Abstract
Epigenetic systems are well known for the roles they play in regulating the differential expression of the same genome in different cell types. However, epigenetic systems can also directly impact genomic integrity by protecting genetic sequences. Using an experimental evolutionary approach, we studied rates of mutation in the fission yeast Schizosaccharomyces pombe strains that lacked genes encoding several epigenetic regulators or mismatch repair components. We report that loss of a functional mismatch repair pathway in S. pombe resulted in the preferential enrichment of mutations in euchromatin, indicating that the mismatch repair machinery preferentially protected genetic fidelity in euchromatin. This preference is probably determined by differences in the accessibility of chromatin at distinct chromatin regions, which is supported by our observations that chromatin accessibility positively correlated with mutation rates in S. pombe or human cancer samples with deficiencies in mismatch repair. Importantly, such positive correlation was not observed in S. pombe strains or human cancer samples with functional mismatch repair machinery.
Keywords: chromatin, chromatin regulation, DNA repair, heterochromatin, mutagenesis mechanism
Introduction
Epigenetic systems are often considered to be biological systems that function beside and beyond the genome (1). Most studies have focused on the mechanisms by which epigenetic regulators achieve differential gene expression without altering the genetic information contained in the DNA sequence. However, an interesting but much less clear question is whether epigenetic systems can directly impact the fidelity of the genetic system and affect the accumulation of DNA mutations.
Mutations continually arise during cell proliferation, development, and evolution and under pathogenic conditions. The mutation rate is controlled by the rate at which errors occur in the DNA sequence and the rate of DNA repair. Therefore, questions related to the impact of epigenetic systems on genetic sequence fidelity can be broken down into two parts: do epigenetic systems affect the rate at which errors occur in the DNA sequence, and/or do they affect the rate of DNA repair?
Upon the occurrence of a DNA synthesis error and a DNA mismatch pair, genetic fidelity is first safeguarded by the 3′ to 5′ exonuclease activity termed proofreading activity of the DNA polymerase (2). For mismatches that escape the surveillance of DNA polymerase proofreading activity, the mismatch repair system is the primary protection mechanism against the fixation of such a replication error or the occurrence of an acquired mutation during DNA replication (3–5). The mismatch repair pathway plays a crucial function in protecting genetic fidelity, because it is tightly associated with DNA replication and ensures that most DNA sequence errors that occur during DNA replication are repaired (3–9).
Analysis of spontaneous mutation rates in various organisms revealed that rates of spontaneous mutation are highly similar across different families within the same order, but they differ substantially among organisms from different orders (10, 11), suggesting that a given organism displays a relatively constant rate of spontaneous mutation. Nevertheless, regional variation in the accumulation of mutations has been reported in several species (12–17). More recently, it has been reported that nucleosome occupancy may impact the accumulation of mutations (18) and that heterochromatic regions tend to accumulate more mutations (15, 17, 19, 20), suggesting that the epigenetic status of different chromatin regions may impact whether a mutation occurs or whether a mutation is repaired.
To directly investigate the impact of epigenetic regulators on the accumulation of mutations and to understand whether either the occurrence of a mutation or the repair of a mutation is predominantly affected by chromatin status, we took advantage of an experimental evolutionary approach by using several Schizosaccharomyces pombe strains that contain deletions of genes encoding key epigenetic regulators or mismatch repair components. We found that loss of the epigenetic players that we tested led to minor increases in the mutation rate. Interestingly, we discovered that the high genetic fidelity that was observed in euchromatic regions was largely caused by preferential protection by the mismatch repair pathway. Defects in the mismatch repair machinery led to profound changes in the mutation landscape in the genome in both S. pombe and human cancers.
Results
Experimental Evolution in Fission Yeast Strains Containing Deletions of Various Epigenetic Regulators
To investigate whether epigenetic regulators affect the fidelity of genetic information, we chose the fission yeast S. pombe as our starting model system for three reasons: 1) S. pombe grows quickly enough to allow an experimental evolutionary approach and to accumulate enough mutations to be useful in analysis within a reasonable period of time; 2) S. pombe has a small haploid genome that can be easily sequenced at sufficient depth to call mutations; 3) S. pombe has a more complicated epigenetic system than the budding yeast Saccharomyces cerevisiae (21–30), which harbors heterochromatin regions marked by histone H3K93 dimethylation (H3K9me2) (23, 27).
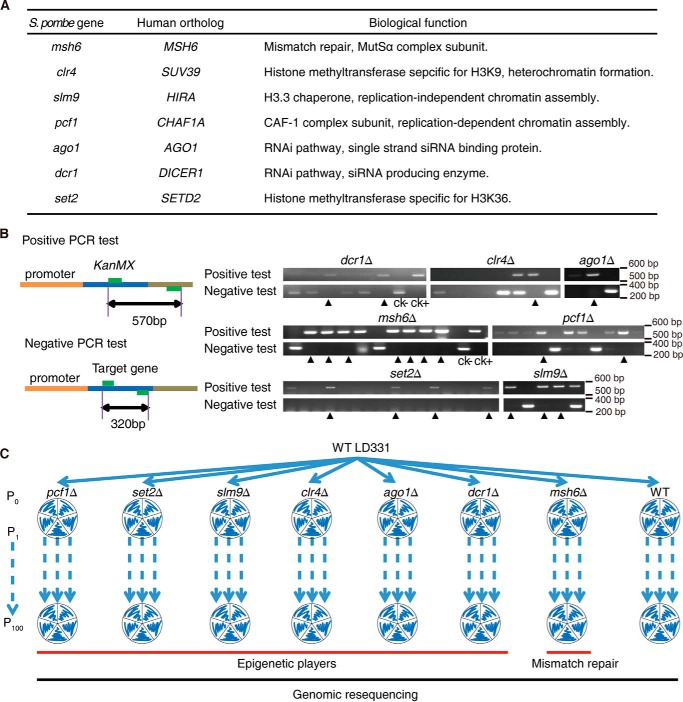
We chose the WT haploid S. pombe strain LD331: h+ as our starting strain and generated KO strains for genes encoding six epigenetic regulators (Clr4, an H3K9-specific histone methyltransferase; Set2, an H3K36-specific histone methyltransferase; Pcf1, a subunit of the chromatin assembly complex CAF1; Slm9, a subunit of the chromatin assembly complex HIRA; Dcr1, an enzyme that generates siRNA and part of the RNAi machinery; Ago1, an siRNA-binding protein and part of the RNAi machinery); and Msh6, a protein that is a key component of the mismatch repair machinery (31) (Fig. 1A). All knock-out strains were verified by PCR (Fig. 1B) and high-throughput sequencing data.
FIGURE 1.
Knock-out strains and experimental scheme. A, list of the genes chosen for analysis. B, PCR validation of the knock-out strains. The KanMX gene replaced the target gene in the genome by using homolog recombination. The arrows indicate positive knock-out clones. C, experimental scheme. Five independent mutation accumulation lines were created for each strain to undergo 100 single-cell bottleneck passages (P0 to P100). Mutations were identified using high-throughput sequencing.
Using an experimental evolutionary approach, all of the above strains were subjected to single cell bottleneck passages with five independent mutation accumulation lines per strain. During each passage, one single colony per line was picked up and streaked onto a new plate of solid YES medium containing full supplements (Fig. 1C). Each mutation accumulation line was numbered according to its colony number at the first passage, and this number was strictly retained only for its progenies to avoid mixing between individual mutation accumulation lines. In each passage, cells were estimated to experience between 19 and 21 mitotic divisions (Table 1). Therefore, at the time point corresponding to the 100th passage (P100), the cells were ∼2,000 generations from their starting point (P0).
TABLE 1.
Generation estimates for single cell bottleneck passages
| Diameter | Cell number | Generation |
|
|---|---|---|---|
| Duplicates | Average | ||
| mm | |||
| 1.0 | 1,050,000 | 20.00 | 19.91 |
| 925,000 | 19.82 | ||
| 1.5–1.7 | 3,175,000 | 21.6 | 21.4 |
| 2,425,000 | 21.21 | ||
| 1.5 | 2,700,000 | 21.36 | 21.2 |
| 2,150,000 | 21.04 | ||
| 0.8 | 525,000 | 19.00 | 19.35 |
| 850,000 | 19.7 | ||
Genomic DNA was extracted from all of the lines at P100 and P0 and subjected to paired-end high-throughput sequencing (PE 100 or PE 125) (Fig. 1C; see “Experimental Procedures” for details). The average coverage of all 48 of the samples (5 at P100 and 1 at P0 for each strain) ranged from 80- to 180-fold across the genome.
