Abstract
Alternative pre-mRNA splicing is a fundamental regulatory process for most mammalian multi-exon genes to increase proteome diversity. Nonsense-mediated mRNA decay (NMD) is a conserved mRNA surveillance mechanism to mitigate deleterious effects caused by gene mutations or transcriptional errors. Coupling alternative splicing and NMD (AS-NMD), in which alternative splicing switches between translational and NMD isoforms, results in fine-tuning overall gene expression to, in turn, expand the functional activities of these two post-transcriptional regulatory processes. AS-NMD is known for maintaining homeostatic expression of many RNA-binding proteins. We further show that AS-NMD is a conserved mechanism among mammals to induce developmental expression of the synaptic scaffold protein PSD-95. Comparing gene sequences between Psd-95 and its ancestral orthologues indicates that AS-NMD regulation of mammalian Psd-95 is a product of selective pressure and that it enforces neural-specific expression of PSD-95 proteins in mammals. Invertebrate homolog of Psd-95 is not subjected to AS-NMD regulation and its protein product does not exhibit neural-specific expression. Given the prevalence of alternative splicing regulation in the mammalian nervous system, neural-specific expression of many other genes could be controlled by AS-NMD in a similar manner. We discuss the implication of these discoveries, as well as the challenges in generalizing the regulation and functional activity of AS-NMD.
1. Introduction
Tissue-specific gene expression is best known to be dictated by transcriptional regulation whereby the targeted tissue is selectively programmed to enhance the promoter activity of the tissue-specific genes. In the nervous system, serving in these roles are many neurogenic transcription factors that induce neural-specific gene expression. The roles of other genetic regulation processes are not as well defined. Recent studies have found that alternative splicing coupled to nonsense-mediated mRNA decay (AS-NMD) is a post-transcriptional regulatory mechanism to fine-tune homeostatic expression of many RNA-binding proteins. We have further shown that AS-NMD mediates developmental induction of mouse synaptic protein PSD-95, a widely used postsynaptic marker for glutamatergic synapses in rodent and human. Although the molecular mechanisms of alternative splicing or NMD have been extensively studied and reviewed, their functional aspects are less clear. Here, we review and discuss the functional roles of AS-NMD regulation in the brain. In light of neural specific expression of PSD-95 protein in human and mouse, we provide further evidence and suggest AS-NMD be a novel posttranscriptional regulatory mechanism to enforce neural specific gene expression as seen for PSD-95.
2. Alternative pre-mRNA splicing
Alternative pre-mRNA splicing is a key regulatory step of mRNA processing that creates different combinations of exons, resulting in multiple transcript variants produced from a single gene locus. The choice of final gene product to be synthesized is primarily governed by special pre-mRNA binding proteins that alter spliceosome assembly at nearby splice sites, leading to changes in the inclusion of exons or exon segments in the final mRNA (Black, 2003; Chen and Manley, 2009). The expression, localization and modification of these splicing regulators are highly regulated to determine splicing outcome during development and in response to environmental cues.
Alternative splicing has two major impacts on gene expression: multiplying the functional outputs of a single pre-mRNA transcript and influencing the use of spliced isoforms. Including or skipping a coding exon often affects protein structures and functions contributing to increased proteome diversity. In contrast, an alternative non-coding or “partial-coding” exon (e.g., exons of 5′ UTR or 3′ UTR) typically harbors cis-regulatory elements that control stability, subcellular localization and translation efficiency of the final spliced isoforms. Thus, with the ability to affect both the meaning and destiny of gene transcripts, alternative splicing is a powerful regulatory invention that enables cells having the same genome to diverge at the molecular level. Alternative splicing regulation or switches of isoform expression are particularly common during development and in cell lineage differentiation. Transcriptomic profiling has identified many tissue-specific splicing events, the largest proportion of which originates from neural tissues (Castle et al., 2008; Pan et al., 2008; Wang et al., 2008). A dozen splicing regulators and their families, including PTBP, RBFOX, NOVA, MBNL, SRRM4, KHDRBS and TDP43, have been implicated to mediate neural-specific splicing events (Darnell, 2013; Raj and Blencowe, 2015; Vuong et al., in press).
Despite the prevalence of alternative splicing in the nervous system, the physiological activity of most alternative splicing events remains poorly understood and presents one of the greatest challenges to the field (Raj and Blencowe, 2015; Zheng and Black, 2013). The first indicator of functional significance for any alternative exon would be the high conservation in sequence and expression. A majority of alternative exons exhibit a bimodal distribution of their sequence conservation (Chen and Zheng, 2008). Those that are not conserved could be newly evolved exons, whereas the existence of conserved exons and flanking introns implicates selective pressure. Indeed, some splicing changes have been characterized in detail and proven essential for neuronal cell biology (Chen et al., 2011; Kim et al., 2012; Krüger et al., 2013; Laurent et al., 2015; Macabuag and Dolphin, 2015; Penn et al., 2012; Sommer et al., 1990; Tan et al., 2012; Wang et al., 2015; Zheng et al., 2012; Zibetti et al., 2010). Nevertheless, a fraction of neural-specific splicing events could be the byproducts of transcriptomic modification toward neuronal lineages and, thus, functionally trivial.
