Abstract
Aim
To investigate the role of p53 gene in cervical carcinogenesis.
Materials and Methods
A total 50 cases and controls were taken after setting exclusion criteria. Venous blood (3 ml) samples were collected in sterile EDTA sterile vials. Both punch biopsy of cervical growth in cases and biopsy from cervix after hysterectomy in controls were performed. Genomic DNA was extracted from tissue and blood using standard protocol of Miller et al. 1994 using chloroform–phenol method. Gene was amplified using specific forward and reverse primers and p53 gene expressions were studied. The present study of p53 gene regulation analyzed the expression of 279-bp bands on 1.5 agarose gel.
Observations
Out of the total 50 samples of cases and controls, we were able to isolate DNA from 38 cases and 28 controls in blood and in 22 cases and 22 controls in tissue. In cases of carcinoma cervix, p53 expression is either downregulated or absent in 71.06 % of cases compared to 50 % of controls in blood and 72.73 % of cases compared to 59.09 % of controls in tissue, but these figures were not statistically significant (p = 0.67 and p = 0.167, respectively). p53 positivity rate was only in 27.78 % of squamous cell cancer and 50 % of adenocarcinoma. Three out of nine patients (33.3 %) with L.N. positive status have p53 gene positivity, whereas 23 % (3 out of 13) with L.N. negative status have p53 gene positivity, which is not significantly associated. In our study, p53 overexpression increases with the various stages of cervical cancer.
Conclusion
In our study, we found that there is the increased frequency of upregulation or overexpression of p53 gene in control in both blood (50 %) and tissue (40.9 %), but this association is statistically nonsignificant. In the present study, there is a lack of relationship between p53 overexpression and prognosis in the cervical cancer patients. However, our study lacked larger sample size which otherwise would have been able to lend support to truly significant findings through much larger combined and comparative datasets.
Keywords: p53 gene expression, Cervical cancer, PCR
Introduction
Cervical cancer is the fourth most common cancer in women. 528,000 new cases were diagnosed worldwide in 2012. Cervical cancer also contributed nearly 8 % of all cancers [1]. This tumor is associated to some specific types of human papillomaviruses (HPV), particularly of types 16, 18, 33, and 42. The peak incidence of the disease occurs in women over 40 years of age; however, the peak incidence for HPV infection is found in women in their twenties; therefore, there is a long latency period between the time of HPV infection and cancer appearance. In the pathogenesis of cervical carcinoma, there are three major components: two of them related to the role of human papilloma viruses (HPV). First, the effects of viral E6 and E7 proteins, and the second, the integration of viral DNA in chromosomal regions associated with the well-known tumor phenotypes. The third factor is the accumulation of cellular genetic damage, not related to HPV, needed for tumor development. HPVs have circular, double-stranded DNA genomes that encode eight genes, of which E6 and E7 have transforming properties. E6/E7 expression promotes chromosomal instability, foreign DNA integration, and other mutagenic events in the cell [2]. The identification of the specific genes involved, and their correlation with specific tumor properties and stages, could improve the understanding and perhaps the management of cervical carcinoma.
p53 protein degradation is induced by virus oncoprotein E6, a determinant step in the cervical HPV-induced oncogenesis. Interestingly, p53 is rarely mutated in early cervical tumors, an exception in cancer genetics. E6 acts as an inactivating mutation. The fact that some p53 mutant proteins are not susceptible to E6-mediated degradation suggests that p53 polymorphisms might be responsible for the variation of the epithelial cell response to the HPV infection.
This study aims
To evaluate the association of p53 gene expression with cervical carcinoma.
To study the effect of p53 gene expression in relation to disease prognosis.
Materials and Methods
This was a case–control study carried out in the Department of Obstetrics and Gynecology, Sir Sunderlal Hospital and Centre of Experimental Medicine and Surgery, The Institute of Medical Sciences, Banaras Hindu University, Varanasi from 2012 to 2013. Institutional Ethical Committee and departmental review board approval was obtained for this study.
Total number of cases and control: 50 each.