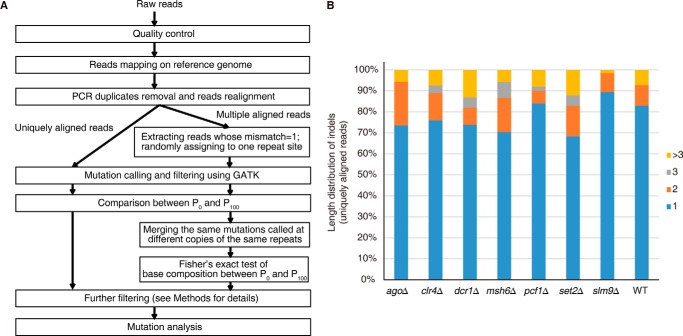
We developed a pipeline (Fig. 2A) for defining the mutations that accumulated during our experimental evolution. Mutations at uniquely aligned reads were relatively easy to call using GATK (32). In contrast, mutations that occurred at multiple aligned reads were more challenging to call and have typically been abandoned during analysis in many previous studies. However, this would compromise the analysis of mutations at heterochromatic regions, which contain more abundant repeats. To improve our analysis in repetitive regions, we first employed PE 100 or PE 125 sequencing to obtain longer reads. Next, we randomly assigned one of the repetitive sites as the target site for multiple aligned reads, and then we excluded those identified mutations that resided exactly at the varied bases among the same group of repeats (Fig. 2A). This approach allowed us to confidently call single-nucleotide substitutions and 1-bp indels in the repetitive regions. However, accurately identifying indels larger than 2 bp remained a challenging task. Nevertheless, based on an analysis of the uniquely aligned reads, we found that 1-bp indels accounted for the majority (>70%) of total indels (Fig. 2B). Therefore, we concentrated our subsequent analysis on single-nucleotide substitutions and 1-bp indels that were called at unique and multiple aligned reads.
FIGURE 2.
The bioinformatics pipeline and length distribution of indels. A, the pipeline used in data analysis for read alignment and mutation identification (see “Experimental Procedures” for details). Uniquely and multiple aligned reads underwent two different procedures for mutation identification. B, length distributions for indels from uniquely aligned reads of the listed strains.
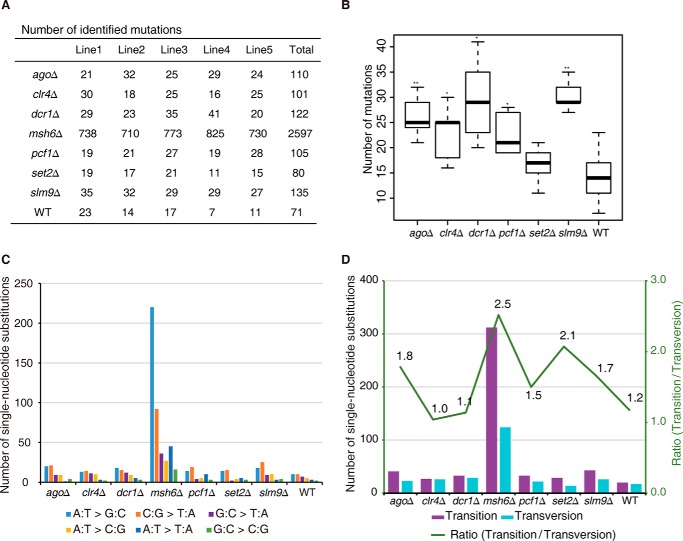
Using the above-described pipeline, we identified a total of 3,321 mutations in all of the P100 samples compared with their own P0 samples (Fig. 3A). To validate our analysis pipeline, we chose one of the mutation accumulation lines (ago1Δ P100-1) and performed Sanger sequencing of the regions surrounding all 23 of the mutations that were called by our analysis pipeline. Among the 23 mutations identified using high-throughput sequencing, only one single-nucleotide substitution at a highly repetitive region was not clarified using Sanger sequencing (Table 2). This single-nucleotide substitution occurred in a region containing 10 different copies in the genome, and it was therefore difficult to validate it using Sanger sequencing. Therefore, we concluded that the vast majority (and potentially all) of the mutations identified by our analysis pipeline were true mutations.
FIGURE 3.
Summary of mutation characteristics in experimental evolution. A, summary of quantity of mutations identified in all samples. B, box plots of the numbers of mutations in the listed strains. p values were calculated using the Wilcoxon Mann-Whitney test (*, p < 0.05; **, p < 0.01). Median, middle bar inside each box; box, first and third quartiles; whiskers, most extreme values within 1.5 times inter-quartile range from the box. C, summary of single-nucleotide substitution types in the listed strains. D, the number of transitions and transversions identified in each strain.
TABLE 2.
Mutation validation for agoΔP100-1
| ID | Chromosome | Position | Base |
Flanking sequence | Single-nucleotide substitution/indel | Copy number in genome | Validation | |
|---|---|---|---|---|---|---|---|---|
| agoΔP0 | agoΔP100-1 | |||||||
| 1 | chr1 | 224 | A | G | gcaagttgtgGtcggccttgc | Single-nucleotide substitution | 2 | True |
| 2 | chr1 | 2224 | T | C | gccaattggtCttgtaagtca | Single-nucleotide substitution | 2 | True |
| 3 | chr1 | 438,086 | C | T | catcattcacCgttagcgact | Single-nucleotide substitution | 1 | True |
| 4 | chr1 | 2,594,351 | T | TA | aaaagcttcaTaacagatgta | Indel | 1 | True |
| 5 | chr1 | 3,789,471 | C | T | aaagaaggtaTattgatgcat | Single-nucleotide substitution | 10 | Not sure |
| 6 | chr1 | 4,455,149 | G | GA | aaattaactaGaaaaaaaaaa | Indel | 1 | True |
| 7 | chr1 | 4,658,949 | T | C | aaacgatttaTcgatatttac | Single-nucleotide substitution | 1 | True |
| 8 | chr1 | 4,667,235 | A | C | ccaaaatttaActgtatggga | Single-nucleotide substitution | 1 | True |
| 9 | chr2 | 960,775 | C | CA | gccgatgcgaCccaggttcga | Indel | 1 | True |
| 10 | chr2 | 1,267,916 | AT | A | caacgaatcaAtttttttttt | Indel | 1 | True |
| 11 | chr2 | 1,635,733 | G | A | agatacgttgAatgttgttgc | Single-nucleotide substitution | 6 | True |
| 12 | chr2 | 1,659,092 | G | GA | ttgtgcttttGaaaaattcta | Indel | 1 | True |
| 13 | chr2 | 1,748,389 | A | C | tgagaaaaaaAaattcaacaa | Single-nucleotide substitution | 1 | True |
| 14 | chr2 | 2,034,216 | C | T | gatatcgaacCggcagaagag | Single-nucleotide substitution | 1 | True |
| 15 | chr2 | 2,088,866 | C | T | tattatagacCcaattattgg | Single-nucleotide substitution | 1 | True |
| 16 | chr2 | 3,058,544 | T | TG | agtacttaagTgggggggggg | Indel | 1 | True |
| 17 | chr2 | 3,725,872 | C | T | gcaggaaaacCggttcaatcg | Single-nucleotide substitution | 1 | True |
| 18 | chr2 | 4,531,749 | C | T | tcttccccccCaccacagcca | Single-nucleotide substitution | 3 | True |
| 19 | chr3 | 826,308 | G | T | atctttaaagGagatttgaat | Single-nucleotide substitution | 1 | True |
| 20 | chr3 | 1,110,963 | TC | T | tttgcgcaacTcctgcttatc | Indel | 10 | True |
| 21 | chr3 | 1,300,027 | G | T | tattttagctGcttttaaagg | Single-nucleotide substitution | 1 | True |
| 22 | chr3 | 2,010,864 | G | GTA | tagaatcatcGtatatatata | Indel | 1 | True |
| 23 | chr3 | 2,448,309 | T | TAA | catatagtttTaaaaaaaaaa | Indel | 1 | True |
Modest Protection of Genetic Fidelity by Epigenetic Regulators
For all eight of the strains tested, the wild-type strain accumulated the lowest number of mutations, and as expected, the mismatch repair-deficient msh6Δ strain accumulated 30-fold more mutations (Fig. 3A). Interestingly, all of the epigenetic regulator-deficient strains that were tested displayed a modest but statistically significant increase in the total number of mutations, with the exception of the set2Δ strain (Fig. 3B). This suggests that these epigenetic regulators may contribute to the protection of genetic fidelity in some degree. However, the total number of mutations accumulated in these lines was too small to allow deeper and more confident analysis, and we reasoned that the use of an experimental evolutionary system at a different scale would be required for such a purpose.