3. Case in Point: AS-NMD regulation of Psd-95
We have recently found that alternative splicing regulation of Psd-95 transcripts by polypyrimidine tract binding proteins PTBP1 and PTBP2 regulate synaptogenesis via controlling expression of PSD-95 protein, a key abundant component of postsynaptic density for glutamatergic synapses (Zheng et al., 2012). PTBP1 and PTBP2 inhibit inclusion of Psd-95 exon 18. Exon 18 is a coding exon; however, instead of modifying protein structure and function, alternative splicing of exon 18 controls overall abundance of PSD-95. Functional PSD-95 proteins are translated from exon 18-plus transcripts. Skipping exon 18 shifts the reading frame, causing earlier translation termination of Psd-95 transcripts at a conserved stop codon in exon 19. Consequently, exon 18-minus transcripts are subject to nonsense-mediated mRNA decay (NMD) without productive translation. Therefore, AS-NMD can serve as an “on-and-off switch” for gene expression independent of transcriptional and miRNA regulation.
AS-NMD is better known as a negative feedback control mechanism to maintain homeostatic expression of RNA-binding proteins (RBP) and splicing regulators (Boutz et al., 2007; Lareau et al., 2007; Ni et al., 2007; Rossbach et al., 2009; Spellman et al., 2007; Valacca et al., 2010). Increase in expression of these factors shifts their own splicing from isoforms that translate normal proteins to isoforms targeted by NMD. Theoretically, non-RBP genes with no ability to control their own splicing can adopt AS-NMD to modulate expression levels as that seen for mouse Psd-95. However, in spite of the widely assumed pervasiveness of AS-NMD regulation in affecting gene abundance, only a few non-RBP genes have been genetically proven to be controlled by AS-NMD in physiological contexts (Hyvönen et al., 2006; Lejeune and Maquat, 2005; McGlincy and Smith, 2008; Zheng et al., 2012). In this regard, AS-NMD regulation of Psd-95 remains a potentially isolated event uniquely associated with mouse neural development
4. Breadth of AS-NMD control over gene expression
Broader use of AS-NMD to control gene expression remains to be understood. Widely used as a bioinformatics rule, transcripts with exon-exon junction 50–55 nucleotides downstream of a premature stop codon are targeted by NMD. Based on this rule, 25–35% of annotated isoforms were predicted to be NMD targets (Lewis et al., 2003). However, some annotated isoforms could have resulted from errors in transcription or mRNA processing or DNA mutations that are not physiologically relevant but are expected to be neutralized by NMD. Indeed, some reports have suggested more limited NMD targeting and showed that most bioinformatics-predicted NMD isoforms were expressed at very low levels, even when NMD factors were depleted (Pan et al., 2006). However, genome-wide profiling of AS-NMD targets after NMD inhibition in this kind of studies may have underestimated the breadth of AS-NMD regulation because many NMD isoforms could be specific to cell types or extracellular cues and, hence, were not readily detectable in certain circumstances.
To investigate the prevalence of AS-NMD as a gene regulatory mechanism, we first need to distinguish genetically regulated splicing from noisy transcriptomic nonsense. Comparative genomics can help in this regard. Most alternative exons are either highly conserved or not conserved at all, exhibiting a bimodal-like distribution of conservation scores (Chen and Zheng, 2008). The conservation of an alternative exon and its flanking introns, as well as its splicing-induced premature stop codons, when taken together, would show the existence of selective pressure to maintain its AS-NMD regulation, which further implies functional relevance. Comparative genomics, on the other hand, has limitation to understanding the use of AS-NMD for speciation and/or individual variability.
Second, we need to better understand the targeting rule of NMD. Computational prediction has faced increasing limitation. NMD has been proposed to be triggered or enhanced by different mechanisms, including downstream exon junction complex (EJC) and long 3′ UTR (Chang et al., 2007; Popp and Maquat, 2013; Rebbapragada and Lykke-Andersen, 2009). The popular 55-nucleotide rule could introduce false NMD targets and miss some true NMD substrates. Further evidence against the generalization of the 55-nucleotide rule has surfaced in recent CLIP-Seq analysis revealing that about 20% of splicing events did not result in EJC deposition and about half of the EJCs bound to regions not associated with exon-exon junctions (Saulière et al., 2012; Singh et al., 2012). In summary, the generality of AS-NMD regulation as a genetic program to control gene expression remains to be scrutinized, even although the primary transcripts of most multi-exon genes undergo alternative splicing.
