Abstract
Functional links between genes can be predicted using phylogenetic profiling, by correlating the appearance and loss of homologs in subsets of species. However, effective genome-wide phylogenetic profiling has been hindered by the large fraction of human genes related to each other through historical duplication events. Here we overcame this challenge by automatically profiling over 30,000 groups of homologous human genes (orthogroups) representing the entire protein-coding genome across 177 eukaryotic species (hOP-profiles). By generating a full pair-wise orthogroup phylogenetic co-occurrence matrix, we derive unbiased genome-wide predictions of functional modules (hOP-modules). Our approach predicts functions for hundreds of poorly characterized genes. The results suggest evolutionary constraints that lead components of protein complexes and metabolic pathways to co-evolve while genes in signaling and transcriptional networks do not. As a proof of principle, we validated two subsets of candidates experimentally for their predicted link to the actin-nucleating WASH complex and cilia/basal body function.
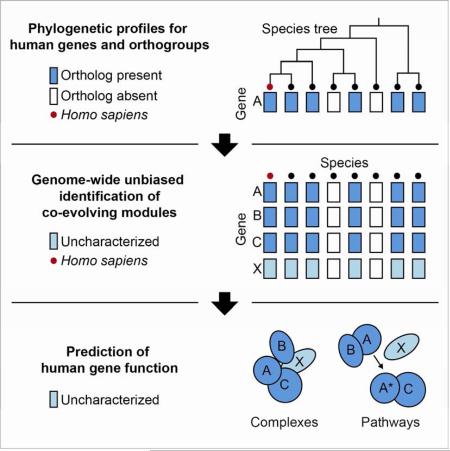
Graphical Abstract
INTRODUCTION
Even though more than a decade has passed since the human genome has been sequenced, the biochemical and cellular function of a large number of human genes remains unknown. Many of these poorly understood genes have been linked to human genetic disorders and are well conserved across a range of eukaryotic species (Domazet-Loso and Tautz, 2008), underscoring their likely relevance to human physiology. However, they often have not been clearly linked to phenotypes (or do not have homologs) in tractable genetic model systems, significantly slowing the rate of discovery. In addition, many have no detectable domain organization or sequence homology to any characterized human genes. We therefore refer to them as ‘refractory genes’. Without reference points for hypothesis-driven experiments, discovery of refractory gene function is left to serendipity or genome-wide functional screens that are often difficult to develop or cannot be performed for processes that are not well understood.
A completely independent approach to predicting gene function was first introduced in bacteria by linking genes based on the joint presence or absence of their orthologs in different species (Pellegrini et al., 1999), defined here as genes with sequence homology derived from a single common ancestor (Gabaldón and Koonin, 2013)(Supplemental Experimental Procedures). This approach, termed phylogenetic profiling, is built on the premise that genes that function together are gained and lost together in evolution. The subsequent extension of phylogenetic profiling to eukaryotic species led to the discovery of cilia genes (Avidor-Reiss et al., 2004), genes linked to Ca2+ influx into mitochondria (Baughman et al., 2011) and small RNA pathway genes (Tabach et al., 2013a). Despite extensive modifications to the original approach (Altenhoff and Dessimoz, 2009; Barker and Pagel, 2005; Bowers et al., 2004; Date and Marcotte, 2003; Kensche et al., 2008; Li et al., 2014), two major challenges have precluded unbiased functional predictions for the human genome. The first is that over half of all human genes are derived from ancestral duplication (Blomme et al., 2006; Cotton and Page, 2005; Zhang, 2003), complicating the one-to-one mapping of orthologs in distant species. This is a critical issue to address, as duplicated genes frequently diverge in function from each other as well as from their ancestor (Conant and Wolfe, 2008). The second major roadblock is that the most sensitive methods for quantifying co-evolution do not scale well with genome size and complexity of the species tree (Barker, Meade, and Pagel 2007; Y. Li et al. 2014).
Aiming to address these challenges and generate a tractable set of global functional predictions, we developed an automated strategy to sequentially assign human genes to hierarchical ‘orthogroups’ of homologous genes with shared ancestry. This enabled us to generate unique phylogenetic profiles for each orthogroup, placing 31406 orthogroups containing 19973 human genes in their evolutionary context across 177 eukaryotic species. We then developed a scoring metric to compare pairs of human orthogroup phylogenetic (hOP) profiles by inferring the number of informative shared losses in a way that accounts for tree topology and noise in homology measurements. This allowed us to create and benchmark a genome-wide human phylogenetic co-occurrence matrix (hOP-matrix) for the first time.
Our main use of the hOP-matrix was to generate clusters in an unbiased fashion, uncovering over a thousand functional modules that vary in size from 2 to over 50 genes (hOP-modules), thereby predicting functions for hundreds of refractory genes. These clusters also reveal unexpected connections between known genes as well as modularity within cellular processes, and enable the exploration of potentially undiscovered biological functions. To emphasize its utility as a discovery tool, we experimentally validated predictions of gene function for two of the identified hOP-modules. Finally, our analysis strongly suggests evolutionary constraints on functional modularity, distinguishing linear metabolic pathways and protein complexes from interlinked signaling and transcriptional regulatory networks. All hOP-profiles, co-occurrence scores and modules can be queried and analyzed on our website (http://web.stanford.edu/group/meyerlab/hOPMAPServer/index.html).
RESULTS
Binary phylogenetic profiles for identification of shared gene function
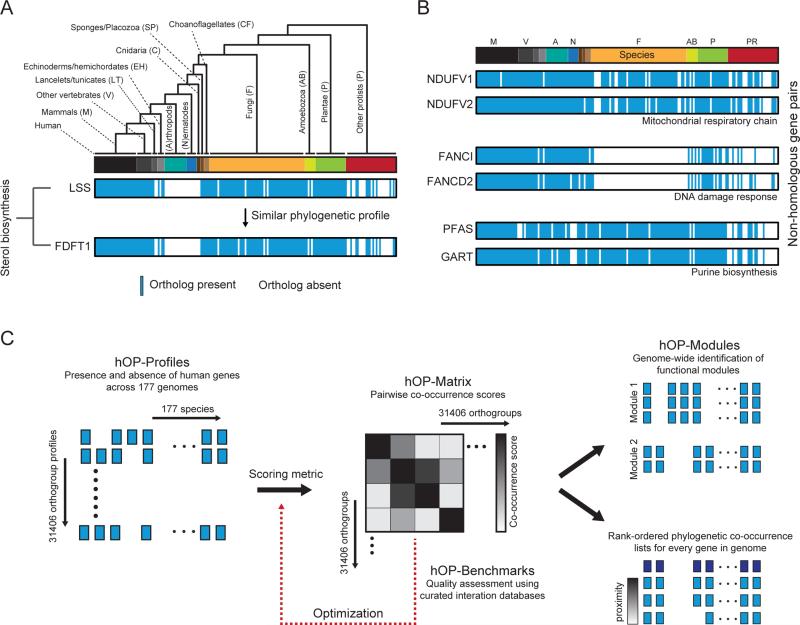
A phylogenetic profile is created by projecting the species tree onto a binary one-dimensional vector with each extant species represented by a single element with a value of 1 if an ortholog is present (177 species, Figure 1A), while keeping closely related species adjacent (Figure S1 and Experimental Procedures). The resulting profile can be used to identify other phylogenetic profiles with similar patterns of presence and absence, exemplified using two ancient genes LSS and FDFT1 (Figure 1A) that highlight the independent loss of the sterol synthesis pathway in the arthropod and nematode lineages (Desmond and Gribaldo, 2009). Additional examples (Figure 1B) highlight gene pairs where correlated phylogenetic profiles are predictive of a shared role in regulating mitochondrial respiration, DNA repair, and purine biosynthesis, respectively. For simplicity, these examples involve genes with no detectable homology to each other or to any other human genes. Our algorithm was designed to extend such a comparative analysis to all human genes and groups of homologous genes (orthogroups) with the robust automation of each step in the generation and unbiased comparison of phylogenetic profiles (Figure 1C).