Preferential Protection against Indels by the Mismatch Repair Complex MutSα
To investigate what kind of mutations were preferentially enriched in the strains containing either a defective mismatch repair pathway or compromised epigenetic regulatory pathways, we first calculated the total number of each type of single-nucleotide substitution. Generally, the total numbers of transitions (C:G > T:A and A:T > G:C) and transversions (G:C > T:A, A:T > C:G, A:T > T:A, and G:C > C:G) were similar in most strains, with the exception of the msh6Δ strain (Fig. 3, C and D). This may not indicate that the mismatch repair machinery preferentially repairs transition type errors; instead, it is more probable that these results are a consequence of transition type errors occurring more frequently during DNA replication than transversion type errors (33).
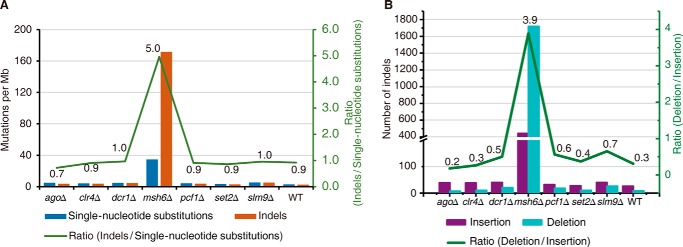
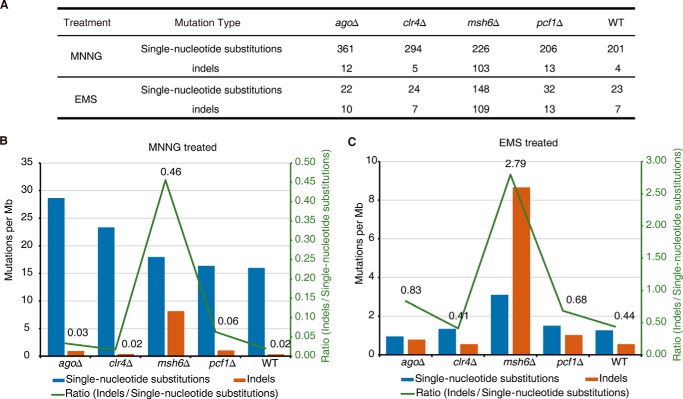
Similar to what was observed in the wild-type strain, the strains lacking epigenetic regulators displayed a ratio of indel/single-nucleotide substitution of ∼0.9 (Fig. 4A), suggesting that they had a minimal impact on this process. In contrast, the msh6Δ strain had a ratio of indel/single-nucleotide substitution of 5 (Fig. 4A). This observation is consistent with findings in previous studies that used budding yeast (34, 35). This may be explained by the fact that the MutSα complex (Msh6 is a subunit of the MutSα complex) preferentially repairs mismatches with looped-out bases.
FIGURE 4.
A mismatch repair-deficient strain displayed a preferentially elevated rate of indels, especially deletions. A, the mutation rate for single-nucleotide substitutions (blue) and indels (orange) in the listed strains. The green line represents the ratio of indels/single-nucleotide substitutions. B, the mutation rate for insertions (violet) and deletions (cyan) in the listed strains. The green line represents the ratio of deletions/insertions.
Interestingly, when we further analyzed the type of indels (insertions or deletions) that accumulated in these strains, we found that most strains, including the wild-type strain, accumulated 2–3-fold fewer deletions than insertions (Fig. 4B). This differs from what was observed in budding yeast S. cerevisiae (36), but it is consistent with a recent fission yeast S. pombe study (37). This is an interesting observation because it suggests that although both insertions and deletions create looped-out structures in double-stranded DNA, they are likely to be differentially sensed by the mismatch machinery, because insertions are associated with looped-out structures at the newly synthesized daughter strand, whereas deletions are associated with looped-out structures at the mother strand. It appears that the fission yeast S. pombe mismatch repair machinery preferentially recognizes the latter. Indeed, the msh6Δ strain accumulated ∼4-fold more deletions than insertions, which reflects a >8-fold change compared with all of the other strains (Fig. 4B), further supporting our notion.
The above observations were obtained from analysis of an experimental evolutionary process in which most of the mutations were classified as spontaneous mutations. We next examined the role of the mismatch repair machinery in protecting the genome against mutations induced by chemical mutagens. We performed chemically induced mutation experiments using mutagens such as methylnitronitrosoguanidine (MNNG) or ethyl methanesulfonate (EMS) in the msh6Δ, ago1Δ, clr4Δ, pcf1Δ, and wild-type strains. MNNG is a mutagen that alkylates the O4 position of thymine or the O6 position of guanine, whereas EMS is an alkylating agent that converts guanine to O-6-ethylguanine (38–40). MNNG did not induce more single-nucleotide substitutions in the msh6Δ strain than in the other strains (Fig. 5A), most likely because MNNG-induced mutations may not be the ideal substrate for the mismatch repair machinery, which primarily functions in proximity to the replication fork to repair DNA replication errors (8, 9, 41–43). Nevertheless, MNNG treatment resulted a robust increase (10–20-fold) in indel accumulation in the msh6Δ strain compared with the other strains (Fig. 5, A and B), which supports our observation that the MutSα complex preferentially targets looped-out structures in double-stranded DNA that may lead to indels if unrepaired. EMS treatment generated fewer mutations (Fig. 5A) in this experimental setup, but it also induced a higher number of indels in the msh6Δ strain than in the other strains (Fig. 5, A and C). Taken together, the above results collectively indicate that the MutSα complex preferentially protects the genome against indels, which is consistent with its known biochemical features (44).
FIGURE 5.
Chemically induced mutation accumulation. A, summary of the mutations in the MNNG- and EMS-treated strains. Each background contained five replicates, and mutations were combined for calculations. B, mutation rate for single-nucleotide substitutions (blue) and indels (orange) in MNNG-treated strains. The green line represents the ratio of indels/single-nucleotide substitutions. C, mutation rate for single-nucleotide substitutions (blue) and indels (orange) in EMS-treated strains. The green line represents the ratio of indels/single-nucleotide substitutions.
Preferential Protection of Genome Fidelity in Euchromatin by the Mismatch Repair Machinery
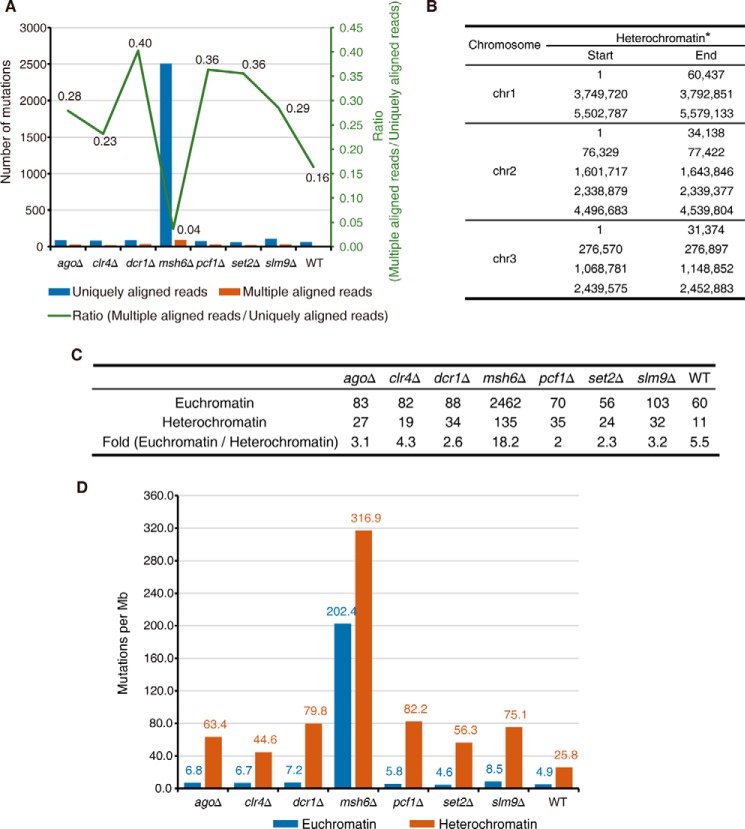
As we described previously, during our analysis of the experimental evolutionary data set, we separated the reads into two groups, the uniquely aligned reads and the multiple aligned reads (Fig. 2A), for technical reasons. Surprisingly, we observed a striking difference in the number of mutations called for each of these two groups of reads. The msh6Δ strain accumulated a much higher number of mutations that were within uniquely aligned reads than all of the other strains, but it had only a much smaller increase in the number of mutations that were within multiple aligned reads (Fig. 6A).
