Abstract
The B-DNA double helix can dynamically accommodate G–C and A–T base pairs in either Watson-Crick or Hoogsteen configurations. Here, we show that G–C+ and A–U Hoogsteen base pairs are strongly disfavored in A-RNA. As a result, N1-methyl adenosine and N1-methyl guanosine, which occur in DNA as a form of alkylation damage, and in RNA as a posttranscriptional modification, have dramatically different consequences. They create G–C+ and A–U Hoogsteen base pairs in duplex DNA that maintain the structural integrity of the double helix, but block base pairing all together and induce local duplex melting in RNA, providing a mechanism for potently disrupting RNA structure through posttranscriptional modifications. The markedly different propensities to form Hoogsteen base pairs in B-DNA and A-RNA may help meet the opposing requirements of maintaining genome stability on one hand, and dynamically modulating the structure of the epitranscriptome on the other.
INTRODUCTION
The Watson-Crick (WC) double helix is the most common structural element in RNA and the dominant structure of genomic DNA. It provides the basis for templated replication, transcription, and translation, and also serves as a scaffold that defines the 3D structure of DNA, RNA, and their protein complexes. The canonical double helices formed by RNA (A-form) and DNA (B-form) differ in several important respects (Fig. 1a). In B-form DNA (B-DNA), the five-membered deoxyribose ring is flexible and favors the C2′-endo sugar pucker (Fig. 1a). In contrast, due to the sugar 2′-hydroxyl group (2′-OH), the sugar in A-RNA is more rigid and adopts an alternative C3′-endo conformation1,2 (Fig. 1a). This in turn brings the oxygen atoms (O5′ and O3′) adjoining sequential nucleotides into closer proximity effectively compressing and rigidifying the A-form helix, widening its helical diameter, and displacing base pairs (bps) away from the helical axis1,3(Fig. 1a). In addition, B-DNA and A-RNA differ considerably with respect to their deformability, with B-DNA being generally more flexible4. For example, B-DNA is more bendable than A-RNA, and this property is of fundamental importance for many biochemical processes including the tight compaction of genome within the nucleus in higher order organisms.
Figure 1.
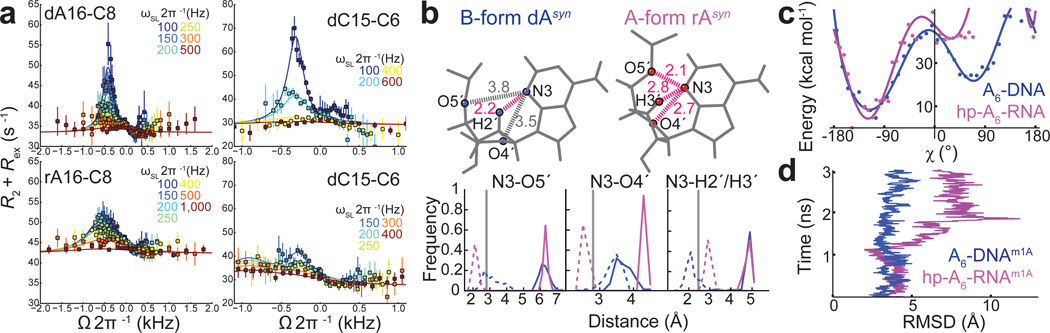
Absence of detectable WC⇄HG exchange in A-RNA by NMR relaxation dispersion. (a) Comparison of A-form RNA (violet) and B-form (blue) DNA double helices. (b) WC and HG bps in dynamic equilibrium in B-DNA. Sites used for RD measurements are highlighted in orange. (c) A6-DNA and hp-A6-RNA duplexes with bps targeted in RD measurements highlighted. (d) Off-resonance RD profiles showing R2+Rex as a function of spin lock offset (Ω 2π−1Hz, where Ω = Ωobs – ωRF) and power (ωSL 2π−1Hz, in insets). Error bars represent experimental uncertainty (one s.d.) estimated from mono-exponential fitting using a Monte-Carlo based method (Methods). Solid line represents a fit to two-state exchange6.
Recently, NMR studies have uncovered a new dynamic property in canonical B-DNA; Watson-Crick (WC) dG–dC and dA–dT bps exist in a dynamic equilibrium with alternative Hoogsteen (HG) bps5,6. A HG bp is created by rotating a WC purine base ≈180° around the glycosidic bond to adopt a syn rather than anti conformation5 (Fig. 1b). The two bases are also brought into closer proximity by ≈2.0–2.5 Å to form a unique set of hydrogen bonds (H-bonds) (Fig. 1b). HG bps exist transiently (lifetimes typically 0.1–1 ms) and in low abundance (populations typically <3%) in naked canonical B-DNA6,7. However, dA–dT and dG–dC+ HG bps can become the dominant configuration (for review see ref. 8) in DNA-protein9 and DNA-small molecule10 complexes where they contribute to DNA recognition, in damaged nucleotides where they contribute to damage accommodation and repair11–13, and in the active sites of translesion synthesis polymerases that use HG pairing to bypass damage during DNA replication14. Purine-purine HG bps have also been shown to play important roles in DNA replication errors and in DNA damage accommodation and repair15,16.
Here, we set out to study WC-HG dynamics in canonical A-RNA duplexes. We show that unlike the canonical B-DNA double helix, rA–rU and rG–rC+ HG bps are strongly disfavored in A-RNA duplexes. As a result, while the DNA double helix can absorb damaged nucleotides incapable of forming WC bps such as N1-methyl deoxyadenosine (m1dA) and N1-methyl deoxyguanosine (m1dG) by forming HG bps; the same methyl marks, N1-methyl adenosine (m1rA) and N1-methyl guanosine (m1rG), acting as a posttranscriptional modification in RNA, block base pairing altogether. This provides a direct mechanism for potently modulating the structure of the epitranscriptome. Our results indicate that HG-dependent DNA biochemical transactions may not be as readily supported in RNA duplexes and identify a unique dynamic property in B-DNA that may help enhance its ability to function as the repository of genetic information.
RESULTS
Absence of conformational exchange in A-RNA
We used NMR spin relaxation in the rotating frame (R1ρ)17–19 to examine whether WC bps in A-RNA duplexes transiently adopt HG bps like in B-DNA. A dynamic equilibrium between a dominant ground state (GS) and short-lived low-abundance ‘excited state’ (ES) conformation can lead to line-broadening of NMR resonances if the conformational exchange occurs on the µs–ms timescale. The R1ρ experiment17 measures this line broadening contribution (Rex) to the transverse relaxation rate (R2) during a relaxation period in which a continuous radiofrequency (RF) field is applied with variable power (ωSL) and frequency (ωRF). The resulting dependence of R2+Rex on ωSL and ωRF, referred to as relaxation dispersion (RD), can be fitted to the Bloch-McConnell equations describing n-site exchange20 to extract exchange parameters of interest, including the population of the ES (pES), the rate constant for conformational exchange (kex = kforward + kbackward), and the difference between the chemical shifts of the ES and GS (Δω = ωES–ωGS).
So far, RD studies have provided evidence for µs–ms conformational exchange in non-coding RNAs involving localized changes in secondary structure in and around non-canonical motifs (reviewed in ref. 18). The RD contributions from such chemical exchange processes can mask the ability to detect WC⇄HG exchange. To hone in on WC⇄HG exchange in A-RNA, we carried out 13C and 15N R1ρ RD experiments21,22 on an RNA duplex (hp-A6-RNA) capped by a stabilizing apical loop lacking non-canonical motifs and containing the same sequence (A6-DNA) for which we first reported transient HG bps in B-DNA6 (Fig. 1c and Supplementary Fig. 1a). We targeted purine-C8, C-C6, G-N1, T-N3 and sugar purine-C1′ sites (highlighted in orange in Fig. 1b), all of which have previously been shown to exhibit significant RD due to WC⇄HG chemical exchange in B-DNA6,7,23 (Supplementary Table 1 and Supplementary Note). In stark contrast to B-DNA, all RD profiles measured in in hp-A6-RNA were flat with no signs of detectable conformational exchange on the µs–ms timescale (Fig. 1d). No RD was observed across a variety of rG–rC and rA–rU WC bps, under low pH conditions (pH = 5.4) that allow optimal RD detection of WC⇄HG exchange in B-DNA6,24, upon increasing the temperature (T = 35°C), and in the presence of 4 mM Mg2+ (at pH = 6.8 and T = 5 or 25°C) (Figs. 1d and Fig. 2b, Supplementary Fig. 1b).
