Abstract
The aim of this study was to apply RAPD technique to analyze the genetic variability among the Iranian CA-MRSA isolates.
The RAPD amplification was implemented on 25 strains isolated from the anterior nares of 410 healthy children using four randomly selected oligonucleotide primers from the stocks available in our laboratory, including the primers 1254, GE6, OLP6 and OLP13 from our stock. The amplified PCR products were detected on a 1.5% agarose gel and subjected to further analysis to establish the band profiles and genetic relationships using the Gel Compar® program.
The Iranian CA-MRSA isolates produced distinct RAPD patterns which varied based on the primer used, however, the primer 1254 revealed highly polymorphic patterns consisting 5 discernable RAPD types (RT), “RT1” (12, 48%), “RT2” (8, 32%), “RT3” (3, 12%), and “RT4 and RT5”, (a single RAPD type each, 4%). Phylogenetic analysis based on RAPD profiles divided most of the CA-MRSA isolates into 2 distinct but related RAPD clusters, a small group and two single unrelated RAPD types.
This study shows that the simple and cost-effective but rather difficult to optimize RAPD fingerprinting could be used to evaluate genetic and epidemiological relationships of CA-MRSA isolates on condition that the patterns are obtained from carefully optimized laboratory tests.
Keywords: staphylococcus aureus, methicillin-resistant staphylococcus aureus, RAPD methods, molecular typing, Iran
1. Introduction
Methicillin resistant S. aureus (MRSA) spread is no longer limited to the hospitals, but has emerged in communities to infect an increasing number of people and in particular children (Green et al., 2012). Nasal carriage of community associated-methicillin resistant S. aureus (CA-MRSA) is a major source of endogenous infection as well as of human to human transmission (David & Daum, 2010). Accurate and rapid typing of S. aureus is crucial to the control of infectious organisms and numerous methods have been described elsewhere (Strandén, Frei, & Widmer, 2003; Mostafa, 2013).
Molecular techniques have been extensively developed for the last 30 years to study the genetic diversity of S. aureus strains and in particular MRSA strains (Mehndiratta & Bhalla, 2012; Vivoni & Moreira, 2005). These techniques have been validated in order to distinguish S. aureus strains for local epidemiologic or outbreak investigation purposes. The molecular approaches such as PCR (Rep-PCR), using primers that target repetitive extragenic palindromic DNA regions (Vecchio et al., 1995), multilocus enzyme electrophoresis (MLEE), a technique that analyzes the electrophoretic mobility of housekeeping enzymes (Boerlin, 1997), restriction fragment length polymorphism (RFLP) and DNA sequence analysis of the coagulase gene isolates (Hookey, Richardson, & Cookson, 1998), the repetitive extragenic palindromic have been applied in order to identify and investigate polymorphism within S. aureus strains. Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) are currently considered the “gold standard” method for epidemiological investigation of MRSA; however, the feasibility of these methods in countries with limited financial and technical resources is a matter of concern (Prevost, Jaulhac, & Piemont, 1992; Murchan et al., 2003; Enright et al., 2000).
The aim of this study was to assess the genetic relationship of nasal carriage CA-MRSA isolates from healthy children in Iran by a more affordable, cost-effective, efficient, quick and easy to assay molecular technique, i.e., RAPD-PCR.
It is noted that the use of this technique has previously been reported in other countries including developing countries and Iran, as suitable for this purpose (Onasanya et al., 2003; Othman, 2010; Fitzgerald et al., 1997; Nikbakht et al., 2008).
2. Materials & Methods
2.1 Isolation and Identification of MRSA
A total of 25 CA-MRSA isolates from anterior nares of 410 healthy preschool children between 2 to 6 years of age without any known risk factors were investigated by the current study. All MRSA isolates were initially identified by conventional microbiological assay and further characterized by PCR amplification of Eap-encoding (eap) and mecA-encoding (mecA) genes as species and MRSA specific markers (Hussain et al., 2008; Jonas et al., 2002).
