Abstract
The primary role of the RNAi machinery is to promote mRNA degradation within the cytoplasm in a microRNA-dependent manner. However, both Dicer and the Argonaute protein family have expanded roles in gene regulation within the nucleus. To further our understanding of this role, we have identified chromatin binding sites for AGO2 throughout the 45S region of the human rRNA gene. The location of these sites was mirrored by the positions of AGO2 cross-linking sites identified via PAR-CLIP-seq. AGO2 binding to the rRNA within the nucleus was confirmed by RNA immunoprecipitation and quantitative-PCR. To explore a possible mechanism by which AGO2 could be recruited to the rRNA, we identified 1174 regions within the 45S rRNA transcript that have the ability to form a perfect duplex with position 2–6 (seed sequence) of each microRNA expressed in HEK293T cells. Of these potential AGO2 binding sites, 479 occurred within experimentally verified AGO2-rRNA cross-linking sites. The ability of AGO2 to cross-link to rRNA was almost completely lost in a DICER knock-out cell line. The transfection of miR-92a-2-3p into the noDICE cell line facilitated AGO2 cross-linking at a region of the rRNA that has a perfect seed match at positions 3–8, including a single G-U base pair. Knockdown of AGO2 within HEK293T cells causes a slight, but statistically significant increase in the overall rRNA synthesis rate but did not impact the ratio of processing intermediates or the recruitment of the Pol I transcription factor UBTF.
Keywords: Argonaute, ChIP-sequencing (ChIP-seq), Dicer, microRNA (miRNA), ribosomal ribonucleic acid (rRNA) (ribosomal RNA), miRNA, par-clip, rRNA
Introduction
The RNAi machinery has many functions in the eukaryotic cell, and aspects of the RNAi molecular mechanism are highly conserved between yeast and humans (1). Essentially, a small RNA is bound by a member of the Argonaute family of proteins and contributes sequence specificity to a larger protein complex. In the cytoplasm, the RNAi machinery uses Watson-Crick base pairing to target the RNA-induced silencing complex to a specific mRNA and facilitate its degradation. A related process is well established in the nucleus of Schizosaccharomyces pombe, where instead of targeting cytoplasmic mRNAs for destruction, a small RNA targets the RNA-induced transcriptional silencing complex to the pericentromeric regions of each chromosome and facilitates the generation of heterochromatin (2, 3).
Work in a chicken-human hybrid cell line supports the possibility that the RNAi machinery is responsible for centromeric chromatin structure in vertebrates as well (4). Indeed, when Dicer is conditionally inactivated, transcription of α-satellite DNA from human chromosome 21 increases. Furthermore, the loss of Dicer results in a loss of siRNAs originating from these repeat regions, a delocalization of HP1, and disruption of mitosis (4). The RNAi machinery is also implicated in the creation and/or maintenance of heterochromatin at various sites throughout the genome, in addition to the centromeric regions. Transfection of a siRNA homologous to the EF1a promoter in human cells silences the endogenous gene (5). In a related study, human Argonaute 1 (AGO1)2 is shown to direct siRNA-mediated gene silencing of both the ccr5 and rassf1a genes by localizing to the promoter regions and inducing heterochromatin formation (6). AGO1 has also been shown to localize to a subset of active promoters (7). However, this same study demonstrated that AGO2 had a distinct nuclear localization and appeared enriched at the nuclear periphery. Studies of a 16-kb region of heterochromatin between the β-globin and folate receptor genes in chicken cells identified an endogenous binding site for chicken Argonaute 2 (cAgo2) (8). The binding of cAgo2 to chromatin at that site required siRNAs homologous to the DNA sequence and expression of Dicer. Knockdown of Dicer or cAGO2 expression caused the chromatin to gain acetylation at histone H4 (H4ac) and histone H3 lysine 9 (H3K9ac). This correlated with an increase in transcription levels and a loss in physical condensation. Recent work has indicated that human AGO proteins can regulate splicing through modulating chromatin structure (9, 10) and can promote gene repression in cis by localizing to nascent tRNA (11). Thus, RNAi-mediated control of gene expression exists also in vertebrate cells but functions in mechanisms distinct from those within the cytoplasm (12, 13). Despite these reports demonstrating a role for RNAi in regulating the chromatin structure of vertebrates, similar studies have shown that the loss of the RNAi machinery impacts chromatin structure indirectly through miRNA biogenesis and post-transcriptional gene regulation (14). Thus, the precise roles for the RNAi machinery in regulating the chromatin structure of higher eukaryotes remain unclear.
The rRNA genes are transcribed by RNA Polymerase I to generate a 45S precursor RNA, which is processed into the 18S, 5.8S, and 28S rRNAs (15). These genes are highly repetitive; ∼400 copies exist in humans. In most human cell types, ∼50% of these gene copies are silenced and heterochromatic (16). A recent study has demonstrated that DICER localizes to the human rRNA genes, although it was found equally distributed between the active and silent loci (17). Here we provide evidence that AGO2 binds to the nascent rRNA. We propose that this interaction is mediated by an interaction between AGO2 and many distinct miRNA. We further demonstrate that a reduction in AGO2 levels causes the overall rRNA gene transcription rate to increase.
