Abstract
Chloroplasts and mitochondria are unique endosymbiotic cellular organelles surrounded by two membranes. Essential metabolic networking between these compartments and their hosting cells requires the exchange of a large number of biochemical pathway intermediates in a directed and coordinated fashion across their inner and outer envelope membranes. Here, we describe the identification and functional characterization of a highly specific, regulated solute channel in the outer envelope of chloroplasts, named OEP40. Loss of OEP40 function in Arabidopsis thaliana results in early flowering under cold temperature. The reconstituted recombinant OEP40 protein forms a high conductance β-barrel ion channel with subconductant states in planar lipid bilayers. The OEP40 channel is slightly cation-selective PK+/PCl− ≈ 4:1 and rectifying (i⃗/i⃖ ≅ 2) with a slope conductance of Ḡmax ≅ 690 picosiemens. The OEP40 channel has a restriction zone diameter of ≅1.4 nm and is permeable for glucose, glucose 1-phosphate and glucose 6-phosphate, but not for maltose. Moreover, channel properties are regulated by trehalose 6-phosphate, which cannot permeate. Altogether, our results indicate that OEP40 is a “glucose-gate” in the outer envelope membrane of chloroplasts, facilitating selective metabolite exchange between chloroplasts and the surrounding cell.
Keywords: chloroplast, glucose transport, ion channel, membrane transport, outer membrane, beta-barrel protein, metabolite transport, solute channel
Introduction
The plant cell is highly compartmentalized, containing at least seven different types of organelles that are involved in metabolism and cellular maintenance. In many cases, metabolic pathways and other cellular processes are not confined to one organelle but require the cooperation of several of them, e.g. in protein secretion or photorespiration. Metabolic networking between compartments thus requires directed and coordinated exchange of biochemical pathway intermediates across organellar membranes (1). Chloroplasts and mitochondria are unique endosymbiotic cellular organelles, which like their prokaryotic ancestors are surrounded by two membranes. The inner membrane of both organelles is derived from the bacterial plasma membrane. Surprisingly, also the outer membrane largely originated from and still resembles the outer membrane of the Gram-negative-like bacterial endosymbiont (2). In the inner chloroplast envelope membrane (IE),5 numerous metabolite transporter proteins were identified and characterized to large detail with respect to their physiological impact and molecular mechanisms (3). These transporters are hydrophobic, polytopic, and mainly α-helical membrane proteins facilitating the exchange of metabolic precursors, intermediates, and final products between plastids and the cytoplasm. In contrast, the characteristic channels of the outer membranes in bacteria, chloroplasts as well as mitochondria, span the membrane in the form of β-strands that are organized to form a barrel-like pore structure (4). Proteins sharing this three-dimensional arrangement were implied in a variety of functions, including protein import (5). In addition, solute pores like the voltage-dependent anion channel (VDAC) in mitochondria (6) or the outer envelope proteins OEP21, OEP24, and OEP37 (7–9) in chloroplasts are essential parts of the outer membrane permeom of these organelles (10, 11). In the outer mitochondrial membrane, so far only VDAC has been identified as the main solute pore. However, it has been shown that ΔVDAC mouse models, in which all three VDAC isoforms VDAC-1, VDAC-2, VDAC-3 have been deleted, were not severely impaired in metabolite exchange between mitochondria and cytosol nor in Ca2+ homeostasis and apoptosis signaling (12). Similarly, yeast VDAC deletion strains were viable and only slightly impaired under harsh growth conditions (13). Thus, other not yet identified metabolite pores are likely present in the outer mitochondrial membrane. In contrast, in the chloroplast outer envelope membrane (OE) four different solute channels have been identified. They can in principle process and transport numerous metabolites (10, 11). Although specific and regulated solute channels are present in the OE, it remained a matter of debate to what extent transport is regulated in vivo (14). Although the analysis of Arabidopsis knock-out lines for the β-barrel channels OEP21, OEP24, and OEP37 did not reveal any visible plant defects under standard growth conditions, the loss of the α-helical OE amino acid channel OEP16.2 showed germination defects and metabolic imbalances in the presence of the seed dormancy hormone abscisic acid (15).
Here, we report the identification and functional characterization of a new member of the β-barrel chloroplast OEP family, namely OEP40, that is selectively permeable to glucose and its phosphorylated derivatives. Remarkably, the OEP40 channel is regulated in its channel properties by trehalose 6-phosphate. Furthermore, loss of OEP40 function in Arabidopsis plants results in an early flowering phenotype at cold temperature. In summary, our results indicate that OEP40 is the “glucose-gate” of the chloroplast OE showing that metabolite transport is regulated at the outer plastid envelope membrane.
Results
OEP40 Is a β-Barrel Protein in the Chloroplast OE
In an attempt to identify porin-type solute channels in the OE membrane of chloroplasts, we separated a highly purified OE membrane fraction from Pisum sativum (pea) chloroplasts by SDS-PAGE in the presence or absence of urea. One of the protein bands, which showed a clear difference in electrophoretic mobility, was further analyzed by mass spectroscopy. The four identified peptides (supplemental Fig. S1) matched pea-expressed sequence tags (16) that are annotated as the Arabidopsis gene At3g57990. At3g57990 represents a single-copy gene, encoding for a protein of 39.8 kDa with an isoelectric point of 10.0, which was named At-OEP40 (supplemental Fig. S1). To analyze protein function, we isolated the cDNA of OEP40 genes from Arabidopsis and pea. Ps-OEP40 (GenBankTM accession number KT361648) corresponds to a polypeptide with a calculated molecular mass of 42.4 kDa and an isoelectric point of 9.3 (supplemental Fig. S1). We confirmed the OE localization by immunoblot on purified pea chloroplast subfractions (Fig. 1A, for antiserum specificity see supplemental Fig. S2). To analyze whether Ps-OEP40 represents an integral membrane protein, pea chloroplast OE membranes were treated with high salt, alkaline pH, urea or detergent and subsequently separated into membrane and soluble fractions. In this assay, Ps-OEP40 behaved like a known β-barrel channel in the chloroplast OE, OEP37 (9, 17), and was only partially or fully solubilized by urea and Triton X-100, respectively (supplemental Fig. S3A). To verify a potential β-barrel structure of OEP40, recombinant At-OEP40 and as control At-OEP37 proteins were refolded in the presence of SDS and 2-methyl-2,4-pentanediol and subsequently subjected to circular dichroism (CD) analysis. CD measurements revealed 35.6% of β-sheet structures in OEP40 (supplemental Fig. S3 B), indicating that OEP40 is likely to form a β-barrel in the OE. These findings were qualitatively confirmed by a bioinformatic analysis (18), predicting a relative content of ∼28% of β-sheets and only ∼11% of α-helices for OEP40. Because OEP40 appears to be a plant-specific protein with no templates in public databases, straightforward homology modeling of its probable structure is not readily possible. However, reliable secondary structure predictions (18–20) propose the presence of 10–12 transmembrane β-sheets at the N-terminal part of OEP40 (supplemental Fig. S3C), similar to predictions for other OEPs (10, 17). Restricted structure modeling (20) suggests a 10 transmembrane β-barrel pore structure within the N-terminal section of the protein as a working model (Fig. 1B).
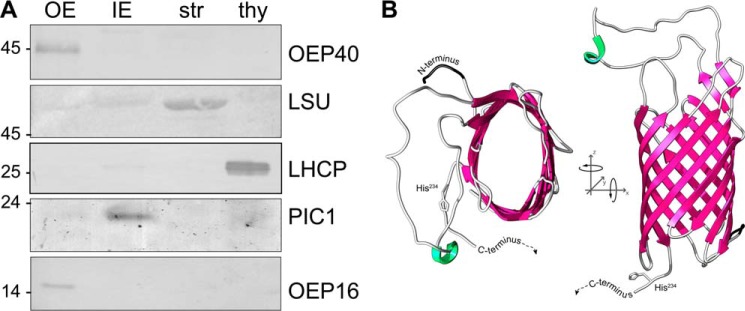
FIGURE 1.
OEP40 forms a β-barrel in the chloroplast OE membrane. A, immunoblot analysis of Ps-OEP40 in chloroplast subfractions. Equal protein amounts (5 μg) of pea chloroplast OE, IE, stroma (str), and thylakoids (thy) were separated by SDS-PAGE and subjected to immunoblot analysis using antibodies directed against recombinant Ps-OEP40. For detection with antisera against marker proteins LSU (str), LHCP (thy), PIC1 (IE), and OEP16.1 (OE), different protein amounts (i.e. 1.0, 0.6, 10.0, and 1.0 μg, respectively) were loaded in each lane. Numbers indicate molecular mass of proteins in kDa. B, three-dimensional structure of a 10-stranded At-OEP40 β-barrel generated by TMBpro. For clarity, the mainly unstructured C terminus starting from His234 (compare supplemental Figs. S1 and S3) is not shown.
OEP40 Forms a High Conductance Channel in Planar Lipid Bilayers
Next, we tested whether OEP40 might also be able to constitute a pore using the planar bilayer technique. Therefore, recombinant Ps-OEP40 from three different expression systems was purified to homogeneity (see supplemental Fig. S2) and reconstituted into liposomes. OEP40-proteoliposomes obtained from the different preparations were then assayed for channel activity using the planar bilayer system (21). After fusion of the OEP40 proteoliposomes with the planar bilayers, channel activity could be detected in all cases. In contrast, mock-control samples did not show any channel activity. Because the basic electrophysiological properties obtained from the individual channel recordings were identical for the three different Ps-OEP40 preparations, we will representatively describe the results obtained with the Ps-OEP40(His6) protein derived from the Escherichia coli expression system.