Figure 1. Strategy for human-centric phylogenetic profiling.
(A) LSS ortholog distribution collapsed into binary phylogenetic vector in accordance with the branching structure of the tree (Figure S1 and Experimental Procedures); vector contains 177 elements with 1 (blue, present) or 0 (white) absent. Individual species are represented with a bar colored according to membership in 13 different branches. The lower profile represents the gene FDFT1, a member of the same cholesterol biosynthetic pathway as LSS. (B) Additional examples of functionally linked gene pairs with correlated phylogenetic profiles. Genes have no detectable homology to each other (BLASTp). The linear ordering of species is represented with a color bar as in (A) with abbreviated labels and without the accompanying species tree, a concise form used in subsequent figures. (C) Schematic illustrating overall workflow for the hOPMAP algorithm. See also Figure S1.
Unbiased generation of phylogenetic profiles for 31406 human orthogroups
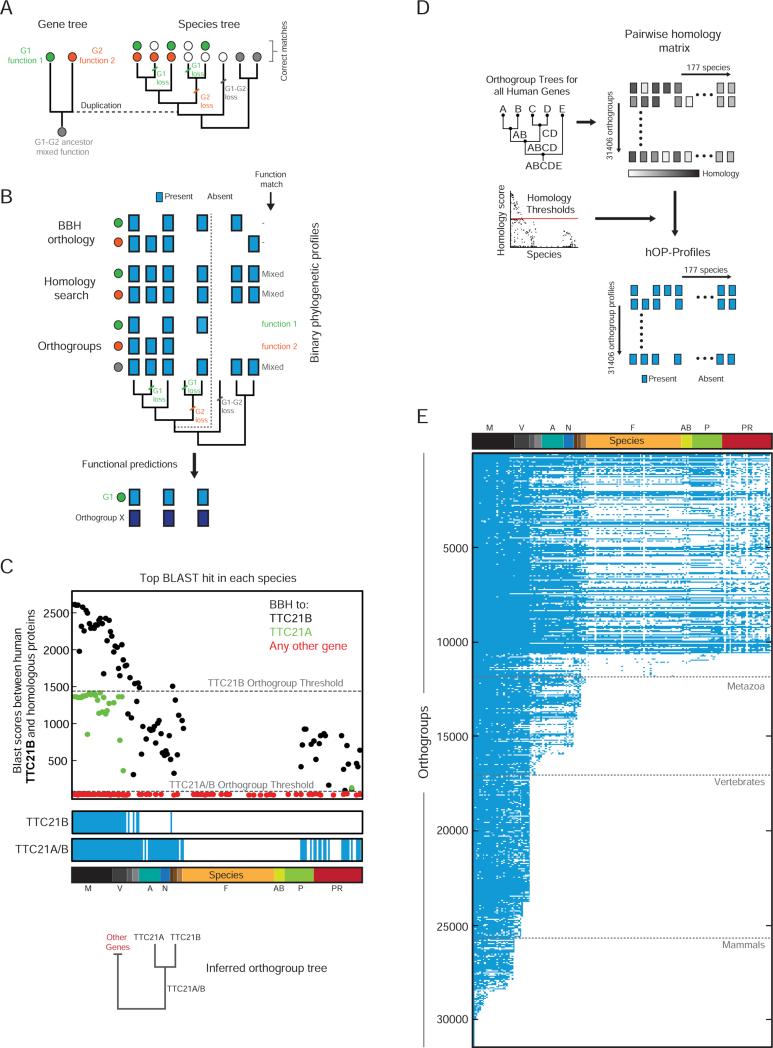
Unlike the examples highlighted in Figure 1, the majority of human genes arose through duplication. Following a duplication event, genes can take different evolutionary trajectories, frequently sharing the functions of the ancestor (subfunctionalization) or acquiring new functions (neofunctionalization) (Figure 2A) (Conant and Wolfe, 2008; Kensche et al., 2008). It is therefore necessary to correctly connect orthologs in each species to either daughter or to a group of homologous human genes (co-orthologs) that reflects the common ancestor (Figure 2A). Assigning human genes to such groups (orthogroups) was a central goal of our analysis. The importance of this step becomes clear in a comparison to other widely used approaches to generating phylogenetic profiles (Figure 2B). The reciprocal best BLAST match criterion (Best Bidirectional Hit or BBH)(Altenhoff and Dessimoz, 2009) applied directly to the daughter genes makes incorrect connections in species that split off before the duplication event (Dalquen and Dessimoz, 2013), while a homology search generates identical profiles for both daughters, linking only to the ancestral function (Li et al., 2014; Tabach et al., 2013a). Separate orthogroup profiles tracking each duplication event provide the most unbiased functional predictions (Figure 2B).
Figure 2. Binary eukaryotic phylogenetic profiles for 31406 human orthogroups.
(A) Schematic to demonstrate the impact of human gene history on the distribution of ortholog functions across species. Genes are represented by filled circles with colors denoting independent functional trajectories. The duplication event is highlighted using a dotted line. (B) A comparison of three different approaches to generating phylogenetic profiles (blue box=present, white box=absent) for the gene family and species tree from (A). The specific functional predictions that each profile can generate are listed to the right. BBH=Best Bidirectional Hit. Dark blue is used to denote a separate phylogenetic profile (Gene X) identified through a similarity search. (C) The top BLAST hit against human TTC21B in each species colored according to a BBH match for the corresponding protein against TTC21B (black), TTC21A (green), or any other human gene (red). Gray lines highlight inferred orthogroup thresholds. Below, resulting phylogenetic profiles (blue/white) and inferred orthogroup tree. See also Figure S2. (D) Extension of algorithm to entire genome. (E) The full hOP-map, containing binary phylogenetic profiles for 31406 orthogroups, listed in decreasing order of estimated first appearance (see Experimental Procedures). Gray lines denote key branch points. See also Figure S2.
For this purpose we needed to obtain unconstrained hierarchical orthogroups defined exclusively by relationships between human genes, on a genome-wide scale. Despite the extensive literature on orthology inference (Kristensen et al., 2011), we could not find a suitable resource meeting these requirements (see Supplemental Experimental Procedures). This fact and the observation that a simple pairwise BBH approach can outperform sophisticated tree-based algorithms, especially when large genomes and many species are involved (Kristensen et al., 2011), led us to develop the modified BBH strategy outlined below. Briefly, we used BLASTp (protein BLAST)(Altschul et al., 1990) bit scores to iteratively join genes into orthogroups and simultaneously identify a score threshold for orthogroup presence (using the longest protein encoded by each gene, see Experimental Procedures). For a given ‘query’ gene or orthogroup (TTC21B in Figure 2C), we used BLAST to identify its closest candidate homolog and the corresponding bit score in each species. We also made use of a reverse-BLAST strategy to define for each of these candidate homologs, its best hit in the human genome (BBH, Figure 2C). We then defined a threshold for identifying a true ortholog of the query as the highest BLAST bit score between the query and any candidate homolog whose BBH in the human genome (‘target’, TTC21A in Figure 2C) was not the query. The target and query were then joined into a new orthogroup and their BLAST scores pooled (TTC21A/B, tree in Figure 2C). This procedure was applied sequentially until the new threshold fell below background levels (see Experimental Procedures), leading to a unique phylogenetic profile for each orthogroup (Figure 2C, Figure S2A). Since other species branching off from the human lineage all evolve their copy of the target (duplicated) gene at approximately the same rate (Figure 2C, after correcting for the higher divergence of a subset of species- such as parasites, see Supplemental Experimental Procedures), this thresholding strategy enables a robust tracking of duplications without the need for explicit models of evolution.
