Abstract
The autoimmune regulator (AIRE) is a transcription factor which is expressed in medullary thymic epithelial cells. It directs the expression of otherwise tissue-specific antigens, which leads to the elimination of autoreactive T cells during development. AIRE is modified post-translationally by phosphorylation and ubiquitylation. In this report we connected these modifications. AIRE, which is phosphorylated on two specific residues near its N terminus, then binds to the F-box protein 3 (FBXO3) E3 ubiquitin ligase. In turn, this SCFFBXO3 (SKP1-CUL1-F box) complex ubiquitylates AIRE, increases its binding to the positive transcription elongation factor b (P-TEFb), and potentiates its transcriptional activity. Because P-TEFb is required for the transition from initiation to elongation of transcription, this interaction ensures proper expression of AIRE-responsive tissue-specific antigens in the thymus.
Keywords: autoimmunity, gene regulation, protein phosphorylation, transcription factor, ubiquitylation (ubiquitination), AIRE, FBXO3, P-TEFb
Introduction
The autoimmune regulator (AIRE)2 is a transcription factor (TF) that regulates central tolerance in the thymus (1–4). In its absence, affected individuals suffer from a congenital autoimmunity, the autoimmune poly-endocrinopathy-candidiasis-ectodermal dystrophy (APECED), also known as the autoimmune polyglandular syndrome type 1 (APS1) (5, 6). Expressed in medullary thymic epithelial cells (mTECs), AIRE directs the expression of otherwise tissue-specific antigens (TSAs). Their presentation to developing T cells allows self-reacting cells to be eliminated. In these cells, AIRE is recruited to the stalled RNA polymerase II (RNAPII) on TSA promoters by the unmodified histone H3K4 and DNA-dependent protein kinase (DNA-PK) (7–10). DNA-PK also phosphorylates AIRE for increased transcriptional activity (11). AIRE binds to the positive transcription elongation factor b (P-TEFb), which modifies RNAPII for elongation and co-transcriptional processing of TSA genes (7, 8, 12). Ectopic expression of AIRE also activates TSA genes in HEK 293T (293T) (13) and mouse 1C6 mTEC cells (8).
Transcriptional activation is a complex process between TFs and RNAPII. Recent evidence suggests that on most promoters, be they transcribed or not, RNAPII is already present; however, it elongates only on active genes (14). Other transcription units can be activated after the recruitment of P-TEFb, which phosphorylates negative elongation factors and the C-terminal domain of RNAPII (15). Many TFs, which contain acidic or amphipathic transcription activation domains (TADs), are also activated by ubiquitylation, especially via phosphorylated degrons that increase their potency and limit their duration of action (16, 17). Previously, we demonstrated that the ubiquitylation of VP16 increased its binding to P-TEFb. Indeed, the TAD and monoubiquitin on VP16 bound to cyclin boxes and the extreme C terminus of cyclin T1 (CycT1), respectively (18). These interactions were not constrained spatially, and besides increased binding they also relieved the autoinhibition of P-TEFb by the C terminus of CycT1 (18).
Of interest, AIRE contains a similar TAD near its C terminus (9). AIRE is also ubiquitylated (19–21). Indeed, an early report suggested that the plant homeodomain 1 (PHD1) in AIRE is a RING domain and that AIRE is itself an E3 ubiquitin ligase (21). Subsequent reports disputed this finding, and the structure of PHD1 proved to be distinct from a RING finger (22). Additionally, the PHD1 lacked E3 ligase activity (19). Nevertheless, the N terminus of AIRE is ubiquitylated (20). In addition, AIRE is phosphorylated on two residues, threonine at position 68 (Thr-68) and serine at position 156 (Ser-156), which when mutated to alanine abrogated AIRE transcriptional activity (11). In this report we discovered the E3 ligase responsible for the ubiquitylation of AIRE. Revealed by our proteomic studies, FBXO3 was found to interact with these phosphorylated Thr-68 and Ser-156 residues, promote the ubiquitylation of AIRE, and lead to its rapid degradation. In addition, this post-translational modification increased interactions between AIRE and P-TEFb and potentiated their transcriptional activity on TSA genes.
Experimental Procedures
Cell Culture and Transfection
HEK 293T (293T) cells and 1C6 mTECs were cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37 °C and 5% CO2. Transfection of plasmid DNA was performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. Small interfering RNA (siRNA) was transfected for 72 h using Lipofectamine RNAiMAX transfection reagent (Life Technologies) according to the manufacturer's instructions. Validated siRNA targeting human FBXO3 was purchased from Qiagen (GeneSolution GS26273) (23), siRNA targeting mouse FBXO3 was purchased from Invitrogen (FBXO3MSS226550), and control siRNA was purchased from Santa Cruz Biotechnology (sc-37007). Mouse thymus samples were collected from C57BL/6J mice from The Jackson Laboratory. Mice were maintained in specific pathogen-free facilities and were handled in accordance with protocols by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
Plasmids and Antibodies
FLAG-AIRE and INSLuc plasmids were described previously (9). Plasmids pSI-AIRE-WT, pSI-AIRE-T68A and pSI-S156A were kind gifts of Pärt Peterson (University of Tartu, Tartu, Estonia). mHis:AIRET68A, mHis:AIRES156Am and mHis:AIRET68AS156A were cloned from pSI-AIRE-T68A and pSI-AIRE-S156A into pcDNA3.1B (−) after the mHis epitope tag. The WT mHis:AIRE and mutant mHis:AIRET68E S156E plasmids were generated by PCR and cloned into pcDNA3.1B (−) as above. v:FBXO3 was a kind gift from Rama Mallampalli (University of Pittsburgh, Pittsburgh). f:FBXO3, h:FBXO3, m:CUL1, and His:Ub were all created by cloning them into pcDNA3.1 vector with the CMV promoter and the appropriate epitope tags. Deletion mutants of AIRE were created by inserting stop codons into f:AIRE using Stratagene XL site-directed mutagenesis kit. h:Ub was created by cloning ubiquitin into pMT2 vector with CMV promoter and 3′ HA tag.
Antibodies used for Western blotting, co-immunoprecipitation, or immunofluorescence were: AIRE (H300, sc-33188, and D17, sc-17986, Santa Cruz), AIRE (phospho Ser-156) antibody (ab78211, Abcam), Cullin 1 (d-5, sc-17775, Santa Cruz), c-Myc (9E10, HRP, ab62928, Abcam), CycT1 (H-245, sc-10750, Santa Cruz), FBXO3 (H300, sc-134722, Santa Cruz), FLAG M2-peroxidase (A8592, HRP, Sigma), FLAG M2-agarose from mouse (A2220, Sigma), GAPDH (6C5, sc-32233, Santa Cruz), His (H3, sc-8036, Santa Cruz), HA (Y-11, sc-805, Santa Cruz), RBX1 (34–2500, Invitrogen), UBC4 (Y20, sc-100617, Santa Cruz), ubiquitin (P4D1, sc-8017, Santa Cruz), and V5-HRP antibodies (R961–25, Life Technologies) as well as rabbit and mouse control IgG (Santa Cruz).
Mass Spectrometry
Immunopurification and mass spectrometry (MS) were carried out as previously described (24). 293T cells transfected with f:AIRE were lysed, and the supernatants were incubated with FLAG M2 affinity gel (Sigma). After extensive washes, bound proteins were eluted and digested using trypsin (Promega). The peptide mixture was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Thermo Scientific Velos Pro ion trap mass spectrometry system equipped with a Proxeon Easy nLC high pressure liquid chromatography and autosampler system. The MS data were searched using a database containing Swiss-Prot human protein sequence for protein identification.
Immunofluorescence Assays
For immunofluorescence assays, 293T cells were cultured on the Collagen I 22-mm round coverslip (354089; Corning) in 6-well plates. Cells were transfected with mHis:AIRE and h:FBXO3. Two days after transfection, cells were washed by PBS and fixed in 4% formaldehyde for 30 min at room temperature. They were then incubated with 0.2% Triton X-100 in PBS for 5 min. Reactions were then blocked for 1 h in PBS containing 5% BSA. Antibodies were diluted in 1% BSA in PBS. The primary anti-His and anti-HA antibodies were added and incubated at 4 °C overnight. Goat anti-mouse secondary antibodies, which were conjugated with green Alexa Fluor 488 dye (56881A; Invitrogen), and donkey anti-rabbit secondary antibodies, which were conjugated with red Alexa Fluor 594 (404239; Invitrogen) were then performed for 2 h at room temperature. DAPI (P36962; Life Technologies) was used for nuclear staining. Images were captured on an Olympus microscope and analyzed using Image-Pro Plus (version 6.2) software.