2.2 RAPD Analysis
Total genomic DNA was obtained using the method of Pitcher as previously described with minor modification for large-scale DNA extractions (Pitcher, Saunders, & Owen, 1989; Shojaei et al., 2000). RAPD-PCR assay was carried out according to procedure outlined by Burucoa, Lhomme, & Fauchere, 1999.
In brief, PCR amplification was performed in a 25 µl total volume containing 1U Taq DNA polymerase, 2.5 µl of 10xPCR buffer, 2µl of the randomly selected primers from our library, i.e., the primers GE6 (5’-CCC GTC AGC A-3’), 1254 (5’-CCG CAG CCA A-3’), OLP6 (5’- GAGGGAAGAG-3’), OLP13 (5’- ACCGCCTGCT-3’) and 50 ng of DNA template. PCR cycling parameters were as follows: an initial hot start at 95ºC followed by4 cycles each consisted of 94ºC for 1 minute, 30ºC for 1 minute, and 72ºC for 2 minutes, followed by 36 cycles of 94ºC for 1 minute, 36ºC for 1 minute, and 72ºC for 2 minutes and a final extension step of 72ºC for 5 minutes. A negative control of the same reaction mixture without DNA and a positive control, containing chromosomal DNA of the reference strain MRSA ATCC 33591 were included in each run. Each isolate was tested under the same conditions at least twice with the selected oligonucleotides. Amplified PCR fragments were subjected to electrophoresis in a 1.5% agarose gel at a constant voltage of 4V/cm and scanned by the Gel Documentation Systems. The software DNA FRAG version 3.03 (Nash, 1991) was used to estimated DNA fragment sizes in the RAPD profiles.
The RAPD profiles were defined based on the number and position of the major bands according to the Dice formula and similarity coefficients for each pair of lanes and a dendrogram generated based on the unweighted pair-group method with averages (UPGMA) by means of the GelCompar 3.1 software (Applied Maths, Kortrijk, Belgium) to estimate the relationships between the isolates.
3. Results
The RAPD analysis with DNA from the CA-MRSA isolates, and the primers OLP6, OLP13 and GE6 was found to yield less informative banding patterns.
However, primers 1254 yielded the best RAPD patterns with regard to number, distribution and intensity of the bands. Consequently, it was selected to be used with all strains for typing analysis. RAPD analysis with the chosen primer generated 5 different patterns. These genotypes assigned to RAPD types 1 to 5 (Figure 1). The RAPD profiles consisted of three to five amplicons ranging from 730 to 1720 base pairs in length. Apart from one isolate, i.e., strain ST34, all other isolates generated some conserved bands that could be detected in the patterns from all isolates, that is, the fragments of 1290, 1410 and 1720 base pairs. The isolate ST34 lacked a 1720 bp fragment (Figure 2). According to our results, RAPD types 1 with 12 isolates (48%) was the most frequently encountered, followed by RAPD type 2 with 8 isolates (32%) as the second, RAPD types 3 (3 isolates) as the third then RAPD types 4 and 5 (each contains 1 isolates), as the fourth and the fifth RAPD types (Figure 2).
Figure 1.
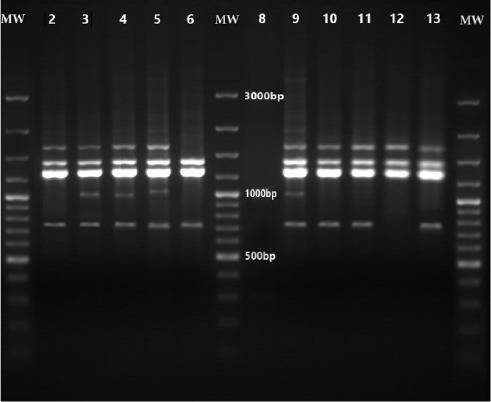
RAPD-PCR assay for CA-MRSA
Lanes: 1, 7 and 14: MW, 100-bp DNA ladder marker, Lanes: 2, 10, 11, 13: RAPD type 1, Lanes: 3, 4: RAPD type 2, Lane 5: RAPD type 4, Lane 6: RAPD type 5, Lane 13: RAPD type 3
Figure 2.

