Abstract
Single nucleotide polymorphisms in the FKBP5 gene increase the expression of the FKBP51 protein and have been associated with increased risk for neuropsychiatric disorders such as major depression and post-traumatic stress disorder. Moreover, levels of FKBP51 are increased with aging and in Alzheimer disease, potentially contributing to disease pathogenesis. However, aside from its glucocorticoid responsiveness, little is known about what regulates FKBP5. In recent years, non-coding RNAs, and in particular microRNAs, have been shown to modulate disease-related genes and processes. The current study sought to investigate which miRNAs could target and functionally regulate FKBP5. Following in silico data mining and initial target expression validation, miR-511 was found to suppress FKBP5 mRNA and protein levels. Using luciferase p-miR-Report constructs and RNA pulldown assays, we confirmed that miR-511 bound directly to the 3′-UTR of FKBP5, validating the predicted gene-microRNA interaction. miR-511 suppressed glucocorticoid-induced up-regulation of FKBP51 in cells and primary neurons, demonstrating functional, disease-relevant control of the protein. Consistent with a regulator of FKBP5, miR-511 expression in the mouse brain decreased with age but increased following chronic glucocorticoid treatment. Analysis of the predicted target genes of miR-511 revealed that neurogenesis, neuronal development, and neuronal differentiation were likely controlled by these genes. Accordingly, miR-511 increased neuronal differentiation in cells and enhanced neuronal development in primary neurons. Collectively, these findings show that miR-511 is a functional regulator of FKBP5 and can contribute to neuronal differentiation.
Keywords: depression, glucocorticoid, microRNA (miRNA), neurodifferentiation, stress, FKBP5
Introduction
FKBP51 (FK506 binding protein 51 kDa) is dysregulated in several diseases, but there is a paucity of data regarding its functional regulation. FKBP51 is an Hsp90 co-chaperone that helps regulate the function of specific Hsp90 clients, such as the glucocorticoid receptor (GR)3 and the microtubule-associated protein Tau. FKBP51 inhibits GR function, leading to delayed hypothalamic-pituitary-adrenal axis feedback and elevated circulating glucocorticoid levels (1–3), a phenomenon observed in major depression (4). In fact, single nucleotide polymorphisms in the FKBP5 gene have been associated with increased risk for depression, as well as other neuropsychiatric disorders including post-traumatic stress disorder (PTSD) (5–7). Mice with a targeted deletion of Fkbp5 display resilience to stress and accelerated hypothalamic-pituitary-adrenal axis reactivity (8, 9). FKBP51 expression is also increased in Alzheimer disease (AD), which is characterized by accumulation of misfolded Tau (10). FKBP51 has been shown to accelerate Tau oligomerization and neurotoxicity in vitro and in vivo, suggesting that it may be contributing to the pathogenesis of AD (11). Therefore, reducing FKBP51 expression is of significant interest in both depression and AD; however, little is known about what genetically regulates FKBP5, aside from its glucocorticoid response element (GRE) responsiveness (12).
The prospect of controlling gene expression through microRNAs (miRNAs) has been an area of intense recent research. miRNAs are non-protein-coding RNAs that bind to the 3′-UTR of specific mRNAs to promote degradation or inhibit translation (13). These endogenous genetic regulators have broad patterns of expression and localization, even within the brain. Moreover, miRNAs have been implicated in several processes associated with disease, including depression and AD. Several studies have shown that miRNAs can regulate neurogenesis, a process believed to be impaired in major depression (14), whereas stress can also differentially affect miRNA expression depending on the type of the stress and area of the brain (15–17). Moreover, miRNA expression is down-regulated in the prefrontal cortex of depressed suicide patients (18), and polymorphisms in miRNA genes can contribute to susceptibility to depression (19). AD brains display miRNA dysregulation as well, providing a common link between miRNAs and FKBP5 (20). However, it is not known which miRNAs target FKBP5 in the brain or whether miRNA regulation can directly alter FKBP51 expression.
The current study sought to determine whether miRNAs could regulate FKBP5 expression as a way to control FKBP51 biology. Multiple programs were initially probed to predict potential miRNA candidates that could target FKBP5. Candidates were then identified that have been linked to diseases associated with FKBP5 dysfunction. This search revealed three miRNAs that could potentially bind to FKBP5: miR-142, miR-340, and miR-511. Experiments in cells demonstrated that each of these miRNAs could regulate FKBP5 expression at the mRNA level, but only miR-511 robustly affected downstream protein levels of FKBP51. Therefore, miR-511 was selected for more in-depth analyses to confirm miRNA-gene binding and functional regulation. Our data reveal that miR-511 definitively targets FKBP5 and regulates neuronal differentiation as predicted by miRNA software programs.
Results
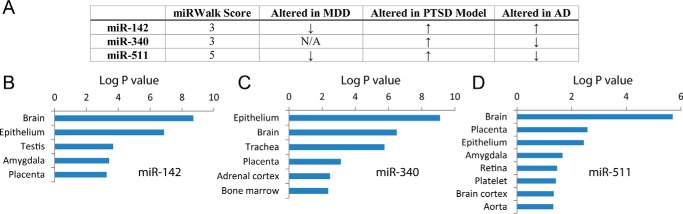
Several freely available atlases and databases have been generated to assist in the prediction of miRNA-target interactions. We employed miRWalk v1.0 to determine which miRNAs were predicted to target human FKBP5 mRNA using a minimum score of 3 of 5 (21) (Fig. 1A). Following this initial search, the literature was explored for any candidate miRNAs that were related to depression, PTSD, or AD, because these are the three major neurological diseases associated with FKBP5. This search revealed three miRNAs with potential binding to FKBP5 and links to these diseases: miR-142, miR-340, and miR-511 (Fig. 1A). Only miR-511 exhibited disease-related expression consistent with a regulator of FKBP5, because it has been found to be down-regulated in major depression and AD and increased in serum from a rat model of PTSD. Expression patterns of the target genes predicted to bind each miRNA were determined by entering the gene targets into the DAVID gene ontology bioinformatics database to evaluate functional gene classifications (22, 23) (Fig. 1, B–D).
FIGURE 1.
MicroRNAs implicated in disease that are predicted to target FKBP5. A, miR-142 and miR-340 were predicted to target FKBP5 by three different programs, whereas miR-511 was predicted to target it by all five programs surveyed using miRWalk v1.0. Arrows indicate up- or down-regulation of each miRNA in major depressive disorder (MDD), a rat model of PTSD, or AD. B–D, the predicted target genes of miR-142 (B), miR-340 (C), and miR-511 (D) were enriched in the brain according to DAVID gene ontology analysis.