Results
AGO2 Binds to the Human rRNA Genes
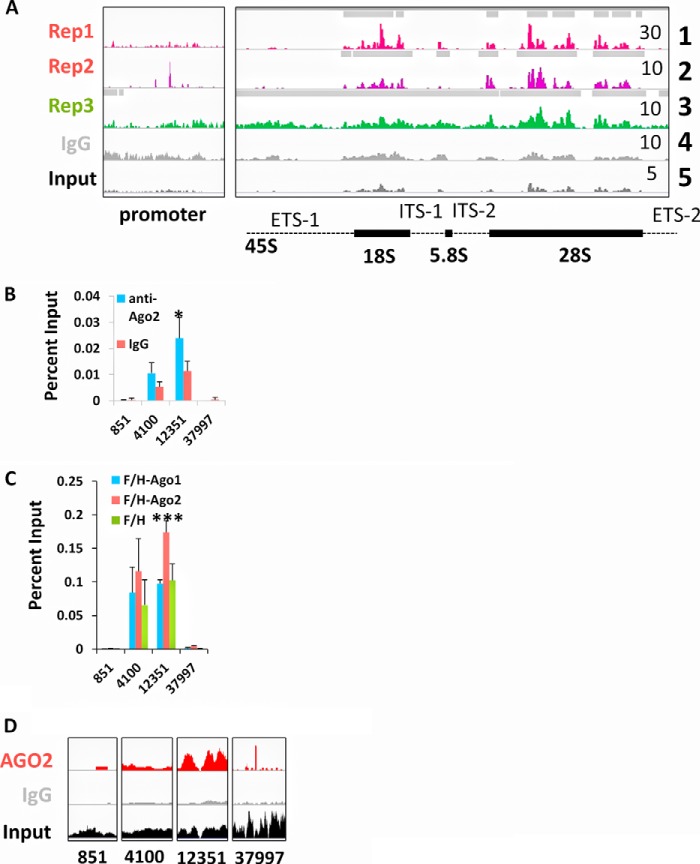
Our recent work indicated that AGO2 interacts with nascent tRNA, which tethers AGO2 in close proximity to most active tRNA genes (11); thus AGO2 can interact with nascent transcripts from both RNA Polymerase II and III (9–11, 18). To investigate if AGO2 had a similar interaction with the sole RNA Polymerase I transcript in human cells, we aligned our previously published ChIP-seq datasets (GEO: GSE68813) to a custom build of human genome hg18, similar to what has been previously described (19). AGO2 binding was detected within the 18S and 28S regions of the consensus rRNA gene from two trials using the anti-AGO2 mAb Millipore clone 9E8.2 (04-642, rep1,2) and one trial using Abcam (ab57113, rep3) (Fig. 1A, tracks 1–3). In both cases, AGO2 was enriched primarily within the 18S and 28S rRNA regions relative to the input control (Fig. 1A). We confirmed the visual identification of enriched regions using the MACS 1.4 peak-calling algorithm (20). We have depicted the regions of MACS-identified peaks as gray bars above each track. We further confirmed the enrichment of endogenous AGO2 relative to IgG by chromatin immunoprecipitation (ChIP)-quantitative PCR (QPCR) (Fig. 1B, n = 7; *, Student's t test p < 0.05). In addition, FLAG/HA-AGO2, but not FLAG/HA-AGO1, was enriched at the rRNA gene relative to empty vector via ChIP-QPCR (Fig. 1C, n = 3; ***, Student's t test, p < 0.001). To provide a comparison between the ChIP-seq and ChIP-QPCR results, we have presented close-up images of the ChIP-seq coverage within the region of each amplicon used in the QPCR (Fig. 1D). These views indicate that the signal of AGO2 ChIP follows the pattern: 12,351 > 4,100 > 3,7997 = 851. It is difficult to quantitatively compare ChIP-QPCR and ChIP-seq data due to differing normalization methods. However, we demonstrate that there is a qualitative agreement between these two complementary methods to measure AGO2 binding to the rRNA gene. These results alleviate concerns that the observed signal is due to incorrect tag alignment.
FIGURE 1.
Human Argonaute 2 localizes to the rRNA genes. A, coverage density (reads per million) of ChIP-seq tags aligned to a consensus sequence for the human rRNA gene. The red tracks represent ChIP-seq done with Millipore anti-AGO2 mAb 9E8.2. The green track shows an identical ChIP-seq performed with Abcam anti-AGO2 mAb ab57116. A negative control ChIP-seq done with IgG is shown in gray. The tag density of the input samples is shown in black. The maximum reads per million for each track is indicated by the number in the upper-right corner of each track. Each scale is linear. A schematic representation of the rRNA gene is shown below the ChIP-seq data: ETS = external transcribed spacer; ITS = internal transcribed spacer. The 45S corresponds to the 45S primary transcript from which the rRNAs are processed. Each of the 3 mature rRNA species is indicated by a blue box (18S, 5.8S, and 28S). All ChIP-seq data were reanalyzed from a previously published AGO2 study (11) and are available within the Gene Expression Omnibus: GSE68813. The horizontal gray bars above each track represent regions called as peaks using MACS version 1.4, false discovery rate < 10% (see “Experimental Procedures”) (20). The track number is listed on the far right for clarity. B, a ChIP-QPCR assay of AGO2 binding to the rRNA gene in HEK293T cells. The y axis indicates the percent input for each immunoprecipitation. A ChIP was done with IgG as a negative control. The alignment of each primer set to the rRNA gene can be matched up using the schematic diagram of the locus. The values shown represent an average and S.D. (n = 7, Student's t test; *, p < 0.05. C, ChIP-QPCR, as described in B but performed in K562 cells transfected with FLAG/HA-AGO2 overexpression construct. Cells transfected with an empty vector were used as the mock control. Each ChIP was done with an anti-FLAG antibody (Sigma F3165). The values represent the average and S.D. n = 3. Student's t test; ***, p < 0.001). D, screen shots showing a zoomed-in view of the ChIP-seq data within the regions of each primer pair that was analyzed in B and C. Each view represents the length of the amplicon (∼100 base pairs).
AGO2 Binds to Nascent rRNA
AGO2 has been widely reported to interact with specific regions of chromatin via an “RNA-tether.” To determine if the interaction between AGO2 and the rRNA gene was mediated by such a mechanism, we analyzed previously published PAR-CLIP-seq data wherein the interactions with the rRNA gene were not investigated (21). To alleviate the concern that any known interaction between AGO2 and the rRNA was due to nonspecific binding/contamination of the AGO2-IP with rRNA, we limited our analysis to cross-linking sites that could be verified computationally. This ability relies on the fact that the modified nucleoside used in PAR-CLIP-seq, 4-thiouridine, induces T to C mutations during the high throughput sequencing process. This phenomenon can be detected using the software program PARAlyzer (22). PARAlyzer detects clusters of T to C mutations that are likely to indicate authentic protein-RNA binding sites. Although the ChIP-seq data were determined within K562 cells, we have focused on publicly available RNA-seq data from HEK293T cells. Our previous work indicated that the interactions between AGO2 and tRNA were conserved between the two cell lines (11).
We used this analysis to demonstrate the existence of a cluster of high confidence AGO2:rRNA binding sites within the 18S and 28S rRNA regions (Fig. 2A, tracks 4–6). Three metrics of this analysis are presented to illustrate AGO2 binding: the total number of converted reads (track 4), the fraction of converted reads (track 5), and the overall PAR-CLIP-seq signal in reads per million (track 6). These same three metrics are shown for the negative control protein HuR, which has much less signal than AGO2 for each metric (Fig. 2A, tracks 7–9).
FIGURE 2.