5. Could AS-NMD be a molecular mechanism to maintain modest protein expression of NMD isoforms
NMD surveillance depends on ribosome scanning or translational activity, which means that at least one protein copy is translated from each recognized NMD isoform transcript. While most studies have focused on mRNA decay of NMD isoforms, few have investigated the protein products of NMD isoforms. These C-terminal truncated proteins could be misfolded and quickly destructed by proteasome, but they could also be stable and functional. The most parsimonious speculation for the functional role of NMD isoforms would be their dominant negative activity, the same reason that explains the necessity of their low abundance. When a NMD isoform has no sign of disrupting the function of the major isoform, it might also have special functions.
Could this be the case for PSD-95? Initial synaptogenesis is prone to errors and intrinsically dynamic before subsequent activity-dependent synapse pruning and stabilization. The exon 18-minus protein, if expressed, would be a “less sticky” scaffolding protein suited to structurally dynamic young post-synaptic density (PSD) in developing dendrites (Marrs et al., 2001). From N- to C-terminus, normal non-NMD PSD-95 proteins have 3 PDZ domains, a SH3 domain and a GK domain that each serves as a scaffolding structure for protein-protein interactions with other post-synaptic proteins. PSD-95 is central to post-synaptic scaffolds because of its ability to multimerize. PSD-95 N-termini are self-linked by “head-to-head” interactions, while the C-termini are bridged by GKAP (DLGAP1) (Christopherson et al., 2003; Hsueh and Sheng, 1999; Kim et al., 1997). The exon 18-minus isoform, if translated, would lose ability to associate with and multimerize via GKAP, but is assumed to keep the N-terminus interactors in contact. Thus, the exon 18-minus isoform would be an ideal candidate to establish young dynamic PSDs in immature neurons.
Exon18-minus proteins could also have functions outside of synapses. Many synaptic proteins including glutamate receptors and PSD-95, are transported in packets along dendrites to synapses (Aoki et al., 2001; El-Husseini et al., 2000; Washbourne et al., 2002). Individual packet is probably too small to include and transport the whole postsynaptic density; however, it may include a fraction of PSD proteins constituting some pre-assembled mini-scaffolds. The exon 18-minus protein could play a role in assembling such mini-scaffolds and/or acting as a mild disruptor to prevent hyperaggregation of these pre-assembled scaffolds in the mobile packets. At the destination synaptic sites, exon 18-minus proteins may then be selectively degraded or exchanged in order for the mini-scaffolds to integrate into the stabilized PSD.
6. AS-NMD as a novel posttranscriptional mechanism to enforce tissue specific gene expression
Since NMD isoforms are not effectively translated into proteins, switching between the translational isoform and the NMD isoform allows alternative splicing to control overall abundance of the transcript and protein. Could AS-NMD thus be used to determine tissue-specific gene expression? This would expand physiological relevance of AS-NMD regulation, at least to PSD-95. We therefore conduct our investigation focusing on Psd-95.
6.1. Human neuronal cell lineage differentiation induces inclusion of PSD-95 exon 18
Neuronal enrichment of PSD-95 protein is conserved between human and mouse. If AS-NMD mediates neuronal specific gene expression of PSD-95, human PSD-95 exon 18 should be regulated in a manner similar to that of mouse orthologue. We monitored exon 18 splicing during the course of human embryonic stem cell (ESC) differentiation into neural progenitor cells (NPC) and neurons.
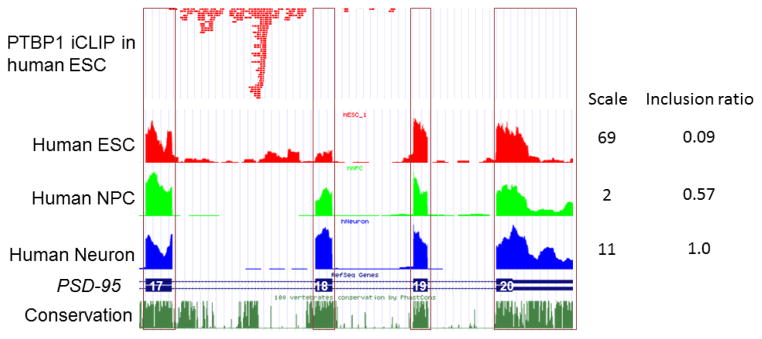
Although PSD-95 protein is one of the best known neural specific proteins, PSD-95 transcripts are readily detectable in human ESC (Figure 1). Exon 18 is, however, only minimally included. On the other hand, intronic reads upstream of exon 18 are relatively abundant in polyA+ RNA-Seq, suggesting inefficient splicing of intron 17 most likely resulting from PTBP1 inhibition in human ESC. Consistently, PTBP1 iCLIP-Seq in human ESC illustrates specific binding of PTBP1 to intron 17 (Figure 1) (Han et al., 2014). Strikingly, the 5′-end of most iCLIP-Seq tags (i.e., the nucleotides immediately succeeding the crosslinked nucleotides) overlay intronic positions conserved among mammals, suggesting that these intronic positions are under selection pressure to preserve PTBP1 binding.