Starting with the 19973 human protein coding genes (NCBI), the algorithm yielded 31406 orthogroups (Figure 2D; Supplemental Experimental Procedures for orthogroup naming conventions) and 31406 matching hOP-profiles (Figure 2E), also available on our website. This analysis generated a comprehensive one-to-one orthology dataset (useful for experiments across multiple species, Figure S2B) and, importantly, confirmed that the majority of human genes can be assigned to gene families (Figure S2C), highlighting both the utility and necessity of an orthogroup-based approach to phylogenetic profiling. We detected a significant fraction of human genes tracing back to early eukaryotes (approximately 25%), with major innovation in both vertebrates and later mammals (>50%, Figure S2D- top)(Blomme et al., 2006). Importantly, our strategy generated phylogenetic profiles for ancestors of many of these vertebrate and mammal-specific genes (Figure S2D- middle), gaining predictive power from the higher inferred loss frequencies in early-branching lineages like the fungi (Figure S2D- bottom). An additional benefit of our approach was the ability to reconstruct gene trees across all gene families, which, though restricted to the human lineage, provide a close match to the literature for well-studied examples (Berg et al., 2001; Boureux et al., 2007) (Figure S2E, S2F, Supplemental Experimental Procedures).
Creation of a genome-wide pair-wise phylogenetic co-occurrence matrix
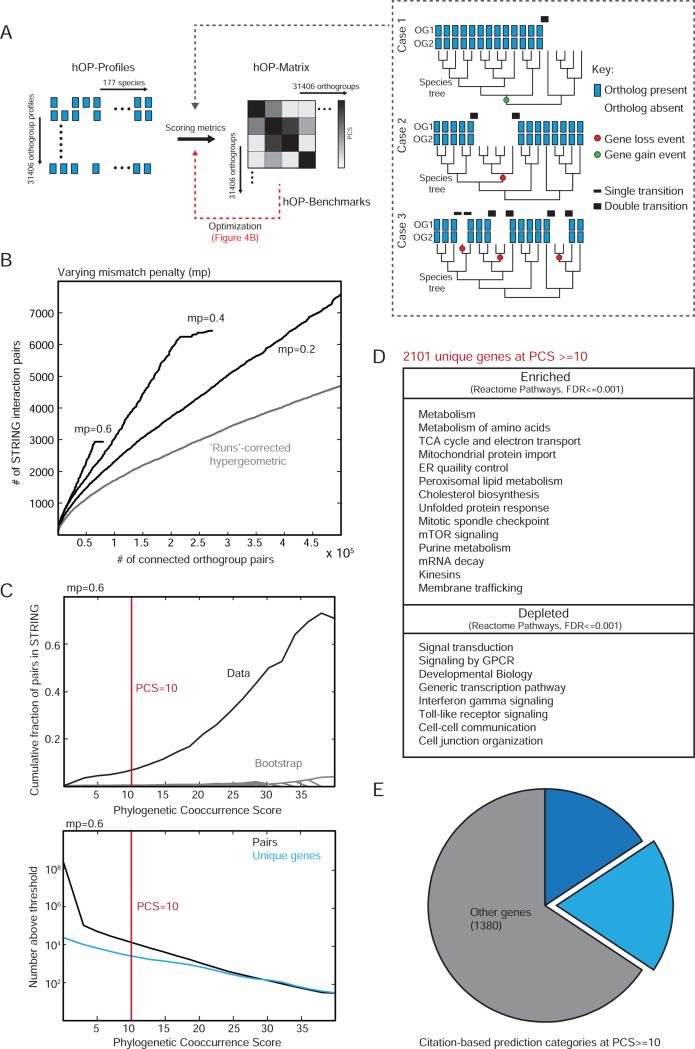
The next major challenge was to define a scoring metric that effectively extracts the predictive value of similarity between pairs of phylogenetic profiles, and then to generate a pairwise phylogenetic co-occurrence matrix between all human orthogroups (hOP-matrix, Figure 3A). Existing methods for comparing phylogenetic profiles fall into two broad categories: linear metrics based on correlation or mutual information and tree-based methods (Kensche et al., 2008). Linear metrics do not account for the interdependence of related species, leading to distorted similarity scores (Figure 3A), while tree-based methods do not scale well and also require the specific fitting of gain/loss models that must also account for noise in homology measurements. A third option, combining the strengths of both approaches, is to use joint binary transitions between presence (1s) and absence (0s) in a pair of linear profiles ordered in accordance with a 1D projection of the species tree (keeping closely related species adjacent) as a proxy for shared loss and gain events (Cokus et al., 2007). Based on the premise that more shared loss events would support a stronger case for co-evolution (Figure 3A), we defined a phylogenetic co-occurrence score (PCS) as the sum of shared transitions weighted to distinguish matches of higher and lower confidence (black bars in Figure 3A) and included a penalty for mismatches (see Experimental Procedures).
Figure 3. Generating a genome-wide phylogenetic co-occurrence matrix.
(A) Schematic for generating a genome-wide pairwise co-occurrence matrix for human orthogroups. The inset highlights 3 pairs of hypothetical orthogroups indistinguishable if independence of species is assumed. However the profiles in case 1 and 2 can be reconciled with likely models of a single gain event (green dot) and a single loss event (red dot) respectively while case 3 involves 3 independent loss events. We used weighted transitions (black bars of varying thickness) to construct the phylogenetic co-occurrence score (PCS, see Experimental Procedures). (B) The PCS was benchmarked against STRING (Experimental Procedures). A range of mismatch penalty (mp) values were compared to each other and the original ‘runs’ algorithm (Cokus et al. 2007) (C) The upper panel shows the cumulative fraction of co-evolving pairs confirmed by STRING for each PCS threshold (black) compared to a random bootstrap (gray), with the lower panel showing the total number of pairs at each corresponding threshold value, at mp=0.6. Red line indicates the threshold used for further analysis. (D) Table highlighting a selected list of enriched and depleted functional terms at PCS >=10 (Experimental Procedures). (E) All genes contained in orthogroups involved in interactions with PCS>=10 were grouped into 3 categories (both poorly studied, citation count <3; one poorly studied, citation count <3 for gene 1 and >10 for gene 2; all other genes; see Supplemental Experimental Procedures for further details). See also Figure S3.
We employed a database of known interactions to optimize and validate the PCS (STRING; see Experimental Procedures). We found that varying the mismatch penalty (mp) value had a strong effect on PCS behavior (Figure 3B), namely that higher stringency improved the known interaction recall (true positive) rate but came at the cost of fewer total interactions recovered (increased false negatives). We found higher stringency (mp=0.6) useful for a genome-wide exploration of functional connections (Figures 4-5), though predictions using a lower penalty value could provide better coverage for specific functional clusters (available on website). We also confirmed that orthogroup-based profiling increases predictive power over a pair-wise BBH approach restricted to genes not assigned to gene families (Figure S3A). Figure 3C plots the cumulative fraction (upper panel) and number of predicted interactions (lower panel) as a function of PCS at mp=0.6, after filters were applied to exclude orthogroups that either appeared too recently or contained too many genes to produce useful functional predictions (Figure S3B and Supplemental Experimental Procedures).
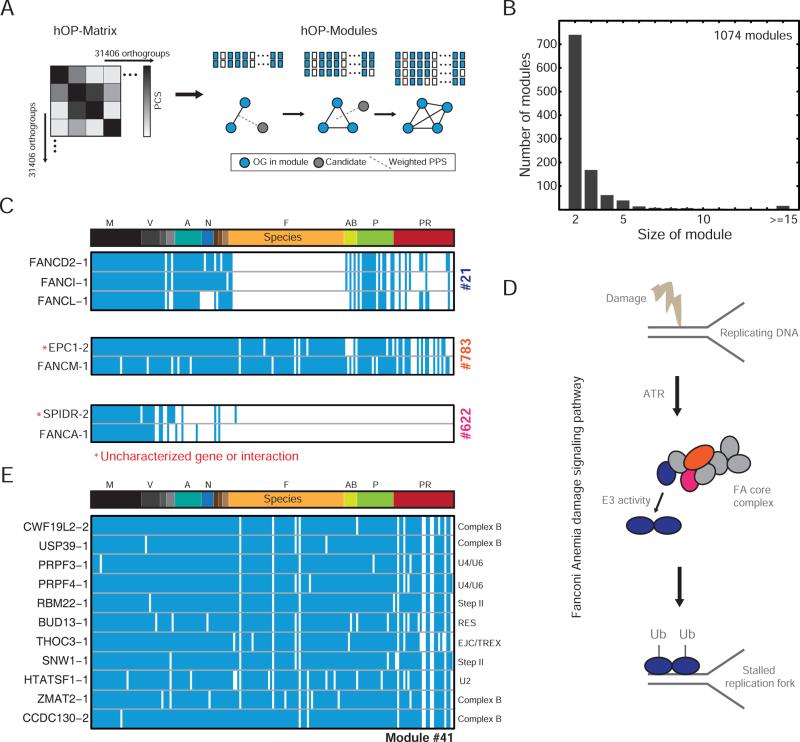
Figure 4. Unbiased clustering of human genes into co-evolving modules.