Human AGO2 binds directly to rRNA within the nucleus. A, the PAR-CLIP-seq of both AGO2 (green) and HuR (gray) (see “Experimental Procedures” for datasets) is shown in comparison to the ChIP-seq alignments from Fig. 1A (58). The top three tracks show the ChIP-seq of AGO2 from Fig. 1 for comparison. For both AGO2 (green) and HuR (gray) the PAR-CLIP-seq data are presented using three methods: the summary of the total number of sequencing reads containing T to C mutations (total T → C), the fraction of the reads containing T to C mutations (fraction T → C), and the overall PAR-CLIP-seq coverage in reads per million. The y axis of each track is in the upper right corner but has different dimensions for each track. The ChIP-seq and PAR-CLIP-seq coverage is in reads per million, the total T → C represents total tags, and the fraction T → C is a fraction ranging from 0 to 1. A schematic of the rRNA gene is shown below the panel of sequencing data. The AGO2 and HuR PAR-CLIP-seq data were reanalyzed from a previous study (58) and are available within the Gene Expression Omnibus: GSE28859. The track numbers are listed to the far right for clarity. B, the same data as presented in A but with each the total T → C and the overall coverage values zoomed in 100-fold. The fraction T → C was zoomed in 5×. The track numbers are listed to the far right for clarity. C, HEK293T cells were lysed, cross-linked, and a RIP-QPCR was carried out as described under “Experimental Procedures.” The y axis indicates the enrichment of each region relative to input. The x axis indicates the location of the primer set within the rRNA gene locus (for the numbered primers, which can be visualized in the schematic of the rRNA gene) or the name of a Pol II gene that was analyzed as a control. Immunoprecipitations were carried out using the antibodies as described under “Experimental Procedures.” Actinomycin D treatment was carried out overnight with (200 ng/ml ActD) before RIP was carried out. The values represent the average and S.D. (n = 3, Student's t test was used to indicate a significant different between AGO2 and HuR; **, p < 0.005; *, p < 0.05). The red asterisks over YY1 indicate that HuR is enriched relative to AGO2. D, to illustrate the significant effect that actinomycin D has on the interactions between AGO2 and the rRNA, the enrichment of AGO2 after actinomycin D treatment was subtracted from the enrichment of AGO2 in untreated control cells. If actinomycin D treatment had no effect, then the values would be zero. A larger bar indicates a greater effect of actinomycin D on the AGO2 enrichment.
The majority of the AGO2 cross-linking sites were found within the 18S and 28S rRNA regions. However, by zooming in 100-fold, we are able to see many AGO2 PAR-CLIP-seq tags within ETS1, ITS-1, ITS-2, and ETS-2. We were unable to detect any such cross-linking sites for HuR (Fig. 2B). These rRNA regions are excised shortly after transcription of the 45S rRNA precursor and suggested to us that perhaps the observed cross-linking between AGO2 and the rRNA gene chromatin could be facilitated by a nuclear interaction between AGO2 and nascent rRNA. To this end we performed a RNA immunoprecipitation (RIP)-QPCR assay in nuclear extract (Fig. 2C). AGO2 was enriched at all regions throughout the rRNA gene, and this enrichment was reduced upon actinomycin D treatment (Fig. 2C, n = 3, Student's t test for AGO2 versus HuR; **, p < 0.005; *, p < 0.05, red asterisks indicate HuR enrichment relative to AGO2). The yy1 gene served as a positive control for HuR binding relative to AGO2, and neither AGO2 nor HuR was significantly enriched within the 3′-UTR of β-actin. Interestingly, AGO2 was also enriched for the 3′-UTR of rpl10a gene.
Actinomycin D treatment inhibits new rRNA synthesis but, due to the long half-life of mature rRNAs, should not affect their abundance in short time frames. Thus, RIP-QPCR in conjunction with actinomycin D treatment can be used to quantify any interactions with nascent or newly transcribed rRNA. To illustrate the specificity of the actinomycin D treatment, we subtracted the enrichment of each gene after treatment from that of control cells (Fig. 2D). Although the enrichment of AGO2 for the Pol II genes did decrease after actinomycin D, the decrease was much greater for each of the rRNA gene regions, indicating that AGO2 binding to the rRNA is sensitive to ongoing Pol I transcription.
AGO2-miRNA Interactions Facilitate AGO2-rRNA Interactions
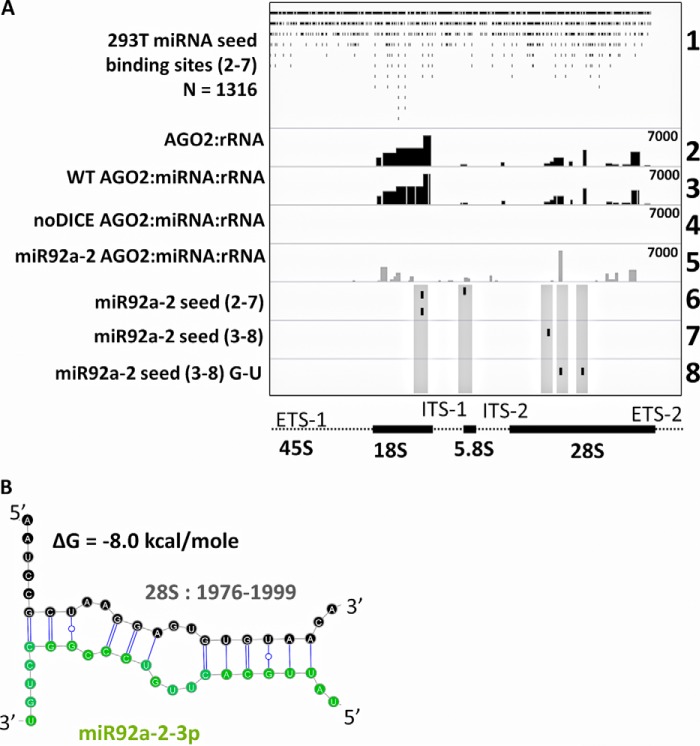
To determine if the binding of AGO2 to the rRNA was related to miRNA base pairing, we modified the high-throughput sequencing alignment program bowtie (23) to align the reverse complement of the seed sequences from miRNA to the rRNA gene (see “Experimental Procedures”). We limited the analysis to miRNAs that were bound by AGO2 in HEK293T cells (see “Experimental Procedures”). We were surprised to find that there were 1,174 regions within the 45S rRNA region that had a perfect match to positions 2–7 of a miRNA (Fig. 3A, track 1). This region, known as the seed sequence, is required to have perfect complementarity to facilitate miRNA interaction with a target sequence (24–28). Additional binding sites existed within the intergenic spacer and from miRNA that were not expressed in HEK293T (data not shown), which suggests that the cell-type specificity to miRNA expression may impart cell-type specific RNAi-mediated control mechanisms to the rRNA gene.