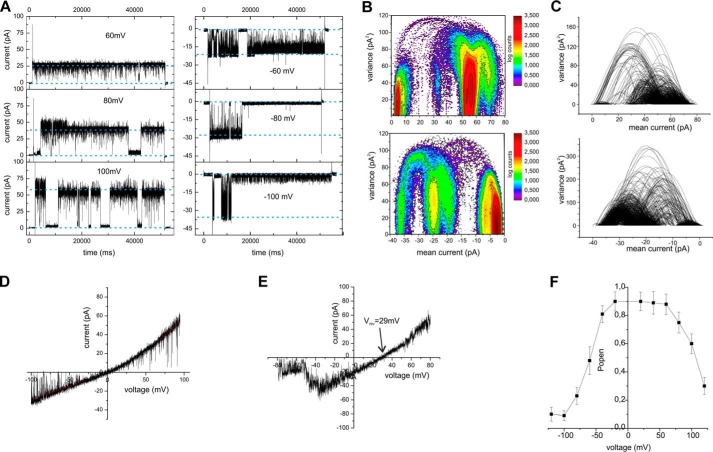
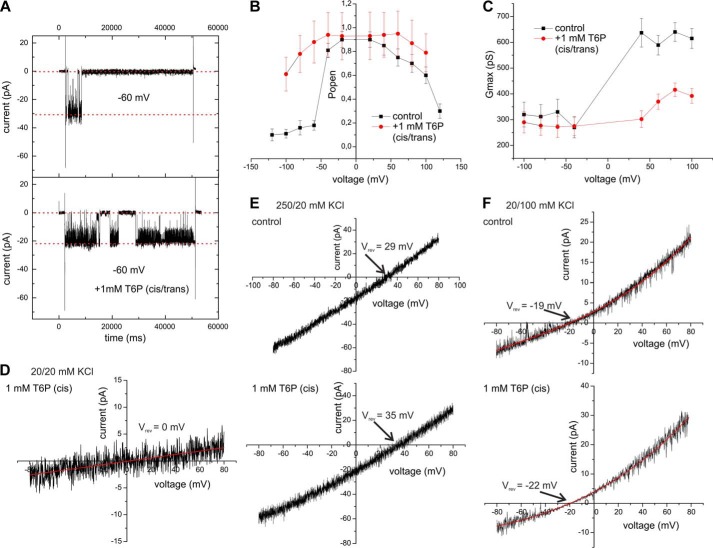
During the application of a voltage gate, the OEP40 channel was mainly open but displayed short flickering gating events at lower membrane potentials (Vm = ±60 mV). At higher membrane potentials (Vm > ±60 mV), the channel closed (Fig. 2A). Mean variance analysis (Fig. 2, B and C) (22) discloses that the OEP40 channel has subconductance states (Ḡmax = ≅692 ± 6 pS, Ḡsub1 ≅550 ± 4 pS, Ḡsub2 ≅325 ± 6 pS at Vm = −100 mV). From the conductance Ḡmax ≅325 pS, a first approximation of the diameter of the restriction zone within the OEP40 channel of dpore ≅1.4 nm may be calculated using the Ohmic model of Hille (23) (for details see supplemental Methods). In voltage ramps the channel also displayed a slightly rectifying current voltage relation with a rectification ratio rrec = i⃗/i⃖ ≈ 1.5 − 2.5 (Fig. 2D). In asymmetric buffers, the channel showed a reversal potential of Vrev = 29.0 ± 2.3 mV (n > 6) (Fig. 2E). Using the Goldman-Hodgkin-Katz (GHK) approach (23), this corresponds to a minor cation selectivity of the OEP40 channel (PK+/PCl− ≈4:1). Additionally we observed a slightly asymmetric open probability with the OEP40 channel more frequently residing in a closed state at negative Vm (Fig. 2F).
FIGURE 2.
Single channel recording from planar bilayers containing reconstituted OEP40. A, single channel recordings in symmetrical buffer conditions (250/250 mm KCl) from bilayers containing a single active OEP40 channel in response to an applied voltage gate of the indicated voltage amplitude (Vm). B, mean variance histogram of the current recording at Vm = 100 mV (top) and Vm = −100 mV (bottom). C, mean variance plot of the current recording at Vm = 100 mV (top) and Vm = −100 mV (bottom). D, current-voltage ramp (−100 to 100 mV, sweep-rate 15 mV/s) of a single open OEP40 channel in symmetrical buffer (see A). E, current-voltage ramp (−80 to 80 mV, sweep-rate 15 mV/s) of a single open OEP40 channel in asymmetrical buffer (250/20 mm KCl). F, voltage-dependent open probability of OEP40 single channels (n = 3).
OEP40 Is Selectively Permeable for Glucose
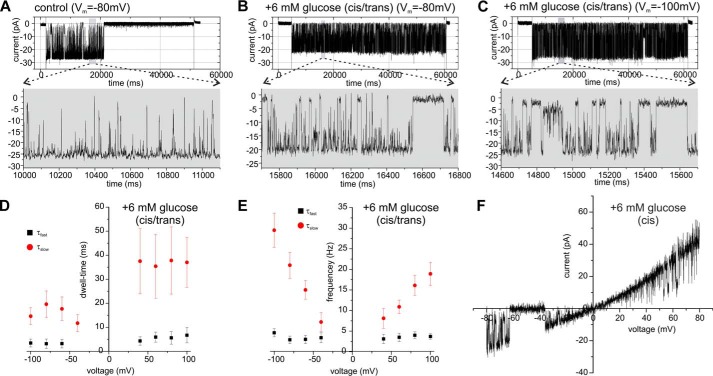
To elucidate transport specificity, we tested the effect of different photosynthetic metabolites, namely glucose, glucose 1-phosphate (Glc-1-P), glucose 6-phosphate (Glc-6-P), maltose, as well as the signaling metabolite trehalose 6-phosphate (Tre-6-P) (24) on the OEP40 channel activity. Because the effects of symmetric addition of glucose, Glc-1-P, and Glc-6-P on the channel gating were identical, representative data are shown in the presence of glucose in Fig. 3. When glucose or the phosphorylated derivatives were added symmetrically to both sides of the bilayer (cis/trans), strong flickering of the channel currents between the fully open and the closed state was observed (Fig. 3, A–C). To acquire information on the details of glucose transport by OEP40, we performed an in-depth mean variance analysis of the glucose-induced gating transitions of OEP40 (22). The concentration- and voltage-dependent flickering presumably represents electrostatic or steric modulation of the current through the pore, caused by partitioning of glucose molecules into, or their transport through the OEP40 channel. The mean variance analysis revealed two classes of gating transitions, “fast” transitions characterized by short dwell times and voltage-independent frequency and “slow” transitions, displaying longer and slightly voltage-dependent dwell times as well as frequency (Fig. 3, D and E). In addition, at a given voltage the reciprocal interevent time between the slow closing events was linearly dependent on the glucose concentration with kon ≅8.58 ± 1.15·10−3 m−1 s−1 at Vm = −80 mV (supplemental Fig. S4). This as well as the pronounced biphasic voltage dependence of the slow dwell times at negative Vm argue in favor of the translocation of glucose through the OEP40 channel (25). Because a complete analysis of the interactions between the OEP40 channel and glucose cannot be performed within the scope of this report, we may in a first approximation interpret the voltage-independent, mean blocking time of τ̄block ≅ 37 ms at positive Vm (see Fig. 3D) as the average transit time for glucose across the channel (25). This would mean that glucose could pass the OEP40 channel at a minimum rate of rtransport ≅27 molecules/s.
FIGURE 3.
Effect of glucose on the single channel activity of reconstituted OEP40. A, single channel recordings in symmetrical buffer (250/250 mm KCl) from a bilayer containing a single active OEP40 channel in response to an applied voltage gate of 60 s duration with the indicated voltage amplitude (Vm). B and C, single channel recordings in symmetrical buffer from the same bilayer as in A but in the presence of 6 mm glucose (cis/trans). D, voltage dependence of the fast (τfast) and slow dwell-times (τslow) of glucose-induced transitions between the fully open and closed state of the OEP40 channel (6 mm glucose, cis/trans). E, voltage dependence of the frequency of τfast and τslow of glucose (6 mm, cis/trans)-induced transitions between the fully open and closed state of the OEP40 channel (n = 3). F, current-voltage ramp (−80 to 80 mV, sweep-rate 15 mV/s) of an OEP40 channel in symmetrical buffer conditions (see A) with the addition of 6 mm glucose only in cis. For a control without glucose, see Figs. 2D and 4C (upper panel).
