesting indeed Caspase-6 played a key role in regulating alternative activation of macrophages. This is a new evidence for non-apoptotic role of Caspase-6.
In nature, there are some atypical macrophages that share some pathological features with Th2, and also they showed similar phenotypes with IL-4/IL-13-induced M2 macrophages, such as TAMs, which also are named as AAMs but do not possess all the features of AAMs (4, 33, 46, 56, 57). In our current research, we obtained a similar conclusion. In TAMs, the expression level and activation of Arginase-1 was highly up-regulated, and the expression levels of IL-10 and IL-1β were enhanced; however, the expression of CD206 was not altered (Fig. 6), suggesting mannose metabolism might be sustained during the activation of TAMs, and the detailed mechanism needs to be further investigated. These results demonstrated an atypical phenotype of TAM, similar to M2 macrophage, but not identical. TAMs, existing in tumor microenvironment, have been shown to play a major role in promoting tumor growth, angiogenesis, immunosuppression, tumor invasion, and migration, etc. (8–10, 37–39, 58, 59). With more and more anti-tumor drugs being developed for clinical application, the roles of TAMs have been studied and showed completely distinct effects toward various drugs, in a case/tumor-dependent manner (59). For instant, doxorubicin and daunomycin can significantly enhance the cytotoxicity of TAMs/macrophages, exhibiting an anti-leukemia efficacy (60); in another case, docetaxel promoted the expansion of cytotoxic M1-like macrophages/TAMs, which further elevated specific T cell responses toward breast tumor cells (61). More drugs, including paclitaxel, gemcitabine, and 5-fluorouracil, can remarkably increase the expression of cathepsin-B/S, a family of proteolytic enzymes expressed on mononuclear macrophages, leading to enhanced tumor infiltration and increased tumor resistance (62). Therefore, effectively targeting TAMs has become a novel approach to increase the anti-tumor efficacy based on current single or combined anti-tumor therapies. For example, DeNardo et al. (63) blocked the migration of TAMs with colony-stimulating factor 1 receptor (CSF1R) signaling antagonists and combined with paclitaxel greatly suppressed primary tumor development and reduced pulmonary metastasis in the murine model. Rolny et al. (64) targeted the polarization of TAMs with histidine-rich glycoprotein by promoting antitumor immune responses and vessel normalization, while improving chemotherapy. Based on the evidence and our above research, we further examined whether and how caspase-6 can regulate the activation of TAMs to further modulate tumor progression.
Our studies here showed that in TAMs, similar to the role in M2, Caspase-6 was also able to regulate the expression levels of the signature molecules of AAMs. More interestingly, Caspase-6 showed a tight association with the expression levels of MMP-2 and MMP-9, which are secreted by TAMs and play important role in tumor infiltration and angiogenesis development (65). Either caspase inhibitor or caspase-6 siRNA can significantly down-regulate the levels of MMP-2 and MMP-9, whereas overexpression of caspase-6 could up-regulate the expression of MMP-2 and MMP-9 (Fig. 8). Thus, we hypothesized that Caspase-6 might have possible effects on the ability of tumor infiltration in TAMs by regulating the expression of MMP-2 and MMP-9. More important, the primary TAMs derived from the breast cancer model (MMTV-PyMT) showed a significant M2 polarization phenotype. Furthermore, in vivo, these activated macrophages in the tumor micro-environment are Caspase-6-dependent, too (Fig. 9). In vitro invasion assays showed that indeed by inhibiting Caspase-6 with either Caspase-6 inhibitor or caspase-6 siRNA suppressed the tumor pro-invasion capacity of TAMs (Fig. 10). Similarly, an inhibitory result was obtained in the medium containing specific neutralizing antibodies that can block MMP-2 or MMP-9 (Fig. 10). Taken together, these results convincingly demonstrated that Caspase-6 could regulate the pro-invasion ability of TAMs through activation of MMP-2 and MMP-9; however, the detailed mechanism needs to be further investigated.
In summary, the data described in this study indicated an essential role of caspase-6 in activation of alternatively activated macrophages; additionally, caspase-6 also controls the tumor pro-invasion capacity of TAMs by regulating the expression of MMP-2 and MMP-9. Given the fact that macrophages affect many physiological or pathological processes and the important role of TAMs in progressing tumors, the current research shed lights on the utilization of Caspase-6 as a potential drug target in clinical application, particularly in developing and improving the efficacy of immunotherapies in cancer treatment. Evidently, every unique clinical approach should fully consider the biological variation on a specific tumor type and individual patient basis to design effective personalized therapies.
Experimental Procedures
Cell Culture and Treatments
RAW264.7 cells, 4T1 cells, and 293T cells (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 units/ml), respectively, in a humidified atmosphere with 5% CO2 at 37 °C. Alternatively, M2-activated macrophages were obtained by stimulating with IL-4 (10 ng/ml) for 24 h. Classically activated macrophages (M1) were obtained by treatment with LPS (100 ng/ml) and IFN-γ (20 ng/ml) for 24 h. TAMs were obtained by stimulation with tumor conditional medium (the supernatant of the 4T1 cells) for 24 h.