FIGURE 6.
The mismatch repair-deficient strain displayed a preferentially elevated rate of mutations in euchromatin. A, number of mutations identified at uniquely (blue) and multiple (orange) aligned reads in the listed strains. The green line represents the ratio of mutations at multiple/uniquely aligned reads. B, genomic distribution of heterochromatin regions defined using H3K9me2 ChIP-seq data. *, heterochromatin at mating type regions is excluded for analysis in this study, because the reference genome of S. pombe is that of an h+ strain that lacks the region between mat2 and mat3. C, summary of the number of mutations in heterochromatin and euchromatin of listed strains. D, mutation rate in euchromatin (blue) and heterochromatin (orange) regions in the listed strains.
Because the initial aim of this study was to examine the role of epigenetic regulation in protecting genome fidelity, this unexpected observation immediately attracted our attention because the multiple aligned reads corresponded to repetitive sequences, which are often enriched in heterochromatin (45–51). Heterochromatic regions in S. pombe are marked by H3K9me2 (23, 27). Therefore, we defined the boundaries of S. pombe heterochromatin and euchromatin using previously published H3K9me2 ChIP-seq data (52) (Fig. 6B) and analyzed the accumulated mutations in heterochromatin and euchromatin (Fig. 6C). Clearly, the msh6Δ strain accumulated a disproportionally high number of mutations in euchromatin in comparison with all other strains (Fig. 6C).
Then we calculated the mutation rate by normalizing the number of mutations to the length of the corresponding genomic regions. The mutation rate was clearly 5–14-fold higher in heterochromatin than in euchromatin in all of the tested strains except for the mismatch repair-deficient msh6Δ strain, in which the difference was reduced to ∼1.5-fold (Fig. 6D). The increased mutation rate in heterochromatin has been reported previously in other organisms (15, 17, 19, 20). Our finding that the loss of mismatch machinery effectively abrogates such trend suggests that the mismatch repair machinery preferentially protects genetic fidelity in euchromatin.
We also noted that although loss of Clr4 is known to disrupt heterochromatic silencing (21), it did not cause a decreased number of mutations within heterochromatin in comparison with the wild-type strain (Fig. 6C), which we believe can be explained by two potential reasons. First, the number of mutations in heterochromatin identified from the wild-type strain (a total of 11) was very small, and it could be hard to expect a further decrease upon loss of Clr4, because at this level, any stochastic changes during the experiments could affect the readout. Second, loss of Clr4 reverts heterochromatin silencing, but it may not fully alter the chromatin structure of heterochromatin. Of note, in mammalian cells lacking both homologues of Clr4 (Suv39h1 and Suv39h2), DAPI-dense regions remained, despite of disruption of heterochromatin silencing (53). This is an indication that the loss of H3K9 methylation does not fully abolish the structure of heterochromatin, and DNA-dense regions could still be formed in the cells.
Loss of Msh6 in S. pombe Altered the Chromatin Distribution of Mutations and Led to a Positive Correlation between Chromatin Accessibility and Mutation Rate
The explanation for the elevated mutation rate in heterochromatin is unclear. Due to the late replicating nature of heterochromatin, it has been proposed that replication occurring at late S phase might be more error-prone during DNA synthesis (54–56). However, this does not explain our observation that the low mutation rate in euchromatin became disproportionally increased by disrupting the mismatch repair machinery (Fig. 6D). We reasoned that preferential protection of euchromatin was probably due to the restricted access of the mismatch repair machinery at the heterochromatic regions. To directly interrogate the relationship between chromatin accessibility and mutation accumulation, we performed DNase I sequencing experiments and cross-examined chromatin accessibility data with our mutation accumulation data.
Currently, due to the large number of repetitive sequences, the reference genome of S. pombe is incomplete, especially at the heterochromatic region, which affects our DNase I sequencing analysis at heterochromatic regions. Therefore, we focused on euchromatin regions and analyzed the relationship between chromatin accessibility and the mutation rate within euchromatin.
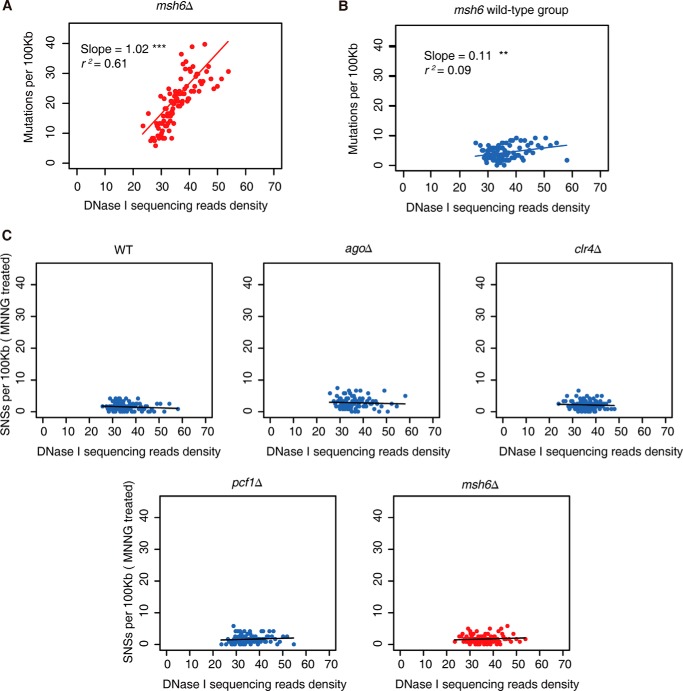
The number of mutations accumulated in individual strains except msh6Δ was insufficient for statistical analysis. Nonetheless, these strains share a similar global DNase I sequencing profile, and we decided to combine these strains as the msh6 wild-type group and pooled their mutations for further analysis. We first divided the yeast genome into 1-kb windows and scored their DNase I sequencing read density. Next we ranked all of these windows according to their read density and merged them into 100 units. We then calculated the mutation rate for each unit and plotted it against the DNase I sequencing read density. It is easily appreciated that the mutation rate displayed a strong positive correlation with read density in msh6Δ strains (r2 = 0.61, slope = 1.02) (Fig. 7A), and such correlation was not observed in the msh6 wild-type group (r2 = 0.09, slope = 0.11) (Fig. 7B).
FIGURE 7.
The relationship between mutation accumulation and chromatin accessibility in fission yeast strains. A and B, correlation analysis of spontaneous mutation rate and DNase I sequencing read density in msh6Δ strain (A) and in the msh6 wild-type group (B) during experimental evolution. C, correlation analysis of mutagen-induced mutation rate (single-nucleotide substitutions per 100 kb) and DNase I sequencing read density during MNNG treatment.
As a control analysis, the single-nucleotide substitutions induced by MNNG that could bypass mismatch repair (Fig. 5A) did not show a positive correlation between DNase I sequencing read density and mutation rate in msh6Δ strains and other tested strains (Fig. 7C).
These data collectively suggest that the function of mismatch repair machinery is regulated by chromatin accessibility, and such regulation is one major player in setting the spontaneous mutation landscape across the genome.