Inclusion criteria: Cases: Patients with histopathologically diagnosed cervical carcinoma. Control: Patients undergoing hysterectomy for some benign conditions without any evidence of HPV infection, PID, active infection, and histopathologically undetermined cervical abnormalities.
Exclusion criteria: Patients with active infections, HPV infections, chronic inflammatory disease, and histopathologically undetermined cervical abnormalities.
Collection and processing of sample: Venous blood (3 ml) was collected in sterile vials containing sterile EDTA and centrifuged within 4 h at 1500 rpm for 15 min at 40 °C. Isolated pellet was transferred into another polypropylene vial and stored at −80 °C until further study.
Punch biopsy of cervical growth in cases and punch biopsy from cervix after hysterectomy in controls were performed after obtaining written consent, and tissue is homogenized using mortar and pestle and stored at −80 °C until further study.
DNA extraction and real-time PCR: genomic DNA was extracted from tissue and blood using standard protocol of Miller et al. 1994 using chloroform–phenol method, and p53 expression were studied using both the forward and reverse primers as respective positive controls. Gene was amplified using specific forward and reverse primers with optimum conditions of the PCR.
Specific forward and reverse primers selected for PCR reaction were as follows:
p53
F = 5′-TGA AGT CTC ATG GAA GCC AGC-3′
R = 5′-GCT CTTT TTC ACC CAT CTA CAG-3′
Total reaction volume of 25 µl containing 50-100 ng of genomic DNA, 20 pmol of each primer, 200 µM of each dNTPs (Fermentas, USA) mixed with Taq buffer (10 mM TrisHCl pH 8.3, 50 mM KCL), 3.0 mM MgCl2, and 3 units of Taq polymerase (New England, Biolabs) was prepared for Taq amplification. Amplification was carried out in a thermal cycler (BioRed, USA). Cycling conditions consisted of 3 min at 94 °C for initial denaturation, followed by 1 min at 60 °C for annealing, followed by 35 cycles, and 7 min at 72 °C for final extension to ensure a complete extension of all PCR products.
Anticontamination Measures
Strict precautions to prevent PCR products’ contamination were enforced.
Statistical Analysis
Statistical analysis was performed using SPSS version-16. The various parameters studied during the observation period were compared using Chi-square test for noncontinuous variables. The critical value of ‘p’ indicating the probability of significant difference was taken as <0.05 for comparison.
Observations
Out of the 50 samples of cases and controls, we were able to isolate DNA from 38 cases and 28 controls in blood and in 22 cases and 22 controls in tissue. In cases of carcinoma cervix, p53 expression is either downregulated or absent in 71.06 % of cases compared to 50 % of controls in blood (Table 1); and 72.73 % of cases compared to 59.09 % of controls in tissue (Table 2), but these are not statistically significant (p = 0.67 and p = 0.167, respectively).
Table 1.
p53 gene expressions in blood in cervical cancer cases and controls
| Expression | Case (n = 38) | Control (n = 28) | ||
|---|---|---|---|---|
| No. | % | No. | % | |
| Up regulation | 11 | 28.94 | 14 | 50.0 |
| Down regulation | 14 | 36.84 | 9 | 32.15 |
| Null (absent) | 13 | 34.22 | 5 | 17.85 |
| Total | 38 | 100 | 28 | 100 |
χ 2 = 3.569, p = 0.67
Table 2.
p53 gene expressions in tissue in cervical cancer cases and controls
| Expression | Case (n = 22) | Control (n = 22) | ||
|---|---|---|---|---|
| No. | % | No. | % | |
| Up regulation | 6 | 27.27 | 9 | 40.91 |
| Down regulation | 9 | 40.91 | 11 | 50.00 |
| Null (absent) | 7 | 31.82 | 2 | 9.09 |
| Total | 22 | 100 | 22 | 100 |
χ 22 = 3.577, p = 0.167
p53 positivity was found only in 27.78 % of squamous cell cancer and in 50 % of adenocarcinoma (Table 3). Three out of nine patients (33.3 %) with L.N. positive status have p53 gene positivity, whereas 23 % (3 out of 13) with L.N. negative status have p53 gene positivity, which is not significantly associated. L.N. positivity indicates poor prognosis (Table 4). In our study, p53 overexpression showed variable increases in different stages of cervical cancer (Table 5).