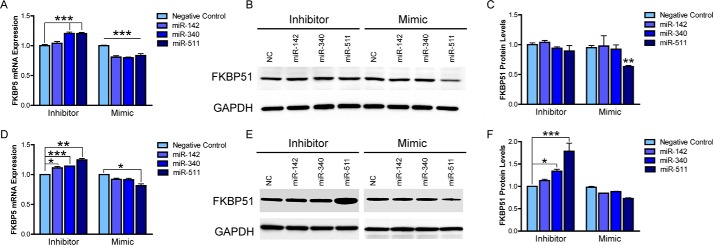
To directly assess whether these miRNAs had a biological effect on FKBP5, mRNA expression of FKBP5 was examined from HEK293T cells following transfection of miRNA mimics or inhibitors. Each of the three miRNA mimics significantly reduced FKBP5 mRNA expression as measured by quantitative RT-PCR, whereas the miRNA inhibitor constructs had the opposite effect on FKBP5 expression for miR-340 and miR-511 but no effect for miR-142 (Fig. 2A). We next examined whether any of these miRNAs affected protein levels of FKBP51. The SDS/PAGE revealed that only the miR-511 mimic had a significant effect on FKBP51 levels (Fig. 2, B and C), indicating a functional impact on FKBP51 biology. Interestingly, no additive effects of the miRNAs were observed when co-transfected (data not shown), suggesting a lack of potentiation on FKBP5.
FIGURE 2.
miR-511 inhibits FKBP5 at the mRNA and protein level. A, miRNA inhibitors and mimics of miR-142, miR-340, and miR-511 were transfected into HEK293T cells, and the mRNA expression of FKBP5 was measured by real time PCR. Inhibitors of miR-142 and miR-511 increased FKBP5 mRNA expression, whereas all three miRNA mimics decreased expression. B, protein levels of FKBP51 were assessed under the same conditions using SDS/PAGE. C, densitometry quantification of the blots in B revealed that only the miR-511 mimic was able to affect protein levels of FKBP51, significantly decreasing them. D–F, experiments were repeated identically in M17 cells, which display 6-fold higher expression of endogenous miR-511. *, p < 0.05; **, p < 0.01; ***, p < 0.001. NC, negative control miRNA.
RT-PCR revealed a ∼6-fold greater expression of miR-511 in M17 cells compared with HEK293T cells (6.056 ± 0.541; data not shown), allowing us to better assess the effects of miRNA inhibition on endogenous FKBP51. As expected, the miRNA inhibitors had a much greater impact on both FKBP5 expression and FKBP51 protein levels in M17 cells, and miR-511 still displayed the most robust effects (Fig. 2, D–F). Therefore, subsequent studies were only performed on miR-511.
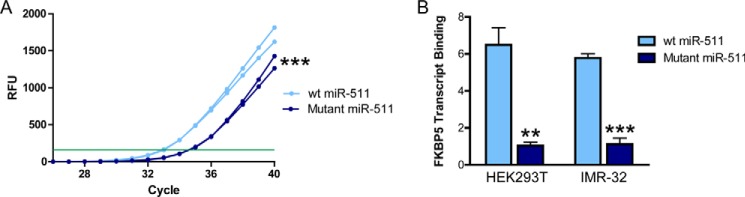
Sequence alignment of miR-511 and the 3′-UTR of FKBP5 revealed high sequence complementarity, further confirming possible miRNA-gene binding (Fig. 3A). We then designed a mutant FKBP5 with an alternate 3′-UTR sequence. The functional effect of WT or mutant FKBP5 was tested by inserting a fragment of FKBP5, surrounding and including the 3′-UTR, into a pMIR-REPORT luciferase reporter, which tests putative miRNA binding sites. Luciferase activity of the mutant FKBP5 was not affected by the miR-511 mimic, whereas activity of WT FKBP5 was decreased (Fig. 3B). To definitively verify that miR-511 was binding to FKBP5, an RNA pulldown assay was performed using biotinylated miR-511 and RT-PCR (Fig. 4A). Both in HEK293T and IMR-32 cells, FKBP5 mRNA was enriched in miR-511 following pulldown, an effect not observed using a mutant miR-511 (Fig. 4B). Together, these findings demonstrate that FKBP5 is a valid target of miR-511. We also examined whether there was a reciprocal relationship between miR-511 and FKBP5 by determining whether elevated levels of FKBP51 could impact miR-511 expression. Using RT-PCR, we did not observe any change in miR-511 expression following overexpression of FKBP5 in HEK293T cells (Vector: 1.00 ± 0.14, FKBP5: 1.10 ± 0.05; p = 0.552), suggesting the regulation is not bidirectional.
FIGURE 3.
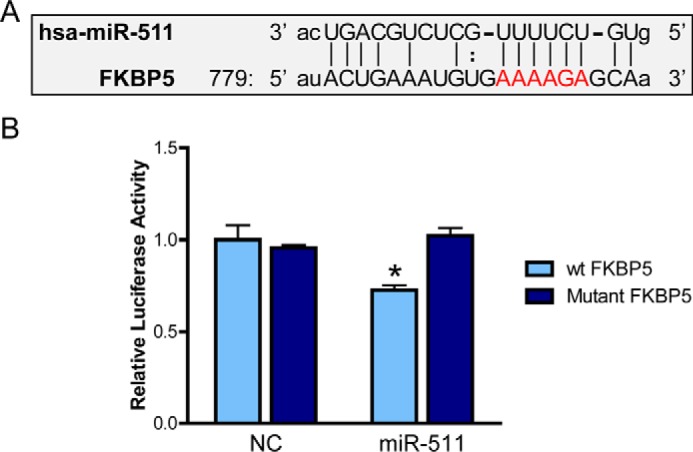
miR-511 represses FKBP5 expression. A, potential miR-511 binding sites in the 3′-UTR of FKBP5 mRNA were discovered using software from microrna.org. The area highlighted in red indicates the sites used for mutagenesis of the seed sequence. B, a fragment of the 3′-UTR of FKBP5 containing the putative miRNA binding sites or the mutant was cloned into a luciferase reporter vector, which was co-transfected with negative control miRNA (NC) or the miR-511 mimic. miR-511 decreased luciferase activity in the wild-type (wt) but not mutant FKBP5 reporter, indicating it could repress FKBP5 expression through direct binding. *, p < 0.05.
FIGURE 4.
miR-511 binds to the 3′-UTR of FKBP5. Biotin-labeled wild-type (wt) or mutant miR-511 mimics were transfected into HEK293T or IMR-32 cells, and the FKBP5 mRNA was detected by RT-qPCR following RNA pulldown. Duplicate RT-PCR amplification curves from IMR-32 cells are shown in A, with the green line denoting the threshold. RFU indicates relative fluorescent units. B, mutant miR-511 did not show any binding to FKBP5 mRNA, whereas WT miR-511 displayed robust binding in both HEK293T and IMR-32 cells. **, p < 0.01; ***, p < 0.001.
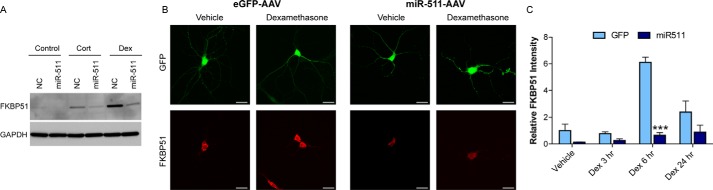
FKBP5 contains GRE binding sites, which lead to glucocorticoid-induced gene up-regulation (24). We initially sought to determine whether miR-511 could suppress glucocorticoid-induced up-regulation of FKBP51 in HeLa cells, which have high levels of endogenous GR. Following treatment with either hydrocortisone or dexamethasone, the miR-511 mimic dramatically decreased the glucocorticoid-induced increase in levels of FKBP51 (Fig. 5A). We further explored the effects of miR-511 overexpression on glucocorticoid-induced FKBP51 up-regulation in primary neurons isolated from WT mice. Neurons were transduced with GFP or miR-511 AAV9 for 10 days and treated with dexamethasone for 3, 6, or 24 h to induce FKBP51 up-regulation prior to examining immunofluorescence (Fig. 5B). As expected, the hormone significantly increased FKBP51 levels at 6 h; overexpression of the miR-511 mimic completely abrogated this increase (Fig. 5C), further validating its ability to target and inhibit FKBP5.
