Abstract
Innate immune responses are important for pathogen elimination and adaptive immune response activation. However, excess inflammation may contribute to immunopathology and disease progression (e.g. inflammation-associated hepatocellular carcinoma). Immune modulation resulting from pattern recognition receptor-induced responses is a potential strategy for controlling immunopathology and related diseases. This study demonstrates that the mycotoxin patulin suppresses Toll-like receptor- and RIG-I/MAVS-dependent cytokine production through GSH depletion, mitochondrial dysfunction, the activation of p62-associated mitophagy, and p62-TRAF6 interaction. Blockade of autophagy restored the immunosuppressive activity of patulin, and pharmacological activation of p62-dependent mitophagy directly reduced RIG-I-like receptor-dependent inflammatory cytokine production. These results demonstrated that p62-dependent mitophagy has an immunosuppressive role to innate immune response and might serve as a potential immunomodulatory target for inflammation-associated diseases.
Keywords: mitochondria, mitophagy, mycotoxin, p62 (sequestosome 1(SQSTM1)), RIG-I-like receptor (RLR), toll-like receptor (TLR), immunomodulation, patulin
Introduction
Innate immune responses are important for pathogen elimination and the activation of adaptive immune responses (1). Pattern recognition receptors, such as Toll-like receptors (TLR), 3 RIG-I-like receptors (RLR), and Nod-like receptors (NLR), recognize pathogen-associated molecular patterns, resulting in cytokine and chemokine production and the induction of other downstream innate immune responses (2). Although innate immune responses are critical to pathogen elimination and immune response activation, the pro-inflammatory responses elicited as a result of pattern recognition receptor signaling can lead to excess inflammation if not controlled and may contribute to disease progression (e.g. Helicobacter pylori-associated gastric cancer (3) and chronic hepatitis virus infection-associated hepatocellular carcinoma (4, 5)). Therefore, controlling immunopathology and related diseases caused by inflammation resulting from pattern recognition receptor-dependent signaling represents a potential treatment strategy (6–8). How to fine tune unwanted inflammation while maintaining protective immune responses intact will be a critical hurdle to overcome.
Hepatic diseases, such as cirrhosis and hepatocellular carcinoma are typically associated with chronic inflammation that can take decades to develop. Activation of proinflammatory cytokines in the liver, such as IL-6/gp130/STAT3, is a critical step in the development of hepatic disease, including hepatocellular carcinoma (9–11). In addition, serum proinflammatory cytokine levels were higher in hepatitis C virus (HCV)-associated hepatocellular carcinoma patients (12). Modulation of these immune responses represents a potential strategy for ameliorating disease progression (7). Therefore, we aimed to identify new targets for immune modulation as a means of reducing proinflammatory cytokine production by screening a library of natural compounds for the treatment of inflammation-related liver diseases. An in vitro screening platform was developed to evaluate natural compounds collected by Microsource Discovery Systems that was composed of two screens: one to evaluate cytokine production activated by LPS/TLR4 pathway signaling and the other to analyze the cytokines triggered by HCV-NS5B-dependent RIG-I/MAVS activation (13). IL-6 production was measured as a readout to evaluate the immune modulatory activity of candidate compounds. This screening process identified patulin that suppressed the activity of both LPS/TLR4 and RIG-I/MAVS activation pathways.
Patulin is a fungal toxin produced by Penicillium and other fugal genera and can be commonly found in rotten crops and fruit (14). With its electrophilic property, patulin reacts with GSH and N-acetylcysteine to form adducts (15). Therefore, patulin treatment could elicit GSH depletion followed by mitochondrial dysfunction and reactive oxygen species elevation in cell (16, 17). Long term treatment with patulin, therefore, leads to the activation of oxidative stress, endoplasmic reticulum stress, and apoptosis (18–22). Patulin also has been linked to skin cancer formation (23) and cytokine dysregulation (24, 25). In this study, we further examined the mechanisms associated with patulin-modulation of the TLR and RLR pathways and identified that patulin disturbed NF-κB-mediated activation and cytokine production through GSH depletion and mitochondrial dysfunction-dependent mitophagy activation. These results reveal an immunosuppressive role for the p62-dependent mitophagy in both TLR- and RLR-dependent innate immune response.