Immunoprecipitations
Immunoprecipitations were performed as described before (24). 293T cells were lysed on ice using lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 1.0% Triton X-100) or radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 0.1% SDS, 1.0% Nonidet P-40, 0.5% sodium deoxycholate) supplement with the protease inhibitor mixture (78430; Thermo) then sonicated with Sonic Dismembrator 100 (Fisher) by 3 cycles of 5 s on and 30 s off at an intensity of 3 and incubated on ice for 30 min. Extracts were then precleared with protein A- or G-Sepharose beads (Invitrogen). After centrifugation, whole cell lysates were incubated with the appropriate primary antibodies or antibody-conjugated beads overnight at 4 °C. After incubations with primary antibodies, lysates were centrifuged, and supernatants were incubated with Protein A- or G-Sepharose beads for 2 h at 4 °C. Beads were washed 3 times using lysis buffer, 5 min every time at 4 °C. Calf intestinal phosphatase (CIP, M0290S, New England BioLabs) treatment was performed by adding 10 units of CIP to whole cell lysates at 37 °C for 30 min. For native immunoprecipitation using mouse thymus, 10 thymi were lysed using pestles (1415–5390; USA Scientific) in 1.5-ml tubes. The lysate was sonicated as above described, and then the lysates were centrifuged and precleared by protein A-Sepharose beads (Invitrogen). Lysates were then incubated with anti-FBXO3 antibodies overnight. Beads were washed 3 times using the lysis buffer. Immunoprecipitated complexes were boiled in SDS sample buffer (Laemmli sample buffer with β-mercaptoethanol) and analyzed by Western blotting.
Chromatin Immunoprecipitations
Chromatin immunoprecipitations (ChIPs) were performed as described previously (9, 24). 293T cells were transfected using Lipofectamine 2000 or Lipofectamine RNAiMAX transfection reagent according to the manufacturer's instructions. 293T cells were harvested, cross-linked with 1% formaldehyde at room temperature for 10 min, and stopped by adding glycine to a final concentration of 125 mm. Cells were lysed in radioimmune precipitation assay buffer and sonicated at power 4 five times for 10 s each by using a Sonic Dismembrator model 100 (Fisher). Lysates were precleared by protein G-Sepharose beads (Invitrogen). Chromatin immunoprecipitations were performed using anti-FLAG beads or anti-His beads. Immunoprecipitated DNA was detected by quantitative PCR using SensiFAST SYBR Lo-ROX Kit (Bioline) on an Mx3005p machine (Stratagene). Precipitated DNA was quantified relative to input and is presented as -fold over IgG control. The following primers were used: KRT14 promoter, forward (5′-GGAAAGTGCCAGACCCGCCC) and reverse (5′-GGAGGGAGGTGAGCGAGCGA); S100A8 promoter, forward (5′-CTGGGCTGCTGGCATCCACT) and reverse (5′-GGCCTGACCACCAATGCAGGG).
In Vivo Ubiquitylation Assay
For in vivo ubiquitylation assays, cells were treated with 1 μm MG132 (M7449; Sigma) for 4 h before harvesting, and then cells were lysed in 1% SDS buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% SDS, 10 mm DTT) and boiled for 30 min. The lysates were centrifuged and diluted 1:10 with lysis buffer (50 mm Tri-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100). The diluted lysates were immunoprecipitated with anti-FLAG beads overnight at 4 °C. Myc, His epitope-tagged AIRE protein was isolated as described above. After SDS-PAGE, the membrane was incubated with the indicated antibodies.
Luciferase Assay
293T cells were transfected with Lipofectamine 2000 to co-express AIRE proteins and INSLuc in 6-well plates. Cells were harvested after 48 h, and luciferase assays were performed using dual luciferase kit (Promega) according to the manufacturer's instructions. Firefly luciferase activity readings of each sample were normalized to total protein in the whole extract to determine relative luciferase activities. Total protein was measured using NanoDrop Lite spectrophotometer (Thermo).
RNA Isolation and RT-PCR
293T cells or 1C6 mTECs were washed by cold PBS and harvested in TRIzol reagent (Life Technologies). RNA was isolated according to the manufacturer's instructions. 2 μg of RNA was reverse-transcribed with SuperScript III Reverse Transcriptase (Life Technologies) and random primers. Levels of expression of AIRE-dependent and -independent genes were determined by real-time PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Real-time PCR was performed using SensiFAST SYBR Lo-ROX Kit (Bioline) on an Mx3005p machine (Stratagene). Relative activations represent the ratio between FBXO3 siRNA knockdown and the siRNA control, which were calculated using the threshold cycles (ΔΔCT) method (10). The Primers used in this study have been published (13, 25–28).
Protein Stability Assay
293T cells were transfected with mHis:AIRE, control siRNA, or siRNA targeting FBXO3 for 48 h, and then cells were treated with translation inhibitor cycloheximide (100 μg/ml, Sigma) (29). Cells were washed by cold PBS and collected at time points 0, 1, 4, and 8 h, then lysed and subjected to Western blotting using GAPDH as the control. Protein signals were quantified by Image J (30).
Bioinformatics
AIRE protein sequences from Homo sapiens, Pan troglodytes, Macaca mulatta, Rattus norvegicus, Mus musculus, and Canis lupus were obtained from NCBI and aligned using MegAlign by Clustal W method (Dnastar, Madison, WI).
Results
AIRE Interacts with FBXO3
To find additional AIRE-interacting proteins, we performed proteomic studies with the FLAG epitope-tagged AIRE protein, which was expressed transiently in 293T cells. Besides many proteins already identified by others (13), we found one E3 ligase, FBXO3 (Table 1). FBXO3 contains 471 residues and migrates with an apparent molecular mass of 55 kDa. It forms a SCF E3 ubiquitin ligase complex with SKP1, Cullin1 (CUL1), and ring-box 1 (RBX1). By ubiquitylating F-box and leucine-rich repeat protein 2 (FBXL2), FBXO3 plays a role in cytokine-mediated inflammation (31). FBXL2 also ubiquitylates, degrades, and inhibits tumor necrosis factor receptor-associated factors (TRAFs). By removing it, FBXO3 increases TRAF signaling (31, 32). FBXO3 also ubiquitylates and degrades homeodomain-interacting protein kinase 2 (HIPK2) and P300 (33). By ubiquitylating and degrading SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1), FBXO3 also increases BMP signaling (34). In all these areas, FBXO3 appears to play an active role in gene expression. However, FBXO3 had not previously been implicated in effects of AIRE.
TABLE 1.
AIRE-interacting proteins identified by MS
Shown is a list of AIRE interacting proteins identified by mass spectrometry. The number of unique peptides is presented. FBXO3 and its number of peptides are written in bold letters.