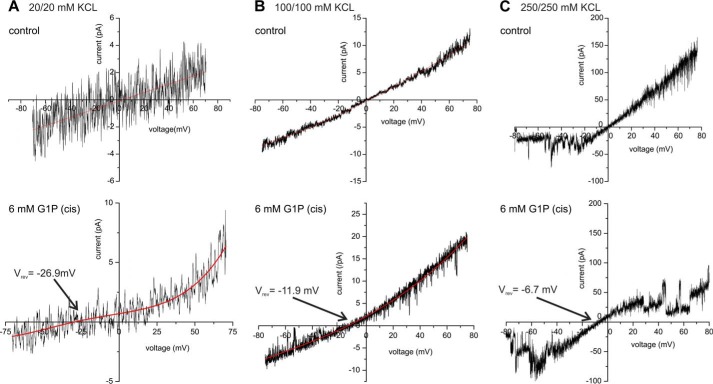
Translocation of the charged glucose derivatives, Glc-1-P and Glc-6-P, can be more easily tested by measuring the effect of asymmetric addition of the respective molecule on the current-voltage relationship of OEP40. Although addition of uncharged glucose to one side of the bilayer does not affect OEP40 (Fig. 3F), we reproducibly observed a shift in the zero current potential of the current-voltage curve upon addition of its charged derivatives, which demonstrates electrogenic transport of Glc-1-P (Fig. 4, A–C) and Glc-6-P (data not shown) through the OEP40 channel. In particular, we investigated the effect of Glc-1-P in the cis compartment on the reversal potential of the OEP40 channel at different symmetrical (cis/trans) concentrations of KCl. According to the GHK expression (see Equations 1–3 in supplemental Methods), it is expected that at low KCl concentrations the shift in the reversal potential (Vrev) at a constant Glc-1-P concentration will be more pronounced than at higher KCl concentrations. Fig. 4, A–C, shows the current-voltage relation of the OEP40 channel in different symmetrical (cis/trans) KCl buffer (20, 100, and 250 mm) with additional 6 mm Glc-1-P in the cis compartment and the respective controls. Using the GHK approximation and assuming that (i) the relative permeability PK+/PCl− = 4:1 does not change in the presence of Glc-1-P and that (ii) K+ and Na+ ions, the latter supplied with the Glc-1-P disodium salt, are equally permeant, we can calculate the relative permeability as PGlc-1-P/PK+/PCl− ≅ 48:4:1, respectively (for details see supplemental Methods). The results of these calculations are summarized in Table 1. From these it is obvious that the conductance values of the OEP40 channel determined from the slope conductance of current-voltage ramps at the low conductance site increase linearly up to an ion concentration of 250 mm K+, revealing that the conductance under these conditions is not saturating. The Glc-1-P-induced change in the reversal potential dropped significantly with increasing KCl concentrations as expected for Glc-1-P being permeant. In line with this, the apparent relative permeability for Glc-1-P, PGlc-1-P (app), as calculated by Equations 1–3 in supplemental Methods, was nearly constant at the different KCl concentrations (see Table 1). In summary, we can conclude that OEP40 is permeable for Glc-1-P and Glc-6-P as well as for uncharged glucose (compare Fig. 3). The disaccharide maltose in contrast did not show any marked effect on the activity of the OEP40 channel (supplemental Fig. S5), indicating that the interaction of the OEP40 channel with glucose is specific.
FIGURE 4.
Effect of Glc-1-P on the single channel activity of reconstituted OEP40. A, current-voltage ramp (−70 to +70 mV, sweep-rate 15 mV/s) of a single open OEP40 channel in the absence (control) and presence of 6 mm Glc-1-P in the cis compartment, which corresponds to the low conductance site of the OEP40 channel (symmetrical 20/20 mm KCl buffer conditions). B, same as A except symmetrical 100/100 mm KCl buffer conditions. C, same as A except symmetrical 250/250 mm KCl buffer conditions.
TABLE 1.
Conductance and Glc-1-P induced zero current potential shift of the OEP40 channel at different ionic strength
Conductance values (G) were calculated from the linear slope conductance of current-voltage sweeps (average from n ≥3 sweeps) at the low conductance site. PGlc-1-P(app) is the relative apparent permeability (PGlc-1-P− = X: PK+ = PNa+ = 4: PCl− = 1) calculated for the tri-ionic conditions using the GHK equation (see supplemental Methods). Because in the OEP40-containing bilayers with 100 mm KCl (cis) and 100 mm NaCl (trans) no zero current shift was observed, we can set PK+ = PNa+.
| cKCl (cis) | cKCl(trans) | cGlc-1-P(cis) | ḡ | Vrev | PGlc-1-P(app) |
|---|---|---|---|---|---|
| mm | mm | mm | pS | mV | |
| 250 | 250 | 0 | 325 ± 38 | 0 | − |
| 250+ | 250 | 6 | 380 ± 23 | −6.7 | 48 |
| 12 Na+ | |||||
| 100 | 100 | 0 | 127 ± 1.8 | 0 | − |
| 100+ | 100 | 6 | 123 ± 3.0 | −11.9 | 47 |
| 12 Na+ | |||||
| 20 | 20 | 0 | 30 ± 4.6 | 0 | − |
| 20+ | 20 | 6 | 26.7 ± 0.6 | −26.9 | 49 |
| 12 Na+ |
We then tested whether Tre-6-P, a known signal of a plant's sucrose status (24), can also permeate the OEP40 channel. Tre-6-P added to both sides of the channel in contrast to glucose and its derivatives barely induced flickering of the OEP40 channel (Fig. 5A). Furthermore, in the presence of Tre-6-P the OEP40 channel more often resided in an open conformation, thus increasing the open probability, especially at negative Vm values (Fig. 5B). However, the channel mainly flickered between channel sub-states (supplemental Fig. S6), thereby reducing the observed maximum conductance Gmax of the OEP40 channel (Fig. 5C). In contrast to our observations with the charged glucose derivative Glc-1-P (see above), addition of 1 mm charged Tre-6-P under symmetrical low KCl concentration (20 mm) to the low conductance cis compartment did not induce a change in the reversal potential (Fig. 5D). Thus, even though the channel would still be large enough to accommodate mono- and disaccharide metabolites, these results argue against a permeability of OEP40 for Tre-6-P. However, in a tri-ionic experiment, where 1 mm Tre-6-P was added under asymmetric buffer conditions to the low conductance cis compartment, the zero current potential shifted slightly toward higher values (Fig. 5, E and F). In these experiments with two different KCl ion gradients across the bilayer, i.e. 250/20 mm cis/trans (Fig. 5E) and 20/100 mm cis/trans (Fig. 5F), we observed in both cases a Tre-6-P-induced shift of the reversal potential from Vrev = 29 ± 1.6 mV to Vrev = 35 ± 2.1 mV (250/20 mm KCl cis/trans) and Vrev = −19 ±1.9 mV to −21 ± 1.3 mV (20/100 mm KCl cis/trans). Using the GHK approach, both results are compatible with a Tre-6-P-induced increase of the selectivity from PK+/PCl− = 4:1 to PK+/PCl− = 6:1 (see analogous calculations in supplemental Methods). The slight increase of the reversal potential, at the given experimental setup, shows that Tre-6-P cannot permeate the OEP40 channel, but rather binds to it and thereby exerts modulating effects on the ion flux through the OEP40 pore. Thus, Tre-6-P appears to be a specific effector of the OEP40 channel.
FIGURE 5.
Effect of Tre-6-P on single channel activity of reconstituted OEP40. A, current recording from a bilayer containing a single active OEP40 channel in response to an applied voltage gate of Vm = −60 mV in the absence (top) and presence of 1 mm Tre-6-P (cis/trans). Symmetrical buffer conditions (250/250 mm KCl). B, voltage-dependent open probabilities (n = 3) of the OEP40 channel in the absence (black square) and presence of 1 mm Tre-6-P (red circle) (cis/trans). C, apparent conductance Gmax obtained from the maximal current amplitudes (Imax) at the given voltages in the absence (black square) and presence of 1 mm Tre-6-P (red circle) (n = 3). D, current-voltage ramp (−80 to 80 mV sweep-rate 15 mV/s) of a single open OEP40 channel in symmetrical buffer (20/20 mm KCl) in the presence of 1 mm Tre-6-P in the cis compartment. E, current-voltage ramps (−80 to 80 mV sweep-rate 15 mV/s) of a single open OEP40 channel in asymmetrical buffer (250/20 mm KCl) in the absence (upper panel) and presence of 1 mm Tre-6-P in cis (lower panel). F, same as E except the asymmetric 20/100 (cis/trans) KCl buffer conditions.
Loss of OEP40 Function in Arabidopsis Results in Early Flowering under Cold Temperature
To assess the function of OEP40 in planta, we analyzed phenotypes of OEP40 T-DNA insertion lines in Arabidopsis (supplemental Fig. S7A). Although the line oep40-1 represents a knockdown with about 30% residual transcripts and proteins, oep40-3 is a true loss-of-function mutant because neither mRNA nor protein was detectable (supplemental Fig. S7, B and C). No obvious phenotypic differences for both mutant lines were observed when compared with wild type under various growth conditions and different length of day, except when plants were grown at 10 °C, a temperature in which plant growth in general is greatly reduced. Here, oep40-1 and oep40-3 both showed an early flowering phenotype (Fig. 6, A and B). We next recorded days to bolting and total leaf numbers, both of which are established parameters to measure differences in flowering time. When grown at 10 °C, both oep40 mutant lines bolted about 5–6 days earlier than wild-type plants (Fig. 6C). This was also reflected by the total leaf numbers recorded for oep40-1 and oep40-3. However, flowering time did not significantly change when plants were grown at a normal temperature (21 °C day/16 °C night, Fig. 6D).