FIGURE 5.
miR-511 suppresses glucocorticoid-induced FKBP51 up-regulation in cells and primary neurons. A, overexpression of the miR-511 mimic in HeLa cells suppressed the increase in endogenous FKBP51 protein levels induced by treatment with hydrocortisone (Cort) or dexamethasone (Dex). B, representative images of primary neurons transduced with eGFP or miR-511 AAV9 for 10 days prior to treatment with vehicle or dexamethasone for 6 h. C, overexpression of miR-511 significantly suppressed FKBP51 levels following 6 h of treatment with dexamethasone. ***, p < 0.001. Scale bar, 30 μm. NC, negative control miRNA.
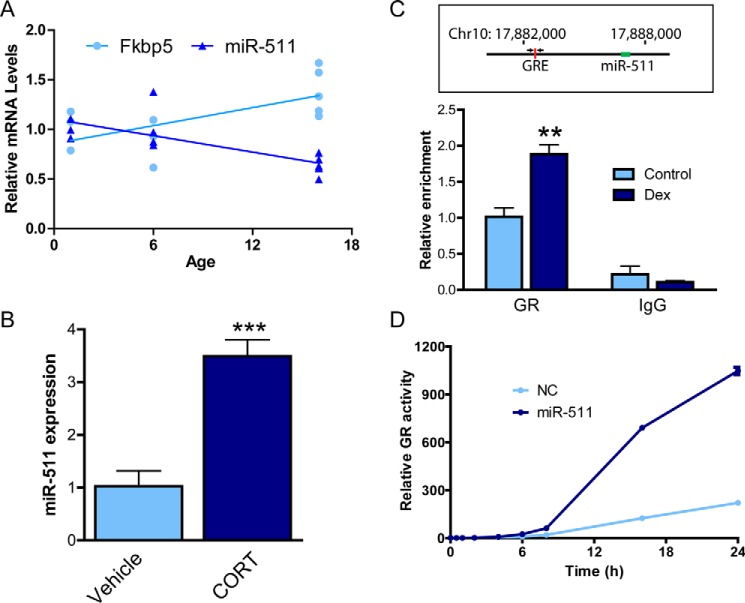
Previous studies have demonstrated that FKBP5 expression increases with age in humans (11). To determine whether murine Fkbp5 expression aligned with these previous findings and to investigate expression of miR-511 with age, RT-PCR was employed to determine mRNA levels in the hippocampus of WT mice at various ages (Fig. 6A). Linear regression analyses revealed a significant increase in Fkbp5 expression and a concomitant decrease in miR-511 expression with age. This decrease in miR-511 levels may partially mediate the age-dependent elevations in FKBP5 expression. Interestingly, peripheral miR-511 expression in the spleen and liver can be up-regulated by glucocorticoids similar to FKBP5 (25). Therefore, we investigated whether miR-511 expression in the brain was affected by chronic glucocorticoid treatment in WT mice. Following corticosterone chronically administered in the drinking water, miR-511 expression was in fact up-regulated in the cortex (Fig. 6B), confirming that GR activity can regulate the miRNA. To explore the mechanism by which GR up-regulates miR-511, we employed Motifmap to search for consensus GRE sequences around the miR-511 gene. This search revealed a GR binding site 4930 bp upstream of the miR-511 gene. Using a ChIP assay, we found that the binding of GR to this specific GRE increased following hormone treatment (Fig. 6C), suggesting that GR can regulate miR-511 expression via direct binding to the GRE adjacent to the miR-511 gene. We next investigated whether this regulation was bidirectional by examining whether miR-511 could alter GR activity. As shown in Fig. 6D, the miR-511 mimic dramatically increased GR activity beginning at 4 h post-treatment. Collectively, these data suggest that there is likely an intricate feedback loop among GR, FKBP51, and miR-511, which may contribute to disease when disrupted.
FIGURE 6.
Reciprocal regulation of miR-511 and glucocorticoid receptor. A, Fkbp5 expression increases, whereas miR-511 expression decreases with age in the hippocampus of wild-type mice. B, chronic corticosterone (CORT) treatment in mice increased expression of miR-511 in the cortex (n = 4). C, schematic depicting the chromosomal locations of a GRE and miR-511 on chromosome 10 (top panel). ChIP of the discrete GRE adjacent to miR-511 revealed increased binding following treatment with dexamethasone (Dex). D, overexpression of the miR-511 mimic increased GRE luciferase reporter activity following treatment with dexamethasone for varying durations. **, p < 0.01; ***, p < 0.001. NC, negative control miRNA.
Because miRNAs tend to regulate hundreds of genes, we employed DAVID gene ontology database mining to determine which functional processes miR-511 was predicted to regulate. Narrowing the results to processes related to the brain, we discovered that neurogenesis, neuronal development, and neuronal differentiation were most prevalent (Fig. 7A). Because neurogenesis has been repeatedly implicated in the pathogenesis of depression, we examined whether miR-511 could affect neuronal differentiation in a cell-based model of neurogenesis. N2a cells, a murine neuroblastoma cell line, were transfected with the miR-511 mimic prior to differentiation with retinoic acid for 3 days (Fig. 7B). miR-511 increased the percentage of differentiated cells (Fig. 7C) and also increased expression of the neuron-specific markers Calbindin 2 and Prox1 (Fig. 7D), suggesting that miR-511 does increase neuronal differentiation and may have similar effects on neurogenesis.
FIGURE 7.