FIGURE 3.
Direct AGO2 binding to the rRNA can be facilitated by specific miRNA. A, top track, the extent of possible interactions between miRNA and the 45S rRNA was determined by aligning the reverse complement of the seed sequences (positions 2–7) of each human miRNA to the rRNA gene. A BED file depicts each potential miRNA binding site. Track 2, a representation of the PARAlyzer-detected cross-linking sites between AGO2 and the rRNA, as shown in Fig. 2. Track 3, an intersection between the top two tracks, which depicts each PARAlyzer-detected AGO2 cross-linking site within the rRNA that also contains perfect Watson-Crick base-pairing potential between the rRNA and at least one human miRNA. Track 4, the same analysis as in the track 3, but analyzed on PAR-CLIP-seq data taken from noDICE, DICER null HEK293T cells. Track 5, the same analysis as in the above two tracks but performed on noDICE cells that were transfected with a miR-92a-2 duplex. Tracks 6–8, potential binding sites for the seed sequence of miR-92a-2-3p when using positions 2–7, 3–8, or 3–8 allowing for G-U base pairs. Each seed sequence binding site is shown by a small black tick mark and is highlighted by a transparent gray box to illustrate the location of the seed sequence relative to the overall PAR-CLIP-seq signal at each site. The noDICE and miR-92a-2 datasets represent a reanalysis of a previously published manuscript (29). The datasets are available within GSE56836, as described under “Experimental Procedures.” The track numbers are listed to the far right for clarity. B, a diagram of the Mfold-predicted (30) base-pairing between miR-92a-2-3p and its binding site within the 28S rRNA region upon transfection of the exogenous duplex. The secondary structure schematic was generated using the Varna software program (59).
To determine if any of these potential miRNA-rRNA interaction sites were actually used to tether AGO2 binding to the rRNA, we detected which of the 1,174 potential seed sequences were found within a PAR-CLIP-seq-verified AGO2 cross-linking site within the rRNA. The cross-linking sites were identified using PARAlyzer (22). There were 479 potential miRNA binding sites that intersected with actual AGO2-rRNA cross-linking sites (Fig. 3A, track 2, AGO2-rRNA cross-linking sites as shown in Fig. 2; track 3, the intersection of those sites with potential miRNA binding sites). We performed the same analysis using PAR-CLIP-seq data taken from HEK293T cells that are homozygous null for DICER (29). The cross-linking of AGO2 to the rRNA was almost completely lost within these “noDICE” cells (Fig. 3A, track 4). Interestingly, as a part of the same study, PAR-CLIP-seq was performed on noDICE cells that were transfected with a miR-92a-2-3p duplex. A PARAlyzer analysis showed a strong enrichment of mutated reads at a single position within the rRNA gene and several weaker positions (Fig. 3A, track 5). To determine if this region corresponded to a potential miR-92a-2-3p binding site, bowtie was used to align the reverse complement of the possible miRNA seed sequences for miR92a-2-3p (Fig. 3A, tracks 6–8). The locations of the potential sites are shown for seed sequences ranging from 2 to 7 (track 6), 3 to 8 (track 7), and 3 to 8, allowing a G to U base pair (track 8). Although the induced AGO2 binding site did not correspond to any sites with a possible 2–7 or 3–8 seed sequence, it did correspond to a site containing a 3–8 seed sequence when a G to U base pair was allowed. Thus, the induced localization of miR-92a-2-3p was distinct from the most likely endogenous location. The miR-92a-2-3p-rRNA duplex (at location 28S: 1976–1999) was estimated to have a ΔG of −8.1 kcal/mol via the Mfold RNA folding algorithm (30) (Fig. 3B).
We next investigated if we could measure changes in the binding of AGO2 to small RNAs derived from the rRNA gene in the presence or absence of DICER. To this end, we analyzed publicly available small RNA-IP-seq data from the same study that generated the noDICE line. Visualization of this data demonstrated that the vast majority of rRNA-derived small RNAs that are loaded into the RNA-induced silencing complex are generated from a small number of sites (Fig. 4 track 1, black asterisks). The binding of AGO2 to small RNAs that are derived from the termini of the processed products of the rRNA has been previously reported (31). Comparison of both the magnitude and location of AGO2-bound rRNA-derived small RNAs did not change when control HEK293T cells were compared with the noDICE cell line (Fig. 4, track 2). A magnification of 100× did not reveal any differences between control and noDICE cell lines (Fig. 4, tracks 3 and 4). Thus, AGO2 is bound to a large number of rRNA-derived small RNAs that are derived in a DICER-independent manner.
FIGURE 4.

The loading of small RNAs derived from the rRNA gene is not affected by DICER knock-out. A BEDGRAPH file is shown summarizing the levels of rRNA-derived small RNA that are bound by AGO2 in both control and noDICE HEK293T cells. The data are shown twice, once above the rRNA gene schematic auto-scaled to 42,000 reads per million and the second time below the schematic zoomed-in 100-fold. Both datasets are available within the Gene Expression Omnibus GSE56836. The track numbers are listed to the far right for clarity.
