Abstract
Corticosteroid-binding globulin (CBG) delivers anti-inflammatory cortisol to inflamed tissues upon elastase-based proteolysis of the exposed reactive center loop (RCL). However, the molecular mechanisms that regulate the RCL proteolysis by co-existing host and bacterial elastases in inflamed/infected tissues remain unknown. We document that RCL-localized Asn347 glycosylation fine-tunes the RCL cleavage rate by human neutrophil elastase (NE) and Pseudomonas aeruginosa elastase (PAE) by different mechanisms. NE- and PAE-generated fragments of native and exoglycosidase-treated blood-derived CBG of healthy individuals were monitored by gel electrophoresis and LC-MS/MS to determine the cleavage site(s) and Asn347 glycosylation as a function of digestion time. The site-specific (Val344-Thr345) and rapid (seconds to minutes) NE-based RCL proteolysis was significantly antagonized by several volume-enhancing Asn347 glycan features (i.e. occupancy, triantennary GlcNAc branching, and α1,6-fucosylation) and augmented by Asn347 NeuAc-type sialylation (all p < 0.05). In contrast, the inefficient (minutes to hours) PAE-based RCL cleavage, which occurred equally well at Thr345-Leu346 and Asn347-Leu348, was abolished by the presence of Asn347 glycosylation but was enhanced by sialoglycans on neighboring CBG N-sites. Molecular dynamics simulations of various Asn347 glycoforms of uncleaved CBG indicated that multiple Asn347 glycan features are modulating the RCL digestion efficiencies by NE/PAE. Finally, high concentrations of cortisol showed weak bacteriostatic effects toward virulent P. aeruginosa, which may explain the low RCL potency of the abundantly secreted PAE during host infection. In conclusion, site-specific CBG N-glycosylation regulates the bioavailability of cortisol in inflamed environments by fine-tuning the RCL proteolysis by endogenous and exogenous elastases. This study offers new molecular insight into host- and pathogen-based manipulation of the human immune system.
Keywords: glycoprotein, glycosylation, molecular dynamics, proteolysis, Pseudomonas aeruginosa (P. aeruginosa), corticosteroid-binding globulin, P. aeruginosa elastase, cortisol, neutrophil elastase, reactive center loop
Introduction
Human corticosteroid-binding globulin (CBG)5 is the main carrier of cortisol in the blood circulatory system. CBG regulates the bioavailability of this anti-inflammatory steroid hormone by binding up to 90% of the entire cortisol pool with high affinity and specificity in a single binding pocket in a temperature-sensitive manner (1, 2). Although produced primarily by hepatocytes (3, 4), human CBG mRNA and protein have been found in smaller amounts in other tissues, including lungs, kidneys, testes, placenta, endometrium, fallopian tube, heart, and some regions of the brain (5–12). The varying circulatory levels of CBG in childhood, puberty, and adulthood and in different physiological states like pregnancy have been documented; for example, the CBG concentration in the blood of healthy individuals was measured to be ∼40 mg/l (13). The circulatory half-life of human CBG has been established to be approximately 6 days, although some forms of CBG may be removed much faster (13), as supported recently by investigation of the human CBG half-life in rabbits (14). The CBG synthesis rate, which from the above can be estimated to be ∼0.1–0.2 mg/h under normal physiology, has been reported to be regulated by the glucocorticoids and IL-6 (15). For instance, during inflammation, CBG acts as a negative acute-phase protein (16), where the level of intact functionally active CBG in circulation is reduced due to the increased rate of proteolytic cleavage of this glucocorticoid carrier with a concomitant cortisol release (17) and an IL-6-driven reduction in CBG synthesis (18, 19). The delivery mechanism of cortisol to target tissues, which is only partially understood, involves 1) interaction of the cortisol-bound CBG with a putative receptor on the membranes of some cortisol-sensitive tissues (i.e. liver, endometrium, placenta, and prostate) (20, 21); 2) direct internalization of an intact cortisol-bound CBG complex in specific tissues (e.g. placental syncytiotrophoblasts) (20); and, the most documented, 3) extracellular elastase-based proteolytic cleavage of the exposed reactive center loop (RCL) of CBG, resulting in cortisol release and cellular uptake of cortisol upon a “stressed-to-relaxed” conformational change of CBG at the site of inflammation (22–27). Readers are directed to a recent review as a source of more in depth coverage of the CBG biology (28). The highly flexible RCL spanning the Glu333–Ile354 region of CBG is the target of human neutrophil elastase (NE), which cleaves Val344-Thr345 to form two complementary fragments: the large (50–55 kDa) N-terminal fragment (CBG-Nt) and the small (5–10 kDa) C-terminal fragment (CBG-Ct) (22). The reduced cortisol affinity of cleaved CBG increases the local concentration of free cortisol, which is beneficial toward resolving inflammation in affected tissues by the anti-inflammatory effects of cortisol (17, 29).
It was recently suggested that in basal, low inflammatory conditions, proteases other than NE may be causing systemic CBG cleavage (30). Both endogenous and exogenous proteases were documented to cleave the RCL and reduce the cortisol affinity of CBG. It was also shown that chymotrypsin cleaves the RCL of CBG at Leu346-Asn347 and Leu348-Thr349, but the biological relevance of this pancreatic protease in the context of the host immune response in inflamed tissues remains unknown (31). In addition, the Thr349-Ser350 and Ser350-Lys351 sites of E. coli-produced (non-glycosylated) recombinant human CBG were reported to be proteolytic target sites, but the responsible protease(s) remains elusive (32). Finally, P. aeruginosa elastase (PAE), the major virulence factor of this opportunistic Gram-negative pathogen and a zinc metalloprotease that is structurally unrelated to the serine proteases NE and chymotrypsin, was shown to cleave the RCL of CBG primarily between Asn347 and Leu348, leading to reduced cortisol affinity and release of the hormone from CBG (33). It was also reported that PAE cleaves several RCL sites (i.e. Thr345-Leu346, Leu346-Asn347, Asn347-Leu348 (main cleavage site), and Leu348-Thr349) (33). Interestingly, PAE and NE are known to co-exist in the inflamed and bacteria-infected respiratory tract of individuals with cystic fibrosis and chronic obstructive pulmonary disease (34). Although growing evidence supports an elastase-driven release of cortisol from CBG as a mechanism for cortisol delivery to inflamed tissues, the molecular basis for the regulation of such a delivery mechanism by endogenous and exogenous elastases remains poorly understood.
The six occupied N-glycosylation sites of mature CBG (383 amino acid residues) create extensive glycoform heterogeneity (4, 5, 35). We recently performed a detailed site-specific glycoprofiling of all six glycosylation sites of blood-derived CBG from healthy individuals (36). The sites displayed different heterogeneous populations of branched, primarily bi- and triantennary, complex glycans displaying α1,6-linked (core) fucose (Fuc) and terminal N-acetylneuraminic acid (NeuAc) residues. It was suggested that CBG carries predominantly α2,3-sialylation, as assessed by LC-MS/MS-based glycomics of free reduced N-glycans at the global (site-nonspecific) level of CBG with and without α2,3-linkage-specific sialidase treatment. In addition, sialylation was found to be an inhibitory feature for the CBG-receptor complex interaction (36). Other lines of evidence suggest that N-glycosylation is a key modulator of CBG functions, including cortisol binding and delivery (e.g. glycosylated CBG was found to bind cortisol with significantly higher affinity and was more temperature-sensitive relative to non-glycosylated CBG) (29). Moreover, upon NE-based RCL cleavage, the cortisol affinity of glycosylated CBG was reduced more than non-glycosylated forms, indicating that CBG glycosylation may yield a more rapid surge of free cortisol at target tissues to thereby facilitate a quicker resolution of inflammation (29). Intriguingly, the C-terminally located Asn347, which is 84.7% occupied by primarily triantennary sialoglycans, is located on the RCL in close proximity to the reported cleavage sites (36). Similarly, other highly occupied glycosylation sites of CBG displaying mainly bi- and triantennary sialoglycans appear to be in relatively close spatial vicinity of the RCL when evaluated on the three-dimensional structure of human CBG (32, 37). However, the site-specific effects of CBG N-glycosylation on the endogenous and exogenous elastase-based RCL proteolysis and cortisol release remain undocumented.
We establish here that CBG uses specific glycan features, in particular at the Asn347 site, to fine-tune the RCL proteolysis by two unrelated and co-existing elastases of host and pathogen origin that may be found in the respiratory tract of bacteria-infected individuals; the endogenous NE and P. aeruginosa-derived PAE. The site-specific modulatory functions of CBG N-glycosylation contribute to advancing our understanding of the complex molecular mechanisms underpinning the competing interests of the host and pathogen that facilitate subtle yet effective manipulation of the human immune response during infection and inflammation.
Results
High NE Specificity of the RCL Cleavage of CBG Irrespective of the Asn347 Glycosylation Status
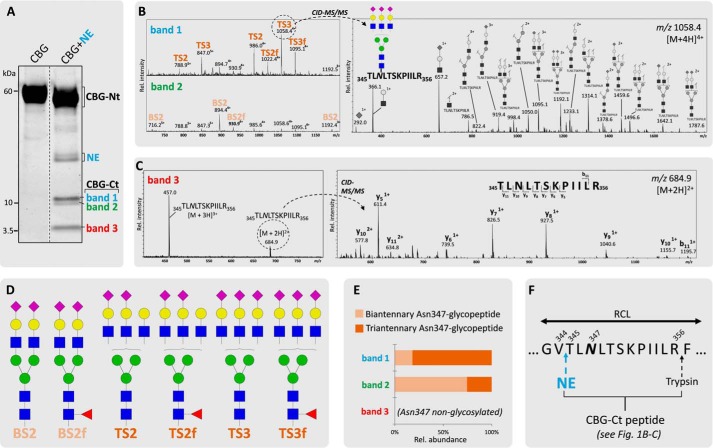
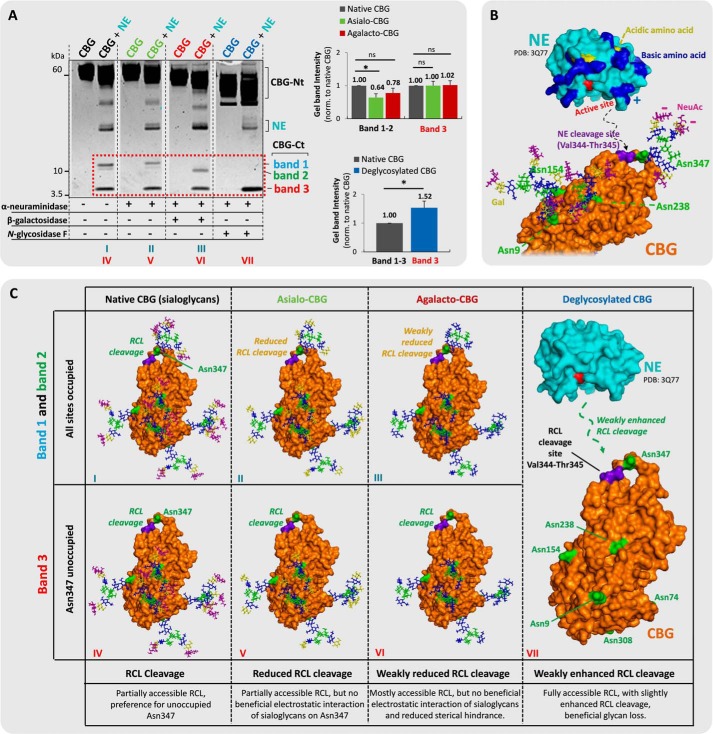
Short (2-min) incubation of human CBG with human neutrophil-derived NE under physiological conditions generated significant RCL cleavage of CBG. Two complementary sets of CBG fragments were formed (i.e. CBG-Ct (bands 1–3; 12, 11, and 5 kDa, respectively) and CBG-Nt (50–55 kDa) fragments) (Fig. 1A. Following in-gel trypsin digestion, multiple variants of the CBG-Ct fragment were identified by the detection of the peptide 345TLNLTSKPIILR356 in an Asn347-glycosylated (bands 1 and 2, Fig. 1B) and in a non-glycosylated (band 3, Fig. 1C) form using collision-induced dissociation (CID)-MS/MS. Thus, the CBG-Ct fragments were a convenient way to monitor the Asn347 glycosylation status of CBG upon RCL cleavage. Consistent with our previous glycoprofiling of native CBG from healthy human blood donors (36), six abundant glycoforms encompassing α1,6 (core)-fucosylated (f) and NeuAc-type α-sialylated (S) biantennary (B) and triantennary (T) complex-type N-glycans were found to occupy Asn347 (Fig. 1D). A few other Asn347 glycoforms of low abundance were observed as reported before (36); however, these trace structures were not included in the further analysis. The Asn347-glycosylated CBG-Ct fragments (bands 1 and 2) displayed predominantly triantennary (81.7%) and biantennary (74.5%) N-glycans, respectively, in agreement with their differential gel migration (Fig. 1E). MS/MS identification of the tryptic 401VMNPVC-term in all CBG-Ct bands confirmed an intact CBG C terminus (data not shown). The data supported a highly specific NE-based RCL cleavage at Val344-Thr345 irrespective of the Asn347 glycosylation status (Fig. 1F).