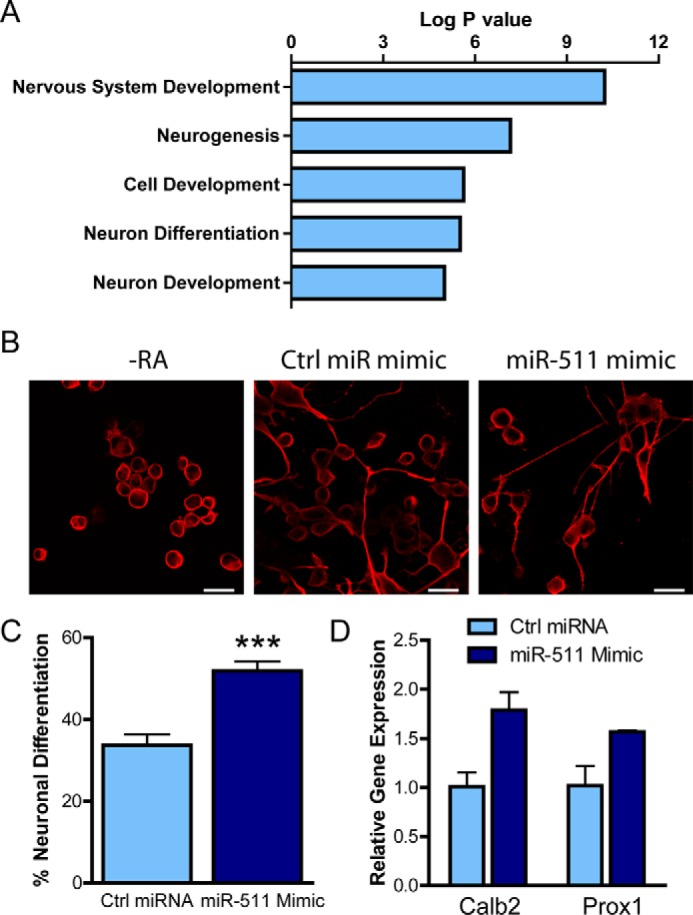
miR-511 increases neuronal differentiation. A, DAVID gene ontology analysis revealed that the target genes of miR-511 were implicated in neuronal development and differentiation. B, N2a cells were differentiated by retinoic acid (RA) for 3 days following transfection with control (Ctrl) miRNA or miR-511 mimics and stained with the neuron-specific marker β3-tubulin. C, the miR-511 mimic significantly increased the percentage of differentiated cells counted. D, the miR-511 mimic increased expression of the neuronal markers Calb2 and Prox1 as measured by real time PCR. ***, p < 0.001. Scale bar, 30 μm.
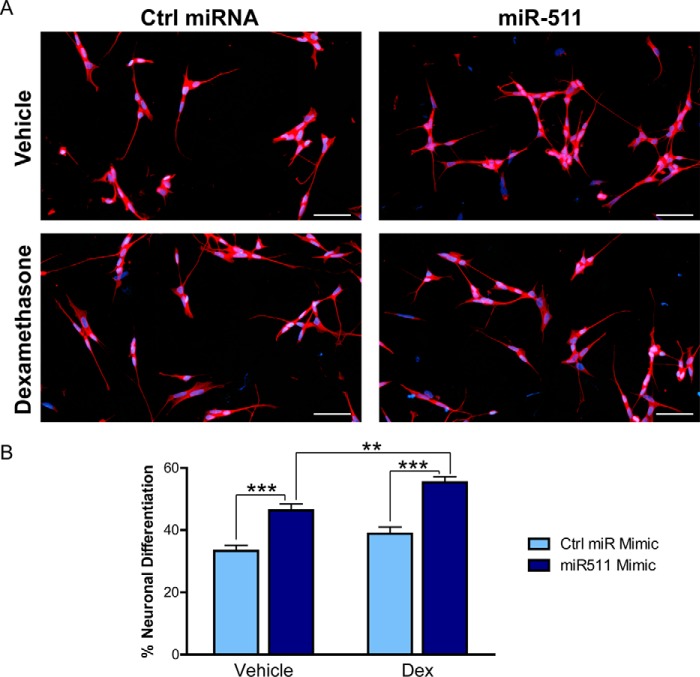
Because of our findings of a positive feedback loop between miR-511 and glucocorticoid responsiveness, we tested whether neuronal differentiation was affected by hormone in combination with miR-511. Using another cell line to further validate the N2a cell results, SH-SY5Y cells were transfected with a control or miR-511 mimic and differentiated with retinoic acid for 3 days. The cells were treated with vehicle or dexamethasone during this period to determine whether hormone could impact the effects of miR-511. Representative images of differentiated cells are displayed in Fig. 8A. Similar to the N2a cells, miR-511 significantly increased neuronal differentiation, and this differentiation was further enhanced by chronic hormone treatment (Fig. 8B), reinforcing the idea of a positive feedback loop between miR-511 and GR activity.
FIGURE 8.
miR-511 and glucocorticoids enhance neuronal differentiation. A, SH-SY5Y cells were transfected with control (Ctrl) miRNA mimic or miR-511 mimic and treated with vehicle or dexamethasone and retinoic acid to induce neuronal differentiation. The cells were stained with β3-tubulin and DAPI prior to imaging. B, the miR-511 mimic significantly increased the percentage of differentiation of the cells, which was further enhanced by chronic dexamethasone (Dex) treatment. **, p < 0.01; ***, p < 0.001. Scale bar, 50 μm.
To further explore the effects of miR-511 on neuronal development and differentiation in a more physiological model, murine primary neurons were transfected with control miRNA or miR-511 plasmids. As shown in Fig. 9A, neuronal development appears to be enhanced by miR-511, with longer processes and neurites observed 3 days following miR-511 transfection. Moreover, markers of late stage neuronal development were significantly up-regulated following miR-511 overexpression, including Calbindin 1 and 2, Prox1, and Rbfox3 (NeuN) (Fig. 9B). Overexpression of miR-511 was confirmed using RT-PCR, wherein a fold increase of 3.89 ± 0.08 (data not shown) was observed. Collectively, these data underscore the role of miR-511 in neuronal development and differentiation, suggesting it could be an important factor in the developing brain.
FIGURE 9.
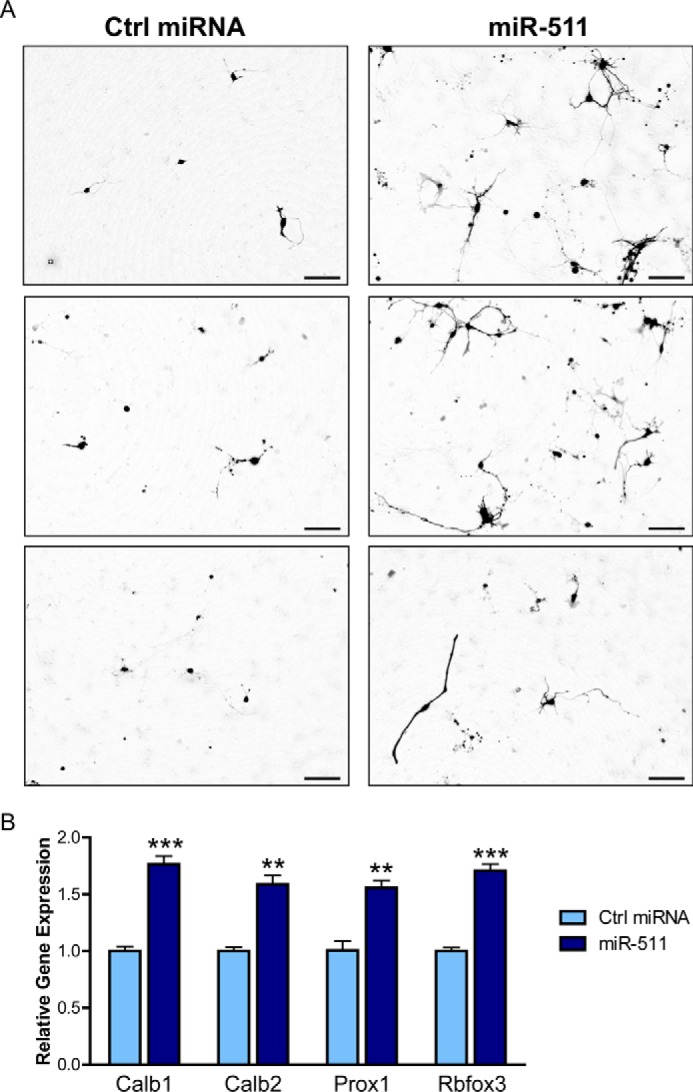
miR-511 increases neuronal development and differentiation in neurons. A, primary neurons were transfected with control (Ctrl) miRNA or miR-511 plasmids on day in vitro 2, and live cell images were obtained 3 days later. B, neurons transfected with miR-511 showed increased expression of late-stage neuronal markers, including Calb1, Calb2, Prox1, and Rbfox3 (also known as NeuN). **, p < 0.01; ***, p < 0.001. Scale bar, 100 μm.