(A) Schematic illustrating the module expansion strategy (see Experimental Procedures). (B) Histogram representing the final sizes of 1074 modules. (C) Modules linked to the Fanconi Anemia (FA) pathway. Red stars indicate predicted interactions. (D) Schematic illustrating aspects of the FA pathway with genes in hOP-modules color-coded according to (C). (E) Module linked to splicing, with each orthogroup annotated to specific splicing complexes. See also Figure S4.
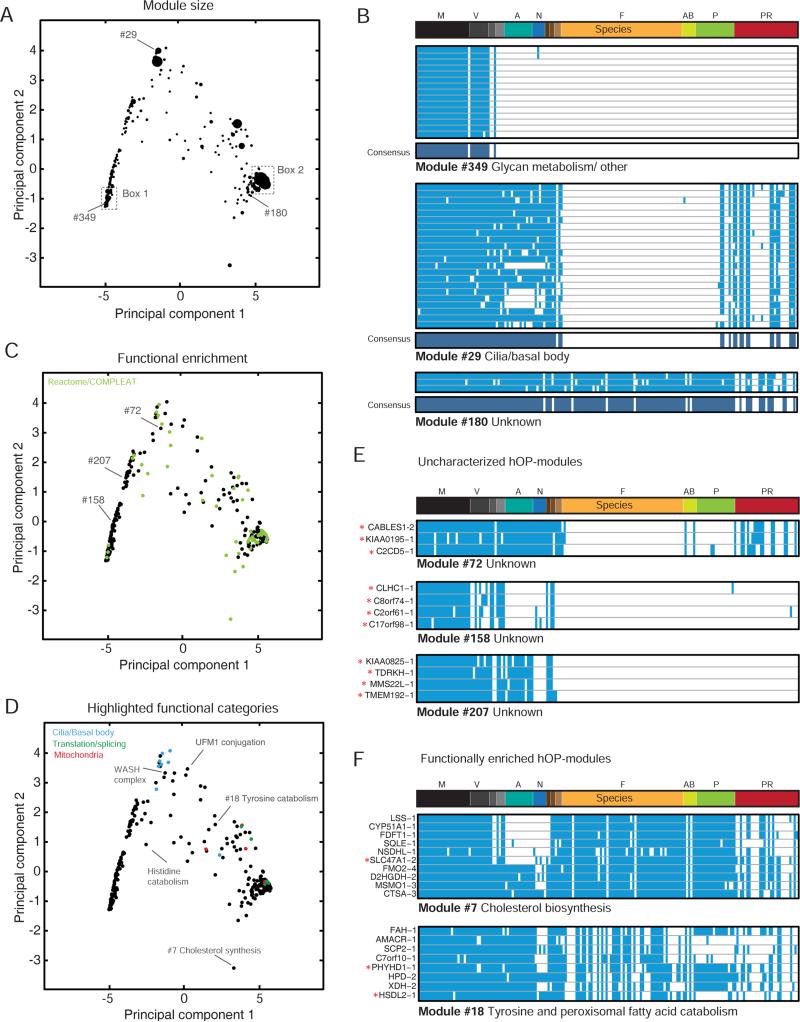
Figure 5. Global analysis of hOP-modules.
(A), (C), (D) The first two principal components obtained from the PCA of consensus profiles corresponding to modules with 3 or more components (334 hOP-modules, Experimental Procedures) plotted against each other superimposing module size (A), functional enrichment (hypergeometric, FDR<0.1) in green (C), or certain highlighted functional categories (D). (B) Examples of modules with consensus profiles, also marked in (A). (E) Examples of uncharacterized hOP-modules, also marked in (C). (F). Examples of functionally enriched hOP-modules, also marked on the map in (D). Red stars indicate refractory genes. See also Figure S4; Table S2 for details of all hOP-modules.
We found that that the strongest co-evolving pairs (2101 unique genes, PCS>= 10) were strongly enriched for large protein complexes, metabolic pathways and some organelles but devoid of genes involved in canonical signaling, immune responses and transcriptional control (Reactome Pathways, see Figure 3D and Table S1), closely mirroring trends in bacteria (Campillos et al., 2006). This observation argues for a generalizable tendency for cellular networks with strong internal coupling (protein complexes and metabolic pathways) to form evolutionary modules over interlinked signaling and transcriptional pathways. Despite these constraints, the finding that the set of strongly co-evolving pairs includes more than 700 genes for which little or nothing is known about its function (Figure 3E and Figure S3C) highlights the power of our approach as a discovery tool. We also noted a strong enrichment for genes involved in monogenic diseases (hypergeometric p-value <1×10−4, Supplemental Experimental Procedures) at the same PCS threshold, highlighting the potential for identifying new functional connections relevant to genetic disorders.
Unbiased genome-wide assignment of co-evolving gene pairs to functional modules
We identified co-evolving modules (hOP-modules) in an unbiased genome-wide approach starting with pairs of proximal orthogroups to which, in sequential steps, a single additional orthogroup was added as the top hit from the weighted PCS average against existing module members (Figure 4A) until a PCS threshold was reached (see Experimental Procedures). The first striking observation was that the size distribution was skewed heavily towards smaller modules with a large number of single pairs failing to pick up additional orthogroups (Figure 4B). These modules often map to one or more sections of highly conserved cellular pathways and multi-protein assemblies (Figure 4C-E), highlighting an unanticipated degree of modularity within these pathways (Figure 4D) and suggesting novel connections (red stars in Figure 4C; for example, SPIDR is a recently identified scaffold protein that has been linked to DNA repair (Wan et al., 2013) but not directly to the Fanconi Anemia pathway). It is worth noting that, in cases like the splicing-related module #41 (Figure 4E) where genes are largely annotated purely based on homology or high-throughput proteomics (Cvitkovic and Jurica, 2013), co-evolution can serve as an independent confirmation of shared function.
When available, orthogonal sources of data helped link the observed evolutionary modularity to its underlying causes. Analyzing a curated compendium of splicing genes (Hegele et al., 2012; Wahl et al., 2009) revealed two clear evolutionary trajectories for ancient splicing genes (Figure S4A), one that is persistent with a few losses in protists, and a second that is characterized by a gradual pruning of the splicing machinery in fungal lineages (Figure S4B-C). These observations parallel previous work on intron losses in fungi (Nielsen et al., 2004) and explain the wide range of complexes observed in hOP-modules enriched for splicing (Figure 4E).