| Gene symbol | # Unique peptides | |
|---|---|---|
| 1 | GCN1L_HUMAN, GCN1-like protein 1 | 44 |
| 2 | LAP2A_HUMAN, lamina-associated polypeptide 2 isoform α | 13 |
| 3 | AIRE_HUMAN, autoimmune regulator | 12 |
| 4 | HNRH1_HUMAN, heterogeneous nuclear ribonucleoprotein H | 11 |
| 5 | FBXO3_HUMAN, F-box protein 3 | 8 |
| 6 | DDX5_HUMAN, probable ATP-dependent RNA helicase DDX5 | 5 |
| 7 | HNRPF_HUMAN, heterogeneous nuclear ribonucleoprotein F | 4 |
| 8 | SMC3_HUMAN, structural maintenance of chromosomes protein 3 | 3 |
| 9 | SF3B3_HUMAN, splicing factor 3B subunit 3 | 3 |
| 10 | TADBP_HUMAN, TAR DNA-binding protein 43 | 2 |
| 11 | SUMO2_HUMAN, small ubiquitin-related modifier 2 precursor | 2 |
| 12 | LANC2_HUMAN, LanC-like protein 2 | 2 |
| 13 | ATR_HUMAN, serine/threonine-protein kinase ATR | 2 |
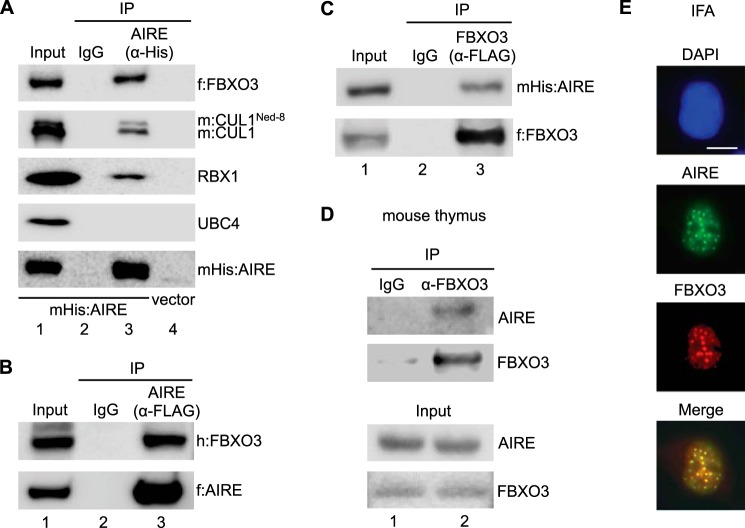
To confirm this association of FBXO3, we co-expressed transiently the Myc, His epitope-tagged AIRE protein (mHis:AIRE) with the FLAG epitope-tagged FBXO3 protein (f:FBXO3) in 293T cells. In addition, because FBXO3 is found in association with CUL1 and RBX1, we co-expressed the Myc epitope-tagged CUL1 (m:CUL1) protein in these cells (33). Anti-His immunoprecipitations were then probed with appropriate antibodies by Western blotting. Presented in Fig. 1A, lane 3, AIRE co-immunoprecipitated with FBXO3, CUL1, and RBX1 but not with UBC4. In the absence of AIRE, none of these proteins was detected (Fig. 1A, vector, lane 4). To confirm this association further, we also co-expressed the FLAG epitope-tagged AIRE protein (f:AIRE) with the HA epitope-tagged FBXO3 protein (h:FBXO3) and found the same association (Fig. 1B, lane 3). FBXO3 could also immunoprecipitate AIRE (Fig. 1C). For this panel, we co-expressed f:FBXO3 and mHis:AIRE and analyzed anti-FLAG immunoprecipitations (Fig. 1C, lane 3). Finally, we isolated 10 thymi from young mice and immunoprecipitated the endogenous FBXO3 protein. These Western blots were then probed with anti-AIRE antibodies. In Fig. 1D, lane 2, the endogenous FBXO3 protein interacted with the native AIRE protein. We conclude that AIRE and FBXO3 interact with each other in transformed cells as well as in the relevant tissue of the host.
FIGURE 1.
AIRE interacts with FBXO3. A, AIRE interacts with the SCFFBXO3 complex. Anti-His antibodies immunoprecipitated Myc, His epitope-tagged AIRE (mHis:AIRE) protein from 293T cell lysates. Western blotting with anti-Myc, anti-FLAG, anti-RBX1, and anti-Cullin1 antibodies revealed the presence of associated proteins in immunoprecipitations. Western blotting with anti-UBC4 antibodies represented a negative control. In the absence of AIRE (vector, lane 4), none of these proteins was detected in our immunoprecipitations. f:FBXO3, FLAG epitope-tagged FBXO3; m:CUL1, Myc epitope-tagged Cullin1; α-, anti. B, AIRE interacts with FBXO3. FLAG epitope-tagged AIRE protein co-immunoprecipitated the HA epitope-tagged FBXO3 (h:FBXO3) protein from 293T cell lysates. AIRE and FBXO3 were detected using anti-FLAG and anti-HA antibodies. C, FBXO3 interacts with AIRE. f:FBXO3 co-immunoprecipitated (IP) mHis:AIRE from 293T cell lysates. D, endogenous FBXO3 and AIRE proteins interact in the mouse thymus. Endogenous FBXO3 and AIRE proteins co-immunoprecipitated from mouse thymus lysates. Input (lower panels, 5% whole thymus lysate) and IgG control are also presented. E, AIRE and FBXO3 co-localize in cells. 293T cells co-expressed Myc, His epitope-tagged AIRE (green), and HA epitope-tagged FBXO3 (red) proteins. AIRE and FBXO3 were detected with anti-His and anti-HA antibodies, respectively. The nuclear staining with DAPI is blue. Scale bar, 10 μm. IFA, immunofluorescence assay.
Finally, immunofluorescence assays revealed that AIRE and FBXO3 co-localize in the same nuclear organelles in 293T cells (Fig. 1E). These subcellular organelles are enriched for splicing factors and represent sites of active transcription (35). Importantly, this intracellular localization is in agreement with all previous reports (33, 35, 36). Thus, not only do AIRE and FBXO3 associate with each other and the rest of the E3 ligase complex, but they also co-localize in areas of active transcription. These findings encouraged us to look more deeply into the role of FBXO3 in regulating the expression of TSA genes.
It is important to note that cells expressing AIRE stop growing and many undergo apoptosis. This cell cycle arrest prevents the continuous cultivation of AIRE-expressing cells in culture. In the thymus, these dying cells are cleared by professional antigen-presenting cells, which present engulfed and processed TSA peptides on major histocompatibility antigens to developing T cells for the elimination of autoreactive cells. This process directs central tolerance in the thymus (3, 4, 37). Nevertheless, the transient expression of AIRE in other epithelial or endothelial cells leads to the activation of TSA genes.
N Terminus of AIRE Binds to FBXO3
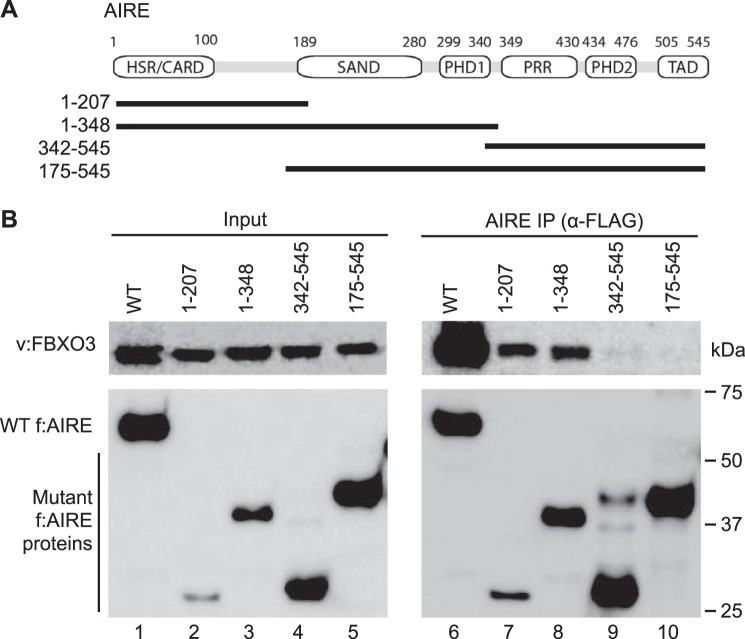
Next, we wanted to map surfaces on AIRE that interact with FBXO3. To this end, we generated several deletion mutant AIRE proteins (Fig. 2A). Below the well described domains of AIRE are depicted fragments that contain N-terminal, middle, and C-terminal sequences of AIRE. The FLAG epitope tag was placed at the N terminus of these truncated AIRE proteins. They were co-expressed with the V5 epitope-tagged FBXO3 (v:FBXO3) protein in 293T cells. Of interest, N-terminal fragments of AIRE were unstable (Fig. 2B, f:AIRE panels, lanes 2, 3, 7, and 8). Nevertheless, only they interacted with v:FBXO3 (Fig. 2B, v:FBXO3 panel, lanes 7 and 8). We conclude that the N-terminal 207 residues in AIRE interact with FBXO3.
FIGURE 2.
N terminus of AIRE binds to FBXO3. A, schematic representation of WT and mutant AIRE proteins. Black lines below the schematic indicate FLAG epitope-tagged deletion mutant AIRE peptides. B, N terminus of AIRE interacts with FBXO3. FLAG epitope-tagged WT and mutant AIRE proteins (f:AIRE) were co-expressed with the V5 epitope-tagged FBXO3 (v:FBXO3) protein in 293T cells. Anti-FLAG immunoprecipitations were performed. The lower panels depict total lysates (lanes 1–5, 3%) and immunoprecipitated (lanes 6–10) WT and mutant AIRE proteins. The upper panels depict the V5 epitope-tagged FBXO3 protein in lysates (lanes 1–5) and co-immunoprecipitations (lanes 6–10) with WT and mutant FLAG epitope-tagged AIRE proteins.