Normalized graph showing the details of RAPD profiles of Iranian nasal carriage CA-MRSA
Dendrogram analysis of RAPD-PCR amplification patterns of the Iranian isolates of CA-MRSA resulted in formation of 2 distinct but related RAPD clusters. RAPD cluster 1 had the highest similarity (89.9%) with RAPD type 4. The remaining 3 isolates occurred in a small group that showed a similarity of 84.5± 4.6% with the other two clusters and RAPD type 4. The RT 5 also formed a rare single line of descent which was distantly related to other isolates with a similarity of 80.2 ± 4.3 % (Figure 3).
Figure 3.
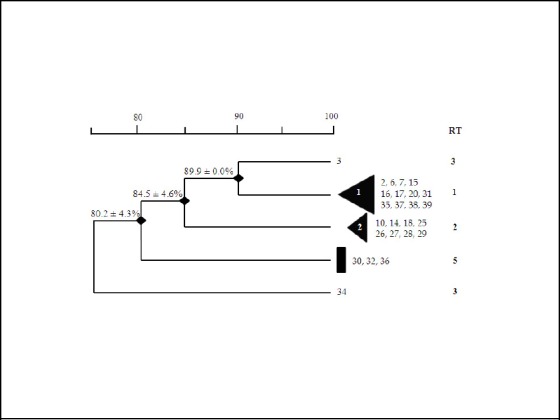
Dendrogram of the nasal carriage CA-MRSA RAPD-PCR DNA fingerprints with the primer 1254; Similarity coefficients are included in the top bar
4. Discussion
Molecular typing methods have been evaluated not only for their ability to discriminate among strains for epidemiologic purposes but also for their potential as taxonomic value. Previous studies have shown that S. aureus is a polymorphic species and has a clonal population structure (Fitzgerald et al., 2001; Feil et al., 2003). Several molecular methods have been used for epidemiological surveillance of MRSA isolates in order to track the distribution, infection source and transmission routes (Tenover et al., 1994; van Belkum et al., 1995; Hookey, Richardson, & Cookson, 1998; Vecchio et al., 1995; Boerlin, 1997). The typing techniques that are most commonly used today for typing of MRSA strains are multilocus sequence typing (MLST) (Enright et al., 2000), pulsed-field gel electrophoresis (PFGE) (Prevost, Jaulhac, & Piemont, 1992; Murchan et al., 2003), spa typing (Frenay et al., 1996), and SCC mec typing (Oliveira & de Lencastre, 2002; Ito et al., 2001).
Most of these methods suffer from the disadvantages that require high expertise, complicated laboratory settings and complex procedures to clearly differentiate the various MRSA. In our study we intended to find the molecular variation of Iranian isolates of CA-MRSA by utilizing a rather simple and cost-effective molecular typing method, namely, random amplified polymorphic DNA (RAPD) analysis (Welsh & McClelland, 1990; Reinoso et al., 2004). Compared with other typing methods for S.aureus strains such as PFGE (Prevost, Jaulhac, & Piemont, 1992), this procedure generates greater polymorphism, is technically friendly and faster, and requires no radioactive materials (Welsh & McClelland, 1990; Reinoso et al., 2004; Linton et al., 1995).