We took advantage of our unbiased approach to ask if a global analysis of hOP-modules would reveal general principles underlying the modular co-evolution of human genes in addition to generating a set of high-confidence functional predictions. We applied principle component analysis (PCA) to ‘consensus profiles’ (Figure 5A-B, Experimental Procedures) for 334 hOP-modules with 3 or more components. The first two principal components (Figure S4D) were dominated by the evolutionary age of hOP-modules and overall loss frequency (examples from Figure 5B define the corners of the PC space in Figure 5A, 5C and 5D). This analysis revealed clear constraints upon the potential range of phylogenetic patterns and also separates out large, poorly-resolved hOP-modules corresponding to mammal-specific (box 1 in Figure 5A) and paneukaryote modules (box 2 in Figure 5A) with a shortage of informative loss events. Analysis of functional enrichment (Reactome/COMPLEAT, Figure 5C) helped identify completely uncharacterized modules (Figure 5E) that may reflect undiscovered cell functions, as well targeted predictions for refractory genes clustering within functionally enriched hOP-modules (Figure 5F). Modules belonging to related functional categories often occupied neighboring areas of the map (Figure 5D). Nevertheless, a large number of consensus profiles were isolated, belonging to modules with unique loss patterns often linked to metabolic functions (Figure 5D, Table S2). Modules occupying sparser areas can be analyzed with a lower threshold to discover more refractory genes without blurring functional boundaries (Figure S4E, S4F). Finally, modules enriched for more than one function may imply novel connections between them, such as Module #18 that is linked to two apparently distinct metabolic pathways, tyrosine/phenylalanine catabolism and the peroxisomal degradation of branched fatty acids (Figure 5F). Interestingly, older studies suggest that these two pathways might be linked (Vamecq and Van Hoof, 1984).
Identifying novel interactors of the WASH complex
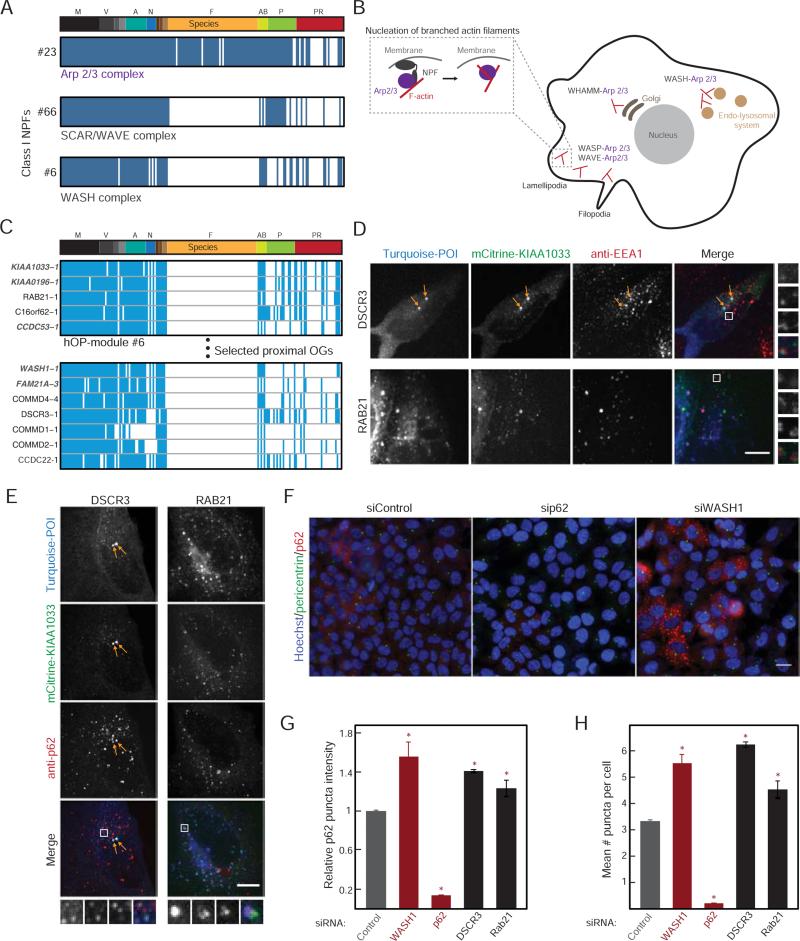
As a first proof of principle, we focused on the regulation of Arp2/3, an ancient multi-protein machine that nucleates actin filaments and organizes them into branched networks (Campellone and Welch, 2010). Arp2/3 can be activated by WASP-WAVE-WASH-family nucleation-promoting factors (Class I NPFs, Figure 6A-B). The Arp2/3 complex as well as the WAVE and WASH nucleation-promoting complexes emerged from our analysis with distinctive loss patterns (Figure 6A) (Kollmar et al., 2012). The WASH complex was of particular interest to us because it is incompletely characterized and we identified promising candidates in its corresponding hOP-module and neighborhood (Figure 6C, Supplemental Experimental Procedures): CCDC22 has been identified in pull-downs with FAM21A (Harbour et al., 2012) and COMM Domain (COMMD) proteins (Starokadomskyy et al., 2013); DSCR3 is an ortholog of the WASH-interacting retromer component VPS26A (Gomez and Billadeau, 2009); the poorly-studied RAB21 localizes to early endosomes (Simpson et al., 2004). The list also provides a close match with very recent predictions from an orthogonal approach (Li et al., 2014).
Figure 6. Investigation of genes with a predicted link to WASH complex function.
(A) Consensus profiles for the Arp2/3 complex, SCAR/WAVE complex and WASH complex (module ID on the left). (B) Schematic highlighting the role of branched actin in various cellular locations and specificity provided by the different Class I NPFs, with inset showing recruitment of Arp2/3 followed by nucleation of a new branch. (C) hOP-profiles from module #6 and extended neighborhood (Supplemental Experimental Procedures) containing WASH complex components (gray) and candidates (red stars). (D), (E) mTurquoise-tagged DSCR3 or RAB21 (blue in merge) were co-expressed with mCitrine-tagged KIAA1033 (green in merge) in HeLa cells also immunostained with an antibody to EEA1 (D, red in merge) or p62 (E, red in merge) and imaged at 63X on a confocal microscope. Images are maximum intensity projections. In the DSCR3 images, arrows highlight characteristic bright co-localizing EEA1-negative foci. In both cases, white boxes represent magnified regions on right and bottom. Scale bar=10 μm. (F) HeLa cells treated with siRNA for 48 hours were subjected to acute stress (10 μM Bortezomib for 30 minutes), and immunostained for pericentrin (green) and p62 (red) and labeled with Hoechst (blue) to mark nuclei. Sample images from 3 treatment conditions obtained at 20X are shown. Scale bar=20 μm. (G) Mean p62 puncta intensity per cell (Supplemental Experimental Procedures) for each condition. Bars represent the mean and standard deviation of two replicates with >1000 cells per replicate from a single experiment (experiments could not be pooled due to the variable fraction of control cells expressing p62, but trends were reproducible across >3 independent experiments). Red stars mark a significant deviation from control (p-value<0.05, estimated using multiple pairwise comparison after one-way ANOVA). (H) The average number of puncta per cell, analyzed as in (G). See also Figure S5.
The WASH complex has been implicated in regulating endosome morphology (Derivery et al., 2009), endosome-to-Golgi transport (Gomez and Billadeau, 2009) and autophagic flux (Xia et al., 2014; Zavodszky et al., 2014) in mammalian cells with data from Dictyostelium (Park et al., 2013) suggesting that the autophagy role might be conserved. To test for such a potential link, we co-expressed fluorescently tagged candidates DSCR3 and RAB21 together with the WASH complex component SWIP/KIAA1033, followed by immunostaining for early endosomes (EEA1) or autophagosomes/aggresomes (p62/SQSTM1). Both DSCR3 and RAB21 colocalized strongly with the WASH complex marker in puncta that partially overlapped with both compartment markers (Figure 6D, 6E). Inspection of larger EEA1-positive rings revealed the WASH marker also in small surface domains, consistent with previous reports (Figure 6D, (Derivery et al., 2009)) while RAB21 was excluded from these domains (Figure 6D, 6E). While multiple COMMD proteins also colocalized strongly with the WASH marker (Figure S5A-B), we found that, unlike the other candidates, they also aggregated when expressed at high levels (data not shown) (Vonk et al., 2014) and did not analyze them further. We confirmed that knockdown of WASH1, which destabilizes the entire complex, led to a visible collapse of the endosomal membrane network (Figure S5C) and also a strong increase in the number and intensity of p62 autophagosome-like puncta visible after acute stress (Figure 6F-6H). Interestingly, knockdown of DSCR3 or RAB21 phenocopied the latter effect (Figure 6F-6H), indicating a likely parallel disruption of autophagic flux. While correlative, these experiments imply a shared function with the WASH complex and provide a starting point for future investigation.