Differential Contributions of Chromatin Accessibility in Human Cancers with or without Mismatch Repair Deficiency
The above data suggest that the mismatch repair machinery preferentially protects open chromatin regions that are highly accessible. Notably, a large number of human cancers are associated with microsatellite instability resulting from deficiency in the mismatch repair pathway (5, 6, 40, 57–60). Recently, it has been revealed that microsatellite instable cancer samples display altered mutation landscape, and mutations arising after the inactivation of mismatch repair are no longer enriched in heterochromatin relative to euchromatin (61). The same study proposed that such a phenomenon might be caused by replication timing or chromatin accessibility (61). These observations, together with our fission yeast data (Fig. 7, A and B) prompted us to investigate whether human cancers with or without microsatellite instability may display distinct patterns of accumulated mutations at genomic regions that display different levels of accessibility within euchromatin.
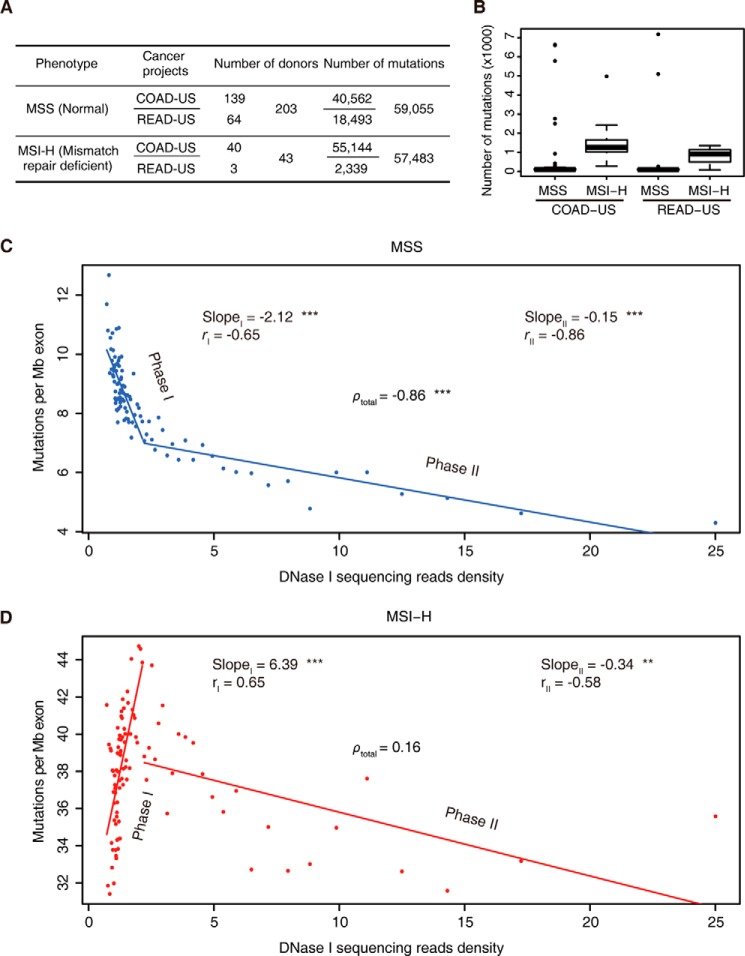
We analyzed a public exon mutation data set of colorectal cancer samples obtained from 246 patients (the International Cancer Genome Consortium database, release 18). Colorectal cancers often display increased mutation rates due to the inactivation of the mismatch repair or other DNA repair pathways (5, 40, 57–60, 62). These colorectal cancer samples were classified into two groups, a microsatellite-stable (MSS) group and a microsatellite highly instable (MSI-H) group, which represent cancer samples with functional or deficient mismatch repair pathways, respectively. Cancer samples in the MSI-H group displayed 5-fold more mutations than the MSS group, on average (Fig. 8, A and B), which is consistent with their deficiencies in the mismatch repair pathway.
FIGURE 8.
The relationship between mutation accumulation and chromatin accessibility in colorectal cancer samples. A, exome mutation summary and donor numbers for colorectal cancers obtained from a public database. Samples were categorized into two groups according to their microsatellite stability status. B, box plots of the numbers of mutations in the indicated cancer samples. Samples were categorized according to their microsatellite stability status. Median, middle bar inside each box; box, first and third quartiles; whiskers, most extreme values within 1.5 times inter-quartile range from the box; points, outliers. C, exonic mutation rate plotted against DNase I sequencing density in MSS cancers. D, exonic mutation rate plotted against DNase I sequencing density in MSI-H cancers.
To validate our hypothesis that the mutation landscape in cancer samples may be influenced by the accessibility of the local chromatin environment, we cross-analyzed the ENCODE DNase I accessibility sequencing data set of the colorectal origin cell line HCT-116 (63) and the aforementioned colorectal cancer exome mutation data set. We modified our fission yeast analysis pipeline slightly, by dividing the human genome into 10-kb windows, and we then selected windows containing at least 50 bp of exonic DNA (63,606 windows in total) for further analysis. We sorted all of these windows according to their DNase I sequencing reads and merged them into 100 units (DNase I sequencing reads from low to high, with units 1–99 containing 637 10-kb windows each and unit 100 containing 543 10-kb windows). We then calculated the number of mutations per megabase of exon length in each unit and plotted this number against the DNase I sequencing read density.
For MSS cancers with a normal mismatch repair pathway, we observed that more mutations occurred within genomic regions with a lower DNase I sequencing read density (ρtotal = −0.86; Fig. 8C). We also noticed that this negative correlation appeared in two distinct linear phases. In genomic regions with DNase I sequencing read densities <2.2 (phase I), a negative correlation with a relatively sharp slope was observed (slope = −2.12, r = −0.65), whereas in genomic regions with DNase I sequencing read densities >2.2 (phase II), a negative correlation with a much flatter slope was observed (slope = −0.15, r = −0.86) (Fig. 8C). We reasoned that in relatively more closed chromatin regions (phase I), chromatin accessibility is probably the major rate-limiting factor that determines the access of DNA repair machineries, and it therefore has a greater impact on the accumulation of mutations. However, the impact of chromatin accessibility becomes less profound in more open chromatin regions (phase II), which are more freely accessible in general.
Importantly, we observed a dramatic change in the mutation landscape in the MSI-H cancer samples. The overall negative correlation between the mutation rate and chromatin accessibility was diminished (ρtotal = 0.16; Fig. 8D). This was primarily caused by changes toward the opposite direction within the closed chromatin regions. Instead of the negative correlation that was observed in the MSS cancer samples, the MSI-H cancer samples displayed a strong positive correlation between mutation rates and chromatin accessibility at closed chromatin regions (phase I) (slope = 6.39, r = 0.65). In contrast, the MSI-H cancer samples displayed much milder changes in the mutation landscape at open chromatin regions (phase II) (slope = −0.34, r = −0.58).
The above results indicate that in fission yeast and human cancer samples, some physical or biological aspect of chromatin structure contributes to the landscape of the mutation accumulation rate, probably involving differential accessibility of the DNA repair machinery to distinct chromatin regions
Discussion
The observation that the chromatin environment can affect the rate at which mutations accumulate and the landscape representing the distribution of mutations across the genome (19, 64–66) has prompted many studies that aim to reveal the mechanisms underlying these relationships. The accumulation of mutations is apparently the consequence of two potential causes, the rate at which DNA sequence errors occur and the DNA repair rate. Because regions with elevated mutation rates appear to be correlated with late replication of DNA sequences (16, 61), it has been proposed that replication timing may be a major contributor to the uneven distribution in mutations observed across the genome, which may imply that late replicating forks are prone to DNA synthesis errors (16). On the other hand, nucleosome occupancy has been reported to reduce the accumulation of mutations, in particular the C to T transitions caused by cytosine deamination that probably occur less frequently in nucleosomal DNA that is protected by histones (18).