Discussion
The area of miRNA research has accelerated exponentially over the last few years with more and more miRNAs being discovered and ascribed gene targets. In the field of depression alone, dozens of miRNAs have been implicated in the disease (19, 26–30). The current study identified and confirmed miR-511 as a regulator of FKBP5, a gene associated with risk for stress-related disorders including major depression and PTSD. FKBP51 has also been linked to AD, with elevated levels observed in AD brains (11). FKBP51 accelerates formation of Tau oligomers and enhances neurotoxicity in a mouse model of AD, suggesting that its inhibition would be beneficial. Interestingly, miR-511 is down-regulated in depression and AD (18, 31) and up-regulated in a rat model of PTSD (32), which could contribute to aberrant processing of FKBP5. Our findings demonstrate that miR-511 is a modulator of FKBP5 and also elucidate a novel role of miR-511 in glucocorticoid signaling and neuronal differentiation.
Therapeutic efforts directed at targeting FKBP51 have been challenging for several reasons, most of all because of the high degree of sequence homology between FKBP51 and other members of the FKBP family (33). Gene therapy may have better success. According to miRWalk and other databases, miR-511 is not a predicted binder of similar FKBPs, such as FKBP4 and FKBP1A. This specificity for FKBP5 could be important for therapeutic development of miR mimics. However, the inherent lack of specificity with which miRNAs operate would almost certainly lead to additional off-target effects. Therefore, the functional processes that a particular miRNA regulates may be more informative regarding its putative effects. Here we showed that a miR-511 mimic enhanced neuronal development and differentiation in two neuroblastoma cell lines and primary neurons, which may be indicative of its ability to regulate neurogenesis as predicted by the DAVID gene ontology analyses. Future studies may be able to determine the effects of miR-511 on neurogenesis in the brain.
It has been previously shown that peripheral miR-511 expression in the spleen and liver can be up-regulated by glucocorticoids (25). The current study confirmed this result in the brain, demonstrating that chronic corticosterone increased miR-511 expression. Our ChIP data provide a mechanism for miR-511 up-regulation by glucocorticoids, revealing that the miR-511 gene is directly downstream of a consensus GRE sequence. Conversely, we found that miR-511 overexpression increased GR activity, indicating a positive feedback loop. It is interesting to consider the consequences of this reciprocal stimulation within the context of a focal GR-centric feedback loop. If stimulating GR up-regulates miR-511, which in turn can repress FKBP51 expression as we have shown here, this would lead to enhanced GR activity and perhaps an altered stress response. Thus, because levels of FKBP51 and miR-511 change with age, the hypothalamic-pituitary-adrenal axis may become dysregulated and lead to diminished ability to adapt to stressors.
Targeting FKBP51 could be beneficial in multiple neuropsychiatric disorders, as well as age-related diseases such as AD. The current study revealed that multiple miRNAs could decrease FKBP5 expression, whereas only miR-511 was able to have a functional impact on FKBP51 protein levels. Gaining a greater understanding of how FKBP5 is regulated will likely be beneficial for potential drug design and development. Using miRNAs as potential biomarkers is also an appealing strategy because more and more effort is being directed at a personalized medicine approach. Our findings that miR-511 expression can be controlled by GR signaling suggest that it is possible that individuals with single nucleotide polymorphisms in FKBP5 may display altered miR-511 expression, which could be detected in serum or cerebrospinal fluid, as has been accomplished in patients with AD or major depression (29, 34). This approach would permit selective targeting of miR-511 when warranted and could potentially enhance regulation of processes implicated in disease such as neurogenesis.
Experimental Procedures
Prediction of miRNAs Target Genes and Functional Annotation
To search for miRNAs that were predicted to target FKBP5, we used the miRWalk v1.0 database with different bioinformatics algorithms (miRanda, miRDB, miRWalk, RNA22, and Targetscan); the miRNAs predicted by at least three algorithms were selected (Fig. 1A). The putative target genes of selected miRNAs were predicted using the same databases and subjected to the DAVID database for gene ontology analysis and functional annotation.
Cell Culture and Transfection
Human embryonic kidney 293T cells (HEK293T), IMR-32 cells (human neuroblastoma), M17 cells (human neuroblastoma), Neuro-2a cells (N2A; mouse neuroblastoma), SH-SY5Y cells (human neuroblastoma), and HeLa cells were purchased from ATCC and cultured in the medium and conditions recommended. Plasmids, miRNA mimics, inhibitors, and controls were transfected or co-transfected with Lipofectamine 2000 according to manufacturer's directions (Invitrogen). See Table 1 for details about miRNA plasmid constructs. To induce differentiation of N2a and SH-SY5Y cells, the cells were treated with retinoic acid (10 μm) for 3 days.
TABLE 1.
List of primers and miRNAs used
Forward and reverse primers are listed for relevant real time PCR experiments and miRNA constructs. NCBI accession numbers are listed for real time PCR studies, whereas restriction enzymes are noted for miR-511 plasmids. For mRNA pulldown assays, wild-type and mutant seed sequences are listed.