FIGURE 6.
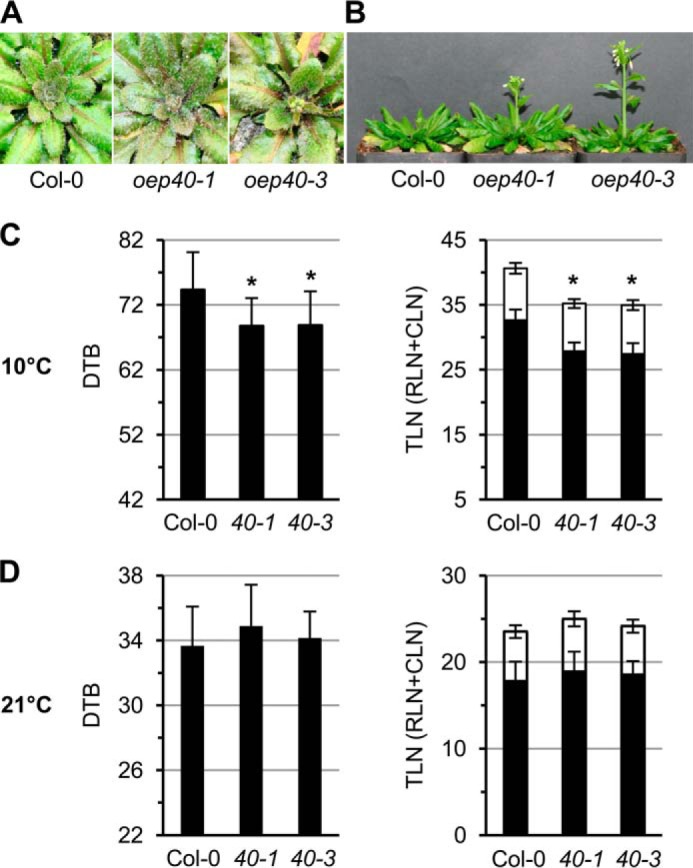
Arabidopsis mutants with reduced OEP40 protein display an early flowering phenotype a low temperature conditions. A and B, representative individuals of 64- and 85-day-old-plants of Col-0 wild-type and oep40-1 and oep40-3 homozygous mutant lines grown at 10 °C. C and D, DTB (left) and TLN (right) were recorded in plant lines described in A, B. Plants were grown at 10 °C in C and at 21 °C (day) and 16 °C (night) in D. Bars depict mean numbers ± S.D. of n = 27 individual plants for each line. Asterisks indicate significant differences in DTB and TLN in oep40 mutants compared with Col-0 (Student's t test, p < 0.05). Right graphs, black bars represent rosette leaf (RLN) and white bars cauline leaf numbers (CLN). Please note that first flowers of oep40-1 and oep40-3 appeared about 8–9 days earlier than for Col-0 wild type.
Transition to flowering in plants is a highly coordinated process influenced by many external factors, like day length or temperature, but also by internal factors such as the plants' age and its sucrose status (26). To determine the transition to flowering also on a morphological and molecular basis, we followed the morphological changes at the shoot apical meristem (SAM) from oep40-1, oep40-3, and wild-type lines over a 3-week time period. We used LEAFY (LFY), a plant-specific transcription factor (27), as an RNA probe to identify emerging floral primordia. About 4 days earlier than in wild type, we detected strong LFY transcript-specific signals in apices of oep40-1 and oep40-3 lines (Fig. 7). In general, effects were slightly more prominent in the knock-out line oep40-3 than in oep40-1 (knockdown). These results correlated well with the morphological changes described above.
FIGURE 7.
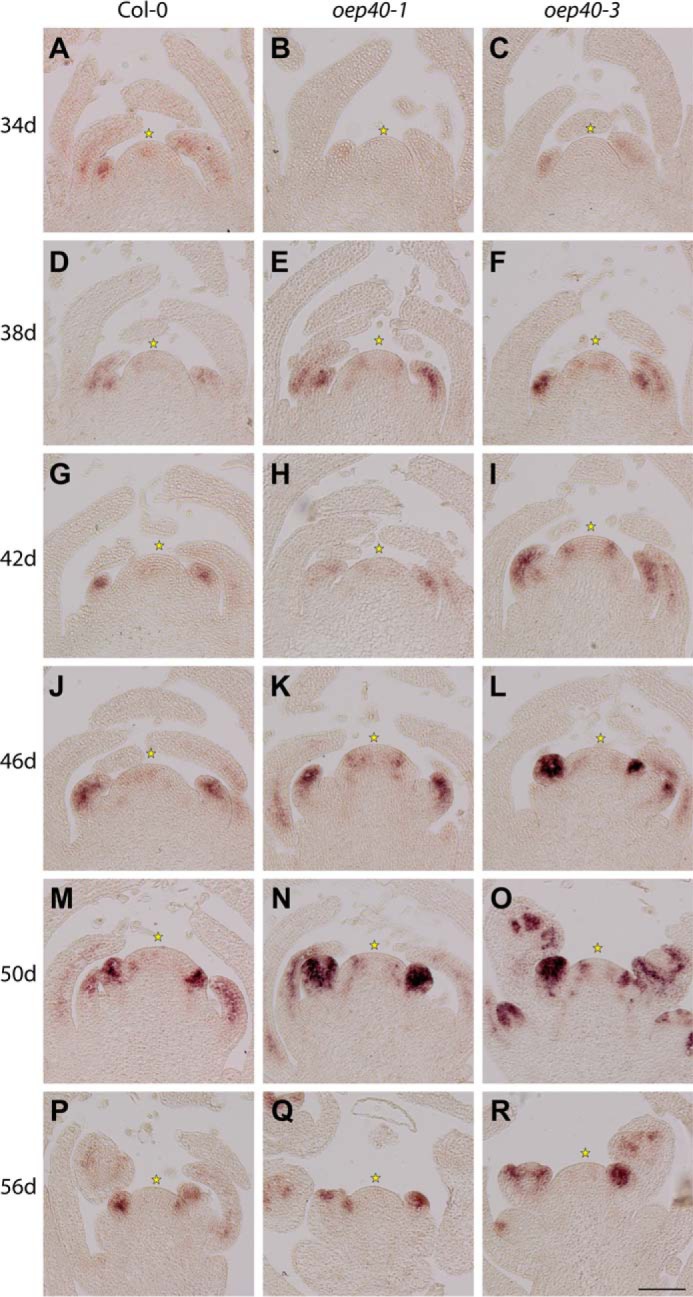
Floral transition occurs earlier in oep40 mutant lines compared with wild type. Emergence of floral primordia was visualized by RNA in situ hybridization using a specific LFY probe on longitudinal sections through apices of Col-0 wild type as well as homozygous oep40-1 and oep40-3 mutant lines. The plants were grown under low temperature conditions (10 °C) and harvested every 2nd day over a period of 3 weeks. Representative images of 34–54-day-old plants are shown as indicated. Asterisks indicate the meristem summit.
Discussion
We have demonstrated that the reconstituted recombinant OEP40 forms a high conductance channel in planar lipid bilayers. CD spectroscopy results of the refolded recombinant protein agree with bioinformatic analysis in classifying OEP40 as a mainly β-sheet protein. Thus, we propose that OEP40 forms a water-filled β-barrel channel pore in the chloroplast OE. The electrophysiological properties of OEP40 show that the channel is permeable for glucose and its phosphorylated derivatives but not for maltose and Tre-6-P. In vivo, glucose, Glc-1-P, and Glc-6-P are major metabolites to be exchanged between the chloroplast and the cytosol (28). Transport across the Arabidopsis chloroplast IE is accomplished by the plastid glucose transporter (29, 30) and Glc-6-P/phosphate translocators (GPT1 and GPT2) (31, 32), respectively. Although Glc-1-P carbon fluxes into chloroplast starch have been measured (33), the corresponding transport proteins are yet unknown. With OEP40, we now were able to identify the first channel protein at the chloroplast OE, specific for transport of glucose and its phosphorylated derivatives. The in vitro OEP40 channel properties match the principal requirement for acting as the regulated glucose gate in the plastid OE. The pore diameter of the restriction site of 1.4 nm is large enough to allow easy permeation of glucose (dglucose ≅0.7 nm), Glc-1-P, or Glc-6-P, and the deduced voltage-independent transit rate of glucose across the OEP40 channel (rtransport ≈ 27 molecules/s) is fast enough to allow efficient transport into or out of the chloroplast. In vivo, the major transport capacity for glucose residues in leaf chloroplasts is export of glucose and maltose molecules for subsequent cytosolic sucrose synthesis during starch breakdown at night (30, 34). Transport of Glc-1-P plays only a minor role for starch synthesis during the day, i.e. via Glc-1-P import, but could also be implicated in night export of phosphorylated glucose (33, 34). Roles for Glc-6-P transport across plastid envelopes are proposed by mutation of IE-intrinsic GPT proteins in non-green tissues, i.e. import for starch synthesis and the oxidative pentose phosphate pathway (35), as well as exchange with the cytosolic hexose-phosphate pool when plastid starch synthesis is impaired (32). Export of glucose and maltose across the IE during starch breakdown at night is mediated by plastid glucose transporter and MEX1 proteins, respectively (30, 34, 36). Interestingly, in vitro OEP40 showed to be impermeable to maltose, and this sugar did also not affect any of the basic OEP40 channel properties. Thus, OEP40 seems to provide in vivo OE specificity for glucose but not maltose fluxes.
