Abstract
Structure-based design of vaccines has been a long-sought goal, especially the iterative optimization used so successfully with structure-based design of drugs. We previously developed a 1st-generation vaccine antigen called DS-Cav1, comprising a pre-fusion-stabilized form of the fusion (F) glycoprotein, which elicited high titers of protective responses against respiratory syncytial virus (RSV) in mice and macaques. Here we report the improvement of DS-Cav1 through iterative cycles of structure-based design that significantly increased the titer of RSV-protective responses. The resultant 2nd-generation “DS2”-stabilized immunogens have F subunits genetically linked, fusion peptide deleted, and interprotomer movements stabilized by an additional disulfide bond. These DS2-immunogens are promising vaccine candidates with superior attributes, such as the absence of a requirement for furin cleavage and increased antigenic stability to heat inactivation. The iterative structure-based improvement described here may have utility in the optimization of other vaccine antigens.
Human respiratory syncytial virus (RSV) infects virtually all children by two years of age and accounts for ~50% of hospitalizations due to respiratory infections in this age group1-4. Maternal antibodies – transferred placentally during the last weeks of pregnancy – provide protective immunity to full-term infants5,6, but this protection wanes ~2-fold each month7-9, with infant hospitalization from RSV peaking in the first five months after birth10. Although maternal antibodies can be transferred after birth, for example, through breast milk, such transference is inefficient and involves primarily IgA. For infants born at <28 weeks gestation, monthly injections of palivizumab (Synagis®), a humanized monoclonal antibody, are recommended11, and this reduces the frequency of hospitalization by ~50%12. Premature infants born between 28 and 34 weeks gestation also have increased risk for severe RSV disease; and while full-term infants without identified risk factors do have reduced rates of severe RSV disease, these full-term infants nevertheless make up 50% of RSV-related hospitalizations10. New interventions are thus needed to reduce the disease burden imposed by RSV infection.
One possibility is maternal immunization. However, assuming roughly a one month half-life, titers would have to be 26-fold (64-fold) above the protective threshold at birth to safeguard infants during their most vulnerable period – and it has been unclear how to induce such very high protective responses. Many RSV vaccine candidates are being developed13-17. Most include the RSV fusion (F) glycoprotein antigen, the target of palivizumab18-20. The F glycoprotein is a type 1 fusion machine that transitions between pre-fusion and post-fusion states to facilitate RSV entry into target cells21. Initially synthesized as an F0 precursor, RSV F0 folds into a trimer, which is activated by furin cleavage into mature pre-fusion F (pre-F), comprising F1 and F2 subunits22. Pre-F is metastable, and spontaneously rearranges into a stable post-fusion state14,23,24. While initial focus has been on postfusion F14,24, many neutralization epitopes are not present on postfusion F25-27, including those for potent antibodies D25, AM22 and 5C428,29, directed against the membrane-distal antigenic site Ø29, and also those for quaternary-specific antibodies MPE8 and AM1426-28. Moreover, the realization that pre-F-specific antibodies determine the magnitude of RSV-neutralizing activity in human sera30 has led to a focus on pre-F. Two 1st-generation pre-F antigens, stabilized through structure-based design, appear especially promising. These are “DS-Cav1”31 a pre-F trimer, stabilized by a disulfide between residues 155 and 290 (“DS”), cavity-filling mutations S190F and V207L (“Cav1”) and an appended C-terminal fibritin-trimerization domain, and “SC-DM”32 also a pre-F trimer, with F2 fused to F1 (“SC”), stabilizing mutations N67I and S215P (“DM”) and an appended fibritin domain. DS-Cav1 induces neutralizing antibody titers ~70- and ~80-fold higher than postfusion F (for subtypes A and B, respectively) in rhesus macaques31, whereas SC-DM induces complete protection against viral challenge in cotton rats and remains primarily in the pre-fusion state for over 50 days at 4°C32. While promising, the duration of infant protection achieved by maternal immunization awaits clinical evaluation, and every 2-fold increase in immunogenicity would potentially extend protection in infants by an additional month. Since RSV-mediated disease may be largely a consequence of obstruction in small airways due to an accumulation of sloughed epithelium, mucus, fibrin and inflammatory debris33, a major goal for an RSV vaccine has been to delay the timing of primary RSV infection until infants are older and airways are larger.
We therefore sought to optimize the immunogenicity of the 1st-generation RSV antigens. Historically, boosting neutralizing activity in humans with pre-existing immunity has been difficult: decades of RSV-vaccine research had previously achieved an increase of only 2-4-fold in RSV-neutralizing titers in healthy adults34-36, and the increase in immunogenicity of pre-F DS-Cav1 over postfusion RSV F was considered one of the top scientific breakthroughs of 201337. How might we increase the immunogenicity of the 1st-generation pre-F antigens?
We hypothesized that each cycle of structure-based design might provide clues to optimize immunogenicity – and that we could explore these clues iteratively. Here, we describe our efforts to improve the immunogenicity of DS-Cav1 through iterative cycles of structure-based design. In each cycle, we used atomic-level design to engineer variants, expressed, and assessed their antigenic and physical properties, determined structures, measured the immunogenicity of select variants, and statistically analyzed results. The statistical analyses involved the “informatics” of design, properties, structure and immunogenicity that we previously postulated31 would provide a direction for improvement of immunogenicity. In total we employed 4 cycles of structure-based engineering, involving the design, expression, and antigenic assessment of hundreds of variants, the crystal structures of 6, and the immunogenicity of 14. The resultant 2nd-generation “DS2” antigens had increased antigenic stability and induced significantly higher RSV-neutralizing activity – an achievement critical to extending and improving vaccine-induced immunity against RSV-mediated disease.
RESULTS
Engineering and properties of vaccine-design cycle 1
In the structure-based design of DS-Cav131, we previously examined the interplay between design, physical and antigenic properties, atomic-level structure, and immunogenicity to obtain insight into the relationship between the antigenic and physical properties of the engineered RSV Fs and the elicitation of RSV-protective responses. Correlation between physical stability of the pre-fusion-specific site Ø and immunogenicity suggested further increases in physical stability might increase immunogenicity31. We therefore sought ways to increase the physical stability of antigenic site Ø29.
Studies with other type 1 fusion proteins suggested that genetically fusing the two subunits of mature pre-F into a single chain might enhance stability32,38,39. We therefore engineered DS-Cav1 variants with F2 fused genetically to F1 and both fusion peptide and pep27 region deleted (deletion of pep27 has been shown to facilitate F trimerization40) (Fig. 1). In particular, for the C terminus of F2 (where we engineered the start of the genetic linkage), we chose two positions: residue 97, at the end of the α1-helix, and residue 105, one residue after the last residue of F2 visible in the election density of the DS-Cav1 structure (Fig. 1a,b). For the N terminus of F1 (where we engineered the end of the genetic linkage), we chose either residue 145, just after the fusion peptide, or residue 151, a glycine in the α2-helix, which might accommodate flexibility (Fig. 1a,b).
Figure 1.
Design, structure, immunogenicity, and informatics of pre-fusion stabilized single-chain RSV F glycoproteins from design cycle 1. (a) RSV F DS-Cav1 structure in ribbon with F2 domain in dark gray and F1 domain in light gray. Close-up of the cleavage site of a single protomer with the fusion peptide colored orange. (b) Single-chain RSV F constructs were designed by replacing both of the furin cleavage sites (R109/E110 and R136/F137) and the fusion peptide with a variable-length linker replacing either residues 97–151 or residues 105–145. DS-Cav1 mutations (DS: disulfide between residues 155 and 290; Cav1: Ser to Phe mutation at residue 190 and Val to Leu at residue 207) were incorporated into all designs with other mutations labeled and indicated by vertical black lines. (c) RSV F sc9 DS-Cav1 structure in ribbon and overlaid with the DS-Cav1 structure (gray). Residues removed in the single chain design or that differed by rmsd > 5 Å highlighted in red. (d) Details of the inter-subunit linker between F2 and F1 (electron density (2Fo-Fc) is shown at 1σ in blue). (e) Serum neutralization titers from mice immunized with RSV F DS-Cav1 single chain variants with optimized linker (blue symbols) and Postfusion F and RSV F DS-Cav1 as controls (gray symbols). Titers from each mouse are shown, with geometric means indicated by horizontal red lines (values shown at the bottom). Each group contained 10 mice except the DS-Cav1 group which included 29 mice. P values <0.0001 (****), <0.01 (**) or <0.05 (*) were determined by two-tailed Mann-Whitney tests. (f) Statistical analysis of design cycle 1. Left: spearman correlation of antigenic and physical properties with neutralization titers for design cycle 1 immunogens (including DS-Cav1). Right: correlation of elution volume with neutralization titer. Pearson correlation for single-chain constructs denoted in green.