FIGURE 1.
Highly specific NE-based RCL cleavage proximal to the Asn347 glycosylation of CBG. A, generation of the two complementary sets of CBG fragments upon NE (light blue) cleavage (i.e. CBG-Ct (band 1, 12 kDa (blue); band 2, 11 kDa (green); band 3, 5 kDa (red)) and CBG-Nt (50–55 kDa) fragments as evaluated by their differential migration on SDS-PAGE. Broken line, non-neighboring gel lanes from the same gel. B, MS-based profiles of the NE-generated Asn347-glycosylated CBG-Ct fragments after trypsin digestion (left). The structures of the glycopeptides were confirmed using CID-MS/MS as exemplified by the fragmentation of a TS3 Asn347 glycopeptide (right) and our previous glycoprofiling of native CBG (36). C, band 3 contained the Asn347-non-glycosylated variant of the CBG-Ct fragment as confirmed by the identification of the non-modified peptide 345TLNLTSKPIILR356. D, overview of the nomenclature and structures of the six most abundant glycans harboring Asn347 (i.e. BS2, BS2f (biantennary (B) glycans in light orange); TS2, TS2f, TS3, and TS3f (triantennary (T) glycans in dark orange)). f, α1,6-linked (core) fucosylation; S, number of NeuAc residues. E, bands 1 and 2 contained primarily tri- and biantennary Asn347 glycans, respectively. F, sequence of a selected region of the exposed RCL of CBG. Blue and black arrows indicate the identified cleavage sites of NE and trypsin, respectively.
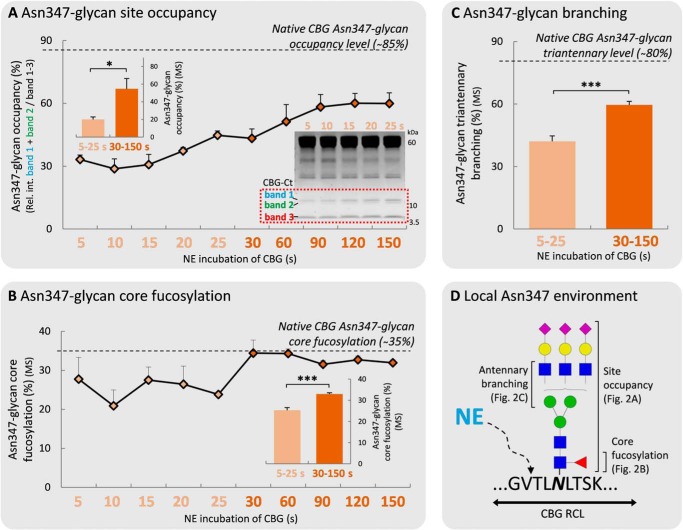
Asn347 Glycan Occupancy, Core Fucosylation, and Branching Reduce the NE-based RCL Cleavage Efficiency
The influence of the Asn347 glycan moiety on the NE-based RCL cleavage was investigated by monitoring three volume-enhancing features (i.e. glycan site occupancy, core fucosylation, and outer antennae GlcNAc branching) of the Asn347 glycan as a function of NE digestion time. The Asn347 glycan occupancy significantly affected the efficiency of the NE-induced RCL cleavage, as evidenced by a significant increase of the site occupancy over the short (5–25 s) (20.0 ± 2.7%) to medium (30–150 s) (54.6 ± 11.7%, p < 0.05) incubation period (Fig. 2A). In addition, the low Asn347 glycan occupancies (∼20–55%) relative to native human CBG (∼85%) (36) supported the strong NE digestion bias toward cleavage of non-glycosylated Asn347 CBG variants within this relatively short digestion window.
FIGURE 2.
The rapid NE-based RCL cleavage of CBG is affected by the Asn347 glycan occupancy, core fucosylation, and outer antennary GlcNAc branching. The relative levels of Asn347 glycan site occupancy (%) (A), core fucosylation (%) (B), and outer antennary GlcNAc branching (measured as the level of triantennary glycans) (%) (C) were measured on the tryptic peptide from the CBG-Ct fragment (345TLNLTSKPIILR356) as a function of NE-based digestion time (0–150 s) of CBG. See Fig. 3A for an extended NE-based incubation of CBG (i.e. 10 min and 4 h). Asn347-specific values of native human CBG based on previously reported data (36) are shown for the three studied glycan features (dotted lines). All data points are plotted as mean ± S.E. (error bars) (n = 3) and based on relative MS abundances except in A, where the occupancies are based the relative gel band intensities (see inset for the 5–25 s gel). The involvement of the three Asn347 glycan features was statistically tested by grouping the early (5–25 s, light orange) and the medium (30–150 s) digestion time points. *, p < 0.05; ***, p < 0.001. D, schematic illustration of the local molecular environment around Asn347 and the three glycan features affecting the NE-based RCL cleavage of CBG at Val344-Thr345.
Core fucosylation of the Asn347 glycans appeared to influence the NE-based proteolysis less, as shown by the only slightly lower levels of core fucosylation of Asn347 glycans over the short (5–25 s) (25.2 ± 1.3%) relative to the medium (30–150 s) (32.9 ± 0.6%, p < 0.05) incubation time scale (Fig. 2B). CBG-Ct fragments generated over the medium incubation time scale showed native-like levels of Asn347-core fucosylation (∼34.9%), illustrating that the digestion preference for non-core fucosylated Asn347 glycans was restricted to the short time scale.
The triantennary Asn347 glycans were found to be significantly underrepresented in the short (5–25 s) (42.1 ± 2.6%) as compared with the medium (30–150 s) (59.6 ± 1.7%) incubation period and compared with native CBG (∼80%) (36) (Fig. 2C). We also investigated these data for a potential bias toward or against higher sialylated Asn347 glycan species but found no different RCL cleavage rates of trisialylated triantennary Asn347 glycoforms relative to bisialylated triantennary Asn347 glycoforms over the investigated time course (data not shown). Together, these data showed that several volume-enhancing Asn347 glycan features (i.e. site occupancy, core fucosylation, and outer antennae GlcNAc branching) temporally affect the rapid NE-induced RCL cleavage efficiency at the proximal Val344-Thr345 cleavage site (Fig. 2D).
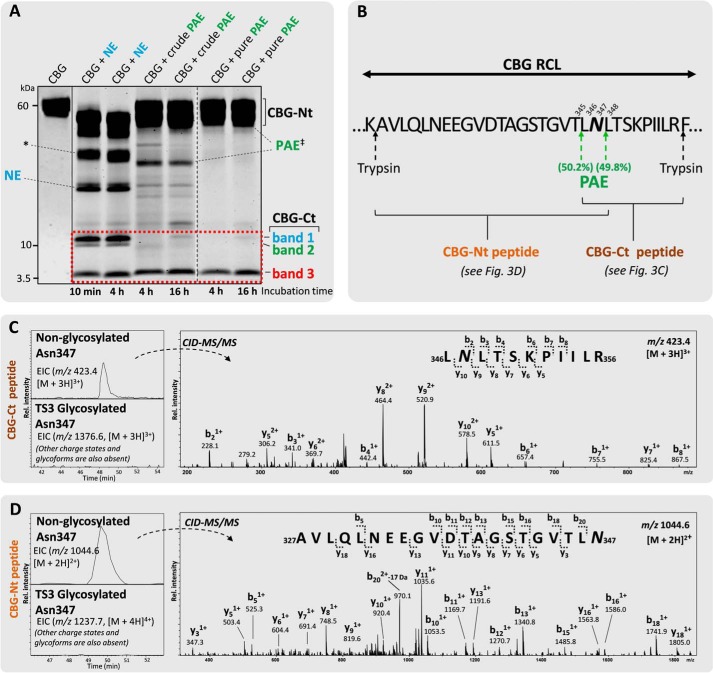
Asn347 Glycosylation Abolishes the Less Efficient PAE-induced RCL Cleavage
The influence of the Asn347 glycan on the RCL cleavage efficiency of the proteolytically less potent PAE from the culture media of virulent P. aeruginosa was then investigated. Surprisingly, even after favorable PAE incubation conditions and extended incubation time (i.e. 1 h (data not shown), 4 h, and 16 h), both crude and pure PAE produced only a single 5-kDa fragment (band 3) corresponding to an Asn347-non-glycosylated CBG-Ct fragment (Fig. 3A). The glycosylated CBG-Ct fragments (bands 1 and 2) that were clearly observed after NE digestion were absent following PAE digestion of CBG. This interesting observation was verified using LC-MS/MS by the identification of the peptide 346LNLTSKPIILR356 (Fig. 3, B and C). The peptide 348LTSKPIILR356 was identified as a second PAE cleavage product (data not shown). By assuming similar ionization efficiency of these two peptide variants, it was estimated that PAE has similar preference for cleaving the RCL at the two sites (i.e. Thr345-Leu346) (50.2%) and Asn347-Leu348 (49.8%) when Asn347 is not occupied. The peptide derived from the complementary CBG-Nt fragment (i.e. 327AVLQLNEEGVDTAGSTGVTLN347) was similarly identified exclusively in the non-glycosylated form by LC-MS/MS, thereby confirming that PAE is unable to cleave the RCL when Asn347 is occupied (Fig. 3D). The 327AVLQLNEEGVDTAGSTGVT345 peptide was also identified, which further confirmed the dual cleavage sites of PAE (data not shown). The other reported, less used, PAE cleavage sites of RCL (i.e. Leu346-Asn347, Leu348-Thr349, Thr349-Ser350, and Ser350-Lys351) (33) were not detected.
FIGURE 3.