Reagents
DMEM and fetal bovine serums (FBS) were supplied by Gibco. Opti-MEM medium was purchased from Invitrogen. Custom DMEM (lacking arginine and lysine), dialyzed fetal calf serum, and stable isotope-containing amino acids l-13C6-lysine and l-13C615N4-arginine were purchased from Pierce. Sequencing grade porcine trypsin was purchased from Promega Corp. (Madison, WI). Solvents for liquid chromatography were purchased as follows: HPLC-grade acetonitrile (Fisher) and formic acid (Sigma). Caspase-6 siRNA (sc-72803) was purchased from Santa Cruz Biotechnology. Caspase-6 inhibitor (Z-VEID-fmk) was purchased from Biovision. Other chemicals were supplied by Sigma, unless otherwise stated. Mouse IL-4, IFN-γ, and M-CSF were purchased from PeproTech.
Animals
All mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and were maintained in a specific pathogen-free facility. Friend virus B-type (FVB)/Nj female mice were crossed with PyMT-positive FVB males. The offspring were genotyped by PCR on a Bio-Rad My-Cycler using the following primers (forward primer, 5′-AACCCGAGTTCTCCAACAG-3′, and reverse primer, 5′-TCAGCAACACAAGGATTTC-3′) to identify MMTV-PyMT-positive female mice. The spontaneously developed mammary tumors in MMTV-PyMT-positive female mice were used in the subsequent experiments. Animal welfare and experimental procedures were carried out in strict compliance with the “Guide for the Care and Use of Laboratory Animals” and the related ethical regulations of Nanjing University.
Preparation of TAMs and PMs
Tumor tissues from sacrificed mice were prepared by mechanical disruption, and the pieces of tumor tissues were incubated in Hanks' buffer with 500 μg/ml collagenase type I and 75 μg/ml DNase I (Sigma) for 40 min at 37 °C with periodic vortexing. Digested tissues were filtered through a 70-μm cell strainer, layered in a 44 and 66% Percoll gradient (Sunshine Bio, China), and centrifuged at 2000 rpm for 25 min without a break. Cells at the interface were collected and analyzed by flow cytometry. Anti-mouse CD11b-PE (eBioscience), anti-mouse MHCII-FITC (eBioscience), and anti-mouse F4/80-APC (eBioscience) antibodies were incubated with the collected cells for 45 min at 4 °C, and CD11b+, F4/80+, and MHCII+ TAMs were sorted by FACS (66).
Peritoneal macrophages were harvested by peritoneal lavage. 5 ml of ice-cold PBS was injected into the peritoneal cavity and extracted after gentle agitation. The peritoneal cell suspension was centrifuged at 1200 rpm for 5 min. After centrifugation, cells were then resuspended in DMEM-complete, and a total of 2 × 107 macrophages were seeded in 100-mm culture dishes in a final volume of 10 ml. After 1 h of incubation, no adherent cells were washed away. Monolayers were assessed by flow cytometry analysis with an anti-mouse F4/80-APC antibody (67).
Preparation of BMDMs
FVB/Nj female mice were killed by cervical dislocation and disinfected with 70% ethanol. Both hind legs were excised, and the femur and tibia were separated by cutting at the knee joint, scraping them clean with a scalpel, and placing into fresh 1× sterile PBS. Both ends of bone were cut, exposing marrow, and we slowly flushed out the bone marrow with 1× sterile PBS using a 1-ml syringe and a 25-gauge needle. We then pipetted the bone marrow cells up and down and passed these cells through a 7-μm nylon cell strainer to bring the cells into single cell suspension, then centrifuged at 1200 rpm for 5 min, and aspirated the suspension. Next, we added 2 ml of ACK lysing buffer and incubated for 5 min, then added 1× sterile PBS to stop the lysis reaction and centrifuged at 1200 rpm for 5 min, aspirated the suspension, and resuspended the bone marrow cells with complete DMEM (with 10% FBS and 100 units/ml penicillin and streptomycin). The bone marrow cells were counted using a hemocytometer, and the cells were plated at 1 × 106 cells/well in a sterile 6-well cell culture plate in 2 ml of complete DMEM. To induce the bone marrow cells differentiated into BMDMs, the bone marrow cells were needed to culture for 6–7 days in the present of M-CSF (20 ng/ml) in a humidified atmosphere with 5% CO2 at 37 °C (67).
In Vitro Stimulation of BMDMs
For transwell co-culture with 4T1 tumor cells, we added the transwell insert to the 6-well plates with BMDMs placed in the bottom, and we added 2 × 105 4T1 cells in the top; meanwhile the untreated BMDMs served as control group. After 72 h of incubation, the BMDMs were collected and used for the subsequent research.