Figure 1.
Psd-95 exon 18 is neural-specific in human cells. Exon 17 to Exon 20 of PSD-95 transcript is shown. PTBP1 binds to the Psd-95 pre-mRNA transcripts in human embryonic stem cells (ESC). PTBP1 iCLIP-Seq tags are enriched in conserved intronic sequences upstream of Psd-95 exon 18. As a result, exon 18 is mostly skipped in ESC. Exon 18 becomes more included as ESCs differentiate into neural progenitor cells (NPC) and eventually neurons.
When human ESCs differentiate to NPCs, exon 18 inclusion level is increased. Because the transcriptomic profiling was not performed at the single-cell level, the elevation of exon 18 read numbers could reflect synchronized induction of exon 18 splicing in all NPCs, or a mixed population of high exon 18 (differentiated) and low exon 18 (undifferentiated) cells. Nevertheless, exon 18 is further increased and fully included in differentiated neurons. These results show that human neuronal cell lineage differentiation induces PSD-95 exon 18 inclusion in a manner similar to mouse orthologue. Only in differentiated neurons is exon 18 completely included, thereby allowing expression of PSD-95 protein.
6.2. Alternative splicing of exon 18 is evolutionarily selected to confer neural specific expression of mammalian PSD-95 protein
Both human and rodent PSD-95 proteins were found to be restricted to neural tissues (Cho et al., 1992; Ohnuma et al., 2000), but their mRNA transcripts are apparently not. ESC expression of PSD-95 transcripts is also seen in mouse (data not shown), suggesting conserved discrepancy between RNA and protein expression of Psd-95. Therefore, neural-specific expression of mammalian PSD-95 protein is controlled at the post-transcriptional level.
The protein products of Dlg1, the “ancestral” orthologue of mammalian Psd-95 gene, are expressed in both neural and non-neural cells of D. melanogaster (Woods and Bryant, 1991). Dlg1, or discs large-1, was initially identified as a tumor suppressor gene whose mutations cause neoplastic overgrowth of the imaginal discs. Different from PSD-95, Dlg1 proteins have been detected in muscle cells and epithelial cells in addition to the nervous system. Dlg1 proteins are often found around the cell border and play a critical role in cellular adhesion and growth (Ogawa et al., 2009). Thus, neuronal specificity cannot be attributed to either Dlg1 RNA or Dlg1 protein.
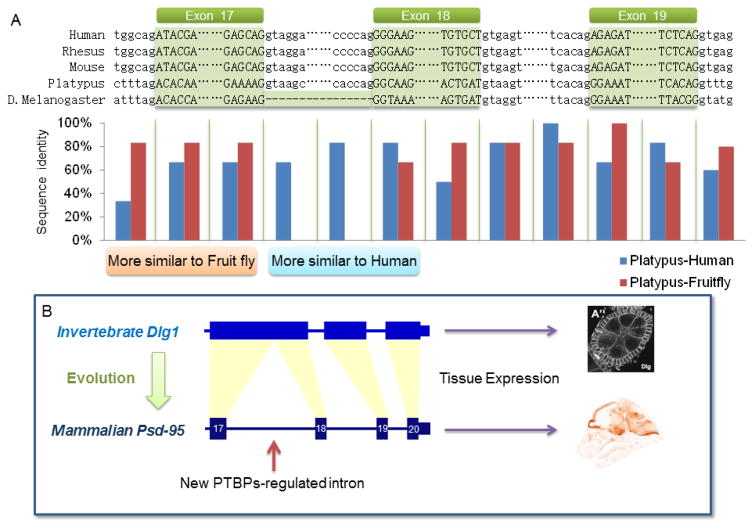
Since neither Dlg1 nor Psd-95 RNA is neural-specifically transcribed, the differential cell-type distribution between mammalian PSD-95 protein and fruit fly Dlg1 protein may be rooted in their distinct pre-RNA structures. To examine this, we compared the exon-intron structures, focusing on sequences from exon 17 to 19, of PSD-95 and its orthologues from various species including human, chimp, orangutan, rhesus, mouse, rat, guinea pig, rabbit, marmoset, dog, cat, horse, cow, elephant, armadillo, opossum, playtypus, frog, zebrafish, and D. melanogaster. Many other species were not included due to incomplete genome annotation or lack of sequence matched to the Psd-95 gene. The segregation of exon 17, 18 and 19 by introns is well conserved with identical or highly similar exon-intron and intron-exon junction sequences among mammals including platypus, one of the extant mammal species most distantly related to humans. The degree of sequence similarity between mammals is consistent with their phylogenetic relationship. We aligned the exon-intron junction sequences, the most important cis-elements in determining splicing functions. Due to space limit, only the representative species are shown (Figure 2A). Platypus sequences appear to be hybrids of Euarchontoglires mammals and fruit fly sequences, in agreement with the phylogenetic distances of these orthologues. In particular, the junction sequence of platypus intron 16-exon 17 and the 3′-end of exon 17 are more similar to the corresponding fruit fly sequences, whereas the 5′-end of exon 18 and intron 17 are more similar to those of human.