Glycan occupancy of Asn347 abolishes the PAE-induced RCL cleavage. A, unlike NE-digested (light blue) CBG (two penultimate left lanes), crudely isolated and pure PAE (green) generated two relatively homogeneous complementary CBG fragments (i.e. a single CBG-Ct (band 3, 5 kDa) and a CBG-Nt (50–55 kDa) fragment under extended incubation (4 and 16 h, right lanes) as evaluated by SDS-PAGE. Undigested native CBG is shown in the far left lane. Broken line, non-neighboring gel lanes from the same gel. Solid line, gel lanes from different gels. B, amino acid residues of the exposed RCL of CBG in the vicinity of the observed PAE cleavage sites (green arrows). The CBG-Nt (gold) and CBG-Ct (brown) peptides covering the Asn347 glycosylation site are shown. C, the presence of 346LNLTSKPIILR356 and 348LTSKPIILR356 (data for the latter not shown) in an Asn347-non-glycosylated form (top EIC, left) and the absence of their corresponding Asn347-glycosylated forms (bottom EIC, left; as exemplified by the absence of an EIC signal for the TS3 Asn347 glycopeptide) in the CBG-Ct fragment was confirmed using CID-MS/MS (right). D, peptides of the complementary CBG-Nt fragments (i.e. 327AVLQLNEEGVDTAGSTGVTLN347 and 327AVLQLNEEGVDTAGSTGVT345 (data for the latter not shown) were also exclusively identified in their non-glycosylated state by LC-MS/MS. No EIC signals were detected in any charge states for any glycoforms of either the CBG-Nt or the CBG-Ct peptides. *, CBG-Nt fragments occurring after extended digestion of CBG with NE that were not further investigated in this study. ‡, crude PAE migrated around 33 kDa as validated by LC-MS/MS analysis, whereas the commercial form of PAE (“pure PAE”) was observed in separate experiments to co-migrate with human CBG around 55–60 kDa (data not shown).
Sialylation of Asn347- and Non-Asn347 Glycans Promotes the PAE- and NE-based RCL Proteolysis
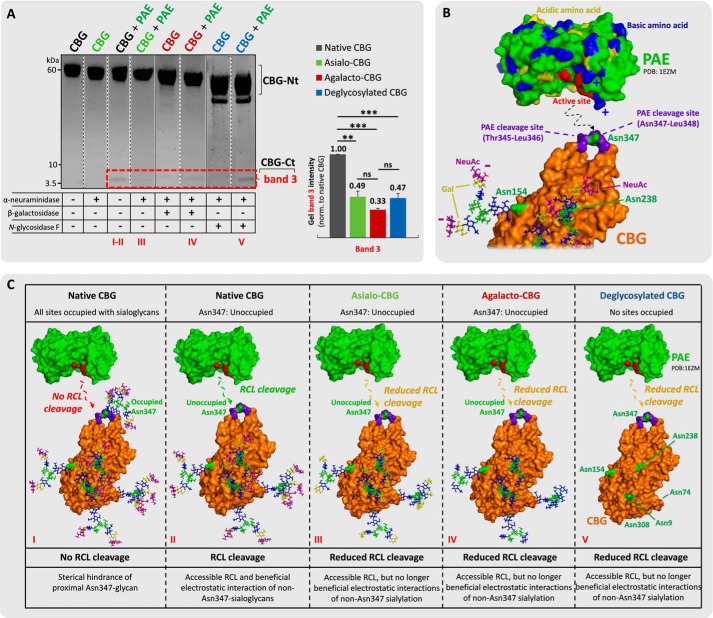
The influence of terminal sialylation of the proximal Asn347 and other non-Asn347 glycans of CBG on the RCL cleavage efficiencies by NE and PAE was then investigated. This was performed using a series of time-fixed digestions of a panel of CBG glycosylation variants consisting of asialo-, agalacto-, and deglycosylated CBG generated by treatment of exoglycosidases and N-glycosidase F under native (conformation-retaining) conditions. Interestingly, the global enzymatic removal of the terminal α-linked NeuAc residues of CBG dramatically reduced the RCL cleavage efficiency of PAE as evaluated by the reduced generation of the CBG-Ct fragment (band 3, 49.1 ± 6.8%) relative to the native CBG (normalized to 100%, p = 1.7 × 10−3) (Fig. 4A). However, global removal of the penultimate β1,4-linked galactose residues (agalacto-CBG, 33.5 ± 1.7%) and the entire N-glycan moieties (deglycosylated CBG, 47.4 ± 5.5%) of CBG did not further change the RCL cleavage efficiency by PAE (p ≥ 0.05). This indicated that sialylation of the non-Asn347 glycans is strongly enhancing the RCL digestion efficiency of PAE. In addition, this data indicated that the RCL digestion efficiency of PAE was rather independent of the galactose residues as well as the presence of the remaining parts of the non-Asn347 glycan structures. By inspecting our glycosylated homology model of uncleaved human CBG, the Asn238 and Asn154 glycans, which naturally comprise biantennary sialoglycans at high occupancies (36), are in relatively close proximity to the RCL. It appears likely that the negatively charged sialic acid residues of these two N-sites may be beneficial for efficient PAE proteolysis, possibly by electrostatic interactions with the highly cationic protein surface surrounding the active site of PAE. This might indeed explain the reduced and non-recoverable RCL cleavage efficiency of PAE on asialo-CBG and further truncated CBG glycosylation analogs (Fig. 4C). Further work is warranted to determine the exact glycosylation site(s) contributing to these effects.
FIGURE 4.
Sialylation of the non-Asn347 glycans is beneficial for PAE-based RCL cleavage. A, incubations of (from left) undigested native CBG, undigested asialo-CBG, pure PAE-digested native CBG, pure PAE-digested asialo-CBG, undigested agalacto-CBG, pure PAE-digested agalacto-CBG, undigested deglycosylated CBG, and pure PAE-digested deglycosylated CBG. Shown is a comparison of the relative gel band intensities of band 3 (red box). Broken line, non-neighboring gel lanes from the same gel. Solid line, gel lanes from different gels. B, three-dimensional structure of PAE (PDB code 1EZM) and RCL-uncleaved CBG (homology model based on uncleaved human TBG, PDB code 2CEO). CBG is presented in a glycoform that allows RCL cleavage by PAE (i.e. all glycosylation sites are occupied except for Asn347). Color coding of monosaccharide residues is in accordance with the established nomenclature by the Consortium for Functional Glycomics (79). NeuAc residues are in purple, galactose residues are in yellow, GlcNAc residues are in blue, fucose residues are in red, and mannose residues are in green. Color coding of the polypeptide chains is as follows. The PAE cleavage sites (Thr345-Leu346 and Asn347-Leu348) on the RCL of CBG are in purple, the basic amino acid residues of PAE are in blue, acidic amino acid residues of PAE are in yellow, the amino acid residues in the active site of PAE are in red, and the Asn glycosylated sites of interest of CBG are in green. The proposed electrostatic interactions and protein docking are schematically illustrated with broken lines. C, proposed interactions of NE and glycosylated variants of CBG, which may explain the differential RCL cleavage efficiencies observed in A; same color coding as in B. **, p < 0.01; ***, p < 0.001; ns, not significant. Data points are represented as mean ± S.E. (error bars), n = 3.
Comparatively less reduction of the RCL digestion efficiency upon global desialylation of CBG was observed for NE (Fig. 5A). Interestingly, the reduced proteolysis was restricted to the Asn347-glycosylated CBG-Ct fragments (bands 1 and 2, 64.2 ± 11.3% versus native CBG, normalized to 100%; p < 0.05), whereas the levels of the Asn347-non-glycosylated CBG-Ct fragment were not altered (band 3, 100.2 ± 13.4% versus native CBG, normalized to 100%; p ≥ 0.05). This indicates that the NeuAc residues of the proximal Asn347 glycan, but not the other CBG N-sites, are beneficial, but ultimately not strictly crucial, for the NE digestion of RCL. The global removal of the galactose residues of CBG showed a recovery of the Asn347-glycosylated CBG-Ct fragments (bands 1 and 2, 77.9 ± 14.7%; p ≥ 0.05) relative to native CBG. In contrast, the levels of the Asn347-non-glycosylated CBG-Ct fragment (band 3, 102.0 ± 13.0%; p ≥ 0.05) remained unchanged relative to native and asialo-CBG. Recovery and indeed enhanced RCL digestion efficiency relative to native CBG (combined bands 1–3, normalized to 100%) were observed with global CBG deglycosylation (153.5 ± 23.3%; p < 0.05). Together, this indicated that only NeuAc residues of Asn347, and not NeuAc residues from the other CBG N-sites, are enhancing the RCL digestion efficiency of NE, whereas the volume-enhancing galactose residues and the underlying glycan cores of Asn347 are antagonistic features reducing its RCL digestion efficiency (Fig. 5, B and C).
FIGURE 5.
Sialylation of Asn347 glycans, not of glycans from other CBG sites, is beneficial for NE-based RCL cleavage. A, incubations of (from left) undigested native CBG (black), NE-digested native CBG, undigested asialo-CBG (green), NE-digested asialo-CBG, undigested agalacto-CBG (red), NE-digested agalacto-CBG, undigested deglycosylated CBG (blue), and NE-digested deglycosylated CBG. Comparisons of the relative gel band intensities (red box) were performed. Broken line, non-neighboring gel lanes from the same gel. B, the three-dimensional structures of NE (PDB code 3Q77) and RCL uncleaved CBG (homology model based on uncleaved human TBG, PDB code 2CEO). CBG is presented with its most abundant glycoforms of the three sites located in proximity to the RCL (i.e. Asn347 displays TS3, and Asn154 and Asn238 both display BS2). The color coding of the monosaccharide residues and amino acid residues is generally as described in the legend to Fig. 4, the NE cleavage site (Val344-Thr345) on the RCL of CBG is in purple. The proposed electrostatic interactions and protein docking to the RCL of CBG are schematically illustrated with broken lines. C, proposed interactions of NE and the glycosylated variants of CBG, which may explain the differential RCL cleavage observed in Fig. 5A. *, p < 0.05; ns, not significant. Data points are represented as mean ± S.E. (error bars), n = 3.
Molecular Dynamics (MD) Simulations of Glycosylated CBG Support an Asn347 Glycan Involvement in the RCL Cleavage
To generate a better understanding of the molecular mechanisms underpinning RCL cleavage, 100-ns MD simulations of abundant glycoforms of CBG were performed. Because the exact triantennary branching of the Asn347 glycans remains unknown (36), both the β1,4- and β1,6-GlcNAc-branched isomers (i.e. TS3f-A and TS3f-B, respectively) were simulated. The simulations did not achieve complete structural convergence but were sufficient to support our hypotheses regarding the relationship between CBG glycoforms and protease activity. Specific interactions formed between the Asn347 glycans and the CBG surface, which persisted throughout the simulation, indicated that they are significantly populated over longer time scales.
Regardless of the glycoform, glycan occupancy at Asn347 significantly perturbed the average conformation adopted by the RCL (root mean square deviation, 2.4 ± 0.3 to 4.1 ± 0.5 Å) relative to unglycosylated RCL (1.5 ± 0.4 Å) (Table 1). The biantennary sialoglycan (BS2) at Asn347 did not interact with CBG and remained projected away from the protein over the entire simulation. This resulted in less perturbation of the RCL position relative to the more branched Asn347 glycoforms. This suggests that the attenuating effects of both glycan occupancy and triantennary branching at Asn347 on the RCL cleavage efficiency (Fig. 2, A and C) result from altered RCL conformations, in addition to the glycan sterically blocking access to the cleavage sites.
TABLE 1.
Asn347 glycosylation, in particular the triantennary glycoforms, causes more perturbation of the average RCL conformation relative to the non-glycosylated RCL structure
Perturbation, which may restrict access of or affect recognition by the RCL-targeting proteases, was measured as the average root mean square deviation (Å) (mean ± S.D.) of the Cα atoms of the RCL over the course of the simulations (t = 100 ns, n = 1,000 sampled time points) of the Asn347 glycoforms of CBG relative to the average structure adopted by the RCL Cα atoms in the non-glycosylated CBG variant.
| CBG Asn347 glycoform | Perturbation of the RCL conformation (RMSD) (mean ± S.D.) |
|---|---|
| Å | |
| Non-glycosylated | 1.5 ± 0.4a |
| BS2 | 2.4 ± 0.3 |
| TS3-A | 4.1 ± 0.5 |
| TS3-B | 3.1 ± 0.4 |
| TS3f-A | 3.1 ± 0.4 |
| TS3f-B | 3.5 ± 0.7 |
| DTS3-A | 3.9 ± 0.4 |
| DTS3-B | 3.5 ± 0.6 |
a This value represents the baseline deviation from the average RCL structure without perturbation by an attached glycan. See the legend to Fig. 1 and supplemental Table S1 for the Asn347 glycoform nomenclature used here.
