Abstract
Efficacy of allergen-specific immunotherapy is often severely impaired by detrimental IgE-mediated side effects of native allergen during vaccination. Here, we present the molecular determinants for IgE recognition of Rhi o 1 and eventually converting the allergen into a hypoallergenic immunogen to restrain health hazards during desensitization. Rhi o 1 is a respiratory fungal allergen. Despite having cross-reactivity with cockroach allergen, we observed that non-cross-reactive epitope predominantly determined IgE binding to Rhi o 1. Denaturation and refolding behavior of the allergen confirmed that its IgE reactivity was not essentially conformation-dependent. A combinatorial approach consisting of computational prediction and a peptide-based immunoassay identified two peptides (44TGEYLTQKYFNSQRNN and 311GAEKNWAGQYVVDCNK) of Rhi o 1 that frequently reacted with IgE antibodies of sensitized patients. Interestingly, these peptides did not represent purely linear IgE epitopes but were presented in a conformational manner by forming a spatially clustered surface-exposed epitope conferring optimal IgE-binding capacity to the folded allergen. Site-directed alanine substitution identified four residues of the IgE epitope that were crucial for antibody binding. A multiple mutant (T49A/Y52A/K314A/W316A) showing 100-fold lower IgE binding and reduced allergenic activity was generated. The TYKW mutant retained T-cell epitopes, as evident from its lymphoproliferative capacity but down-regulated pro-allergic IL-5 secretion. The TYKW mutant induced enhanced focusing of blocking IgG antibodies specifically toward the IgE epitope of the allergen. Anti-TYKW mutant polyclonal IgG antibodies competitively inhibited binding of IgE antibodies to Rhi o 1 up to 70% and suppressed allergen-mediated histamine release by 10-fold. In conclusion, this is a simple yet rational strategy based on epitope mapping data to develop a genetically modified hypoallergenic variant showing protective antibody response for immunotherapeutic applications.
Keywords: allergen, epitope mapping, immunoglobulin E (IgE), immunotherapy, mutagenesis, vaccine, asthma, mold allergy
Introduction
IgE-mediated allergic sensitization to indoor allergens is a serious health problem with worldwide prevalence. Fungal spores and fragments are major contributors of indoor allergens (1, 2). Rhizopus sp. is a common airborne fungus that elicits hypersensitive reactions in the respiratory tract upon inhalation (3, 4). For several years, growing investigations have supported the association of particular species of Rhizopus with bronchial asthma and other types of allergy (5–10). In India, emerging clinical and immunological studies have suggested the involvement of Rhizopus oryzae allergens in type I allergy (11–13). Rhi o 1 is a major allergen (aspartic protease, molecular mass 44 kDa) of R. oryzae, which causes respiratory sensitization to mold allergic patients (14). This allergen has been officially recognized and listed in the database of the International Union of Immunological Societies. Recombinant Rhi o 1 (rRhi o 1) was identified as a potential candidate for component-resolved diagnosis of mold allergy. In addition to diagnosis, purified allergens are also used for provocative vaccination in allergen-specific immunotherapy, which is considered as the long lasting and disease modifying approach in allergy treatment (15). A disadvantage for allergen-specific immunotherapy with a native allergen is the chance of severe side effects, such as anaphylaxis. This can be overcome by strategic modification of allergen-encoding genes, in which the allergen will evolve into a “safe hypoallergenic vaccine.” Various molecular strategies have been adopted to formulate safe vaccines against common environmental allergens (16–20). However, possibly there has been no report on a hypoallergenic vaccine against fungal allergens. The present study has been conducted for Rhi o 1 due to its close association with the prevalence and severity of the fungal allergy, especially among Indian patients. Our overall aim was to generate a genetically modified Rhi o 1 with reduced allergenicity but unaltered immunogenicity. We assumed that this would be beneficial regarding safe administration and mounting protective antibody response against the original allergen. As a first step, mapping of the IgE epitope and its critical residues was performed. We observed the low degree of immunological relevance of an earlier reported (14) cross-reactive IgE epitope shared by Rhi o 1 and the cockroach allergen Bla g 2. Instead, we identified two novel IgE-binding peptides that together constitute a major epitope of Rhi o 1. This epitope was predominantly involved in the immunorecognition of this allergen. Subsequently, based on epitope mapping data, we developed a mutated hypoallergenic molecule that strongly induced blocking IgG antibody response against WT Rhi o 1.
Results
IgE Antibody Reactivity Pattern of Rhi o 1 and Bla g 2 among R. oryzae-sensitized Patients
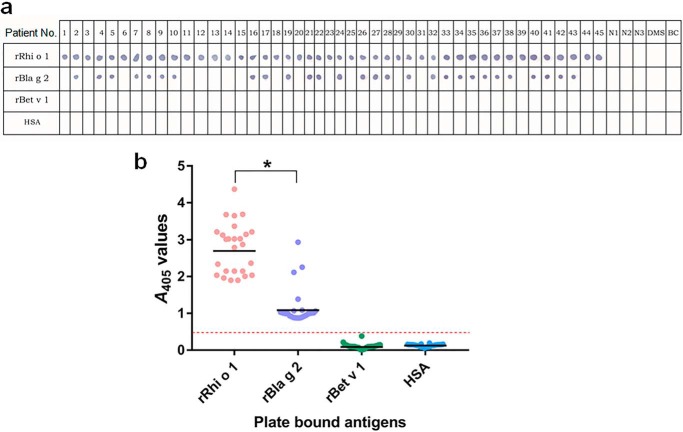
Co-sensitization to multiple allergens having conserved molecular surfaces can often give rise to unwanted cross-reactivity of low clinical importance. Here, we studied the significance of an earlier reported (14) cross-reactivity between rRhi o 1 and rBla g 2 by using sera from a population of 45 R. oryzae-sensitized patients. As observed in an IgE dot blot (Fig. 1a), 17 of 45 patients (38%) showed strong reactions with rRhi o 1 but not with rBla g 2. Hence, these 17 patients had no detectable IgE antibodies cross-reacting between rRhi o 1 and rBla g 2. The remainder of the 28 patients were found to react positively with rRhi o 1 as well as rBla g 2. However, in ELISA with these 28 patients' sera (Fig. 1b), the intensity of IgE antibody binding to rRhi o 1 was on average ∼2.45-fold higher (p < 0.001) than to rBla g 2. Such a frequently strong IgE binding to Rhi o 1 suggests that IgE recognition of this allergen principally depends on non-cross-reactive epitope(s) and is not significantly influenced by the presence of cross-reactive epitope in these two less conserved allergens (28% sequence identity).
FIGURE 1.
IgE reactivity of Rhi o 1 and Bla g 2 among R. oryzae allergic patients. a, nitrocellulose strips were dotted with recombinant allergens and negative controls (rBet v 1 and HSA). IgE immunoblotting was done with 45 patients' sera (1–45), three healthy sera (N1–N3), a dust mite-allergic serum (DMS), and buffer control (BC). 28 of 45 sera displayed IgE reactivity to rRhi o 1 as well as rBla g 2. b, in ELISA, these 28 sera were used to determine the bound IgE level (A values; y axis) to the allergens (x axis). In the scatter plot, the horizontal bar represents a geometric mean of A values against each antigen. *, p < 0.001. Dotted line, cut-off level of IgE binding.
IgE Recognition of rRhi o 1 under Denaturing Conditions
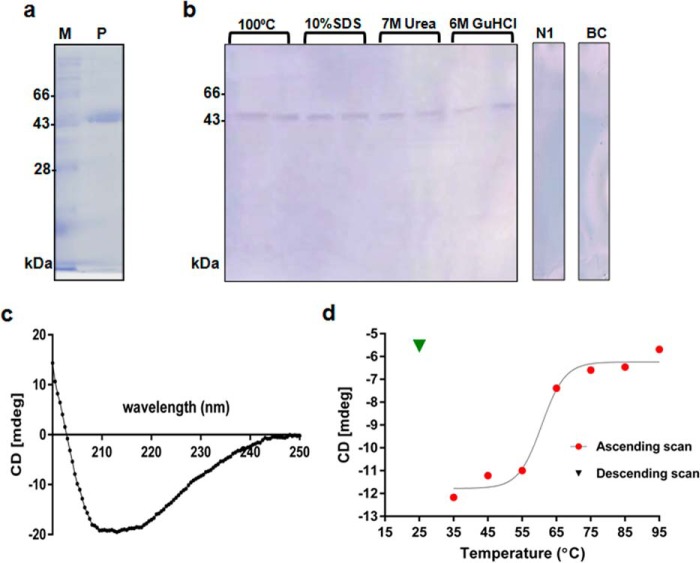
We tested the IgE-binding ability of rRhi o 1 after being treated with four denaturing agents (boiling temperature, anionic detergent, and chaotropes). Under any denaturing condition, rRhi o 1 was able to react with IgE antibodies (Fig. 2, a and b). Therefore, IgE binding to Rhi o 1 may not essentially depend on its native folds.
FIGURE 2.
IgE antibody binding to denatured Rhi o 1. a, Ni-NTA-purified rRhi o 1 in 12% SDS-PAGE (lane P) and molecular mass marker (lane M). b, IgE-Western blotting of rRhi o 1 under four denaturing conditions using a pool of patients' sera, healthy serum (N1), and buffer control (BC). c, CD spectrum of rRhi o 1 at 20 °C. d, thermal denaturation curve showing gradual changes in ellipticity signal at 215 nm (CD millidegrees at λmax; y axis) due to rRhi o1 unfolding with increasing temperatures (ascending scan) from 35 to 95 °C (x axis). Heat-denatured rRhi o 1 did not fold back to native conformation (no increase in ellipticity signal) after cooling down (descending scan) from 95 to 25 °C.