Table 3.
p53 gene expressions in tissue in cervical cancer cases and their correlation to histopathology
| Histopathology | Case (n = 22) | p53 positive | p53 negative | |||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Squamous cell cancer | 18 | 81.8 | 5 | 27.78 | 13 | 72.22 |
| Adenocarcinoma | 2 | 9.1 | 1 | 50 | 1 | 50 |
| Adenosquamous | 2 | 9.1 | 0 | 0 | 2 | 100 |
| Total | 22 | 100 | 6 | 27.27 | 16 | 72.7 |
χ 2 = 1.273, p = 0.529
Table 4.
p53 gene expressions in tissue in cervical cancer cases and their correlation to lymph node status
| Histopathology | Lymph node mets | |||
|---|---|---|---|---|
| Positive (n = 9) | Negative (n = 13) | |||
| p53 positivity | p53 negative | p53 positive | p53 negative | |
| Squamous cell cancer (n = 18) | 2 | 6 | 3 | 7 |
| Adenocarcinoma (n = 2) | 1 | 0 | 0 | 1 |
| Adenosquamous (n = 2) | 0 | 0 | 0 | 2 |
Table 5.
p53 gene overexpressions with respect to stages of carcinoma cervix
| Stages | Cases (n) | p53 overexpression | Percentage |
|---|---|---|---|
| Stage I | 5 | 1 | 20 |
| Stage II | 10 | 2 | 20 |
| Stage III | 4 | 1 | 25 |
| Stage IV | 3 | 2 | 66 |
| Total | 22 | 6 | 27.27 |
Discussion
p53 is a tumor suppressor protein and transcription factor present in a latent state in all cells and activated by various stresses such as hypoxia, free radicals, DNA damage, and UV light. Some of the first molecular explanations for the malignant potential of the high-risk HPV types, 16 and 18, came with the observations that their E6 and E7 oncoproteins could interact with the key cellular tumor suppressors, p53 and pRB. p53 is a multifunctional transcriptional modulator and inducer of apoptosis. In response to DNA damage, nucleotide depletion, or hypoxia, it becomes activated by acetylation and phosphorylation, functioning as a nuclear transcription factor to induce genes involved in cell-cycle inhibition or apoptosis, or it can induce apoptosis more directly by interacting with proteins in the cytoplasm at mitochondrial sites. Similarly, pRB is also a key regulator, interaction of which with the members of the E2F family of transcription factors regulates both cell-cycle progression and apoptosis. Both p53 and pRB mutations are common in many types of cancer, but occur very rarely in early-stage cervical cancers, inviting the speculation that their functional abrogations by the E6 and E7 oncoproteins from high-risk mucosal HPVs are functionally equivalent to mutations in other cancer types to some extent. The E7 proteins encoded by the high-risk type HPVs, such as HPV 16 and HPV 18, bind Rb with a much higher affinity compared to those encoded by the low-risk type HPVs, such as HPV 6 and HPV 11. E7 disrupts the interaction between Rb and E2F, resulting in the release of E2F factors in their transcriptionally active forms. The E7-mediated conversion of E2Fs to their activator forms stimulates replication and cell division [3]. Subsequently, it was shown that high-risk, but not low-risk, E7 could induce the proteasome-mediated degradation of pRB.
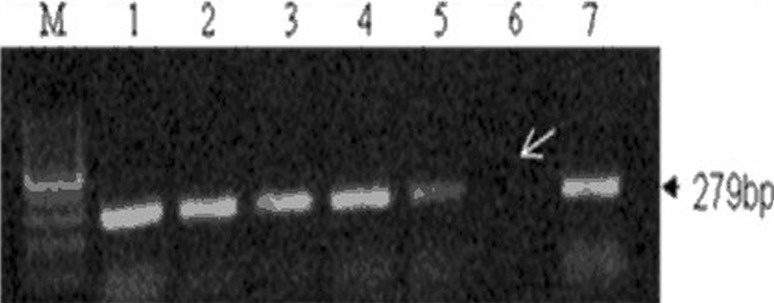
The present study of p53 gene regulation analyzed the expression of 279-bp bands on 1.5 agarose gel as shown in Fig. 1. The p53 activity was analyzed in terms of expressions in three different forms: 1. complete disappearance (null) of 279 bands, 2. overexpression, or 3. regression (underexpression) compared with controls. Interestingly, in our study, we found that there is the increased frequency of upregulation or overexpression of p53 gene in control both in blood (50 %) and tissue samples (40.9 %), but this association is statistically nonsignificant. These observations had the possibility of a predisposive effect due to the small sample size of this study.