In this study, we attempted to directly investigate the contribution of epigenetic regulators to the control of the mutation rate using an experimental evolutionary system that allowed us to compare different S. pombe strains that contained deletions of various epigenetic regulators. In human cells, H3K36 methylation facilitates mismatch repair by recruiting human MSH6 (67). In our study, set2Δ strain displayed no significant change of mutation rate. This could be explained by the fact that fission yeast S. pombe MSH6 lacks a PWWP domain that preferentially associates with methylated H3K36. We did observe a modest increase in mutation rates in strains lacking several other epigenetic regulators (Fig. 3, A and B), suggesting that loss of these epigenetic regulators, including components of the RNAi machinery and histone methyltransferase and chaperones, may affect the protection of genome fidelity. However, we also would like to point out that the observed level of change was relatively subtle, and the number of accumulated mutations was too small for further analysis. Because of these reasons, our observation may even be affected by stochastic events that occurred during the experiment procedure. We think that future investigations on a much greater scale may help to pinpoint the roles of these epigenetic regulators in the regulation of genome fidelity.
On the other hand, we observed a much more profound role of chromatin in genome protection. An elevated mutation rate at heterochromatin has been observed previously in various organisms (15, 16, 19, 35). A number of reasons have been proposed as potential explanations, including error-prone late replication, chromatin structure-mediated DNA sequence protection, chromatin accessibility for DNA repair, etc. The dramatically elevated mutation rate and its preferential distribution in euchromatin observed in the fission yeast S. pombe msh6Δ strain (Fig. 6, C and D) provide direct evidence that the elevated mutation rate in heterochromatin is primarily due to preferential protection in euchromatin under wild-type conditions, whereas error-prone DNA synthesis during late replication may only be a minor contributor. Furthermore, we observed that the spontaneous mutation rate displayed a strong positive correlation with chromatin accessibility only when mismatch repair machinery was impaired, both in fission yeast (Fig. 7A) and in human cancer samples (Fig. 8D). These observations argue that chromatin accessibility-regulated mismatch repair is one major mechanism for an elevated mutation rate in heterochromatin regions. Although we cannot rule out contributions from potential error-prone DNA synthesis in late replicating heterochromatic regions, we identify that restricted access of the mismatch repair machinery is one important contributor to the elevated mutation rate in these regions.
One interesting question is why chromatin can play a role in regulating mismatch repair, a process that largely accompanies DNA replication, during which chromatin structure must be unpacked. Of note, unpacking of chromatin structure during DNA replication is a transient event coupled with the passage of the replication fork. DNA exists in the context of chromatin immediately before and immediately after the passage of the replication fork (68–71). Thus, we reason that before the passage of the replication fork, chromatin accessibility can regulate the local concentration of mismatch machinery and therefore impact subsequent repair efficiency. Indeed, in mammalian cells, H3K36 methylation, a chromatin mark, facilitates the recruitment of mismatch repair machinery and promotes repair efficiency (67), which is a good example of the role of chromatin in facilitating mismatch repair during replication.
We hypothesize that the preferential protection of genetic fidelity in euchromatin has two beneficial roles. First, mutations in actively transcribed open chromatin regions may cause deleterious effects to cells and should therefore be preferentially suppressed. Second, the increased mutation rate observed in heterochromatin regions may allow the origin of new regulatory elements or even new genes during evolution. Indeed, it has been reported that many newborn enhancers in humans are derived from Alu repeat sequences (72), which are often embedded within less accessible genomic regions (73–78). Finally, this study also provides direct evidence indicating a regulatory role for epigenetic information in the control of genetic fidelity in addition to its well understood role in the regulation of transcription.
Experimental Procedures
S. pombe Knock-out Strains
In this study, all knock-out strains (Fig. 1A) were derived from the haploid wild-type strain LD331: h+. Knock-out strains were generated according to the methods described previously (79). DNA fragments containing the KanMX6 cassette for recombination were amplified from the pFA6a-KanMX6 plasmid using long primers that covered the upstream or downstream flanking sequences of the target genes. All mutant strains were verified by positive and negative PCR tests, as shown in Fig. 1B. For positive tests, the upstream primers were designed within the KanMX cassette, and the downstream primers were designed according to sequences downstream of the stop codon of the target genes. For negative tests, both primers were designed within the corresponding target genes. These knock-out strains were also confirmed by subsequent high-throughput sequencing data.
Mutation Accumulation Lines and Estimation of the Number of Cell Generations
All 40 mutation accumulation lines were selected from single P0 colonies of eight strains. They were then continuously passed using a single-colony bottleneck culture approach on solid YES medium containing full supplements (80). To minimize variation in cell generation across all of the strains, we consistently chose colonies that were ∼1 mm in diameter for passage.
To estimate the number of cell generations in each passage, we randomly picked two colonies of each size, counted the cell numbers, and then estimated the number of cell divisions or generations. Cells went through ∼20 generations in each passage under our culture conditions (Table 1).
DNA Library Preparation and Illumina® Sequencing
Genomic DNA samples were sonicated into fragments with average length of 500 bp using a Covaris® M220 sonicator, and then sequencing libraries were prepared using KAPA® Hyper Prep kits (KK8504), according to the manufacturer's instructions. We also designed 24 paired-end sequencing adapters with barcodes to multiplex the sequencing experiments.
Forty experimentally evolved lines (five lines per strain for the ago1Δ, clr4Δ, dcr1Δ, msh6Δ, pcf1Δ, set2Δ, slm9Δ, and WT strains), eight corresponding P0 strains, 15 MNNG-treated lines (five lines per strain for the clr4Δ, msh6Δ, and WT strains), and three corresponding untreated strains were sequenced using an Illumina® HiSeq 2000 sequencer for PE 100 sequencing with an average insertion size of 500 bp. Two additional WT P0 strains were sequenced using an Illumina® HiSeq 2000 sequencer for PE 50 sequencing with an average insertion size of 200 bp. Ten lines (5 lines/strain for the ago1Δ and pcf1Δ strains) were treated with MNNG, with their corresponding untreated strains used as controls, and 25 lines (5 lines/strain for the ago1Δ, clr4Δ, msh6Δ, pcf1Δ, and WT strains) were treated with EMS, and their corresponding untreated strains were used as controls. All strains were sequenced using an Illumina® HiSeq 2500 sequencer for PE 125 sequencing with an average insertion size of 500 bp.
Mutation Identification
Contaminated adapters were removed using Cutadapt (81). Then we used SolexaQA (82) (version 3.1.3) to filter the sequencing data. For each read, the longest contiguous segment with a quality score of >20 was retained. Reads were aligned to the fission yeast reference genome (NCBI Schizosaccharomyces_pombe_uid127) using BWA (83). Potential PCR duplicates were removed using samtools (version 0.1.18) (84). Then local realignment around indels was performed using IndelRealigner in the Genome Analysis Toolkit (GATK version 3.2.0) (32). UnifiedGenotyper in GATK was used with the parameters “-mbq 35 -ploidy 1 -stand_call_conf 30 -stand_emit_conf 10 -minIndelFrac 0.7” to call mutations. VariantFiltration in GATK was used with the parameters “DP < 5.0 QD<2 FS>60 MQ<40 MQRankSum< −12.5 ReadPosRankSum < −8.0” for single-nucleotide substitutions and with the parameters “DP < 5.0 QD < 2 FS > 200 ReadPosRankSum < −20.0” for INDEL to filter mutations. For single-nucleotide substitutions, the frequency of the mutant allele was required to be >75%. In addition, all mutations resulting in variations within the adjacent 10 bp were removed. Then evolved strains were compared with P0 to identify mutations. For multiple aligned reads, we chose those with a mismatch = 1 and randomly assigned one site for each mismatch. Then we set the MAPQ to 40 and subjected them to mutation calling and filtering with GATK. The same settings were applied for mutation calling in uniquely aligned reads. When the same mutations were found in different copies of one given repeat group, they were merged. For each mutation that was identified in the multiple aligned reads, the base compositions of the P0 and P100 (or untreated and chemically treated) samples at the corresponding mutation site were computed, and we retained those mutations with a p value of <0.05 (Fisher's exact test). If opposite single-nucleotide substitutions were identified, both of them were discarded.
Mutation Validation
We chose ago1Δ P100-1 as the validation sample. From it, 23 mutations were identified using our data analysis pipeline. PCR primers were designed using Primer3 (libprimer3 release 2.3.6) (85, 86), and DNA fragments containing mutation sites were amplified from genomic DNA. The amplified products were cloned for Sanger sequencing.