Thermal Denaturation and Refolding Behavior of rRhi o 1
CD spectrum of rRhi o 1 (Fig. 2c) at 20 °C represented a correctly folded protein with predominantly β-sheet contents (56%), and the minimum was obtained at 215 nm (λmax). The thermal denaturation curve of rRhi o 1 based on negative ellipticity signals at 215 nm during ascending scan (from 35 to 95 °C) is shown in Fig. 2d. Transition temperature (Tm) was observed at ∼60 °C, at which unfolding took place. The protein lost most of its native folds at ∼95 °C. After cooling down (descending scan), there was no significant increase in the CD signal at 215 nm, indicating that rRhi o 1 did not refold upon cooling. This result led us to hypothesize that rRhi o 1 remained denatured on the membrane during Western blotting, and IgE binding under this condition could only be possible principally with continuous sequences.
Two IgE Antibody-reactive Peptides of Rhi o 1
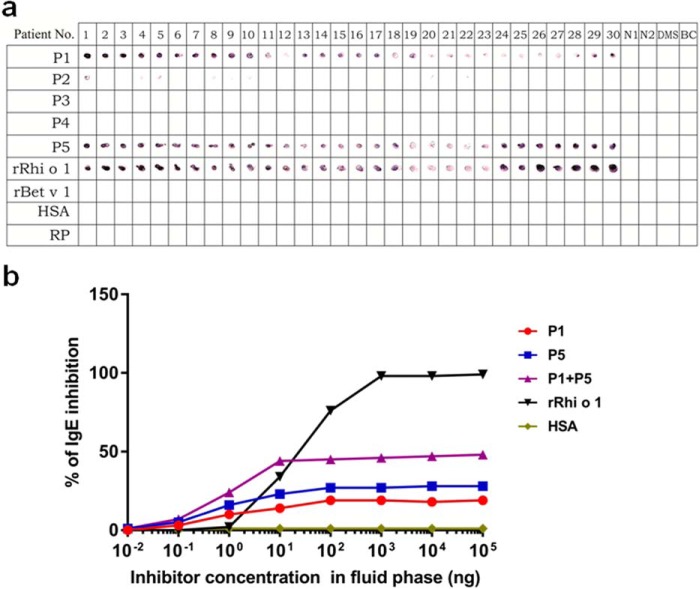
The IgE epitope was identified by experimental validation of the results generated by computational prediction as summarized in Table 1. Two B-cell epitope prediction servers were simultaneously used that predicted five 16-mer peptides (P1–P5) as high scoring antigenic regions of Rhi o 1. These peptides were chemically synthesized for confirmatory immunoassays. For preliminary screening of IgE reactivity, five synthetic peptides were immobilized as plate-bound antigens, and ELISA was performed with pooled sera. The threshold value was set as 15 times higher A405 than negative controls (i.e. human serum albumin (HSA)4 and random peptide with mean A405 values of 0.055 ± 0.003). P1 and P5 displayed >15 times higher IgE binding than negative controls. The highest IgE binding was observed for P5 (A405 = 1.090 ± 0.011) followed by P1 (A405 = 0.883 ± 0.030). P2, P3, and P4 displayed on average >20 times lower IgE binding than positive control (i.e. intact rRhi o 1 with A405 = 2.041 ± 0.014). The ELISA data were cross-verified by dot blotting using 30 individual sera. As shown in Fig. 3a, all of the sera reacted with only P1 and P5 as strongly as they did with intact rRhi o 1. The intensity of IgE reactivity was conspicuously high, suggesting that these two peptides constituted the immunodominant IgE epitope of Rhi o 1.
TABLE 1.
Identification of IgE antibody-binding peptides of Rhi o 1 allergen
Critical residues selected for mutagenesis are also listed. IgE binders are shown with asterisks. Prediction score, confidence score of ABCpred server (threshold >0.5); Apos., A405 value of positive control (intact rRhi o1); Apep., A405 value of individual peptides (P1–P5); Aneg., A405 value of negative controls (HSA and random peptide); ELISA results (A405 values of bound IgEs) are the means of triplicate determinations with variations <5%.
| Peptide no. | Sequence of putative B-cell epitopes predicted by the servers | Prediction score | IgE binding (ELISA) data |
Mutations in critical residues | |
|---|---|---|---|---|---|
| Apos./Apep. | Apep./Aneg. | ||||
| *P1 | 44TGEYLTQKYFNSQRNN | 0.96 | 2.3 | 16.0 | T49A/Y52A |
| P2 | 90EIEIGTPPQPFTVVFD | 0.95 | 22.0 | 1.8 | |
| P3 | 252EGDIHWSDVRRKGYWE | 0.95 | 25.5 | 1.7 | |
| P4 | 244GGVDEDHFEGDIHWSD | 0.89 | 29 | 1.4 | |
| *P5 | 311GAEKNWAGQYVVDCNK | 0.87 | 1.9 | 19.8 | K314A/W316A |
FIGURE 3.
Immunoscreening of IgE-binding peptides. a, nitrocellulose strips were dotted with synthetic peptides (P1–P5), positive control (intact rRhi o 1), and negative controls (rBet v 1, HSA, and random peptide (RP)). IgE-immunoblotting was done with 30 patients' sera (1–30), two healthy sera (N1 and N2), a dust mite allergic serum (DMS), and buffer control (BC). b, competitive ELISA, in which plate-bound rRhi o 1 was exposed to a sera pool preincubated with increasing doses (x axis) of peptides (test inhibitors), rRhi o 1 (autoinhibitor), and HSA (non-inhibitor). Antibody binding strength of individual peptides (P1 or P5) or in combination (P1 + P5; equimolar mix) is represented by their respective percentages of IgE inhibition (y axis) to immobilized rRhi o 1.
The IgE-binding Capacity of P1 and P5
In a competitive ELISA, the antibody binding strength of the IgE-reactive peptides (in fluid phase) was quantified as a measure of their respective capacity to inhibit IgE binding to intact rRhi o 1 (in solid phase). Peptides were preincubated with IgE antibodies from a pool of six patients' sera. These sera were reactive to only rRhi o 1 and not to rBla g 2. By using these sera, we precluded the interference of immunologically insignificant IgE epitope responsible for cross-reactivity between Rhi o 1 and Bla g 2. This, in turn, helped us to understand the precise contribution of P1 and P5 to determine the immunoreactivity of Rhi o 1. As shown in Fig. 3b, individual peptide (either P1 or P5) did not attain the IC50 value (i.e. 50% IgE inhibition to immobilized rRhi o 1) at its highest concentration (105 ng in 100 μl of serum (i.e. 1 mg/ml)). Maximum IgE inhibitions by P1 and P5 were ∼19 and ∼28%, respectively. Interestingly, complete IgE inhibition (i.e. ∼99%, as displayed by autoinhibitor rRhi o 1) was not observed even when these two peptides were simultaneously used (P1 + P5) as fluid phase inhibitors. Both of the peptides collectively displayed a maximum IgE binding inhibition to rRhi o 1 of up to ∼48%. Similar IgE inhibition patterns were observed when peptides were preincubated with individual patients' serum instead of pooled sera (supplemental Fig. S1). Hence, P1 and P5, as free peptides in the fluid phase, were unable to capture the full repertoire of Rhi o 1-specific free IgE antibodies of the patients' sera. Consequently, IgE binding to plate-bound rRhi o 1 was only partially inhibited. This partial inhibition can be explained by the fact that these two linear peptides might not represent explicitly linear IgE epitopes. It also suggested the likely presence of particular IgE antibodies in the patients' sera specifically directed toward the topological conformations of P1 and P5 on the native folds of Rhi o 1 protein.
Spatial Clustering of P1 and P5 on the Allergen Surface
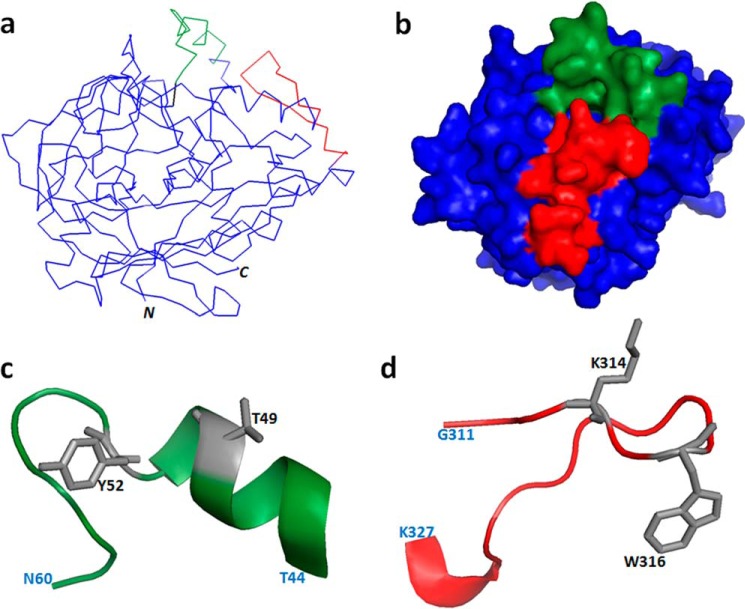
Based on the homology model of Rhi o 1, peptides P1 and P5 were mapped on the solvent-accessible surface of the allergen, which formed an electrostatically active molecular domain responsible for IgE binding (Fig. 4, a and b). In this domain, P1 and P5 assumed helix coil and coil helix conformations, respectively. P1 and P5 were two independent sequences distantly located on N- and C-terminal ends of Rhi o 1, respectively. However, in the folded conformation of the tertiary structure, these peptides were found to come in proximity, forming a contiguous antigenic cluster. In this cluster, the side chain-accessible surface area of these two peptides comprised up to 2181.19 Å2 (i.e. 12% of the total solvent-accessible surface area of Rhi o 1 model).
FIGURE 4.
Mapping of IgE epitope. IgE-reactive peptides P1 (green) and P5 (red) mapped on Rhi o 1 homology model (blue) represented as α-carbon backbone structure (a) and space-filled model (b). Shown are enlarged schematic representations of the conformation of P1 (c) and P5 (d) on a Rhi o 1 three-dimensional model. Critical residues (gray side chains) and terminal residues (light blue) are labeled.