We synthesized forty-five constructs with a variety of linkers (Supplementary Table 1) and tested the expression of these in a 96-well microplate-formatted transient transfection format31. Five of these constructs expressed at levels which allowed characterization of their antigenic and physical properties (Table 1,2). Of these, sc9 DS-Cav1, comprising a GS linkage between residues 105 and 145, showed 4-fold enhanced expression and recognition by a number of pre-fusion specific antibodies (Fig. 1c). Notably, the single-chain variants showed altered mobility in size-exclusion chromatography (SEC) (Supplementary Fig. 1) and many displayed reduced tolerance to physical extremes (Table 2). However, sc9 DS-Cav1 did show increased ability to retain antigenic site Ø after incubation at 70°C, as assessed by binding to the site Ø-specific antibody D25. Specifically, 39% of sc9 DS-Cav1 was still active, whereas site Ø on the parent DS-Cav1 was completely lost, upon 1 h incubation at 70°C (Table 2).
Table 1.
Antigenic characteristics of single-chain RSV F glycoproteins stabilized in the pre-fusion state (design cycles 1 and 2)
| Antibody KD value (nM) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site Ø | Site II | Quaternary epitope | |||||||||
| Construct | Linker sequence | C-terminus | Oligomeric state | Elution volume (mL)† | Yield (mg/L) | D25 | AM22 | 5C4 | Mota$ | MPE8 | AM14 |
|
Cycle 1
| |||||||||||
| DS-Cav1 | N.A. | Foldon | Trimer | 61.17 | 3.5 | 0.15 | <0.01 | 13 | 0.04 | 0.5 | 3.3 |
| sc4 DS-Cav1 | M97 GSGGNGIGLG G151 | Foldon | Trimer | 59.46 | 3.4 | N. B. | N. B. | N. B. | 7.6 | N. B. | N. B. |
| sc8 DS-Cav1 | M97 GSGNVGLGG G151 | L512C, L513C | Monomer and Trimer | 59.97/72.88 | 1.0 | 7.4 | 3.9 | N. B | <0.01 | 0.5 | 1330 |
| sc9 DS-Cav1 | M97QSTPATNNGS G145 | Foldon | Trimer* | 55.77 | 18.5 | 0.8 | <0.01 | 31.3 | 13.7 | 0.8 | 33.1 |
| sc10 DS-Cav1 | M97QSTPATNNGSGSAIASG151 | S491 | Monomer and Trimer | 64.02/74.71 | 0.5 | 0.3 | 0.3 | 38.1 | 22.4 | 0.8 | N. B. |
| sc11 DS-Cav1 | M97QSTPATNNGSGSAIASG151 | A514 | Monomer | 72.14 | 1.0 | 0.7 | 1.6 | 58.8 | 29.3 | 1.6 | N. B. |
|
Cycle 2 | |||||||||||
| sc9-9 DS-Cav1 | M97QSTPAT G145 | Foldon | Trimer | 62.87 | 17.1 | 0.2 | 2.8 | 17.9 | 1.0 | <0.01 | 3.3 |
| sc9-10 DS-Cav1 | M97QSTPAT GS G145 | Foldon | Trimer | 61.59 | 17.8 | 0.8 | <0.01 | 26.2 | 9.7 | 0.8 | 6.6 |
| sc9-19 DS-Cav1 | M97QSTPAT GGSGGSG G145 | Foldon | Trimer* | 58.87 | 16.1 | 0.8 | 0.06 | 22.6 | 16.7 | 1.0 | 25.6 |
| sc9-24 DS-Cav1 | N97SALSAT GS G145 | Foldon | Trimer* | 60.75 | 14.5 | 0.8 | 0.3 | 20.8 | 15.1 | 0.8 | 25.1 |
| sc9-41 DS-Cav1 | M97QST G G145 | Foldon | Trimer* | 59.60 | 12.3 | N. B. | N. B. | N. B. | 0.04 | N. B. | N. B. |
| sc9-42 DS-Cav1 | A97QST G G145 | Foldon | Trimer* | 62.19 | 11.2 | N. B. | N. B. | N. B. | 5.9 | N. B. | N. B. |
| sc9 DS-Cav1 105Q | M97QSTPATNQGS G145 | Foldon | Trimer* | 56.65 | 13.5 | 0.7 | <0.01 | 28.8 | 16.3 | 0.8 | 46.2 |
Elution volume was determined using size-exclusion column HiLoad 16/60 Superdex 200, GE.
Motavizumab (Mota).
N.A.: not applicable.
Elution volume by gel filtration was slightly lower than DS-Cav1.
N.B.: no binding observed at 1000 nM Fab concentration
Table 2.
Physical stability of single-chain RSV F glycoproteins stabilized in the pre-fusion state (design cycles 1–4)
| Physical stability (fractional D25 reactivity) | |||||||
|---|---|---|---|---|---|---|---|
| Temp (°C) | pH | Freeze Thaw | Osmolality (mM) | ||||
| Construct | 50 | 70 | 3.5 | 10.0 | 10 | 3000 | |
| DS-Cav1 | 0.9 | 0.0 | 0.8 | 0.5 | 0.1 | 0.9 | 1.0 |
|
Cycle 1 | |||||||
| sc8 DS-Cav1 | 1.1 | 0.1 | 0.4 | 0.5 | 0.1 | 0.2 | 0.1 |
| sc9 DS-Cav1 | 0.4 | 0.4 | 0.2 | 0.1 | 0.2 | 0.1 | 0.1 |
| sc10 DS-Cav1 | 0.3 | 0.3 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 |
| sc11 DS-Cav1 | 0.2 | 0.1 | 0.5 | 0.2 | 0.0 | 0.1 | 0.1 |
|
Cycle 2 | |||||||
| sc9-10 DS-Cav1 | 0.9 | 0.9 | 1.5 | 0.7 | 0.2 | 1.4 | 0.7 |
| sc9-19 DS-Cav1 | 0.9 | 0.9 | 0.7 | 0.8 | 0.04 | 0.6 | 0.5 |
| sc9-24 DS-Cav1 | 0.9 | 0.9 | 0.7 | 0.7 | 0.0 | 0.5 | 0.5 |
|
Cycle 3 | |||||||
| sc9-10 DS-Cav1 A149C Y458C | 1.0 | 0.8 | 2.3 | 0.7 | 0.3 | 0.7 | 0.6 |
| sc9-10 DS-Cav1 N183GC N428C | 1.0 | 0.5 | 2.6 | 0.7 | 0.3 | 0.9 | 0.4 |
|
Cycle 4 | |||||||
| sc9-10 DS-Cav1 A149C Y458C S46G E92D S215P K465Q | 1.0 | 0.9 | 2.7 | 0.7 | 0.1 | 0.8 | 0.7 |
| sc9-10 DS-Cav1 A149C Y458C S46G N67I E92D S215P K465Q | 1.0 | 0.9 | 2.7 | 0.4 | 0.1 | 0.7 | 0.8 |
| sc9-10 DS-Cav1 A149C Y458C S46G S215P K465Q | 1.0 | 0.9 | 2.7 | 0.5 | 0.2 | 0.8 | 0.7 |
| sc9-10 DS-Cav1 N183GC N428C S46G N67I E92D S215P K465Q | 1.0 | 0.1 | 1.0 | 0.2 | 0.0 | 0.3 | 0.2 |
| sc9-10 DS-Cav1 Q98C Q361C L95M I221M R429K | 1.0 | 0.8 | 1.7 | 0.3 | 0.1 | 0.3 | 0.3 |
| sc9-10 DS-Cav1 Q98C Q361C S46G E92D L95M S215P I217P I221M R429K K465Q | 1.0 | 0.9 | 2.7 | 0.7 | 0.1 | 0.8 | 0.7 |
The fractional value refers to D25 reactivity retained after exposure to various physical extremes as compared to initial D25 binding level measured by Octet biolayer interferometry.
Structures, immunogenicities, and informatics of cycle 1
To assess the effects of the sc9 alteration on DS-Cav1, we crystallized and determined the structure of sc9 DS-Cav1 at 2.98 Å resolution (Table 3). The structure of sc9 DS-Cav1 was similar to the parent DS-Cav1, with a root-mean-square deviation (rmsd) of 0.6 Å for all residues excluding 105-151 (Fig. 1c). The fusion peptide deletion did not induce any substantial movements in the neighboring region (Fig. 1d, left), where the new connection between 105 and 145 showed partially ordered electron density (Fig. 1d, right). Thus the structural effects of fusing F2 to F1 and removing the fusion peptide on the structure of DS-Cav1 were minimal, localized almost entirely to the linker region.
Table 3.