Results
Patulin Blocked LPS-dependent Cytokine and NO Production
To determine whether patulin possessed immunosuppressive properties, we examined its effects on IL-6 production following LPS stimulation of mouse leukemic monocyte macrophage (RAW264.7) cells. RAW264.7 cells were treated with LPS (1 μg/ml) in the presence of various doses of patulin (1–100 μm). As shown in Fig. 1A (left), LPS-dependent IL-6 expression was significantly reduced in the presence of 1 μm patulin and completely blocked at a concentration of 5 μm. NO production was also completely inhibited in the presence of 5 μm patulin (Fig. 1A, middle). To determine whether patulin treatment resulted in any cytotoxicity effects, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to monitor cell viability. As shown in Fig. 1A (right), patulin treatment at 1 μm did not significantly affect MTT activity; however, MTT activity was slightly reduced to 80% when cells were incubated with 5–20 μm patulin, and activity did not drop significantly until 50 μm patulin was used, suggesting that patulin suppressed LPS-dependent proinflammatory cytokine and NO production by RAW264.7 cells in vitro.
FIGURE 1.
Patulin blocked LPS-dependent cytokine secretion and NO production. A, mouse leukemic monocyte macrophages (RAW264.7 cells) were treated with various concentrations of patulin in the presence or absence of LPS (1 μg/ml) for 24 h (n = 3). IL-6 and NO levels in culture supernatants were measured by ELISA and the Griess reaction, respectively. Cell viability was determined using the MTT assay. B, cytokine mRNA expression profiles of cells 2 h after LPS treatment (100 ng/ml) were evaluated by quantitative RT-PCR (n = 3). C, cytokine production after 24-h LPS treatment (100 ng/ml) was measured by ELISA (n = 6). D, to stimulate inflammasome activity, J774A.1 cells pretreated with or without LPS (500 ng/ml) for 4 h were stimulated with 5 mm ATP for 30 min. The cells were then washed with PBS and incubated in low serum medium for an additional 4 h. Intracellular (immature) and extracellular (mature) forms of IL-1β and caspase-1 were monitored by immunoblotting. The statistical significance was evaluated by Student's t test. *, p < 0.05; **, p < 0.01. Error bars, S.E.
Proinflammatory cytokine and type I IFN mRNA and protein expression levels following LPS (100 ng/ml) stimulation were evaluated by real-time PCR (Fig. 1B) and ELISA (Fig. 1C) in the presence or absence of patulin. LPS-induced IL-6, TNFα, IL-1β, and IFNβ mRNA expression were reduced in the presence of patulin treatment. IL-6, IL-1β, and IFNβ protein expression upon LPS stimulation were more sensitive to patulin treatment compared with TNFα expression. The maturation process of IL-1β protein from pro-IL-1β is inflammasome/caspase-1-dependent, and the activation of inflammasome usually needs a second signal to trigger the self-activation of procaspase-1 zymogen by proteolytic cleavage (26). Although the IL-1β mRNA was up-regulated by LPS stimulation in RAW267.4 cells, IL-1β protein was under the ELISA detection limit. It has been shown that RAW264.7 macrophage is defective in caspase-1 activation (27), so the J774.1 mouse macrophage was used to evaluate the effect of patulin on inflammasome activation. The expression of pro-IL-1β and NLRP3, a component of the inflammasome, was induced inside the LPS-primed J774.1 cells, but the induction was reduced in the presence of patulin (Fig. 1D). Upon extracellular ATP stimulation for inflammasome activation, mature IL-1β (p19) and caspase-1 (p20) were detected in the culture supernatant of control cells but were blocked in the supernatant of patulin-treated cells, which could be due to the reduction of pro-IL-1β and NLRP3 expression. TNFα secretion, an inflammasome-independent process, was not disturbed dramatically by patulin treatment.