Critical Residues of IgE Epitopes and Design of Mutagenesis
The rationale behind identifying the critical residues in each of the two IgE-reactive peptides of the epitope was to change selected amino acid residue(s) showing (i) high surface accessibility as well as antigenic propensity values and (ii) reported involvement in antigen-antibody interaction, as evident from published literature. Four residues (two from each peptide) were selected as potential targets of mutations as listed in Table 1, and their respective locations on IgE epitope are shown in Fig. 4, c and d. Polar residues (Thr49) tend to cluster in the antigen-antibody interface to increase binding affinity (21), whereas charged residues (Lys314) reside in the edge and exclude water from the interior of the antigen-antibody interface, thereby stabilizing electrostatic interaction (22). Aromatic residues (Tyr52 and Trp316) with bulky side chains participate in networks of cooperative interactions with hydrophobic, charged, and hydrogen bond donor/acceptor groups of interacting antibody (23). Mutation of these selected residues to alanine in silico, resulted in a reduction of antigenicity and surface accessibility index (data not shown). In ELISA and immuno-dot blotting, synthetic mutant peptides containing alanine substitutions (mP1 and mP5) displayed significantly (p < 0.001) reduced reactivity with IgE antibody (Fig. 5, a and b). This observation implicated the crucial role of these residues in mediating the interaction between IgE antibodies and the corresponding epitope. To study the effect of these mutations on IgE binding to an intact allergen, sequential point substitutions were induced in identified IgE-reactive regions of the intact Rhi o 1. Two double mutants (P1-mut and P5-mut, each with two substitutions on P1 and P5, respectively) and a multiple mutant (TYKW-mut with four substitutions on both of the peptides) were generated. Purified mutant proteins (Fig. 6a) had a folding pattern similar to that of WT rRhi o 1, as evident from their CD spectra (supplemental Fig. S2).
FIGURE 5.
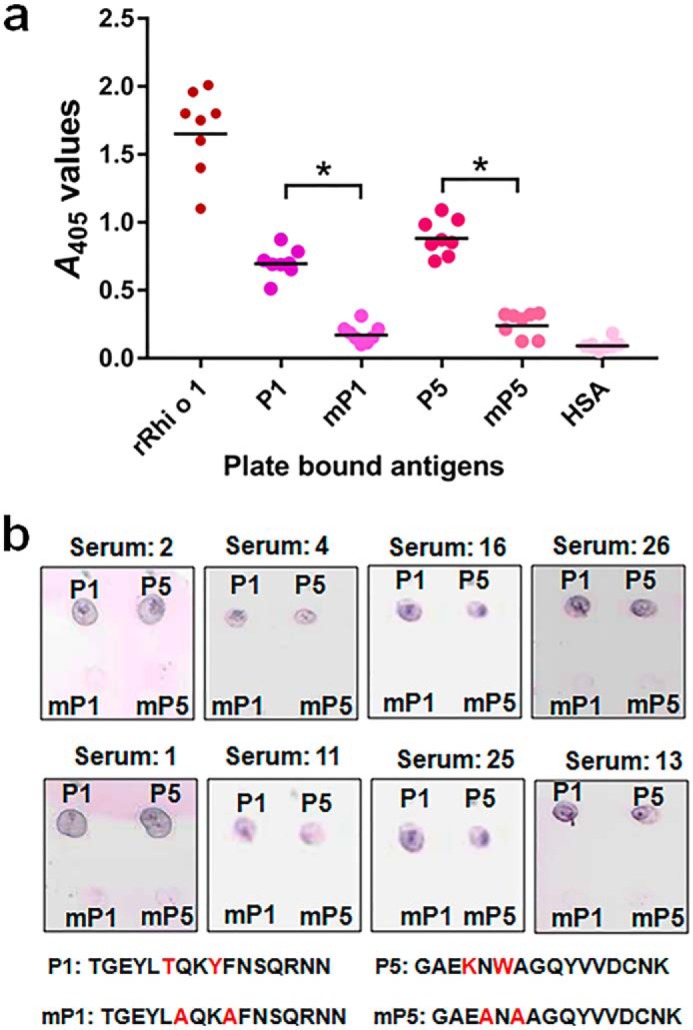
Critical residues of IgE epitope. a, in ELISA, synthetic mutant peptides (mP1 and mP5) with selected residues changed into alanine displayed significantly reduced (*, p < 0.001) IgE binding (A values; y axis) from patients' sera (n = 8) as compared with corresponding equimolar WT peptides. Equimolar rRhi o 1 and HSA were used as positive and negative controls, respectively. Horizontal bars in the scatter plot represent geometric means of A values against each antigen (x axis). b, reconfirmation of reduced IgE binding by immuno-dot blotting with these eight sera. Sequences of original and mutant peptides are shown with changed residues marked in red.
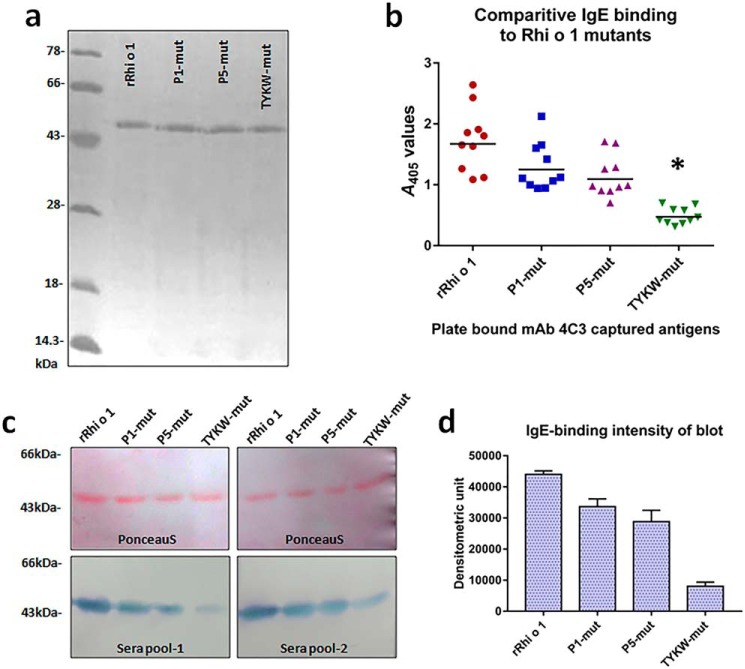
FIGURE 6.
IgE reactivity of Rhi o 1 mutants. a, purified rRhi o 1 and its mutants in silver-stained 12% SDS-PAGE. b, scatter plot showing reactivity of mAb4C3-captured Rhi o 1 and its mutants (x axis) to IgE antibody (A values; y axis) from patients' sera (n = 10) in sandwich ELISA. Horizontal bars, geometric means of A values against each antigen. *, p < 0.001 (i.e. significant difference in IgE binding to TYKW-mut versus WT rRhi o 1). c, top, confirmation of equal loading (2 μg) of rRhi o 1 and its mutants on PVDF membrane by Ponceau S staining followed by IgE immunoblotting (bottom) with two sera pools. d, comparison of IgE-reactive band intensity (mean of densitometric units ± S.D. (error bars); y axis) of rRhi o 1 and mutants (x axis) in immunoblots with patients' sera pools (n = 2).
Reduction in IgE Binding to Rhi o 1 Mutants
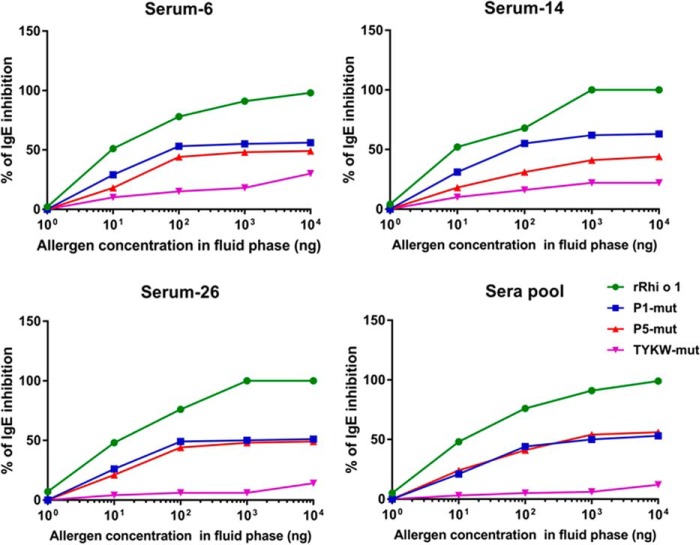
Mutants were variously tested for any change in the IgE-binding capacity as compared with WT rRhi o 1 by comparative immunoassays using patients' sera. For sandwich ELISA, rRhio 1 mutants were first presented in a defined orientation on mAb4C3 used as a capture antibody, followed by the addition of patients' sera. The idea of presenting Rhi o 1 on mAb4C3 was taken from a recently published work on Bla g 2 (24). The mAb4C3 was originally developed against a conformational IgE epitope of Bla g 2. Interestingly, it was observed in our earlier study (14) that Rhi o 1 also had a 4C3 binding site, and this site overlapped with a conformational IgE epitope of this allergen. In the present study, the 4C3 binding site remained intact because there was no mutation induced on it. Therefore, mAb4C3 binding to Rhi o 1 would exclude IgE binding to the corresponding epitope. It would help to understand the effect of mutations on IgE antibody binding to the rest of the allergen. As shown in Fig. 6b, all of the three mutants displayed a decrease in IgE binding to different extents. The highest percentage of reduction (∼93%) was observed for TYKW-mut as compared with WT rRhi o 1. In the next step, equal amounts of rRhi o 1 and its mutants were IgE-immunoblotted with two sets of pooled sera (Fig. 6c). Band densitometric analysis of IgE binding intensity (Fig. 6d) revealed the TYKW-mut displaying the highest percentage (∼88%) of reduction in IgE binding, which was in agreement with the results of the ELISA experiment. Next, we performed competitive ELISA to compare the capacity of three mutants (as fluid phase inhibitors) to inhibit IgE binding to plate-bound rRhi o 1 (Fig. 7). In all of the four sera, it was observed that TYKW-mut did not reach an IC50 value at a concentration as high as 104 ng in 100 μl of each serum (i.e. 100 μg/ml). On the contrary, autoinhibitor rRhi o 1 exhibited ∼99% inhibition at only 10 μg/ml concentration. Hence, TYKW-mut among the three mutants displayed >100-fold reduction in competitive IgE-binding capacity. These results also indicated the combined role of P1 and P5 in constituting a dominant IgE epitope on Rhi o 1 protein. Finally, we selected TYKW-mut to study further any change in its allergenic activity because it had a maximum number of mutations and consequently the highest reduction in IgE-binding capacity.