AGO2 Regulates rRNA Synthesis Rate
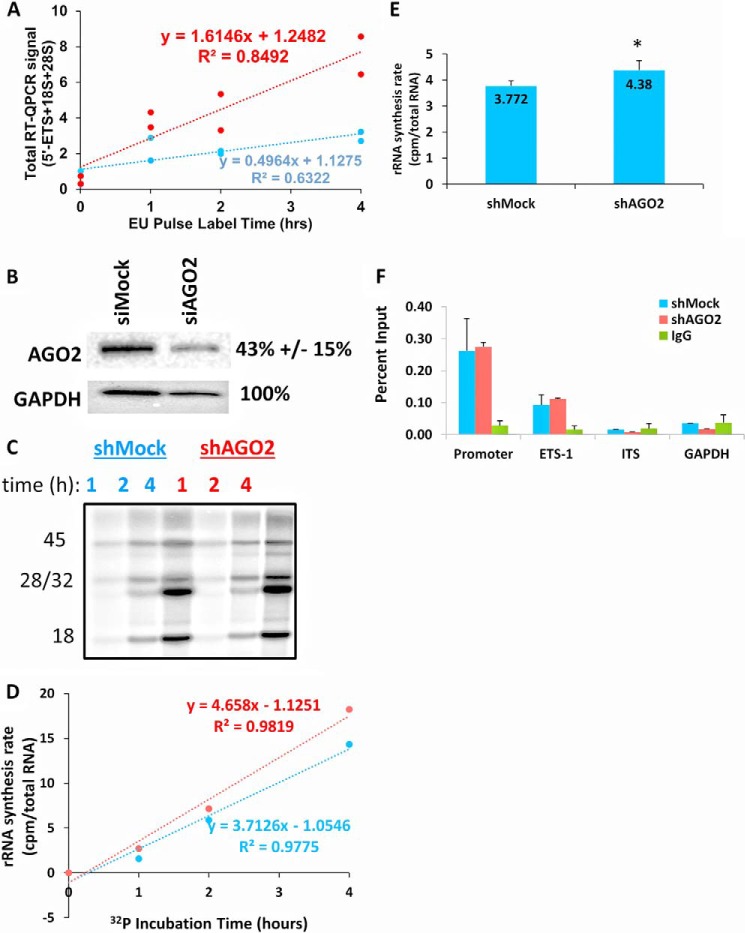
To investigate if AGO2 binding to rRNA had any measurable effect on the synthesis of rRNA, we utilized Click-iT® metabolic labeling to isolate all nascent RNA from either siMock- or siAGO2-treated K562 cells (see “Experimental Procedures”). The two treatments were incubated with ethylene uridine (EU) for 0, 2, or 4 h followed by isolation of nascent RNA, which was then analyzed by RT-QPCR using primers for the 5′-ETS, 18S, and 28S, each of which was normalized to GAPDH. The total signal from nascent rRNA was then plotted as a function of time for both siMock and siAGO2 cells (Fig. 5, A and B). There was a significant increase in the rate of incorporation of EU (slopes = 1.6 versus 0.5, n = 8, p = 0.00424 two-way analysis of covariance) in siAGO2-treated K562 cells.
FIGURE 5.
AGO2 knockdown increases the apparent synthesis rate of rRNA. A, K562 cells were metabolically labeled with EU in both siMock- and siAGO2-treated cells for the indicated length of time (see “Experimental Procedures”). RT-QPCR was used to quantify total rRNA, normalized to GAPDH, at each time point. The time points were fitted to a linear regression to determine the rate of incorporation of EU in each treatment (1.61 versus 0.496, p = 0.00424, two-way analysis of covariance). B, a Western blot indicating the ability to knockdown AGO2 expression using the Silencer® Select siRNA (“Experimental Procedure”). A representative blot is shown, and the percentage of AGO2 knockdown is indicated to the right of the blot. In each case, AGO2 levels were normalized to GAPDH (n = 4, Student's t test). C, shMock and shAGO2 HEK293T cells were metabolically labeled with 32P for the indicated lengths of time, and total RNA was harvested, gel-electrophoresed, and imaged in a Typhoon scanner (see “Experimental Procedures”). The gel was also stained with SYBR Gold to normalize for total RNA levels. The identity of each radioactive RNA band was indicated based on its size and is indicated to the left of the gel. D, the total counts of the 45S, 32S, 28S, and 18S radioactive bands were summed, normalized to total RNA (image J software, NIH), and plotted as function of incubation time. The best fit line was determined by Microsoft Excel 2013, and the slope, y-intercept, and χ2 of the line are shown (n = three biological replicates, and three time points per replicate; *, p < 0.05, Student's t-test). E, a summary of the difference in total 32P incorporation between shMock- and shAGO2-treated HEK293T cells after 4 h of labeling (n = 5). F, A UBTF ChIP-QPCR (n = 3) was performed in both shMock and shAGO2 cells. The amplicons indicate the region of the rRNA gene that was amplified: promoter = −57, ETS1 = 851, IGS = 37997. The primer sequences were all previously reported (11).
The incorporation of EU is a sensitive method for detecting changes to nascent RNA (32). A drawback of using modified nucleosides is that they may inhibit polymerase activity (33), and a multistep process is required to isolate and analyze nascent RNA (see “Experimental Procedures”). To generate independent confirmation of the effect of AGO2 on the overall rRNA synthesis rate, we repeated the metabolic labeling with 32P in our previously established (11) shMock and shAGO2 knockdown system in HEK293T cells (Fig. 5C). Total RNA was electrophoresed, each rRNA species was quantified using the Typhoon phosphorimaging, and the sum intensity of each of the following rRNA species was plotted as a function of time: 45S, 32S, 28S, and 18S (Fig. 5D, “Experimental Procedures”). A representative experiment is shown for clarity (Fig. 5, C and D). We were able to reproducibly determine that the rRNA synthesis rate was increased 15% in shAGO2 relative to shMock cells after 4 h of labeling (Fig. 5E, n = 5, Student's t test; *, p < 0.05). We did not detect any change in the ratios of the 45S precursor to the mature species (28S and 18S), demonstrating that AGO2 did not affect processing of the rRNA (Fig. 5C). This increase in rRNA synthesis was not concomitant with an increase in the binding of UBTF to the rRNA gene (Fig. 5F), which suggests that the change in the “observable” rRNA synthesis rate was not due an increase in transcription initiation.
Discussion
Here we report the observation that AGO2 can be cross-linked to the chromatin of the rRNA gene (Fig. 1). These chromatin binding sites are largely coincident with sites of direct contact between AGO2 and the rRNA (Fig. 2). The interaction of AGO2 with the rRNA is drastically reduced when cells are treated briefly with actinomycin D, which impairs Pol I transcription (34, 35). This drug treatment would not affect the overall ribosomal levels, as their half-lives have been estimated to be up to 5 days (36). Thus, we conclude that interactions between AGO2 and the rRNA occurs at least in part within the nucleus.
One caveat to this study is the comparisons between K562 and HEK293T cell lines. Our previous work has indicated that the roles of AGO2 at tRNA genes appear to be conserved within the two cell lines, and there is no evidence to the contrary with regard to the rRNA gene. However, our model, as with most models describing the interaction between AGO2 and nascent transcripts, indicates a direct interaction with the RNA that allows for chromatin cross-linking. Although there are likely specific interactions between AGO2 and chromatin-binding proteins that occur within the rRNA gene, these are not likely to be possible in the absence of a strong interaction between AGO2 and the rRNA. In addition, the AGO2-RNA interaction generates a stronger signal-to-noise ratio than the AGO2-chromatin interaction. For these reasons we chose to focus on the interactions between AGO2 and both miRNA and rRNA as opposed to furthering an analysis of the AGO2-chromatin interaction. We were able to demonstrate that AGO2 knockdown causes an increase in rRNA synthesis rate in both cell lines through the use of two distinct knockdown methods. However, it must be mentioned that AGO2 knockdown is likely to impact many components of cellular physiology, and it is difficult to know exactly how these changes impact rRNA synthesis. Interestingly, AGO2 knockdown had a much stronger effect on rRNA synthesis in K562 cells than in HEK293T cells despite similar levels of knockdown. This result suggests that the nucleolus of each cell line may have a unique level of susceptibility to miRNA-mediated regulation. However, this could also be due to the wide range of possible indirect effects caused by bulk AGO2 knockdown. Future work will be directed at isolating the nucleolar-specific function of AGO2.