Double-phosphorylated AIRE Binds Better to FBXO3
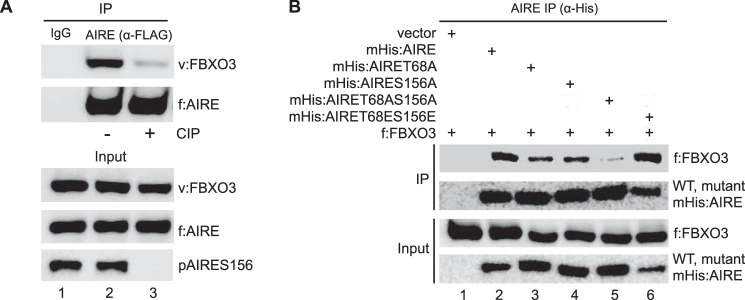
As mentioned in the Introduction, the N terminus of AIRE contains two residues that are phosphorylated by DNA-PK for full activity (11). They are Thr-68 and Ser-156 and are contained in sequences that bind to FBXO3. To determine if the phosphorylation of AIRE is required for binding to FBXO3, we first treated cell lysates with CIP before the immunoprecipitation. As presented in Fig. 3A, lane 3, the dephosphorylated AIRE protein bound only weakly to FBXO3. The removal of the phosphate group was confirmed with specific antibodies against the phosphorylated Ser-156 residue in AIRE (Fig. 3A, pAIRES156 panel, lane 3). Next, we changed Thr-68 and Ser-156 to alanine, which cannot be phosphorylated, and the phosphomimetic glutamic acid, creating the mutant mHis:AIRET68A, mHis:AIRES156A, mHis:AIRET68AS156A, and mHis:AIRET68E S156E proteins. Whereas the mutant mHis:AIRET68ES156E protein bound FBXO3 as well as its wild type (WT) counterpart (Fig. 3B, lanes 2 and 6), these interactions were diminished greatly with the mutant mHis:AIRET68AS156A protein (Fig. 3B, lanes 2 and 5). Single mutant AIRET68A or AIRES156A proteins bound FBXO3 5-fold less well than the WT AIRE protein (Fig. 3B, lanes 2–4). Thus, AIRE binds to FBXO3 via phosphorylated Thr-68 and Ser-156 residues, and both are required for optimal interactions between these two proteins.
FIGURE 3.
Doubly phosphorylated AIRE binds better to FBXO3. A, CIP treatment decreases interactions between AIRE and FBXO3. f:AIRE proteins were co-expressed with the V5 epitope-tagged FBXO3 (v:FBXO3) protein in 293T cells. Cell extracts were treated with CIP (lane 3) before anti-FLAG immunoprecipitations (IP). Upper and lower panels contain specific immunoprecipitations and inputs (3%), respectively. Antibody detecting the phosphorylated Ser-156 site in AIRE input (pAIRES156, 3%) and IgG control are also presented. B, progressive alanine substitutions of Thr-68 and Ser-156 in AIRE diminish interactions with FBXO3. 293T cells expressed WT (lane 2) or mutant mHis:AIRET68A (lane 3), mHis:AIRES156A (lane 4), mHis:AIRET68AS156A (lane 5), and mHis:AIRET68ES156E (lane 6) proteins. Cell lysates were incubated with anti-His antibodies. Input (lower panels, 3%) and no AIRE (lane 1) controls are also presented.
FBXO3 Promotes the Ubiquitylation of AIRE
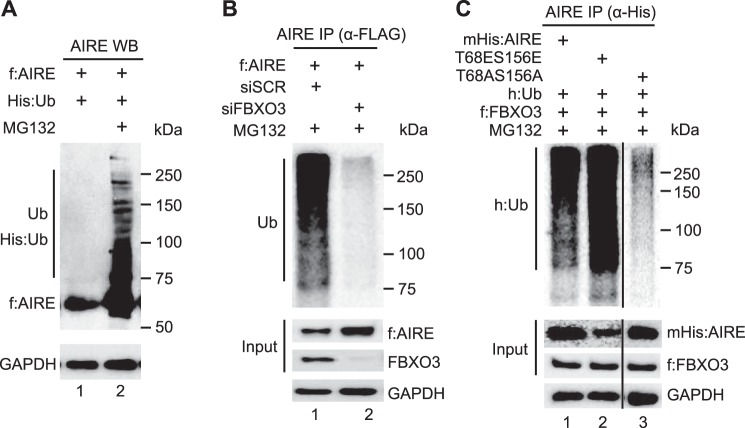
FBXO3 is an E3 ubiquitin ligase (23, 31, 33, 34). To determine if it is responsible for the ubiquitylation of AIRE, we co-expressed f:AIRE and the His epitope-tagged ubiquitin (His:Ub) in 293T cells. These cells were also exposed to DMSO (control) or MG132, which is a proteasome inhibitor. As expected, in its absence the ubiquitylated AIRE protein was degraded, and little to no ubiquitylation was observed on AIRE (Fig. 4A, lane 1). In contrast, in the presence of MG132, AIRE ubiquitylation became apparent with the endogenous FBXO3 protein (Fig. 4A, lane 2). In Fig. 4B, we observed that the depletion of FBXO3 with siFBXO3 RNA reduced AIRE ubiquitylation (Fig. 4B, lane 2). Finally, we examined if Thr-68 and Ser-156 residues contribute to this ubiquitylation of AIRE. As presented in Fig. 4C (upper, h:Ub panel, lane 2), when these residues were changed to the phosphomimetic glutamic acid the ubiquitylation of AIRE was increased. In contrast, when they were mutated to alanine, the mutant mHis:AIRET68AS156A protein had decreased ubiquitylation (Fig. 4C, h:Ub panel, lane 3). Thus, the phosphorylation of Thr-68 and Ser-156 not only allows for the binding to FBXO3 but also leads to the ubiquitylation of AIRE.
FIGURE 4.
FBXO3 promotes ubiquitylation of AIRE. A, AIRE is ubiquitylated in cells. 293T cells co-expressed the His epitope-tagged ubiquitin and f:AIRE in the presence of DMSO or MG132. Proteins were detected with anti-FLAG antibodies by Western blotting. Ub depicts the polyubiquitylated AIRE protein. GAPDH served as a loading control. B, depletion of FBXO3 reduces greatly the ubiquitylation of AIRE. 293T cells expressed f:AIRE with control siRNA (scrambled, lane 1) or siRNA targeting FBXO3 (siFBXO3, lane 2). Lysates were immunoprecipitated (IP) with anti-FLAG antibodies followed by detection with indicated antibodies by Western blotting. Input proteins (f:AIRE and FBXO3) and the loading control (GAPDH) are presented in the lower three panels. C, the mutant mHisAIRET68AS156A protein was only weakly ubiquitylated. 293T cells co-expressed HA epitope-tagged ubiquitin (h:Ub), f:FBXO3, WT mHis:AIRE, mutant mHis:AIRET68ES156E, and mutant mHis:AIRET68AS156A proteins for 48 h. Anti-His antibodies immunoprecipitated AIRE. Western blotting was performed with anti-Myc, anti-HA, and anti-FLAG antibodies. Input proteins (mHis:AIRE and f:FBXO3) and the loading control (GAPDH) are presented in the lower three panels.