| Method | Name | Forward | Reverse | Accession number/restriction enzymes |
|---|---|---|---|---|
| Real time PCR | hFKBP5 | caagaagtttgcagagcaggat | cactgggactcttccctcctt | NM_004117 |
| GAPDH | tcctgcaccaccaactgctta | cacagtcttctgggtggcagt | NM_002046 (human); NM_008084 (mouse) | |
| mFkbp5 | gtacaacaaagccgtggagtg | gccctgttctgaggattgact | NM_010220 | |
| mProx1 | agctataccgagccctcaacat | ccaggaaggatcaacatctttg | NM_008937 | |
| mRbfox3 | agtgaccagtttcccctaccc | cagcaccataaaatccatcctg | NM_001039167 | |
| mCalb1 | ctccgcgcactctcaaacta | tgcagctcctttccttccag | NM_009788 | |
| mCalb2 | tttcagggtatgaagctgacctc | tgacactcttcctgtaggtggtg | NM_007586 | |
| AAV9 | mmiR-511 | GAGAAGCTTcccttgcattttcttctcttca | ACTACCGGTaggctctgatgatggacttcct | HindIII/AgeI |
| GR ChIP | GRE-511 | TGCAGTGAGTGGAGATTGAGC | AGAAACAACACGGACCTCAGC | |
| Luciferase reporter | miR-511 | CTAGTAAGTTCTGAGATACTGAAATGTGAAAAGAGCAATCAGAATTGTA | AGCTTACAATTCTGATTGCTCTTTTCACATTTCAGTATCTCAGAACTTA | SpeI/HindIII |
| miR-511-Mut | CTAGTAAGTTCTGAGATACTGAAATGTGCCCCTGGCAATCAGAATTGTA | AGCTTACAATTCTGATTGCCAGGGGCACATTTCAGTATCTCAGAACTTA | SpeI/HindIII | |
| mRNA pulldown | miR-511-Bio | GUGUCUUUUGCUCUGCAGUCA-biotin | GUGUCUUUUGCUCUGCAGUCA-biotin | RNA |
| miR-511-Mut-Bio | GUGAGAAAAGCUCUGCAGUCA-biotin | GUGAGAAAAGCUCUGCAGUCA-biotin | RNA |
RNA Isolation and Quantitative PCR
Total RNA was isolated from cells, primary neurons, or mouse brain using TRIzol reagent according to manufacturer's protocol (Invitrogen). cDNA was prepared through reverse transcription using the iScript cDNA synthesis kit (Bio-Rad), and qPCR was conducted using SYBR Green PCR Master Mix (Applied Biosystems) using the primers listed in Table 1. Triplicate PCR reactions were run and GAPDH mRNA expression was analyzed for each sample for normalization. The data were analyzed according to the 2−ΔΔCt method (35).
For miR-511 expression analysis, total RNA including small RNA was isolated from cells, primary neurons or mouse tissue using the miRNeasy micro kit (Qiagen). 10 ng of RNA was used for reverse transcription using TaqMan® MicroRNA reverse transcription kit (Applied Biosystems) and hsa-miR-511 or mmu-miR-511 specific primers. qPCR was performed using TaqMan PCR master mix (Bio-Rad) and specific probes and primers in triplicate. The expression values of miR-511 were normalized to RUNU6B.
Western Blot
The cells were lysed with radioimmune precipitation assay buffer with a protease inhibitor mixture (Sigma) and 1 mm PMSF. 40 μg of total protein was subjected to SDS-PAGE using 4–15% gradient gels and then transferred to PVDF membranes (Amersham Biosciences). After blocking with 7% nonfat milk, the membrane was incubated with rabbit anti-FKBP5 polyclonal antibody (1:1000; Cell Signaling) or mouse anti-GAPDH (1:1000; Meridian Life Science) at room temperature for 1 h, followed by incubation with secondary antibodies (1:1000, Southern Biotech). Blots were developed using ECL (Pierce) on a LAS-4000 mini imager (GE Healthcare). Densitometry was performed using Scion Image, and duplicate samples of FKBP51 were normalized to GAPDH.
Dual Luciferase Reporter Assay
The putative miR-511 binding site in the 3′-UTR of FKBP5 and its flanking sequence (10 bp per side) were synthesized by Sigma (the sequence of the oligonucleotides is listed in Table 1) and annealed and inserted into the pMIR-Report vector (Ambion). The fragment that replaced the seed sequence of miR-511 binding sites was inserted and served as the mutant control. HEK293T cells were plated in 24-well plates and transfected with 400 ng of the reporter plasmids, 20 ng of pRL-CMV (Promega), and the miR-511 mimic or control at a final concentration of 20 nm using Lipofectamine 2000 (Life Technologies). Following 40 h of incubation, the cells were harvested with Glo lysis buffer (Promega) and subjected to a luciferase reporter assay using the Dual-Luciferase reporter assay kit (Promega). Firefly luciferase activities were normalized to Renilla luciferase activities. Each experiment was repeated at least three times in duplicate.
To measure GR activity, the GRE reporter (200 ng) was co-transfected with a GR cDNA plasmid (200 ng), pRL-CMV (20 ng), and miR-511 mimic or control (20 nm) in HEK293T cells, which were growing in phenol red-free DMEM (Life Technologies) with 10% charcoal-stripped FBS (Life Technologies). At different time points, dexamethasone was added at a final concentration of 10 nm, and the cells were harvested and subjected to Dual-Luciferase assay as described above.
mRNA Pulldown Assay
To validate the binding of miR-511 to the mRNA of FKBP5, an mRNA pulldown assay was conducted as described (36). Briefly, the biotinylated miR-511 or mutant miR-511 (constructed by replacing the seed sequence as shown in Table 1) was transfected into HEK293T or IMR-32 cells. The cells were fixed with 1% formaldehyde 40 h later (Sigma) at room temperature for 15 min and stopped with 0.2 m glycine. After washing with TBS, the cells were lysed with lysis buffer (50 mm HEPES, pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton, 0.1% sodium deoxycholate), and genomic DNA was digested with RNase-free DNase I (NEB). The miRNA-mRNA complex was pulled down by streptavidin beads (Dynabeads M-280 Streptavidin), washed three times with washing buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.15 mm LiCl), and eluted with elution buffer (0.1 m NaHCO3, 1% SDS) and heat treatment for 10 min at 85 °C. The proteins were digested with the addition of protease K and incubation at 65 °C for 2 h was performed to fully reverse the cross-linkages. RNA was purified with TRIzol, and the mRNA level of FKBP5 was measured with RT-qPCR.
AAV9 Construction and Production
The mouse genomic DNA including mmu-miR-511 and its flanking sequence (about 150 bp upstream and downstream) was amplified using Phusion high fidelity DNA polymerase (NEB) with the primers listed in Table 1. After digestion, the fragment was subcloned into the 3′-UTR of GFP in the pTR12.1-MCS vector, which contains the short hybrid CMV chicken β-actin promoter as described (37). The sequence-confirmed, endotoxin-free plasmid was co-transfected with helper plasmids pFdelta6 and pAAV9 into HEK293T cells (ATCC, Manassas, VA). The resulting recombinant virus was harvested by three cycles of freeze-thaw and purified using an iodixanol gradient as previously described (38) and concentrated to 200 μl. A Sybr-green-based real time PCR was used to determine the viral titer and is expressed as vector genomes/ml. The primary neurons were infected with equivalent titers of AAV9, with eGFP-AAV9 serving as control.
Primary Neurons
Primary cortical neurons were isolated from E16 WT mouse brains according to previous established protocols (39). Briefly, the brains were extracted, meninges were removed, and cortices were placed in ice-cold isotonic buffer (137 mm NaCl, 5 mm KCl, 0.2 mm NaH2PO4, 0.2 mm KH2PO4, 5.5 mm glucose, and 6 mm sucrose, pH 7.4). Following washes, cortices were minced, digested in trypsin, triturated, and resuspended in DMEM supplemented with 10% FBS, penicillin/streptomycin, and amphotericin B. The DMEM was exchanged 24 h later for Neurobasal medium supplemented with GlutaMAX and B27 supplement (Life Technologies). For experiments investigating the effects of miR-511 on FKBP51 levels, primary neurons were transduced on DIV4 with 2 μl of miR-511-eGFP or eGFP AAV in PBS at 1013 vector genomes/ml. On DIV18, neurons were treated with dexamethasone (0.5 μm) for 3, 6, or 24 h prior to fixation with 4% paraformaldehyde. For experiments examining the role of miR-511 in neuronal development and differentiation, primary neurons were transfected on DIV2 with control miRNA or miR-511 plasmids. In vivo cell imaging was performed on DIV5 using a Cytation 3 cell imaging multimode reader (BioTek Instruments Inc., Winooski, VT) with a 10× objective, and neurons were then harvested for qPCR as described above.