In contrast to glucose and phosphorylated derivatives, the disaccharide Tre-6-P is not transported by OEP40, although the pore size of about 1.4 nm would allow permeation. However, Tre-6-P in vitro interacts with OEP40 presumably by binding to the channel. Therefore, it probably induces topological changes of the pore through which the channel properties, such as gating, the open probability, and to a small extent also its selectivity, are modulated in a complex way. Even though in the presence of Tre-6-P, the apparent maximum current through OEP40, and its permeability for anions were slightly reduced, the increased open probability of the channel might result in an increased average permeability for glucose derivatives. Thus, Tre-6-P might be involved in regulation of the transport of glucose derivatives through OEP40.
Recently, Tre-6-P emerged as signaling metabolite within the plant cell, affecting leaf sucrose and starch accumulation during the day and regulating starch breakdown at night, thereby controlling cytosolic availability of sucrose, the major transport sugar in vascular plants (24, 37, 38). In addition, Tre-6-P signaling is necessary for induction of flowering, by integrating responses to changing day length (photoperiod) and high carbohydrate levels with the photoperiod pathway in leaves, as well as fine-tuning flowering time through the age pathway in the SAM (39). Corresponding to night-time chloroplast starch breakdown, export of glucose residues and subsequent cytosolic sucrose synthesis via Glc-6-P, Glc-1-P intermediates is initiated (34). Furthermore, diurnal changes of leaf Tre-6-P concentrations are similar to alterations in starch content (39) and therefore might stimulate chloroplast export of glucose and Glc-1-P and Glc-6-P molecules via OEP40 in particular at the beginning of the night when starch breakdown is activated. Furthermore, it was shown only very recently that an induced Tre-6-P increase leads to a mild reduction in sucrose and no change in starch but elevated glucose levels during the daytime (40). Thus, for the day period the latter might provide a link between Tre-6-P modulation of OEP40 function in glucose transport. Because OEP40 is embedded in the chloroplast OE membrane, the channel protein is ideal for integration of cytosolic signals, i.e. via Tre-6-P modulation. Please note that in vitro Tre-6-P concentrations for modulation of OEP40 are in the low millimolar range (0.2–1 mm), but physiological Tre-6-P concentrations in plant cells are between 1 and 15 μm (41). However, the latter rise significantly under abiotic stress (see below) and are likely to be subject to subcellular variation, e.g. might peak locally at the chloroplast OE. Furthermore, in vitro OEP40 is isolated from its natural environment, which might explain discrepancies.
Interestingly, low levels of OEP40 transcripts can be found throughout Arabidopsis development and tissues (supplemental Fig. S8 A), but in isolated SAM cells OEP40 mRNA amounts are similar when compared with Tre-6-P-synthase (TPS1 (39)) or the LFY transcription factor for floral meristem identity (supplemental Fig. S8B). Because Tre-6-P levels significantly rise in the SAM during the transition to flowering (39), OEP40 could more specifically respond to Tre-6-P regulation in SAM cells and thereby be involved in modulating glucose, Glc-1-P, and Glc-6-P fluxes important for floral induction. This is supported by the fact that both oep40-1 and oep40-3 are early flowering. Furthermore, in young leaf primordia, glucose has been described to be involved in juvenile to adult phase transition via the miR156/SPL signaling pathway (42, 43), and in root meristem tissue, glucose promotes cell proliferation via the so-called target-of-rapamycin pathway (44). Thus, in general the balance of cellular hexose sugar levels seems to be implicated in regulation of developmental processes.
A flowering phenotype of OEP40 loss-of-function mutants, however, was only detected at cold temperature (i.e. 10 °C), indicating that modulation of chloroplast OE fluxes of glucose derivatives becomes important under stress conditions. Surprisingly, the cold-responsive early flowering phenotype already appeared when OEP40 proteins were reduced (i.e. in oep40-1 knockdown lines), pointing to a potential flowering-associated fine-tuning function of hexose metabolite threshold levels and/or involvement of OEP40 in signaling, e.g. as target of Tre-6-P. Furthermore, the overall profiles of soluble metabolites (sugars, organic acids) in total leaf tissue did not change in cold-grown oep40 mutants when compared with wild type (data not shown), which might argue for a cell type-specific function of OEP40 within the SAM (see above).
Cold treatment of Arabidopsis plants leads to rapid metabolic changes, such as accumulation of cryoprotective solutes, i.e. amino acids and sugars (45). In response to cold, starch breakdown is initiated and maltose, glucose, as well as sucrose molecules accumulate. Corresponding to the rising sucrose content, Tre-6-P levels increase up to 55-fold (46, 47) and thus are implicated in cold temperature-associated regulation as well. Interestingly, a block in chloroplast maltose export via MEX1 (maltose translocator) leads to cold stress-like metabolic profiles already at normal temperatures, including maltose accumulation (48). Thus, elevated maltose levels in plastids are suggested to be involved in the cold response. Because OEP40 permeates glucose, but not maltose molecules, chloroplast export of the latter in oep40 loss-of-function mutants might be increased, because glucose export is blocked, and all sugar residues from starch degradation have to be transported as maltose molecules. Therefore, the changed chloroplast/cytosol glucose/maltose ratios could play a role in the observed early flowering phenotype. Cold-treated chloroplasts, besides raffinose, also showed glucose uptake and hexose accumulation (49, 50), implicating that when OEP40 function is impaired, deregulated hexose distribution between chloroplasts and cytosol could lead to induction of flowering. Moreover, modulation of a vacuolar glucose exporter affects freezing tolerance (51), indicating that tight control of the sugar balance between cytosol and organelles is crucial in response to cold. Because temperature control of flowering at the molecular level is only starting to be understood (52), involvement of signaling processes addressed by OEP40 function are still speculative.
However, hexokinase, which converts glucose to Glc-6-P during sucrose synthesis, has been described as a site of glucose sensing, affecting plant growth and development as well as flowering (53–55). Because hexokinase is also attached to the chloroplast OE (56), this enzyme might be directly addressed by changed glucose and Glc-6-P fluxes through OEP40 in the response to cold stress and/or the presence of Tre-6-P. In summary, the presence of the OEP40 glucose gate, together with the previously characterized solute channels in the chloroplast OE membrane (10), adds a new important mosaic part to the emerging picture of the plastid outer membrane as an important player in the regulation of metabolite and solute exchange between the organelle and the cell.
Experimental Procedures
Plant Material and Growth Conditions
Experiments were performed on Arabidopsis thaliana ecotype Columbia 0 (Col-0, Lehle seeds, Round Rock, TX). T-DNA insertion lines oep40-1 (SAIL_266_D10) and oep40-3 (SAIL_759_A01) were obtained from Nottingham Arabidopsis Stock Center, Nottingham, UK. To synchronize germination, all seeds were kept at 4 °C in darkness for 2 days. Seedlings for transcript analysis (see supplemental Fig. S7B) were grown on plates (0.3% Gelrite medium (Roth, Karlsruhe, Germany); 1% d-sucrose; 0.53 MS salts, pH 5.7). Mature plants for immunoblots (supplemental Fig. S7C) and all plants for phenotyping (Figs. 6 and 7) were grown in soil (Stender Substrate A210, Stender AG, Schermbeck, Germany). Plant growth occurred in growth chambers with a long day 16-h light (21 °C; 100 μmol photons·m−2·s−1) and 8-h dark (16 °C) cycle. For growth under cold temperature conditions, 7-day-old seedlings grown in soil at 21/16 °C (day/night) were transferred to a 16-h light and 8-h dark cycle at constant 10 °C. Pea plants (P. sativum L., cv. arvica, Bayrische Futtersaatbau GmbH, Ismaning, Germany) were grown in sand in the greenhouse under a 16-h light (220 μmol·m−2·s−1)/8-h dark regime at 21 °C.
Genotyping and Phenotyping of OEP40 Arabidopsis Mutant Lines
Genomic DNA of the T-DNA insertion lines oep40-1 and oep40-3 was screened by PCR genotyping. To identify plants with T-DNA insertion in both At-OEP40 alleles (homozygous), combinations of gene-specific primers that flank the predicted insertion sites with each other and with T-DNA-specific left border primers (supplemental Table S1) were used. Positions and orientations of T-DNA inserts and oligonucleotide primers in oep40-1 and oep40-3 are shown in supplemental Fig. S7A. To verify insertion sites, PCR-genotyping products were sequenced. To analyze flowering phenotypes of oep40-1, oep40-3 mutants, and Col-0 wild type, we determined days to bolting (DTB; i.e. days until the inflorescence had elongated to 0.5 cm), and total leaf number (TLN; i.e. sum of rosette and cauline leaves).
RNA in Situ Hybridization
Tissue embedding and RNA in situ hybridizations were carried out as described in Wahl et al. (39). Briefly, RNA in situ hybridizations were performed on longitudinal sections through oep40 mutant and Col-0 wild-type apices of plants grown under cold temperature conditions from 34 to 56 days after transfer to the growth chamber. Samples were fixed, dehydrated, and embedded into wax using an automated embedding system (ASP300S, Leica, Wetzlar, Germany). Sections of 8 μm thickness were prepared using a rotary microtome (Leica RM2265; Leica, Wetzlar, Germany). Probes were synthesized with the DIG RNA labeling kit (Roche Applied Science, Mannheim, Germany) on PCR products of the LFY gene cDNA (39). Histological sections were imaged with an Olympus BX-61 microscope equipped with a DC View III digital camera.