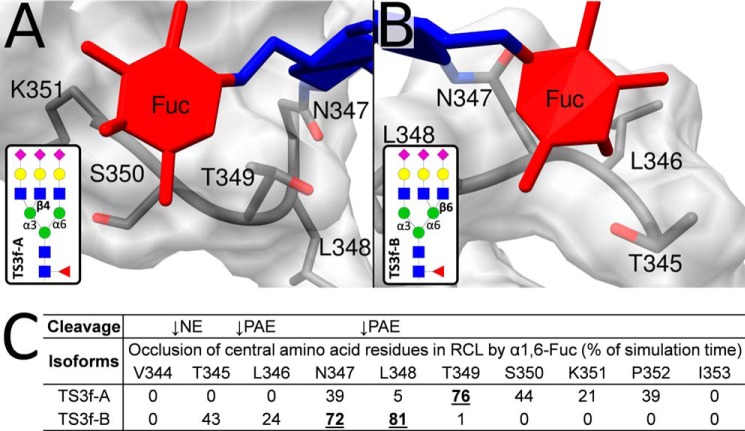
It is intuitive that the proximal core fucose of Asn347 glycans would affect the RCL proteolysis. Consistent with the experimental data (Fig. 2B), the MD simulations supported that core fucosylation reduces access to central amino acid residues in the RCL. Interestingly, the core fucose of the TS3f-A and TS3f-B isomers formed sustained interactions with, and thereby sterically masked, different regions of the RCL (Fig. 6, A and B). The TS3f-A isomer (Man 3′-arm GlcNAc branching) occluded the Asn347–Pro352 region involving the two PAE cleavage sites, whereas the TS3f-B isomer (Man 6′-arm GlcNAc branching) occluded Thr345–Leu348, which contains both the NE and PAE cleavage sites (Fig. 6C). Upon close reexamination of our existing glycome (site-nonspecific) LC-MS/MS data of CBG (36), it is estimated that triantennary A-isomers (∼80–85%) dominate over the B-isomers (∼15–20%) in CBG (data not shown). This may explain the limited, but still significant, influence of the core fucosylation of Asn347 glycans on the NE cleavage of the RCL. However, a detailed linkage and branch analysis of the Asn347 glycoforms is required to confirm the exact glycan structures at this site.
FIGURE 6.
MD simulations indicate that core fucosylation of Asn347 glycans occludes central amino acid residues of RCL. The α1,6-linked core fucose of the TS3f-A (Man 3′-arm GlcNAc branching) (A) and TS3f-B (Man 6′-arm GlcNAc branching) (B) isomers formed prolonged interactions with two different regions of the RCL. C, the interactions generated by the TS3f-B isomer resulted in significant occlusion of both the NE and PAE proteolytic sites as measured by the reduced solvent accessibility of the Thr345-Leu348 region generated by the core fucose residue over the simulation time. In contrast, the core fucose-centric interactions generated by the supposedly more abundant TS3f-A isomer influenced only the PAE digestion sites by occluding the Asn347–Pro352 region. Amino acid residues occluded for >50% of the simulation time are in boldface type and underlined. See Fig. 7 and Table 2 for the RCL occlusions provided by the entire Asn347 glycan moieties, not just their core fucose residues. See the legend to Fig. 1 and supplemental Table S1 for the nomenclature used for the Asn347 glycoforms.
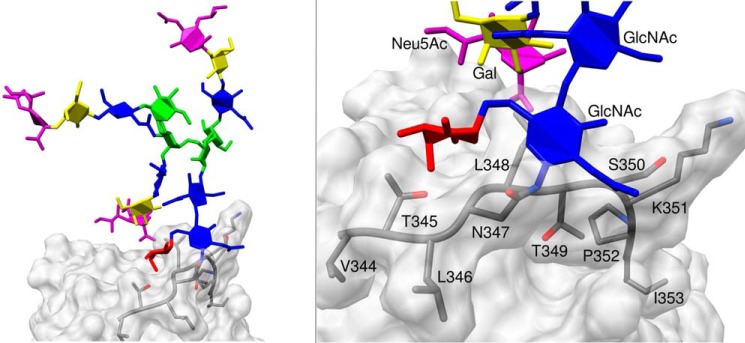
The MD simulations also permitted a deeper examination of the average shielding effect of the RCL generated by the entire Asn347 glycan structure during the simulation (Table 2). Irrespective of the occupying Asn347 glycoform, it was predominantly the PAE sites that were affected by the presence of a glycan in this position, which formed sustained contacts with the CBG surface (Fig. 7). The negligible steric hindrance of the NE digestion site by the Asn347 glycans on the RCL is in strong agreement with the experimental observations.
TABLE 2.
Proportion of simulation time (%) that individual RCL residues are shielded by the Asn347 glycan
Amino acid residues occluded >50% of the simulation time are in bolface type and underlined. These heavily occluded residues were predominantly located in proximity to the two PAE cleavage sites rather than the NE cleavage site (indicated with arrows). See legend for Fig. 1 and supplemental Table S1 for the Asn347 glycoform nomenclature used here.
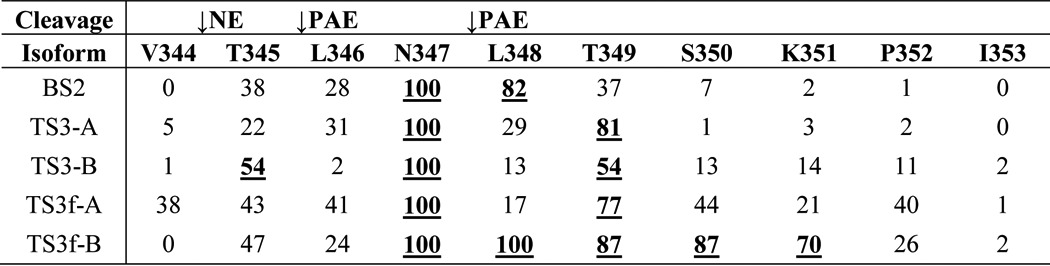
FIGURE 7.
MD simulations suggest that Asn347 glycans form sustained interactions to the RCL backbone of CBG that occlude primarily the PAE digestion sites. Shown is an example of Asn347 glycan-CBG protein surface interaction as demonstrated by the prolonged contact of the chitobiose core of TS3f-B to the PAE digestion sites of the RCL (left), in particular the Asn347–Lys351 region; see enlarged region (right) for details. See also Table 2 for details on the reduction of solvent accessibilities of TS3f-B and other Asn347 glycoforms over the MD simulation time. See the legend to Fig. 1 and supplemental Table S1 for the nomenclature used for the Asn347 glycoforms.
Abundant Secretion of the Inefficient PAE of Virulent P. aeruginosa May Trigger Slow Cortisol Release for Sustained Host Immune Suppression and Avoidance of Growth Inhibition
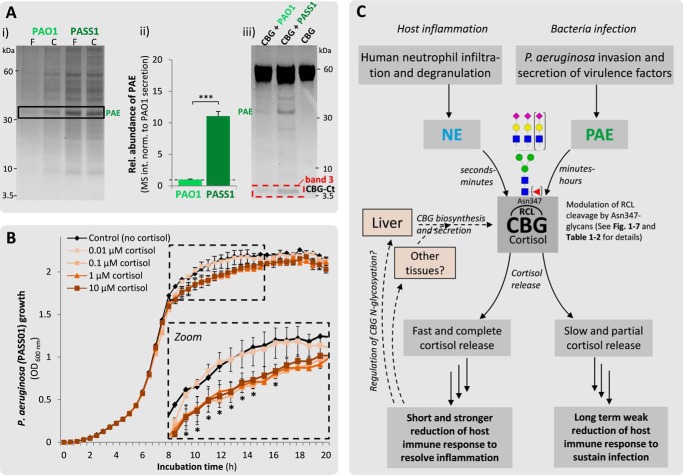
We observed that PAE, a well known virulence factor in cystic fibrosis and chronic obstructive pulmonary disease (34), is among the most abundant proteins secreted by P. aeruginosa (Fig. 8A, i). The cystic fibrosis-derived virulent PASS1 strain clearly secreted more PAE into the culture media than the laboratory PAO1 wound strain over the same period as evaluated by their gel band intensities on SDS-PAGE. A virulence-dependent expression of PAE was also observed in the cell lysates of P. aeruginosa, showing significantly higher intracellular PAE levels in PASS1 compared with in the same number of PAO1 bacteria (11.1 ± 0.7-fold, p = 9.4 × 10−6) as measured using LC-MS/MS (Fig. 8A, ii). Only the virulent PASS1 resulted in the RCL cleavage as evaluated by the generation of a CBG-Ct fragment when equivolume culture media from the same number of the two bacterial strains grown until their mid-log phase under identical conditions were incubated with CBG (Fig. 8A, iii). The differential RCL cleavage efficiency appeared to be reflecting the difference in PAE secretion, but strain-specific differences of the PAE structure and/or activity cannot be ruled out.
FIGURE 8.
High PAE secretion and cortisol concentration-dependent growth inhibition of virulent P. aeruginosa may explain the relatively weak RCL potency of PAE. A, when cultured overnight, virulent P. aeruginosa (PASS1) isolated from the sputum of a cystic fibrosis patient showed, relative to the equivalent number of (or equivolume media from) the laboratory PAO1 wound strain, the following: a higher PAE secretion as evaluated by gel band intensities of filtered (F) and crude (C) culture media (i); significantly higher intracellular PAE levels as evaluated by intensity-based LC-MS/MS analysis of bacteria cell lysates (ii); and a higher capacity for generating the CBG-Ct fragments, indicating enhanced RCL cleavage (iii). ***, p < 0.001 (mean ± S.D. (error bars), n = 3). B, the PASS1 growth profile was monitored over time (0–20 h) without (black) and with various concentrations of cortisol (0.01–10 μm, various shadings of brown) covering the physiological cortisol range in humans. No significant differences were observed for the 0.01 μm cortisol relative to the untreated control (p ≥ 0.05 for all time points), whereas a minor concentration-dependent bacteriostatic growth inhibition was observed for the 0.1–10 μm cortisol relative to the 0.01 μm cortisol concentration (p < 0.05 for time points marked with an asterisk in the expanded inset). All data points are presented as mean ± S.E. (error bars) (n = 3). C, proposed simplistic model of the relationship between the human host inflammation and bacterial infection pathways in the context of NE- and PAE-induced cortisol release upon RCL cleavage. The potent NE acts preferentially on non-glycosylated Asn347 variants or CBG glycoforms carrying Asn347 glycans that are sialylated, non-core-fucosylated, and biantennary branched on the second-to-minute time scale, yielding a substantial release of cortisol from hepatocyte- and potentially extrahepatocyte-derived CBG present at the site of inflammation (24). In contrast, the less potent PAE acts exclusively on non-glycosylated Asn347 CBG variants during the longer (minutes-to-hours) incubation period and benefits significantly from the sialylation of non-Asn347 glycans. This rather inefficient proteolysis may generate a slow release of cortisol of only a subset of CBG-cortisol complexes, which may be below the bacteriostatic concentration threshold but perhaps still sufficient to reduce the host immune response by the anti-inflammatory effects of cortisol, thereby possibly creating a favorable infection environment of P. aeruginosa. The comparative faster and more substantial release of cortisol by NE, on the contrary, would, in principal, create a more rapid resolution of the inflammation and possibly generate a significant bacteriostatic effect and a more hostile environment for bacterial infection. See Fig. 1 and supplemental Table S1 for the nomenclature used for the Asn347 glycoforms.