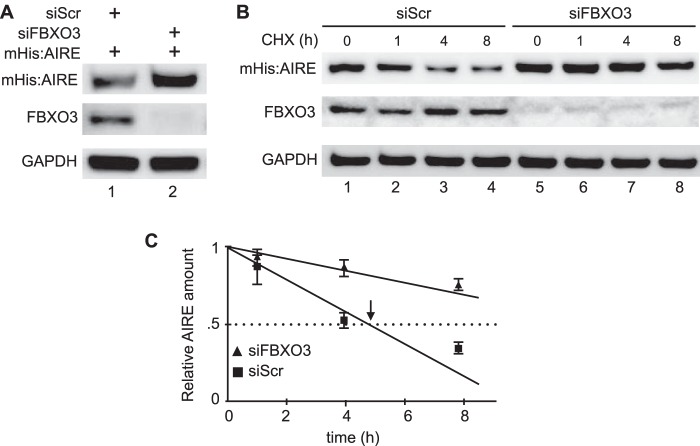
FBXO3 Destabilizes AIRE
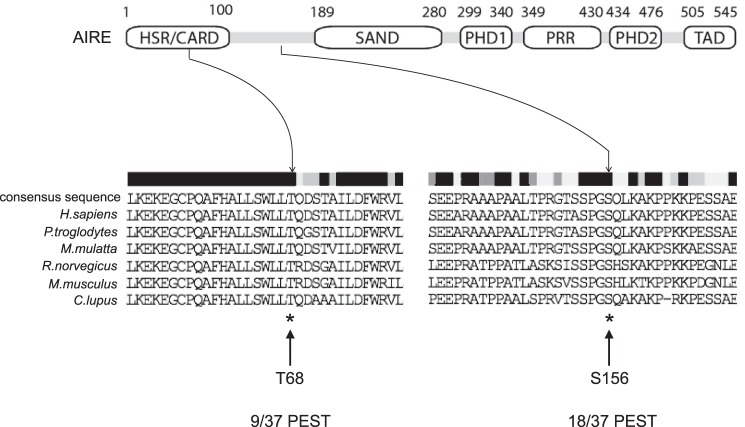
As inferred from Fig. 4, the ubiquitylated AIRE protein could be destabilized. To determine the rate of decay of AIRE in the presence and absence of FBXO3, we performed steady state and decay experiments. When the endogenous FBXO3 was depleted with siRNA, levels of AIRE increased (Fig. 5A, top panel, lane 2). To distinguish between increased synthesis and decreased rates of decay, we also treated cells with cycloheximide and followed the expression of mHis:AIRE by Western blotting. With endogenous FBXO3, the half-life of AIRE was 5 h. In the absence of FBXO3, this half-life was increased 2–3-fold (Fig. 5, B and C). Thus, FBXO3 not only ubiquitylates but also destabilizes the AIRE protein in cells. Because Thr-68 and Ser-156 are found in conserved sequences rich in proline, glutamic acid, serine, and threonine residues (PEST) in vertebrates (Fig. 6), we conclude that they are contained in degrons.
FIGURE 5.
FBXO3 destabilizes AIRE. A, knockdown of FBXO3 increases amounts of steady state AIRE protein. 293T cells co-expressed mHis:AIRE, control (scrambled) siRNA (siScr) or FBXO3 siRNA (siFBXO3). Whole cell lysates were probed with anti-Myc or anti-FBXO3 antibodies by Western blotting. GAPDH served as a loading control. B, knockdown of FBXO3 stabilizes AIRE. Cells were transfected with mHis:AIRE, control siRNA (siScr), or siRNA targeting FBXO3 (siFBXO3) for 48 h. 100 μg/ml cycloheximide (CHX) was added to cells, and the disappearance of AIRE was followed by Western blotting with specific antibodies (anti-Myc, anti-FBXO3, and anti-GAPDH) for the indicated times (0, 1, 4, and 8 h). C, densitometric analysis of mHis:AIRE Western blot from panel B. Protein bands were quantified using ImageJ software, and AIRE protein half-life was calculated. Bars represent the S.E. of three independent experiments (S.E., n = 3).
FIGURE 6.
Thr-68 and Ser-156 are conserved across species. Comparisons of conserved Thr-68 and Ser-156 residues in AIRE between species. Sequences from H. sapiens, P. troglodytes, M. mulatta, R. norvegicus, M. musculus, C. lupus, and Danio rerio are presented. The number of PEST residues surrounding Thr-68 and Ser-156 is indicated on the bottom. White to black bars indicate increased conservation of amino acids in these FBXO3 proteins.
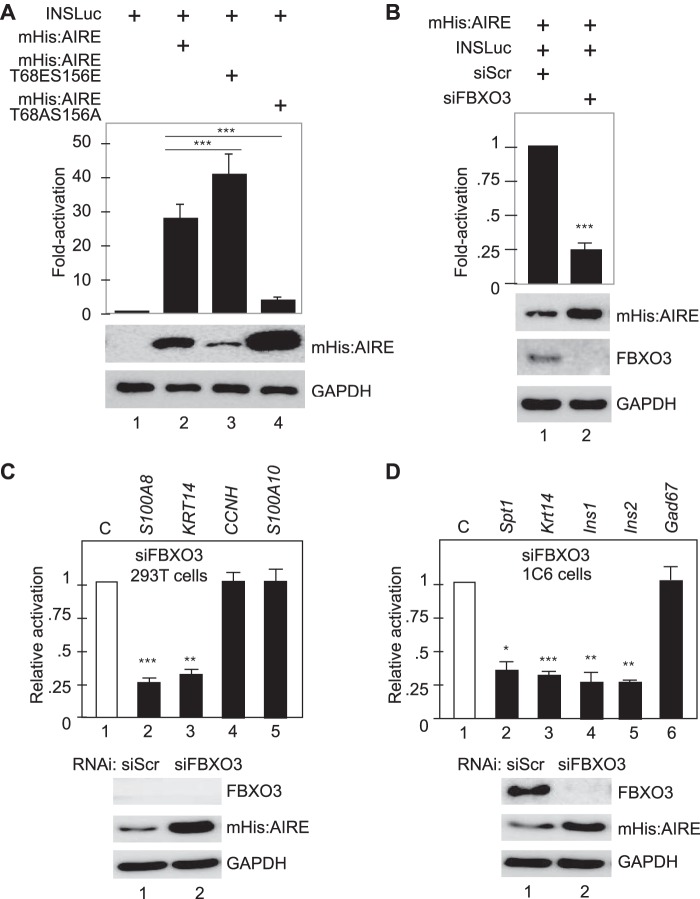
FBXO3 Is Required for AIRE Transcriptional Activity
There is extensive literature on TADs, which exist in proteins that contain degrons and function better when ubiquitylated. They include cJun, cFos, cMyc, p53, and VP16 (16–18). To determine if AIRE also acts in this fashion, we co-expressed mHis:AIRE and the human insulin promoter linked to the luciferase reporter gene (INSLuc) plasmid target in 293T cells. The insulin gene is a bona fide target of AIRE and is expressed in AIRE-expressing mTECs (38). First, we investigated transcriptional activity of the WT and two mutant AIRE proteins on INSLuc. As presented in Fig. 7A, whereas the WT mHis:AIRE and mutant mHis:AIRET68ES156E proteins activated INSLuc 25- and 37.5-fold, respectively, the mutant mHis:AIRET68AS156A only increased the luciferase activity 5-fold. Of note, levels of AIRE proteins also paralleled their relative degradation such that the mutant mHis:AIRET68AS156A achieved the highest levels of expression in these cells (Fig. 7A, mHis:AIRE panel). Next, we repeated these report assays in WT or FBXO3-depleted cells (by siFBXO3 RNA). As presented in Fig. 7B (bar 2), the depletion of FBXO3 resulted in a 4-fold decreased activation of the human insulin promoter. Note also that the levels of the WT mHis:AIRE protein increased in these cells (Fig. 7B, mHis:AIRE panel, lane 2). We conclude that the ubiquitylation of AIRE is important for its transcriptional activity on a validated target promoter.
FIGURE 7.