Isolation of OEP40 cDNA Molecules
For amplification of At-OEP40 and Ps-OEP40, RT-PCR was performed using Arabidopsis and pea seedling cDNA as template. Oligonucleotide primers were designed according to At3g57990, accessible at the ARAMEMNON database (57) for At-OEP40, and a pea EST contig sequence (16), respectively. The corresponding cDNA molecules were 1101 bp (At-OEP40) and 1167 bp (Ps-OEP40) long. The coding sequence for Ps-OEP40, including a 189-bp 3′UTR, is deposited at GenBankTM, accession number KT361648. For primer sequences see supplemental Table S1 and for amino acid sequences see supplemental Fig. S1.
Protein Extraction and Immunoblot Analysis
Pea chloroplasts isolated from leaf tissue of 10-day-old pea plantlets were sub-fractionated into OE and IE membranes, stroma, and thylakoids as described (58). For total membrane protein extracts from Arabidopsis, leaves were ground in liquid nitrogen and subsequently mixed with an equal volume of urea/buffer (50 mm Tris/HCl, pH 8, 0.2 mm EDTA, 6 m urea). Samples were briefly mixed and rotated for 10 min at 20 °C to solubilize proteins. After centrifugation (16,000 × g, 10 min, 20 °C), total membrane proteins were resuspended in 200 μl of SDS buffer (50 mm Tris/HCl, pH 8, 0.2 mm EDTA, 1% SDS) and solubilized as described above. After a second centrifugation, the membrane protein-containing supernatant was recovered. At-OEP40 and Ps-OEP40 antisera were raised in rabbit (Pineda Antibody Service, Berlin, Germany) against the purified recombinant proteins (see below and supplemental Fig. S2). Antisera for marker proteins were produced as described previously (59, 60). For immunoblot analysis the appropriate amounts of organellar or total cellular proteins were separated by SDS-PAGE, transferred to PVDF membranes, and subjected to immunoblot analysis using primary antisera in 1:250 to 1:5000 dilutions in TTBS buffer (100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.2% Tween 20; 0.1% BSA). Nonspecific signals were blocked by 3% skim milk powder and 0.1% BSA. Secondary anti-rabbit IgG alkaline phosphatase antibodies (Sigma) were diluted 1:30,000. Blots were stained using the alkaline phosphatase reaction with 0.3 mg/ml nitro blue tetrazolium and 0.16 mg/ml bromochloroindolyl phosphate in 100 mm Tris, pH 9.5, 100 mm NaCl, 5 mm MgCl2.
Purification of Recombinant At-OEP40, Ps-OEP40 Proteins
The cDNA of At-OEP40 and Ps-OEP40 was subcloned via NcoI/XhoI into the pET21d plasmid vector (Novagen/Merck, Darmstadt, Germany), resulting in a C-terminal fusion of Ps-OEP40 with a polyhistidine tag (Ps-OEP40(His6)). For At-OEP40, fusion with the His6 tag was avoided by integration of a C-terminal stop codon (for oligonucleotide primers see supplemental Table S1). The generated plasmids were used for overexpression after transforming E. coli BL21(DE3) cells (Novagen/Merck). Overexpression was induced by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside to rapidly growing cells (OD600 of 0.4–0.6). After 3 h of growth at 37 °C, cells were harvested at 6000 × g for 10 min at 4 °C and resuspended in Tris/NaCl buffer (50 mm Tris/HCl, pH 8, 200 mm NaCl, 5 mm β-MeSH). Cells were twice broken in a microfluidizer processor (Microfluidics, Westwood, MA), and bacterial gDNA was degraded using ultrasonification. Samples were centrifuged at 20,000 × g for 30 min at 4 °C, and the resulting inclusion body (IB) pellet was washed twice with detergent buffer (20 mm Tris/HCl, pH 7.5, 200 mm NaCl, 1% deoxycholic acid, 1% Nonidet P-40, 5 mm β-MeSH), followed by centrifugation at 12,000 × g for 10 min, 4 °C. IBs were further cleaned by repeated washing with Triton X-100 (20 mm Tris/HCl, pH 7.5, 0.5% Triton X-100, 5 mm β-MeSH) and finally Tris buffer (200 mm Tris/HCl, pH 8, 10 mm DTT).
For generation of antiserum, cleaned rec_Ps-OEP40(His6) IBs were dissolved in buffer G (50 mm NaPi, pH 8, 100 mm NaCl, 2 mm β-MeSH, 6 m guanidinium chloride). Proteins were purified via their C-terminal His6-tag (compare supplemental Fig. S2, A and B) using Ni-NTA-Sepharose (GE Healthcare) and were eluted with 50–500 mm imidazole in buffer A (50 mm NaPi, pH 8, 100 mm NaCl, 2 mm β-MeSH, 8 m urea). For rec_At-OEP40, IBs were resuspended by rotation for 1 h at 20 °C in SDS buffer (50 mm Tris/HCl, pH 8, 100 mm NaCl, 1% SDS). After centrifugation at 20,000 × g for 15 min at 20 °C, rec_At-OEP40 was present in the supernatant. Because of the lack of an affinity tag, 7 mg of protein were further separated by SDS-PAGE and purified by electroelution. For this, the respective band was excised from the gel, placed into a dialysis tubing (size exclusion 14 kDa), and electroeluted at 16 mA overnight in SDS buffer.
For analysis in planar lipid bilayers, we first exchanged a methionine residue at position 19 of Ps-OEP40/pET21d into isoleucine using site-directed mutagenesis to avoid occurrence of a second translational start. IBs containing rec_Ps-OEP40M19I(His6) were prepared after overexpression in E. coli as described above. After the second wash with Triton buffer, IBs were resuspended for 1 h (20 °C) in 15 ml of urea buffer (50 mm Tris/HCl, pH 8, 100 mm NaCl, 6 m urea). After centrifugation at 20,000 × g for 15 min at 20 °C, rec_Ps-OEP40M19I(His6) was present in the supernatant and bound to Ni-NTA-Sepharose (GE Healthcare) in the presence of 5 mm imidazole overnight at 20 °C. After centrifugation at 1000 × g for 2 min (20 °C), the pelleted Ni-NTA beads were applied onto a 2.5-ml column (MoBiTec Gmbh, Goettingen, Germany) and rec_Ps-OEP40M19I(His6) protein was eluted with 50–500 mm imidazole in urea buffer (see supplemental Fig. S2, A–C).
Recombinant Control Proteins (Mock Samples)
To exclude contaminations with E. coli membrane proteins in planar lipid bilayers, we produced control rec_PsOEP40-M19I(His6) protein by two independent in vitro translation systems. (i) For translation in a cell-free eukaryotic system, the TnT® Coupled Reticulate Lysate System (Promega, Madison, WI) was used according to the manufacturers' instructions. Thereby Ps-OEP40-M19I/pET21d and a control reaction without plasmid DNA were transcribed using T7 RNA polymerase and translated in the coupled non-radiolabeled system for 120 min at 30 °C (see supplemental Fig. S2D). Both reactions were centrifuged at 80,000 × g for 15 min at 4 °C, and proteins in supernatants (containing rec_PsOEP40-M19I(His6) and empty control, respectively) were subsequently analyzed in planar lipid bilayers. Presence of rec_Ps-OEP40-M19I(His6) proteins in the respective samples was confirmed by immunoblots (see supplemental Fig. S2D). (ii) To in vitro translate the Ps-OEP40-M19I(His6) protein using a plant wheat germ lysate, Ps-OEP40-M19I/pET21d was transcribed by T7 RNA polymerase and translated in the absence of radiolabeled amino acids as described previously (61). Subsequently, translation reactions were supplemented with lysis buffer (30 mm Tris/HCl, pH 7.9, 300 mm NaCl, 0.5% SDS, 5 mm imidazole), incubated at 55 °C for 3 min, and purified by 200 μl of Ni-NTA-Sepharose on mobicol minispin columns (MoBiTec Gmbh, Goettingen, Germany). After elution with 30 mm Tris/HCl, pH 7.9, 300 mm NaCl, 0.5% SDS, 300 mm imidazole, fractions of eight translation reactions were pooled and concentrated to 100 μl using an Amicon® Ultra 0.5-ml filter unit (Merck KGaA, Darmstadt, Germany). The concentrated fraction was separated via a 12.5% SDS gel and reversibly stained by zinc imidazole, and the band corresponding to rec_Ps-OEP40-M19I(His6), as well as a second control band, was excised from the gel. After destaining, the excised protein bands were used for reconstitution and lipid bilayer measurements, and the presence or absence of rec_Ps-OEP40-M19I(His6) was confirmed by immunoblotting (see supplemental Fig. S2E).