Figure 2.
Intron 17 contributes to neuron-specific expression of PSD-95 protein in mammals. (A) Sequence alignment of human, rhesus, mouse and playtypus Psd-95 and its ancestral orthologue D. melanogaster Dlg1. Introns are lowercase. Exons are uppercase and highlighted by the green blocks corresponding to human exon 17, 18 and 19 sequentially. Only the intron-exon and exon-intron junction sequences are shown. Nucleotides between the shown junctions are represented by dots. A gap in D. melanogaster Dlg1 is represented by dashes. Sequences are segmented by the vertical lines in the lower panel to calculate sequence identity between platypus and human or fruit fly for each segment. (B) Therefore, Dlg1, the “ancestral” orthologue of Psd-95, does not have an intron homologous to mammalian intron 17. Dlg1 is expressed in many cells, including epithelial cells, as shown by. Mammalian Psd-95 has intron 17 whose splicing is regulated by PTBP1 and PTBP2. As a result, PSD-95 expression is restricted to the brain. The image of PSD-95 expression is from GENESAT.
Most interestingly, the orthologous exon 17 and 18 are not separated in the D. melanogaster Dlg1 gene (Figure 2). The 3′-end of orthologous exon 17 immediately precedes the 5′-end of orthologous exon 18 at the DNA/pre-mRNA level, indicating that D. melanogaster has no “intron 17”. As a result, fruit fly Dlg1 pre-mRNA does not undergo alternative splicing regulation similar to that seen in mammalian PSD-95 transcripts. Also, the fruit fly Dlg1 spliced isoform is not subject to NMD regulation like that seen for PSD-95 exon 18-minus isoform.
The insertion of intron 17 may thus be an evolutionary event resulting in the neural-specific expression of PSD-95 (Figure 2B). PTBP1 iCLIP-Seq clearly shows specific binding to intron 17 (Figure 1) at the conserved positions. This new intron, when bound to PTBP1, has introduced a new layer of regulation, i.e., AS-NMD regulation, to the PSD-95 gene that does not exist in its fruit fly orthologue. Mammalian PSD-95 genes and fruit fly Dlg1 both inherit non-neuronal transcriptional activity, but their post-transcriptional regulation differs significantly. D. melanogaster Dlg1 has no intron 17 and, hence, no regulation by PTB proteins. In mammals, however, exon 18 is widely repressed because PTBP1 is ubiquitously expressed outside the nervous system (Keppetipola et al., 2012). As a result, PSD-95 protein is not expressed in non-neuronal tissues as seen in fruit fly Dlg1. In terminally differentiated rodent neurons that have significantly downregulated PTBP1 and PTBP2, PSD-95 protein is then allowed to express. Therefore, mouse PSD-95 expression is restricted to neural tissues in part as a result of new intron 17 regulated by PTB proteins. Do human PTBP1 and PTBP2 also exhibit temporal expression similar to mouse during brain development to regulate PSD-95 splicing and expression?