Data collection and refinement statistics
| Protein | sc9 DS-Cav1 (5K6B) |
sc9-10 DS-Cav1 (5K6C) |
sc9-19 DS-Cav1 (5K6F) |
sc9-24 DS-Cav1 (5K6G) |
sc9-10 DS-Cav1 A149C Y458C (5K6H) |
sc9-10 DS-Cav1 A149C Y458C S46G N67I E92D S215P K465Q (5K6I) |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P4132 | P4132 | P4132 | P4132 | P4132 | P4132 |
| Cell dimensions | ||||||
| a, b, c (Å) | 171.3, 171.3, 171.3 | 171.2, 171.2, 171.2 | 168.9, 168.9, 168.9 | 168.2, 168.2, 168.2 | 169.1, 169.1, 169.1 | 170.7, 170.7, 170.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–2.98 (3.21–3.09; 3.09–2.98) | 50–3.58 (3.86–3.71, 3.71–3.58) | 50–2.55 (2.75–2.64, 2.64–2.55) | 50–2.90 (3.12–3.00, 3.00–2.90) | 50.0–2.65 (2.74–2.65) | 50–2.92 (3.02–2.92) |
| R merge | 10.9 | 8.2 | 15.0 | 14.9 | 11.8 | 14.0 |
| I / σI | 11.5 (1.8, 1.0) | 11.3 (1.67, 1.0) | 16.2 (2.5, 1.3) | 13.8 (1.9, 1.0) | 8.2 (1.87) | 11.57 (2.25) |
| Completeness (%) | 97.3 (99.7, 89.2) | 91.5 (94.1, 91.2) | 100.0 (100.0, 49.6) | 100.0 (100.0, 99.9) | 79.4 (66.3) | 98.0 (94.2) |
| Redundancy | 4.3 (4.0, 2.7) | 5.9 (5.5, 3.6) | 19.9 (14.7, 7.3) | 11.6 (10.7, 8.8) | 3.2 (1.9) | 5.4 (3.2) |
| Refinement | ||||||
| Resolution (Å) | 50–2.98 | 50–3.58 | 50–2.55 | 50–2.90 | 50.0–2.65 | 50.0–2.92 |
| No. reflections | 17652 | 9773 | 12401 | 18563 | 19565 | 18357 |
| Rwork / Rfree | 19.4/23.7 | 25.3/28.9 | 19.1/25.3 | 27.2/31.1 | 22.7/27.1 | 22.3/25.9 |
| No. atoms | ||||||
| Protein | 3522 | 3488 | 3461 | 3435 | 3433 | 3462 |
| Carbohydrate | 19 | – | – | – | – | 54 |
| Water | – | – | – | – | – | – |
| B–factors | ||||||
| Protein | 111.3 | 108.3 | 100.8 | 92.9 | 72.5 | 82.3 |
| Carbohydrate | 181 | – | – | – | – | 131 |
| Water | – | – | – | – | – | – |
| R.m.s. deviation | ||||||
| Bond lengths (Å) | 0.009 | 0.002 | 0.010 | 0.010 | 0.004 | 0.002 |
| Bond angles (°) | 1.26 | 0.63 | 1.42 | 1.31 | 0.815 | 0.514 |
Values in parentheses are for highest-resolution shells. One crystal was used to measure the data for each crystal structure.
To measure the immunogenicity of the various single-chain DS-Cav1 molecules, we immunized CB6F1/J mice with 10 μg RSV F combined with 50 μg polyinosinic-polycytidylic acid adjuvant at weeks 0 and 3 intra-muscularly, and evaluated the ability of week 5 sera to prevent RSV infection of HEp-2 cells. Notably all titers elicited by these single-chain antigens were lower than those elicited by DS-Cav1 (Fig. 1e). Variants sc8 DS-Cav1 and sc11 DS-Cav1 had especially low immunogenicity, whereas sc9 DS-Cav1, the single-chain with highest immunogenicity, had a geometric mean titer one third that of DS-Cav1.
The increased physical stability of sc9 DS-Cav1 at 70°C did not yield an increase in immunogenicity, likely indicating other factors with more substantial effects on immunogenicity had been altered between DS-Cav1 and sc9 DS-Cav1. We analyzed the immunogenicity of the five characterized single-chain DS-Cav1 constructs, sc4, sc8, sc9, sc10 and sc11, in the context of their (i) antigenicity toward site-Ø antibodies (D25, AM22, 5C4), motavizumab, and quaternary antibodies (MPE8 and AM14), and (ii) physical properties such as elution volume, expression yield, and physical stability (D25 reactivity retained after exposure to various physical extremes) (Fig. 1f). No single property significantly correlated with immunogenicity. However, we noticed that the property that has the largest absolute correlation with immunogenicity was elution volume (anti-correlates, Fig. 1f) (Pearson correlation r=−0.976, P=0.0042 for single-chain constructs). Although the relationship between elution volume and immunogenicity likely has an optimum as opposed to monotonically increasing or decreasing and the number of data points was small, we nevertheless selected sc9 DS-Cav1 for further optimization based on it’ antigenicity and it also having the highest immunogenicity of the single-chain constructs.
Structure-based optimization for vaccine-design cycle 2
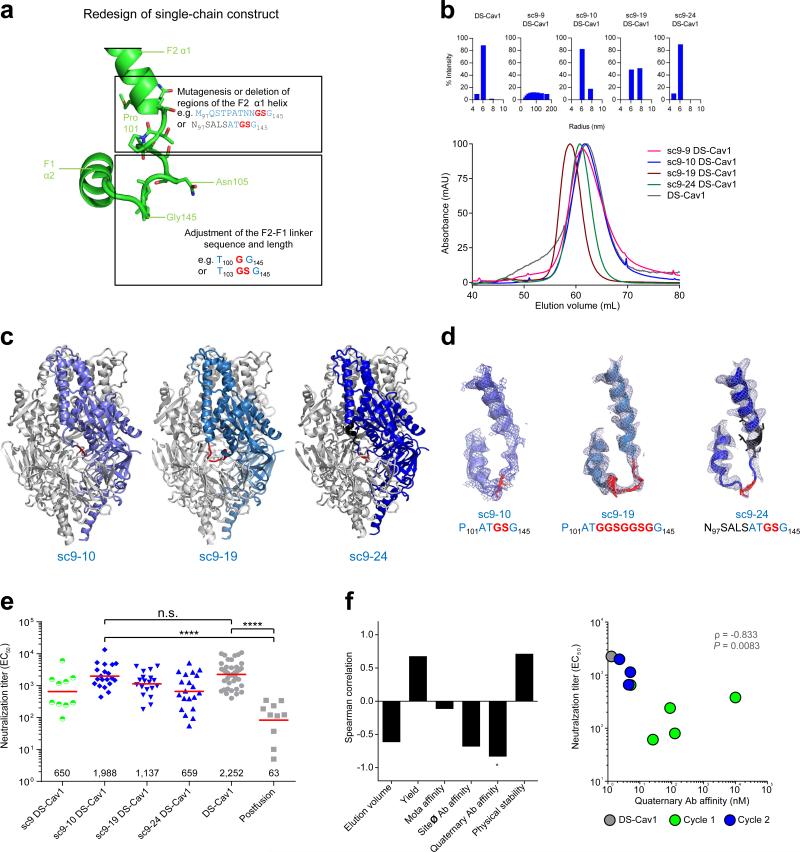
As differences in the linker between F2 and F1 subunits appeared to affect immunogenicity, we further varied this linker and also altered residues in the F2 α-1 helix (Fig. 2A). We synthesized 45 variants of sc9 DS-Cav1 (Supplementary Table 1) and tested the expression of these in the 96-well microplate transient transfection format31. Most of these constructs expressed at levels which allowed characterization of their antigenic and physical properties (Table 1, 2, Fig. 2b). Of these, one construct, sc9-10 DS-Cav1 (which differs from sc9 DS-Cav1 by having residues 104 and 105 removed), had a SEC-elution volume similar to that of DS-Cav1 (Fig. 2b), and displayed improved affinity (from 33.1 nM to 6.6 nM) to the pre-fusion quaternary-specific antibody AM14 (Table 1). Constructs sc9-41 and sc9-42 with shortened F2-linkers did not bind to pre-fusion specific antibodies despite containing DS-Cav1 stabilizing mutations indicating that subtle changes in the F2-F1 linker sequence and length can have substantial impact on antigenicity. sc9-10 had a similarly improved affinity over sc9 to antibody AM14 as did sc9-9, however sc9-9 was not chosen for further improvement because it had a suboptimal SEC profile and was poly-disperse as assessed by dynamic light scattering (DLS).
Figure 2.
Design, properties, structure, immunogenicity, and informatics of single-chain RSV F with altered F2-F1 linkers from design cycle 2. (a) Design of altered F2-F1 linkers. (b)Representative dynamic light scattering profiles and gel filtration chromatograms of single-chain RSV F glycoprotein variants with altered linkers. A representative gel filtration profile for each protein is shown as assessed on a Superdex-200 16/60 column (120 ml column volume). sc9-10 DS-Cav1 exhibited similar elution profiles characteristic as DS-Cav1. (c) Structures of single-chain RSV F glycoproteins with optimized F2-F1 linkers. (d) Structural details of the F2-F1 linker with the linker region in red, mutations in black, and electron density in blue. (e) Serum neutralization titers from mice immunized with RSV F DS-Cav1 single chain variants with optimized linker (blue symbols) and Postfusion F and RSV F DS-Cav1 as controls (gray symbols). Immunization titers from sc9 DS-Cav1 (green symbols) are shown for reference. Titers from each mouse are shown, with geometric means indicated by horizontal red lines (values shown at the bottom). Each group contained 10 mice except the sc9-10 DS-Cav1 (20 mice), sc9-19 DS-Cav1 (19 mice), sc9-24 DS-Cav1 (20 mice) and DS-Cav1 (29 mice) groups. P values <0.0001 (****) or <0.001 (***) were determined by two-tailed Mann-Whitney tests. (f)Statistical analysis of design cycles 1–2. Left: spearman correlation of antigenic and physical properties with neutralization titers for variants from design cycles 1 and 2 (including DS-Cav1). Correlations with adjusted P value (Bonferroni correction) of less than 0.05 were marked with “*”. Right: correlation of quaternary antibody antigenicity with neutralization titer. “10,000” was used for antigenicity with “N.B.” values.