Figure 2.
Lack of detectable exchange across diverse RNA sequence and structural contexts. (a) Secondary structures with bps showing no detectable RD highlighted in red. (b) Off-resonance RD profiles for the highlighted bps with error bars representing experimental uncertainty (one s.d.) estimated from mono-exponential fitting using a Monte-Carlo based method (Methods).
To broaden the search for WC⇄HG exchange in A-RNA duplexes, we carried out additional RD measurements over a wide range of conditions (pH = 5.4–8.4 and T = 5–35°C) for another ten rG–rC and seven rA–rU bps embedded in distinct sequence and structural contexts in three additional RNA molecules, including a GC-rich hairpin (hp-gcGU), elongated duplex (E-gc), the transactivation response element (TAR) and a mutant form of TAR (TAR-UUCGGU) that is impaired from undergoing secondary structure chemical exchange25 (Fig. 2a). In all cases we did not detect any signs of RD (Fig. 2b and Supplementary Fig. 1b). These results, together with our studies25–27 reporting flat RD profiles for RNA WC bps near non-canonical motifs and mismatches (wtTAR and P5abc, Fig. 2a) and for the reverse wobble rGsyn–rU mispairs in apical loops28 stand in striking contrast to canonical duplex DNA in which we have robustly observed WC⇄HG exchange in all 35 dA–dT and dG–dC bps examined to date in a wide variety of positional and sequence contexts in eight different duplexes that have varying lengths and stabilities6,7.
The lack of detectable WC⇄HG exchange in A-RNA could in principle result from small differences between the WC and HG NMR chemical shifts (Δω < 0.5 p.p.m. for carbon chemical shifts). However, based on density functional theory calculations (DFT)6,29 and a survey of syn purine base chemical shifts in the Biological Magnetic Resonance Data Bank30, it is highly unlikely that such a large transformation in base pairing would result in such small changes in chemical shifts for the different sugar (C1′) and base (C8, C6 and N1/N3) sites targeted for RD measurements (Supplementary Note). The absence of RD is unlikely to be due to the exchange rate falling outside the detection limits of the RD experiment given that flat profiles are observed over a wide range of temperatures and pH conditions (Fig. 2b and Supplementary Fig. 1b) known to significantly alter the WC⇄HG exchange rate in B-DNA6,7.
A more likely explanation is that HG bps are energetically disfavored in A-RNA duplexes, and have an abundance that falls below the detection threshold of the RD experiment (population < 0.01%). Indeed, a survey of X-ray structures of RNA duplexes in the Protein Data Bank (PDB)31 failed to identify a single rG–rC+ or rA–rU HG bp within continuous A-RNA duplexes out of a total of 123,935 rG–rC and rA–rU bps (Methods); while in sharp contrast, a similar survey conducted recently on B-DNA duplexes32 identified 54 dG–dC+ or dA–dU HG bps out of a much smaller set of 51,485 bps. The survey identified a single rA–rU HG bp (for example, PDBID: 1GID33) within an RNA duplex that fell well outside the A-form structural context being surrounded by a bulge and internal loop. The survey did identify several examples of long-range rG–rC+ and rA–rU HG bps forming tertiary contacts; rG–rC+ and rA–rU HG bps in triplexes and reverse rA–rU HG bps within duplexes typically near rG–rA mismatches where purines adopt anti rather than syn conformation, as well as several examples of HG mispairs in A-RNA duplexes (e.g. rGsyn–rGanti, and rGsyn–rAanti) (Supplementary Note).
m1A and m1G modified A-RNA
If rG–rC+ and rA–rU HG bps are indeed thermodynamically disfavored in A-RNA, they should prove more difficult to trap using chemical modifications known to stabilize dG–dC+ and dA–dT HG bps in B-DNA6. We therefore examined whether HG bps could be stably trapped in A-RNA duplexes using m1rA and m1rG. These modified bases block WC pairing because of steric collisions with the methyl group and because the methylation knocks out one of the WC H-bonds (Fig. 3a). Both m1dA and m1dG occur in DNA due to alkylation damage12,13. In B-DNA, m1dA and m1dG are accommodated as m1dA–dT and m1dG–dC+ HG bps6,11,34 (Fig. 3a), which can in turn be recognized and repaired by damage repair enzymes12,13. m1rA and m1rG can also occur as a form of alkylation damage in RNA but they are also highly conserved post-transcriptional modifications in transfer and ribosomal RNAs that play critical structural and functional roles often by blocking WC base pairing35–39. m1rG and m1rA have been shown to induce duplex-to-hairpin transitions in palindromic RNA sequences where the modified base favors an unpaired conformation within apical loops40,41. Recent genome-wide studies have shown m1rA to be a dynamic reversible eukaryotic messenger RNA (mRNA) modification that can potentially play roles in epitranscriptomic regulation42,43.
Figure 3.
m1A and m1G do not form HG bps and disrupt A-RNA structure. (a) N1-methylated purines trap HG bps in B-DNA. NMR chemical shift probes of HG bps are in orange and of purine methylation state in cyan. Arrows indicate characteristic HG NOE cross-peaks. (b) Duplexes containing m1A or m1G (turquoise circles). syn or anti purines deduced by NMR are shown as open and filled letters, respectively. HG and partially melted bps as deduced by NMR are indicated using open and dashed lines, respectively. Residues showing significant chemical shift perturbations or line-broadening due to m1A or m1G are colored orange and grey, respectively. (c) m1A or m1G induced purine-C1′ chemical shift perturbations (Δω = ωmodified – ωunmodified) in A-RNA (violet) and B-DNA (blue). Shown for comparison are Δω = ωHG – ωWC measured for transient dA–dT HG bps by RD (“RD”) in unmodified DNA duplexes (error bars showing one s.d.) and computed for adenine residues using DFT (Methods). (d) NOESY H1′–H8 cross-peaks showing syn purine bases in B-DNA but not A-RNA. Shown for reference is the cytosine base H5–H6 NOE with inter-atomic distance ≈2.5 Å. (e) 1D 1H spectra showing the imino/amino resonances expected for HG type H-bonds in A6-DNAm1A and A6-DNAm1G but not in methylated RNA at 5°C and 15°C, respectively. (f) Example showing m1G induced loss of a WC imino resonance (highlighted in a circle) in A6-RNA but not A6-DNA in 2D NMR spectra. (g) Example downfield shifted carbon chemical shifts induced by m1A. (h) Free energy (ΔΔG) and enthalpy (ΔΔH) destabilization due to m1A and m1G in DNA (blue) and RNA (violet) duplexes measured by UV melting experiments with error bars, one s.d. (n = 3 independent measurements) (Methods and Supplementary Table 2).
In prior studies6, we showed that m1dA16 and m1dG10 form stable m1dA16–dT9 and m1dG10–dC15+ HG bps stabilized by unique H-bonds in A6-DNA while minimally impacting neighboring WC bps as judged based on observation of HG-specific chemical shifts, Nuclear Overhauser effect spectroscopy (NOESY) cross-peaks, and imino resonances (highlighted in Fig. 3a). In contrast, we did not observe any NMR evidence for HG bps or syn purine bases in the corresponding A6-RNA duplex containing m1rA16 or m1rG10 (Fig. 3b – e and Supplementary Fig. 2). This was the case despite the fact that A6-DNA and A6-RNA duplexes have very similar thermodynamic stabilities. Rather, the rA-C1′ chemical shifts falls in a region that is consistent with A-form helical residues (Fig. 3c and Supplementary Note). We also observe continuous NOE distance-based connectivity between the m1rA and its preceding residue that are consistent with an anti conformation for the purine base (Supplementary Fig. 2). These data, together with the absence of strong H1′–H8 NOEs expected for syn base (Fig. 3d) and imino and amino resonances indicative of H-bonding (Fig. 3e) suggest that in A6-RNA, m1rA adopts a predominantly unpaired anti conformation although we cannot rule out transient formation of syn base conformations. The resonances belonging to m1rG in A6-RNA were broadened out of detection suggesting extensive conformational exchange at the µs–ms timescale with no NMR evidence for HG pairing given the absence of downfield-shifted rC-H4 (Supplementary Fig. 1 and Fig. 3e). However, we cannot exclude micro-to-millisecond exchange between syn and anti conformations for the m1rG base since the resonances are broadened out of detection24.