FIGURE 7.
Comparative IgE-binding capacity of rRhi o 1 mutants by competitive ELISA. Plate-bound mAb4C3-captured rRhi o 1 was exposed to three individual sera and one pooled serum separately preincubated with increasing doses (x axis) of mutants and rRhi o 1 (autoinhibitor). TYKW-mut displayed the lowest percentage of IgE inhibition (y axis) to immobilized rRhi o 1.
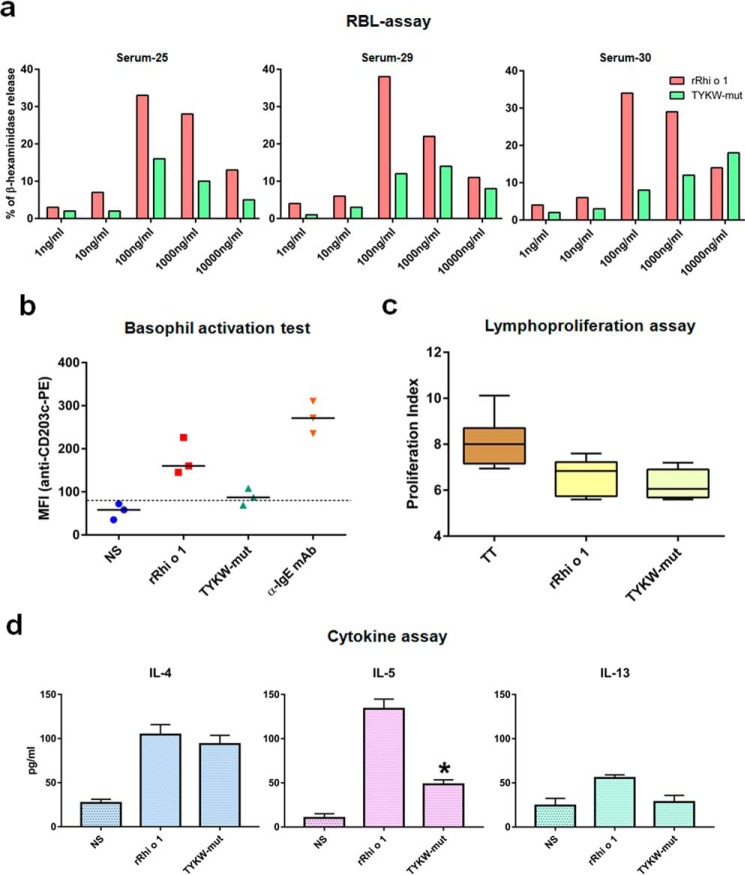
TYKW-mut Displayed Reduced Capacity of Basophil Activation
In a degranulation assay, IgE-sensitized rat basophil leukemia (RBL) cells were challenged with different concentrations of allergens (Fig. 8A). In the case of rRhi o 1, typical dose-dependent degranulation was observed. The highest release (∼35%) of β-hexaminidase took place at 100 ng/ml allergen, and then a gradual decrease in degranulation was observed at higher concentrations. On the other hand, 2–3 times lower inflammatory release was observed at corresponding concentrations of the TYKW-mut. Only for one serum (number 30) did TYKW-mut show maximum degranulation up to 18% at its highest concentration (10,000 ng/ml). In addition to the mediator release assay, we also analyzed the expression level of CD203c, a surface marker of basophil activation, by flow cytometry (Fig. 8b). Basophils from three patients displayed on average 3.2- and 4.9-fold stimulation of CD203c expression upon challenge with rRhi o 1 and anti-IgE, respectively, as compared with non-stimulated control. In the case of TYKW-mut challenge, the stimulation was ∼2 times lower than rRhi o 1 and almost at the threshold level of non-stimulated expression. Because of this reduced allergenic activity, TYKW-mut was considered as a hypoallergenic derivative of Rhi o 1.
FIGURE 8.
Allergenic activity and T-cell reactivity of TYKW-mut. a, mediator release (percentage; y axis) from RBL cells IgE-sensitized with three different sera and challenged with different concentrations (x axis) of rRhi o 1 or TYKW-mut. b, scatter plot shows CD203c expression (mean fluorescence intensity; y axis) on basophils from patients (n = 3) induced with different stimuli or buffer (NS) in x axis. Horizontal bar, geometric mean of mean fluorescence intensities for each stimulus; dotted line, cut-off level of expression. c, PBMCs from patients (n = 6) were stimulated with either allergens or tetanus toxoid (TT; positive control) (x axis). The lymphocyte proliferation index (y axis) is shown as box-whisker plots with median and interquartile ranges. d, cytokine concentrations (mean ± S.D. (error bars); y axis) in PBMC culture supernatants of the patients (n = 4) stimulated with antigens or medium (NS) (x axis). *, p < 0.001 for IL-5 induction by TYKW-mut versus rRhi o 1.
TYKW-mut Retained T-cell Responses
Despite having diminished allergenicity, TYKW-mut exhibited T-cell reactivity, as analyzed by the lymphoproliferative response and Th2 cytokine profile in the allergen-stimulated peripheral blood mononuclear cell (PBMC) cultures. As illustrated in Fig. 8c, the proliferation index of TYKW-mut was almost similar to that of rRhi o 1 (p value = 0.4778 (i.e. insignificant difference)) in the MTT assay. The Th2 cytokine profile of stimulated cultures is shown in Fig. 8d. Stimulation with rRhi o 1 resulted in increased production of three pro-allergic cytokines as compared with non-stimulated cultures. We observed the significantly lower level of secreted IL-5 in TYKW-mut-stimulated culture as compared with rRhi o 1. No significant difference was observed in IL-4 and IL-13 levels between the two antigens.
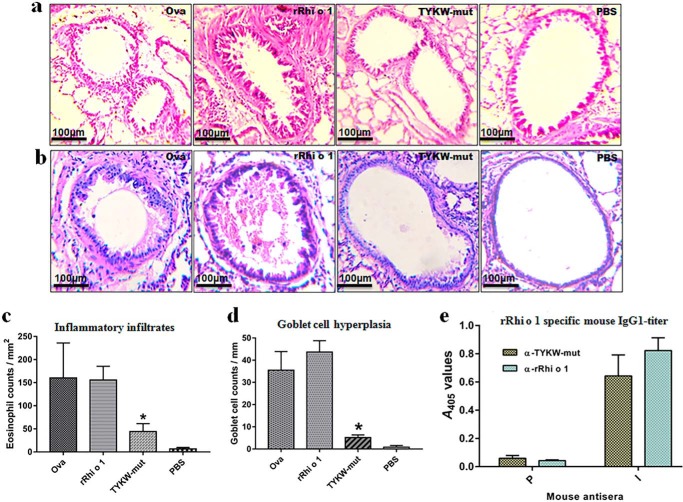
TYKW-mut Elicited a Less Allergic Reaction in Vivo but Induced Rhi o 1-specific IgG Responses in Animal Models
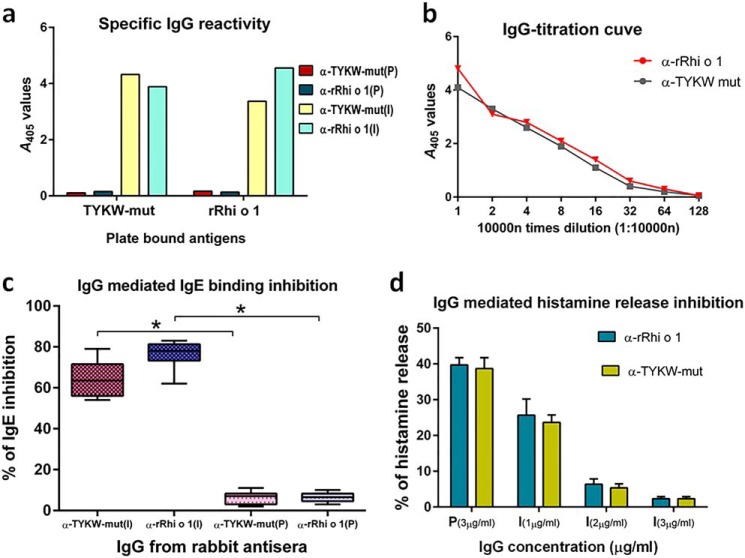
Lung histopathology of rRhi o 1-challenged mice revealed severe allergic inflammation characterized by extensive infiltration of eosinophils in peribronchial tissues and hyperplasia of mucous-secreting goblet cells on the inner linings of bronchial basement membranes. In comparison with rRhi o 1, the TYKW-mut-challenged lungs displayed histological evidence of less pronounced allergic reactions, as illustrated in Fig. 9, a–d. Immunization of animals with either TYKW-mut or rRhi o 1 gave rise to the corresponding antiserum designated as α-TYKW-mut and α-rRhi o 1, respectively. As shown in Fig. 9e, mouse α-rRhi o 1 had high specific IgG1 titer against rRhi o 1. Interestingly, α-TYKW-mut IgG1 also recognized plate-bound rRhi o 1 in ELISA. For further investigations, which required a larger volume of antisera, immunization in a rabbit model was performed. As shown in Fig. 10a, IgGs from α-TYKW-mut and α-rRhi o 1 rabbit antisera were able to cross-recognize the plate-bound rRhi o 1 and TYKW-mut, respectively, with almost identical specificity. In a comparative titration experiment (Fig. 10b), α-TYKW-mut rabbit antiserum was found to contain a high titer of IgG specific for rRhi o 1 (recognized even at 1:160,000 dilution), and these titers under different dilutions were nearly identical to that of α-rRhi o 1.