AGO2 has been shown to interact with long RNA (>24 nucleotides) directly (11, 37). However, the primary mechanism by which AGO2 is recruited to specific loci is through base-pairing between a miRNA and target. To investigate if this conserved pathway was responsible for our observed AGO2 interactions, we aligned the seed sequence from each human miRNA and demonstrated >1000 regions within the 45S rRNA that were potential base-pairing sites (Fig. 3). When these data were compared with experimentally determined cross-linking regions between AGO2 and the rRNA, >400 potential miRNA binding sites were within 30 nucleotides of an actual AGO2-rRNA cross-link (Fig. 3). The cross-linking of AGO2 to the rRNA was reduced to background levels in the absence of DICER, and the transfection of a single miRNA, miR-92a-2-3p, created a strong AGO2 cross-linking site within the 28S region of the rRNA that had base-pairing potential with the miR-92a-2-3p. Thus, we have demonstrated that the binding of AGO2 to the rRNA is dependent upon the presence of DICER and at least one miRNA.
This work represents the first observation of specific interactions between AGO2, miRNA, and rRNA. There have been numerous reports suggesting a connection between the RNAi pathway and nucleolar regulation. DICER has been shown to reside within the nucleolus and bind to both active and inactive copies of the rRNA gene (17). The microprocessor complex is required for the normal processing of the 45S rRNA precursor (38). Many miRNAs have been isolated within the nucleolus (39–45). The function of AGO2 and the RNAi machinery within the nucleolus remains unclear. The complexity of interactions that impinge on Pol I transcription combined with the large number of probable AGO2-miRNA binding sites within the rRNA will make determining a functional role for this process difficult. Interestingly, the lack of any change in AGO2-bound small RNAs derived from the rRNA region in the absence of DICER indicates that either (a) AGO2 does not cleave the rRNA into small RNA fragments that can be loaded into AGO2, or (b) the abundance of these putative small RNAs is too low to affect the overall distribution of AGO2-bound rRNA-derived small RNA. An analysis of the rRNA synthesis rate within the noDICE cells may seem prudent. However, the massive physiological consequences of prolonged DICER knock-out would render any result difficult to interpret. Future work will be directed at developing an assay to detect the entire set of miRNAs that are actually bound to the rRNA gene at any given time. Once detected, the binding sites for these miRNAs can be altered using genome editing. This approach will allow measurement of the specific cause and effect of each AGO2-miRNA-rRNA interaction as well as the sum total of all such interactions. Although this may be a technically challenging research problem, the investment may be well worth it as many miRNA have been implicated as markers for cancer. It is always assumed that the mechanism of action relevant to cancer is related to the well-known role of miRNA in cytoplasmic RNAi. However, cancer cells are often associated with aberrant nucleoli (46), and a Pol I inhibitor is currently in stage I trials as an anti-cancer compound (47, 48). Thus, the connection between miRNA and cancer may be related to this poorly understood role in the nucleolus.
Experimental Procedures
Cell Culture and Transfections
All experiments were carried out in K562 and HEK293T cells. Culturing conditions were carried out essentially as previously described (8) FLAG/HA-AGO2 constructs were acquired through Addgene (49). shRNA constructs were acquired from GE Dharmacon (GIPZ Lentiviral #RHS4531-EG271611). Transfections of both shRNAs and FLAG/HA constructs were performed using Calfectin reagent (Signagen), according the manufacturer's instructions. Briefly, HEK293T cells were grown to ∼25% confluency. Cells were given fresh growth media (DMEM with 50% FBS and 5 ml pen/strep). The vectors (1 μg) were added to DMEM (100 μl/ml of media) without antibiotics or serum. Next, 2.5 μl of Calfectin was added per 1 ml of media and incubated for 10 min at room temperature. The pGIPZ/Calfectin mixture was added to the cell culture, swirled, and incubated 4 h, after which the growth medium was replaced, and the cells were cultured for the indicated times (at least 48 h). Transfections of siMock (Thermo Fisher Scientific catalog #AM4635) and siAGO2 siRNA (Thermo Fisher Scientific Silencer® Select, catalog #4392420, siRNA ID = s25931) were carried out as described (8).
ChIP/RIP-QPCR
Cells were lysed in LB50 buffer (10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1% Triton X-100, 1 mm EDTA) supplemented with 1× Halt protease inhibitor mixture (Thermo Scientific) and RNaseOUT (Invitrogen). The nuclei were then pelleted at 5000 × g and then resuspended in 5 ml of 1× PBS. Formaldehyde was added to a final concentration of 1.2%, and cells were rotated at room temperature for 15 min. Cross-linking was quenched by adding glycine to a final concentration of 0.125 m. Nuclei were pelleted, washed once with 1× PBS, and then resuspended in radioimmune precipitation assay buffer. Nuclei were then sonicated in a Bioruptor 300 (Diagenode) for 60 cycles of 30 s on and 30 s off. Sonicated product was spun at 17,000 × g for 15 min to remove the insoluble fraction, and the supernatant was saved. The immunoprecipitation was performed as previously described (60) with the following exceptions. After the immunoprecipitation and final wash, beads were resuspended in Tris-EDTA elution buffer (1 × Tris-EDTA, 1% SDS, RNaseOUT (Invitrogen), 4 μg Proteinase K) incubated at 45 °C for 1 h. For ChIP, supernatant was isolated via phenol-chloroform extraction, ethanol precipitation, and resuspension in 200 μl of water. For RNA, supernatant was harvested and resuspended in TRIzol. QPCR or RT-QPCR was carried out as previously described (8).