Our results indicated that RAPD fingerprinting can classify isolates of MRSA into clusters through which the relationship of strains can be evaluated. Previous studies has documented that the polymorphism generated by RAPD is almost the same as that yielded by RFLP (Linton et al., 1995). However, RAPD is almost incomparable in terms of simplicity, fastness and low-cost conditions with most other molecular typing methods (Burucoa, Lhomme, & Fauchere, 1999; Welsh & McClelland, 1990). In addition, a smaller volume of total DNA is required than for methods such as hybridization based methods (Welsh & McClelland, 1990; Linton et al., 1994). This is of obvious significance in terms of cost analysis as, in our experience; DNA isolation from staphylococcus aureus is almost unlikely without using a rather expensive enzyme, that is, lysostaphin (Enright et al., 2000). Although RAPD is simple and useful for epidemiological analysis, optimization of the PCR conditions is very important for reliability and reproducibility of the polymorphic patterns (Burucoa, Lhomme, & Fauchere, 1999). For instance in our experiment it was highly crucial to optimize the amount of DNA in each RAPD assay (50 ng of DNA) to ensure that non-specific bands were not present.
In fact our rather long involvement in utilization of RAPD as a favorite typing techniques for developing countries has shown that, even with rigid precaution and standardization, it is rather difficult to obtain reproducible patterns with RAPD assay and as far as our experiences concern it is of high practical significance to carry out the RAPD experiment in duplicate for all isolates in order to acquire the true profile differences for the studied strains.
In our investigation the RAPD application in CA-MRSA genotype analysis based on primer 1254, resulted in 5 RAPD types for 25 isolates. We classified CA-MRSA strains as either cluster (containing >3 members), small group (2 or 3 members) or unique, i.e., single isolates. Of 25 CA-MRSA isolates, 20 (80%) grouped into two clusters, i.e., 1 &2, and 3 isolates (12%) classified into a small group, and 2 isolates (8%) had a unique RAPD profile. The RAPD types 1 and 2 that encompassed 80% of all isolates prevailed. From this finding it can be inferred that RT 1 and RT2 are the major prevalent strains circulating in our community. Whether these strains have a particular feature that facilitates their colonization and adaptation capacity within Iranian children is a question that remains to be answered by further studies including more advance molecular typing assay such as MLST.
5. Conclusion
Taking into account the fact that advances in the development of more accurate and reproducible molecular methods have offered new typing choices for S.aureus strains, such as PFGE, MLST and spa typing, our data indicated that a simple, rapid and cost-efficient but rather difficult to optimize RAPD-PCR can be successfully utilized to find out the distributional and epidemiological relationship of CA-MRSA isolates provided that the technique is implemented under careful reproducibility condition and in particular duplication of PCR runs to obtain valid results.
Acknowledgments
We would like to thank Isfahan University of Medical Sciences for sponsoring our project and Nosocomial Infection Research Center, and Infectious Diseases and Tropical Medicine Research center for technical support.
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- Boerlin P. Applications of multilocus enzyme electrophoresis in medical microbiology. J Microbiol Methods. 1997;28:221–231. http://dx.doi.org/10.1016/S0167-7012(97)00010-9 . [Google Scholar]