6.3. Conserved temporal expression of PTBP1 and PTBP2 in developing human brains
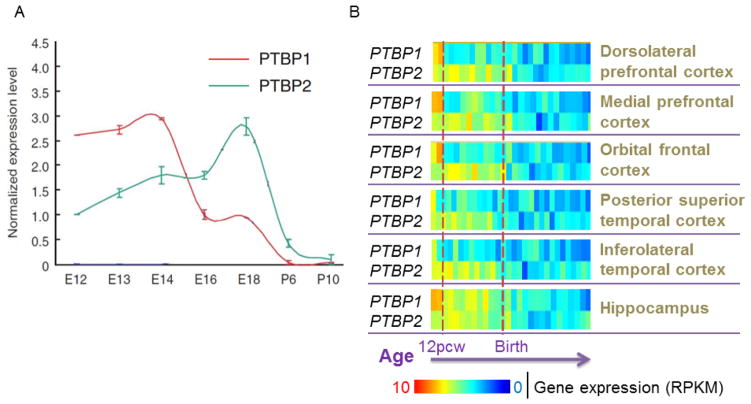
During mouse brain development PTBP1 and PTBP2 are sequentially expressed and sequentially downregulated to control Psd-95 splicing. PTBP1 is abundantly expressed in neural stem cells (NSCs) and progenitor cells (NPCs) (Boutz et al., 2007). When NPCs start to differentiate into neurons, PTBP1 is suppressed. Reduction in PTBP1 induces the expression of PTBP2 protein (Boutz et al., 2007; Spellman et al., 2007). PTBP2 expression peaks in immature neurons and subsequently wanes right before synaptogenesis (Zheng et al., 2012). In developing mouse neocortex, PTBP1 starts to decrease as early as E14, while PTBP2 continues to increase until birth. Both proteins are significantly reduced postnatally (Figure 3A).
Figure 3.
Sequential expression of PTBP1 and PTBP2 during development is conserved between mouse and human in various brain regions. (A) Temporal expression patterns of PTBP1 and PTBP2 in developing mouse neocortices (Zheng et al., 2012). PTBP1/2 protein expression is first normalized to internal control GAPDH. The normalized time-serial expression is then normalized to the expression level at E16 for PTBP1 and E12 for PTBP2. (B) Temporal expression patterns of PTBP1 and PTBP2 in various human brain regions at 8, 9, 12, 13, 16, 17, 19,21, 24, 25,26, 35, 37 pcw (post- conception week), 4 months, 10 months, as well as 1, 2, 3, 4, 8, 11, 13, 15, 18, 19, 21, 23, 30, 36, and 37 years of age. Each color block represents one specific age.
To probe PTBP1 and PTBP2 gene expression in developing human brains, we turned to the BrainSpan project that has profiled the transcriptome of multiple postmortem cortical and subcortical structures spanning pre- and postnatal development. The rudimentary structures and major compartments of the human brain are established around eight post-conception weeks (pcw) (Stiles and Jernigan, 2010). From then on throughout the prenatal period, both cortical and subcortical structures undergo rapid growth and expansion, while neurons are born, migrate, and shape their elaborative morphology. Brain development continues after birth during childhood and adolescence until adult ages. During this extended postnatal period, neurons make extensive connections that simultaneously undergo activity-dependent pruning, and various types of glia cells populate throughout the brain. We analyzed PTBP1 and PTBP2 expression from 8 pcw to 37 years of age. Brain areas were segregated to test the possibility of area-specific expression patterns.
Various human brain regions are almost indistinguishable with respect to the developmental expression patterns of PTBP1 or PTBP2, which also closely mirror those of mouse orthologues, respectively (Figure 3B). PTBP1 is generally expressed at higher levels between 8 and 12 pcw and consistently lower after 12 pcw, except in hippocampus. PTBP2 expression increases when PTBP1 is initially downregulated. Later, both human PTBP1 and PTBP2 are expressed at a much lower level postnatally than prenatally. The conserved expression patterns of PTBP1 and PTBP2 between human and mouse suggest that these two proteins play fundamental roles in neural development. Since PTBP1 and PTBP2 are the major alternative splicing regulators of Psd-95 exon 18 during brain development, their developmental expression levels determine the dynamics of exon 18 splicing, as seen in the ESC differentiation model (Figure 1), leading to developmental induction and neural-specific expression of PSD-95 protein.
Conclusions and future perspectives
By directly influencing the overall abundance of translational isoforms, AS-NMD is a regulatory mechanism to control gene expression independent of transcriptional and miRNA-mediated posttranscriptional controls. Many RNA-binding proteins harness AS-NMD mechanism to maintain homeostatic expression. Here, we have further shown that AS-NMD regulation of Psd-95 RNA is evolutionarily selected to enforce neural-specific expression of PSD-95 protein and that AS-NMD regulation of Psd-95 is conserved between human and mouse and probably among all mammals. PSD-95 thereby serves as an example of AS-NMD regulation being a molecular mechanism used to convert tissue-specific splicing to tissue-specific protein expression.
Tissue-specific transcriptional regulators selectively enhance expression of target genes in the host tissues and cell types. AS-NMD regulation, on the other hand, diminishes transcript abundance in the surrounding tissues; as a result, the spared tissue stands out exhibiting higher abundance of the transcripts. AS-NMD thus provides a new layer of regulation complexity in addition to much more extensively studied transcriptional and miRNA-mediated posttranscriptional gene regulation. This kind of mechanism, that we term selective repression at the posttranscriptional level, may be particularly suited to control tissue-specific expression of genes with pervasive transcription activity, e.g., Psd-95. PSD-95 shares a promoter with VLCAD (very long chain acyl-CoA dehydrogenase) that encodes a protein catalyzing the first step of mitochondrial long-chain fatty acid β-oxidation (Zhang et al., 2003). Because VLCAD is a ubiquitously-expressed housekeeping gene, the shared promoter might have contributed to the non-neuronal transcriptional activity of PSD-95. Since the nervous system expresses many tissue-specific genes and displays proportionally most tissue-specific splicing events, it is conceivable that many brain-specific genes are controlled via AS-NMD as that seen for Psd-95.