FIGURE 9.
Allergic reaction and immunogenic response of TYKW-mut. Representative lung histology after H&E (a) and PAS staining (b) from four groups of intranasally challenged BALB/c mice (n = 6/group). Shown are eosinophil counts (c) in the peribronchial area (mm2) and PAS-positive goblet cell counts (d) along the length (mm) of bronchial basement membrane (mean of cell counts ± S.D. (error bars); y axis) from different treatment groups (x axis). *, p < 0.001 in TYKW-mut versus rRhi o 1. e, ELISA showing plate-bound rRhi o 1-specific IgG1 titers (mean of A values ± S.D.; y axis) in antisera (x axis) from mice (n = 3) immunized with TYKW-mut or rRhi o 1. P and I, preimmune and immune sera, respectively.
FIGURE 10.
Anti-TYKW-mut IgG-antibodies with blocking effects on Rhi o 1 allergen. P and I, rabbit preimmune and immunized sera, respectively. a, ELISA showing plate-bound antigen (x axis) specificity of IgG (A values; y axis) in rabbit preimmune and immunized sera (α-rRhi o 1 and α-TYKW-mut). b, titration curve showing rRhi o 1-specific IgG-titers (A values; y axis) in two antisera at different dilutions (x axis). c, percentage of inhibition of patients' IgE antibody binding (y axis) to rRhi o 1 by purified IgG (x axis) from immunized or preimmune (negative control) sera. Percentages were determined from 10 patients' sera and are represented as box-whisker plots with medians and interquartile ranges. *, p < 0.001 for IgE inhibition by immunized versus corresponding preimmune IgG. d, preincubation of rRhi o 1 with increasing concentrations (x axis) of purified anti-serum IgGs resulted in a dose-dependent reduction in histamine release (mean of percentages ± S.D. (error bars); y axis) from granulocytes of patients (n = 3). Uninhibited release was observed with preimmune IgGs.
IgG against TYKW-mut Specifically Blocked IgE Binding to Rhi o 1
Purified IgG from α-TYKW-mut rabbit antiserum was used to study its inhibitory effect on IgE binding to rRhi o 1. In an ELISA inhibition assay (Fig. 10c), it was observed that preincubation of plate-bound rRhi o 1 with α-TYKW-mut IgG resulted in the blocking of IgE epitopes, and the subsequent addition of patients' sera did not allow IgE binding to rRhi o 1. The extent of IgE inhibition was calculated to be >70% on average (ranging from 54 to 79% in 10 patients). In a reciprocal experiment, purified IgG from α-rRhi o 1 serum (positive control) inhibited IgE binding to rRhi o 1 up to 83% on average. As a negative control, IgG from corresponding preimmune serum did not inhibit IgE binding (<5%) to Rhi o 1. In addition to IgE inhibition, α-TYKW-mut IgG also displayed inhibitory effects on the allergenic activity of rRhi o 1. Different concentrations of α-TYKW-mut IgGs were used to saturate the IgE epitope(s) of rRhi o 1 to various extents. The resulting rRhi o 1 with blocked IgE epitopes displayed reduced activation of patients' granulocytes and consequently a low level of histamine release (Fig. 10d). For comparison, we used 3 μg/ml IgG purified from preimmune sera, which resulted in efficient histamine release of up to 39%. On the contrary, a 3 μg/ml concentration of α-TYKW-mut IgGs used for allergen preincubation resulted in 4% release (i.e. ∼10-fold reduction). A similar pattern in histamine release inhibition was observed with rRhi o 1 preincubated with α-rRhi o 1 IgG (positive control).
Discussion
The present study describes an epitope mapping-based approach to develop a genetically modified, hypoallergenic Rhi o 1 with adequate immunogenicity as an aid for safe vaccination. It was achieved by introducing molecular mutations in only four critical residues of IgE epitope and without changing the overall fold of the allergen to protect a functional repertoire of non-IgE-binding B-cell epitopes as well as T-cell epitopes.
The foremost strategy was to generate precise information on the major antigenic regions of Rhi o 1 responsible for IgE binding. Antibody recognition of many inhalant allergens with globular structures was reported to be conformation-dependent (25). However, several respiratory fungal allergens, such as Asp f 2, Alt a 1, Asp f 1, Asp f 13, Pen n 18, Pen c 13, and Cur l 3, were identified as having linear IgE epitopes (26–32). In the case of Rhi o 1, IgE recognition was observed under denaturing conditions. In earlier studies, birch allergen Bet v 1, despite having conformational IgE epitopes (33, 34), displayed IgE binding even under denaturing conditions of Western blotting (35). This discrepancy was experimentally explained (36) by the fact that Bet v 1 was able to regain partially its native folds from a fully denatured state. However, rRhi o 1, once denatured, did not fold back even when the denaturing conditions were removed (i.e. irreversible denaturation). Therefore, we presumed that binding sites for IgE antibody on denatured Rhi o 1 were probably linear. Our presumption was empirically confirmed through identification of two IgE-reactive peptides of Rhi o 1 and the critical residues mediating antibody interaction. Loss-of-function mutations in these two peptides resulted in a decrease in IgE affinity by 2–3 orders of magnitude. This implied the presence of a limited number of high affinity IgE-binding sites on this allergen and consequently the oligoclonal nature of anti-Rhi o 1 IgE antibody as also evident for other allergens (24, 33). These two IgE-reactive peptides of Rhi o 1 were not conserved in other allergen sequences (data not shown) and therefore probably non-cross-reactive in nature. Our study sustains the lower clinical relevance of cross-reactivity between moderately conserved allergens (37). Therefore, epitope information generated in this study may be sufficient for the molecular diagnosis of Rhi o 1 allergy and may not be compromised due to its cross-reactivity with Bla g 2.
Interestingly, these two IgE-reactive peptides did not form purely linear IgE epitopes as quantified from their low IgE-binding capacity as free peptides in solution. The IgE-binding capacity of the peptides (P1, P5, and P1 + P5) was sufficiently lower than that of the corresponding intact proteins (P5-mut with functional P1, P1-mut with functional P5, and WT rRhi o 1, respectively). Hence, efficient IgE binding to these peptides was thought to be facilitated by their native conformations and positions on the folded structure of Rhi o 1. Reassuringly, these two peptides were found to form a cluster of antigenic surfaces, which gave rise to a localized IgE epitope on Rhi o 1. Therefore, individual peptides, despite independently binding to IgE antibody, were presented together in a conformational manner on the extended structure of Rhi o 1. Such a steric orientation in the epitope is effective for IgE cross-linking and effector cell activation (38). Degranulation can occur when allergens are recruited on the effector cell surface by specific interaction between receptor-bound IgE antibodies and corresponding epitopes of different affinities. This interaction can be disrupted by targeted changing of epitope residues, which in turn hampers the IgE affinity of the allergen. In this study, the failure of TYKW-mut to activate patients' basophils can be explained by such a mechanism of affinity disruption. Therefore, reduced allergenicity of TYKW-mut (in vitro and in vivo) can be attributed to non-functional mutations in the IgE epitope.
Allergen extracts and purified allergens are the active ingredients used to manufacture most of the clinically available allergy vaccines (39). Considering the risk of IgE-mediated side effects, these hazardous ingredients are currently being replaced by non-IgE-reactive allergen fragments. However, a broader use of this technology is limited due to the poor immunogenicity of the fragments, which may result in major shortcomings, such as inconvenient application to patients, weak induction of blocking antibody, and less specificity. These disadvantages can be overcome by protecting the three-dimensional structure of the allergen within the engineered vaccine (40). Such a strategy can concomitantly increase the specific immunogenicity of a hypoallergenic molecule. In TYKW-mut, the IgE reactivity was interfered with by rationally introducing only four substitutions located at discrete points. These subtle mutations did not result in a disruption of native folds of the allergen, and therefore, most of the antigenic regions were preserved. The preservation of T-cell epitopes in TYKW-mut was evident from its mitogenic effect on lymphocytes. This may help the mutant to exert preferential tolerance in allergen-specific CD4+ T-cells and to confer T-cell help necessary for an immunogenic response. Additionally, TYKW-mut modulated Th2 priming by down-regulating prototype IL-5 secretion. This indicated that the TYKW-mut had the immunotherapeutic potential of skewing the T-cell response from pro-allergic Th2 to possibly a tolerogenic phenotype while retaining ∼99% of the WT allergen sequences. The most important mechanism of immunotherapy is the induction of allergen-specific blocking IgG antibodies (15, 39). Remarkably, TYKW-mut, despite carrying mutations on the IgE epitopes, was able to induce a high titer of IgG antibodies specifically directed (comparable with anti-rRhi o 1 IgG) toward the IgE epitope of Rhi o 1. Binding of this polyclonal IgG to Rhi o 1 competitively inhibited patients' IgE binding by a mechanism of either directly blocking the IgE epitopes or creating a steric hindrance for IgE binding. Another beneficial feature of blocking IgG antibodies was the allergen neutralization, associated with the suppression of allergenic activity (17). This happened due to reduced histamine release as a result of ineffective IgE cross-linking on granulocytes. Taken together, the information generated from antigenic and functional analysis of TYKW-mut may be of potential consideration for therapeutic applications. In conclusion, the present findings have brought us closer to the development of a safe and efficient vaccine candidate for the treatment of mold allergy, especially for Indian patients.
Experimental Procedures
Human Subjects
Residual sera and clinical records of 45 mold allergy patients with elevated serum IgE against R. oryzae antigenic extract (20.91 ± 10.51 kilo units (KUA)/liter) were collected with prior consent (for details, see supplemental Table S1). Sera from one dust mite-allergic patient and three healthy volunteers were also collected for negative controls. Approval was obtained from the institutional ethics committee.