FBXO3 increases AIRE transcriptional activity. A, mutations of Thr-68 and Ser-156 influence AIRE transcriptional activity. The relative luciferase activity of INSLuc for WT mHis:AIRE (lane 2) and mutant mHis:AIRET68ES156E (lane 3) or mHis:AIRET68AS156A proteins (lane 4) is presented as -fold activation above levels of the empty plasmid vector. Levels of mHis:AIRE and GAPDH (as internal control) proteins are given below the bar graph. B, knockdown of FBXO3 decreases AIRE transcriptional activity. Presented is the relative luciferase activity of INSLuc, which is induced by AIRE, after FBXO3 knockdown (lane 2) or control (lane 1). Values are presented as -fold activation above levels of control siRNA. Levels of mHis:AIRE, FBXO3, and GAPDH (as internal control) proteins are given below the bar graph. C, FBXO3 knockdown affects the expression of AIRE-dependent genes in 293T cells. RT-quantitative PCR analysis was performed on four candidate genes after FBXO3 knockdown in 293T cells. 293T cells co-expressed mHis:AIRE with control (siScr) or FBXO3 targeting siRNAs (siFBXO3) for 72 h before harvesting. Bottom panels present levels of FBXO3, AIRE, and GAPDH (as the internal control) proteins. D, FBXO3 knockdown affects AIRE-dependent genes in the mTECs. RT-quantitative PCR analysis was performed on five candidate genes after FBXO3 knockdown in 1C6 mTECs. 1C6 cells co-expressed mHis:AIRE with control (siScr) or FBXO3 targeting (siFBXO3) siRNAs for 72 h before harvesting. Values and Western blots are as in panel C. For panels A–D, bars represent S.E., n = 3. Student's t test was preformed to measure the significance of the data (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
FBXO3 Increases AIRE Transcriptional Activity
To validate our findings with a transiently expressed plasmid target, we also performed studies on host cell genes in 293T cells (Fig. 7C) and continuously growing 1C6 cells (Fig. 7D). Although isolated as mTECs, continuously growing 1C6 cells no longer express the endogenous AIRE protein. However, when AIRE is introduced into these cells, it activates TSA genes (8, 13). Levels of expression were determined by RT-quantitative PCR. In 293T cells, the ectopically expressed mHis:AIRE activated two AIRE-responsive genes, S100A8, and KRT14 but not two AIRE-unresponsive genes, CCNH and S100A10 (Fig. 7C). When FBXO3 was depleted in these cells by siFBXO3 RNA, the expression of S100A8 and KRT14 was decreased up to 4-fold (Fig. 7C, bars 2 and 3), whereas that of CCNH and S100A10 remained unchanged (Fig. 7C, bars 4 and 5). Levels of mHis:AIRE also increased upon the depletion of FBXO3 (Fig. 7C, mHis:AIRE panel, lane 2). The same situation was observed in 1C6 cells. In these cells, we investigated the expression of Spt1, Krt14, Ins1, Ins2, and Gad67 genes, of which the first four were AIRE-responsive. Upon the depletion of FBXO3, a similar reduction in the expression of AIRE-responsive genes, but not of Gad67, was observed in these cells (Fig. 7D, bars 2–6). Again, the levels of mHis:AIRE increased upon the absence of FBXO3. Thus, transcriptional activity of AIRE depends upon its post-translational modification also on genes in chromatin.
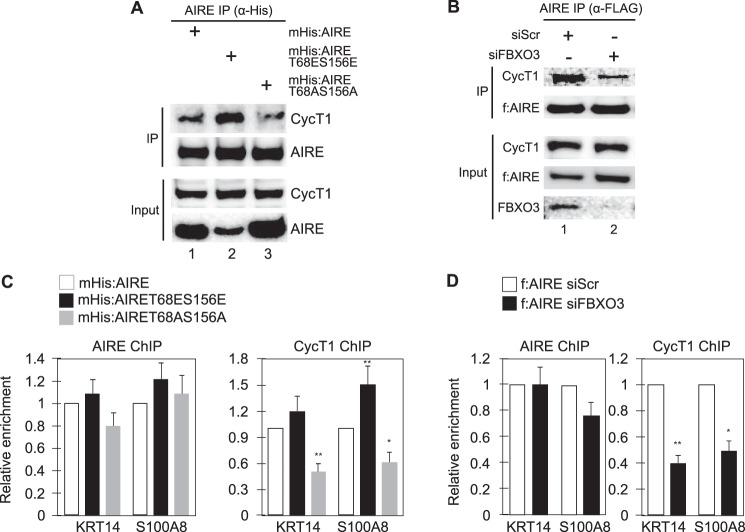
Ubiquitylation of AIRE Increases the Binding and Recruitment of P-TEFb to Target Genes
The ubiquitylation of TFs increases the recruitment of the proteasome and P-TEFb to transcription units (16–18). Although the recruitment of the 19S rather than the complete 26S proteasome suffices, this assembly is thought to help clean up and degrade histones and other misfolded proteins that are generated during active transcription (16, 17). In addition, we found that ubiquitylated TFs also bind better to CycT1, which together with CDK9 (cyclin-dependent kinase 9) forms P-TEFb (18). To determine if this ubiquitylation of AIRE also increases its interactions with P-TEFb, we performed binding studies between f:AIRE and the endogenous CycT1 protein. Indeed, mHis:AIRE and the mutant mHis:AIRET68ES156E proteins bound to CycT1 better than the mutant mHis:AIRET68AS156A protein (Fig. 8A, left top, CycT1 panel, lanes 1–3). Densitometric analyses revealed that the WT mHis:AIRE protein bound CycT1 ∼3-fold better than the mutant mHis:AIRET68AS156A protein and that the mutant mHis:AIRET68ES156E protein bound CycT1 ∼5-fold better than its WT counterpart (Fig. 8A). In addition, f:AIRE bound to CycT1 4-fold better in the presence of FBXO3 than in its absence (Fig. 8B, top, CycT1 panel, lanes 1 and 2). This improved binding should increase the transcriptional activity of AIRE.
FIGURE 8.
Ubiquitylation of AIRE increases the binding and recruitment of P-TEFb to target genes. A, mutations of Thr-68 and Ser-156 influence the AIRE interaction with P-TEFb. 293T cells expressed WT mHis:AIRE or mutant mHis:AIRE proteins for 48 h. Whole cell lysates were immunoprecipitated (IP) with anti-His antibodies followed by Western blotting with indicated antibodies. B, knockdown of FBXO3 decreases interactions between AIRE and P-TEFb. f:AIRE was expressed in 293T cells with control (siScr) or siFBXO3 (siFBXO3) RNAs for 72 h before harvesting. Total cell lysates were incubated with anti-FLAG beads. AIRE, the endogenous CycT1 and FBXO3 proteins, were examined with indicated antibodies. The top panels contain immunoprecipitated CycT1 and f:AIRE proteins. The lower panels contain 3% input CycT1, f:AIRE, and FBXO3 proteins. C and D, interactions between AIRE and P-TEFb on DNA are decreased by deleterious mutations in AIRE and the depletion of FBXO3. C, presented are ChIPs of AIRE and CycT1 at promoters of TSA genes (KRT14 and S100A8) in cells expressing WT and mutant mHis:AIRE proteins. D, presented are ChIPs of AIRE and CycT1 at the promoter of TSA genes (KRT14 and S100A8) in control or FBXO3-depleted cells. Bars represent the S.E. of three independent experiments (S.E., n = 3). Student's t test was preformed to measure the significance of the data (*, p < 0.05; **, p < 0.01).
To this end, we also examined the recruitment of P-TEFb to AIRE-responsive genes and performed ChIPs on KRT14 and S100A8 promoters (Fig. 8, C and D). Anti-His immunoprecipitations revealed no difference in the amounts of WT mHis:AIRE and mutant mHis:AIRET68ES156E or mHis:AIRET68 AS156A proteins at these promoters (Fig. 8C, left panel, all bars). In contrast, levels of CycT1 were attenuated with the mutant mHis:AIRET68AS156.
A protein (Fig. 8C, right panel, gray bars). The same situation was observed with the depletion of FBXO3 using siFBXO3 RNA (Fig. 8D, left and right panels, black bars). Because AIRE does not travel with elongating RNAPII, we did not investigate the body or 3′ ends of these genes (8). We conclude that the ubiquitylation of AIRE increases the binding between AIRE and CycT1 and the recruitment of P-TEFb to AIRE-responsive promoters.
Discussion
In this study we identified and validated FBXO3 as the E3 ubiquitin ligase that modifies AIRE post-translationally and increases its activity on TSA genes. After mass spectrometry, further binding studies indicated that N-terminal sequences in AIRE bind to FBXO3. FBXO3 was also found to promote the ubiquitylation of AIRE. Moreover, two phosphorylated residues were required for this binding. Dephosphorylating AIRE and mutating Thr-68 and Ser-156 residues to alanine abrogated the binding between AIRE and FBXO3. It also stabilized the AIRE protein and diminished its transcriptional activity. Mutating the same residues to glutamic acid, which is phosphomimetic, not only destabilized AIRE but also increased its transcriptional activity. Of note, depleting FBXO3 in 293T and 1C6 cells attenuated the ability of AIRE to activate TSA genes. Finally, we found that AIRE phosphorylation is important for its binding to FBXO3 and its ubiquitylation. AIRE ubiquitylation increased its interactions with CycT1 and recruited more P-TEFb to AIRE-responsive genes. P-TEFb is required to modify stalled RNAPII and transcription complexes for efficient elongation and co-transcriptional processing of target genes. We conclude that FBXO3 is the E3 ligase that modifies AIRE for its transcriptional activity and for central tolerance in the thymus.