Compared to A6-DNA, m1A and m1G also induced more significant structural perturbations in A6-RNA (Fig. 3b and Supplementary Table 2). We do not observe some of the imino resonances belonging to WC bps neighboring the modified site (Fig. 3f), which suggests loss of H-bonds and the melting of these base pairs. The modifications also induced more extensive chemical shift perturbations (Fig. 3g and Supplementary Fig. 1c; highlighted in orange in Fig. 3b) and line broadening (Supplementary Fig. 1c; highlighted in grey in Fig. 3b) in the sugar and base resonances that extend to the partner strand. The direction of the perturbations is consistent with deviations from a helical conformation (Supplementary Fig. 1c). The perturbations were particularly pronounced for m1rG, which broadened all imino resonances out of detection at 35°C, consistent with significant melting of the entire duplex (Supplementary Fig. 1d). Thus, HG bps are so sufficiently disfavored in A-RNA that m1rA and m1rG prefer to adopt predominantly non-helical conformations that disrupt the duplex structure.
Similar results were obtained in GC-rich (gcm1A) and scrambled (A2m1A)6 (B.S., H.Z., Y.X., H.M.A., unpublished) duplexes with m1A consistently forming HG bps or adopting a syn conformation in B-DNA but not in A-RNA (Fig. 3b,d and Supplementary Fig. 3 and Supplementary Fig. 4) and with the modification more significantly perturbing the structure of A-RNA as compared to B-DNA (Fig. 3b,g). The structural perturbations induced by m1rA varied with sequence, and were either distributed across many WC bps (A6-RNA and A2-RNA) or more severe but localized to the modified and partner base (gc-RNA) (Fig. 3b and Supplementary Fig. 1c and Supplementary Fig. 3). In all cases we did not observe any evidence for m1rA or m1rG induced duplex-to-hairpin transition based on spectral overlays with the unmodified counterparts (Supplementary Fig. 1c and Supplementary Fig. 3).
We corroborated the more portent destabilization of A-RNA as compared to B-DNA duplexes by m1A and m1G using UV melting experiments. m1dA destabilized A6-DNA, A2-DNA, and gc-DNA duplexes by ΔΔG = 1.8–3.4 kcal mol−1 (Fig. 3h) in good agreement with the relative stability of transient HG bps measured by NMR RD (2.1–4.3 kcal mol−1) and prior UV-melting studies of m1dA containing DNA duplexes (≈2 kcal mol−1)44. By comparison, m1rA and m1rG destabilized the corresponding A-RNA duplexes by a larger amount ΔΔG = 4.3–6.5 kcal mol−1. Interestingly, this greater destabilization is comparable to the relative stability of the base-opened state45. This suggests that in A-RNA, the modification results in a conformation similar to the base-opened state, consistent with the NMR evidence for local melting. Greater destabilization (by ≈1.1–4.7 kcal mol−1) of A-RNA as compared to B-DNA was robustly observed across different duplex and hairpin contexts in the presence or absence of Mg2+ and with the destabilization being principally enthalpic (Fig. 3h, and Supplementary Table 3). The potent m1rA induced destabilization of duplex RNA is highly significant considering recent studies showing it to be a dynamic mRNA modification with roles in post-transcriptional gene regulation42,43. For comparison, the other well studied mRNA modification46 N6-methyladenosine (m6A) which affects mRNA localization, stability, translation, and splicing destabilizes A-RNA by only 0.5–1.7 kcal mol−1 47.
The more potent m1A and m1G destabilization of A-RNA as compared to B-DNA is unlikely to be due to differences in steric contacts involving the methyl group in a HG bp configuration (Supplementary Note). While the positive charge on m1rA may affect stacking and H-bonding interactions, significant destabilization is also observed with the neutral m1rG, and the m1A destabilization is greater for A-RNA as compared to B-DNA (Fig. 3h). Rather, the greater destabilization observed in A-RNA is likely due to the higher energetic cost of forming HG bps in A-RNA as compared to B-DNA.
Why are HG bps disfavored in A-RNA?
Why are HG bps so strongly disfavored in RNA as compared to DNA duplexes? The HG bp could in principle be disfavored in RNA due to the sugar 2′-OH at the purine residue. The 2′-OH helps bias the sugar pucker toward the C3′-endo conformation (Fig. 1a) due to unfavorable steric contacts between O2′and O3′ and electronic effects involving the 2′-OH group2,48. This in turn disfavors the syn purine base conformation even in nucleosides49 and single-stranded polynucleotides50 due to unfavorable base-sugar steric contacts (N3–H3′ and N3–O4′). The syn purine conformation may also destabilize water-bridged interactions involving the 2′-OH and N3 of the anti purine base51. To examine whether the mere presence of a 2′-OH group on the ribose moiety of the flipping purine base is sufficient to suppress WC⇄HG exchange, we carried out R1ρ RD experiments on site or strand specifically labeled A6-DNA duplexes containing a single ribonucleotide, rA16 or rG10 (Supplementary Fig. 5a,b and Methods). These RD measurements were also of interest given that single ribonucleotides are frequently incorporated in DNA during replication and can have important biological consequences through mechanisms that are not fully understood52.
Both rA16 and rG10 formed the expected rA16–dT9 and rG10–dC15 WC bps53 and exhibited RD consistent with WC⇄HG exchange (Fig. 4a). The lower Rex contribution observed for the rA16 and rG10 substituted samples relative to the unmodified DNA duplex can be attributed to ≈4-fold faster exchange rate (kex = 2325 s−1 versus 595 s−1) in the case of rA16 and a combination of slightly smaller Δω (1.8 versus 2.1 p.p.m.) and transient HG population (0.8% versus 1.3%) in the case of rG10 (Fig. 4a, Supplementary Table 4 and Supplementary Fig. 5c). Neither rA16 nor rG10 significantly impacted the abundance of the transient HG bps relative to the unmodified A6-DNA duplex (Fig. 4a and Supplementary Table 4) indicating that the purine sugar 2′-OH group alone cannot account for the lack of observable WC⇄HG exchange in A-RNA duplexes. We confirmed these findings by analyzing A6-DNA duplexes containing N1-methylated single ribonucleotide, m1rA16 or m1rG10. In both cases, we observe stably formed m1rA16–dT9 and m1rG10–dC15+ HG bps (Supplementary Fig. 5b). These data suggest that the destabilization of HG bps requires the broader A-form RNA helical context.
Figure 4.
Source of HG instability in A-RNA. (a) Comparison of RD profiles measured in A6-DNArA, A6-DNArG, and A6-DNA. Error bars correspond to one s.d. estimated from mono-exponential fitting using a Monte-Carlo based method (Methods). Note that the larger R2 value in A6-DNArA A16-C8 as compared to A6-DNA likely reflects decreased flexibility in rA16. Exchange parameters are shown in Supplementary Fig. 4b. (b) Inter-atomic distances (in Å) with unfavorable steric contacts in pink when rotating the purine base 180° around the glycoside bond in WC bps derived from idealized B-DNA and A-RNA duplexes (Methods) to adopt a syn conformation. Shown below are corresponding distance distributions in WC bps derived from X-ray structures of A-RNA (total 146) and B-DNA (total 159) duplexes before (solid line) and following (dashed line) 180° rotation of the purine base. The inter-atomic cut-off distance (grey line) was defined based on the van der Waals radii. (c) Relative interaction energy versus χ-angle from biased MD trajectories of A6-DNA (blue) and hp-A6-RNA (violet). (d) Simulation time (ns) versus the global RMSD (Methods) for single A6-DNAm1A and hp-A6-RNAm1A trajectories depicting the destabilization of the RNA strand within the time of the simulation.
Next, we examined whether there were unique steric clashes that could disfavor syn purine bases within the compact A-RNA helix context that are absent in the more capacious B-form DNA helix. Indeed, flipping the purine base around the glycosidic χ-angle through a range of angles (160°–200°) that span syn base conformations found in RNA helices (Supplementary Note) resulted in greater steric clashes in A-RNA as compared to B-DNA. The additional base-sugar (N3–H3′ and N3–O4′) and base-backbone (N3–O5′) clashes observed in A-RNA arise due to both the C3′-endo sugar pucker and unique phosphodiester backbone conformation at the syn purine residue (Fig. 4b).