To provide atomic level details, we crystallized and determined the structures of sc9-10, sc9-19, and sc9-24 versions of DS-Cav1 at 3.6, 2.6, and 2.9 Å resolution respectively (Table 3). All of these structures were highly similar to the parent sc9 DS-Cav1 (Fig. 2c), though with important differences in their F2-F1 linkers as expected from the sequence changes in this region (Fig. 2d). The sc9-10 and sc9-19 constructs had improved electron density in the linker region compared to sc9, while sc9-24 electron density was similar to sc9.
As before, we measured the immunogenicity of the various single-chain DS-Cav1 molecules by immunizing CB6F1/J mice at weeks 0 and 3. Importantly, the titers of sc9-10 DS-Cav1 at week 5 (geometric mean of 1988) were statistically indistinguishable from those of DS-Cav1 (Fig. 2e). Notably, statistical analysis indicated quaternary antibody affinity correlated significantly with improved immunogenicity (P=0.0083, Fig. 2f), suggesting reduced protomer flexibility might lead to improved neutralizing titers. For the 3rd cycle of structure-based design, we thus sought to reduce protomer flexibility even further, by introducing interprotomer disulfide linkages.
Structure-based optimization for vaccine-design cycle 3
We analyzed the structure of sc9-10 DS-Cav1 for interprotomer positions where disulfide linkages might be introduced (Fig. 3a). We synthesized 17 variants of sc9-10 with introduced interprotomer disulfides (Supplementary Table 1) and tested the expression of these in the 96-well microplate-formatted transient transfection format. Four of these constructs expressed at levels which allowed characterization by reducing and non-reducing SDS-PAGE (Fig. 3b); in the absence of reducing agent, all of these constructs ran as disulfide-linked oligomers of over 150 kDa without detectable dimer or monomer populations. In the presence of reducing agent, all four ran as monomers of ~60 kDa.
Figure 3.
Design, structure, immunogenicity, and informatics of single-chain RSV F glycoproteins with interprotomer disulfides (DS2) from design cycle 3. (a) Crystal structure of RSV sc9-10 DS-Cav1 trimer in ribbon. Insets show close-ups of Cycle 3 designs for residues mutated to Cys to form interprotomer disulfides (mutations labeled and in stick representation). (b) Engineered single-chain RSV F glycoproteins with interprotomer disulfides by SDS-PAGE (uncropped form is shown in Supplementary Fig. 1c). (c) Negative-stain EM class-averaged images of RSV F DS2 variants. (d) Structure of single-chain RSV F glycoproteins sc9-10 with interprotomer disulfide A149C Y458C shown in close-up with electron density. (e) Serum neutralization titers of 10 mice immunized with sc9-10 DS-Cav1 A149C Y458C and 15 mice immunized with sc9-10 DS-Cav1 N183GC N428C. Titers from each mouse are shown as individual purple symbols, with geometric means indicated by horizontal red lines (values shown at the bottom). Sera titers from 20 mice immunized with sc9-10 DS-Cav1 (blue symbols) and 39 mice immunized with RSV F DS-Cav1 (gray symbols) are shown for reference. P values <0.0001 (****) or <0.001 (***) were determined by two-tailed Mann-Whitney tests. (f)Statistical analysis of design cycles 1–3. Left: spearman correlation of each physical property with neutralization titers for design cycle 1, 2, and 3 variants (including DS-Cav1). Correlations with adjusted P value (Bonferroni correction) of less than 0.05 were marked with “*”. Right: correlation of quaternary antibody antigenicity and physical stability with neutralization titer. Constructs containing a mutation at residue 183 were not included in the correlation for quaternary antibody antigenicity as N183 is part of the AM14 epitope. “10,000” was used for antigenicity with “N.B.” values.
The sc9-10 DS-Cav1 A149C Y458C variant expressed at 1 mg/L, the N183GC N428C variant at only ~0.1 mg/L (but showed a typical pre-F structure by negative stain-EM (Fig. 3c)), and the T100C S362C and the T369C T455C variants at quantities too low for characterization. Disulfide bonds also formed when tested with two-residue variation around the T100C S362C disulfide bond location (e.g. 98C 361C). Both sc9-10 DS-Cav1 A149C Y458C and N183GC N428C variants showed increased physical stability, being 6-fold and 19-fold more stable, respectively, than DS-Cav1 at 60°C (Table 2, Supplementary Fig. 2, Supplementary Table 2).
We crystallized and determined the structure of sc9-10 DS-Cav1 A149C Y458C at 2.65 Å resolution (Table 3). The structure of sc9-10 DS-Cav1 A149C Y458C was similar to the parent sc9-10 DS-Cav1 (Fig. 3d). Notably, we observed strong electron density between residue 149 and 458 consistent with the formation of a disulfide bond. The overall quaternary structure, moreover, was maintained between sc9-10 and sc9-10 DS-Cav1 A149C Y458C, with low rmsd for protomer (0.9Å) and for the entire trimer (1.0 Å).
To measure the immunogenicity of the 2nd-generation DS-Cav1 with interprotomer disulfides (which we named “DS2” immunogens, for their 2nd-interprotomer disulfides), we immunized CB6F1/J mice intra-muscularly as described for earlier variants (Fig. 3e). Notably, two DS2 immunogens, sc9-10 DS-Cav1 A149C Y458C and N183GC N428C, showed a significant increase in RSV-neutralizing titer, 3–4-fold higher than that of DS-Cav1. When we analyzed statistically the cumulative data on immunogenicity versus various properties from design cycles 1–3, both quaternary antibody affinity (P=0.0016) and physical stability (P=0.0037) significantly correlated with immunogenicity (Fig. 3f), indicating the importance of appropriate quaternary organization as well as improved physical stability to the elicitation of RSV-neutralizing antibodies.
Structure-based optimization for vaccine-design cycle 4
The heightened immunogenicity of DS2 immunogens with interprotomer disulfides indicated interprotomer stabilization might be a general way to increase the elicitation of RSV-protective responses. However, the low expression levels of many of the designed interprotomer variants revealed expression to be a roadblock. For design cycle 4, we thus strove to combine interprotomer disulfides A149C Y458C and N183GC N428C (Fig. 3) with other mutations that might additionally stabilize the pre-fusion state or increase expression – including mutations N67I and S215P (identified by Krarup and colleagues32) (Supplementary Fig. 3) as well as variant strains of RSV such as the Long strain and subtype-B strains such as CH1853741 (uniprot P13843) (Fig. 4, Supplementary Table 1). We synthesized 396 constructs and assessed their antigenicity and level of expression in the 96-well microplate transient transfection format. Six of these constructs expressed at levels which allowed characterization of their antigenic and physical properties (Table 4). Expression levels appeared to be heightened by the presence of the mutants, S46G, K465Q and S215P32 as well as by mutations L95M, I221M, and R429K. Importantly, all of these DS2-stabilized constructs maintained increased physical stability at 60°C, which ranged from 6-7-fold for the A149C Y458C-based constructs to 13–19-fold for the N183GC N428C-based constructs (Supplementary Fig. 2, Supplementary Table 2).
Figure 4.
Design, structure, immunogenicity, and informatics from design cycle 4 comprising combinations of interprotomer disulfides (DS2) and other mutations. (a) Design cycle 4 involving interprotomer disulfides combined with other variants highlighted here in orange and detailed in Table S1. (b) Phylogenetic tree of strains tested. (c) Structures of single-chain RSV F glycoproteins sc9-10 DS-Cav1 A149C Y458C S46G E92D S215P K465Q with details of surface mutations shown in insets. (d) Serum neutralization titers from mice immunized with RSV F DS-Cav1 single-chain variants (n=10). Titers from each mouse are shown as individual orange symbols, with geometric means indicated by horizontal red lines. Titers from mice immunized with designs based on sc9-10 DS-Cav1 A149C Y458C are shown with differently formatted circle symbols. Designs based on sc-9-10 DS-Cav1 N183GC N428C are shown with orange square symbols, and designs based on sc9-10 DS-Cav1 Q98C Q361C with orange triangle symbols. These latter two designs were based on the RSV CH18537 and Long strains. Sera titers from 39 mice immunized with RSV F DS-Cav1 (gray symbols) are shown for reference. P values <0.0001 (****), <0.01 (**) or <0.05 (*) were determined by two-tailed Mann-Whitney tests. (e) Statistical analysis of design cycles 1–4. Left: spearman correlation of antigenic and physical properties with neutralization titers for variants from design cycles 1–4 (including DS-Cav1). Correlations with adjusted P value (Bonferroni correction) of less than 0.05 are marked with “*”. Right: correlation of quaternary antibody antigenicity and physical stability with neutralization titer. Constructs containing mutations at residue 183 were not included in the correlation for quaternary antibody antigenicity as N183 is part of the AM14 epitope. “10,000” was used for antigenicity with “N.B.” values.