Reconstitution of OEP40 into Proteoliposomes
For reconstitution, the purified protein in urea buffer (see above) was supplemented with 2% SDS. Preformed liposomes (50 mg/ml, l-α-phosphatidylcholine from egg in 100 mm NaCl, 10 mm MOPS/Tris, pH 7.0) were solubilized with 80 mm Mega9 and added to a protein/lipid ratio between 1:50 (w/w). Detergent and urea were then removed by dialysis initially for 2 h at room temperature and subsequently overnight at 4 °C in 5 liters of buffer (100 mm NaCl, 10 mm MOPS/Tris, pH 7.0) with the inclusion of Calbiosorb adsorbent into the dialysis cup for efficient SDS removal. As mock samples, IBs from untransformed E. coli cells and reticulocyte lysate translations without Ps-OEP40 templates subjected to the above purification steps were used. These mock samples did not show any channel activity at all (n = 5).
Single Channel Recordings from Planar Lipid Bilayers and Data Analysis
Planar lipid bilayer measurements were performed as described previously (62). If not explicitly stated otherwise, symmetric conditions (250 mm KCl, 10 mm MOPS/Tris, pH 7.0) were used in cis and trans compartments. 250 mm KCl, 10 mm MOPS/Tris, pH 7.0, in cis and 20 mm KCl, 10 mm MOPS/Tris, pH 7.0, in trans was used as standard asymmetric conditions. Sugars were purchased from Sigma as α-d-glucose, α-d-glucose 1-phosphate disodium salt hydrate (Glc-1-P), d-glucose 6-phosphate sodium salt (Glc-6-P), and trehalose 6-phosphate dipotassium salt (Tre-6-P), respectively. Single channel currents were recorded using a current amplifier (Gene Clamp 500, Axon Instruments) with a CV-5-1G Headstage (Axon Instruments) and filtered with the inbuilt 4-pole Bessel low pass filter at 5 kHz. For data acquisition at a sampling rate of 50 kHz, a personal computer equipped with a Digidata 1200 (Axon Instruments) and Clampex 9 software was used. Voltage ramps were conducted with a rate of 7.5 or 15 mV/s. The denomination cis and trans corresponds to the half-chambers of the bilayer unit. Reported membrane potentials are always referred to the trans compartment. Channel insertion into the bilayer occurred mostly (≥70%) unidirectionally as deduced from their rectification characteristics. Those channels integrated in reverse orientation were replotted for analysis. Gating transition amplitudes were analyzed and subsequently filtered for dwell times exceeding 0.1 ms (corresponding to five times the sampling interval) to exclude incompletely resolved gating transitions from the analysis (63). Single channel analysis was performed essentially as described in detail in Ref. 21. In brief, we used the robust mean variance approach (22) that allows rigorous unbiased analysis of single channel recordings. For this, we developed a Mathlab-based program (64) that allows the automatic current amplitude and channel state dwell time analysis of single channel recordings (21).
Circular Dichroism Analysis
Recombinant At-OEP40/pET21d and At-OEP37/pET14b proteins were refolded according to Ref. 65. Therefore, IBs (see above) were solubilized in SDS buffer (120 mm SDS, 10 mm NaPi, pH 7.5, 200 mm NaF) by rotation for 1 h at 20 °C. After centrifugation at 20,000 × g for 15 min at 20 °C, an equal volume of MPD buffer (3 m 2-methyl-2,4-pentanediol, 10 mm NaPi, pH 7.5, 200 mm NaF) was added to the resuspended proteins in the supernatant. Refolding of proteins occurred by rotation of samples for 24 h at 20 °C. For CD analysis according to Ref. 66, rec_At-OEP40 and rec_At-OEP37 samples were diluted to a final concentration of 0.065 and 0.042 mg/ml with 10 mm NaPi, pH 7.5. CD measurements in 10 consecutive scans from 190 to 260 nm were performed with a J-810 spectropolarimeter (Jasco Inc., Easton, MD). Data were analyzed by the DICHROWEB analysis webserver (67), using SELCON3, CONTIN, and CDSSTR programs.
Bioinformatic Analysis of the Putative OEP40 Topology and Structure
Sequence-based structure prediction of OEP40 from Arabidopsis (UniProtKB, Q9M2P9) was performed using different algorithms as follows: PSIPRED (18), SPP (Scratch Protein Predictor) (68), PRED-TMBB (19), TMBpro (20), and I-TASSER (69). A putative model of the tertiary structure predicted by TMBpro was illustrated using the UCSF Chimera modeling system (70).
Author Contributions
K. P., A. H., J. S., and R. W. designed the study, analyzed the data, and wrote the paper. A. H. and B. H. performed electrophysiological analysis of OEP40. A. Schock and I. J. isolated Arabidopsis and pea OEP40 proteins, performed immunoblot analysis, and characterized mutant phenotypes. H. P.-O. supplied additional immunoblots. V. W. and A. Schlereth contributed to phenotyping of early flowering and performed in situ hybridization of shoot apical meristems. P. B. carried out bioinformatic prediction of OEP40 structure and topology. T. A. G. and K. P. established and analyzed circular dichroism spectroscopy for OEP40 proteins. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Karl Mayer (Ludwig-Maximilians-University Munich) and Christin Abel (Max Planck Institute of Molecular Plant Physiology) for excellent technical assistance; John Lunn (Max Planck Institute of Molecular Plant Physiology) and Tabea Mettler/Andreas Weber (Heinrich Heine-University of Düsseldorf) for metabolite analysis; and the group of Mark Stitt (Max Planck Institute of Molecular Plant Physiology) for hosting A. Schock.
The authors declare that they have no conflicts of interest with the contents of this article.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KT361648

This article contains supplemental Figs. S1–S8, Methods, and Table S1.
- IE
- chloroplast inner envelope membrane
- VDAC
- voltage-dependent anion channel
- OEP
- outer envelope proteins
- OE
- chloroplast outer envelope membrane
- Col-0
- A. thaliana ecotype Columbia 0
- DTB
- days to bolting
- TLN
- total leaf number
- Glc-1-P
- glucose 1-phosphate
- Glc-6-P
- glucose 6-phosphate
- Tre-6-P
- trehalose 6-phosphate
- GHK
- Goldman-Hodgkin-Katz
- SAM
- shoot apical meristem
- Ni-NTA
- nickel-nitrilotriacetic acid
- IB
- inclusion body
- pS
- picosiemens
- MeSH
- mercaptoethanol.
References
- 1. Philippar K., and Soll J. (2007) in Plant Solute Transport (Yeo A. R., and Flowers T. J., eds) pp. 133–192, Blackwell Publishing, Ltd., Oxford, UK [Google Scholar]
- 2. Block M. A., Douce R., Joyard J., and Rolland N. (2007) Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth. Res. 92, 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weber A. P., and Linka N. (2011) Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62, 53–77 [DOI] [PubMed] [Google Scholar]
- 4. Zeth K., and Thein M. (2010) Porins in prokaryotes and eukaryotes: common themes and variations. Biochem. J. 431, 13–22 [DOI] [PubMed] [Google Scholar]
- 5. Bredemeier R., Schlegel T., Ertel F., Vojta A., Borissenko L., Bohnsack M. T., Groll M., von Haeseler A., and Schleiff E. (2007) Functional and phylogenetic properties of the pore-forming β-barrel transporters of the Omp85 family. J. Biol. Chem. 282, 1882–1890 [DOI] [PubMed] [Google Scholar]
- 6. Colombini M. (2012) VDAC structure, selectivity, and dynamics. Biochim. Biophys. Acta 1818, 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bölter B., Soll J., Hill K., Hemmler R., and Wagner R. (1999) A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J. 18, 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pohlmeyer K., Soll J., Grimm R., Hill K., and Wagner R. (1998) A high-conductance solute channel in the chloroplastic outer envelope from pea. Plant Cell 10, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goetze T. A., Philippar K., Ilkavets I., Soll J., and Wagner R. (2006) OEP37 is a new member of the chloroplast outer membrane ion channels. J. Biol. Chem. 281, 17989–17998 [DOI] [PubMed] [Google Scholar]
- 10. Duy D., Soll J., and Philippar K. (2007) Solute channels of the outer membrane: from bacteria to chloroplasts. Biol. Chem. 388, 879–889 [DOI] [PubMed] [Google Scholar]
- 11. Breuers F. K., Bräutigam A., and Weber A. P. (2011) The plastid outer envelope–a highly dynamic interface between plastid and cytoplasm. Front. Plant Sci. 2, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baines C. P., Kaiser R. A., Sheiko T., Craigen W. J., and Molkentin J. D. (2007) Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death 1. Nat. Cell Biol. 9, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blachly-Dyson E., Song J., Wolfgang W. J., Colombini M., and Forte M. (1997) Multicopy suppressors of phenotypes resulting from the absence of yeast VDAC encode a VDAC-like protein. Mol. Cell. Biol. 17, 5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bölter B., and Soll J. (2001) Ion channels in the outer membranes of chloroplasts and mitochondria: open doors or regulated gates? EMBO J. 20, 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pudelski B., Schock A., Hoth S., Radchuk R., Weber H., Hofmann J., Sonnewald U., Soll J., and Philippar K. (2012) The plastid outer envelope protein OEP16 affects metabolic fluxes during ABA-controlled seed development and germination. J. Exp. Bot. 63, 1919–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franssen S. U., Shrestha R. P., Bräutigam A., Bornberg-Bauer E., and Weber A. P. (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schleiff E., Eichacker L. A., Eckart K., Becker T., Mirus O., Stahl T., and Soll J. (2003) Prediction of the plant β-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci. 12, 748–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41, W349–W357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bagos P. G., Liakopoulos T. D., Spyropoulos I. C., and Hamodrakas S. J. (2004) PRED-TMBB: a web server for predicting the topology of β-barrel outer membrane proteins. Nucleic Acids Res. 32, W400–W404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Randall A., Cheng J., Sweredoski M., and Baldi P. (2008) TMBpro: secondary structure, β-contact and tertiary structure prediction of transmembrane β-barrel proteins. Bioinformatics 24, 513–520 [DOI] [PubMed] [Google Scholar]