RNA Isolation and Quantitative Real Time PCR
Total RNA was isolated from macrophages with TRIzol reagent (Invitrogen), and the protocol was followed according to the manufacturer's instructions, and then RNA was quantified with Nanodrop. First strand cDNA was synthesized from 1.5 μg of total RNA using the Transgene first strand cDNA kit. The primer sequences of arginase-1, CD206, IL-1β, IL-10, TNF-α, IFN-γ, MMP-2, MMP-9, VEGF, caspase-6, gnpnat1, ahnak, plec1, Sptbn1, ubr4, vps13c, slc16a1, map4, hmox1, and soat1 were synthesized by GenScript (Table 2). Quantitative-PCR assays were carried out on the CFX96TM real time PCR detection system (Bio-Rad), using the quantitative-PCR kit (Roche Applied Science). The comparative threshold method for relative quantification was used, and the results are expressed as fold change. The expression of the measured genes in each sample was normalized to actin expression.
TABLE 2.
Primers used in real time quantitative PCR
| Gene name | Accession no. | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) |
|---|---|---|---|
| Actin | NM_007393.5 | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| arginase-1 | NM_007482.3 | CTCCAAGCCAAAGTCCTTAGAG | AGGAGCTGTCATTAGGGACATC |
| IL-10 | NM_010548.2 | GCTCTTACTGACTGGCATGAG | CGCAGCTCTAGGAGCATGTG |
| CD206 | NM_008625.2 | CTCTGTTCAGCTATTGGACGC | CGGAATTTCTGGGATTCAGCTTC |
| caspase-6 | NM_009811.4 | GGAAGTGTTCGATCCAGCCG | GGAGGGTCAGGTGCCAAAAG |
| MMP-2 | NM_008610.3 | CAAGTTCCCCGGCGATGTC | TTCTGGTCAAGGTCACCTGTC |
| MMP-9 | NM_013599.4 | CTGGACAGCCAGACACTAAAG | CTCGCGGCAAGTCTTCAGAG |
| VEGF | NM_001025250.3 | GCACATAGAGAGAATGAGCTTCC | CTCCGCTCTGAACAAGGCT |
| TNF-α | NM_001278601.1 | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| IL-1β | NM_008361.4 | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| gnpnat1 | NM_019425.2 | ATGAAACCCGATGAAACTCCC | GCCTCAAAACCAAGCCTTCTC |
| ahnak | NM_001039959.2 | GGTCCCAAATTCAGGATGCCT | GGGCCTTCAAAATCCAGACCT |
| plec1 | NM_011117.2 | GGCTGAAGGTAGTTCCAATGG | GTGCCTCTGAGCCTTGATAAG |
| sptbn1 | NM_175836.2 | CCACCTTGCGAGAGTGTCC | GTCCCTTAGTGGGTTTAGGCA |
| ubr4 | NM_001160319.1 | GGGACGCCACCTTCTAACAG | TTCAGAGTGTTCGTCTCCAGC |
| vps13c | NM_177184.3 | GAAGCTAAAGTAAAAGCCCACGA | ACACATCAGAGGTGTTGACAATG |
| slc16a1 | NM_009196.4 | TGTTAGTCGGAGCCTTCATTTC | CACTGGTCGTTGCACTGAATA |
| map4 | NM_001205330.1 | CAGTCTTGTGGATGCGTTGAC | TTCCCGGTTTTCTCATCACCA |
| hmox1 | NM_010442.2 | AAGCCGAGAATGCTGAGTTCA | GCCGTGTAGATATGGTACAAGGA |
| soat1 | NM_009230.3 | GAAGGCTCACTCATTTGTCAGA | GTCTCGGTAAATAAGTGTAGGCG |
Arginase-1 Activity Assay
After exposure to treatments, cells in 12-well cell culture plates were harvested and lysed using a lysis buffer (50 mm Tris-HCl, 0.1 mm EDTA, and EGTA, pH 7.5, containing protease inhibitors). The lysate was centrifuged at 14,000 × g (10 min, 4 °C), and we then collected the supernatant for assaying the arginase-1 activity. 25 μl of this lysate were taken and mixed with 20 μl of 25 mm Tris-HCl, pH 7.5 and 5 μl of 10 mm MnCl2, and the enzyme was activated by heating for 3 min at 56 °C. Then, 25 μl of 0.5 m l-arginine was added and hydrolyzed at 37 °C for 90 min. After the reaction was terminated by 200 μl of H2SO4/H3PO4/H2O, 25 μl of 2-isonitrosopropiophenone was used to react with product urea. Then 200 μl of 95% ethanol was added, followed by the measurement on a spectrophotometer at 540 nm. The optical density values were normalized based on protein concentrations of the lysates.