Categorization of Euchromatin and Heterochromatin Regions
The heterochromatin regions in fission yeast S. pombe were defined using previously published H3K9me2 ChIP-Seq data (52). The reads were aligned using Bowtie (87), and the peaks were called using MACS2 (88) with the parameter “-q 1e-6”. Then peaks within 15 kb were merged to define the heterochromatin regions.
DNase I Sequencing
Nuclei from S. pombe were prepared as described (89), with some modifications. Yeast cells were collected, washed, and then suspended with 5 ml of lysis solution (5 mm 2-mercaptoethanol, 1 m sorbitol) per g of cells (wet weight). Cells were digested for 40 min at 28 °C with 8 mg/ml Zymolase 20T (MP Biomedicals). The spheroplasts were washed with 1 m sorbitol and then resuspended in 4 ml of Ficoll buffer (18% (w/v) Ficoll, 20 mm KH2PO4, pH 6.8, 1 mm MgCl2, 0.25 mm EGTA, and 0.25 mm EDTA). After 13,000 × g centrifugation, the nuclear pellet was washed by DNase I buffer (15 mm Tris-HCl, pH 7.5, 75 mm NaCl, 3 mm MgCl2, 0.05 mm CaCl2, and 1 mm 2-mercaptoethanol). The nuclear pellet was suspended in 2.4 ml of DNase I buffer/g of starting cells (wet weight). Aliquots of 200 μl each were digested with 10 units/ml DNase I (Beyotime Biotechnology) at 37 °C for 5 min. Digestion was stopped with 1% SDS and 10 mm EDTA, pH 8.0. Then 5% (v/v) proteinase K (20 mg/ml) was added and incubated overnight. 1 m NaClO4 was added for maxima isolating DNA from chromatin, and DNA samples were treated with RNase A (10 mg/ml). A DNase I sequencing library was prepared similar to the above mentioned genomic sequencing library, with one modified step of size selection for DNA fragments between 160 and 400 bp.
The data set supporting the results of this article is publically available at the Sequence Read Archive of NCBI under the accession number SRP065655. All barcodes have been removed in our deposited sequencing data.
Correlation Analysis between Mutation Rates and DNA Accessibility in Colorectal Cancers
The somatic mutations called from colorectal cancer exomes were downloaded from the International Cancer Genome Consortium database (release 18). Exomes with 50 or more mutations were retained, including 179 samples in the COAD-US project and 67 samples in the READ-US project. The microsatellite instability data for these cancer samples were downloaded from the NCI, National Institutes of Health, TCGA Data Portal. The DNA accessibility data for the HCT-116 cell line were downloaded from two DNase I hypersensitivity data files in ENCODE.
We divided the human genome into 10-kb windows. Only windows with at least 50 bp of exonic DNA were retained. For each window, the mutation rate and DNase I sequencing read densities were calculated. Then the windows were sorted according their DNase I sequencing read densities and divided into 100 units. The total mutation rate and the mean DNase I sequencing read density were calculated for each unit.
Author Contributions
L. S. performed the fission yeast experimental evolution experiments. Y. Zhang performed the bioinformatics analysis. Z. Z. participated in the bioinformatics analysis. Y. Zheng assisted with the experiments. L. D. supervised the fission yeast experiments. The experiments were designed by L. S. and B. Z. The manuscript was written by L. S., Y. Zhang, and B. Z. All authors read the manuscript and provided suggestions.
This work was supported by Chinese Ministry of Science and Technology Grant 2015CB856200. This work was also supported by China National Science Foundation Grant 31401114 (to Z. Z.), Grant 31501059 (to Y. Zhang), and Grants 31425013, 31530037, and 31521002 (to B. Z.); Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB08010103; and the Howard Hughes Medical Institute International Early Career Scientist Program. The authors declare that they have no conflicts of interest with the contents of this article.
- H3K9
- histone H3 Lys-9
- H3K9me2
- H3K9 dimethylation
- MNNG
- methylnitronitrosoguanidine
- EMS
- ethyl methanesulfonate
- MSS
- microsatellite-stable
- MSI-H
- microsatellite highly instable
- PE
- paired-end.
References
- 1. Allis C. D., Jenuwein T., and Reinberg D. (2007) Epigenetics, p. 16, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Shevelev I. V., and Hübscher U. (2002) The 3′ 5′ exonucleases. Nat. Rev. Mol. Cell Biol. 3, 364–376 [DOI] [PubMed] [Google Scholar]
- 3. Lahue R. S., Au K. G., and Modrich P. (1989) DNA mismatch correction in a defined system. Science 245, 160–164 [DOI] [PubMed] [Google Scholar]
- 4. Iyer R. R., Pluciennik A., Burdett V., and Modrich P. L. (2006) DNA mismatch repair: functions and mechanisms. Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 5. Li G. M. (2008) Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 [DOI] [PubMed] [Google Scholar]
- 6. Jiricny J. (2006) The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 7. Fukui K. (2010) DNA mismatch repair in eukaryotes and bacteria. J. Nucleic Acids 10.4061/2010/260512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hombauer H., Campbell C. S., Smith C. E., Desai A., and Kolodner R. D. (2011) Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147, 1040–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hombauer H., Srivatsan A., Putnam C. D., and Kolodner R. D. (2011) Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science 334, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drake J. W. (1991) Spontaneous mutation. Annu. Rev. Genet. 25, 125–146 [DOI] [PubMed] [Google Scholar]
- 11. Drake J. W., Charlesworth B., Charlesworth D., and Crow J. F. (1998) Rates of spontaneous mutation. Genetics 148, 1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolfe K. H., Sharp P. M., and Li W. H. (1989) Mutation rates differ among regions of the mammalian genome. Nature 337, 283–285 [DOI] [PubMed] [Google Scholar]
- 13. Smith N. G., Webster M. T., and Ellegren H. (2002) Deterministic mutation rate variation in the human genome. Genome Res. 12, 1350–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaffney D. J., and Keightley P. D. (2005) The scale of mutational variation in the murid genome. Genome Res. 15, 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hodgkinson A., and Eyre-Walker A. (2011) Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12, 756–766 [DOI] [PubMed] [Google Scholar]
- 16. Lang G. I., and Murray A. W. (2011) Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol. Evol. 3, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Makova K. D., and Hardison R. C. (2015) The effects of chromatin organization on variation in mutation rates in the genome. Nat. Rev. Genet. 16, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X., Chen Z., Chen H., Su Z., Yang J., Lin F., Shi S., and He X. (2012) Nucleosomes suppress spontaneous mutations base-specifically in eukaryotes. Science 335, 1235–1238 [DOI] [PubMed] [Google Scholar]
- 19. Schuster-Böckler B., and Lehner B. (2012) Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507 [DOI] [PubMed] [Google Scholar]
- 20. Polak P., Karlić R., Koren A., Thurman R., Sandstrom R., Lawrence M. S., Reynolds A., Rynes E., Vlahoviček K., Stamatoyannopoulos J. A., and Sunyaev S. R. (2015) Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., and Cranston G. (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218–233 [DOI] [PubMed] [Google Scholar]
- 22. Ekwall K., Javerzat J. P., Lorentz A., Schmidt H., Cranston G., and Allshire R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269, 1429–1431 [DOI] [PubMed] [Google Scholar]
- 23. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., and Grewal S. I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 [DOI] [PubMed] [Google Scholar]
- 24. Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., and Moazed D. (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 [DOI] [PubMed] [Google Scholar]
- 25. Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S. I., and Moazed D. (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanoh J., Sadaie M., Urano T., and Ishikawa F. (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 15, 1808–1819 [DOI] [PubMed] [Google Scholar]
- 27. Yamada T., Fischle W., Sugiyama T., Allis C. D., and Grewal S. I. (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20, 173–185 [DOI] [PubMed] [Google Scholar]
- 28. Garcia J. F., Dumesic P. A., Hartley P. D., El-Samad H., and Madhani H. D. (2010) Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 24, 1758–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun S., Garcia J. F., Rowley M., Rougemaille M., Shankar S., and Madhani H. D. (2011) The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Egel R. (2013) The Molecular Biology of Schizosaccharomyces pombe: Genetics, Genomics and Beyond, pp. 149–169, Springer, Berlin [Google Scholar]
- 31. Tornier C., Bessone S., Varlet I., Rudolph C., Darmon M., and Fleck O. (2001) Requirement for Msh6, but not for Swi4 (Msh3), in Msh2-dependent repair of base-base mismatches and mononucleotide loops in Schizosaccharomyces pombe. Genetics 158, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., Hanna M., McKenna A., Fennell T. J., Kernytsky A. M., Sivachenko A. Y., Cibulskis K., et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang D., Kreutzer D. A., and Essigmann J. M. (1998) Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. 400, 99–115 [DOI] [PubMed] [Google Scholar]