The motivation for P. aeruginosa secreting high amounts of “inefficient” PAE, causing inefficient CBG RCL cleavage and thereby sub-optimal cortisol release from the host, was studied by monitoring the growth of the virulent PASS1 strain over 20 h with 4 orders of magnitude of free cortisol concentrations (0.01–10 μm) in the culture media. The physiological level of free cortisol in humans is 0.1–1 μm (38). The lowest cortisol concentration (0.01 μm) neither promoted nor inhibited the growth of PASS1 relative to the control grown under identical conditions without cortisol supplement (p ≥ 0.05 for all time points) (Fig. 8B). However, the higher cortisol concentrations (0.1–10 μm) had an equal suppressive effect on the PASS1 growth in the 8–16 h period (p < 0.05 for the indicated time points). In other words, the higher cortisol concentration (10 μm) did not suppress the PASS1 growth more than the 0.1 μm cortisol concentration, indicating a saturation of the bacteriostatic effect of cortisol within the physiological range. Any bacteriostatic effects of cortisol were absent in the beginning (0–8 h) and less pronounced in the end (16–20 h) of the studied growth period. Taken together, the data suggest that the high secretion of the relatively weakly potent PAE of virulent P. aeruginosa strains facilitates a limited cortisol release from the host upon slow RCL cleavage, which may be below the bacteriostatic concentration but sufficient to achieve sustained suppression of the host immune system to promote bacterial infection (Fig. 8C).
Discussion
CBG is a central protein in inflammation due to its transport of corticosteroids in circulation and its ability to deliver such anti-inflammatory stress hormones in an accurate and timely way to the required tissues. Although not fully resolved, one aspect of the delivery mechanism that is rather well established is the NE-driven proteolysis of the RCL leading to cortisol release upon a so-called stress-to-relaxed conformational change of CBG at the neutrophil-rich inflammatory site (22, 24). As demonstrated here by time-based digestion assays and structural analysis of the resulting glycoform fragments and supported by MD simulations, the digestion efficiency of the highly potent and cleavage site-specific (Val344-Thr345) endogenous NE is modulated in an intriguingly complex manner by the Asn347 glycosylation of CBG. The predominant bi- and trivalent NeuAc-type sialylation of the Asn347 glycans was found to be highly beneficial for NE digestion, possibly by enabling electrostatic attraction to the arginine-rich surface of NE (39) upon docking onto the RCL. With respect to the RCL cleavage rate, no additional benefits of trivalent over bivalent sialylation of the Asn347 glycan were observed, indicating that the orientation and the position of the anionic charges around the RCL are more important than the sheer number of negative charges. The MD simulations indicated that some Asn347 sialoglycoforms form interactions with the CBG protein surface (data not shown). However, further work is warranted to investigate whether these interactions promote RCL digestion by NE.
In contrast, other volume-enhancing neutral features of the Asn347 glycans, including the site occupancy, core fucosylation, outer antennary β-GlcNAc branching, and β-galactosylation, were found to antagonize the RCL digestion efficiency of NE. It is anticipated that these inhibitory glycan features are exerting their action by steric hindrance by forming hydrophilic (hydrogen bonding from hydroxyl groups) and/or hydrophobic (from N-acetyl groups of GlcNAc and NeuAc) interactions with the CBG surface, thereby masking the digestion sites of the RCL. The presence of core fucosylation of N-glycans has previously been shown to inhibit the proximal proteolysis of polypeptides by serine proteases (40). None of the five other CBG N-glycans (at sites Asn9, Asn74, Asn154, Asn238, and Asn308) displayed any detectable modulation of the NE digestion efficiency of the RCL, which can be rationalized from the relative small size of NE (mature polypeptide, 25.5 kDa) and the significant distance of the digestion site to the “neighboring” Asn154 (∼44 Å) and Asn238 (∼34 Å) of uncleaved CBG (data not shown, but see Figs. 4 and 5 for model). Despite the presence of these multiple inhibitory features of the Asn347 glycans, NE yielded significant CBG fragments in the second-to-minute time scale. Thus, the Asn347 glycosylation does not prevent the NE-based cleavage of CBG but instead fine-tunes the digestion rate and, thus, is able to manipulate the speed at which cortisol is released into inflamed tissues. Consequently, the Asn347 glycosylation may be viewed as an anti-inflammatory regulator.
Unlike NeuAc-type α-sialylation and β-galactosylation, which forms the terminal and penultimate glycoepitopes, respectively, of all native CBG glycoforms on Asn347, it is interesting to note that the other regulatory Asn347 glycan features are partial modifications (i.e. the Asn347 site occupancy is ∼85%, core fucosylation is ∼35%, and triantennary GlcNAc branching is ∼70% as evaluated from an “average” CBG population derived from a pool of healthy donors) (36). The degree of expression of these glycan features on CBG may be manipulated by hepatic cells or extrahepatic tissues as required to alter the NE-based RCL digestion rate and the cortisol release from CBG (e.g. during inflammation or bacterial infection). Both cellular factors (e.g. levels of glycosylation enzymes and nucleotide sugar donors) (41) and protein features (e.g. glycosylation site accessibility (42) and subcellular localization/trafficking rate) (41, 43) may contribute to the regulation of the N-glycosylation of a protein. Specifically, the global glycosylation pattern of hepatocyte-derived CBG has indeed been shown, in vitro, to be regulated by glucocorticoids (44); however, it still needs to be assessed whether altered cortisol levels affect the secretion and glycosylation pattern of hepatocyte-derived CBG and/or potentially the expression of extrahepatic CBG displaying different glycoforms and activities in a feedback mechanism. In relation to this, it would be interesting to map the natural variation of the Asn347 glycosylation of blood- and tissue-derived CBG in healthy donors and to compare these profiles with those of individuals undergoing local/systemic inflammation and bacterial infection to further test the observations described herein. Understanding the molecular basis for rapid tissue-specific cortisol delivery may have great utility in the rational design of CBG glycoforms with enhanced cleavage characteristics as a new type of therapeutics that can be directed to inflammatory tissues (e.g. in septic shock, prostatic hyperplasia, and rheumatoid arthritis) (45–47).
The immune modulation caused by other endogenous and exogenous proteases upon CBG cleavage is even less studied. PAE produced by virulent P. aeruginosa was shown to cleave the RCL of CBG, causing a diminished cortisol binding capacity (33). Importantly, other pathogens (i.e. Burkholderia cenocepacia and Staphylococcus aureus) did not produce RCL-cleaving proteases, suggesting that this ability may be unique to P. aeruginosa (33). Supporting these observations, the RCL of CBG was here shown to be a proteolytic target of PAE; two equally preferred cleavage sites (Thr345-Leu346 and Asn347-Leu348) were identified. PAE is the most secreted protein of P. aeruginosa, and despite its poor proteolytic activity (acting on a minutes-to-hours time scale) relative to NE, the total proteolytic capacity of PAE may still be significant, considering its abundance. In contrast to NE, the action of PAE was affected by the glycans on multiple glycosylation sites of CBG. Specifically, PAE digestion was shown to benefit from the sialylation of glycans on sites other than Asn347. Further investigation needs to specify whether these promoting effects are generated directly by electrostatic attractions of the neighboring Asn154 and Asn238 to the larger (relative to NE) PAE molecule (mature polypeptide, 33.1 kDa) upon RCL docking or whether the sialic acid residues are indirectly facilitating a more cleavage-susceptible conformation of CBG by engaging in charge-based interactions with its polypeptide backbone. Despite the lack of any direct evidence, we cannot rule out the possibility that negative charges from intra- or intermolecular sources other than sialylation of CBG, including glycosaminoglycans and phosphorylation, may similarly play a yet to be explored role in RCL modulation. It is also of interest to note that although the CBG sialylation appears to be a promoting feature of both NE and PAE digestion, we have previously shown that these negatively charged terminal glycoepitopes are antagonistic features reducing the interaction of CBG to yet to be thoroughly characterized putative cell surface receptors (36). Thus, the same glycosylation features may display multiple, diverse, and even opposite modulatory functions.
Interestingly, PAE-induced RCL cleavage was restricted to the subpopulation of CBG molecules lacking Asn347 glycosylation, amounting to only ∼15% of native CBG molecules (36). This suggests that PAE can only access a small proportion of the CBG-bound cortisol in a biological context, which may, however, be sufficient for promoting long term inhibition of the host immune response and thus allow the bacteria to gain a growth advantage by initiating and sustaining colonization. P. aeruginosa is an opportunistic pathogen that is known to establish chronic infection of immunocompromised individuals (e.g. in the respiratory tract of cystic fibrosis and chronic obstructive pulmonary disease patients) by having evolved refined strategies for long term infection, such as quorum sensing and biofilm formation (48–50). Because the release of free cortisol delays the production of pro-inflammatory factors designed to combat bacterial invasion, the PAE-induced RCL cleavage of CBG may also be viewed as one such infection strategy that enhances the chances of this opportunistic pathogen to evade the host immune system. The low potency of PAE may be crucial to avoid triggering a more complete release of cortisol, which appears to be detrimental to the growth of virulent P. aeruginosa, as demonstrated here. The weak bacteriostatic effect of low micromolar cortisol concentrations and the underlying suppressive mechanisms on the growth of P. aeruginosa need to be further investigated by orthogonal approaches.
The RCL of human α1-antitrypsin (A1AT, UniProtKB, P01009), another member of the serpin superfamily, is also a target of PAE (51). This is important because A1AT, unlike CBG, shows serpin-inhibitory activity by irreversibly binding and inactivating NE (and other serine proteases) and thereby reducing its CBG-cleaving abilities (52). Thus, the PAE-induced cleavage of A1AT may result in reduced NE inactivation and consequently an increased NE-based CBG digestion and cortisol release. Similar to NE, A1AT is present at the site of inflammation due to its localized production and secretion from leukocytes (53). Future work will reveal whether glycosylation is involved in the A1AT-based inhibition of NE and whether this inhibition is as potent as the well described NE inhibition displayed by a number of chemical inhibitors (e.g. N-benzoylindazole derivatives (54) as well as acylating and carbamylating agents and inhibitors based on other mechanisms (55)) and other protein/peptide-based inhibitors, such as cyclotides (56). Interestingly, the exposed RCLs of serpins generally do not share a common amino acid sequence and harbor no conserved N-glycosylation sequon motifs (NX(S/T), where X is not proline) (57). For example, no RCL-localized glycosylation sites similar to Asn347 of CBG were identified for A1AT and the structurally related thyroxine-binding globulin (TBG). It could be speculated that the RCL of especially inhibitory serpins encompassing the majority of the members within this superfamily lacks glycosylation sites because the binding and inactivation of proteases through the RCL may be compromised by the presence of a glycan in this region. However, it remains to be investigated whether any of the more than 70 serpin members, in addition to CBG, harbor an RCL glycosylation site, which may similarly modulate their inhibitory/non-inhibitory functions (58). The lack of serpin sequence conservation between species is attributed to the adaptive changes necessary to escape pathogenic species-specific elastases (57) and to accommodate for species-specific hormonal and physiological differences (59). In this context, the Asn347 glycosylation of the RCL of human CBG may have evolved as an additional level of fine control of and/or protection from digestion by exogenous and endogenous elastases, considering the diverse environments of CBG (i.e. produced in liver, circulates in blood, and migrates to inflammatory and steroid-sensitive tissues). The RCL glycosylation of CBG may regulate its spatial and temporal cleavage and thus be crucial to ensure accurate cortisol delivery.