Recombinant Allergens and Monoclonal Antibody
Recombinant Rhi o 1 was expressed in E. coli BL21(DE3)Rosetta and purified using an Ni-NTA column (Qiagen), as described earlier (14). rBet v 1 (an unrelated pollen allergen for negative control) was from Dr. Margarete Focke-Tejkl (Medical University of Vienna). rBla g 2 and mAb4C3 were from Dr. Anna Pomes (Indoor Biotechnologies, Charlottesville, VA).
IgE Immuno-dot Blotting
Nitrocellulose Strips were dotted with 0.4 μg each of rRhi o 1, rBla g 2, rBet v 1, and HSA. After blocking with 3% BSA, strips were then exposed to 45 sera in 1:10 dilution at 4 °C overnight. Bound IgEs were detected with 1:1000 diluted alkaline phosphatase-conjugated mouse monoclonal anti-human IgE (Sigma, A3076) and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate (Abcam).
IgE-ELISA
Polystyrene plates (Maxisorp; Nunc) were coated with 5 ng/μl of either rRhi o 1, rBla g 2, rBet v 1, or HSA. Wells were blocked with 1% BSA and then exposed to 28 sera at a 1:10 dilution overnight at 4 °C. Bound IgEs were detected with 1:1000 diluted alkaline phosphatase-conjugated mouse monoclonal anti-human IgE (Sigma, A3076) and one-step para-nitro phenyl phosphate substrate (Thermo Scientific). Absorbance was taken at 405 nm.
IgE-Western Blotting
Around 1 μg of rRhi o 1 was treated for denaturation with either 10% SDS, prolonged heating, or 7 m urea or 6 m guanidine-HCl. Treated allergens were then subjected to Western blotting (41) with 1:10 diluted pooled sera, and bound IgEs on blots were detected as described above.
Circular Dichroism Spectroscopy
The CD spectra of 5 μm rRhi o 1 were recorded in a Jasco Corp. J-815 CD spectropolarimeter at 20 °C within a wavelength range of 200–250 nm. In a separate experiment, the CD spectra were recorded at temperatures ranging from 35 to 95 °C with every 10 °C increase by a step scan procedure (heating rate, 60 °C/h; scan speed, 50 nm/min). The spectrum was also recorded after cooling down the system at 25 °C. Changes in millidegree ellipticity during thermal denaturation were studied by generating a melting curve, which was fitted with a sigmoidal function. The transition temperature was determined from the inflection point of the curve using Origin version 5.0 software.
B-cell Epitope Prediction
The amino acid sequence of Rhi o 1 was given as input in two B-cell epitope prediction servers, such as artificial neural network-based ABCpred (42) and physicochemical parameters-based BCEPRED (43). Peptides with residues above a threshold value of 2.38 in all of the seven scales of BCEPRED were selected. The ABCpred server was also run (threshold >0.5) to compare and verify the results of BCEPRED analysis.
Immunoscreening of Peptides
Peptides corresponding to predicted B-cell epitopes were either chemically synthesized using an Endeavor 90-II peptide synthesizer (AAPTec, Louisville, KY) as described (44) or purchased from GL Biochem (Shanghai, China). For ELISA, 5 μm each of synthetic peptides (P1–P5), rRhi o 1, HSA, and a random irrelevant oligopeptide were reconstituted in binding buffer (40 mm Na2CO3, 60 mm NaHCO3, 10 mm CaCl2, 10 mm DTT, pH 9.6) and separately coated on polystyrene plates. Wells were exposed to a pool of five sera at 1:10 dilution for overnight at 4 °C. For dot blotting, a 5 μm concentration of each of the five peptides and control antigens was dotted onto nitrocellulose strips. Strips were exposed to 30 sera at a 1:10 dilution. Bound IgEs in ELISA and dot blots were detected.
Competitive ELISA with Peptides
For this assay, we selected six sera (patients 11–15 and 39), which contained high titers (A405 > 2.00) of reaginic IgEs against rRhi o 1 but no reactivity (A405 < 0.10) against rBla g 2. These sera were pooled and separately preincubated with an equal volume of 10-fold serial dilutions of rRhio 1, synthetic peptides (P1, P5, or P1 + P5), and HSA, respectively, overnight at 4 °C. About 1 ng/μl plate-bound rRhi o 1 was then exposed to these preincubated serum (1:10, v/v), and bound IgEs were detected. The percentage of inhibition of IgE binding to rRhi o 1 was calculated as follows.
 |
Structural Bioinformatics
A homology model of Rhi o 1 reported previously (14) was used to map IgE-binding peptides using PyMOL version 1.8.2.0 software. Residue-wise solvent-accessible surface area and the percentage of relative solvent accessibility were calculated by the POPS version 1.6.4 (45) and SARpred (46) servers, respectively.
Critical Residues of IgE Epitope
The antigenic role of each residue in IgE epitope mapped on the three-dimensional model of Rhi o 1 was carefully examined, and their involvement in immunorecognition was reviewed from established literature. Based on this analysis, we labeled individual residues as probable mediators of IgE interaction. These selected residues were changed into alanine, and corresponding mutant peptides were synthesized. Synthetic mutant peptides along with WT peptides (for comparison) were tested for IgE reactivity using eight representative sera by ELISA and dot blotting.
Site-directed Mutagenesis
Critical residues of the IgE epitope were changed into alanine by base pair substitution in the Rhi o 1 cDNA using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). Mutant proteins were expressed in BL21(DE3) with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 5 h and purified using an Ni-NTA column (Qiagen). The TYKW-mut was purified from the inclusion bodies under denaturing conditions. For solubilization, denatured TYKW-mut was brought to 0.003 mg/ml concentration by rapidly diluting in refolding buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 5% glycerol, 0.5 mm oxidized glutathione, 3 mm reduced glutathione) at 4 °C. The refolded TYKW-mut protein was then concentrated up to 0.5 mg/ml. CD spectra of the three purified mutants were recorded.
IgE Reactivity of the Mutants by Sandwich ELISA and Western Blotting
ELISA plates were coated with 1 ng/μl anti-Bla g 2 mAb4C3 at 37 °C for 3 h followed by washing to remove the unbound mAbs. After blocking the plate with 1% BSA, a 10 ng/μl concentration of either rRhi o 1 or its mutants was added and incubated at 37 °C for 5 h. The mAb4C3-captured antigens were exposed to 10 patients' sera at 1:10 dilution. For Western blotting, equal loading of 2 μg (Bradford-quantified) of rRhi o 1 and the mutants on PVDF membrane was confirmed by Ponceau S staining. After destaining and blocking, two such membranes were separately incubated with two pools (1:10, v/v) of patients' sera. Each pool represented an equal mixture of sera from three patients. Bound IgEs in ELISA and blots were detected. The IgE-reactive band intensities were determined by QuantityOne software (Bio-Rad). The percentage of reduction in IgE binding in ELISA and Western blotting was calculated as follows.
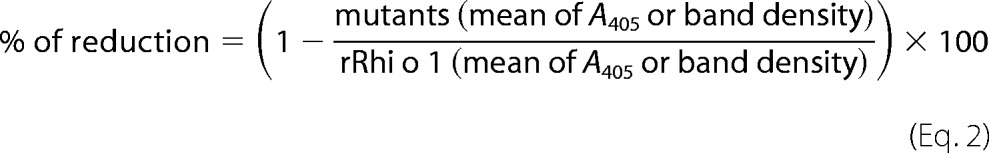 |
Competitive ELISA with Mutants
Serially increasing (10-fold) doses of rRhio 1 or its mutants were separately preincubated with three individual and one pooled (mix of five) sera for overnight at 4 °C. Preincubated samples of sera (1:10, v/v) were then added to plate-bound mAb4C3-captured rRhi o 1, and bound IgEs were detected. The percentage of IgE binding inhibition was calculated as described under “Competitive ELISA with Peptides.”
Degranulation Assay
RBL-2H3 cells expressing the human FCER1 receptor was cultured in RPMI medium supplemented with 10% FCS, 4 mm l-glutamine, 2 mm sodium pyruvate, 10 mm HEPES, and 1% penicillin/streptomycin. Cells were sensitized passively with IgE from three different patients' sera (diluted 1:10 in media) and incubated overnight at 37 °C, 95% humidity, and 5% CO2. After washing with Tyrode's buffer containing 0.1% BSA (pH 7.2), cells were challenged with either rRhi o 1 or TYKW-mut in five different concentrations diluted in Tyrode's buffer with 50% D2O and incubated at 37 °C for 1 h. For spontaneous release, cells were incubated with sera but without any challenge. For the complete release, cells were lysed with 10% Triton X-100. Released β-hexaminidase was estimated by incubating with 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide in 0.1 m citrate buffer (pH 4.5) for 30 min. Reactions were stopped by adding glycine buffer (200 mm l-glycine and 200 mm NaCl, pH 10.5), and fluorescence was measured in a microplate reader (λex = 360 nm; λem = 465 nm). The percentage of release was calculated as follows.
 |
Flow Cytometry
The CD203c-based basophil activation test was performed as described (47). Briefly, 100 μl of heparinized blood from each of three R. oryzae-sensitized patients were stimulated with 100 ng/ml (optimized from the RBL assay) of either rRhi o 1 or TYKW-mut and incubated for 45 min at 37 °C. Instead of allergens, 1 μg/ml anti-human IgE (Sigma) and PBS were used as positive and negative control, respectively. Staining was done with 10 μl of phycoerythrin-conjugated anti-CD203c mAb97A6 (BioLegend) for 30 min in the dark at room temperature. Erythrocytes were removed by RBC lysis solution (eBioscience), and cells were analyzed in a FACS Verse flow cytometer (BD Biosciences). Basophils were detected based on forward-scatter and side-scatter characteristics. The level of CD203c expression was represented as a measure of the mean fluorescence intensities.
MTT Assay and Cytokine Assay
Around 2 × 106 PBMCs from each of six patients were cultured as described (18) and stimulated for 7 days with a 10 μg/ml concentration of either endotoxin-free allergens, tetanus toxoid (Medline India), or medium only. Lymphocyte proliferation was determined using the Vybrant® MTT cell proliferation assay kit (Thermo Scientific) and expressed as the proliferation index (stimulated A570/unstimulated A570). Secreted Th2 cytokines in PBMC culture supernatants (unstimulated, rRhi o 1, and TYKW-mut) of four patients were estimated using the RayBio® human IL-4, IL-5, and IL-13 ELISA kit (RayBiotech, Inc.).