Patulin Blocked Cytokine Production Induced by Various TLR Ligands
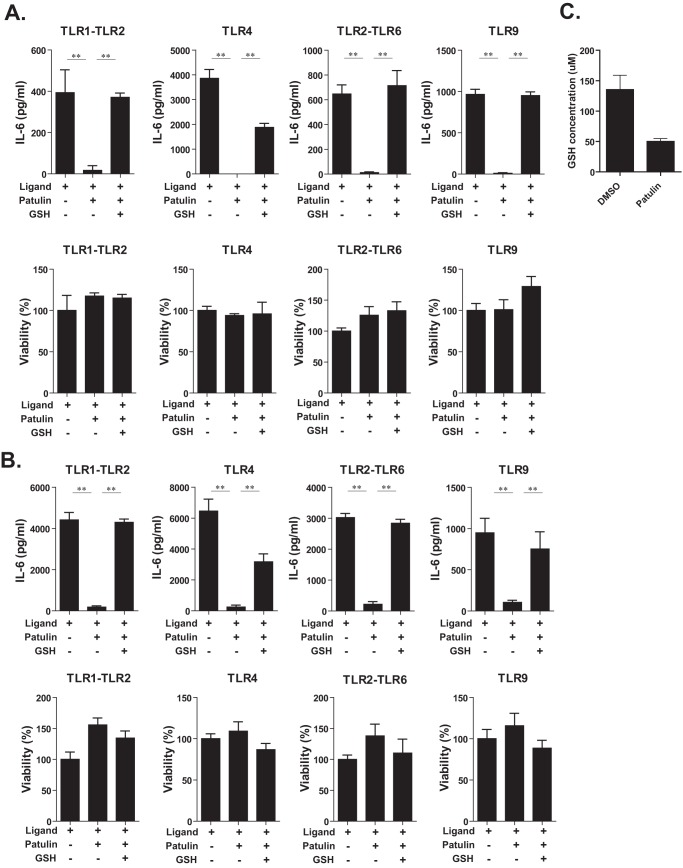
Because patulin suppressed LPS/TLR4-dependent IL-6 production, we next examined whether patulin could block cytokine production resulting from ligation to other TLR ligands. Pam3CSK and FSL-1 are synthetic lipoproteins recognized by TLR1-TLR2 (28) and TLR2-TLR6 (29) receptor complexes, respectively. CpG ODN is a synthetic oligodeoxynucleotide that is a ligand for TLR9, resulting in activation signals (30, 31). As shown in Fig. 2A, the ligands for TLR1-TLR2, TLR4, TLR2-TLR6, and TLR9 induced IL-6 production by RAW267.4 macrophage cells with different intensities. Surprisingly, IL-6 production induced by these different ligands was suppressed by patulin treatment (1 μm). Because agonists specific for TLR3, TLR5, and TLR7 did not induce production of IL-6 in RAW267.4 macrophages, the impact of patulin on signaling via these pattern recognition receptors remains undefined.
FIGURE 2.
Patulin blocked IL-6 production induced by other TLR ligands. A, RAW264.7 cells were pretreated with patulin (1 μm) in combination with GSH (1 mm) for 2 h and then stimulated with various TLR ligands for 16 h in the presence of patulin and GSH (n = 6). Pam3CSK4 (0.2 μg/ml), LPS (0.1 μg/ml), FSL1 (10 ng/ml), or ODN1862 (0.1 μg/ml) were used as TLR1-TLR2, TLR4, TLR2-TLR6, and TLR9 ligands, respectively. B, peritoneal macrophages harvested from thioglycollate-stimulated C57BL/6 mice were treated as described in A (n = 6). C, total GSH level in cell lysate prepared from RAW264.7 cells treated with patulin (1 μm) or DMSO (vehicle control) for 8 h was measured (n = 3). Error bars, S.E.
It has been shown previously that patulin treatment could elicit GSH depletion followed by mitochondrial dysfunction (16, 17). We therefore speculated that the inhibitory effects of patulin on proinflammatory cytokine production might be due to the ability of patulin to deplete GSH and affect mitochondrial function. When high concentrations of GSH (1 mm) were added to the culture medium, the inhibitory effect of patulin on IL-6 production resulting from ligation to various TLR ligands was reversed (Fig. 2A). To verify whether primary macrophages were also sensitive to patulin, peritoneal macrophages were stimulated with TLR agonists in the presence or absence of patulin. As shown in Fig. 2B, patulin also suppressed TLR-dependent IL-6 production in primary macrophages that was also reversed by the addition of GSH to the culture medium as described above. Cell viability of both RAW264.7 and primary macrophages was not affected significantly by exposure to either patulin or GSH (Fig. 2, A and B). To ensure that patulin indeed depleted GSH in RAW264.7 cells, intracellular GSH concentration at 8 h after patulin treatment was measured, and the result showed that >50% of GSH was depleted by patulin in cells (Fig. 2C). Taken together, these data suggest that patulin depleted the GSH level and inhibited TLR-dependent cytokine production in macrophages.