Although the ability of glycans to protect their carrier proteins from proteolytic degradation has been known for many years (60, 61), still very few examples have been reported, in particular, in biological systems where the regulatory role of the individual glycan features have been documented (62). The fine control provided by the Asn347 glycosylation of CBG regulating the RCL proteolysis by important endogenous and exogenous proteases as reported here represents a fascinating example of the extremely complex molecular regulation underpinning the immune system. This work has demonstrated the central role of CBG glycosylation in providing an efficient immune response upon challenge and has brought us closer to understanding the molecular basis underpinning the regulation of these processes in the context of inflammation and bacterial infection.
Experimental Procedures
Materials
Human CBG (UniProtKB, P08185) was purified from pooled sera of healthy donors to >90% purity as assessed by SDS-PAGE (Affiland (Liège, Belgium), product code hCBGs). We have previously performed deep structural analysis of this source of CBG and confirmed high purity and structural and functional integrity of the protein (36). Human NE (UniProtKB, P08246) was isolated from blood-derived resting human neutrophils to >96% purity (Lee Biosolutions (Maryland Heights, MO), product code 342-40). Chromatographically purified PAE (here called “pure” PAE) (UniProtKB, P14756) was from a pathogenic P. aeruginosa strain (Elastin Products (Owensville, MO), product code PE961). The cystic fibrosis-derived (PASS1) (63) and laboratory wound (PAO1) P. aeruginosa strains, from which PAE was also derived, were kindly provided by Prof. Ian Paulsen (Macquarie University) (63). Cortisol (product code H0888) was from Sigma-Aldrich. Broad specificity Arthrobacter ureafaciens α2,3/6/8/9-linkage-specific neuraminidase (product code 480716, recombinant in E. coli) and β1,4-linkage-specific Streptococcus pneumoniae galactosidase (product code 345806, recombinant in E. coli) were from Calbiochem. Flavobacterium meningosepticum peptide:N-glycosidase F (recombinant in E. coli) was from Roche Diagnostics. Ultra-high quality water was used for all experiments. All chemicals and LC solvents were from Sigma and Merck, respectively, unless otherwise stated.
Bacteria Culture and Crude Isolation and Identification of PAE
PASS1 and PAO1 were cultured on Luria-Bertani agar plates and incubated overnight at 37 °C as described (64). Cell colonies were transferred to 5 ml of LB medium and incubated overnight at 37 °C, and the culture media containing PAE were collected. The culture media were centrifuged at 5,000 × g, and the abundantly expressed PAE was crudely separated from other more acidic molecular components in the supernatant fraction as described previously (33). In brief, the supernatant was mixed with strong anion exchange resin (Q-Sepharose, GE Healthcare) equilibrated in 0.02 m sodium phosphate, pH 7.7. End-over-end mixing was performed for 30 min at room temperature. The unbound fraction containing the PAE was collected after 5 min of centrifugation at 3,000 rpm; this fraction is called “filtered” PAE. For some experiments, PAE was used directly from the culture media (labeled “crude” PAE) because it showed largely the same purity on SDS-PAGE and activity as the crudely isolated PAE (data not shown). For the identification of PAE, a fraction of the culture medium was analyzed by SDS-PAGE. The intense band around 35 kDa was excised, reduced, alkylated, and in-gel-digested using trypsin. The resulting peptides were extracted, desalted, and analyzed by LC-MS/MS (for details, see below). The processed peptide data were searched against the UniProtKB protein database containing proteins from a compilation of P. aeruginosa strains using Mascot with the following search criteria: Cys carbamidomethylation as a fixed modification and Met oxidation and Asn/Gln deamidation as variable modifications. A maximum two missed tryptic cleavages were allowed. The remainder of the PAE media was aliquoted and stored at −20 °C until used.
Cortisol-enriched P. aeruginosa Growth Assay
The PASS1 strain was cultured on Luria-Bertani agar plates and incubated overnight at 37 °C (64). A cell colony was transferred to 5 ml of LB medium and incubated overnight at 37 °C. The bacteria were separated from the culture medium by centrifugation at 4,000 × g for 20 min. The cell pellet was washed twice with 1 ml of PBS and centrifuged at 4,000 × g for 5 min. The cells were then diluted with LB medium to approximately A600 nm = 0.1 as measured using a biophotometer (BMG Labtech). PASS1 bacteria were grown in triplicates in the four specified cortisol concentrations (i.e. 10, 1, 0.1, and 0.01 μm) and without cortisol (control) in LB media in a total volume of 200 μl in 96-well plates. LB medium was used for the blank measurement. The 96-well plate was placed in a biophotometer and incubated overnight at 37 °C. A600 nm was measured every 30 min for 16 h. The plate was automatically shaken before each measurement.
Exoglycosidase and N-Glycosidase F Treatment of CBG
Asialylated CBG (asialo-CBG) was generated using α2,3/6/8/9-specific neuraminidase (0.02 milliunits/μg CBG) for 24 h at 37 °C. After 6 h, the sample was split into multiple vials to generate more glycosylation variants of CBG. Agalactosylated CBG (agalacto-CBG) and deglycosylated CBG variants were generated by adding β1,4-galactosidase (0.2 milliunits/μg CBG and N-glycosidase F (1 unit/μg CBG) to the respective vials and incubation for another 18 h at 37 °C. All experiments were performed in 50 mm sodium phosphate buffer, pH 6.5. Reactions were stopped by freezing the glycosylation variants at −80 °C, where they remained stored until use.
NE- and PAE-based Digestion of CBG
Native CBG was incubated with NE in 50 mm sodium acetate, 0.6 m sodium chloride, pH 5.5, at an enzyme/substrate ratio of 1:25 (w/w) for 5 s, 10 s, 15 s, 20 s, 25 s, 30 s, 60 s, 90 s, 120 s, 150 s, 10 min, and 4 h at room temperature. In addition, human CBG was incubated with pure PAE in 50 mm Tris-HCl, 10 mm CaCl2, pH 7.5, at an enzyme/substrate ratio of 1:5 (w/w) for 1, 4, and 16 h or crude PAE (see details above) using 5 μg of CBG to 15 μl of culture medium (containing ∼1 and ∼2 μg of PAE secreted from the PAO1 and PASS1 strains, respectively, as evaluated by SDS-polyacrylamide gel band intensity) for 1 h at 37 °C. The glycosylation variants of CBG were incubated with NE for 2 min at room temperature or with pure PAE for 4 h at 37 °C with the same w/w ratios as indicated above. The digestion reactions were quenched by instantly boiling the samples in NuPAGE LDS sample buffer (Life Technologies, Inc.) containing 50 mm dithiothreitol and 500 mm iodoacetamide for 5 min. Intact CBG and CBG fragments were separated on 8–12% NuPAGE gels (Life Technologies) using a MES (1×) buffer (Life Technologies) for 40 min at 180 V. Gels were stained using Coomassie Brilliant Blue. The gel band intensities were measured using ImageJ software as described (65). All NE and PAE digests of native and glycosylated variants of CBG were performed in triplicates.
In-gel Digestion and Isolation of CBG Peptides
Gel bands corresponding to undigested intact CBG (55–60 kDa), the large CBG-Nt fragment (50–55 kDa), and the small CBG-Ct fragment (5–10 kDa) were excised from the gels, diced, and washed with 100 mm NH4HCO3 (aqueous) and 50% (v/v) acetonitrile. In-gel digestion was performed using modified porcine trypsin (Promega) (100 ng/gel band) overnight in 50 mm NH4HCO3 (aqueous), pH 6.8, at 37 °C. CBG peptides were extracted from the gel pieces using washing cycles of 100% acetonitrile and 0.1% (v/v) formic acid (aqueous). The resulting peptide mixtures were desalted using C18 stage tips (Thermo Scientific) according to the manufacturer's instructions, dried, and redissolved in water before LC-MS/MS. The peptide mixtures derived from intact CBG and CBG-Nt fragments were analyzed individually. The multiple bands corresponding to CBG-Ct fragments (band 1–3; see Fig. 1A) were either analyzed separately (for band-specific analysis) or pooled and analyzed together (for quantitative glycoprofiling) using LC-MS/MS.
Liquid Chromatography and Tandem Mass Spectrometry (LC-MS/MS)
Peptides were separated on a ProteCol C18 column (300-μm inner diameter, 10-cm length, 3-μm particle size, 300-Å pore size, SGE Analytical Science (Melbourne, Australia)) with an attached 0.5-μm Peek filter (Fisher Scientific (Pittsburgh, PA)). The flow rate provided by an Ultimate 3000 HPLC (Dionex) was 5 μl/min. The column was equilibrated with 100% solvent A (i.e. 0.1% (v/v) formic acid (aqueous) before sample load followed by a linear gradient of up to 50% solvent B (i.e. 0.1% (v/v) formic acid in acetonitrile) over 70 min. The column was then washed using 90% solvent B for 5 min and reequilibrated in 100% solvent A for 15 min. The LC column was conjugated to an HCT three-dimensional ion trap (Bruker Daltonics). Peptides were analyzed using electrospray ionization CID-MS/MS in positive polarity mode. Throughout the analysis, an MS full scan (m/z 200–2,200, scan speed 8,100 m/z/s) was followed by data-dependent CID fragmentation of the two most abundant signals in the full scan. Single LC-MS/MS injections for each of the three digestion replicates of all time points were performed. The instrument was calibrated before analysis using a multipoint calibrant (Agilent). The mass discrepancies were generally better than 0.2 Da for the precursor and fragment ions.
LC-MS/MS Data Handling and Glycopeptide Characterization
Tryptic and semitryptic (partially NE- or PAE-cleaved) CBG peptides and glycopeptides were manually identified by de novo sequencing from the acquired LC-MS/MS data using Compass DataAnalysis version 4.0 (Bruker Daltonics) as described (36, 66). Tryptic Asn347-glycosylated (bands 1 and 2) and non-glycosylated (band 3) CBG-Ct peptides eluted in a cluster (50–53 min) and were identified based on their monoisotopic masses, LC retention times, and CID-MS/MS fragmentation. The relevant CBG-Nt peptides were similarly identified as described above. Our previous site-specific N-glycan profiling of human CBG served as a template for the characterization (36). Thus, only the glycan monosaccharide compositions, N-glycan site occupancies, and peptide identities were assigned. The Asn347 glycoprofiles and the Asn347 site occupancy were calculated from the relative areas of the extracted ion chromatograms (EICs) for all observed charge states of the relevant precursors (67, 68).
Statistics
The significance of the experiments was assessed by one- or two-tailed Student's t tests, where p < 0.05 (as indicated by an asterisk) was chosen as the minimum confidence supporting a rejection of the null hypothesis. Stronger significance was indicated by double asterisks (p < 0.01) and triple asterisks (p < 0.001). The sample number (n) is given for the individual experiments. Data points are presented as the mean and the S.D. or S.E. (see the figure legends for details).
Three-dimensional Structures of CBG, NE, and PAE
The experimentally determined three-dimensional structures of human NE (PDB code 3Q77, 1.86 Å), PAE (PDB code 1EZM, 1.50 Å), and relaxed (RCL-cleaved) human CBG (PDB code 4BB2, 2.48 Å) were obtained from the Protein Data Bank. The latter was used as a template to generate a homology model of stressed (RCL uncleaved) CBG in Modeler (69). Cleaved TBG (UniProtKB, P05543) (PDB code 2XN7, 1.43 Å) is a close structural homolog of cleaved CBG with similar conformational epitopes despite the modest (∼45%) sequence identity (32). We used uncleaved TBG (PDB code 2CEO, 2.80 Å) to generate the RCL region (amino acids 341–349) of uncleaved CBG. The unresolved amino acids 1–12 and 96–100 of cleaved CBG were generated using default settings in Modeler.