- Burucoa C, Lhomme V, Fauchere J. L. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pyloriisolates:Experimental results and meta-analysis. J Clin Microbiol. 1999;37(12):4071–1080. doi: 10.1128/jcm.37.12.4071-4080.1999. PMID:10565934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M. Z, Daum R. S. Community-associated methicillin-resistant Staphylococcus aureus:Epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. PMID:20610826. http://dx.doi.org/10.1128/CMR.00081-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M. C, Day N. P, Davies C. E, Peacock S. J, Spratt B. G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. PMID:10698988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil S. J, Smith J. M, Murphy M, Spratt B. G, Moore C. E, Day N. P. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185(11):3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. PMID:12754228. http://dx.doi.org/10.1128/JB.185.11.3307-3316.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald J. R, Sturdevant D. E, Mackie S. M, Gill S. R, Musser J. M. Evolutionary genomics of Staphylococcus aureus:Insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. ProcNatlAcadSci. 2001;98(15):8821–8826. doi: 10.1073/pnas.161098098. PMID:11447287. http://dx.doi.org/10.1073/pnas.161098098 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald J, Meaney W, Hartigan P, Smyth C, Kapur V. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997;119(2):261–269. doi: 10.1017/s0950268897007802. PMID:9363026. http://dx.doi.org/10.1017/S0950268897007802 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenay H. M, Bunschoten A. E, Schouls L. M, van Leeuwen W. J, Vandenbroucke-Grauls C. M, Verhoef J, Mooi F. R. Molecular typing of methicillin resistant Staphylococcus aureuson the basis of protein a gene polymorphism. Eur J Clin Microbiol Infect Dis. 1996;15:60–64. doi: 10.1007/BF01586186. PMID:8641305. http://dx.doi.org/10.1007/BF01586186 . [DOI] [PubMed] [Google Scholar]
- Green B. N, Johnson C. D, Egan J. T, Rosenthal M, Griffith E. A, Evans M. W. Methicillin-resistant Staphylococcus aureus:An overview for manual therapists. J Chiropr Med. 2012;11(1):64–76. doi: 10.1016/j.jcm.2011.12.001. PMID:22942844. http://dx.doi.org/10.1016/j.jcm.2011.12.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookey J. V, Richardson J. F, Cookson B. D. Molecular Typing of Staphylococcus aureusBased on PCR Restriction Fragment Length Polymorphism and DNA Sequence Analysis of the Coagulase Gene. J Clin Microbio. 1998;36(4):1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. PMID:9542942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Eiff C. V, Sinha B, Joost I, Herrmann M, Peters G, Becker K. EapGene as Novel Target for Specific Identification of Staphylococcus aureus. J Clin Microbiol. 2008;46(2):470–476. doi: 10.1128/JCM.01425-07. PMID:18094124. http://dx.doi.org/10.1128/JCM.01425-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. PMID:11302791. http://dx.doi.org/10.1128/AAC.45.5.1323-1336.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas D, Speck M, Daschner F. D, Grundmann H. Rapid PCR-Based Identification of Methicillin-Resistant Staphylococcus aureusfrom Screening Swabs. J Clin Microbiol. 2002;40(5):1821–1823. doi: 10.1128/JCM.40.5.1821-1823.2002. PMID:11980967. http://dx.doi.org/10.1128/JCM.40.5.1821-1823.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton C. J, Jalal H, Leeming J. P, Millar M. R. Rapid discrimination of Mycobacterium tuberculosis strains by random amplified polymorphic DNA analysis. J Clin Microbiol. 1994;32(9):2169–2174. doi: 10.1128/jcm.32.9.2169-2174.1994. PMID:7814542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton C. J, Smart A. D, Leeming J. P, Jalal H, Telenti A, Bodmer T. Comparison of random amplified polymorphic DNA with restriction fragment length polymorphism as epidemiological typing methods for Mycobacterium tuberculosis. J Mol Pathol. 1995;48:133–135. doi: 10.1136/mp.48.3.m133. PMID:16695993. http://dx.doi.org/10.1136/mp.48.3.M133 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehndiratta P. L, Bhalla P. Typing of Methicillin resistant Staphylococcus aureus:A technical review. Indian J Med Microbiol. 2012;30(1):16–23. doi: 10.4103/0255-0857.93015. PMID:22361755. http://dx.doi.org/10.4103/0255-0857.93015 . [DOI] [PubMed] [Google Scholar]
- Mostafa S. I. Molecular typing of methicillin resistant Staphylococcus aureus by spa gene polymorphism. AJMR. 2013;7(9):755–759. [Google Scholar]