Immunohistochemistry and Imaging
Cells and primary neurons were permeabilized using 0.1% Triton X-100 for 10 min and blocked in 5% goat serum in PBS for 30 min. To examine neuronal differentiation, the cells were incubated in a class III β-tubulin (Tuj1) antibody (1:500; BioLegend, San Diego, CA) for 1 h at room temperature. The neurons were incubated overnight at 4 °C with primary antibody directed against FKBP51 (mouse FKBP51E antibody at 1:100; gift from Marc B. Cox). Following washes, Alexa Fluor secondary antibody was added for 90 min at room temperature (1:500), and coverslips were mounted onto slides using Prolong Gold (Life Technologies).
Confocal imaging was performed using an Olympus FluoView confocal microscope as previously described (40) to capture z-stack images at 60× magnification for primary neurons. To examine FKBP51 intensity, the analyze particles function on ImageJ software (National Institutes of Health) was utilized to measure integrated density of the positive staining. Six images were captured for each sample and then averaged; each condition was performed in triplicate. For differentiation experiments, the cells were counted by blinded experimenters, and the percentage of differentiation was determined.
Mice
Male and female mice from an FVB/SVEV background (Jackson Laboratories, Bar Harbor, ME) were used to investigate expression of Fkbp5 and miR-511. All animal studies were approved by the University of South Florida Institutional Animal Care and Use Committee and carried out in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals. The mice were group housed under a 12-h light-dark cycle (lights on at 06:00) and permitted ad libitum access to food and water.
Chronic Glucocorticoid Treatment
For glucocorticoid treatment, 4.5-month-old mice were chronically administered 0.1 mg/ml corticosterone (Sigma) in 1% ethanol in tap water ad libitum through leak-proof sipper tubes for ∼10 weeks and harvested at 6 months of age. The brains were rapidly removed following overdose with sodium pentobarbital, and the cortices were dissected out and flash frozen.
Chromatin Immunoprecipitation
GR binding sites around the miR-511 gene were searched by Motifmap (41), and the primers were designed using DNA around the binging site (−150 to +150 bp). ChIP analyses were conducted as described previously (42). In brief, HEK293T cells were transfected with human GR cDNA in charcoal-stripped medium for 40 h and treated with vehicle or 10 nm dexamethasone for 4 h. The cells were harvested and cross-linked, and sonicated chromatin was pulled down with 5 μg of GR (Cell Signaling) or IgG antibody (as a control). After washing and de-cross-linking, the DNA was purified with phenol chloroform, and the enrichment of specific DNA was analyzed by RT-PCR with the primers listed in Table 1.
Statistics
Statistical significance for each analysis was determined with Student's t tests, one- or two-way analysis of variance with Tukey, or Bonferroni post tests to compare groups or linear regression where appropriate. All figures and statistics were generated using GraphPad Prism software; each graph represents the mean ± S.E.
Author Contributions
D. Z. designed experiments, collected and analyzed most of the data, and contributed to the writing of the manuscript. J. J. S. designed experiments, collected and analyzed data, and contributed to the writing of most of the manuscript. L. J. B., A. L. D., and X. W. collected and analyzed data. C. A. D. conceived and designed the experiments and contributed to the writing of the manuscript.
This work was supported by the National Institutes of Mental Health Grant R01 MH103848 and National Institutes of Neurological Disease and Stroke R01 Grant NS073899. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GR
- glucocorticoid receptor
- AD
- Alzheimer disease
- miRNA
- microRNA
- PTSD
- post-traumatic stress disorder
- GRE
- glucocorticoid response element
- CORT
- corticosterone
- AAV
- adeno-associated virus
- qPCR
- quantitative PCR.
References
- 1. O'Leary J. C. 3rd, Dharia S., Blair L. J., Brady S., Johnson A. G., Peters M., Cheung-Flynn J., Cox M. B., de Erausquin G., Weeber E. J., Jinwal U. K., and Dickey C. A. (2011) A new anti-depressive strategy for the elderly: ablation of FKBP5/FKBP51. PLoS One 6, e24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wochnik G. M., Rüegg J., Abel G. A., Schmidt U., Holsboer F., and Rein T. (2005) FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 280, 4609–4616 [DOI] [PubMed] [Google Scholar]
- 3. Denny W. B., Prapapanich V., Smith D. F., and Scammell J. G. (2005) Structure-function analysis of squirrel monkey FK506-binding protein 51, a potent inhibitor of glucocorticoid receptor activity. Endocrinology 146, 3194–3201 [DOI] [PubMed] [Google Scholar]
- 4. Gillespie C. F., and Nemeroff C. B. (2005) Hypercortisolemia and depression. Psychosom. Med. 67, S26–S28 [DOI] [PubMed] [Google Scholar]
- 5. Binder E. B., Salyakina D., Lichtner P., Wochnik G. M., Ising M., Pütz B., Papiol S., Seaman S., Lucae S., Kohli M. A., Nickel T., Künzel H. E., Fuchs B., Majer M., Pfennig A., et al. (2004) Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat. Genet. 36, 1319–1325 [DOI] [PubMed] [Google Scholar]
- 6. Binder E. B., Bradley R. G., Liu W., Epstein M. P., Deveau T. C., Mercer K. B., Tang Y., Gillespie C. F., Heim C. M., Nemeroff C. B., Schwartz A. C., Cubells J. F., and Ressler K. J. (2008) Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA 299, 1291–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klengel T., Mehta D., Anacker C., Rex-Haffner M., Pruessner J. C., Pariante C. M., Pace T. W., Mercer K. B., Mayberg H. S., Bradley B., Nemeroff C. B., Holsboer F., Heim C. M., Ressler K. J., Rein T., et al. (2013) Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 16, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sabbagh J. J., O'Leary J. C. 3rd, Blair L. J., Klengel T., Nordhues B. A., Fontaine S. N., Binder E. B., and Dickey C. A. (2014) Age-associated epigenetic upregulation of the FKBP5 gene selectively impairs stress resiliency. PLoS One 9, e107241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartmann J., Wagner K. V., Liebl C., Scharf S. H., Wang X. D., Wolf M., Hausch F., Rein T., Schmidt U., Touma C., Cheung-Flynn J., Cox M. B., Smith D. F., Holsboer F., Müller M. B., et al. (2012) The involvement of FK506-binding protein 51 (FKBP5) in the behavioral and neuroendocrine effects of chronic social defeat stress. Neuropharmacology 62, 332–339 [DOI] [PubMed] [Google Scholar]