The homology model of uncleaved CBG was glycosylated with the predominant glycoforms (i.e. for Asn9, Asn154, Asn238, and Asn308, all BS2; for Asn74, TeS4; and for Asn347, TS3, based on our previous report (36) using Glycam-web). An asialo version of the fully glycosylated CBG was also generated. In addition, a series of CBG glycoforms with varying Asn347 glycoforms (i.e. BS2, TS3, TS3f, and DTS3) was generated with no glycans occupying the other sites (supplemental Table S1). Because the triantennary CBG glycoforms remain incompletely determined (36), isomers were built with β1,4/6-GlcNAc branching on the α1,3- or α1,6-Man arm (70) (denoted as the TS3-A and TS3-B isomer, respectively). A single rotamer per glycan was selected that directs the glycan antennae away from the protein; the glycan interconverts between different rotamers during the MD simulation. The gt rotamer for the ω torsion angle was selected for α1,6-Fuc and for outer α1,6-linkages in triantennary structures; the gg rotamer was used for Manα1,6Manβ. For α2,3-linkages, the anti-rotamer of the ϕ torsion angle was selected. The χ1, χ2, ψ, and ϕ torsion angles for the GlcNAc-Asn linkage were selected based on the average Asn-conjugated rotamers in the Protein Data Bank (71) and adjusted to avoid any steric overlap with the protein. The structures were visualized and inspected using UCSF Chimera version 1.10.1, VMD version 1.9.2, and PyMOL version 1.7.6.
Energy Minimization and MD Simulation
All simulations were performed with the CUDA implementation of PMEMD (72, 73) in the Amber14 suite. The GLYCAM06-j (74) and Amber14SB parameters (75) were employed. Berendsen barostats with a time constant of 1 ps was used for pressure regulation, and a Langevin thermostat with a collision frequency of 2 ps−1 was used for temperature regulation. A non-bonded interaction cut-off of 8 Å was employed. Long range electrostatics were treated with the particle mesh-Ewald method (76). Covalent bonds involving hydrogen were constrained with the SHAKE algorithm, allowing time steps of 2 fs (77). Hydrogen atoms were added to each structure. The protonation states of the ionizable side chains were assigned by tleap. The structures were placed in a periodic box of ∼20,000 TIP5P waters (78) to provide an 8-Å buffer between the glycan and the edge of the periodic box. Energy minimization of all atoms was performed for 20,000 steps (10,000 steepest descent and 10,000 conjugant gradients). The energy-minimized structures were equilibrated at 300 K under nPT conditions for 400 ps with 5 kcal/mol/Å2 Cartesian restraints on the solute heavy atoms. This was followed by a 1-ns equilibration and then a 100-ns production MD simulation with molecular tumbling prevented by restraining the Cα atoms of residues Met1, Ile43, Ile83, His84, and Gln85 (5 kcal/mol/Å2). The simulations were insufficiently converged to allow statistical evaluation of the glycoform conformational populations but provided support for the experimental data.
Author Contributions
Z. S.-B. and M. T.-A. conceived the experimental design and hypotheses. Z. S.-B., V. V., and O. C. G. performed the experiments. Z. S.-B., O. C. G., V. V., R. J. W., and M. T.-A. analyzed data. R. J. W., N. H. P., and M. T.-A. supplied reagents/analytical tools/expertise. Z. S.-B., O. C. G., V. V., R. J. W., N. H. P., and M. T.-A. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Karthik Kamath and Dr. Jodie Abrahams for assistance with LC-MS/MS data collection and interpretation. This research was facilitated through access to the Australian Proteomics Analysis Facility (APAF).
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
- CBG
- corticosteroid-binding globulin
- CBG-Ct
- corticosteroid-binding globulin C-terminal fragment
- CBG-Nt
- corticosteroid-binding globulin N-terminal fragment
- A1AT
- α1-antitrypsin
- CID
- collision-induced dissociation
- EIC
- extracted ion chromatogram
- GlcNAc
- N-acetylglucosamine
- MD
- molecular dynamics
- NE
- neutrophil elastase
- NeuAc
- N-acetylneuraminic acid
- PAE
- P. aeruginosa elastase
- PAO1
- wound-derived P. aeruginosa laboratory strain
- PASS1
- cystic fibrosis-derived P. aeruginosa strain
- RCL
- reactive center loop
- TBG
- thyroxine-binding globulin
- Fuc
- fucose
- Man
- mannose
- PDB
- Protein Data Bank.
References
- 1. Siiteri P. K., Murai J. T., Hammond G. L., Nisker J. A., Raymoure W. J., and Kuhn R. W. (1982) The serum transport of steroid hormones. Recent Prog. Horm. Res. 38, 457–510 [DOI] [PubMed] [Google Scholar]
- 2. Cameron A., Henley D., Carrell R., Zhou A., Clarke A., and Lightman S. (2010) Temperature-responsive release of cortisol from its binding globulin: a protein thermocouple. J. Clin. Endocrinol. Metab. 95, 4689–4695 [DOI] [PubMed] [Google Scholar]
- 3. Khan M. S., Aden D., and Rosner W. (1984) Human corticosteroid binding globulin is secreted by a hepatoma-derived cell line. J. Steroid Biochem. 20, 677–678 [DOI] [PubMed] [Google Scholar]
- 4. Hammond G. L., Smith C. L., and Underhill D. A. (1991) Molecular studies of corticosteroid binding globulin structure, biosynthesis and function. J. Steroid Biochem. Mol. Biol. 40, 755–762 [DOI] [PubMed] [Google Scholar]
- 5. Hammond G. L., Smith C. L., Goping I. S., Underhill D. A., Harley M. J., Reventos J., Musto N. A., Gunsalus G. L., and Bardin C. W. (1987) Primary structure of human corticosteroid binding globulin, deduced from hepatic and pulmonary cDNAs, exhibits homology with serine protease inhibitors. Proc. Natl. Acad. Sci. U.S.A. 84, 5153–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Misao R., Iwagaki S., Sun W. S., Fujimoto J., Saio M., Takami T., and Tamaya T. (1999) Evidence for the synthesis of corticosteroid-binding globulin in human placenta. Horm. Res. 51, 162–167 [DOI] [PubMed] [Google Scholar]
- 7. Misao R., Hori M., Ichigo S., Fujimoto J., and Tamaya T. (1994) Corticosteroid-binding globulin mRNA levels in human uterine endometrium. Steroids 59, 603–607 [DOI] [PubMed] [Google Scholar]
- 8. Miska W., Peña P., Villegas J., and Sánchez R. (2004) Detection of a CBG-like protein in human Fallopian tube tissue. Andrologia 36, 41–46 [DOI] [PubMed] [Google Scholar]
- 9. Schäfer H. H., Gebhart V. M., Hertel K., and Jirikowski G. F. (2015) Expression of corticosteroid-binding globulin CBG in the human heart. Horm. Metab. Res. 47, 596–599 [DOI] [PubMed] [Google Scholar]
- 10. Sivukhina E. V., Jirikowski G. F., Bernstein H. G., Lewis J. G., and Herbert Z. (2006) Expression of corticosteroid-binding protein in the human hypothalamus, co-localization with oxytocin and vasopressin. Horm. Metab. Res. 38, 253–259 [DOI] [PubMed] [Google Scholar]
- 11. Perrot-Applanat M., Racadot O., and Milgrom E. (1984) Specific localization of plasma corticosteroid-binding globulin immunoreactivity in pituitary corticotrophs. Endocrinology 115, 559–569 [DOI] [PubMed] [Google Scholar]
- 12. Jirikowski G. F., Pusch L., Möpert B., Herbert Z., and Caldwell J. D. (2007) Expression of corticosteroid binding globulin in the rat central nervous system. J. Chem. Neuroanat. 34, 22–28 [DOI] [PubMed] [Google Scholar]
- 13. Heyns W., and Coolens J. L. (1988) Physiology of corticosteroid-binding globulin in humans. Ann. N.Y. Acad. Sci. 538, 122–129 [DOI] [PubMed] [Google Scholar]
- 14. Lewis J. G., Saunders K., Dyer A., and Elder P. A. (2015) The half-lives of intact and elastase cleaved human corticosteroid-binding globulin (CBG) are identical in the rabbit. J. Steroid Biochem. Mol. Biol. 149, 53–57 [DOI] [PubMed] [Google Scholar]
- 15. Perogamvros I., Ray D. W., and Trainer P. J. (2012) Regulation of cortisol bioavailability: effects on hormone measurement and action. Nat. Rev. Endocrinol. 8, 717–727 [DOI] [PubMed] [Google Scholar]
- 16. Verhoog N., Allie-Reid F., Vanden Berghe W., Smith C., Haegeman G., Hapgood J., and Louw A. (2014) Inhibition of corticosteroid-binding globulin gene expression by glucocorticoids involves C/EBPβ. PLoS One 9, e110702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nenke M. A., Rankin W., Chapman M. J., Stevens N. E., Diener K. R., Hayball J. D., Lewis J. G., and Torpy D. J. (2015) Depletion of high-affinity corticosteroid-binding globulin corresponds to illness severity in sepsis and septic shock; clinical implications. Clin. Endocrinol. 82, 801–807 [DOI] [PubMed] [Google Scholar]
- 18. Emptoz-Bonneton A., Crave J. C., LeJeune H., Brébant C., and Pugeat M. (1997) Corticosteroid-binding globulin synthesis regulation by cytokines and glucocorticoids in human hepatoblastoma-derived (HepG2) cells. J. Clin. Endocrinol. Metab. 82, 3758–3762 [DOI] [PubMed] [Google Scholar]
- 19. Tsigos C., Kyrou I., Chrousos G. P., and Papanicolaou D. A. (1998) Prolonged suppression of corticosteroid-binding globulin by recombinant human interleukin-6 in man. J. Clin. Endocrinol. Metab. 83, 3379. [DOI] [PubMed] [Google Scholar]
- 20. Strel'chyonok O. A., and Avvakumov G. V. (1991) Interaction of human CBG with cell membranes. J. Steroid Biochem. Mol. Biol. 40, 795–803 [DOI] [PubMed] [Google Scholar]
- 21. Hryb D. J., Khan M. S., Romas N. A., and Rosner W. (1986) Specific binding of human corticosteroid-binding globulin to cell membranes. Proc. Natl. Acad. Sci. U.S.A. 83, 3253–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pemberton P. A., Stein P. E., Pepys M. B., Potter J. M., and Carrell R. W. (1988) Hormone binding globulins undergo serpin conformational change in inflammation. Nature 336, 257–258 [DOI] [PubMed] [Google Scholar]
- 23. Lin H. Y., Underhill C., Gardill B. R., Muller Y. A., and Hammond G. L. (2009) Residues in the human corticosteroid-binding globulin reactive center loop that influence steroid binding before and after elastase cleavage. J. Biol. Chem. 284, 884–896 [DOI] [PubMed] [Google Scholar]
- 24. Hammond G. L., Smith C. L., Underhill C. M., and Nguyen V. T. (1990) Interaction between corticosteroid binding globulin and activated leukocytes in vitro. Biochem. Biophys. Res. Commun. 172, 172–177 [DOI] [PubMed] [Google Scholar]
- 25. Klieber M. A., Underhill C., Hammond G. L., and Muller Y. A. (2007) Corticosteroid-binding globulin, a structural basis for steroid transport and proteinase-triggered release. J. Biol. Chem. 282, 29594–29603 [DOI] [PubMed] [Google Scholar]
- 26. Qi X., Loiseau F., Chan W. L., Yan Y., Wei Z., Milroy L. G., Myers R. M., Ley S. V., Read R. J., Carrell R. W., and Zhou A. (2011) Allosteric modulation of hormone release from thyroxine and corticosteroid-binding globulins. J. Biol. Chem. 286, 16163–16173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis J. G., and Elder P. A. (2011) Corticosteroid-binding globulin reactive centre loop antibodies recognise only the intact natured protein: elastase cleaved and uncleaved CBG may coexist in circulation. J. Steroid Biochem. Mol. Biol. 127, 289–294 [DOI] [PubMed] [Google Scholar]