- Murchan S, Kaufmann M. E, Deplano A, de Ryck R, Struelens M, Zinn C. E, Cookson B. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus:A single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41(4):1574–1585. doi: 10.1128/JCM.41.4.1574-1585.2003. PMID:12682148. http://dx.doi.org/10.1128/JCM.41.4.1574-1585.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash J. H. E. DNA frag, Version 3.03. Ottawa, Ontario, Canada: Institute for Biological Sciences, National Research Council of Canada; 1991. [Google Scholar]
- Nikbakht M, Nahaei M, Akhi M, Asgharzadeh M, Nikvash S. Molecular fingerprinting of meticillin-resistant Staphylococcus aureus strains isolated from patients and staff of two Iranian hospitals. J Hosp Infect. 2008;69(1):46–55. doi: 10.1016/j.jhin.2008.01.034. PMID:18358563. http://dx.doi.org/10.1016/j.jhin.2008.01.034 . [DOI] [PubMed] [Google Scholar]
- Oliveira D. C, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. PMID:12069968. http://dx.doi.org/10.1128/AAC.46.7.2155-2161.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onasanya A, Mignouna H, Thottappilly G. Genetic fingerprinting and phylogenetic diversity of Staphylococcus aureus isolates from Nigeria. AJB. 2003;2(8):246–250. http://dx.doi.org/10.5897/ajb2003.000-1051 . [Google Scholar]
- Othman H. E. RAPD-PCR Markers in Genetic Relationship and Phylogenetic Diversity Analysis of Staphylococcus aureus Isolated from Dr. Khalid and Rizgary General Hospitals. J Koya Univer. 2010;(17):85–100. [Google Scholar]
- Pitcher D, Saunders N. A, Owen R. J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;l8:151–158. http://dx.doi.org/10.1111/j.1472-765X.1989.tb00262.x . [Google Scholar]
- Prevost G, Jaulhac B, Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. PMID:1572985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinoso E, Bettera S, Frigerio C, DiRenzo M, Calzolari A, Bogni C. RAPD-PCR analysis of Staphylococcus aureus strains isolated from bovine and human hosts. Microbiol Res. 2004;159(3):245–255. doi: 10.1016/j.micres.2004.04.002. PMID:15462524. http://dx.doi.org/10.1016/j.micres.2004.04.002 . [DOI] [PubMed] [Google Scholar]
- Shojaei H, Magee J. G, Freeman R, Yates M, Horadagoda N. U, Goodfellow M. Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic Mycobacterium isolated from an elephant. Int J Syst Evol Microbiol. 2000;50(5):1817–1820. doi: 10.1099/00207713-50-5-1817. PMID:11034492. http://dx.doi.org/10.1099/00207713-50-5-1817 . [DOI] [PubMed] [Google Scholar]
- Strandén A, Frei R, Widmer A. F. Molecular Typing of Methicillin-Resistant Staphylococcus aureus Can PCR Replace. Pulsed-Field Gel Electrophoresis? J Clin Microbiol. 2003;41(7):3181–3186. doi: 10.1128/JCM.41.7.3181-3186.2003. PMID:12843061. http://dx.doi.org/10.1128/JCM.41.7.3181-3186.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Pfaller M. A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32(2):407–415. doi: 10.1128/jcm.32.2.407-415.1994. PMID:7908673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, Koeleman Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbial. 1995;33(6):1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. PMID:7650182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio V. G, Petroziello J. M, Gress M. J, McCleskey F. K, Melcher G. P, Crouch H. K, Lupski J. R. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J ClinMicrobiol. 1995;33:2141–2144. doi: 10.1128/jcm.33.8.2141-2144.1995. PMID:7559964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivoni A. M, Moreira B. M. Application of molecular techniques in the study of Staphylococcus aureus clonal evolution-A Review. MemInst Oswaldo Cruz. 2005;100(7):693–698. doi: 10.1590/s0074-02762005000700001. PMID:16410952. http://dx.doi.org/10.1590/S0074-02762005000700001 . [DOI] [PubMed] [Google Scholar]
- Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. PMID:2259619. http://dx.doi.org/10.1093/nar/18.24.7213 . [DOI] [PMC free article] [PubMed] [Google Scholar]