Pulse-Chase Labeling
Click-iT®
Labeling was performed as described by manufacturer (Thermo Fisher Scientific). Briefly, 3 ml of ∼1 million K562 cells per ml were grown in the presence of 200 μm Ethylene uridine (EU) for the indicated length of time. RNA was harvested, biotinylated, and biotinylated RNA was affinity purified according to manufacturer's instructions. RT-QPCR on affinity purified RNA was performed using primers: 851, 4100, 12351, and GAPDH as previously described (11). The fold-increase in signal over time for each rRNA primer pair (851, 4100, and 12351) was normalized by the fold-increase to GAPDH. The sum of the three rRNA primers was then plotted for each time point. The best fit line and ordinary least squares analysis was calculated in Microsoft Excel. The Two-way Analysis of covariance testing was performed using R studio and the built-in lm() function.
32Phosphorous
Cells were grown to confluency, washed with 1X PBS at room temperature, and then incubated for 1 h in phosphate free media. 32Pi was added to a final concentration of 20 μCi/ml. Cells were harvested 60, 120, and 240 min after the addition of 32Pi in 500 μl of TRIzol reagent, and RNA was harvested according to the manufacturer's instructions (Life Technologies). RNA was then electrophoresed on a 1% Formaldehyde/MOPS agarose gel, stained with SYBR Gold (Invitrogen), and visualized using Bio-Rad Gel Doc system. The gel was then transferred to a Nylon membrane overnight, and then exposed to Phosphor screen (GE) and quantified using a Typhoon Trio + imager (GE).
Data Analysis
Bioinformatics
High throughput sequencing FASTQ data were aligned to a custom human genome (hg18 + single rRNA gene copy as an extra chromosome) using bowtie2 with default parameters (50). These parameters map each individual read once to the best possible mapping location. In the instance of a tie between two or more mapping locations, the read will be randomly assigned to one of the tied locations. The output SAM file was converted into the BAM format using the SAMtools view option (51). For all ChIP-seq data, PCR duplicates were removed, meaning that all identical reads were collapsed to one single read. This is a common and established processing step for ChIP-seq data (52–54). BAM data were converted to BED format using the BEDtools bamToBed option (55).
Visualization
For meta-gene analyses, the coverage of each nucleotide across each feature (small nucleolar RNAs or miRNAs) was determined using coverageBed against a BED file of the hg18 small nucleolar RNA database taken from UCSC table browser, with the −d option of the BEDTools Suite (55). For all figures, the coverage was aligned at either the 5′ or 3′ position of the feature, as indicated within each figure legend, and scaled according to reads per million mapped reads. Average coverage across all features was generated by the BEDtools groupBy tool. The groupBy output was visualized using a line graph in Microsoft Excel. Binding profile heatmaps were generated essentially as previously described, except without binning the genes (56). Briefly, the coverage of each nucleotide across the features was determined as described above, and outputs were converted to data matrices using a custom script and then visualized using conditional formatting options in Microsoft Excel 2013. Each heatmap is scaled as described within each figure legend.
Data Acquisition
All acquired data were taken from the Gene Expression Omnibus (57). AGO2 ChIP-seq was taken from GSE68813 (11). WT AGO2 PAR-CLIP-seq was a concatenation of datasets SRR189784 and SRR189785, and HuR PAR-CLIP-seq was a concatenation of datasets SRR189777 and SRR189778, available within the Gene Expression Omnibus: GSE28859 (58). The noDICE and miR-92a-2p PAR-CLIP-seq experiments, which were analyzed using PARAlyzer (22), were taken from datasets SRR1241607 and SRR1241613, respectively. The sRIP-seq data were taken from SRR1241606 (WT) and SRR1241607 (noDICE). These datasets were a reanalysis of data from Bogerd et al. (29) and are available within the Gene Expression Omnibus at GSE56836.
Author Contributions
B. L. A., J. L. W., M. S. J. R., G. M. L., and K. E. G. performed the experiments and assisted with the manuscript preparation. K. E. G. and G. F. designed the experiments and prepared the manuscript.
Acknowledgments
We thank Harold Smith (NIDDK high throughput sequencing core) and Michael Crowley (Heflin Genomics Center at UAB) for operation of the Illumina Hi-Seq and Kevin Brick (NIDDK) for assistance in analyzing bioinformatic data. We also thank Bryan Cullen for kindly providing the noDICE cell line.
This work was supported in part by the National Institutes of Health, NIDDK Intramural Research Program. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- AGO1
- Argonaute 1
- cAgo2
- chicken Argonaute 2
- miRNA
- microRNA
- EU
- ethylene uridine
- QPCR
- quantitative-PCR
- RIP
- RNA immunoprecipitation.
References
- 1. Grewal S. I. (2010) RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 20, 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., and Martienssen R. A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 [DOI] [PubMed] [Google Scholar]
- 3. Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., and Grewal S. I. (2002) Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 [DOI] [PubMed] [Google Scholar]
- 4. Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., and Oshimura M. (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6, 784–791 [DOI] [PubMed] [Google Scholar]
- 5. Morris K. V., Chan S. W., Jacobsen S. E., and Looney D. J. (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305, 1289–1292 [DOI] [PubMed] [Google Scholar]
- 6. Kim D. H., Villeneuve L. M., Morris K. V., and Rossi J. J. (2006) Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 13, 793–797 [DOI] [PubMed] [Google Scholar]
- 7. Huang V., Zheng J., Qi Z., Wang J., Place R. F., Yu J., Li H., and Li L. C. (2013) Ago1 interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 9, e1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giles K. E., Ghirlando R., and Felsenfeld G. (2010) Maintenance of a constitutive heterochromatin domain in vertebrates by a Dicer-dependent mechanism. Nat. Cell Biol. 12, 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alló M., Buggiano V., Fededa J. P., Petrillo E., Schor I., de la Mata M., Agirre E., Plass M., Eyras E., Elela S. A., Klinck R., Chabot B., and Kornblihtt A. R. (2009) Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724 [DOI] [PubMed] [Google Scholar]
- 10. Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J. C., Mathieu J., Hamiche A., Ait-Si-Ali S., Muchardt C., Batsché E., and Harel-Bellan A. (2012) Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 19, 998–1004 [DOI] [PubMed] [Google Scholar]
- 11. Woolnough J. L., Atwood B. L., and Giles K. E. (2015) Argonaute 2 binds directly to tRNA genes and promotes gene repression in cis. Mol. Cell. Biol. 35, 2278–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gagnon K. T., Li L., Chu Y., Janowski B. A., and Corey D. R. (2014) RNAi factors are present and active in human cell nuclei. Cell Rep. 6, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zamudio J. R., Kelly T. J., and Sharp P. A. (2014) Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell 156, 920–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanellopoulou C., Muljo S. A., Dimitrov S. D., Chen X., Colin C., Plath K., and Livingston D. M. (2009) X chromosome inactivation in the absence of Dicer: Proc. Natl. Acad. Sci. U.S.A. 106, 1122–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McStay B., and Grummt I. (2008) The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131–157 [DOI] [PubMed] [Google Scholar]