Predicting cilia/basal body components
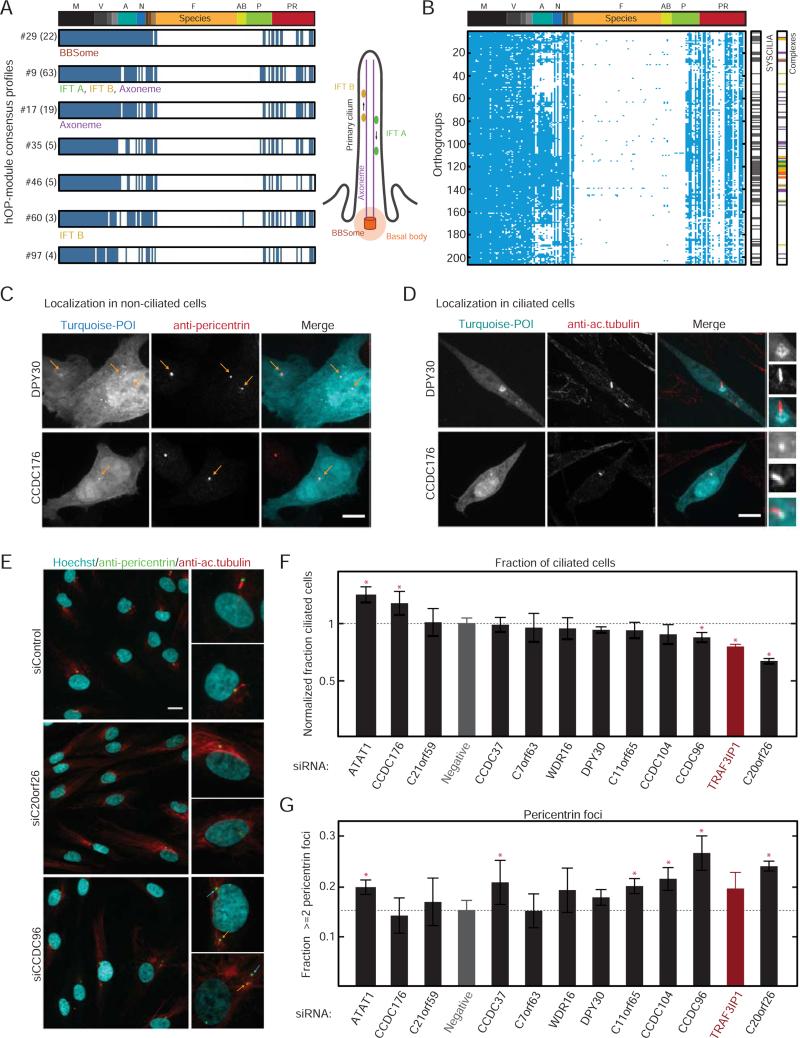
We noted that several hOP-modules were enriched for cilia function (Figure 5D). Cilia are highly specialized organelles that date back to the first eukaryotic ancestor, providing motility and signaling specificity to a wide range of organisms and cell types. Despite their importance, cilium and associated basal body (CBB) genes have been lost in multiple lineages (e.g. fungi), or diverged significantly (e.g. insects), giving rise to the striking loss patterns observed in Figure 7A (Carvalho-Santos et al., 2011). Given the success of previous phylogenetic profiling studies (Avidor-Reiss et al., 2004; Li et al., 2004), we conducted a genome-wide analysis with lower PCS stringency to identify human cilia/basal body genes (Supplemental Experimental Procedures). We clustered this ‘super-module’ based on losses in species with diverged or no cilia (Carvalho-Santos et al., 2011). Orthogroups belonging to core cilia components (BBSome and interflagellar transport complexes A and B) clustered together in this analysis (Figure 7B, 206 unique genes). While these genes overlapped strongly with a recently curated ‘gold standard’ ciliome (Figure 7B, 88/206) and a more expansive database of cilia genes (91/206), our analysis identified a sizeable number of poorly characterized candidates (Table S3).
Figure 7. Investigation of genes with a predicted role in cilia/basal body function.
(A) Consensus profiles for hOP-modules (labels: #ID (no. of orthogroups)) enriched for genes involved in CBB function. (B) Ciliome (206 hOP-profiles) identified through module expansion (Experimental Procedures). “SYSCILIA” and “Complexes” bars indicate overlap with respective lists. (C) Localization of transiently expressed candidate proteins (tagged with mTurquoise, cyan in merge) at centrosomes (antibody to pericentrin, red in merge) in HeLa cells. Yellow arrows highlight locations of centrosomal foci. Scale bar=10 μm. Images are maximum intensity projections of confocal stacks acquired at 63X. (D) Localization of the same candidates (cyan) in NIH3T3 cells with primary cilia marked by an antibody to acetylated tubulin (red), Boxes highlight ciliary region, zoom to the right. Imaged as in (C). Scale bar=10 μm. (E) Serum-starved Hs68 cells subjected to 60 hours of siRNA treatment were stained with Hoechst (cyan in merge) and labeled with pericentrin (green) and acetylated tubulin antibodies (red), followed by wide-field imaging at 20X. Boxes correspond to zoom on right. Yellow arrows highlight centrosome with associated cilium, blue arrows highlight additional centrosomal foci. Scale bar=20 μm. (F) The normalized fraction of cells with a detectable cilium following siRNA treatment, averaged over 4 replicates pooled from multiple experiments for each condition with >1000 cells per replicate (Experimental Procedures); scrambled siRNA (“Negative”) in gray, positive control TRAF3IP1 in red. Red stars highlight significant difference from negative control (p-value <0.05, multiple pairwise comparison following one-way ANOVA) (G) The fraction of cells with 2 or more detectable pericentrin foci, averaged, ordered and assessed for significance as in (F). See also Figure S6.
We selected 12 of these candidate genes for experimental validation (Table S4, see Supplemental Experimental Procedures), using the recently identified IFTA component TRAF3IP1 (Berbari et al., 2011) as a positive control. For the subset we were able to express with a fluorescent tag (Supplemental Experimental Procedures), we measured their colocalization with antibodies labeling pericentrin and/or acetylated tubulin in non-ciliated and ciliated cells. All candidates except for one localized at least partially to the centrosome and/or basal body (Figure 7C, 7D, and Figure S6A-B) with a strong correlation between the two cell lines. Notably, the DPY30 protein localized to a diffuse area around the basal body (Figure 7D, upper panel) in ciliated cells. We further assayed the effect of knocking down each candidate gene on cilia and basal body organization in serum-starved primary human fibroblasts (Figure 7E-7G). We used antibodies against acetylated tubulin and pericentrin to label the primary cilium and centrosome respectively, and custom image analysis scripts (Figure S6C, Experimental Procedures) to quantify defects in cilia formation/disassembly (the fraction of ciliated cells at steady state) and centrosome duplication/separation (the number of pericentrin foci per cell at steady state). Depletion produced a cilia phenotype for 3 and an apparent centrosome defect for 6 candidates (Figure 7G). It should be noted that the genes with no phenotype in this assay might be specifically associated with motile cilia function (Table S4). These survey experiments suggest that most of the tested genes are linked to cilia/basal body function, highlighting our capacity to systematically generate experimentally tractable predictions.
DISCUSSION
Our study shows that the hOP-profiles, -lists and -modules that we derived provide a starting point to answer important questions about human gene function and modularity. First, simply exploring the phylogenetic profiles and orthogroup trees can help develop a better understanding of the evolutionary roots of individual genes or gene families, or to identify suitable model organisms to investigate relevant orthologs. Second, the rank-ordered lists for known genes and orthogroups (available online) can be used to discover functions of refractory genes that are as of yet uncharacterized or incompletely understood. Third, exploration of the clustered hOP-modules allows one to learn about the modular architecture of specific cell functions of interest, and perhaps even the discovery of novel cellular functions. When analyzed more globally, the hOP-modules enable the derivation of general principles: for example, that evolution often operates on distinct sub-modules and sub-complexes at surprisingly small scales. Furthermore, the depletion of signaling and transcriptional networks compared to the strong enrichment for large protein complexes, structural modules, and linear metabolic pathways in our analysis highlights the role of single gene plasticity over module gain and loss in the evolution of interlinked regulatory networks. Within these evolutionary constraints, functional linkages can be confidently inferred for over 10% of the genome using the currently available species.