- 10. Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., and Binder L. I. (1986) Abnormal phosphorylation of the microtubule-associated protein Tau (Tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blair L. J., Nordhues B. A., Hill S. E., Scaglione K. M., O'Leary J. C. 3rd, Fontaine S. N., Breydo L., Zhang B., Li P., Wang L., Cotman C., Paulson H. L., Muschol M., Uversky V. N., Klengel T., et al. (2013) Accelerated neurodegeneration through chaperone-mediated oligomerization of Tau. J. Clin. Invest. 123, 4158–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paakinaho V., Makkonen H., Jääskeläinen T., and Palvimo J. J. (2010) Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol. Endocrinol. 24, 511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iwakawa H. O., and Tomari Y. (2015) The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 25, 651–665 [DOI] [PubMed] [Google Scholar]
- 14. Eisch A. J., and Petrik D. (2012) Depression and hippocampal neurogenesis: a road to remission? Science 338, 72–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dwivedi Y., Roy B., Lugli G., Rizavi H., Zhang H., and Smalheiser N. R. (2015) Chronic corticosterone-mediated dysregulation of microRNA network in prefrontal cortex of rats: relevance to depression pathophysiology. Transl. Psychiatry 5, e682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smalheiser N. R., Lugli G., Rizavi H. S., Zhang H., Torvik V. I., Pandey G. N., Davis J. M., and Dwivedi Y. (2011) MicroRNA expression in rat brain exposed to repeated inescapable shock: differential alterations in learned helplessness vs. non-learned helplessness. Int. J. Neuropsychopharmacol. 14, 1315–1325 [DOI] [PubMed] [Google Scholar]
- 17. Smalheiser N. R., Zhang H., and Dwivedi Y. (2014) Enoxacin elevates microRNA levels in rat frontal cortex and prevents learned helplessness. Front. Psychiatry 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smalheiser N. R., Lugli G., Rizavi H. S., Torvik V. I., Turecki G., and Dwivedi Y. (2012) MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One 7, e33201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He Y., Zhou Y., Xi Q., Cui H., Luo T., Song H., Nie X., Wang L., and Ying B. (2012) Genetic variations in microRNA processing genes are associated with susceptibility in depression. DNA Cell Biol. 31, 1499–1506 [DOI] [PubMed] [Google Scholar]
- 20. Femminella G. D., Ferrara N., and Rengo G. (2015) The emerging role of microRNAs in Alzheimer's disease. Front. Physiol. 6, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dweep H., Sticht C., Pandey P., and Gretz N. (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 [DOI] [PubMed] [Google Scholar]
- 22. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 23. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Binder E. B. (2009) The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 34, S186–S195 [DOI] [PubMed] [Google Scholar]
- 25. Puimège L., Van Hauwermeiren F., Steeland S., Van Ryckeghem S., Vandewalle J., Lodens S., Dejager L., Vandevyver S., Staelens J., Timmermans S., Vandenbroucke R. E., and Libert C. (2015) Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol. Med. 7, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Camkurt M. A., Acar S., Coskun S., Gunes M., Gunes S., Yilmaz M. F., Gorur A., and Tamer L. (2015) Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J. Psychiatr. Res. 69, 67–71 [DOI] [PubMed] [Google Scholar]
- 27. Wang X., Sundquist K., Hedelius A., Palmér K., Memon A. A., and Sundquist J. (2015) Circulating microRNA-144–5p is associated with depressive disorders. Clin. Epigenetics 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song M. F., Dong J. Z., Wang Y. W., He J., Ju X., Zhang L., Zhang Y. H., Shi J. F., and Lv Y. Y. (2015) CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J. Affect. Disorders 178, 25–31 [DOI] [PubMed] [Google Scholar]
- 29. Wan Y., Liu Y., Wang X., Wu J., Liu K., Zhou J., Liu L., and Zhang C. (2015) Identification of differential microRNAs in cerebrospinal fluid and serum of patients with major depressive disorder. PLoS One 10, e0121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Issler O., Haramati S., Paul E. D., Maeno H., Navon I., Zwang R., Gil S., Mayberg H. S., Dunlop B. W., Menke A., Awatramani R., Binder E. B., Deneris E. S., Lowry C. A., and Chen A. (2014) MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83, 344–360 [DOI] [PubMed] [Google Scholar]
- 31. Lau P., Bossers K., Janky R., Salta E., Frigerio C. S., Barbash S., Rothman R., Sierksma A. S., Thathiah A., Greenberg D., Papadopoulou A. S., Achsel T., Ayoubi T., Soreq H., Verhaagen J., et al. (2013) Alteration of the microRNA network during the progression of Alzheimer's disease. EMBO Mol. Med. 5, 1613–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balakathiresan N. S., Chandran R., Bhomia M., Jia M., Li H., and Maheshwari R. K. (2014) Serum and amygdala microRNA signatures of posttraumatic stress: fear correlation and biomarker potential. J. Psychiatr. Res. 57, 65–73 [DOI] [PubMed] [Google Scholar]
- 33. Schmidt M. V., Paez-Pereda M., Holsboer F., and Hausch F. (2012) The prospect of FKBP51 as a drug target. Chem. Med. Chem. 7, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 34. Burgos K., Malenica I., Metpally R., Courtright A., Rakela B., Beach T., Shill H., Adler C., Sabbagh M., Villa S., Tembe W., Craig D., and Van Keuren-Jensen K. (2014) Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. PLoS One 9, e94839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 36. Hassan T., Smith S. G., Gaughan K., Oglesby I. K., O'Neill S., McElvaney N. G., and Greene C. M. (2013) Isolation and identification of cell-specific microRNAs targeting a messenger RNA using a biotinylated anti-sense oligonucleotide capture affinity technique. Nucleic Acids Res. 41, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mah C., Sarkar R., Zolotukhin I., Schleissing M., Xiao X., Kazazian H. H., and Byrne B. J. (2003) Dual vectors expressing murine factor VIII result in sustained correction of hemophilia A mice. Hum. Gene Ther. 14, 143–152 [DOI] [PubMed] [Google Scholar]
- 38. Zolotukhin S., Potter M., Zolotukhin I., Sakai Y., Loiler S., Fraites T. J. Jr., Chiodo V. A., Phillipsberg T., Muzyczka N., Hauswirth W. W., Flotte T. R., Byrne B. J., and Snyder R. O. (2002) Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28, 158–167 [DOI] [PubMed] [Google Scholar]
- 39. Katnik C., Guerrero W. R., Pennypacker K. R., Herrera Y., and Cuevas J. (2006) Sigma-1 receptor activation prevents intracellular calcium dysregulation in cortical neurons during in vitro ischemia. J. Pharmacol. Exp. Ther. 319, 1355–1365 [DOI] [PubMed] [Google Scholar]
- 40. Abisambra J. F., Jinwal U. K., Blair L. J., O'Leary J. C. 3rd, Li Q., Brady S., Wang L., Guidi C. E., Zhang B., Nordhues B. A., Cockman M., Suntharalingham A., Li P., Jin Y., Atkins C. A., et al. (2013) Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation. J. Neurosci. 33, 9498–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Daily K., Patel V. R., Rigor P., Xie X., and Baldi P. (2011) MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics 12, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Decker K. F., Zheng D., He Y., Bowman T., Edwards J. R., and Jia L. (2012) Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res. 40, 10765–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]