Flow Cytometry Analysis
RAW264.7 cells were plated in 12-well plastic vessels at a density of 3 × 105 cells per well in DMEM with 10% FBS and treatment with or without IL-4 for 12 and 24 h. The cells were analyzed on a FACSCalibur cytometer using CellQuest software (BD Biosciences). Results were analyzed by FlowJo. The statistics presented are based on 10,000 events gated on the population of interest. The phycoerythrin-conjugated anti-mouse CD206 (M1) and corresponding isotype control were purchased from eBioscience.
Apoptosis was detected by staining cells with FITC-conjugated annexin-V and propidium iodide (KeyGEN, BioTECH) and analysis by flow cytometry. Cells staining negative for annexin-V and propidium iodide were defined as viable cells. Phagocytosis was detected by challenging cells with latex beads (BD Biosciences) for 4 h at 37 °C. Following this, unbound beads were removed by washing three times with cold PBS. Results were analyzed by flow cytometry.
SILAC Labeling and Sample Processing
Cells were grown in two different SILAC media, including either “light” isotopes of l-12C6-lysine and l-12C6 14N4-arginine (Lys-0/Arg-0) or “heavy” isotopes of l-13C6-lysine and l-13C615N4-arginine (Lys-6/Arg-10) (ThermoFisher Scientific) for at least five rounds of cell division. Subsequently, the cells in the “light” condition were stimulated with IL-4 for 24 h, whereas the cells in the “heavy” condition were retained as controls. After stimulating with IL-4 for the indicated times in SILAC medium, cells were harvested and lysed in the buffer containing 7 m urea, 2 m thiourea, 4% w/v CHAPS, 65 mm DTT, 1% (v/v) protease inhibitor mixture (Sigma), and 0.1 volume of the mixture of deoxyribonuclease I (1 mg/ml) and ribonuclease A (0.25 mg/ml) incubated for 30 min on ice. The lysates were dissolved with repeated vortex and ultrasonication, followed by centrifugation at 13,000 × g at 4 °C for 10 min to remove insoluble substances.
The protein concentration was determined by a two-dimensional quantitative kit according to the manufacturer's protocol (GE Healthcare). Proteins of “light group” and “heavy group” were mixed at 1:1 for a total of100 μg and were separated by one-dimensional 12.5% SDS-polyacrylamide gel, and the gels were fixed and stained with Coomassie Brilliant Blue. One gel lane was excised into 32 slices. The slices were then cut into 1-mm3 pieces and destained in 50% acetonitrile with 25 mm ammonium bicarbonate solution, before being dehydrated in 100% acetonitrile and dried. The in-gel proteins were reduced by incubation with 10 mm DTT for 40 min at 56 °C, followed by alkylation with 55 mm iodoacetamide for 80 min in the dark. After washing and dehydrating, the proteins were digested with 8 ng/μl sequencing grade trypsin (Promega) at 37 °C overnight. The peptides were extracted from gel pieces with 0.1% trifluoroacetic acid and 50% acetonitrile for 120 min twice, and the extracts were dried in a vacuum centrifuge (ThermoFisher Scientific).
LC-MS/MS Analysis and Statistical Analysis
Peptides were re-dissolved in 10 μl of 0.2% formic acid and injected into a fused silica emitter via auto-sampler with a commercial C18 reverse phase column, before being eluted with a nano-flow liquid chromatography system (Micro-Tech Scientific). These procedures used 30-min linear gradients from 5 to 30% and 5-min linear gradients from 30 to 45% of acetonitrile in 0.1% formic acid at a constant flow rate of 500 nl/min. Eluted peptides were sprayed into a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific Inc) via a nano-electrospray ion source. Raw MS spectra were processed using the MaxQuant software (version1.4.1.2) that did peak list generation, quantitation, and data filtration and presentation (28, 29). The derived peak lists were searched by the Andromeda search engine against a database combining 50,798 proteins from the Uniprot mouse protein sequence database (updated in April, 2013) supplemented with frequently observed contaminants and concatenated with reversed copies of all sequences. The data were acquired in a data-dependent mode to automatically switch between the full-scan MS (from m/z 300 to 2000) and the MS/MS acquisition. The main parameters were set as follows: the protein modifications were carbamidomethylation (fixed), oxidation (variable), and protein N-terminal acetylation (variable); the enzyme specificity was set to trypsin; the maximum missed cleavages were set to 2; the initial mass deviation of precursor ion and fragment ions were up to 10 ppm, and MS/MS tolerance was 0.5 Da; the minimum required peptide length was set to 7 amino acids; and the maximum false discovery rate of proteins was set at 1%. For protein identification, at least one peptide was required to be considered as unique in the database. The protein ratios were calculated as from the medians of all SILAC-normalized peptide pair ratios that belonged to the peptides in this protein. Statistical analysis of data and t tests were performed with Perseus (version 1.4.1.3) (30).