Table 4.
Antigenic characteristics of single-chain RSV F glycoproteins stabilized in the pre-fusion state (design cycles 3 and 4)
| Antibody KD value (nM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Site Ø |
Site II | Quaternary epitope |
|||||||
| Construct | Strain | Oligomeric state | Yield (mg/L) | D25 | AM22 | 5C4 | Mota | MPE8 | AM14 |
|
Cycle 3
| |||||||||
| sc9-10 A DS-Cav1 A149C Y458C | A2 | Trimer | 0.8 | 0.2 | 0.1 | 32.1 | 0.4 | 0.1 | 6.3 |
| sc9-10 DS-Cav1 N183GC N428C# | A2 | Trimer | 0.1 | 0.2 | 0.2 | 14.0 | 0.3 | 0.1 | 787# |
|
Cycle 4 | |||||||||
| sc9-10 DS-Cav1 A149C Y458C S46G E92D S215P K465Q | A2 | Trimer | 10.8 | 0.2. | 0.3 | 314.6 | 0.6 | 0.2 | 15.9 |
| sc9-10 DS-Cav1 A149C Y458C S46G N67I E92D S215P-K465Q | A2 | Trimer | 14.0 | 0.6 | 0.2 | 126.9 | 0.1 | 0.2 | 1.0 |
| sc9-10 DS-Cav1 A149C Y458C S46G S215P K465Q | A2 | Trimer | 10.4 | 0.1 | 0.5 | 92.2 | 0.4 | 0.4 | 35.1 |
| sc9-10 DS-Cav1 N183GC N428C S46G N67I E92D S215P K465Q | A2 | Trimer | 5.1 | 0.2 | 0.4 | 79.7 | 0.6 | 0.2 | 2210 |
| sc9-10 DS-Cav1 Q98C Q361C L95M I221M R429K | CH18537 | Trimer | 1.0 | 0.1 | 0.1 | 100.9 | 0.2 | 0.2 | 18.5 |
| sc9-10 DS-Cav1 Q98C Q361C S46G E92D L95M S215P I217P I221M R429K K465Q | Long (VR26) | Trimer | 1.0 | 0.1 | 0.1 | 35.8 | 11.1 | 0.2 | 33.9 |
N183 is a contact residue for antibody AM14.
To provide insight into the increased expression associated with the S46G, K465Q and S215P mutants, we determined and solved the structure of the DS2 immunogen, sc9-10 A149C Y458C S46G K465Q S215P E92D (Fig. 4c) at 2.92 Å resolution (Table 3). Notably, none of the crystallographically visualized alterations, S46G, E92D, S215P and K465Q, resulted in changes in backbone movements of over 1 Å (Fig. 4c). We examined the local structure (Fig. 4c) of the mutants S46G, E92D, and K465Q and observed several of these mutations to reduce unfavorable interactions with proximal residues: S46 directly points towards another hydroxyl, E92 clashes with N254 (which hydrogen bonds to D92), and K465 clashes electrostatically with a neighboring lysine. Meanwhile, as noted previously32, S215P destabilizes the formation of an elongated helix found in the RSV F-postfusion form.
To measure the impact of these mutations on the immunogenicity of these DS2 immunogens, we immunized CB6F1/J mice (Fig. 4d). Notably, four of the DS2-stabilized RSV F trimers showed 2.5–4-fold increased elicitation of RSV-neutralizing responses relative to DS-Cav1. All of the increased titers involved variants with the triple mutant S46G, K465Q and S215P in the A2 strain. RSV F variants from the CH18537 and Long RSV strains containing these or other mutations also showed both homologous and heterologous neutralization titers (Supplementary Fig. 4). To understand factors that related to immunogenicity, we again used statistical analyses, incorporating information from cycles 1-4 (Fig. 4e). Similar to the analyses using cycles 1-3 data, both quaternary antibody affinity (P<0.0001) and physical stability (P<0.0001) significantly correlated with immunogenicity (Fig. 4e), again suggesting that appropriate quaternary organization and improved physical stability can result in increased elicitation of RSV-neutralizing antibodies.
To provide additional insight into factors that might improve immunogenicity, we assessed sera reactivity to DS-Cav1 as well as to D25, AM14, motavizumab, and MPE8 antigenic sites from select immunization groups across design cycles 1 to 4 (Supplementary Fig. 5). Antibody competition for binding to DS-Cav1 using D25, AM14, motavizumab, and MPE8 significantly reduces the sera reactivity to pre-fusion stabilized F by at least two-thirds suggesting much of the response to be either site Ø-directed or quaternary specific. Overall, sera reactivity agreed well with immunogenicity, with the more stabilized DS2 immunogens eliciting increased sera reactivity to DS-Cav1 as well as to the antigenic sites recognized by antibodies D25, AM14, motavizumab, and MPE8.
Discussion
By employing iterative cycles of structure-based vaccine design, we identified 2nd-generation DS2-F glycoprotein immunogens with interprotomer disulfides that induced ~4-fold higher neutralizing activity than DS-Cav1 after two immunizations of RSV-naïve CB6F1/J mice. We previously observed titers in these mice and in rhesus macaques to be correlated (r=0.9975, P=0.0451)31, and would thus expect DS2 immunogens to also have increased immunogenicity in primates. Whether the immunogenicity of these DS2 immunogens is sufficient to induce titers in pregnant women beyond the critical 64-fold protective threshold needed to protect infants during the first 6 months of life, however, will need to be answered through clinical trials. Relevant to this, we note that maternal immunization may not require durability, just high magnitude during the last weeks of gestation, and – in the case of neutralizing activity – more is better. The DS2 RSV F immunogens containing interprotomer disulfides such as RSV F sc9-10 DS-Cav1 A149C Y458C are thus likely to extend the duration and to increase the protection from RSV.
We were surprised by the substantial increase in immunogenicity of sc9-10 versus sc9 (Fig. 2e), from which it differed by only two amino acids, and we thus sought further characterization of the difference between these two immunogens. Small angle X-ray scattering (SAXS) characterization of sc9 and sc9-10 yielded distinct scattering patterns (Supplementary Fig. 6a). The Kratky plots (Supplementary Fig. 6b) showed that at high q, sc9-10 converged to near zero, as expected for a compact molecule. In contrast, the sc9 Kratky plot plateaued near its maximum, consistent with a substantially flexible or unfolded molecule42. The estimated Porod exponent (Px) for sc9 and sc9-10 were 4.0 and 2.1, respectively, also consistent with sc9-10 being compact and sc9 occupying a larger volume (Supplementary Fig. 6c)43. Altogether, these data indicate substantial conformational flexibility of sc9, not apparent in the sc9 crystal structure, suggesting solution characterization may be more appropriate for analyses of flexibility.
While the 2nd-generation DS2 RSV F immunogens showed significant increases in stability at higher temperature relative to DS-Cav1, other single-chain variants were less stable at physical extremes of pH and osmolality. Notably the correlation between physical stability and increased elicitation of RSV-protective responses was only significant when variants with interprotomer disulfides were included (Figs. 3e and 4e). The increased stability of interprotomer disulfide-stabilized immunogens likely results from the inhibition by the interprotomer disulfides of conformational rearrangements required to access the post-fusion state. Moreover, different unfolding pathways may be inhibited by each interprotomer disulfide, as each showed characteristic increases in stability (~5-fold for A149C Y458C and ~20-fold for N183GC N428C) (Table 2, Supplementary Fig. 2, Supplementary Table 2). It remains to be seen whether combinations of interprotomer disulfides can be expressed and whether these will be further increased in stability. We note that in this study, we used phosphate-buffered saline as a nominal formulation; when we increased salt concentrations to the 350 nM NaCl used in our previously published stability assessments31, the 2nd-generation immunogens showed greater tolerance of physical extremes (Table 2). Also, we were concerned by the extensive loss of activity resulting from 10 rounds of freeze-thaw, but found we could limit freeze-thaw damage by the introduction of 10% glycerol (Supplementary Table 3). While in vivo data on RSV F stability is not available, we note that the introduction of the “DS” mutation in the viral context also results in increased immunogenicity. The link between increased stability of pre-F and increased immunogenicity thus appears to be applicable to both protein subunit and live-attenuated vaccine contexts44.
We previously tested 16 disulfide variants including A149C Y458C, and 3 variants of N183C during our 1st-generation stabilization of mature RSV F, but all of these failed to express31. This failure may relate to structural rearrangements of pre-fusion RSV F between mature and immature states. Specifically, mature pre-F buries the C terminus of F2 and the N terminus of F1, both of which are exposed in immature pre-F (the trimeric uncleaved F0), as furin must have access to these sites to perform the cleavage required for F activation. If interprotomer disulfides were incompatible with trimeric uncleaved F0, then their formation (and expression) might require folding directly into the mature pre-fusion state, as we appear to have achieved with the sc9-10 version of DS-Cav1. We note that interprotomer disulfides do form with influenza A hemagglutinin45, however, influenza A hemagglutinin does not show the extensive pre-fusion structural rearrangements of fusion machines from either RSV or HIV-131,46.