AIRE is expressed in the small subpopulation of mTECs in the thymus. Thus, it is impossible to collect enough mTECs for mechanistic studies. In addition, AIRE-expressing cells stop growing, and most undergo apoptosis. For continuous growth in culture, mTECs, such as 1C6 cells, lose the expression of the endogenous AIRE protein. In our study we expressed AIRE transiently in 293T and 1C6 cells, which is an approach that has been validated by others (13, 19, 21, 39). Nevertheless, because FBXO3 is expressed in all tissues and all cells, we could study the endogenous protein and perform its knockdowns. Indeed, the endogenous FBXO3 protein ubiquitylated AIRE efficiently (Fig. 4). Because of its deleterious effects in cells, levels of transiently expressed AIRE protein approximate those of the endogenous AIRE protein in the thymus, thus further validating these cell culture models for our studies (9, 10). In this context, it is also likely that the ubiquitylation of AIRE ensures the optimal expression of TSA genes before cells stop growing, thus promoting their ability to orchestrate the process of central tolerance in the thymus.
Our studies extend and confirm work on the role of Thr-68 and Ser-156, which are phosphorylated by DNA-PK for optimal transcriptional activity of AIRE (11). These residues are located in a sequence rich in PEST, which could contribute to the rapid degradation of AIRE (Fig. 6) (40). Previously, we and others found that DNA-PK and unmodified histone H3K4 were required to recruit AIRE to TSA genes (10, 26, 41). DNA-PK is found at transcription start sites because of DNA breaks created during initiation of transcription and topoisomerase relaxation of chromatin. We also found other members of FBXO3-E3 ligase complex in our immunoprecipitations, which include CUL1 and RBX1 (Fig. 1A). Other components of the SCFFBXO3 include SKP1 and an E2 neddylation enzyme that modifies CUL1 for activity (42). An interesting aside to our studies is that we created a mutant AIRE protein with increased activity. This mutant AIRET68ES156E protein was constitutively more active than the WT AIRE protein. It warrants expression in transgenic mice, which should become more tolerant to self. It is also possible that this mutant AIRET68ES156E protein could abrogate autoimmunity in yet other mice, such as the NOD mouse (43).
A recent report suggested that bromodomain containing 4 (BRD4) bridges interactions between AIRE and P-TEFb (44). BRD4 bound to acetylated lysines in the HSR/CARD domain at the very N terminus of AIRE. However, this mapping differs from studies of an APECED patient's mutation, which identified the TAD at the extreme C terminus of the protein (9). In VP16, a similar TAD and ubiquitin bind to cyclin boxes and the extreme C terminus of CycT1, respectively (18). A critical finding of this new report was decreased expression of TSA genes after the repeated administration of I-BET151, which is a BET bromodomain inhibitor, to young mice (44). However, these compounds also increase levels of hexamethylene bisacetamide inducible 1 (HEXIM1) that suppress the kinase activity of P-TEFb, which would decrease the expression of TSA genes (45). Of interest, another study found that rather than acetylation, it is the deacetylation by Sirt1 that is essential for AIRE transcriptional activity (46). Nevertheless, we and others found that AIRE is destabilized upon the phosphorylation of Thr-68 and Ser-156 (11). Upon this post-translational modification, we found that FBXO3 binds and ubiquitylates AIRE. Because TAD and ubiquitin bind cooperatively to different surfaces on CycT1, they increase the recruitment of P-TEFb to target genes (18). It is also possible that BRD4, ubiquitin, and TAD all cooperate or act individually in different contexts to recruit more P-TEFb to RNAPII to optimize effects of AIRE on TSA genes.
In addition to its effects on AIRE, this SCFFBXO3 complex has been reported to increase proinflammatory responses by eliminating FBXL2, which inhibits TRAF proteins (31, 32). A more recent study found an association between FBXO3 and SMURF1, which is a Nedd4 family member (34). By degrading SMURF1, it stabilized intermediates in BMP signaling cascades, which include SMAD1/2 and JunB, thus increasing the responsiveness of target genes. FBXO3 can also degrade HIPK2 and p300 as well as p62 during IFN-mediated suppression of the Rift Valley fever virus (RVFV) (23). These other effects of FBXO3 could complicate knock-out studies in transgenic mice, which would have to be controlled for yet additional published phenotypes. Nevertheless, we were able to observe direct transcriptional effect of AIRE on TSA genes in 293T and 1C6 cells, and they were specific to transcription units that had been validated previously by whole genome and targeted studies from several laboratories (25, 47). It is also possible that these global effects might render mutations in or inactivation of FBXO3 more deleterious to the organism than those found in AIRE in APECED/APS1 patients. Thus, it would be of interest to investigate severe cases of APECED/APS1 or of other congenital autoimmunities for possible mutations in or around the FBXO3 gene.
Author Contributions
W. S. and K. Z. performed all the experiments. W. S., K. Z., K. F., and B. M. P. analyzed the data and wrote the manuscript.
Acknowledgments
We thank Dr. Zhiyong Yang for the mouse thymus sample, Pärt Peterson and Rama Mallampalli for reagents, and members of the Peterlin laboratory for helpful discussions and comments on the manuscript.
This work was supported by a grant from the Nora Eccles Treadwell Foundation. The authors declare that they have no conflict of interest with the contents of this article.
- AIRE
- autoimmune regulator
- FBXO3
- F-box protein 3
- P-TEFb
- positive transcription elongation factor b
- TF
- transcription factor
- APECED
- autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy
- TSA
- tissue-specific antigen
- DNA-PK
- DNA-dependent protein kinase
- RNAPII
- RNA polymerase II
- CycT1
- cyclin T1
- TAD
- transcription activation domain
- PHD1
- plant homeodomain 1
- mTEC
- medullary thymic epithelial cell
- CUL1
- Cullin1
- RBX1
- ring-box 1
- FBXL2
- F-box and leucine-rich repeat protein 2
- HIPK2
- homeodomain-interacting protein kinase 2
- SMURF1
- SMAD-specific E3 ubiquitin protein ligase 1
- PEST
- proline, glutamic acid, serine, and threonine residues
- CIP
- calf intestinal phosphatase
- TRAF
- tumor necrosis factor receptor-associated factor
- BRD4
- bromodomain containing 4.