We experimentally validated our computational predictions by focusing on predicted refractory human genes associated with the cilia/basal body or the WASH complex. Localization and siRNA analysis suggested that both tested sets have roles linked to cilia/basal body and WASH-associated functions, highlighting the potential of our approach to provide experimentally tractable predictions for a significant fraction of the remaining dark matter of the human protein-coding genome (comprising more than 6000 poorly studied genes, using the metric defined in this study, Figure S3C).
Despite the large number of algorithms devoted to orthology inference (Altenhoff and Dessimoz, 2009; Kristensen et al., 2011), and a recent study highlighting the utility of an orthogroup-based approach in identifying gene gains and losses in fungi (Wapinski et al., 2007), orthogroup-based phylogenetic profiling has not yet, to our knowledge, been applied to a systematic analysis of eukaryotic species. The lack of a suitable orthology resource for this application led us to develop a modified BBH strategy (using all-against-all BLAST scores) to define hierarchical orthogroups and threshold phylogenetic profiles. As highlighted in Figure 2B and in earlier studies (Kensche et al., 2008), the alternative strategies of using a pair-wise BBH criterion, or simply comparing genes based on homology scores (Tabach et al., 2013b) are only suitable for singleton genes with ancient origins, that by our estimate and others (Zhang, 2003) make up less than half the genome.
A second critical step in our analysis was the generation of a genome-wide pairwise cooccurrence matrix that enables direct optimization and benchmarking against known functional interactions. The metric we used was inspired by a previously introduced “runs” algorithm that aligned species in a linear profile according to evolutionary proximity and used transitions in the linear vector as a proxy for loss events (Cokus et al., 2007). Such an analysis has a number of advantages over full tree-based inference: it does not require training sets and scales easily with genome size and tree complexity. These advantages are highlighted in comparison to a recent tree-based algorithm that learns from and expands existing human pathways (Li et al., 2014). While effective for large modules with good training sets (such as the ciliome or mitochondrial modules), this algorithm could not be extended to all human genes or orthogroups, and due to its seed-based approach, is unable to identify very small or poorly characterized modules. As tree-based algorithms continue to develop, however, we expect that it will be eventually feasible to replace our linear metric with explicit and more accurate models of gene gain and loss.
Finally, it is useful to point out that increasing the number of species and a better annotation of their genomes would allow for a significant extension of the predictive power of phylogenetic profiling. This provides a strong argument for a 1000 eukaryotic genome challenge: a large effort in high quality sequencing and annotation, particularly in the currently poorly covered evolutionary side-branches. Our algorithm is specifically designed to scale with this anticipated increase in species coverage with a negligible increase in computational requirements or error rates.
In summary, our study presents an unbiased genome-wide strategy for a species-centered phylogenetic loss analysis that we show has practical utility in discovering functions of refractory genes as well as for understanding how specific functional modules evolve. We show that phylogenetic profiling provides fundamental insights into human gene function and into the modular logic of how our cells are built and adapt over the course of evolution.
EXPERIMENTAL PROCEDURES
Projection of species tree
We obtained the coarse branching relationships between the 177 species from the NCBI Taxonomy browser (September 2013) in consensus with the current literature (Burki, 2014). The tree was projected in one dimension starting with the human genome at the leftmost extreme, and moving through 13 primary branches relative to the direct human lineage. Within smaller poorly resolved sub-branches, we reordered species according to their local proximity to neighbors based on gene content (shared presence of orthologs across all human genes), starting with the highest number of conserved human genes on the left (final species ordering in Figure S1). This local reordering allowed us to minimize spurious transitions (apparent loss events) in the linear vector (Cokus et al., 2007).
Generation of hOP-profiles
The species used for phylogenetic profiling were restricted to high-quality verified RefSeq genomes to prevent propagation of annotation errors. We first identified the top-scoring homolog (BLASTp bit scores) for the longest protein encoded by each human gene (19973 unique genes, NCBI, March 2013) in each fully annotated genome (177 eukaryotic species, NCBI, March 2013), as well as the reciprocal best match in the human genome for each contributing protein in another species (BBH). In the first step of our iterative algorithm, the third highest BLASTp bit score (Supplemental Experimental Procedures) against a human gene (query) belonging to a gene with a different human BBH match (target) was used to combine the target and query into a single orthogroup and simultaneously set the threshold for orthogroup presence. BLASTp scores for each new orthogroup were pooled and the algorithm iterated until a bit score threshold (50) was reached or the orthogroup size exceeded 100, resulting in 31406 orthogroups for 19973 genes. The orthogroup thresholds were then applied to a matrix of the top BLASTp bit scores (corrected for species evolving significantly faster than their neighbors) against each orthogroup to generate 31406 binary phylogenetic profiles (available in File S1, see Supplemental Experimental Procedures for details).
Generation of the hOP-matrix
A tree-aware score to compare pairs of hOP-profiles was defined as the linear sum of shared transitions between presence and absence with a positive weight for transitions involving doublets (00→11 and 11→00) and a (negative) penalty for mismatches. This score was calculated for each pair of profiles to generate a 31406*31406 sparse co-occurrence matrix. The STRING database of protein-protein interactions was used to benchmark the score and optimize the weight and penalty factors (maximizing the number of identified STRING interactions as a fraction of the total number of co-evolving pairs). Orthogroups first appearing in the vertebrate branch and orthogroups containing more than 4 genes (poor information content) were excluded for the global analyses of pairs and modules, leaving a total of 14412 orthogroups (scores in File S1, see Supplemental Experimental Procedures for details).
Benchmarking and enrichment statistics
We used the STRING protein-protein interaction database (http://string-db.org/, version 9.1, (Franceschini et al., 2013)) to optimize and benchmark the co-occurrence metrics between hOP-profiles. P-values for functional enrichment were estimated using the hypergeometric test and reported using the false discovery rate (FDR). Terms and associated annotations were obtained from the Reactome database (http://www.reactome.org/, version 48 downloaded 07/ 2014 and the COMPLEAT database (http://www.flyrnai.org/compleat/, downloaded 06/2014, (Vinayagam et al., 2013)). See Supplemental Experimental Procedures for details.
Generation of hOP-modules
Agglomerative modules were created in a stepwise fashion starting with high-scoring seed pairs of orthogroups (PCS>=5 for full coverage of the map). At each step, the top-scoring orthogroup from the weighted average of co-occurrence scores between existing module components and the rest of the co-occurrence matrix was added to the module, and the iterative process halted at PCS=5. To maximize the growth opportunity for high-confidence co-evolving seed pairs, modules were created sequentially starting with the strongest seed pair and module components removed from the general pool. See Table S2 and Supplemental Experimental Procedures for additional details.
Consensus profiles and PCA
Binary consensus profiles were generated for each hOP-module by assigning a 1 to each species with an ortholog in 50% or more of the member hOP-profiles and a 0 otherwise. The 334 consensus profiles corresponding to hOP-modules with 3 or more components were then subjected to a principal component analysis (PCA) implemented using inbuilt MATLAB functions. The first two principal components were used for all subsequent analysis.