Could the iterative structure-based optimization described here have utility in the improvement of other vaccine immunogens? We previously showed how structure-based design could be used to stabilize the pre-fusion closed conformation of HIV-146), and it should be possible to stabilize the pre-fusion forms of other type 1 fusion proteins, including those from paramyxovirus pathogens47 such as human metapneumovirus48 and parainfluenza virus49. Whether the iterative optimization described here can be applied to other fusion proteins will likely depend on the relationship between the pre-fusion stability of these other fusion glycoproteins and their immunogenicity.
In creating a matrix of information31,50 by which to improve immunogenicity, we utilized not only measurements of immunogenicity but also of antigenic and physical properties. To these we added a design dimension, allowing us to sample the information matrix so that statistical analyses could identify, from experimental observations, a direction by which to improve RSV-protective responses. In the four cycles described here, structure provided both a means to design variants as well as to understand their properties. Overall we believe that, in the same way that structure-based drug design allows for the iterative optimization of lead therapeutic compounds (e.g. see Klebe, 200051), the informatics of design, properties, structure, and immunogenicity described here to achieve DS2 RSV F immunogens should allow for the iterative optimization of other lead vaccine antigens.
Online Methods
Protein expression and purification
RSV F mutations were made by site-directed mutagenesis using the Stratagene Quik-change procedure. RSV F variants were expressed by transient transfection of Expi293F cells using 293Fectin (Invitrogen). Cell culture supernatants were harvested 5 days post transfection and centrifuged at 10,000 g to remove cell debris. The supernatants were sterile-filtered, and RSV F proteins were purified by nickel and streptactin-affinity chromatography followed by size-exclusion chromatography. Final yield was calculated based on at least two purification runs. The nickel and streptactin purification tags were removed for animal immunization and crystallization studies. To remove the purification tags, proteins were digested with 2 U/mg restriction-grade thrombin (Novagen) overnight at 4 °C and then purified by a second round of size-exclusion chromatography in PBS buffer. The Expi293F cells were authenticated at the source. Mycoplasma testing was performed through Hoechst DNA staining.
Screening of pre-fusion-stabilized RSV F constructs
Pre-fusion RSV F variants were derived from the RSV F (+) Foldon construct, which consists of RSV F residues 1–513 with a C-terminal T4 fibritin trimerization motif (foldon), thrombin site, 6x His-tag, and StreptagII or the modified RSV F DS-Cav1 construct31. A 96-well microplate-formatted transient gene expression approach was used to achieve high-throughput expression of various RSV F proteins as described previously52. Briefly, 24 h prior to transfection HEK 293T cells were seeded in each well of a 96-well microplate at a density of 2.5×105 cells/ml in expression medium (high glucose DMEM supplemented with 10 % ultra-low IgG fetal bovine serum and 1x-non-essential amino acids), and incubated at 37°C, 5% CO2 for 20 h. Plasmid DNA and TrueFect-Max (United BioSystems, MD) were mixed and added to the growing cells, and the 96-well plate incubated at 37°C, 5% CO2. One day post transfection, enriched medium (high glucose DMEM plus 25% ultra-low IgG fetal bovine serum, 2x nonessential amino acids, 1x glutamine) was added to each well, and the 96-well plate was returned to the incubator for continuous culture. On day five, post transfection, supernatants with the expressed RSV F variants were harvested and tested by ELISA for binding to D25, AM22, 5C4, and motavizumab antibodies using Ni2+-NTA microplates. After incubating the harvested supernatants at 4° C for one week, ELISAs were repeated. The HEK 293T cells were authenticated at the source. Mycoplasma testing was performed through Hoechst DNA staining.
RSV F antigenic characterization
A fortéBio Octet Red384 instrument was used to measure binding kinetics of RSV F foldon-removed variants to antibodies that target the pre-fusion form5,19,27,29,53. All assays were performed with agitation set to 1,000 rpm in phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin (BSA) to minimize nonspecific interactions. The final volume for all solutions was 50 μl/well. Assays were performed at 30°C in tilted black 384-well plates (Geiger Bio-One). Ni-NTA sensor tips (fortéBio) were used to load his-tagged proteins for 300 s to capture. Biosensor tips were then equilibrated for 90 s in PBS + 1% BSA prior to measuring association with antigen binding fragments (Fabs) in solution (0.007 μM to 0.5 μM) for 300 s; Fabs were then allowed to dissociate for 300 s-1200 s depending on the observed dissociation rate. Parallel correction to subtract systematic baseline drift was carried out by subtracting the measurements recorded for a loaded sensor incubated in PBS + 1% BSA. Data analysis and curve fitting were carried out using Octet software, version 8.0. Experimental data were fitted with the binding equations describing a 1:1 interaction. Global analyses of the data sets assuming reversible binding (full dissociation) were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for at least four concentrations used in each experiment.
Physical stability of RSV F variants
To assess the physical stability of the pre-fusion conformation of designed RSV F glycoproteins under various stress conditions, we treated the proteins with a variety of pharmaceutically relevant stresses such as extreme pH, high temperature, low and high osmolality, and repeated freeze/thaw cycles while at a concentration of 50 μg/ml. The physical stability of treated RSV F variants was evaluated by the preservation of antigenic site Ø after treatment as assessed by the site Ø-specific antibody D25.
In pH treatments, the RSV F glycoprotein solution was adjusted to pH 3.5 and pH 10 with appropriate buffers exchange and incubated at room temperature for 60 minutes and subsequently neutralized to pH 7.4. Temperature treatments were carried out by incubating the RSV F glycoprotein solutions at 50°C and 70°C for 60 minutes in a PCR cycler with heated lid. In osmolality treatments, RSV F glycoprotein solutions originally containing 137 mM NaCl in PBS buffer were either diluted with 2.5 mM Tris buffer (pH 7.5) to an osmolality of 10 mM NaCl or adjusted with 4.5 M MgCl2 to a final concentration of 3.0 M MgCl2. Protein solutions were incubated for 60 minutes at room temperature and then returned to PBS buffer and concentrated to 50 μg/ml. The freeze/thaw treatment was carried out by repeatedly freezing RSV F glycoprotein solutions in liquid nitrogen and thawing at 37°C ten times. The 10 cycles of freeze/thaw treatment were also used to assess some RSV F variants which were supplemented with 5 or 10% glycerol. All RSV F glycoproteins were diluted to 50 μg/ml with PBS buffer, and their ability to bind D25 Fab was measured with an Octet instrument using the protocol described above. Each measurement was carried out at least twice. The degree of physical stability is reported as the ratio of steady state D25-binding level before and after stress treatment.
Negative stain electron microscopy
Samples were adsorbed to freshly glow-discharged carbon-film grids, rinsed twice with buffer and stained with freshly made 0.75% uranyl formate. Images were recorded on an FEI T20 microscope with a 2k × 2k Eagle CCD camera at a pixel size of 1.5 Å. Image analysis and 2D averaging was performed with Bsoft54 and EMAN55.
Dynamic light scattering
A DynaPro Plate Reader II (Wyatt Technology Corp., Santa Barbara, CA) was used to perform dynamic light scattering analysis at 830 nm to measure the approximate size and size distributions of RSV F immunogens. Protein samples were concentrated to 1 mg/ml in PBS and were centrifuged at 13,000 rpm for 40 minutes at 4°C immediately prior to analysis. Samples of 30 μl each were read in black clear bottom 384 well plates (Corning 3540, NY) at 25°C, with 10 data acquisitions per sample. Data was analyzed using Dynamics software v7.1.7 (Wyatt Technology Corp., Santa Barbara, CA).
Mouse immunizations
All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH, under animal protocol 13–454, and all animals were housed and cared for in accordance with local, state, federal, and institute policies in an American Association for Accreditation of Laboratory Animal Care (AAALAC)-accredited facility at the NIH. Hybrid mice that were the first filial offspring of a cross between BALB/cJ females (C) and C57BL/6J males (B6) (The Jackson Laboratory) known as CB6F1/J at ages 6 weeks to 12 weeks were intramuscularly injected with RSV F immunogens at week 0 and week 3. At least ten mice were used per immunization group. Researchers carrying out the immunizations were blinded to the immunogens. All mice used in these studies were female, there was no randomization of animals. Using our previous immunization results with DS-Cav1 and Poly I:C56 which gave a neutralization geometric mean titer of 1765 with a standard deviation of 1589, if the sample size was 10 and the neutralization titer was ≥4000 the statistical power was 99.4%. The frozen RSV F variant immunogen proteins were thawed on ice and mixed with 5-fold w/w poly I:C (Invivogen) adjuvant (i.e. 10 μg RSV F, 50 μg Poly I:C per animal per immunization), with injections taking place within 1 h of immunogen:adjuvant preparation. No adverse effect from immunization was observed. Blood was collected at least three days before immunization, and at week two, week five and week seven post initial immunization.