Fig. 1.
PCR based DNA analysis of p53 gene showing overexpression in lane 1, underexpression in lane 5 and complete disappearance of 279bp amplicon in lane 6 in carcinoma cervix
Hunt et al. study [4] showed p53 gene product was overexpressed in 17.1 % (14/82) of all carcinomas and also in areas of cervical intraepithelial neoplasia grade III adjacent to invasive squamous carcinoma. Inactivation of wild-type p53 gene product represents the most common genetic alteration in human carcinogenesis, and its overexpression, as detected by immunohistochemistry, has been proposed to indicate a worsened prognosis in some malignancies [5, 6]. Loss of p53 function is believed to play an important role in the pathogenesis of carcinomas of the uterine cervix, where it has been shown that the p53 gene product may be inactivated in three principal ways: (1) after somatic point mutation; (2) after loss of heterozygosity; and (3) following human papillomavirus (HPV) infection, as the viral oncoprotein E6 is known to bind to, stabilize, and ultimately degrade wild-type p53.
In the present study, p53 upregulation was found in 28.94 % of cases in blood and 27.27 % of tissue. p53 overexpression was present in 25.5 % of cases in Oka et al.’s study [7], 33.6 % in Hanprasertpong J et al.’s study [8], and 14.0 % in Busby-Earle et al.’s study [9].
p53 Expression and Histopathology of Cervical Cancer
In our study, p53 positivity was present in 27.78 % of cases with squamous cell cancer (n = 18), in 50 % of cases with adenocarcinoma (n = 2), and 0 % of cases with adenosquamous carcinoma (n = 2). The results are different from Hunt et al.’s [4] study in which p53 positivity was present in 22.7 % of cases with squamous cell cancer (n = 22), in 14.3 % of cases with adenocarcinoma (n = 35), and 16 % of cases with adenosquamous carcinoma (n = 25). Helland et al.’s study [10] showed p53 positivity in 44 % of cases with adenocarcinoma (n = 9) and 57 % of cases with adenosquamous carcinoma (n = 7). In Holm et al.’s study [11] and Bosari et al.’s [12] study, rates of positivity were 11 % (n = 36) and 100 % (n = 1), respectively, in adenocarcinoma.
Lymph node status helps in predicting the prognosis of carcinoma of cervix. Our study tried to correlate the positivity of p53 gene with lymph node status. 33.3 % with L.N. positive status , whereas 23 % with L.N. negative status have p53 gene positivity, which is not significant. Hunt et al.’s study [4] showed the corresponding figures as 33 and 36 %, respectively. There was no relation between p53 overexpression and prognosis in cervical cancer [4, 8]. The present study shows lack of relationship between p53 overexpression and prognosis in the cervical cancer patients.
At various stages, p53 overexpression increased from 20.1 % in stage IB to 60 % in stage IV, according to Bremer et al. [13]. In our study also, p53 overexpressions showed variable increases with different stages, i.e., 20 % in both stage I and stage II, 25 % in stage III, and 66 % in stage IV, but were not statistically significant.
Conclusion
Interestingly, In our study, we found that frequency of upregulation or overexpression of p53 gene increased in control in both blood (50 %) and tissue (40.9 %). Although this association is statistically nonsignificant, this study’s findings will, however, help in meta-analysis studies done on p53 gene expression in carcinoma cervix. The study had the possibility of a predisposive effect due to its small sample size or unknown environmental factor. In most of the earlier studies on cervical cancers, the function of p53 is downregulated and perhaps failed to regulate the process of carcinogenesis. The present study shows lack of relationship between p53 overexpression and prognosis in the cervical cancer patients. Traditional parameters such as tumor size and lymph node status still seem to be the most important determinants of prognosis in the carcinoma of the uterine cervix. However, our study lacked larger sample size, which otherwise would have been able to lend support to truly significant findings through much larger combined and comparative datasets.