To identify genes that display tissue-specific expression via AS-NMD regulation genome-wide, tissue-specific splicing need to be correlated with protein expression, followed by a search for possible NMD regulation. Recent technological improvement on global profiling of tissue-based proteome will expedite the discovery (Kim et al., 2014). However, proteome profiling techniques, e.g., mass spectrometry, have not reached the same level of throughput, resolution and sensitivity as that provided by next-generation sequencing. Until that happens, examples of tissue-specific protein expression via AS-NMD regulation will most likely need to be confirmed individually.
To understand the broader impact of AS-NMD regulation for neural development, it will be necessary to comprehensively catalogue genetically programmed AS-NMD targets and investigate possible physiological activities of those conserved NMD isoforms. Both tasks are challenging because understanding the regulation and functional consequences of each alternative splicing event presents a substantial question of its own. While changes in exon inclusion that happen within known functional protein sites can be predicted to cause drastic changes, the underlying biology of most alterations in spliced isoform expression is unknown.
Materials and methods
Next-generation sequencing data analysis
Gene expression data of developing human brains by RNA-Seq were from Brainspan (http://brainspan.org/). Transcriptomic RNA-Seq data of human ESC, NPC and Neurons were from ENCODE (ENCFF000FCX, ENCFF585VNQ, ENCFF509QEV). Gene expression was estimated by GPSeq (Srivastava and Chen, 2010).
Sequence comparison of PSD-95 and its orthologues
Sequences PSD-95 gene and its orthologs in human, chimp, orangutan, rhesus, mouse, rat, guinea pig, rabbit, marmoset, dog, cat, horse, cow, elephant, armadillo, opossum, playtypus, frog, zebrafish, and D. melanogaster were directly obtained from NCBI Gene portal if annotated or via BLAT search if unannotated. To obtain gene sequences of unannotated orthologues (e.g., Psd-95 gene in platypus), we used Mutliz alignment of UCSC Genome Browser to identify the orthologous exonic sequences which were then BLAT back to organism genomes to obtain neighboring intronic sequences. All sequences were then aligned by their exon-intron structures. Sequence identity of Psd-95 genes between any two species was calculated by taking the ratio between the exactly matched and total nucleotides within the compared sequence fragments.
Highlights.
Alternative splicing is most prevalent in the nervous system while the physiological activity of most alternative splicing events remains to be understood.
Coupling alternative splicing and NMD (AS-NMD), in which alternative splicing switches between translational and NMD isoforms, results in fine-tuning overall gene expression to, in turn, expand the functional activities of these two post-transcriptional regulatory processes.
Determining the breadth of AS-NMD control over gene expression needs separation of genetically regulated splicing events from noisy transcriptomic nonsense as well as a better defined NMD targeting rule.
AS-NMD might be a molecular mechanism to maintain modest protein expression of NMD isoforms.
AS-NMD regulation of mammalian Psd-95 is a product of selective pressure and enforces neural-specific expression of PSD-95 proteins in mammals. PSD-95 thereby serves as an example of AS-NMD regulation being a molecular mechanism used to convert tissue-specific splicing to tissue-specific protein expression.