Bioinformatics Analysis
In the experiment comparing control and alternatively activated macrophages, the quantified proteins were divided into three groups corresponding to the cutoffs of 1.50 and 0.667 of the normalized heavy/light ratios. Fold changes of ≥1.50 were considered as up-regulation, whereas ≤0.667 (reciprocal of 1.50)-fold changes were considered as down-regulation, and values between 0.667 and 1.5 indicated no change. The classification and enrichment analysis of up- and down-regulated proteins for gene ontology biological process (GOBP) and KEGG were performed by Genecodis3 system ( (31), in which the hypergeometric test was employed for enrichment. The categories that were enriched at least in one of the quantiles with a p < 0.05 were filtered for hierarchical clustering.
Lentiviral Plasmids and Macrophage Infection
HEK293T cells were transiently transfected with Fugw3-GFP (control vector), Fugw3-caspase-6 (caspase-6 overexpressed vector) plus RRE, REV, and VsVg envelopes to obtain lentiviral particles using Lipofectamine 2000 (Invitrogen). Culture supernatants were collected after transfection for 72 h, and the obtained lentiviral particles were used to infect RAW264.7 cells.
Small Interfering RNA (siRNA) Transfection
Cells were plated in 6-well plates 12 h before transfection with scramble siRNA or siRNA oligonucleotides against caspase-6 (sc-72803, Santa Cruz Biotechnology). We used Lipofectamine 2000 reagent (Invitrogen) and Opti-MEM according to the manufacturer's recommendations. Treatments were taken 24 h after siRNA transfection.
Western Blotting
RAW264.7 cells were harvested, and proteins were extracted by whole cell lysis or a nuclear protein extraction kit purchased from Beyotime. Protein concentration was determined by BCA reagent from Pierce. Lysates were resolved on 12.5% SDS-polyacrylamide gels, transferred to PVDF membranes (Roche Applied Science), and then probed with primary antibodies. Rabbit anti-pro-caspase-6 (catalog no. 9762) and rabbit anti-cleaved Caspase-6 (catalog no. 9761) were purchased from Cell Signaling Technology. Anti-MMP-2 and MMP-9 were supported by Bioworld Co.; anti-β-actin antibody and anti-GAPDH antibody were purchased from Kangcheng Co. Membranes were then washed in TBST and exposed for 2 h to a 1:3000 dilution of species-specific secondary antibody. Signals were detected by chemiluminescence using Amersham ECL reagents (GE Healthcare).
Cytokine Assays by Specific Enzyme-linked Immunosorbent Assay (ELISA)
IL-10 in supernatants was quantified using standard sandwich ELISAs according to instruction manuals. IL-10 concentrations were expressed in nanograms/ml, as calculated from calibration curves from serial dilutions of murine recombinant standards (eBioscience) in each assay. The sensitivity of both IL-10 assays was 20 pg/ml.
Caspase-6 Activity Assay
After exposure to treatments, cells were harvested and lysed using a lysis buffer (50 mm Tris-HCl, 0.1 mm EDTA, and EGTA, pH 7.5, containing protease inhibitors). The lysate was centrifuged at 14,000 × g (10 min, 4 °C), and we then collected the supernatant for assaying the caspase-6 activity with the Caspase-6 activity assay kit (Beyotime Biotechnology). We took 50 μl of this lysate and added 50 μl of 2× reaction buffer (pre-added 10 mm DTT) and 5 μl of 1 mm VEID-AFC and incubated for 2 h at 37 °C. Then, the fluorescence intensity was measured at excitation/emission (400 nm/505 nm) in a macroplate spectrophotometer. The optical density values were normalized based on protein concentrations of the lysates.
Induced NOS Activity Assay
The activity of iNOS was measured using 105 cells by the nitric oxide synthase assay kit purchased from Beyotime with the employment of the iNOS selective inhibitor 1400W also provided by Beyotime.
Cell Invasion Assay
The ability of breast cancer cells (4T1) to migrate through Matrigel-coated filters was measured using insert cell culture chambers (NUNC) with polycarbonate membranes (8.0-μm pore size, Millipore) coated with 100 μl of v/v 10% Matrigel (BD Biosciences) on the top side of the membrane. The upper surface of the matrix was challenged with 20,000 4T1 cells, and cells were kept in serum-free medium containing 0.1% BSA. The lower chamber was planted macrophages and contained medium supplemented with 10% serum in the presence or absence of Z-VEID-fmk or MMP-2/MMP-9 antibodies. After 24 h, the cells were fixed and stained with 0.1% crystal violet solution. Cells and Matrigel on the upper surface of the membrane were removed carefully with a cotton swab. After a phosphate-buffered saline rinse, three times, microscopic images were taken as qualitative results, or the crystal violet on these cells was resolubilized into 10% acetic acid for absorption spectroscopic analysis at 600 nm to generate quantitative data.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5.0 software. Error bars indicated the standard deviation (S.D.) around the average data point, unless otherwise indicated. Differences were analyzed by using the non-parametric Mann-Whitney analysis (two-tailed). p values of plotted data ≤ 0.05 were considered statistically significant.