Patulin Suppressed LPS-induced NF-κB Activation
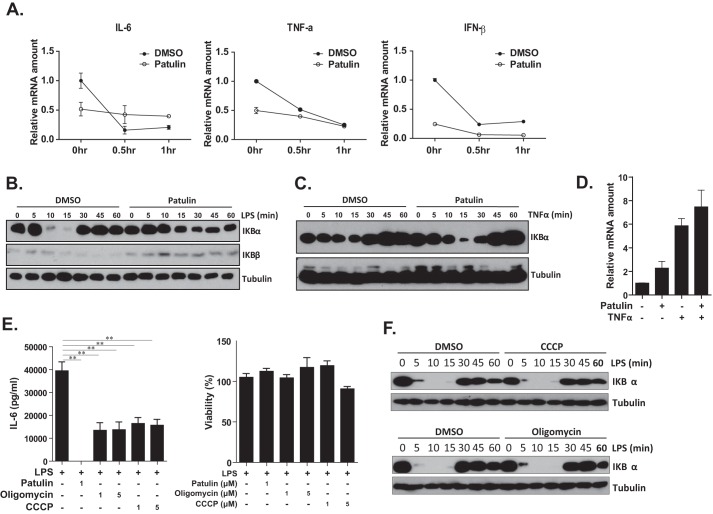
Because it was possible that the reduced levels of LPS-dependent cytokine mRNA observed following patulin treatment could be due to rapid mRNA degradation, actinomycin D was used to block de novo mRNA synthesis to determine the cytokine mRNA half-life. Data presented in Fig. 3A show that the half-life (t½) of IL-6, TNFα, and IFNβ was <30 min in control cells but was longer than 1 h in patulin-treated cells, suggesting that RNA stability might not contribute to patulin-mediated cytokine suppression. TLR-dependent proinflammatory cytokine production is mainly induced following NF-κB activation (32). To further understand the mechanism by which patulin suppressed cytokine production, we evaluated whether patulin modulated LPS-dependent NF-κB activation. In a resting state, NF-κB bound to I-κBα is trapped inactive in the cytoplasm. Upon stimulation (e.g. LPS treatment), TLR4 activation leads to I-κBα degradation, resulting in the release of NF-κB and its translocation into the nucleus, where it can activate cytokine transcription (32). As shown in Fig. 3B, I-κBα was degraded in 10 min post-LPS exposure, returning to baseline levels within 30 min in control cells. In the presence of patulin, however, I-κBα degradation was reduced between 10 and 30 min post-LPS treatment, suggesting that TLR4-dependent signal transduction was affected by patulin. To examine whether patulin disturbed NF-κB activation triggered by other stimuli, RAW264.7 cells were exposed to TNFα in the presence of patulin. As shown in Fig. 3, C and D, I-κBα degradation and IL-6 mRNA expression were not significantly suppressed by patulin treatment, suggesting that patulin disturbed NF-κB activation specifically induced by TLR but not the TNF receptor.
FIGURE 3.