To further examine the energetics of the WC⇄HG transition, we carried out biased MD simulations on the A6-DNA duplex and hp-A6-RNA hairpin, as well as a 3′→5′ inverted sequence of the hp-A6-RNA hairpin. A bias was applied on dA16 or rA16 starting in a WC bp configuration to force purine base flipping and a transition to a target HG configuration (Methods and Supplementary Movie 1, 2). The computed mean interaction energy (averaged over an ensemble of biased trajectories) as a function of the χ-angle along the WC⇄HG transition (Methods) reveals a clear two state transition in the case of B-DNA, consistent with previous results6, whereas in the cases of A-RNA the resultant HG bp is significantly destabilized relative to its WC bp counterpart; for A-RNA, the energy profile in the syn region has much higher relative energies than in the case of DNA (Fig. 4c). In accord with this energetic destabilization, the simulations reveal that flipping the purine base in A-RNA is accompanied by major structural disruption of the surrounding base pairs (Supplementary Movie 2), consistent with m1rA induced NMR chemical shift perturbations, which were more pronounced for residues 3′ to the modified nucleotide (Supplementary Fig. 1c). The extent of the disruption for the neighboring bp was far less significant in B-DNA (Supplementary Movie 1).
We corroborated these findings using unbiased MD simulations, which began with a HG bp embedded in various duplex and hairpin contexts (Methods). The HG H-bonding remained stable during the course of the simulation in the case of B-DNA, B-DNA containing a single rA, and B-DNA containing m1dA (Supplementary Table 5 and Supplementary Movie 3–5). In contrast, for A-RNA strong disruption of N7---H3-N3 HG H-bond between A16 and U9 was observed in cases of the hp-A6-RNA hairpin (Supplementary Table 5). In ≈35% of the trials in the case of 3′→5′ sequence hp-A6-RNA, the HG bp even transitioned rapidly after equilibration back to a WC bp. Strikingly, in the case of m1rA embedded in A-RNA, the HG bp caused melting of the A-form helix (Fig. 4d and Supplementary Movie 6).
Taken together, these results indicate that HG bps are disfavored in the more compact A-RNA helix due to steric contacts that are difficult to alleviate without substantially perturbing the A-form helix structure.
DISCUSSION
Duplex B-DNA can stably accommodate dA–dT and dG–dC+ HG bps which can in turn play roles in sequence-specific DNA recognition, damage induction and repair, and in DNA replication. In contrast, our results indicate that rA–rU and rG–rC+ HG bps are so unstable in the more compressed A-RNA that melting is preferred over the HG bp conformation. It remains to be seen whether the greater instability of HG bps in A-RNA as compared to B-DNA extends to purine-purine HG mispairs (Supplementary Note), which play important roles in replication14,54 and translation errors55, mismatch repair8, as well in translational reprogramming56,57.
The markedly different stability of the A–T/U and G–C+ HG bp in RNA and DNA duplexes provides a basis for achieving opposing functions at the genome and transcriptome levels (Fig. 5a). If DNA did not have a capacity to form HG bps, and instead behaved like RNA, lesions such as m1dA and m1dG that block canonical WC base pairing could greatly destabilize the double helix and potentially cause genomic instability (Fig. 5a). The ability to form HG bps therefore endows DNA with an additional layer of chemical stability over its RNA counterpart that goes beyond resistance to hydrolysis due to the absence of the sugar 2′-OH group. On the other hand, the greater instability of HG bps in A-RNA gives rise to a chemical switch in the form of m1rA and m1rG that can potently modulate RNA structure (Fig. 5a). While it has long been recognized that m1A and m1G can modulate the structure and function of tRNA, rRNA, and other non-coding RNAs37–39,58, this functionality hinges on the unique instability of HG bps in A-RNA uncovered in this work.
Figure 5.
Different propensities to form HG bps in B-DNA and A-RNA enable contrasting roles at the genome and transcriptome level. (a) In DNA, m1dA or m1dG damage is absorbed as HG bps that can be recognized by repair enzymes (in red). Had B-DNA lacked the ability to form HG bps, damage could result in duplex melting and genomic instability. In RNA, post-transcriptional modifications resulting in m1rA and m1rG block both WC and HG pairing, melting or modulating RNA secondary structure to favor functional states or effect epigenetic regulation. Had A-RNA had the ability to form HG, the m1rA and m1rG would form HG bps and potentially fail to more significantly alter RNA structure and function. (b) Highly conserved m1rA9 in human mitochondrial tRNALys blocks rA–rU WC base pairing and stabilizes native tRNA structure in which m1rA9 is in a single strand58. The m1rA9 modification would not stabilize native tRNA structure if it were simply absorbed as a HG bp. (c) Highly conserved m1rG37 next to the anti-codon loop37 blocks base pairing between m1rG37 and the first rC in the codon and prevents +1 frameshifting in tRNAPro, which could occur if m1rG37 formed stable HG bp with rC. (d) Proposed mechanism for m1rA enhanced translation through destabilization of secondary structure in the 5′ UTR of mRNA.
For example, m1rA9 has been shown to stabilize the native structures of human mitochondrial tRNAs by blocking helical rA–rU WC bps that would otherwise stabilize alternative secondary structures58 (Fig. 5b). Likewise, m1rG37 next to the anti-codon loop, which is highly conserved in most tRNAs that read the CNN codon, has been shown to prevent +1 frameshifting by blocking base-pairing between G37 and the first rC in the codon sequence37,39 (Fig. 5c). If RNA behaved like DNA, such posttranscriptional modifications would simply create HG bps and fail to block base pairing and have their intended functional consequence (Fig. 5).
m1rA was recently shown to be a reversible mRNA modification in eukaryotic cells, from yeast to mammals, that can respond to changes in physiological conditions42,43. It is enriched in the 5′ UTR near start codons and was shown to promote translation through mechanisms that are not yet understood42,43. The formation of stable mRNA secondary structure around start codons has been shown to reduce translational efficiency59,60. Although it is unclear whether these m1rA modifications target adenine nucleotides involved in WC base pairing, it is possible that m1rA enhances translation in part by destabilizing secondary structure at the 5′ UTR near the start codons. Indeed, based on our results, m1rA should also be capable of stabilizing alternative RNA secondary structures that feature bulged adenosines even if they are disfavored by as much as ≈5 kcal mol−1 in the absence of the modification. At the same time, placement of m1rA in an unpaired bulged conformation can make it accessible to demethylases for achieving efficient reversible control at the epitranscriptomic level (Fig. 5d). Further studies are needed to test this proposed mechanism for m1A-enhanced translation.
ONLINE METHODS
Sample Preparation
NMR buffer
All RNA and DNA samples were buffer exchanged at least three times using a centrifugal concentrator (EMD Millipore) until containing >99.9% of the desired buffer, which unless stated otherwise, consisted of 15mM sodium phosphate, 25 mM NaCl, 0.1 mM EDTA with pH 5.4 or 6.8 and 10% D2O.
Uniformly 13C/15N labeled RNA and DNA samples
hp-A6-RNA and single-strands of the E-gc and TAR-UUCGGU were prepared using in vitro transcription with uniformly 13C/15N-labeled ribonucleotide triphosphates (Cambridge Isotope Laboratories), T7 polymerase (Takara Mirus Bio Inc.) and synthetic DNA templates (Integrated DNA Technologies, Inc.), purified by 20% (w/v) denaturing polyacrylamide gel electrophoresis (PAGE) and electro-eluted into 20 mM Tris buffer (pH = 8) followed by ethanol precipitation as described previously26. The uniformly labeled T6-strand in A6-DNArG and uniformly labeled A6-DNA were prepared by the primer-extension approach61 using uniformly 13C/15N-labeled deoxyribonucleotide triphosphates (Silantes) as previously described6.
m1A and m1G containing oligonucleotides
Oligonucleotides were purchased from Keck Oligo Synthesis Resource (W.M. Keck Foundation) with Glen-Pak™ DNA/RNA cartridge purification (A6-RNAm1A, A2-RNAm1A, gc-RNAm1A, A6-DNAm1G, A6-DNAm1rA, gc-DNAm1A, hp-A6-RNAm1A, hp-A6-DNAm1A, hp-A6-DNAm1G, hp-gc-RNAm1A, and hp-gc-DNAm1A) Midland Certified Reagents with reverse-phase (RP) HPLC purification (A6-DNAm1A and A2-DNAm1A), and GE Healthcare Dharmacon Inc. with RP-HPLC purification (A6-RNAm1G, A6-DNAm1rG and hp-A6-RNAm1G). To minimize Dimroth rearrangement of m1A into m6A62,63, all DNA and RNA oligonucleotides containing m1A were synthesized and deprotected using the UltraMild protocol (http://www.glenresearch.com/Technical/TB_UltraMild_Deprotection.pdf; Glen Research Corporation).