- 28. Meyer E. J., Nenke M. A., Rankin W., Lewis J. G., and Torpy D. J. (2016) Corticosteroid-binding globulin: a review of basic and clinical advances. Horm. Metab. Res. 48, 359–371 [DOI] [PubMed] [Google Scholar]
- 29. Chan W. L., Carrell R. W., Zhou A., and Read R. J. (2013) How changes in affinity of corticosteroid-binding globulin modulate free cortisol concentration. J. Clin. Endocrinol. Metab. 98, 3315–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nenke M. A., Holmes M., Rankin W., Lewis J. G., and Torpy D. J. (2016) Corticosteroid-binding globulin cleavage is paradoxically reduced in α-1 antitrypsin deficiency: implications for cortisol homeostasis. Clin. Chim. Acta 452, 27–31 [DOI] [PubMed] [Google Scholar]
- 31. Lewis J. G., and Elder P. A. (2014) The reactive centre loop of corticosteroid-binding globulin (CBG) is a protease target for cortisol release. Mol. Cell. Endocrinol. 384, 96–101 [DOI] [PubMed] [Google Scholar]
- 32. Gardill B. R., Vogl M. R., Lin H. Y., Hammond G. L., and Muller Y. A. (2012) Corticosteroid-binding globulin: structure-function implications from species differences. PLoS One 7, e52759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simard M., Hill L. A., Underhill C. M., Keller B. O., Villanueva I., Hancock R. E., and Hammond G. L. (2014) Pseudomonas aeruginosa elastase disrupts the cortisol-binding activity of corticosteroid-binding globulin. Endocrinology 155, 2900–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henke M. O., John G., Rheineck C., Chillappagari S., Naehrlich L., and Rubin B. K. (2011) Serine proteases degrade airway mucins in cystic fibrosis. Infect. Immun. 79, 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Avvakumov G. V., and Hammond G. L. (1994) Glycosylation of human corticosteroid-binding globulin. Differential processing and significance of carbohydrate chains at individual sites. Biochemistry 33, 5759–5765 [DOI] [PubMed] [Google Scholar]
- 36. Sumer-Bayraktar Z., Kolarich D., Campbell M. P., Ali S., Packer N. H., and Thaysen-Andersen M. (2011) N-Glycans modulate the function of human corticosteroid-binding globulin. Mol. Cell. Proteomics 10.1074/mcp.M111.009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou A., Wei Z., Stanley P. L., Read R. J., Stein P. E., and Carrell R. W. (2008) The S-to-R transition of corticosteroid-binding globulin and the mechanism of hormone release. J. Mol. Biol. 380, 244–251 [DOI] [PubMed] [Google Scholar]
- 38. Melby J. C., and Spink W. W. (1958) Comparative studies on adrenal cortical function and cortisol metabolism in healthy adults and in patients with shock due to infection. J. Clin. Invest. 37, 1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hansen G., Gielen-Haertwig H., Reinemer P., Schomburg D., Harrenga A., and Niefind K. (2011) Unexpected active-site flexibility in the structure of human neutrophil elastase in complex with a new dihydropyrimidone inhibitor. J. Mol. Biol. 409, 681–691 [DOI] [PubMed] [Google Scholar]
- 40. Deshpande N., Jensen P. H., Packer N. H., and Kolarich D. (2010) GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J. Proteome Res. 9, 1063–1075 [DOI] [PubMed] [Google Scholar]
- 41. Rudd P. M., and Dwek R. A. (1997) Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 32, 1–100 [DOI] [PubMed] [Google Scholar]
- 42. Thaysen-Andersen M., and Packer N. H. (2012) Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 22, 1440–1452 [DOI] [PubMed] [Google Scholar]
- 43. Lee L. Y., Lin C. H., Fanayan S., Packer N. H., and Thaysen-Andersen M. (2014) Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front. Immunol. 5, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mihrshahi R., Lewis J. G., and Ali S. O. (2006) Hormonal effects on the secretion and glycoform profile of corticosteroid-binding globulin. J. Steroid Biochem. Mol. Biol. 101, 275–285 [DOI] [PubMed] [Google Scholar]
- 45. Sprung C. L., Annane D., Keh D., Moreno R., Singer M., Freivogel K., Weiss Y. G., Benbenishty J., Kalenka A., Forst H., Laterre P. F., Reinhart K., Cuthbertson B. H., Payen D., Briegel J., and CORTICUS Study Group (2008) Hydrocortisone therapy for patients with septic shock. N. Engl. J. Med. 358, 111–124 [DOI] [PubMed] [Google Scholar]
- 46. Chan W. L., Zhou A., and Read R. J. (2014) Towards engineering hormone-binding globulins as drug delivery agents. PLoS One 9, e113402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nenke M. A., Lewis J. G., Rankin W., McWilliams L., Metcalf R. G., Proudman S. M., and Torpy D. J. (2016) Reduced corticosteroid-binding globulin cleavage in active rheumatoid arthritis. Clin. Endocrinol. 48, 359–371 [DOI] [PubMed] [Google Scholar]
- 48. Kamath S., Kapatral V., and Chakrabarty A. M. (1998) Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol. Microbiol. 30, 933–941 [DOI] [PubMed] [Google Scholar]
- 49. Lee J., and Zhang L. (2015) The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ciofu O., Hansen C. R., and Høiby N. (2013) Respiratory bacterial infections in cystic fibrosis. Curr. Opin. Pulm. Med. 19, 251–258 [DOI] [PubMed] [Google Scholar]
- 51. Rapala-Kozik M., Potempa J., Nelson D., Kozik A., and Travis J. (1999) Comparative cleavage sites within the reactive-site loop of native and oxidized α1-proteinase inhibitor by selected bacterial proteinases. Biol. Chem. 380, 1211–1216 [DOI] [PubMed] [Google Scholar]
- 52. Beatty K., Bieth J., and Travis J. (1980) Kinetics of association of serine proteinases with native and oxidized α-1-proteinase inhibitor and α-1-antichymotrypsin. J. Biol. Chem. 255, 3931–3934 [PubMed] [Google Scholar]
- 53. Brantly M. (2002) α1-Antitrypsin: not just an antiprotease: extending the half-life of a natural anti-inflammatory molecule by conjugation with polyethylene glycol. Am. J. Respir. Cell Mol. Biol. 27, 652–654 [DOI] [PubMed] [Google Scholar]
- 54. Crocetti L., Schepetkin I. A., Cilibrizzi A., Graziano A., Vergelli C., Giomi D., Khlebnikov A. I., Quinn M. T., and Giovannoni M. P. (2013) Optimization of N-benzoylindazole derivatives as inhibitors of human neutrophil elastase. J. Med. Chem. 56, 6259–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Groutas W. C., Dou D., and Alliston K. R. (2011) Neutrophil elastase inhibitors. Expert Opin. Ther. Pat. 21, 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Craik D. J., Cemazar M., and Daly N. L. (2006) The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 9, 251–260 [PubMed] [Google Scholar]
- 57. Hill R. E., and Hastie N. D. (1987) Accelerated evolution in the reactive centre regions of serine protease inhibitors. Nature 326, 96–99 [DOI] [PubMed] [Google Scholar]
- 58. Law R. H., Zhang Q., McGowan S., Buckle A. M., Silverman G. A., Wong W., Rosado C. J., Langendorf C. G., Pike R. N., Bird P. I., and Whisstock J. C. (2006) An overview of the serpin superfamily. Genome Biol. 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vashchenko G., Das S., Moon K. M., Rogalski J. C., Taves M. D., Soma K. K., Van Petegem F., Foster L. J., and Hammond G. L. (2016) Identification of avian corticosteroid-binding globulin (SerpinA6) reveals the molecular basis of evolutionary adaptations in SerpinA6 structure and function as a steroid-binding protein. J. Biol. Chem. 291, 11300–11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bernard B. A., Yamada K. M., and Olden K. (1982) Carbohydrates selectively protect a specific domain of fibronectin against proteases. J. Biol. Chem. 257, 8549–8554 [PubMed] [Google Scholar]
- 61. Rutledge E. A., and Enns C. A. (1996) Cleavage of the transferrin receptor is influenced by the composition of the O-linked carbohydrate at position 104. J. Cell. Physiol. 168, 284–293 [DOI] [PubMed] [Google Scholar]
- 62. Hane M., Matsuoka S., Ono S., Miyata S., Kitajima K., and Sato C. (2015) Protective effects of polysialic acid on proteolytic cleavage of FGF2 and ProBDNF/BDNF. Glycobiology 25, 1112–1124 [DOI] [PubMed] [Google Scholar]
- 63. Penesyan A., Kumar S. S., Kamath K., Shathili A. M., Venkatakrishnan V., Krisp C., Packer N. H., Molloy M. P., and Paulsen I. T. (2015) Genetically and phenotypically distinct Pseudomonas aeruginosa cystic fibrosis isolates share a core proteomic signature. PLoS One 10, e0138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luria S. E., and Burrous J. W. (1957) Hybridization between Escherichia coli and Shigella. J. Bacteriol. 74, 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sumer-Bayraktar Z., Nguyen-Khuong T., Jayo R., Chen D. D., Ali S., Packer N. H., and Thaysen-Andersen M. (2012) Micro- and macroheterogeneity of N-glycosylation yields size and charge isoforms of human sex hormone binding globulin circulating in serum. Proteomics 12, 3315–3327 [DOI] [PubMed] [Google Scholar]
- 67. Thaysen-Andersen M., Mysling S., and Højrup P. (2009) Site-specific glycoprofiling of N-linked glycopeptides using MALDI-TOF MS: strong correlation between signal strength and glycoform quantities. Anal. Chem. 81, 3933–3943 [DOI] [PubMed] [Google Scholar]
- 68. Stavenhagen K., Hinneburg H., Thaysen-Andersen M., Hartmann L., Varón Silva D., Fuchser J., Kaspar S., Rapp E., Seeberger P. H., and Kolarich D. (2013) Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 48, 627–639 [DOI] [PubMed] [Google Scholar]
- 69. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., and Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stanley P., Schachter H., and Taniguchi N. (2009) N-Glycans. in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E., eds) pp. 101–114, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 71. Petrescu A. J., Milac A. L., Petrescu S. M., Dwek R. A., and Wormald M. R. (2004) Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14, 103–114 [DOI] [PubMed] [Google Scholar]
- 72. Salomon-Ferrer R., Götz A. W., Poole D., Le Grand S., and Walker R. C. (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 [DOI] [PubMed] [Google Scholar]
- 73. Götz A. W., Williamson M. J., Xu D., Poole D., Le Grand S., and Walker R. C. (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., and Woods R. J. (2008) GLYCAM06: a generalizable biomolecular force field: carbohydrates. J. Comput. Chem. 29, 622–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Case D. A., Babin V., Berryman J. T., Betz R. M., Cai Q., Cerutti D. S., Cheatham T. E. III, Darden T. A., Duke R. E., Gohlke H., Goetz A. W., Gusarov S., Homeyer N., Janowski P., Kaus J., et al. (2014) The FF14SB force field. In AMBER 14 Reference Manual, AMBER 14, University of California, San Francisco [Google Scholar]
- 76. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089 [Google Scholar]
- 77. Ryckaert J.-P., Ciccotti G., and Berendsen H. J. C. (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 78. Mahoney M. W., and Jorgensen W. L. (2000) A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 112, 8910–8922 [Google Scholar]
- 79. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.