- 16. Schlesinger S., Selig S., Bergman Y., and Cedar H. (2009) Allelic inactivation of rDNA loci. Genes Dev. 23, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sinkkonen L., Hugenschmidt T., Filipowicz W., and Svoboda P. (2010) Dicer is associated with ribosomal DNA chromatin in mammalian cells. PloS One 5, e12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alló M., Agirre E., Bessonov S., Bertucci P., Gómez Acuña L., Buggiano V., Bellora N., Singh B., Petrillo E., Blaustein M., Miñana B., Dujardin G., Pozzi B., Pelisch F., et al. (2014) Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc. Natl. Acad. Sci. U.S.A. 111, 15622–15629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zentner G. E., Saiakhova A., Manaenkov P., Adams M. D., and Scacheri P. C. (2011) Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 39, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A. C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., and Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Corcoran D. L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R. L., Keene J. D., and Ohler U. (2011) PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yilmazel B., Hu Y., Sigoillot F., Smith J. A., Shamu C. E., Perrimon N., and Mohr S. E. (2014) Online GESS: prediction of miRNA-like off-target effects in large-scale RNAi screen data by seed region analysis. BMC Bioinformatics 15, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sudbery I., Enright A. J., Fraser A. G., and Dunham I. (2010) Systematic analysis of off-target effects in an RNAi screen reveals microRNAs affecting sensitivity to TRAIL-induced apoptosis. BMC Genomics 11, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okamura K., Liu N., and Lai E. C. (2009) Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell 36, 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klinghoffer R. A., Magnus J., Schelter J., Mehaffey M., Coleman C., and Cleary M. A. (2010) Reduced seed region-based off-target activity with lentivirus-mediated RNAi. RNA 16, 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joseph T. T., and Osman R. (2012) Thermodynamic basis of selectivity in guide-target-mismatched RNA interference. Proteins 80, 1283–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bogerd H. P., Whisnant A. W., Kennedy E. M., Flores O., and Cullen B. R. (2014) Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20, 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z., Ender C., Meister G., Moore P. S., Chang Y., and John B. (2012) Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40, 6787–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ierusalimsky V. N., and Balaban P. M. (2016) RNA synthesis and turnover in the molluscan nervous system studied by Click-iT method. Brain Res. 1633, 139–148 [DOI] [PubMed] [Google Scholar]
- 33. Burger K., Mühl B., Kellner M., Rohrmoser M., Gruber-Eber A., Windhager L., Friedel C. C., Dölken L., and Eick D. (2013) 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 10, 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kramer B. (1980) The effect of actinomycin D on the nucleolus and on pigment synthesis in pigment cells of Xenopus laevis: an ultrastructural study. J. Anat. 130, 809–820 [PMC free article] [PubMed] [Google Scholar]
- 35. Zatsepina O. V., Voit R., Grummt I., Spring H., Semenov M. V., and Trendelenburg M. F. (1993) The RNA Polymerase I-specific transcription initiation factor UBF is associated with transcriptionally active and inactive ribosomal genes. Chromosoma 102, 599–611 [DOI] [PubMed] [Google Scholar]
- 36. McElhone M. J., O'Connor P. J., and Craig A. W. (1971) The stability of rat liver ribonucleic acid in vivo after methylation with methyl methanesulphonate or dimethylnitrosamine. Biochem. J. 125, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tan G. S., Garchow B. G., Liu X., Yeung J., Morris J. P. 4th, Cuellar T. L., McManus M. T., and Kiriakidou M. (2009) Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 37, 7533–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liang X. H., and Crooke S. T. (2011) Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 39, 4875–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts T. C. (2014) The MicroRNA biology of the mammalian nucleus. Mol. Ther. Nucleic Acids 3, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Politz J. C., Zhang F., and Pederson T. (2006) MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. U.S.A. 103, 18957–18962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Politz J. C., Hogan E. M., and Pederson T. (2009) MicroRNAs with a nucleolar location. RNA 15, 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pickering B. F., Yu D., and Van Dyke M. W. (2011) Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J. Biol. Chem. 286, 44095–44103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z. F., Liang Y. M., Lau P. N., Shen W., Wang D. K., Cheung W. T., Xue C. J., Poon L. M., and Lam Y. W. (2013) Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials. PloS One 8, e70869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bai B., Yegnasubramanian S., Wheelan S. J., and Laiho M. (2014) RNA-Seq of the nucleolus reveals abundant SNORD44-derived small RNAs. PloS One 9, e107519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bai B., Liu H., and Laiho M. (2014) Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Bio. 4, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hein N., Hannan K. M., George A. J., Sanij E., and Hannan R. D. (2013) The nucleolus: an emerging target for cancer therapy. Trends Mol. Med. 19, 643–654 [DOI] [PubMed] [Google Scholar]
- 47. Haddach M., Schwaebe M. K., Michaux J., Nagasawa J., O'Brien S. E., Whitten J. P., Pierre F., Kerdoncuff P., Darjania L., Stansfield R., Drygin D., Anderes K., Proffitt C., Bliesath J., Siddiqui-Jain A., Omori M., Huser N., Rice W. G., and Ryckman D. M. (2012) Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 3, 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. (2014) CX-5461 inhibits RNA Pol I in blood cancers. Cancer Discov. 4, OF5. [DOI] [PubMed] [Google Scholar]
- 49. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., and Tuschl T. (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 [DOI] [PubMed] [Google Scholar]
- 50. Langmead B., and Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bailey T., Krajewski P., Ladunga I., Lefebvre C., Li Q., Liu T., Madrigal P., Taslim C., and Zhang J. (2013) Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput. Biol. 9, e1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Landt S. G., Marinov G. K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B. E., Bickel P., Brown J. B., Cayting P., Chen Y., DeSalvo G., Epstein C., Fisher-Aylor K. I., Euskirchen G., et al. (2012) ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quinlan A. R., and Hall I. M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang W., Lee Y. H., Jones A. E., Woolnough J. L., Zhou D., Dai Q., Wu Q., Giles K. E., Townes T. M., and Wang H. (2014) The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat. Commun. 5, 3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., and Zavolan M. (2011) A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 8, 559–564 [DOI] [PubMed] [Google Scholar]
- 59. Darty K., Denise A., and Ponty Y. (2009) VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnson D. S., Mortazavi A., Myers R. M., and Wold B. (2007) Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 [DOI] [PubMed] [Google Scholar]