- 21. Harsman A., Bartsch P., Hemmis B., Kruger V., and Wagner R. (2011) Exploring protein import pores of cellular organelles at the single molecule level using the planar lipid bilayer technique. Eur. J. Cell Biol. 90, 721–730 [DOI] [PubMed] [Google Scholar]
- 22. Patlak J. B. (1993) Measuring kinetics of complex single ion channel data using mean-variance histograms. Biophys. J. 65, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hille B. (2001) Ionic Channels of Excitable Membranes, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 24. Lunn J. E., Delorge I., Figueroa C. M., Van Dijck P., and Stitt M. (2014) Trehalose metabolism in plants. Plant J. 79, 544–567 [DOI] [PubMed] [Google Scholar]
- 25. Movileanu L., Schmittschmitt J. P., Scholtz J. M., and Bayley H. (2005) Interactions of peptides with a protein pore. Biophys. J. 89, 1030–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blümel M., Dally N., and Jung C. (2015) Flowering time regulation in crops–what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129 [DOI] [PubMed] [Google Scholar]
- 27. Siriwardana N. S., and Lamb R. S. (2012) The poetry of reproduction: the role of LEAFY in Arabidopsis thaliana flower formation. Int. J. Dev. Biol. 56, 207–221 [DOI] [PubMed] [Google Scholar]
- 28. Schäfer G., and Heber U. (1977) Glucose transport into spinach chloroplasts. Plant Physiol. 60, 286–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weber A., Servaites J. C., Geiger D. R., Kofler H., Hille D., Gröner F., Hebbeker U., and Flügge U. I. (2000) Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12, 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cho M. H., Lim H., Shin D. H., Jeon J. S., Bhoo S. H., Park Y. I., and Hahn T. R. (2011) Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol. 190, 101–112 [DOI] [PubMed] [Google Scholar]
- 31. Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A., and Flügge U.-I. (1998) Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 10, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kunz H. H., Häusler R. E., Fettke J., Herbst K., Niewiadomski P., Gierth M., Bell K., Steup M., Flügge U. I., and Schneider A. (2010) The role of plastidial glucose 6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol. 12, 115–128 [DOI] [PubMed] [Google Scholar]
- 33. Fettke J., Malinova I., Albrecht T., Hejazi M., and Steup M. (2011) Glucose-1-phosphate transport into protoplasts and chloroplasts from leaves of Arabidopsis. Plant Physiol. 155, 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stitt M., and Zeeman S. C. (2012) Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292 [DOI] [PubMed] [Google Scholar]
- 35. Niewiadomski P., Knappe S., Geimer S., Fischer K., Schulz B., Unte U. S., Rosso M. G., Ache P., Flügge U. I., and Schneider A. (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17, 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niittylä T., Messerli G., Trevisan M., Chen J., Smith A. M., and Zeeman S. C. (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303, 87–89 [DOI] [PubMed] [Google Scholar]
- 37. Tsai A. Y., and Gazzarrini S. (2014) Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci. 5, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yadav U. P., Ivakov A., Feil R., Duan G. Y., Walther D., Giavalisco P., Piques M., Carillo P., Hubberten H. M., Stitt M., and Lunn J. E. (2014) The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 65, 1051–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J. E., Stitt M., and Schmid M. (2013) Regulation of flowering by trehalose 6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707 [DOI] [PubMed] [Google Scholar]
- 40. Figueroa C. M., Feil R., Ishihara H., Watanabe M., Kölling K., Krause U., Höhne M., Encke B., Plaxton W. C., Zeeman S. C., Li Z., Schulze W. X., Hoefgen R., Stitt M., and Lunn J. E. (2016) Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 85, 410–423 [DOI] [PubMed] [Google Scholar]
- 41. Lunn J. E., Feil R., Hendriks J. H., Gibon Y., Morcuende R., Osuna D., Scheible W. R., Carillo P., Hajirezaei M. R., and Stitt M. (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang L., Xu M., Koo Y., He J., and Poethig R. S. (2013) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2, e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu S., Cao L., Zhou C. M., Zhang T. Q., Lian H., Sun Y., Wu J., Huang J., Wang G., and Wang J. W. (2013) Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife 2, e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiong Y., McCormack M., Li L., Hall Q., Xiang C., and Sheen J. (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaplan F., Kopka J., Sung D. Y., Zhao W., Popp M., Porat R., and Guy C. L. (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 50, 967–981 [DOI] [PubMed] [Google Scholar]
- 46. Nunes C., O'Hara L. E., Primavesi L. F., Delatte T. L., Schluepmann H., Somsen G. W., Silva A. B., Fevereiro P. S., Wingler A., and Paul M. J. (2013) The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 162, 1720–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carillo P., Feil R., Gibon Y., Satoh-Nagasawa N., Jackson D., Bläsing O. E., Stitt M., and Lunn J. E. (2013) A fluorometric assay for trehalose in the picomole range. Plant Methods 9, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Purdy S. J., Bussell J. D., Nunn C. P., and Smith S. M. (2013) Leaves of the Arabidopsis maltose exporter1 mutant exhibit a metabolic profile with features of cold acclimation in the warm. PLoS ONE 8, e79412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schneider T., and Keller F. (2009) Raffinose in chloroplasts is synthesized in the cytosol and transported across the chloroplast envelope. Plant Cell Physiol. 50, 2174–2182 [DOI] [PubMed] [Google Scholar]
- 50. Nägele T., and Heyer A. G. (2013) Approximating subcellular organisation of carbohydrate metabolism during cold acclimation in different natural accessions of Arabidopsis thaliana. New Phytol. 198, 777–787 [DOI] [PubMed] [Google Scholar]
- 51. Klemens P. A., Patzke K., Trentmann O., Poschet G., Büttner M., Schulz A., Marten I., Hedrich R., and Neuhaus H. E. (2014) Overexpression of a proton-coupled vacuolar glucose exporter impairs freezing tolerance and seed germination. New Phytol. 202, 188–197 [DOI] [PubMed] [Google Scholar]
- 52. Capovilla G., Schmid M., and Posé D. (2015) Control of flowering by ambient temperature. J. Exp. Bot. 66, 59–69 [DOI] [PubMed] [Google Scholar]
- 53. Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., Hwang I., Jones T., and Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336 [DOI] [PubMed] [Google Scholar]
- 54. Smith A. M., and Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ. 30, 1126–1149 [DOI] [PubMed] [Google Scholar]
- 55. Smeekens S., Ma J., Hanson J., and Rolland F. (2010) Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 274–279 [DOI] [PubMed] [Google Scholar]
- 56. Wiese A., Gröner F., Sonnewald U., Deppner H., Lerchl J., Hebbeker U., Flügge U., and Weber A. (1999) Spinach hexokinase I is located in the outer envelope membrane of plastids. FEBS Lett. 461, 13–18 [DOI] [PubMed] [Google Scholar]
- 57. Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., and Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Waegemann K., Eichacker S., and Soll J. (1992) Outer envelope membranes from chloroplasts are isolated as right-side-out vesicles. Planta 187, 89–94 [DOI] [PubMed] [Google Scholar]
- 59. Duy D., Wanner G., Meda A. R., von Wirén N., Soll J., and Philippar K. (2007) PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19, 986–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Philippar K., Geis T., Ilkavets I., Oster U., Schwenkert S., Meurer J., and Soll J. (2007) Chloroplast biogenesis: the use of mutants to study the etioplast-chloroplast transition. Proc. Natl. Acad. Sci. U.S.A. 104, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fellerer C., Schweiger R., Schöngruber K., Soll J., and Schwenkert S. (2011) Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol. Plant 4, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 62. Hinnah S. C., Wagner R., Sveshnikova N., Harrer R., and Soll J. (2002) The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys. J. 83, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McManus O. B., Blatz A. L., and Magleby K. L. (1987) Sampling, log binning, fitting, and plotting durations of open and shut intervals from single channels and the effects of noise. Pflügers Arch. 410, 530–553 [DOI] [PubMed] [Google Scholar]
- 64. Schmedt D. (2010) Peptide–Translocation through Tom40: Dynamic Interaction and Transport of CoxIV. Ph.D. thesis, University of Osnabrück [Google Scholar]
- 65. Roussel G., Perpète E. A., Matagne A., Tinti E., and Michaux C. (2013) Towards a universal method for protein refolding: the trimeric β barrel membrane Omp2a as a test case. Biotechnol. Bioeng. 110, 417–423 [DOI] [PubMed] [Google Scholar]
- 66. Kelly S. M., Jess T. J., and Price N. C. (2005) How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139 [DOI] [PubMed] [Google Scholar]
- 67. Whitmore L., and Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 68. Cheng J., Randall A. Z., Sweredoski M. J., and Baldi P. (2005) SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 33, W72–W76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.