Viruses and cells
Viral stocks were prepared and maintained as previously described57. Recombinant mKate-RSV expressing prototypic subtype A (strain A2) F genes and the Katushka fluorescent protein were constructed as reported by Hotard et al., 201258. HEp-2 cells were maintained in Eagle's minimal essential medium containing 10% fetal bovine serum (10% EMEM), supplemented with glutamine, penicillin and streptomycin.
RSV neutralization assays
Sera were distributed as four-fold dilutions from 1:10 to 1:163,840, mixed with an equal volume of recombinant mKate-Respiratory syncytial virus expressing prototypic F genes from subtype A (strain A2) and the Katushka fluorescent protein, and incubated at 37°C for 1 h. Next, 50 μl of each serum dilution/virus mixture was added to HEp-2 cells, which had been seeded at a density of 2.4×104 in 30 μl MEM (minimal essential medium) in each well of 384-well black optical bottom plates, and incubated for 20–22 h before spectrophotometric analysis at 588 nm excitation and 635 nm emission (SpectraMax Paradigm, Molecular Devices, CA). The IC50 for each sample was calculated by curve fitting and non-linear regression using GraphPad Prism (GraphPad Software Inc., CA).
Protective threshold was calculated as follows: clinical administration of palivizumab (Synagis) at 15 mg/kg, leads to patient sera levels at trough of ~40 μg/ml. This serum concentration provides protection in infants from severe disease and protection in cotton rats from RSV infection. In the neutralization assay described above, 40 μg/ml of palivizumab yields an EC50 of 100.
Crystallization and X-ray data collection of pre-fusion-stabilized RSV F proteins
Crystals of RSV F single-chain RSV F pre-fusion variants were grown by the vapor diffusion method in hanging drops at 20°C by mixing 1 μl of RSV F with 1 μl of reservoir solution. Crystals of RSV F sc9 DS-Cav1 and sc9-24 DS-Cav1 grew in sodium acetate, pH 5.5, lithium sulfate, magnesium sulfate, Peg 400 conditions. Specifically, sc9 DS-Cav1 crystals grew in 0.1 M sodium acetate pH 5.5, 1.09 M Li2SO4, 0.12 M MgSO4, 3.33% (w/v) PEG 400 and sc9-24 DS-Cav1 crystals grew in 0.1 M sodium acetate pH 5.5, 1.82 M Li2SO4, 0.1 M MgSO4, 5% (w/v) PEG 400. sc9 DS-Cav1 crystals were crystals were transferred to a solution of mother liquor supplemented with 1.0 M Li2SO4, and flash frozen in liquid nitrogen while sc9-24 DS-Cav1 crystals could be flash frozen in mother liquor without additional Li2SO4 addition. sc9-10 DS-Cav1 crystals grew in 0.1 M HEPES pH 7.5, 0.19 M (NH4)2SO4, 11% iso-propanol, 17% (w/v) PEG 8,000 and were transferred to a solution containing mother liquor supplemented with 30% glycerol prior to cryocooling in liquid nitrogen. sc9-19 DS-Cav1 crystals grew in 0.1 M Na citrate pH 5.6, 15% iso-propanol, 17% (w/v) PEG 4,000, and were transferred to mother liquor supplemented with 15% 2R-3R-butanediol prior to storage in liquid nitrogen. sc9-10 DS-Cav1 A149C Y458C crystals grew in 0.1 M phosphate-citrate, pH4.2, 0.12 M NaCl, 9.5% (w/v) PEG 8,000 and were also transferred to mother liquor supplemented with 15% 2R-3R-butanediol prior to storage in liquid nitrogen. The sc9-10 DS-Cav1 A149C Y458C S46G N67I E92D S215P K465Q crystals grew in 0.1M Na cacodylate, pH 6.5, 0.2M ZnAc, 17% (w/v) PEG 8,000 and were frozen in mother liquor supplemented with 22% ethylene glycol. All X-ray diffraction data were collected at a wavelength of 1.00 Å at the SER-CAT beamline ID-22.
Structure determination, refinement and analysis of pre-fusion-stabilized RSV F
X-ray diffraction data were integrated and scaled with the HKL2000 suite59, and molecular replacement solutions were obtained by PHASER60 using the RSV F DS-Cav1 structure (PDB ID: 4MMU31) as a search model. Manual model building was carried out using COOT61, and refinement was performed in PHENIX62. Final data collection and refinement statistics are presented in Table 3. Ramachandran plots indicated that for all structures >93% of residues were in the most favored regions, <2% of residues in disallowed regions. Antigenic site Ø rmsd calculations were based on residues 61–71 and 194–219 which were within 10 Å of the D25 antibody in the RSV F-D25 complex structure.
Correlation of Immunogenicity with different properties
The correlation between immunogenicity and the following properties were measured using Spearman correlation: elution volume, expression yield, antigenicity toward site-Ø antibodies (geometric mean of D25, AM22, and 5C4 KDs), Motavizumab (KD), quaternary antibodies (geometric mean of MPE8 and AM14 KDs), and physical stability (average D25 reactivity retained after exposure to seven different physical extremes). The correlation is considered significant if the adjusted P-values based on Bonferroni correction is less than 0.05 (equivalent to P<0.05/6=0.00833).
Sera antigenic analysis
Mouse sera from a subset of immunization groups from the iterative optimization cycles were assessed for binding to DS-Cav1 pre-F. Antigenic epitope targeting was assessed using the fortéBio Octet HTX instrument by competition using antigenic site Ø antibody D25, quaternary targeting antibodies MPE8 and AM14, as well as motavizumab. Week 5 sera were diluted 1:300 in 1% BSA/PBS. Anti-penta His (HIS1K) sensor tips obtained from fortéBio were equilibrated in PBS prior to assay. RSV F DS-Cav1 trimeric protein at 20 μg/ml in 1% BSA/PBS was loaded onto HIS1K biosensors for 300s. The sensor tips were subsequently equilibrated in 1% BSA/PBS for 60s followed by capture of antibodies D25, AM14, MPE8 or Mota at 50 μg/ml for 300s. HIS1K tips loaded with DS-Cav1/D25, /AM14, /MPE8 or /Mota were equilibrated for 180s in 1% BSA/PBS followed by a serum association step for 300s and a subsequent dissociation step for an additional 60s. A parallel assay was performed in the absence of antibodies D25, AM14, Mota or MPE8 to obtain serum response in the absence of competing antibodies. Naïve pre-bleed sera response for each group of immunogens was used as a reference. All experiments were performed in duplicate and the average of the two data sets was analyzed using Octet and GraphPad Prism 6 software.
SEC-SAXS
Data were collected on beamline 4-2 of the Stanford Synchrotron and Radiation Lightsource (SSRL) using a focused 11-keV X-ray beam to irradiate a thin-wall quartz capillary cell placed at 2.5 m upstream of a Rayonix MX 225HE detector as described previously 3. 50 μL of SC9 or SC9-10 (1 mg/mL) were injected onto a high resolution Sepharose 200 column (GE Healthcare) with a flow rate of 50 μL/min in 20 mM Na3PO4 (pH 7.4), 150 mM NaCl, 0.02% NaN3, 1 mM EDTA. Guinier fits, real-space transformation, Porod-Debye, and volume-of-correlation analyses were performed in ScÅtter (www.bioisis.net/tutorial). Measurements were repeated at least twice.
Supplementary Material
Acknowledgments
We thank M. Moore and A. Hotard, Department of Pediatrics, Emory University, and Children's Healthcare of Atlanta, Atlanta, Georgia, USA, for engineered RSVs used in neutralization assays and protocols. We thank J. Stuckey for assistance with figures, and L. Shapiro and members of the Structural Biology Section, the Structural Bioinformatics Core Section, the Virology Core Section of the Virology Laboratory and the Viral Pathogenesis Laboratory, Vaccine Research Center, NIAID, NIH for discussions and comments on the manuscript. Funding was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health for MGJ, BZ, LO, MC, G-YC, AD, W-PK, Y-TL, EJR, YT, YY, ISG, CRL, MP, MS, CS, GBES-J, PVT, JGVG, JRM, BSG, and PDK, and by the Gates Foundation Global Health Vaccine Accelerator Platform (GH-VAP) # OPP1126258 for MG and KKL. This project has been funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. Leidos Biomedical Research, Inc., as support in the form of salaries for authors YT and UB. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory by MG and KKL, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source by MGJ and PDK, was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
Footnotes
Author contributions M.G.J., B.Z., B.S.G., and P.D.K conceived, designed, and coordinated the study; M.G.J., B.Z., L.O., G.-Y.C., and P.D.K wrote and revised the manuscript and figures; M.G.J., B.Z., L.O., M.C., A.D., W.-P.K., Y.-T.L., E.J.R., Y.T., Y.Y., M.G., C.R.L., M.S., G.B.E.S.-J., P.V.T., J.G.V.G., and U.B., performed experiments; M.G.J., B.Z., I.S.G., M.P., C.S., B.S.G., and P.D.K. designed stabilized pre-fusion F immunogens. G.-Y.C., and P.D.K. carried out bioinformatics analysis. M.G.J., B.Z., L.O., M.C., G.-Y.C., M.G., K.K.L., J.R.M., B.S.G., and P.D.K analyzed data. L.O., M.C., G.-Y.C., A.D., W.-P.K., Y.-T.L., E.J.R., Y.T., and Y.Y., contributed equally to this study. All authors read and approved the manuscript.