References
- 1. Peterson P., Org T., and Rebane A. (2008) Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat. Rev. Immunol. 8, 948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zumer K., Saksela K., and Peterlin B. M. (2013) The mechanism of tissue-restricted antigen gene expression by AIRE. J. Immunol. 190, 2479–2482 [DOI] [PubMed] [Google Scholar]
- 3. Metzger T. C., and Anderson M. S. (2011) Control of central and peripheral tolerance by Aire. Immunol. Rev. 241, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mathis D., and Benoist C. (2009) Aire. Annu. Rev. Immunol. 27, 287–312 [DOI] [PubMed] [Google Scholar]
- 5. Finnish-German APECED Consortium (1997) An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17, 399–403 [DOI] [PubMed] [Google Scholar]
- 6. Nagamine K., Peterson P., Scott H. S., Kudoh J., Minoshima S., Heino M., Krohn K. J., Lalioti M. D., Mullis P. E., Antonarakis S. E., Kawasaki K., Asakawa S., Ito F., and Shimizu N. (1997) Positional cloning of the APECED gene. Nat. Genet. 17, 393–398 [DOI] [PubMed] [Google Scholar]
- 7. Giraud M., Yoshida H., Abramson J., Rahl P. B., Young R. A., Mathis D., and Benoist C. (2012) Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 109, 535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oven I., Brdicková N., Kohoutek J., Vaupotic T., Narat M., and Peterlin B. M. (2007) AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell. Biol. 27, 8815–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Žumer K., Plemenitaš A., Saksela K., and Peterlin B. M. (2011) Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription. Nucleic Acids Res. 39, 7908–7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Žumer K., Low A. K., Jiang H., Saksela K., and Peterlin B. M. (2012) Unmodified histone H3K4 and DNA-dependent protein kinase recruit autoimmune regulator to target genes. Mol. Cell. Biol. 32, 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liiv I., Rebane A., Org T., Saare M., Maslovskaja J., Kisand K., Juronen E., Valmu L., Bottomley M. J., Kalkkinen N., and Peterson P. (2008) DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim. Biophys. Acta 1783, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giraud M., Jmari N., Du L., Carallis F., Nieland T. J., Perez-Campo F. M., Bensaude O., Root D. E., Hacohen N., Mathis D., and Benoist C. (2014) An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc. Natl. Acad. Sci. U.S.A. 111, 1491–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abramson J., Giraud M., Benoist C., and Mathis D. (2010) Aire's partners in the molecular control of immunological tolerance. Cell 140, 123–135 [DOI] [PubMed] [Google Scholar]
- 14. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., and Young R. A. (2010) c-Myc regulates transcriptional pause release. Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peterlin B. M., and Price D. H. (2006) Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 [DOI] [PubMed] [Google Scholar]
- 16. Geng F., Wenzel S., and Tansey W. P. (2012) Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81, 177–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muratani M., and Tansey W. P. (2003) How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 18. Kurosu T., and Peterlin B. M. (2004) VP16 and ubiquitin: binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr. Biol. 14, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 19. Akiyoshi H., Hatakeyama S., Pitkänen J., Mouri Y., Doucas V., Kudoh J., Tsurugaya K., Uchida D., Matsushima A., Oshikawa K., Nakayama K. I., Shimizu N., Peterson P., and Matsumoto M. (2004) Subcellular expression of autoimmune regulator is organized in a spatiotemporal manner. J. Biol. Chem. 279, 33984–33991 [DOI] [PubMed] [Google Scholar]
- 20. Meloni A., Meloni A., Fiorillo E., Corda D., Incani F., Serra M. L., Contini A., Cao A., and Rosatelli M. C. (2010) DAXX is a new AIRE-interacting protein. J. Biol. Chem. 285, 13012–13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uchida D., Hatakeyama S., Matsushima A., Han H., Ishido S., Hotta H., Kudoh J., Shimizu N., Doucas V., Nakayama K. I., Kuroda N., and Matsumoto M. (2004) AIRE functions as an E3 ubiquitin ligase. J. Exp. Med. 199, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bottomley M. J., Stier G., Pennacchini D., Legube G., Simon B., Akhtar A., Sattler M., and Musco G. (2005) NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J. Biol. Chem. 280, 11505–11512 [DOI] [PubMed] [Google Scholar]
- 23. Kainulainen M., Habjan M., Hubel P., Busch L., Lau S., Colinge J., Superti-Furga G., Pichlmair A., and Weber F. (2014) Virulence factor NSs of rift valley fever virus recruits the F-box protein FBXO3 to degrade subunit p62 of general transcription factor TFIIH. J. Virol. 88, 3464–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eifler T. T., Shao W., Bartholomeeusen K., Fujinaga K., Jäger S., Johnson J. R., Luo Z., Krogan N. J., and Peterlin B. M. (2015) Cyclin-dependent kinase 12 increases 3′ end processing of growth factor-induced c-FOS transcripts. Mol. Cell. Biol. 35, 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Org T., Rebane A., Kisand K., Laan M., Haljasorg U., Andreson R., and Peterson P. (2009) AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum. Mol. Genet. 18, 4699–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Org T., Chignola F., Hetényi C., Gaetani M., Rebane A., Liiv I., Maran U., Mollica L., Bottomley M. J., Musco G., and Peterson P. (2008) The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee M., Ji H., Furuta Y., Park J. I., and McCrea P. D. (2014) p120-catenin regulates REST and CoREST, and modulates mouse embryonic stem cell differentiation. J. Cell Sci. 127, 4037–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seiden-Long I. M., Brown K. R., Shih W., Wigle D. A., Radulovich N., Jurisica I., and Tsao M. S. (2006) Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene 25, 91–102 [DOI] [PubMed] [Google Scholar]
- 29. Chen Z. W., Liu B., Tang N. W., Xu Y. H., Ye X. Y., Li Z. M., Niu X. M., Shen S. P., Lu S., and Xu L. (2014) FBXL5-mediated degradation of single-stranded DNA-binding protein hSSB1 controls DNA damage response. Nucleic Acids Res. 42, 11560–11569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y., Zhou Z., Alimandi M., and Chen C. (2009) WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer. Oncogene 28, 2948–2958 [DOI] [PubMed] [Google Scholar]
- 31. Chen B. B., Coon T. A., Glasser J. R., McVerry B. J., Zhao J., Zhao Y., Zou C., Ellis B., Sciurba F. C., Zhang Y., and Mallampalli R. K. (2013) A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat. Immunol. 14, 470–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mallampalli R. K., Coon T. A., Glasser J. R., Wang C., Dunn S. R., Weathington N. M., Zhao J., Zou C., Zhao Y., and Chen B. B. (2013) Targeting F box protein Fbxo3 to control cytokine-driven inflammation. J. Immunol. 191, 5247–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shima Y., Shima T., Chiba T., Irimura T., Pandolfi P. P., and Kitabayashi I. (2008) PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol. Cell. Biol. 28, 7126–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D., Xie P., Zhao F., Shu J., Li L., Zhan Y., and Zhang L. (2015) F-box protein Fbxo3 targets Smurf1 ubiquitin ligase for ubiquitination and degradation. Biochem. Biophys. Res. Commun. 458, 941–945 [DOI] [PubMed] [Google Scholar]
- 35. Björses P., Pelto-Huikko M., Kaukonen J., Aaltonen J., Peltonen L., and Ulmanen I. (1999) Localization of the APECED protein in distinct nuclear structures. Hum. Mol. Genet. 8, 259–266 [DOI] [PubMed] [Google Scholar]
- 36. Rattay K., Claude J., Rezavandy E., Matt S., Hofmann T. G., Kyewski B., and Derbinski J. (2015) Homeodomain-interacting protein kinase 2, a novel autoimmune regulator interaction partner, modulates promiscuous gene expression in medullary thymic epithelial cells. J. Immunol. 194, 921–928 [DOI] [PubMed] [Google Scholar]
- 37. Laan M., and Peterson P. (2013) The many faces of aire in central tolerance. Front. Immunol. 4, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anderson M. S., Venanzi E. S., Klein L., Chen Z., Berzins S. P., Turley S. J., von Boehmer H., Bronson R., Dierich A., Benoist C., and Mathis D. (2002) Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 39. Su M. A., Giang K., Zumer K., Jiang H., Oven I., Rinn J. L., Devoss J. J., Johannes K. P., Lu W., Gardner J., Chang A., Bubulya P., Chang H. Y., Peterlin B. M., and Anderson M. S. (2008) Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest. 118, 1712–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rogers S., Wells R., and Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368 [DOI] [PubMed] [Google Scholar]
- 41. Koh A. S., Kuo A. J., Park S. Y., Cheung P., Abramson J., Bua D., Carney D., Shoelson S. E., Gozani O., Kingston R. E., Benoist C., and Mathis D. (2008) Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 105, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skaar J. R., Pagan J. K., and Pagano M. (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Unanue E. R. (2014) Antigen presentation in the autoimmune diabetes of the NOD mouse. Annu. Rev. Immunol. 32, 579–608 [DOI] [PubMed] [Google Scholar]
- 44. Yoshida H., Bansal K., Schaefer U., Chapman T., Rioja I., Proekt I., Anderson M. S., Prinjha R. K., Tarakhovsky A., Benoist C., and Mathis D. (2015) Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc. Natl. Acad. Sci. U.S.A. 112, E4448–E4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bartholomeeusen K., Xiang Y., Fujinaga K., and Peterlin B. M. (2012) Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 287, 36609–36616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chuprin A., Avin A., Goldfarb Y., Herzig Y., Levi B., Jacob A., Sela A., Katz S., Grossman M., Guyon C., Rathaus M., Cohen H. Y., Sagi I., Giraud M., McBurney M. W., Husebye E. S., and Abramson J. (2015) The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nature Immunology 16, 737–745 [DOI] [PubMed] [Google Scholar]
- 47. Waterfield M., Khan I. S., Cortez J. T., Fan U., Metzger T., Greer A., Fasano K., Martinez-Llordella M., Pollack J. L., Erle D. J., Su M., and Anderson M. S. (2014) The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat. Immunol. 15, 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]