- 34. Zanders S., Ma X., Roychoudhury A., Hernandez R. D., Demogines A., Barker B., Gu Z., Bustamante C. D., and Alani E. (2010) Detection of heterozygous mutations in the genome of mismatch repair defective diploid yeast using a Bayesian approach. Genetics 186, 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Serero A., Jubin C., Loeillet S., Legoix-Né P., and Nicolas A. G. (2014) Mutational landscape of yeast mutator strains. Proc. Natl. Acad. Sci. U.S.A. 111, 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Romanova N. V., and Crouse G. F. (2013) Different roles of eukaryotic MutS and MutL complexes in repair of small insertion and deletion loops in yeast. PLoS Genet. 9, e1003920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farlow A., Long H., Arnoux S., Sung W., Doak T. G., Nordborg M., and Lynch M. (2015) The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics 201, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loveless A. (1969) Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature 223, 206–207 [DOI] [PubMed] [Google Scholar]
- 39. Sega G. A. (1984) A review of the genetic effects of ethyl methanesulfonate. Mutat. Res. 134, 113–142 [DOI] [PubMed] [Google Scholar]
- 40. Modrich P., and Lahue R. (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65, 101–133 [DOI] [PubMed] [Google Scholar]
- 41. Branch P., Aquilina G., Bignami M., and Karran P. (1993) Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature 362, 652–654 [DOI] [PubMed] [Google Scholar]
- 42. Kat A., Thilly W. G., Fang W. H., Longley M. J., Li G. M., and Modrich P. (1993) An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 90, 6424–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koi M., Umar A., Chauhan D. P., Cherian S. P., Carethers J. M., Kunkel T. A., and Boland C. R. (1994) Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 54, 4308–4312 [PubMed] [Google Scholar]
- 44. Tian L., Gu L., and Li G. M. (2009) Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSα and MutSβ determine their differential mismatch binding activities. J. Biol. Chem. 284, 11557–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clarke L., and Baum M. P. (1990) Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol. Cell. Biol. 10, 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takahashi K., Murakami S., Chikashige Y., Niwa O., and Yanagida M. (1991) A large number of tRNA genes are symmetrically located in fission yeast centromeres. J. Mol. Biol. 218, 13–17 [DOI] [PubMed] [Google Scholar]
- 47. Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., and Yanagida M. (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3, 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steiner N. C., Hahnenberger K. M., and Clarke L. (1993) Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol. Cell. Biol. 13, 4578–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ngan V. K., and Clarke L. (1997) The centromere enhancer mediates centromere activation in Schizosaccharomyces pombe. Mol. Cell. Biol. 17, 3305–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., Basham D., Bowman S., Brooks K., Brown D., Brown S., et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880 [DOI] [PubMed] [Google Scholar]
- 51. Pidoux A. L., and Allshire R. C. (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 12, 521–534 [DOI] [PubMed] [Google Scholar]
- 52. Yu R., Jih G., Iglesias N., and Moazed D. (2014) Determinants of heterochromatic siRNA biogenesis and function. Mol. Cell 53, 262–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., and Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 54. Koren A., Polak P., Nemesh J., Michaelson J. J., Sebat J., Sunyaev S. R., and McCarroll S. A. (2012) Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 91, 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lawrence M. S., Stojanov P., Polak P., Kryukov G. V., Cibulskis K., Sivachenko A., Carter S. L., Stewart C., Mermel C. H., Roberts S. A., Kiezun A., Hammerman P. S., McKenna A., Drier Y., Zou L., et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stamatoyannopoulos J. A., Adzhubei I., Thurman R. E., Kryukov G. V., Mirkin S. M., and Sunyaev S. R. (2009) Human mutation rate associated with DNA replication timing. Nat. Genet. 41, 393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Leach F. S., Nicolaides N. C., Papadopoulos N., Liu B., Jen J., Parsons R., Peltom−aki P., Sistonen P., Aaltonen L. A., Nyström-Lahti M., et al. (1993) Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 58. Li G. M., and Modrich P. (1995) Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 92, 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Peltomäki P. (2001) Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 10, 735–740 [DOI] [PubMed] [Google Scholar]
- 60. Peltomäki P. (2001) DNA mismatch repair and cancer. Mutat. Res. 488, 77–85 [DOI] [PubMed] [Google Scholar]
- 61. Supek F., and Lehner B. (2015) Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521, 81–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goodfellow P. J., Buttin B. M., Herzog T. J., Rader J. S., Gibb R. K., Swisher E., Look K., Walls K. C., Fan M. Y., and Mutch D. G. (2003) Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc. Natl. Acad. Sci. U.S.A. 100, 5908–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Prendergast J. G., Campbell H., Gilbert N., Dunlop M. G., Bickmore W. A., and Semple C. A. (2007) Chromatin structure and evolution in the human genome. BMC Evol. Biol. 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sasaki S., Mello C. C., Shimada A., Nakatani Y., Hashimoto S., Ogawa M., Matsushima K., Gu S. G., Kasahara M., Ahsan B., Sasaki A., Saito T., Suzuki Y., Sugano S., Kohara Y., et al. (2009) Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science 323, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Michaelson J. J., Shi Y., Gujral M., Zheng H., Malhotra D., Jin X., Jian M., Liu G., Greer D., Bhandari A., Wu W., Corominas R., Peoples A., Koren A., Gore A., et al. (2012) Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li F., Mao G., Tong D., Huang J., Gu L., Yang W., and Li G. M. (2013) The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153, 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ramachandran S., and Henikoff S. (2016) Transcriptional regulators compete with nucleosomes post-replication. Cell 165, 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Blumenthal A. B., Kriegstein H. J., and Hogness D. S. (1974) The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38, 205–223 [DOI] [PubMed] [Google Scholar]
- 70. McKnight S. L., and Miller O. L. Jr. (1977) Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12, 795–804 [DOI] [PubMed] [Google Scholar]
- 71. Gasser R., Koller T., and Sogo J. M. (1996) The stability of nucleosomes at the replication fork. J. Mol. Biol. 258, 224–239 [DOI] [PubMed] [Google Scholar]
- 72. Su M., Han D., Boyd-Kirkup J., Yu X., and Han J. D. (2014) Evolution of Alu elements toward enhancers. Cell Rep. 7, 376–385 [DOI] [PubMed] [Google Scholar]
- 73. Korenberg J. R., and Rykowski M. C. (1988) Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53, 391–400 [DOI] [PubMed] [Google Scholar]
- 74. Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 75. Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., Gocayne J. D., Amanatides P., Ballew R. M., Huson D. H., Wortman J. R., et al. (2001) The sequence of the human genome. Science 291, 1304–1351 [DOI] [PubMed] [Google Scholar]
- 76. Price A. L., Eskin E., and Pevzner P. A. (2004) Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 14, 2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Peng J. C., and Karpen G. H. (2009) Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5, e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hormozdiari F., Alkan C., Ventura M., Hajirasouliha I., Malig M., Hach F., Yorukoglu D., Dao P., Bakhshi M., Sahinalp S. C., and Eichler E. E. (2011) Alu repeat discovery and characterization within human genomes. Genome Res. 21, 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Forsburg S. L., and Rhind N. (2006) Basic methods for fission yeast. Yeast 23, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Barrick J. E., and Lenski R. E. (2013) Genome dynamics during experimental evolution. Nat. Rev. Genet. 14, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 82. Cox M. P., Peterson D. A., and Biggs P. J. (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koressaar T., and Remm M. (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291 [DOI] [PubMed] [Google Scholar]
- 86. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., and Rozen S. G. (2012) Primer3: new capabilities and interfaces. Nucleic Acids Res. 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Feng J., Liu T., Qin B., Zhang Y., and Liu X. S. (2012) Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Almer A., and Hörz W. (1986) Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 5, 2681–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]