Cell culture, siRNA and expression constructs, antibodies
HeLa, NIH3T3, and Hs68 cells were cultured in 10%FBS/high-glucose DMEM/PSG medium (GIBCO). For siRNA experiments or transient transfections, cells were cultured on 96-well plastic-bottom (Corning 3904) or glass-bottom (In Vitro Scientific, P96-1.5H-N) plates. siRNA reagents were obtained from Qiagen (pools of 3). Expression constructs were derived from the human ORFeome collection (version 5.1, http://horfdb.dfci.harvard.edu/hv5/) or from PCR. Antibodies against p62/SQSTM1 (mouse, used at 1:400, BD Biosciences 610832), acetylated tubulin (mouse, used at 1:3000, Sigma T6793), pericentrin (rabbit, used at 1:800, Abcam ab4448), and EEA1 (mouse, used at 1:400, BD Biosciences 610457) were employed for immunofluorescence. See Supplemental Experimental Procedures for detailed protocols and Table S5 for siRNA specifications.
Imaging and image analysis
High-resolution images were acquired out on a custom-assembled spinning disk confocal system built around an inverted Zeiss Axiovert 200M microscope, equipped with 442 nm, 514 and 593.5 nm lasers, appropriate excitation and emission filters, and a CCD camera (CoolSNAP HQ, Photometrics/Roper Scientific). A 63X 1.2 NA Plan-APOCHROMAT Zeiss water-immersion objective was used and images acquired using μManager (Edelstein et al., 2010). High-throughput imaging was carried out on an integrated ImageXpress (Molecular Devices) high-content analysis system with a 20X 0.75 NA Plan Apo objective (Nikon). All image analysis was carried out using custom-written MATLAB routines (see Supplemental Experimental Procedures for a description of the algorithms employed).
Supplementary Material
Highlights.
Correlated evolutionary histories predict functions for human genes
Reconstructing shared ancestry extends predictive ability to gene families
Scalable scoring metric leads to unbiased genome-wide functional predictions
Predictions are experimentally tractable and a subset were validated in cell lines
ACKNOWLEDGEMENTS
We thank Arnold Hayer, Amy Winans, Steve Cappell and Chad Liu for critical reading of the manuscript, Arnold Hayer for providing GATEWAY destination vectors, Mingyu Chung for a MATLAB nuclear segmentation routine and members of the Meyer lab for helpful discussions and feedback. This work was supported by NIH grants GM063702 and GM030179, the Stanford Center for Systems Biology, and the Stanford Graduate Fellowship in Science and Engineering (G.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
G.D. and T.M. conceived, developed and implemented the algorithm. S.R.C. and T.M. developed a precursor to the algorithm. G.D. performed secondary and statistical analyses. G.D. carried out experiments. A.J. developed the web server. A.S. developed experimental protocols. T.M. supervised the project. G.D., S.R.C., A.J. and T.M. wrote the paper.
eTOC
Dey et al. extend phylogenetic profiling, the analysis of shared genetic evolutionary history, to create an unbiased global map of functional modules in the human genome. In addition to experimentally tractable predictions for hundreds of uncharacterized genes, this resource also suggests underlying principles of evolutionary modularity in human cellular networks.
REFERENCES
- Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding Cilia FunctionDefining Specialized Genes Required for Compartmentalized Cilia Biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Barker D, Pagel M. Predicting functional gene links from phylogenetic-statistical analyses of whole genomes. PLoS Comput. Biol. 2005;1:e3. doi: 10.1371/journal.pcbi.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20. doi: 10.1093/bioinformatics/btl558. [DOI] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Kin NW, Sharma N, Michaud EJ, Kesterson RA, Yoder BK. Mutations in Traf3ip1 reveal defects in ciliogenesis, embryonic development, and altered cell size regulation. Dev. Biol. 2011;360:66–76. doi: 10.1016/j.ydbio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers PM, Cokus SJ, Eisenberg D, Yeates TO. Use of logic relationships to decipher protein network organization. Science. 2004;306:2246–2249. doi: 10.1126/science.1103330. [DOI] [PubMed] [Google Scholar]
- Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 2014;6:a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M, von Mering C, Jensen LJ, Bork P. Identification and analysis of evolutionarily cohesive functional modules in protein networks. Genome Res. 2006;16:374–382. doi: 10.1101/gr.4336406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S, Mizutani S, Pellegrini M. An improved method for identifying functionally linked proteins using phylogenetic profiles. BMC Bioinformatics 8 Suppl. 2007;4:S7. doi: 10.1186/1471-2105-8-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cotton JA, Page RDM. Rates and patterns of gene duplication and loss in the human genome. Proc. Biol. Sci. 2005;272:277–283. doi: 10.1098/rspb.2004.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovic I, Jurica MS. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res. 2013;41:D132–D141. doi: 10.1093/nar/gks999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalquen DA, Dessimoz C. Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol. Evol. 2013;5:1800–1806. doi: 10.1093/gbe/evt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date SV, Marcotte EM. Discovery of uncharacterized cellular systems by genome-wide analysis of functional linkages. Nat. Biotechnol. 2003;21:1055–1062. doi: 10.1038/nbt861. [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Desmond E, Gribaldo S. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 2009;1:364–381. doi: 10.1093/gbe/evp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol. Biol. Evol. 2008;25:2699–2707. doi: 10.1093/molbev/msn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, Seaman MNJ. Recruitment of the endosomal WASH complex is mediated by the extended “tail” of Fam21 binding to the retromer protein Vps35. Biochem. J. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Kensche PR, van Noort V, Dutilh BE, Huynen MA. Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. J. R. Soc. Interface. 2008;5:151–170. doi: 10.1098/rsif.2007.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Lbik D, Enge S. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res. Notes. 2012;5:88. doi: 10.1186/1756-0500-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen DM, Wolf YI, Mushegian AR, Koonin EV. Computational methods for Gene Orthology inference. Brief. Bioinform. 2011;12:379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC. Comparative Genomics Identifies a Flagellar and Basal Body Proteome that Includes the BBS5 Human Disease Gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Li Y, Calvo SE, Gutman R, Liu JS, Mootha VK. Expansion of Biological Pathways Based on Evolutionary Inference. Cell. 2014;158:213–225. doi: 10.1016/j.cell.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CB, Friedman B, Birren B, Burge CB, Galagan JE. Patterns of intron gain and loss in fungi. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev. Cell. 2013;24:169–181. doi: 10.1016/j.devcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JC, Griffiths G, Wessling-Resnick M, Fransen JAM, Bennett H, Jones AT. A role for the small GTPase Rab21 in the early endocytic pathway. J. Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- Starokadomskyy P, Gluck N, Li H, Chen B, Wallis M, Maine GN, Mao X, Zaidi IW, Hein MY, McDonald FJ, et al. CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling. J. Clin. Invest. 2013;123:2244–2256. doi: 10.1172/JCI66466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Billi AC, Hayes GD, Newman M. a, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013a;493:694–698. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Golan T, Hernández-Hernández A, Messer AR, Fukuda T, Kouznetsova A, Liu J-G, Lilienthal I, Levy C, Ruvkun G. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol. Syst. Biol. 2013b;9:692. doi: 10.1038/msb.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamecq J, Van Hoof F. Implication of a peroxisomal enzyme in the catabolism of glutaryl-CoA. Biochem. J. 1984;221:203–211. doi: 10.1042/bj2210203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayagam A, Hu Y, Kulkarni M, Roesel C, Sopko R, Mohr SE, Perrimon N. Protein complex-based analysis framework for high-throughput data sets. Sci. Signal. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk WIM, Kakkar V, Bartuzi P, Jaarsma D, Berger R, Hofker MH, Klomp LWJ, Wijmenga C, Kampinga HH, van de Sluis B. The Copper Metabolism MURR1 domain protein 1 (COMMD1) modulates the aggregation of misfolded protein species in a client-specific manner. PLoS One. 2014;9:e92408. doi: 10.1371/journal.pone.0092408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wan L, Han J, Liu T, Dong S, Xie F, Chen H, Huang J. Scaffolding protein SPIDR/KIAA0146 connects the Bloom syndrome helicase with homologous recombination repair. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10646–10651. doi: 10.1073/pnas.1220921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Xia P, Wang S, Huang G, Du Y, Zhu P, Li M, Fan Z. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014;24:943–958. doi: 10.1038/cr.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MNJ, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC. Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 2014;5:3828. doi: 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.