Acknowledgments
This work was supported by grants from NIH (R00MH096807) and CUBRI to S.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoki C, Miko I, Oviedo H, Mikeladze-Dvali T, Alexandre L, Sweeney N, Bredt DS. Electron microscopic immunocytochemical detection of PSD-95, PSD-93, SAP-102, and SAP-97 at postsynaptic, presynaptic, and nonsynaptic sites of adult and neonatal rat visual cortex. Synapse. 2001;40:239–257. doi: 10.1002/syn.1047. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin C-H, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chen B-S, Thomas EV, Sanz-Clemente A, Roche KW. NMDA receptor-dependent regulation of dendritic spine morphology by SAP102 splice variants. J Neurosci. 2011;31:89–96. doi: 10.1523/JNEUROSCI.1034-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zheng S. Identify alternative splicing events based on position-specific evolutionary conservation. PLoS One. 2008;3:e2806. doi: 10.1371/journal.pone.0002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Sweeney NT, Craven SE, Kang R, El-Husseini AE-D, Bredt DS. Lipid- and protein-mediated multimerization of PSD-95: implications for receptor clustering and assembly of synaptic protein networks. J Cell Sci. 2003;116:3213–3219. doi: 10.1242/jcs.00617. [DOI] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Craven SE, Chetkovich DM, Firestein BL, Schnell E, Aoki C, Bredt DS. Dual palmitoylation of PSD-95 mediates its vesiculotubular sorting, postsynaptic targeting, and ion channel clustering. J Cell Biol. 2000;148:159–172. doi: 10.1083/jcb.148.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Stoilov P, Linares AJ, Zhou Y, Fu X-D, Black DL. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS Comput Biol. 2014;10:e1003442. doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y-P, Sheng M. Requirement of N-terminal Cysteines of PSD-95 for PSD-95 Multimerization and Ternary Complex Formation, but Not for Binding to Potassium Channel Kv1.4. J Biol Chem. 1999;274:532–536. doi: 10.1074/jbc.274.1.532. [DOI] [PubMed] [Google Scholar]
- Hyvönen MT, Uimari A, Keinänen TA, Heikkinen S, Pellinen R, Wahlfors T, Korhonen A, Närvänen A, Wahlfors J, Alhonen L, Jänne J. Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. RNA. 2006;12:1569–1582. doi: 10.1261/rna.39806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang TC, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Wong ACY, Power JM, Tadros SF, Klugmann M, Moorhouse AJ, Bertrand PP, Housley GD. Alternative splicing of the TRPC3 ion channel calmodulin/IP3 receptor-binding domain in the hindbrain enhances cation flux. J Neurosci. 2012;32:11414–11423. doi: 10.1523/JNEUROSCI.6446-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger JM, Favaro PD, Liu M, Kitlinska A, Huang X, Raabe M, Akad DS, Liu Y, Urlaub H, Dong Y, Xu W, Schlüter OM. Differential roles of postsynaptic density-93 isoforms in regulating synaptic transmission. J Neurosci. 2013;33:15504–15517. doi: 10.1523/JNEUROSCI.0019-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, Steen H, Shi Y. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell. 2015;57:957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabuag N, Dolphin AC. Alternative Splicing in CaV2.2 Regulates Neuronal Trafficking via Adaptor Protein Complex-1 Adaptor Protein Motifs. Journal of Neuroscience. 2015;35:14636–14652. doi: 10.1523/JNEUROSCI.3034-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs GS, Green SH, Dailey ME. Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat Neurosci. 2001;4:1006–1013. doi: 10.1038/nn717. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CWJ. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ohta N, Moon W, Matsuzaki F. Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J Cell Sci. 2009;122:3242–3249. doi: 10.1242/jcs.050955. [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Kato H, Arai H, Faull RL, McKenna PJ, Emson PC. Gene expression of PSD95 in prefrontal cortex and hippocampus in schizophrenia. Neuroreport. 2000;11:3133–3137. doi: 10.1097/00001756-200009280-00019. [DOI] [PubMed] [Google Scholar]
- Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Penn AC, Balik A, Wozny C, Cais O, Greger IH. Activity-mediated AMPA receptor remodeling, driven by alternative splicing in the ligand-binding domain. Neuron. 2012;76:503–510. doi: 10.1016/j.neuron.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW-L, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Rossbach O, Hung L-H, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Chen L. A two-parameter generalized Poisson model to improve the analysis of RNA-seq data. Nucleic Acids Res. 2010;38:e170. doi: 10.1093/nar/gkq670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GMY, Yu D, Wang J, Soong TW. Alternative Splicing at C Terminus of CaV1.4 Calcium Channel Modulates Calcium-dependent Inactivation, Activation Potential, and Current Density. J Biol Chem. 2012;287:832–847. doi: 10.1074/jbc.M111.268722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol. 2010;191:87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S. The Neurogenetics of Alternative Splicing. Nat Rev Neurosci. n.d doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Telese F, Tan Y, Li W, Jin C, He X, Basnet H, Ma Q, Merkurjev D, Zhu X, Liu Z, Zhang J, Ohgi K, Taylor H, White RR, Tazearslan C, Suh Y, Macfarlan TS, Pfaff SL, Rosenfeld MG. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci. 2015;18:1256–1264. doi: 10.1038/nn.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Bennett JE, McAllister AK. Rapid recruitment of NMDA receptor transport packets to nascent synapses. Nat Neurosci. 2002;5:751–759. doi: 10.1038/nn883. [DOI] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Ding JH, Yang BZ, He GC, Roe C. Characterization of the bidirectional promoter region between the human genes encoding VLCAD and PSD-95. Genomics. 2003;82:660–668. doi: 10.1016/s0888-7543(03)00211-8. [DOI] [PubMed] [Google Scholar]
- Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013;29:442–448. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–8. S1. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]