Author Contributions
P. S. supervised the project. P. S., Y. Y., and Q. S. designed the experiments, analyzed the data, and wrote the manuscript. Y. Y., Q. W., and J. J. performed the proteome experiments and data analysis. Y. Y., B. C., X. L., and L. L. performed all other experiments. Y. Y., B. C., and Y. H. analyzed the data. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Edward Kalmykov (Dept. of Biochemistry, University of Texas Health Science at San Antonio) for editing the language of this manuscript. We also thank Dr. Yan Lu (Nanjing University, China) for the scientific suggestions on this project.
This work was supported by National Natural Science Foundation of China Projects 81473220, 81273527, and 81421091 and by National Key Research and Development Program no. 2016YFA0500301. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- TAM
- tumor associated macrophage
- AAM
- alternatively activated macrophage; M
- SILAC
- stable isotope labeling with amino acids in cell culture
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- BMDM
- bone marrow-derived macrophage
- L
- light
- H
- heavy
- iNOS
- induced nitric-oxide synthase
- PPAR
- peroxisome proliferator-activated receptor
- GOBP
- GO biological process
- PM
- peritoneal macrophage
- MMTV
- murine mammary tumor virus.
References
- 1. Ginhoux F., and Jung S. (2014) Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 [DOI] [PubMed] [Google Scholar]
- 2. Murray P. J., and Wynn T. A. (2011) Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mills C. D., Kincaid K., Alt J. M., Heilman M. J., and Hill A. M. (2000) M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164, 6166–617310843666 [Google Scholar]
- 4. Mantovani A., Sozzani S., Locati M., Allavena P., and Sica A. (2002) Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 [DOI] [PubMed] [Google Scholar]
- 5. Gordon S., and Martinez F. O. (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 [DOI] [PubMed] [Google Scholar]
- 6. Nucera S., Biziato D., and De Palma M. (2011) The interplay between macrophages and angiogenesis in development, tissue injury and regeneration. Int. J. Dev. Biol. 55, 495–503 [DOI] [PubMed] [Google Scholar]
- 7. Mantovani A., Biswas S. K., Galdiero M. R., Sica A., and Locati M. (2013) Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 229, 176–185 [DOI] [PubMed] [Google Scholar]
- 8. Qian B.-Z., and Pollard J. W. (2010) Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Squadrito M. L., and De Palma M. (2011) Macrophage regulation of tumor angiogenesis: implications for cancer therapy. Mol. Aspects Med. 32, 123–145 [DOI] [PubMed] [Google Scholar]
- 10. Obeid E., Nanda R., Fu Y. X., and Olopade O. I. (2013) The role of tumor-associated macrophages in breast cancer progression (review). Int. J. Oncol. 43, 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bingle L., Brown N. J., and Lewis C. E. (2002) The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196, 254–265 [DOI] [PubMed] [Google Scholar]
- 12. Clear A. J., Lee A. M., Calaminici M., Ramsay A. G., Morris K. J., Hallam S., Kelly G., Macdougall F., Lister T. A., and Gribben J. G. (2010) Increased angiogenic sprouting in poor prognosis FL is associated with elevated numbers of CD163+ macrophages within the immediate sprouting microenvironment. Blood 115, 5053–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heusinkveld M., and van der Burg S. H. (2011) Identification and manipulation of tumor associated macrophages in human cancers. J. Transl. Med. 9, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laoui D., Movahedi K., Van Overmeire E., Van den Bossche J., Schouppe E., Mommer C., Nikolaou A., Morias Y., De Baetselier P., and Van Ginderachter J. A. (2011) Tumor-associated macrophages in breast cancer: distinct subsets, distinct functions. Int. J. Dev. Biol. 55, 861–867 [DOI] [PubMed] [Google Scholar]
- 15. Orth K., Chinnaiyan A. M., Garg M., Froelich C. J., and Dixit V. M. (1996) The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J. Biol. Chem. 271, 16443–16446 [PubMed] [Google Scholar]
- 16. Hirata H., Takahashi A., Kobayashi S., Yonehara S., Sawai H., Okazaki T., Yamamoto K., and Sasada M. (1998) Caspases are activated in a branched protease cascade and control distinct downstream processes in Fas-induced apoptosis. J. Exp. Med. 187, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J., and Yuan J. (2008) Caspases in apoptosis and beyond. Oncogene 27, 6194–6206 [DOI] [PubMed] [Google Scholar]