Assessing purity of m1A and m1G containing oligonucleotides
Samples were assessed using 20% denaturing PAGE, MALDI Mass Spectrometry, Liquid Chromatography-Mass Spectrometry (LC-MS) and NMR spectroscopy. For hairpin constructs hp-gc-RNAm1A and hp-A6-RNAm1A, we obtained evidence for incomplete base deprotection during synthesis based on observation of additional imino proton and acetyl group (the N4 protecting group on the cytosine) resonances and NOE talk between the two. Evidence for the acetyl groups was also obtained by quantitative mass spectrometry (LC-MS). We suspect that incomplete deprotection arises due to formation of stable secondary structure in these hairpin constructs during the UltraMild deprotection step. These impurities could be effectively eliminated by synthesizing individual single strands of duplex versions of the hairpin sequence (gc-RNAm1A and A6-RNAm1A).
In all cases, the NMR chemical shifts of the N1-methyl group and base moieties (A-C2, N1C, N1H) were consistent with m1A with no evidence for Dimroth rearrangements62, which lead to formation of m6A (Supplementary Fig. 1c and Supplementary Fig. 3). In particular, we observed ≈4 p.p.m. upfield shift in m1A-C2 (Supplementary Fig. 1c and Supplementary Fig. 3) consistent with base protonation, which is expected for m1A but not m6A. This, as well as the observation of two amino protons (H61 and H62) with distinct chemical shifts involved in HG H-bonding in DNA (Fig. 3e and Supplementary Fig. 2) indicates a major positively charged amine tautomer rather than a neutral imine tautomeric form64. Nevertheless, we cannot rule out the existence of the neutral imine tautomeric form transiently and/or in low-abundance.
Unmodified oligonucleotides
Unmodified RNA oligonucleotides were synthesized using an in-house MerMade 6 Oligo Synthesizer employing 2′-TBDMS RNA phosphoramidites (ChemGenes) on 1 µmol standard synthesis columns (1000 Å) from BioAutomation using the option to leave the final 4,4′-dimethoxytrityl (DMT), the 5′-protection group on for the cartridge purification. The oligonucleotide was cleaved from each 1 µmol column using ≈1 mL ammonia methylamine (1:1 ratio of 30% ammonium hydroxide and 30% methylamine) followed by 2-hour incubation at room temperature to allow base deprotection. The solution was then subjected to airflow until complete evaporation, leaving the desired product oligonucleotide as dried crystals. The crystals were then dissolved in 115 µL DMSO, mixed with 60 µL TEA and 75uL TEA"l35;3HF, and incubated at 65°C for 2.5 hours for 2′-deprotection. The reaction was quenched using Glen-Pak™ RNA quenching buffer and loaded onto Glen-Pak™ RNA cartridges (Glen Research Corporation) for purification following the online protocol (http://www.glenresearch.com/Technical/GlenPak_UserGuide.pdf). Samples were ethanol precipitated and exchanged into NMR buffer. Unmodified DNA oligonucleotides were purchased from Integrated DNA Technologies with standard desalting.
Site-specifically 13C/15N-labeled samples
The A6 strand of A6-DNArA16 containing C8-13C/15N-labeled adenosine was synthesized using an in-house Solid-phase Oligonucleotide Synthesizer (BioAutomation MerMade 6), C8-13C/15N-labeled adenosine phosphoramidite (see below), and unlabeled DNA phosphoramidites (ChemGenes) using 1 µmol scale 1000 Å CPG DNA columns (BioAutomation). The synthesized oligonucleotides were cleaved and deprotected as described above for unlabeled RNA oligonucleotides and purified with the Glen-Pak™ RNA cartridge (Glen Research Corporation) followed by ethanol precipitation and exchange into the desired NMR buffer.
Synthesis of 8-13C-adenosine phosphoramidite
The 8-13C-adenine nucleobase was synthesized according to a published procedure65. The protection of the exocyclic amino group with a benzoyl moiety and the conversion to the 5′-O-DMT-2′-O-TOM-protected 8-13C-adenosine 3′-O-phosphoramidite was accomplished according to published procedures66,67. A detailed description on the chemical synthesis of 8-13C-purine RNA phosphoramidite building blocks will soon be published elsewhere.
NMR experiments
Resonance assignment
NMR data were collected on an 800 MHz Varian DirectDrive2 spectrometer equipped with a triple resonance HCN cryogenic probe; 700 Bruker Avance III spectrometer equipped with a triple-resonance HCN cryogenic probe; and 600 MHz Varian Inova NMR spectrometer equipped with a Bruker HCPN cryogenic probe. Data were processed and analyzed using NMRpipe68 and SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco), respectively. Resonances were assigned using conventional 2D HSQC, HMQC, NOESY and HCN experiments.
Chemical shift perturbations (CSPs) induced by m1A or m1G for each residue (Δωresidue) were calculated using equation (1)69 from the average Euclidean distance of all measured CSP (ΔωC, ΔωN, and ΔωH):
| (1) |
where γi is the gyromagnetic ratio of the ith nucleus (C, H or N), N is the total number of CSPs measured for each residue, and Δωi is the difference in chemical shifts (in p.p.m.) for the ith nucleus between the m1A or m1G modified and unmodified duplex. Residues with Δωresidue≥0.1 p.p.m. are highlighted on the duplexes in Fig. 3b and Supplementary Fig. 2 and Supplementary Fig. 4. An average CSP (Δωavg) was calculated for each duplex by averaging Δωresidue for two base pairs above and below the modified bp.
13C and 15N R1ρ relaxation dispersion
13C and 15N R1ρ RD experiments were performed at 600 MHz (14.1 T) and 700 MHz (16.4 T) Bruker spectrometers as previously described6,21,23 using spinlock powers (ωSL 2π−1 Hz) and offset frequencies (Ω 2π−1 Hz) listed in Supplementary Table 1. Magnetization of the spins of interest was allowed to relax under an applied spinlock for the following durations: [0–120 ms] for N1/N3 in hp-A6-RNA and E-gc; [0–60 ms] for C8/C1′ in hp-A6-RNA, E-gc, TAR-UUCGGU, A6-DNA, A6-DNArA and A6-DNArG.
Analysis of R1ρ data
Fitting of 13C and 15N R1ρ data
Experimental R1ρ relaxation rate constants were calculated by fitting peak intensities versus relaxation delay durations to a single exponential decay21. Uncertainty in the fitted R1ρ values (one s.d.) were derived using a Monte-Carlo method18. R1ρ data were fitted to simulated R1ρ values given by the solution to the Bloch–McConnell (BM) equations20 at each given Ω and ωSL combination. Residual sum of squares were minimized using a bounded least-squares algorithm70 to give best-fit exchange parameters. The uncertainty in the chemical exchange parameters was calculated as the standard error of the fit18. A 2-state exchange model was used to fit the R1ρ RD profiles of A6-DNA, A6-DNArA and A6-DNArG with the initial magnetization aligned either along the effective field of the ground (for slow exchange with kex Δω−1 < 1) or average (for fast exchange with kex Δω−1 ≥ 1) state71. For the dA16-C8 RD data measured in A6-DNA at low temperatures, both protocols yielded acceptable fits but resulted in different exchange parameters given the slower exchange rate (Supplementary Table 4). The exchange parameters obtained from average alignment protocol were selected based on a van’t Hoff analysis6 (Supplementary Fig. 5d). For the dC15-C6 RD data measured in A6-DNArG, a 3-state chemical exchange model without minor exchange with average alignment was statistically favored over 2-state models (Supplementary Fig. 5c). In all cases, Akaike information criterion (AIC)72 and Bayesian information criterion (BIC)72 were used to select the models.