Animal Experiments
Each group of six female BALB/c mice was intranasally instillated (0.5 μg/mouse) with either rRhi o 1 or TYKW-mut or ovalbumin (positive control) or PBS (negative control) following a protocol described previously (48). H&E and PAS staining of lung histology were done to quantify the eosinophilic infiltrates and mucous gland hyperplasia, respectively, as described (49). For raising antisera, three BALB/c mice (10 μg of antigens/mouse) and one New Zealand White rabbit (200 μg of antigens/rabbit) were immunized with either rRhi o 1 or TYKW-mut adsorbed to Freund's adjuvants following protocols described previously (50). About 3–4 mg of IgG were purified from rabbit sera (preimmune and immune) using a PierceTM Protein A IgG purification kit.
Specificity and Titer of Antiserum IgGs
Plate-bound 10 ng/μl rRhi o 1 or TYKW-mut was exposed to either 1:100 diluted mouse or 1:10,000 diluted rabbit antisera. Mouse IgG1s and rabbit IgGs were detected with alkaline phosphatase-conjugated rat monoclonal SB77e anti-mouse IgG1 H&L (1:1000, v/v) (Abcam, ab99602) and alkaline phosphatase-conjugated mouse monoclonal anti-rabbit IgG (γ-chain-specific) (1:50,000, v/v) (Sigma, A2556) respectively. Specific IgG titers in two rabbit antisera (α-rRhi o 1 and α-TYKW-mut) were compared by ELISA-titration (49), in which immobilized 10 ng/μl rRhi o1 was exposed to gradually increasing dilutions of antisera. A titration curve was prepared with A405 values of bound IgG against each dilution.
IgG-mediated Blocking of Patients' IgE Binding to rRhi o 1
Purified rRhi o 1 (50 ng/μl)-coated ELISA plates were exposed to purified IgG antibodies (1.0–1.26 μg/μl) from either of the two rabbit antisera and incubated for 1 h at 37 °C. As a negative control, an equal concentration of IgG from corresponding preimmune serum was added. After washing five times, plates were exposed to 10 different patients' sera (1:10, v/v) overnight at 4 °C, and bound IgEs were detected. The percentage of IgE inhibition was calculated as follows.
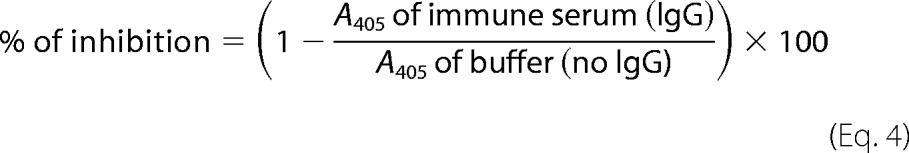 |
IgG-mediated Histamine Release Inhibition
About 100 ng/ml rRhi o 1 was preincubated at 37 °C for 60 min with different concentrations (1–3 μg/ml) of purified IgGs from rabbit antisera. Isolated granulocytes from each of three patients were resuspended in stimulation buffer (25 mm HEPES, 1% BSA, pH 7.4) and then challenged with preincubated allergens. Released histamines were quantified using an EIA histamine assay kit (Immunotech, Beckman Coulter), and the percentage of release was calculated as described (51).
Statistical Analysis
We compared the significance (p < 0.001) of changes in (i) IgE binding to rRhi o 1 versus rBla g 2, mutant peptides versus WT peptides, mutant proteins versus rRhi o 1; (ii) T-cell and IL response of TYKW-mut versus rRhi o 1; (iii) inflammations in lung histology of TYKW-mut- versus rRhi o 1-treated mice; and (iv) IgE inhibition by antiserum IgG versus preimmune IgG by paired Student's t test using GraphPad Prism version 6.
Author Contributions
The entire study was conceived and the experiments were designed by G. S. and S. G. B. G. S. was responsible for peptide assays, mutagenesis, mutant purification, immunological assays, and functional assays of the mutants under the supervision of S. G. B. K. J. performed the entire animal experiments. A. D. performed clinical diagnosis of patients and estimation of antibody titers in patient sera. T-cell assays and flow cytometry were jointly performed by G. S. and A. D. S. S. was responsible for epitope prediction, structural bioinformatic studies, and statistical analysis. G. S. and S. G. B. jointly wrote the paper. All of the authors read and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Jadab Kr. Ghosh, Chanchal Chakraborty, Kaberi Ghosh, and Soumyo Shubhra Gupta (Bose Institute) for technical help.
This work was supported in part by a grant from the Department of Science and Technology, Government of India (Institutional Plan Project-1, Bose Institute). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1 and Figs. S1 and S2.
- HSA
- human serum albumin
- PAS
- periodic acid-Schiff
- PBMC
- peripheral blood mononuclear cell
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Osborne N. J., Thornton C. R., and Sharpe R. A. (2015) Indoor fungal exposure and allergic respiratory disease. Curr. Allergy Asthma Rep. 15, 71. [DOI] [PubMed] [Google Scholar]
- 2. Twaroch T. E., Curin M., Valenta R., and Swoboda I. (2015) Mold allergens in respiratory allergy: from structure to therapy. Allergy Asthma Immunol. Res. 7, 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karlsson-Borgå A., Jonsson P., and Rolfsen W. (1989) Specific IgE antibodies to 16 widespread mold genera in patients with suspected mold allergy. Ann. Allergy 63, 521–526 [PubMed] [Google Scholar]
- 4. O'Connell M. A., Pluss J. L., Schkade P., Henry A. R., and Goodman D. L. (1995) Rhizopus-induced hypersensitivity pneumonitis in a tractor driver. J. Allergy Clin. Immunol. 95, 779–780 [DOI] [PubMed] [Google Scholar]
- 5. Klarić M. Š., Varnai V. M., Calušić A. L., and Macan J. (2012) Occupational exposure to airborne fungi in two Croatian sawmills and atopy in exposed workers. Ann. Agric. Environ. Med. 19, 213–219 [PubMed] [Google Scholar]
- 6. Færden K., Lund M. B., Mogens Aaløkken T., Eduard W., Søstrand P., Langård S., and Kongerud J. (2014) Hypersensitivity pneumonitis in a cluster of sawmill workers: a 10-year follow-up of exposure, symptoms, and lung function. Int. J. Occup. Environ. Health 20, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gioulekas D., Damialis A., Papakosta D., Spieksma F., Giouleka P., and Patakas D. (2004) Allergenic fungi spore records (15 years) and sensitization in patients with respiratory allergy in Thessaloniki-Greece. J. Investig. Allergol. Clin. Immunol. 14, 225–231 [PubMed] [Google Scholar]
- 8. Zhang Y., Chen J., Chen Y., Dong J., Wei Q., and Lou J. (2005) Environmental mycological study and allergic respiratory disease among tobacco processing workers. J. Occup. Health 47, 181–187 [DOI] [PubMed] [Google Scholar]
- 9. Devars D. M. M., Gratacap M., Malinvaud D., Grenouillet F., and Bonfils P. (2015) An uncommon cause of allergic fungal sinusitis: Rhizopus oryzae. Ear Nose Throat J. 94, E17–E20 [PubMed] [Google Scholar]
- 10. Matsuse H., Tsuchida T., Fukahori S., Kawano T., Nishino T., Fukushima C., and Kohno S. (2013) Dissociation between sensitizing and colonizing fungi in patients with allergic bronchopulmonary aspergillosis. Ann. Allergy Asthma Immunol. 111, 190–193 [DOI] [PubMed] [Google Scholar]
- 11. Chowdhary A., Agarwal K., Kathuria S., Gaur S. N., Randhawa H. S., and Meis J. F. (2014) Allergic bronchopulmonary mycosis due to fungi other than Aspergillus: a global overview. Crit. Rev. Microbiol. 40, 30–48 [DOI] [PubMed] [Google Scholar]
- 12. Agarwal K., Gaur S. N., and Chowdhary A. (2015) The role of fungal sensitisation in clinical presentation in patients with chronic obstructive pulmonary disease. Mycoses 58, 531–535 [DOI] [PubMed] [Google Scholar]
- 13. Sircar G., Chakrabarti H. S., Saha B., and Gupta-Bhattacharya S. (2012) Identification of aero-allergens from Rhizopus oryzae: an immunoproteomic approach. J. Proteomics 77, 455–468 [DOI] [PubMed] [Google Scholar]
- 14. Sircar G., Saha B., Mandal R. S., Pandey N., Saha S., and Gupta Bhattacharya S. (2015) Purification, cloning and immuno-biochemical characterization of a fungal aspartic protease allergen Rhi o 1 from the airborne mold Rhizopus oryzae. PLoS One 10, e0144547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valenta R., Campana R., Focke-Tejkl M., and Niederberger V. (2016) Vaccine development for allergen-specific immunotherapy based on recombinant allergens and synthetic allergen peptides: lessons from the past and novel mechanisms of action for the future. J. Allergy Clin. Immunol. 137, 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gafvelin G., Parmley S., Neimert-Andersson T., Blank U., Eriksson T. L., van Hage M., and Punnonen J. (2007) Hypoallergens for allergen-specific immunotherapy by directed molecular evolution of mite group 2 allergens. J. Biol. Chem. 282, 3778–3787 [DOI] [PubMed] [Google Scholar]
- 17. Focke-Tejkl M., Weber M., Niespodziana K., Neubauer A., Huber H., Henning R., Stegfellner G., Maderegger B., Hauer M., Stolz F., Niederberger V., Marth K., Eckl-Dorna J., Weiss R., Thalhamer J., et al. (2015) Development and characterization of a recombinant, hypoallergenic, peptide-based vaccine for grass pollen allergy. J. Allergy Clin. Immunol. 135, 1207–1217.e1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marth K., Breyer I., Focke-Tejkl M., Blatt K., Shamji M. H., Layhadi J., Gieras A., Swoboda I., Zafred D., Keller W., Valent P., Durham S. R., and Valenta R. (2013) A nonallergenic birch pollen allergy vaccine consisting of hepatitis PreS-fused Bet v 1 peptides focuses blocking IgG toward IgE epitopes and shifts immune responses to a tolerogenic and Th1 phenotype. J. Immunol. 190, 3068–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bohle B., Breitwieser A., Zwölfer B., Jahn-Schmid B., Sára M., Sleytr U. B., and Ebner C. (2004) A novel approach to specific allergy treatment: the recombinant fusion protein of a bacterial cell surface (S-layer) protein and the major birch pollen allergen Bet v 1 (rSbsC-Bet v 1) combines reduced allergenicity with immunomodulating capacity. J. Immunol. 172, 6642–6648 [DOI] [PubMed] [Google Scholar]