- 18. Guo H., Albrecht S., Bourdeau M., Petzke T., Bergeron C., and LeBlanc A. C. (2004) Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am. J. Pathol. 165, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., et al. (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 20. Uribe V., Wong B. K., Graham R. K., Cusack C. L., Skotte N. H., Pouladi M. A., Xie Y., Feinberg K., Ou Y., Ouyang Y., Deng Y., Franciosi S., Bissada N., Spreeuw A., Zhang W., et al. (2012) Rescue from excitotoxicity and axonal degeneration accompanied by age-dependent behavioral and neuroanatomical alterations in caspase-6-deficient mice. Hum. Mol. Genet. 21, 1954–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruchaud S., Korfali N., Villa P., Kottke T. J., Dingwall C., Kaufmann S. H., and Earnshaw W. C. (2002) Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 21, 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rust C., Wild N., Bernt C., Vennegeerts T., Wimmer R., and Beuers U. (2009) Bile acid-induced apoptosis in hepatocytes is caspase-6-dependent. J. Biol. Chem. 284, 2908–2916 [DOI] [PubMed] [Google Scholar]
- 23. Wong B. K., Ehrnhoefer D. E., Graham R. K., Martin D. D., Ladha S., Uribe V., Stanek L. M., Franciosi S., Qiu X., Deng Y., Kovalik V., Zhang W., Pouladi M. A., Shihabuddin L. S., Hayden M. R. (2015) Partial rescue of some features of Huntington disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol. Dis. 76, 24–36 [DOI] [PubMed] [Google Scholar]
- 24. Zheng T. S., Hunot S., Kuida K., Momoi T., Srinivasan A., Nicholson D. W., Lazebnik Y., and Flavell R. A. (2000) Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat. Med. 6, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe C., Shu G. L., Zheng T. S., Flavell R. A., and Clark E. A. (2008) Caspase-6 regulates B cell activation and differentiation into plasma cells. J. Immunol. 181, 6810–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi H., Nolan A., Naveed B., Hoshino Y., Segal L. N., Fujita Y., Rom W. N., and Weiden M. D. (2011) Neutrophils activate alveolar macrophages by producing caspase-6-mediated cleavage of IL-1 receptor-associated kinase-M. J. Immunol. 186, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ong S.-E., and Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., and Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 30. Cox J., and Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carmona-Saez P., Chagoyen M., Tirado F., Carazo J. M., and Pascual-Montano A. (2007) GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 8, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 33. Biswas S. K., Gangi L., Paul S., Schioppa T., Saccani A., Sironi M., Bottazzi B., Doni A., Vincenzo B., Pasqualini F., Vago L., Nebuloni M., Mantovani A., and Sica A. (2006) A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-κB and enhanced IRF-3/STAT1 activation). Blood 107, 2112–2122 [DOI] [PubMed] [Google Scholar]
- 34. Qian X., Zhang J., and Liu J. (2011) Tumor-secreted PGE2 inhibits CCL5 production in activated macrophages through cAMP/PKA signaling pathway. J. Biol. Chem. 286, 2111–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Solinas G., Schiarea S., Liguori M., Fabbri M., Pesce S., Zammataro L., Pasqualini F., Nebuloni M., Chiabrando C., Mantovani A., and Allavena P. (2010) Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J. Immunol. 185, 642–652 [DOI] [PubMed] [Google Scholar]
- 36. Biswas S. K., and Mantovani A. (2010) Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11, 889–896 [DOI] [PubMed] [Google Scholar]
- 37. Bingle L., Lewis C. E., Corke K. P., Reed M. W., and Brown N. J. (2006) Macrophages promote angiogenesis in human breast tumour spheroids in vivo. Br. J. Cancer 94, 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DeNardo D. G., Barreto J. B., Andreu P., Vasquez L., Tawfik D., Kolhatkar N., and Coussens L. M. (2009) CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qian B.-Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L. R., Kaiser E. A., Snyder L. A., and Pollard J. W. (2011) CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang J., and Tsirka S. E. (2005) Neuroprotection by inhibition of matrix metalloproteinases in a mouse model of intracerebral haemorrhage. Brain 128, 1622–1633 [DOI] [PubMed] [Google Scholar]
- 41. Vandooren J., Van den Steen P. E., and Opdenakker G. (2013) Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit. Rev. Biochem. Mol. Biol. 48, 222–272 [DOI] [PubMed] [Google Scholar]
- 42. Farina A. R., and Mackay A. R. (2014) Gelatinase B/MMP-9 in tumour Pathogenesis and Progression. Cancers 6, 240–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kalhori V., and Törnquist K. (2015) MMP2 and MMP9 participate in S1P-induced invasion of follicular ML-1 thyroid cancer cells. Mol. Cell Endocrinol. 404, 113–122 [DOI] [PubMed] [Google Scholar]
- 44. McKenzie A. N., Culpepper J. A., de Waal Malefyt R., Brière F., Punnonen J., Aversa G., Sato A., Dang W., Cocks B. G., and Menon S. (1993) Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc. Natl. Acad. Sci. U.S.A. 90, 3735–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abramson S. L., and Gallin J. (1990) IL-4 inhibits superoxide production by human mononuclear phagocytes. J. Immunol. 144, 625–630 [PubMed] [Google Scholar]