Analysis of chemical shift and NOESY data
Chemical shifts and NOESY cross-peaks were used to characterize WC versus HG bps. The NOESY cross-peaks unique to HG bps include strong intra-nucleotide H1′–H8 NOE for syn purine, (i)A-H2–(i-1)H1′/H2′ and (i)A-H2–(i-1)H6/H8 for syn adenosine, and H8–H3, A-H6/C+-H4–H3, (i)H3–(i+1)/(i-1)H1/H3, (i)A-H6/C+-H4–(i+1)/(i-1)H1/H3 NOEs for connectivity involving imino or amino protons in both G-C and A-U/T HG bps6,73,74. Absence of the canonical sequential (i-1)H1′–(i)H8 NOE is also expected for syn purines due to the base flip. As described previously6, the HG bps are also characterized by a unique set of chemical shifts relative to WC bps including downfield shifted purine-C8 and purine-C1′, protonated cytosine-C6 (≈3 p.p.m.) and upfield shifted protonated cytosine-C5, guanine-N1 and thymine-N3 (≈1–2 p.p.m.). The C1′ chemical shifts is also sensitive to sugar pucker. In A-RNA, deviations from the A-form C3′-endo toward C2′-endo sugar pucker leads to an upfield shift (≈4 p.p.m.)75,76. In addition, deviations from the A-form conformation due to loss of stacking and bulging out of nucleotides results in a downfield shift in the base C6/C8 and cytosine-C5 and upfield shift on sugar-C1′18.
Density functional theory geometry optimizations and CS calculations
Density functional theory (DFT) calculations77 using Gaussian 09c (Gaussian, Inc.) were performed as previously described6 to compute chemical shifts for WC and HG bps in A-RNA and B-DNA. In all cases, protons were added using PyMOL (https://www.pymol.org/) and the phosphate backbone truncated leaving only the nucleoside motifs for each bp6. Calculations were performed on rA–rU HG bp obtained from snapshots of rA16–rU9 bp in biased MD simulation of hp-A6-RNA, rA–rU HG bp from the X-ray structure of P4-P6 domain of the Group I intron RNA (PDBID: 1L8V78), tertiary rG–rC+ HG bp in the structure of 23S ribosomal RNA-protein complex (PDBID: 3U5679) and rGsyn–rGanti mispair in duplex RNA structure (PDBID: 3CZW80). Reference rA–rU or rG–rC WC bps were taken from MD snapshot or from the same X-ray structures used to obtain HG bps (Supplementary Fig. 5e). Two runs of geometry optimizations were carried out using the B3LYP functional with 3–21G and 6–311+G(2d,p) basis sets, respectively, with all heavy atoms (C, N and O) frozen. Carbon chemical shifts were computed using the GIAO method within the B3LYP/6–311+G(2d,p) basis set6 on the converged configuration after the second run of optimization. The isotropic carbon chemical shift (ω13C) was referenced to that of TMS (ωTMS = 182.4656 p.p.m.), which was optimized and computed at the same level of theory. The carbon chemical shifts computed for reference WC bps were subtracted from those computed for HG bps to yield chemical shift changes upon HG formation (Δω = ωHG-ωWC).
Analysis of UV melting data
The UV absorbance at 260nm (A260) as a function of temperature was measured on a Shimadzu UV-3600 UV-Vis-NIR spectrophotometer using the 8-cell sample holder with a Fisher Isotemp Refrigerated Circulator to regulate sample temperature. All DNA and RNA oligonucleotides were diluted directly from NMR samples using the same NMR buffer (15mM phosphate, 25mM NaCl, 0.1mM EDTA at pH = 5.4 or 6.8) unless stated otherwise and triplicate measurements were carried out for each oligonucleotide simultaneously using a sample volume of 125 µL in each cell and an additional reference cell containing the same amount of buffer. The temperature was varied between 5°C and 90°C at a ramping rate of 1 °C min−1.
The melting temperature (Tm) and enthalpy (ΔH) for duplex association and hairpin folding was obtained by fitting the melting curves to equation (2) and equation (3)81, respectively,
| (2) |
| (3) |
T is the measured temperature and f is the fraction of the remaining unmelted duplex or folded hairpin over the total concentration of duplex or hairpin (CT), which is proportional to the measured A260.
The thermodynamic parameters ΔG and ΔS were calculated using equation (4) for duplex association and equation (5) for hairpin folding, respectively.
| (4) |
| (5) |
The fitting was carried out using nonlinear model fitting with Mathematica 10.0 (Wolfram Research). Errors in Tm and ΔH represent the standard deviation (one s.d.) from the triplicate measurements. The destabilization effects due to m1A and m1G in DNA or RNA were calculated by taking the difference in free energy for folding i.e. ΔΔG=ΔGmod−ΔGunmod, ΔΔH=ΔHmod−ΔHunmod and ΔΔS=ΔSmod−ΔSunmod.
Since the Dimroth rearrangement can occur for m1A in both DNA and RNA under basic conditions62 and high temperatures63, melting experiments were repeated for all m1A containing duplexes when restricting the temperature to <65°C and when using both neutral (pH = 6.8) and acidic (pH = 5.4) conditions. These control experiments yielded reproducible melting curves and fitted thermodynamic parameters at neutral or acidic pH conditions that are within experimental error (Supplementary Table 3). 1H 1D NMR spectra recorded for the A6-DNAm1A sample following melting showed insignificant changes and no evidence for Dimroth rearrangements (data not shown).
Steric analysis and survey of HG bps in RNA
A–T/U and G–C WC bps were obtained from idealized B-DNA and A-RNA helices built using 3DNA82 (Fig. 4b). 146 and 159 WC bps surrounded by at least one WC bp above and below were obtained from high resolution (< 2 Å) X-ray structures of A-RNA and B-DNA duplexes, respectively in the PDB. Purine bases were flipped around the glycosidic bond and inter-atomic distances measured using an in-house python script. Note that proximity of the exocyclic amino group on guanine to the phosphate group during the base flip was not considered a steric clash given the potential for H-bonding. The survey of HG bps in RNA was carried out following the same protocol reported for B-DNA32. Briefly, all RNA X-ray structures with resolution ≤ 3 Å were downloaded from the PDB on August 31 2014. The same in-house program was used to identify rA–rU and rG–rC bps in RNA structures using three HG criteria (H-bonding, constricted C1′–C1′ distance and syn purine)32. Redundancies defined as bps that are surrounded with the same sequence contexts and from same RNA or RNA-protein/ligand complexes were removed by manual inspection as described previously32. The survey identified a single rA–rU HG bp that reoccurs in four distinct X-ray structures of the P4-P6 domain of the Tetrahymena thermophila group 1 intron RNA (PDBID: 1GID33, 1l8V78, 1HR283, and 2R8S84). The RNA HG survey also identified several examples of long-range HG bps forming tertiary contacts, HG bps in triplexes, and reverse rA–rU HG bps within duplexes typically near rG–rA mismatches.
Biased and Unbiased Molecular Dynamics Simulations
Structure Generation for MD Simulation
hp-A6-RNA, hp-A6-RNA 3′→5′, and A6-DNA helices were built using make-na85 with all bases in WC conformation. In the case of hp-A6-RNA, a duplex structure was generated using make-na and the UUCG loop attached and annealed using the CHARMM simulation package86. Structures with HG conformation at A16 were created by rotating along the glycosidic bond angle χ by 180°.
Unbiased MD equilibrium simulations
All structures were simulated using constant temperature MD with CHARMM36 forcefield87 and a generalized Born molecular volume (GBMV) implicit solvent88; parameters for m1A were taken from Xu et al 89. Integration used a velocity-Verlet algorithm with a timestep of 1 fs. The cutoff for non-bonded list generation was 21 Å, the cutoff for non-bonded interactions was 18 Å, and the onset of switching for non-bonded interactions occurred at 16 Å. The SHAKE algorithm was used to constrain the covalent bonds to hydrogen atoms involved. Each structure was heated to 300.0 K with harmonic constraints on all non-hydrogen atoms, heating occurred in1 ps increments of 1.0 K for a total of 300 ps steps, followed by 200 ps equilibration at 300.0 K. Harmonic constraints were then gradually removed during a sequence of 4 reductions for 50 ps each. Unbiased production-run simulations were then run for 3 ns without constraints for each system. Ten independent simulations with hp-A6-RNA and A6-DNArA with A16 in HG conformation were produced from independent conformations obtained during the heating and equilibration method described above. A6-DNA in HG was repeated twice.
Global RMSD was calculated from the single 3 ns trajectories of m1A starting in HG for both hp-A6-RNA and A6-DNA,
| (6) |
in which ri(t) is the instantaneous coordinate of an atom and rR is the position of the reference structure.
H-bond presence was evaluated using CHARMM’s COOR HBOND module for each trajectory with cutoff distance and angle of 3.6 Å, and 120° following Goldsmith et al90.