Figures
Structure figures were prepared using PyMOL ((http://www.pymol.org/). Neutralization data and correlation figures were generated using GraphPad Prism (http://www.graphpad.com/scientific-software/prism/). Phylogenetic trees were generated using Mega6.
Accession codes Coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 5K6B, 5K6C, 5K6F, 5K6G, 5K6H, and 5K6I.
Competing financial interests The NIH has filed patents on the use of pre-fusion-stabilized RSV F glycoproteins.
References
- 1.Hall CB, et al. The Burden of Respiratory Syncytial Virus Infection in Young Children. N. Engl. J. Med. 2009;360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shay DK, et al. Bronchiolitis-Associated Hospitalizations Among US Children, 1980-1996. JAMA. 1999;282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 3.Thompson WW, et al. Mortality Associated With Influenza and Respiratory Syncytial Virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 4.Nair H, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. The Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Berg JP, Westerbeek EA, van der Klis FR, Berbers GA, van Elburg RM. Transplacental transport of IgG antibodies to preterm infants: a review of the literature. Early Hum Dev. 2011;87:67–72. doi: 10.1016/j.earlhumdev.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Hobbs JR, Davis JA. Serum gamma-G-globulin levels and gestational age in premature babies. Lancet. 1967;1:757–9. doi: 10.1016/s0140-6736(67)91369-4. [DOI] [PubMed] [Google Scholar]
- 7.Leuridan E, et al. Early waning of maternal measles antibodies in era of measles elimination: longitudinal study. BMJ. 2010;340:c1626. doi: 10.1136/bmj.c1626. [DOI] [PubMed] [Google Scholar]
- 8.Sarvas H, Seppala I, Kurikka S, Siegberg R, Makela O. Half-life of the maternal IgG1 allotype in infants. J Clin Immunol. 1993;13:145–51. doi: 10.1007/BF00919271. [DOI] [PubMed] [Google Scholar]
- 9.Lee MS, et al. Half-life of human parainfluenza virus type 3 (hPIV3) maternal antibody and cumulative proportion of hPIV3 infection in young infants. J Infect Dis. 2001;183:1281–4. doi: 10.1086/319690. [DOI] [PubMed] [Google Scholar]
- 10.Hall CB, et al. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics. 2013;132:e341–8. doi: 10.1542/peds.2013-0303. [DOI] [PubMed] [Google Scholar]
- 11.Palivizumab: new indication Moderate reduction in hospitalisation rate. Prescrire Int. 2004;13:213–6. [PubMed] [Google Scholar]
- 12.The IMpact-RSV Study Group Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 13.Glenn GM, et al. A Randomized, Blinded, Controlled, Dose-Ranging Study of a Respiratory Syncytial Virus Recombinant Fusion (F) Nanoparticle Vaccine in Healthy Women of Childbearing Age. J Infect Dis. 2016;213:411–22. doi: 10.1093/infdis/jiv406. [DOI] [PubMed] [Google Scholar]
- 14.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proceedings of the National Academy of Sciences. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modjarrad K, Giersing B, Kaslow DC, Smith PG, Moorthy VS. WHO consultation on Respiratory Syncytial Virus Vaccine Development Report from a World Health Organization Meeting held on 23-24 March 2015. Vaccine. 2016;34:190–7. doi: 10.1016/j.vaccine.2015.05.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karron RA, Buchholz UJ, Collins PL. Live-attenuated respiratory syncytial virus vaccines. Curr Top Microbiol Immunol. 2013;372:259–84. doi: 10.1007/978-3-642-38919-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham BS, Modjarrad K, McLellan JS. Novel antigens for RSV vaccines. Curr Opin Immunol. 2015;35:30–8. doi: 10.1016/j.coi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollack P, Groothuis JR. Development and use of palivizumab (Synagis): a passive immunoprophylactic agent for RSV. J Infect Chemother. 2002;8:201–6. doi: 10.1007/s10156-002-0178-6. [DOI] [PubMed] [Google Scholar]
- 19.Young J. Development of a potent respiratory syncytial virus-specific monoclonal antibody for the prevention of serious lower respiratory tract disease in infants. Respir Med. 2002;96(Suppl B):S31–5. doi: 10.1053/rmed.2002.1298. [DOI] [PubMed] [Google Scholar]
- 20.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176:1215–24. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 21.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–19. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 22.Bolt G, Pedersen LØ, Birkeslund HH. Cleavage of the respiratory syncytial virus fusion protein is required for its surface expression: role of furin. Virus Research. 2000;68:25–33. doi: 10.1016/s0168-1702(00)00149-0. [DOI] [PubMed] [Google Scholar]
- 23.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110:11133–8. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magro M, et al. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012;109:3089–94. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corti D, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–43. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 27.Gilman MS, et al. Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein. PLoS Pathog. 2015;11:e1005035. doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLellan JS, et al. Structure of RSV Fusion Glycoprotein Trimer Bound to a Prefusion-Specific Neutralizing Antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngwuta JO, et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med. 2015;7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–8. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krarup A, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod Pathol. 2007;20:108–19. doi: 10.1038/modpathol.3800725. [DOI] [PubMed] [Google Scholar]
- 34.Langley JM, et al. A dose-ranging study of a subunit Respiratory Syncytial Virus subtype A vaccine with and without aluminum phosphate adjuvantation in adults > or =65 years of age. Vaccine. 2009;27:5913–9. doi: 10.1016/j.vaccine.2009.07.038. [DOI] [PubMed] [Google Scholar]
- 35.Paradiso PR, et al. Safety and immunogenicity of a subunit respiratory syncytial virus vaccine in children 24 to 48 months old. Pediatr Infect Dis J. 1994;13:792–8. doi: 10.1097/00006454-199409000-00008. [DOI] [PubMed] [Google Scholar]
- 36.Tristram DA, Welliver RC, Hogerman DA, Hildreth SW, Paradiso P. Second-year surveillance of recipients of a respiratory syncytial virus (RSV) F protein subunit vaccine, PFP-1: evaluation of antibody persistence and possible disease enhancement. Vaccine. 1994;12:551–6. doi: 10.1016/0264-410x(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 37.Cohen J. 2013 Runners-Up. In vaccine design, looks do matter. Science. 2013;342:1442–3. doi: 10.1126/science.342.6165.1442-a. [DOI] [PubMed] [Google Scholar]
- 38.Georgiev IS, et al. Single-Chain Soluble BG505.SOSIP gp140 Trimers as Structural and Antigenic Mimics of Mature Closed HIV-1 Env. J Virol. 2015;89:5318–29. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma SK, et al. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–50. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Begona Ruiz-Arguello M, et al. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298:317–26. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- 41.Coates HV, Kendrick L, Chanock RM. Antigenic differences between two strains of respiratory syncytial virus. Proc Soc Exp Biol Med. 1963;112:958–64. doi: 10.3181/00379727-112-28221. [DOI] [PubMed] [Google Scholar]
- 42.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 43.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–71. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang B, et al. Enhanced Neutralizing Antibody Response Induced by Respiratory Syncytial Virus Prefusion F Protein Expressed by a Vaccine Candidate. J Virol. 2015;89:9499–510. doi: 10.1128/JVI.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godley L, et al. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–45. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 46.Kwon YD, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–31. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong JJ, Paterson RG, Lamb RA, Jardetzky TS. Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form. Proc Natl Acad Sci U S A. 2016;113:1056–61. doi: 10.1073/pnas.1523303113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen X, et al. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat Struct Mol Biol. 2012;19:461–3. doi: 10.1038/nsmb.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012;109:16672–7. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nabel GJ. Philosophy of science. The coordinates of truth. Science. 2009;326:53–4. doi: 10.1126/science.1177637. [DOI] [PubMed] [Google Scholar]
- 51.Klebe G. Recent developments in structure-based drug design. J Mol Med (Berl) 2000;78:269–81. doi: 10.1007/s001090000084. [DOI] [PubMed] [Google Scholar]
- 52.Pancera M, et al. N332-Directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS One. 2013;8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLellan JS, et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat. Struct. Mol. Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 56.Stewart-Jones GB, et al. A Cysteine Zipper Stabilizes a Pre-Fusion F Glycoprotein Vaccine for Respiratory Syncytial Virus. PLoS One. 2015;10:e0128779. doi: 10.1371/journal.pone.0128779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graham BS, Perkins MD, Wright PF, Karzon DT. Primary respiratory syncytial virus infection in mice. J Med Virol. 1988;26:153–62. doi: 10.1002/jmv.1890260207. [DOI] [PubMed] [Google Scholar]
- 58.Hotard AL, et al. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology. 2012;434:129–36. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W. Methods Enzymol. Vol. 276. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 60.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.