- 20. Chan S. L., Ong S. T., Ong S. Y., Chew F. T., and Mok Y. K. (2006) Nuclear magnetic resonance structure-based epitope mapping and modulation of dust mite group 13 allergen as a hypoallergen. J. Immunol. 176, 4852–4860 [DOI] [PubMed] [Google Scholar]
- 21. Neuvirth H., Raz R., and Schreiber G. (2004) ProMate: a structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 338, 181–199 [DOI] [PubMed] [Google Scholar]
- 22. Kringelum J. V., Nielsen M., Padkjær S. B., and Lund O. (2013) Structural analysis of B-cell epitopes in antibody:protein complexes. Mol. Immunol. 53, 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pedraza-Escalona M., Becerril-Luján B., Agundis C., Domínguez-Ramírez L., Pereyra A., Riaño-Umbarila L., and Rodríguez-Romero A. (2009) Analysis of B-cell epitopes from the allergen Hev b 6.02 revealed by using blocking antibodies. Mol. Immunol. 46, 668–676 [DOI] [PubMed] [Google Scholar]
- 24. Woodfolk J. A., Glesner J., Wright P. W., Kepley C. L., Li M., Himly M., Muehling L. M., Gustchina A., Wlodawer A., Chapman M. D., and Pomés A. (2016) Antigenic determinants of the bilobal cockroach allergen Bla g 2. J. Biol. Chem. 291, 2288–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pomés A. (2009) Relevant B cell epitopes in allergic disease. Int. Arch. Allergy Immunol. 152, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banerjee B., Greenberger P. A., Fink J. N., and Kurup V. P. (1999) Conformational and linear B-cell epitopes of Asp f 2, a major allergen of Aspergillus fumigatus, bind differently to immunoglobulin E antibody in the sera of allergic bronchopulmonary aspergillosis patients. Infect. Immun. 67, 2284–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kurup V. P., Vijay H. M., Kumar V., Castillo L., and Elms N. (2003) IgE binding synthetic peptides of Alt a 1, a major allergen of Alternaria alternata. Peptides 24, 179–185 [DOI] [PubMed] [Google Scholar]
- 28. Kurup V. P., Banerjee B., Murali P. S., Greenberger P. A., Krishnan M., Hari V., and Fink J. N. (1998) Immunodominant peptide epitopes of allergen, Asp f 1 from the fungus Aspergillus fumigatus. Peptides 19, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 29. Chow L.. P., Liu S. L., Yu C. J., Liao H. K., Tsai J. J., and Tang T. K. (2000) Identification and expression of an allergen Asp f 13 from Aspergillus fumigatus and epitope mapping using human IgE antibodies and rabbit polyclonal antibodies. Biochem. J. 346, 423–431 [PMC free article] [PubMed] [Google Scholar]
- 30. Yu C. J., Chen Y. M., Su S. N., Forouhar F., Lee S. H., and Chow L. P. (2002) Molecular and immunological characterization and IgE epitope mapping of Pen n 18, a major allergen of Penicillium notatum. Biochem. J. 363, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J. C., Chiu L. L., Lee K. L., Huang W. N., Chuang J. G., Liao H. K., and Chow L. P. (2012) Identification of critical amino acids in an immunodominant IgE epitope of Pen c 13, a major allergen from Penicillium citrinum. PLoS One 7, e34627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma V., Singh B. P., Gaur S. N., Pasha S., and Arora N. (2009) Bioinformatics and immunologic investigation on B and T cell epitopes of Cur l 3, a major allergen of Curvularia lunata. J. Proteome Res. 8, 2650–2655 [DOI] [PubMed] [Google Scholar]
- 33. Spangfort M. D., Mirza O., Ipsen H., Van Neerven R. J., Gajhede M., and Larsen J. N. (2003) Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J. Immunol. 171, 3084–3090 [DOI] [PubMed] [Google Scholar]
- 34. Hecker J., Diethers A., Schulz D., Sabri A., Plum M., Michel Y., Mempel M., Ollert M., Jakob T., Blank S., Braren I., and Spillner E. (2012) An IgE epitope of Bet v 1 and fagales PR10 proteins as defined by a human monoclonal IgE. Allergy 67, 1530–1537 [DOI] [PubMed] [Google Scholar]
- 35. Schöll I., Kalkura N., Shedziankova Y., Bergmann A., Verdino P., Knittelfelder R., Kopp T., Hantusch B., Betzel C., Dierks K., Scheiner O., Boltz-Nitulescu G., Keller W., and Jensen-Jarolim E. (2005) Dimerization of the major birch pollen allergen Bet v 1 is important for its in vivo IgE-cross-linking potential in mice. J. Immunol. 175, 6645–6650 [DOI] [PubMed] [Google Scholar]
- 36. Laffer S., Hamdi S., Lupinek C., Sperr W. R., Valent P., Verdino P., Keller W., Grote M., Hoffmann-Sommergruber K., Scheiner O., Kraft D., Rideau M., and Valenta R. (2003) Molecular characterization of recombinant T1, a non-allergenic periwinkle (Catharanthus roseus) protein, with sequence similarity to the Bet v 1 plant allergen family. Biochem. J. 373, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mueller G. A., Pedersen L. C., Glesner J., Edwards L. L., Zakzuk J., London R. E., Arruda L. K., Chapman M. D., Caraballo L., and Pomés A. (2015) Analysis of glutathione S-transferase allergen cross-reactivity in a North American population: relevance for molecular diagnosis. J. Allergy Clin. Immunol. 136, 1369–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flicker S., Steinberger P., Ball T., Krauth M. T., Verdino P., Valent P., Almo S., and Valenta R. (2006) Spatial clustering of the IgE epitopes on the major timothy grass pollen allergen Phl p 1: importance for allergenic activity. J. Allergy Clin. Immunol. 117, 1336–1343 [DOI] [PubMed] [Google Scholar]
- 39. Valenta R., Ferreira F., Focke-Tejkl M., Linhart B., Niederberger V., Swoboda I., and Vrtala S. (2010) From allergen genes to allergy vaccines. Annu. Rev. Immunol. 28, 211–241 [DOI] [PubMed] [Google Scholar]
- 40. Holm J., Gajhede M., Ferreras M., Henriksen A., Ipsen H., Larsen J. N., Lund L., Jacobi H., Millner A., Würtzen P. A., and Spangfort M. D. (2004) Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses. J. Immunol. 173, 5258–5267 [DOI] [PubMed] [Google Scholar]
- 41. Ghosh D., Mueller G. A., Schramm G., Edwards L. L., Petersen A., London R. E., Haas H., and Gupta Bhattacharya S. (2014) Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin Cat r 1. J. Biol. Chem. 289, 21374–21385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saha S., and Raghava G. P. S. (2006) Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65, 40–48 [DOI] [PubMed] [Google Scholar]
- 43. Saha S., and Raghava G. P. S. (2004) BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. Lecture Notes Comput. Sci. 3239, 197–204 [Google Scholar]
- 44. Ganguly H. K., Kaur H., and Basu G. (2013) Local control of cis-peptidyl–prolyl bonds mediated by CH···π interactions: the Xaa-Pro-Tyr motif. Biochemistry 52, 6348–6357 [DOI] [PubMed] [Google Scholar]
- 45. Cavallo L., Kleinjung J., and Fraternali F. (2003) POPS: a fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res. 31, 3364–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garg A., Kaur H., and Raghava G. P. S. (2005) Real value prediction of solvent accessibility in proteins using multiple sequence alignment and secondary structure. Proteins 61, 318–324 [DOI] [PubMed] [Google Scholar]
- 47. Hauswirth A. W., Natter S., Ghannadan M., Majlesi Y., Schernthaner G. H., Sperr W. R., Bühring H. J., Valenta R., and Valent P. (2002) Recombinant allergens promote expression of CD203c on basophils in sensitized individuals. J. Allergy Clin. Immunol. 110, 102–109 [DOI] [PubMed] [Google Scholar]
- 48. Chen J. C., Chuang J. G., Su Y. Y., Chiang B. L., Lin Y. S., and Chow L. P. (2011) The protease allergen Pen c 13 induces allergic airway inflammation and changes in epithelial barrier integrity and function in a murine model. J. Biol. Chem. 286, 26667–26679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lanckacker E. A., Tournoy K. G., Hammad H., Holtappels G., Lambrecht B. N., Joos G. F., and Maes T. (2013) Short cigarette smoke exposure facilitates sensitisation and asthma development in mice. Eur. Respir. J. 41, 1189–1199 [DOI] [PubMed] [Google Scholar]
- 50. Linhart B., Focke-Tejkl M., Weber M., Narayanan M., Neubauer A., Mayrhofer H., Blatt K., Lupinek C., Valent P., and Valenta R. (2015) Molecular evolution of hypoallergenic hybrid proteins for vaccination against grass pollen allergy. J. Immunol. 194, 4008–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saha B., Sircar G., Pandey N., and Gupta Bhattacharya S. (2015) Mining novel allergens from coconut pollen employing manual de novo sequencing and homology-driven proteomics. J. Proteome Res. 14, 4823–4833 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.