Biased MD Simulations
The protocols for minimization, heating, and solvation were identical to those used for the unbiased simulations. The biased molecular dynamics method91 implemented in the CHARMM package was used to force conformational transitions between WC and HG states using a biasing potential W (ρ(t)) applied according to equation (7),
where
| (7) |
ρ(t) is a collective distance between the instantaneous (rij) and the reference structure , and α the strength of the half-harmonic bias. In all cases, biases were placed between pairs of atoms that share a H-bond in the target structure, ensuring that the adenine base would not only perform the roughly 180° flip, but also form the definitive H-bonding structure of the desired WC or HG configuration. After the biased trajectories were generated, they were post-processed in CHARMM, outputting the χ-angle dependence of the relative interaction energy value in the absence of the bias. Only successfully flipping trajectories were used, resulting in 40 trajectories for A6-DNA, 24 for hp-A6-RNA, and 25 for hp-A6-RNA 3′→5′. The relative interaction energy was calculated for the base pair that includes the flipping base as well as the base pairs above and below the flipping base. Angle-energy pairs were binned into 50 bins and the mean of the energy was evaluated within each bin. Plots of relative interaction energy as a function of the χ-angle were thus generated.
Supplementary Material
Acknowledgments
We thank M. Juen (University of Innsbruck, Austria), N. Orlovsky, Y. Xue, A. Shakya, A. Rangadurai, E. Szymanski, members of the Al-Hashimi lab, and T. Mustoe (UNC-Chapel Hill) for assistance and critical input. We acknowledge technical support and resources from the Duke Magnetic Resonance Spectroscopy Center, the Duke Compute Cluster and the Shared Materials Instrumentation Facility at Duke University. This work was supported by an NIH grant (R01GM089846 to I.A. and H.M.A) and (P26550 and P28725 to C.K.) from Austrian Science Fund (FWF). H.Z. acknowledges support from the China Scholarship Council.
Footnotes
AUTHOR CONTRIBUTIONS H.Z., E.N.N. and H.M.A. conceived the project and experimental design. H.Z. prepared NMR samples with assistance from I.J.K and E.N.N, as well as performed NMR experiments and analyzed NMR data with assistance from I.J.K and B.S. H.Z. performed DFT calculations and modeling of steric analysis. H.Z., I.J.K and E.N.N. performed the structure-based survey of RNA Hoogsteen base pairs. I.A., G.G., J.M. performed and analyzed the molecular dynamics simulations. C.W. and C.K. prepared the 13C-C8-adenosine phosphoramidite. H.Z. and T.B. carried out the UV melting experiments and performed the data analysis. H.M.A., H.Z. and I.A. wrote the manuscript with critical input from I.J.K., B.S., E.N.N, G.G., J.M., T.B., C.H.W. and C.K.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Neidle S. Principles of nucleic acid structure. Academic Press; 2010. [Google Scholar]
- 2.Brameld KA, Goddard WA. Ab initio quantum mechanical study of the structures and energies for the pseudorotation of 5′-dehydroxy analogues of 2′-deoxyribose and ribose sugars. J. Am. Chem. Soc. 1999;121:985–993. [Google Scholar]
- 3.Seeman NC, Rosenberg JM, Suddath FL, Kim JJP, Rich A. RNA double-helical fragments at atomic resolution: I. The crystal and molecular structure of sodium adenylyl-3′,5′-uridine hexahydrate. J. Mol. Biol. 1976;104:109–144. doi: 10.1016/0022-2836(76)90005-x. [DOI] [PubMed] [Google Scholar]
- 4.Lipfert J, et al. Double-stranded RNA under force and torque: Similarities to and striking differences from double-stranded DNA. Proceedings of the National Academy of Sciences. 2014;111:15408–15413. doi: 10.1073/pnas.1407197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoogsteen K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Crystallographica. 1959;12:822–823. [Google Scholar]
- 6.Nikolova EN, et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvey HS, Gottardo FL, Nikolova EN, Al-Hashimi HM. Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nat Commun. 2014;5:4786. doi: 10.1038/ncomms5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolova EN, et al. A historical account of Hoogsteen base-pairs in duplex DNA. Biopolymers. 2013;99:955–968. doi: 10.1002/bip.22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitayner M, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quigley GJ, et al. Non-Watson-Crick G.C and A.T base pairs in a DNA-antibiotic complex. Science. 1986;232:1255–1258. doi: 10.1126/science.3704650. [DOI] [PubMed] [Google Scholar]
- 11.Lu L, Yi C, Jian X, Zheng G, He C. Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system. Nucleic Acids Res. 2010;38:4415–4425. doi: 10.1093/nar/gkq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 13.Chen F, et al. Adaptive response enzyme AlkB preferentially repairs 1-methylguanine and 3-methylthymine adducts in double-stranded DNA. Chem. Res. Toxicol. 2016 doi: 10.1021/acs.chemrestox.5b00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarova AV, Kulbachinskiy AV. Structure of human DNA polymerase iota and the mechanism of DNA synthesis. Biochemistry (Moscow) 2012;77:547–561. doi: 10.1134/S0006297912060016. [DOI] [PubMed] [Google Scholar]
- 15.Kouchakdjian M, et al. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H–dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H–dG(syn)•dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 16.Kuchino Y, et al. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 17.Palmer AG., III A dynamic look backward and forward. J. Magn. Reson. 2016 doi: 10.1016/j.jmr.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue Y, et al. In: Methods Enzymol. Vol. Woodson Sarah A, Allain Frédéric HT, editors. Vol. 558. Academic Press; 2015. pp. 39–73. [Google Scholar]
- 19.Sekhar A, Kay LE. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12867–12874. doi: 10.1073/pnas.1305688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trott O, Palmer AG., III Theoretical study of R1ρ rotating-frame and R2 free-precession relaxation in the presence of n-site chemical exchange. J. Magn. Reson. 2004;170:104–112. doi: 10.1016/j.jmr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R1ρ NMR Spectroscopy. J. Am. Chem. Soc. 2009;131:3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 22.Korzhnev DM, Orekhov VY, Kay LE. Off-Resonance R1ρ NMR studies of exchange dynamics in proteins with low spin-lock fields: an application to a Fyn SH3 domain. J. Am. Chem. Soc. 2005;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 23.Nikolova EN, Gottardo FL, Al-Hashimi HM. Probing transient Hoogsteen hydrogen bonds in canonical duplex DNA using NMR relaxation dispersion and single-atom substitution. J. Am. Chem. Soc. 2012;134:3667–3670. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolova EN, Goh GB, Brooks CL, Al-Hashimi HM., III Characterizing the protonation state of cytosine in transient G•C Hoogsteen base pairs in duplex DNA. J. Am. Chem. Soc. 2013;135:6766–6769. doi: 10.1021/ja400994e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Dethoff EA, Al-Hashimi HM. Invisible RNA state dynamically couples distant motifs. Proceedings of the National Academy of Sciences. 2014;111:9485–9490. doi: 10.1073/pnas.1407969111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Visualizing transient low-populated structures of RNA. Nature. 2012;491:724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue Y, Gracia B, Herschlag D, Russell R, Al-Hashimi HM. Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat Commun. 2016;7 doi: 10.1038/ncomms11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salmon L, et al. Modulating RNA alignment using directional dynamic kinks: application in determining an atomic-resolution ensemble for a hairpin using NMR residual dipolar couplings. J. Am. Chem. Soc. 2015;137:12954–12965. doi: 10.1021/jacs.5b07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghose R, Marino JP, Wiberg KB, Prestegard JH. Dependence of 13C chemical shifts on glycosidic torsional angles in ribonucleic acids. J. Am. Chem. Soc. 1994;116:8827–8828. [Google Scholar]
- 30.Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H, et al. New insights into Hoogsteen base pairs in DNA duplexes from a structure-based survey. Nucleic Acids Res. 2015;43:3420–3433. doi: 10.1093/nar/gkv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cate JH, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Zhan Y, Fenn D, Chi LM, Lam SL. Effect of 1-methyladenine on double-helical DNA structures. FEBS Lett. 2008;582:1629–1633. doi: 10.1016/j.febslet.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Dunn D. The occurence of 1-methyladenine in ribonucleic acid. Biochim. Biophys. Acta. 1961;46:198–200. doi: 10.1016/0006-3002(61)90668-0. [DOI] [PubMed] [Google Scholar]