- 46. Martinez F. O., Helming L., and Gordon S. (2009) Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 27, 451–483 [DOI] [PubMed] [Google Scholar]
- 47. Mosser D. M. (2003) The many faces of macrophage activation. J. Leukocyte Biol. 73, 209–212 [DOI] [PubMed] [Google Scholar]
- 48. Van Dyken S. J., and Locksley R. M. (2013) Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu. Rev. Immunol. 31, 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vats D., Mukundan L., Odegaard J. I., Zhang L., Smith K. L., Morel C. R., Wagner R. A., Greaves D. R., Murray P. J., and Chawla A. (2006) Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., and Chawla A. (2007) Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Odegaard J. I., Ricardo-Gonzalez R. R., Red Eagle A., Vats D., Morel C. R., Goforth M. H., Subramanian V., Mukundan L., Ferrante A. W., and Chawla A. (2008) Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metab. 7, 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liao X., Sharma N., Kapadia F., Zhou G., Lu Y., Hong H., Paruchuri K., Mahabeleshwar G. H., Dalmas E., Venteclef N., Flask C. A., Kim J., Doreian B. W., Lu K. Q., Kaestner K. H., et al. (2011) Krüppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 121, 2736–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pello O. M., De Pizzol M., Mirolo M., Soucek L., Zammataro L., Amabile A., Doni A., Nebuloni M., Swigart L. B., Evan G. I., Mantovani A., and Locati M. (2012) Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 119, 411–421 [DOI] [PubMed] [Google Scholar]
- 54. Ishii M., Wen H., Corsa C. A., Liu T., Coelho A. L., Allen R. M., Carson W. F. 4th., Cavassani K. A., Li X., Lukacs N. W., Hogaboam C. M., Dou Y., and Kunkel S. L. (2009) Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., Honma K., Matsuyama T., Yui K., Tsujimura T., Standley D. M., et al. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 [DOI] [PubMed] [Google Scholar]
- 56. Gal A., Tapmeier T. T., and Muschel R. J. (2012) Plasticity of tumor associated macrophages in a metastatic melanoma model in the mouse. Cancer Res. 72, 40222127927 [Google Scholar]
- 57. Wang T., Ge Y., Xiao M., Lopez-Coral A., Azuma R., Somasundaram R., Zhang G., Wei Z., Xu X., Rauscher F. J. 3rd., Herlyn M., and Kaufman R. E. (2012) Melanoma-derived conditioned media efficiently induce the differentiation of monocytes to macrophages that display a highly invasive gene signature. Pigment Cell Melanoma Res. 25, 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lewis C. E., and Pollard J. W. (2006) Distinct role of macrophages in different tumor microenvironments. Cancer Res. 66, 605–612 [DOI] [PubMed] [Google Scholar]
- 59. De Palma M., and Lewis C. E. (2013) Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell 23, 277–286 [DOI] [PubMed] [Google Scholar]
- 60. Mantovani A., Polentarutti N., Luini W., Peri G., and Spreafico F. (1979) Role of host defense mechanisms in the antitumor activity of adriamycin and daunomycin in mice. J. Natl. Cancer Inst. 63, 61–66 [PubMed] [Google Scholar]
- 61. Kodumudi K. N., Woan K., Gilvary D. L., Sahakian E., Wei S., and Djeu J. Y. (2010) A novel chemoimmunomodulating property of docetaxel: suppression of myeloid-derived suppressor cells in tumor bearers. Clin. Cancer Res. 16, 4583–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bruchard M., Mignot G., Derangère V., Chalmin F., Chevriaux A., Végran F., Boireau W., Simon B., Ryffel B., Connat J. L., Kanellopoulos J., Martin F., Rébé C., Apetoh L., and Ghiringhelli F. (2013) Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat. Med. 19, 57–64 [DOI] [PubMed] [Google Scholar]
- 63. DeNardo D. G., Brennan D. J., Rexhepaj E., Ruffell B., Shiao S. L., Madden S. F., Gallagher W. M., Wadhwani N., Keil S. D., Junaid S. A., Rugo H. S., Hwang E. S., Jirström K., West B. L., and Coussens L. M. (2011) Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1, 54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rolny C., Mazzone M., Tugues S., Laoui D., Johansson I., Coulon C., Squadrito M. L., Segura I., Li X., Knevels E., Costa S., Vinckier S., Dresselaer T., Åkerud P., De Mol M., et al. (2011) HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Cancer Cell 19, 31–44 [DOI] [PubMed] [Google Scholar]
- 65. Carmeliet P., and Jain R. K. (2000) Angiogenesis in cancer and other diseases. Nature 407, 249–257 [DOI] [PubMed] [Google Scholar]
- 66. Franklin R. A., Liao W., Sarkar A., Kim M. V., Bivona M. R., Liu K., Pamer E. G., and Li M. O. (2014) The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Colegio O. R., Chu N.-Q., Szabo A. L., Chu T., Rhebergen A. M., Jairam V., Cyrus N., Brokowski C. E., Eisenbarth S. C., Phillips G. M., Cline G. W., Phillips A. J., and Medzhitov R. (2014) Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.