NF-κB activation suppression and mitochondrial dysfunction may contribute to immunosuppressive activity of patulin. A, RAW264.7 cells were treated with patulin (1 μm or DMSO control) for 2 h and then exposed to LPS (100 ng/ml) for 30 min. Cells were then exposed to actinomycin D (Act D; 10 μg/ml) to block de novo mRNA transcription. Cells were then collected at various time points after actinomycin D treatment, and proinflammatory cytokine mRNA in treated cells was quantified using quantitative RT-PCR. B, RAW264.7 cells were pretreated with patulin (1 μm) or DMSO for 2 h and stimulated with LPS (100 ng/ml). Treated cells were collected at different time points post-LPS stimulation and subjected to immunoblotting with anti-IκBα and anti-tubulin antibodies. C, mouse TNFα (10 ng/ml) treatment was used to replace LPS stimulation as described in B. D, RAW264.7 cells were treated with patulin (or DMSO) for 2 h and then exposed to TNFα (10 ng/ml) for 6 h, and IL-6 mRNA expression was measured by quantitative RT-PCR. E, the effects of mitochondrial inhibitors, CCCP and oligomycin, on LPS-dependent IL-6 production were evaluated in RAW264.7 in parallel with patulin treatment (n = 6). F, CCCP and oligomycin (1 μm) were used to replace patulin for pretreatment before LPS stimulation as described in B. Error bars, S.E.
Mitochondrial Dysfunction Led to Immunosuppression
It has been demonstrated that patulin may have various effects on cells, including GSH depletion and mitochondrial depolarization (17). Because mitochondrial dysfunction occurs following patulin treatment, we considered the possibility that mitochondrial function might be important in TLR-dependent cytokine production. Therefore, the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which can depolarize the mitochondrial proton gradient, and oligomycin, an inhibitor of mitochondrial ATP synthase (33), were used to interfere with mitochondrial function. LPS/TLR4-dependent IL-6 production was inhibited by both CCCP and oligomycin treatment of RAW264.7 cells, suggesting that mitochondrial function is important for TLR-dependent innate immune response (Fig. 3E). The CCCP and oligomycin treatment did not affect cell viability significantly (Fig. 3E). Because CCCP and oligomycin did not affect LPS/TLR4-dependent NF-κB activation (Fig. 3F), we concluded that mitochondrial dysfunction might be involved in TLR-dependent cytokine production at the post-transcriptional level, such as translation.
Patulin Enhanced p62 Expression and Mitophagy Formation
Dysfunctional mitochondria are removed from cells by mitophagy through a priming process mediated by parkin, ubiquitin, and p62/sequestosome-1 (SQSTM1) (34, 35). Because mitochondrial dysfunction might be induced by patulin in the context of inflammation suppression, we examined whether p62 expression was affected by patulin treatment using quantitative RT-PCR. As shown in Fig. 4A, p62 mRNA expression was induced in RAW264.7 cells following patulin treatment and was further enhanced by LPS stimulation compared with levels observed in control cells. Replenishment of GSH level limited patulin-dependent p62 mRNA induction (Fig. 4A). p62 expression at the protein level was also induced by patulin treatment (Fig. 4B). Expression levels of the autophagy marker LC3 were also slightly increased following patulin treatment (Fig. 4B). To confirm that p62 was translocated to the mitochondria after patulin treatment, patulin-treated cells were subjected to mitochondrial fractionation, and p62 expression was examined by immunoblotting. As shown in Fig. 4C, p62 was present mainly in the cytosolic fraction of control cells, but following patulin treatment, some p62 translocated into the mitochondrial fraction, suggesting that patulin-dependent mitochondrial dysfunction might induce p62 expression and activation of mitophagy.
FIGURE 4.
Patulin induced p62 expression and mitophagy activation. A, RAW264.7 cells were pretreated with patulin (or DMSO) in combination with GSH (1 mm) for 2 h and exposed to LPS for 4 h. The expression of the p62 autophagy marker was evaluated by quantitative RT-PCR (n = 3). B, p62 protein expression was examined by immunoblotting with specific antibodies. C, RAW264.7 cells were treated with patulin for 8 h and subjected to mitochondrial fractionation. p62, AIP, and tubulin protein expression in cytosolic and mitochondrial fractions were evaluated by immunoblotting. D, autophagy inhibitor 3-MA was used in combination with patulin in LPS-stimulated RAW264.7 cells (n = 6). E, RAW264.7 cells were exposed to patulin (1 μm), CCCP (5 μm), oligomycin (5 μm), or patulin (1 μm) in combination with 3-MA (10 μm). p62 protein expression was examined by immunoblotting. Error bars, S.E.