- 36.Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA. 2010;16:1317–1327. doi: 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagervall TG, Tuohy TMF, Atkins JF, Björk GR. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 1993;232:756–765. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 38.Agris PF. In: Prog. Nucleic Acid Res. Mol. Biol. Vol. Cohn Waldo E, Moldave Klvle, editors. Vol. 53. Academic Press; 1996. pp. 79–129. [DOI] [PubMed] [Google Scholar]
- 39.Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 40.Micura R, et al. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 2001;29:3997–4005. doi: 10.1093/nar/29.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tianming Yang WQAC, Xiangrui Mai, Shui Zou, Esther CY. Woon. A methylation-switchable conformational probe for sensitive and selective detection of RNA demethylase activity. Chem. Commun. 2016 doi: 10.1039/c6cc01045h. [DOI] [PubMed] [Google Scholar]
- 42.Dominissini D, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol advance online publication. 2016 doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Lam SL. Effect of 1-methyladenine on thermodynamic stabilities of double-helical DNA structures. FEBS Lett. 2009;583:1548–1553. doi: 10.1016/j.febslet.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Huang Y, Weng X, Russu IM. Enhanced base-pair opening in the adenine tract of a RNA double helix. Biochemistry. 2011;50:1857–1863. doi: 10.1021/bi1014997. [DOI] [PubMed] [Google Scholar]
- 46.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roost C, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015;137:2107–2115. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uesugi S, Miki H, Ikehara M, Iwahashi H, Kyogoku Y. A linear relationship between electronegativity of 2′-substituents and conformation of adenine nucleosides. Tetrahedron Lett. 1979;20:4073–4076. [Google Scholar]
- 49.Haschemeyer AEV, Rich A. Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J. Mol. Biol. 1967;27:369–384. doi: 10.1016/0022-2836(67)90026-5. [DOI] [PubMed] [Google Scholar]
- 50.Olson WK. Syn-Anti effects on the spatial configuration of polynucleotide chains. Biopolymers. 1973;12:1787–1814. doi: 10.1002/bip.1973.360120808. [DOI] [PubMed] [Google Scholar]
- 51.Sundaralingam M, Pan B. Hydrogen and hydration of DNA and RNA oligonucleotides. Biophys. Chem. 2002;95:273–282. doi: 10.1016/s0301-4622(01)00262-9. [DOI] [PubMed] [Google Scholar]
- 52.Williams JS, Kunkel TA. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu W-J, et al. How a low-fidelity DNA polymerase chooses non-Watson-Crick from Watson-Crick incorporation. J. Am. Chem. Soc. 2014;136:4927–4937. doi: 10.1021/ja4102375. [DOI] [PubMed] [Google Scholar]
- 55.Topal MD, Fresco JR. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976;263:289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- 56.Fernandez IS, et al. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimsey I, Al-Hashimi HM. Increasing occurrences and functional roles for high energy purine-pyrimidine base-pairs in nucleic acids. Curr. Opin. Struct. Biol. 2014;24:72–80. doi: 10.1016/j.sbi.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helm M, Giegé R, Florentz C. A Watson−Crick base-pair-disrupting methyl group (m1A9) Is sufficient for cloverleaf folding of human mitochondrial tRNALys . Biochemistry. 1999;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- 59.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 61.Zimmer DP, Crothers DM. NMR of enzymatically synthesized uniformly 13C15N–labeled DNA oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3091–3095. doi: 10.1073/pnas.92.8.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macon JB, Wolfenden R1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry. 1968;7:3453–3458. doi: 10.1021/bi00850a021. [DOI] [PubMed] [Google Scholar]
- 63.Timofeev EN, et al. Oligodeoxynucleotides containing 2′-deoxy-1-methyladenosine and Dimroth rearrangement. Helv. Chim. Acta. 2007;90:928–937. [Google Scholar]
- 64.Pivovarov VB, Stepanian SG, Reva ID, Sheina GG, Blagoi YP. Infrared spectra and the structure of 1-methyladenine in an argon matrix and solutions. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 1995;51:843–853. [Google Scholar]
- 65.Longhini AP, et al. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wenter P, Reymond L, Auweter SD, Allain FHT, Pitsch S. Short, synthetic and selectively 13C–labeled RNA sequences for the NMR structure determination of protein-RNA complexes. Nucleic Acids Res. 2006;34:e79–e79. doi: 10.1093/nar/gkl427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wunderlich CH, et al. Synthesis of (6–13C)pyrimidine nucleotides as spin-labels for RNA dynamics. J. Am. Chem. Soc. 2012;134:7558–7569. doi: 10.1021/ja302148g. [DOI] [PubMed] [Google Scholar]
- 68.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 69.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Branch M, Coleman T, Li Y. A subspace, interior, and conjugate gradient method for large-scale bound-constrained minimization problems. SIAM Journal on Scientific Computing. 1999;21:1–23. [Google Scholar]
- 71.Bothe JR, Stein ZW, Al-Hashimi HM. Evaluating the uncertainty in exchange parameters determined from off-resonance R1ρ relaxation dispersion for systems in fast exchange. J. Magn. Reson. 2014;244:18–29. doi: 10.1016/j.jmr.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wagenmakers E-J, Farrell S. AIC model selection using Akaike weights. Psychonomic Bulletin & Review. 11:192–196. doi: 10.3758/bf03206482. [DOI] [PubMed] [Google Scholar]
- 73.Sklenar V, Felgon J. Formation of a stable triplex from a single DNA strand. Nature. 1990;345:836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]
- 74.de los Santos C, Rosen M, Patel D. NMR studies of DNA (R+)n•(Y-)n•(Y+)n triple helices in solution: imino and amino proton markers of T•A•T and C•G•C+ base-triple formation. Biochemistry. 1989;28:7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]
- 75.Fürtig B, Richter C, Bermel W, Schwalbe H. New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J. Biomol. NMR. 2004;28:69–79. doi: 10.1023/B:JNMR.0000012863.63522.1f. [DOI] [PubMed] [Google Scholar]
- 76.Fonville JM, et al. Chemical shifts in nucleic acids studied by density functional theory calculations and comparison with experiment. Chemistry - A European Journal. 2012;18:12372–12387. doi: 10.1002/chem.201103593. [DOI] [PubMed] [Google Scholar]
- 77.Xu X-P, Au−Yeung SCF. Investigation of chemical shift and structure relationships in nucleic acids using NMR and density functional theory methods. The Journal of Physical Chemistry B. 2000;104:5641–5650. [Google Scholar]
- 78.Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proceedings of the National Academy of Sciences. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crystal structure of ribosomal protein tthl1 in complex with 80nt 23s rna from thermus thermophilus FAU. Tishchenko SV, Nikonova EY, Kostareva OS, Gabdulkhakov AG, Sarskikh AV, Piendl W, Nikonov SV, Garber MB, Nevskaya NA. [Google Scholar]
- 80.Rypniewski W, Adamiak DA, Milecki J, Adamiak RW. Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex. RNA. 2008;14:1845–1851. doi: 10.1261/rna.1164308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crothers DM, Bloomfield VA, Tinoco I. Nucleic acids: structures, properties, and functions. University science books Sausalito; 2000. [Google Scholar]
- 82.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA-solvent Interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 84.Ye J-D, et al. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proceedings of the National Academy of Sciences. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Macke TJ, Case DA. Molecular Modeling of Nucleic Acids Vol. 682 ACS Symposium Series. Vol. 24. American Chemical Society; 1997. pp. 379–393. Ch. [Google Scholar]
- 86.Brooks BR, et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Denning EJ, Priyakumar UD, Nilsson L, MacKerell AD. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. J. Comput. Chem. 2011;32:1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee MS, Feig M, Salsbury FR, Brooks CL. New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. J. Comput. Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 89.Xu Y, Vanommeslaeghe K, Aleksandrov A, MacKerell AD, Nilsson L. Additive CHARMM force field for naturally occurring modified ribonucleotides. J. Comput. Chem. 2016;37:896–912. doi: 10.1002/jcc.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goldsmith G, Rathinavelan T, Yathindra N. Selective preference of parallel DNA triplexes Is due to the disruption of Hoogsteen hydrogen bonds caused by the severe nonisostericity between the G*GC and T*AT Triplets. PLoS One. 2016;11:e0152102. doi: 10.1371/journal.pone.0152102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paci E, Karplus M. Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations. J. Mol. Biol. 1999;288:441–459. doi: 10.1006/jmbi.1999.2670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.