Dr. Garima
has passed her MBBS and MS (Obs and Gynaec) both from BHU, Varanasi. Currently she is working as Senior Resident in the Department of Obstetrics and Gynaecology at UCMS and GTB Hospital, Delhi. Her field of interest includes high-risk pregnancy and infertility.
Compliance with Ethical Standards
Conflict of interest
No conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Ethical approval
Institutional ethical clearance was taken. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institution and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
Garima is Senior Resident, Sulekha Pandey is Professor, L. K. Pandey is Professor, A. K. Saxena Professor, Nidhi Patel is Senior Resident in the BHU, Varanasi, Uttar Pradesh, India
References
- 1.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon: International Agency for Research on Cancer; 2013.
- 2.Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev Med Virol. 2006;16:83–97. doi: 10.1002/rmv.488. [DOI] [PubMed] [Google Scholar]
- 3.Yim E-K, Park J-S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37(6):319–324. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt CR, Hale RJ, Buckley CH, et al. p53 expression in carcinoma of the cervix. J Clin Pathol. 1996;49(12):971–974. doi: 10.1136/jcp.49.12.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildirim M, Kaya V, Demirpence O, et al. Prognostic significance of p53 in gastric cancer: a meta-analysis. Asian Pac J Cancer Prev. 2015;16(1):327–332. doi: 10.7314/APJCP.2015.16.1.327. [DOI] [PubMed] [Google Scholar]
- 6.Bansal A, Gupta A, Saxena S. Correlation of p53 immunoexpression with DNA ploidy and apoptotic index in subsets of prostate cancer: a marker reiterated in progression and recurrence of prostate cancer. South Asian J Cancer. 2015;4(2):88–90. doi: 10.4103/2278-330X.155693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oka K, Nakano T, Arai T. P53CM1 expression is not associated with prognosis in uterine cervical carcinoma. Cancer. 1993;72:160–163. doi: 10.1002/1097-0142(19930701)72:1<160::AID-CNCR2820720130>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 8.Hanprasertpong J, Tungsinmunkong K, Chichareon S, et al. Correlation of P53 and Ki-67 (MIB-1) expressions with clinicopathological features and prognosis of early stage cervical squamous cell carcinomas. J Obstet Gynaecol Res. 2010;36(3):572–580. doi: 10.1111/j.1447-0756.2010.01227.x. [DOI] [PubMed] [Google Scholar]
- 9.Busby-Earle RMC, Steel CM, Williams ARW, et al. P53 mutations in cervical carcinogenesis—low frequency and lack of correlation with human papillomavirus status. Br3r Cancer. 1994;69:732–737. doi: 10.1038/bjc.1994.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helland A, Holm R, Kristensen G, Kaern J, Karlsen F, Trope C, et al. Genetic alterations of the TP53 gene, P53 protein expression and HPV infection in primary cervical carcinomas. J Pathol. 1993;171:105–114. doi: 10.1002/path.1711710207. [DOI] [PubMed] [Google Scholar]
- 11.Holm R, Skomedal H, Helland A, et al. Immunohistochemical analysis of P53 protein overexpression in normal, premalignant and malignant tissues of the cervix uteri. J Pathol. 1993;169:21–26. doi: 10.1002/path.1711690105. [DOI] [PubMed] [Google Scholar]
- 12.Bosari S, Roncalli M, Viale G, et al. P53 immunoreactivity in inflammatory and neoplastic diseases of the uterine cervix. J Pathol. 1993;169:425–430. doi: 10.1002/path.1711690407. [DOI] [PubMed] [Google Scholar]
- 13.Bremer GL, Tieboschb AT, van der Putten HW, et al. P53 tumor suppressor gene protein expression in cervical cancer: relationship to prognosis. Eur J Obstet Gynecol Reprod Biol. 1995;63(1):55–59. doi: 10.1016/0301-2115(95)02225-V. [DOI] [PubMed] [Google Scholar]