Mitophagy represents a selective form of autophagy used to clear dysfunctional mitochondria following linkage of the adaptor protein p62 to damaged mitochondria forming the autophagosome (36). We therefore considered whether autophagy was directly involved with patulin-mediated immunosuppression. Using the autophagy inhibitor 3-methyladenine (3-MA) (37) to block patulin-dependent mitophagy, we next examined the impact of LPS stimulation on IL-6 production. As shown in Fig. 4D, 3-MA treatment partially restored patulin-induced immunosuppression, suggesting that mitophagy has a suppressive effect on inflammation. CCCP and oligomycin have been used to activate p62-dependent mitophagy (38), but patulin treatment was a more potent p62 activator (Fig. 4E).
The intracellular localization of p62 was examined by immunofluorescent staining demonstrating the presence of p62-containing speckles in the cytoplasm following patulin treatment (Fig. 5A). These p62-containing speckles gradually appeared in the cytoplasm as early as 2 h after patulin treatment, becoming larger after longer incubation times (Fig. 5B). Patulin-treated cells were also stained with the mitochondrial marker AIF to define the spatial correlation between p62 bodies and mitochondria. As shown in Fig. 5B, the staining intensity of the line plot showed that p62-containing dots were closely associated with AIF staining signals at 2 h after patulin treatment. At later time points (4 and 8 h after patulin treatment), some of the p62-containing bodies were still closely associated with AIF, but others were distant from the mitochondria. To test whether p62 expression is sufficient to trigger large p62-containing dot formation, p62-GFP expressing RAW264.7 cells were examined by a fluorescence microscope. As shown in Fig. 5C, p62-GFP formed small dots in cytoplasm and was accumulated in large complexes after 8-h LPS or patulin treatment. In contrast, GFP protein in RAW264.7 cells was distributed in both cytoplasm and nucleus and did not form any unique pattern upon LPS or patulin treatment. The results suggested that both LPS and patulin activate mitophagy to modulate inflammation.
FIGURE 5.
Patulin and LPS treatment induced p62-containing complex formation. A, RAW264.7 cells were exposed to patulin for 8 h and stained with Mitotracker dye and an anti-p62 antibody. Stained cells were examined by confocal microscopy. B, RAW264.7 cells exposed to patulin were stained for p62 and the mitochondrial marker AIF with specific antibodies, and the nuclei were counterstained with DAPI. The line plot quantifications of p62, AIF, and DAPI fluorescent intensities of individual cells were analyzed. C, p62-GFP- or GFP-expressing RAW264.7 cells were stimulated with LPS (100 ng/ml) or patulin (1 μm) for 8 h and stained with Mitotracker and DAPI to locate mitochondria and nuclei. The images were obtained by fluorescence microscopy.
Patulin Increased Nrf2 Accumulation and p62-TRAF6 Interaction
p62 transcription is regulated by transcription factor Nrf2 (39), and p62 can further promote Nrf2 accumulation through the inactivation of Keap1-dependent Nrf2 degradation as a positive loop (40). To understand how p62 was activated by patulin in RAW264.7 cells, the intracellular Nrf2 protein level after patulin treatment was examined by immunoblotting (Fig. 6A). Both p62 and Nrf2 were present in the detergent-insoluble fraction as described previously (41), and the accumulation of p62 and Nfr2 was enhanced upon LPS and patulin treatment, suggesting that Nrf2 accumulation might participate in patulin-dependent p62 expression. p62 is a multimodule adaptor protein participating in various signaling pathways by interacting with different binding partners and polyubiquitin chains (42). TRAF6, an ubiquitin E3 ligase involved in TLR activation, also interacts with p62 (43, 44). We speculated that p62-TRAF6 interaction might be enhanced by patulin treatment and, therefore, disturb TLR signaling. As shown in Fig. 6B, the interaction of p62-TRAF6 can be observed in Myc-tagged TRAF6- and p62-GFP-overexpressing 293T cells by co-immunoprecipitation and was further enhanced in the presence of patulin. In addition to exogenous p62-GFP and Myc-TRAF6 pulled down by GFP antibody, endogenous p62 and ubiquitin-positive molecules were also co-precipitated from the complex upon patulin treatment. When the proteasome activity was blocked by MG132, more p62 and ubiquitinated proteins in complex with TRAF6 were observed (Fig. 6B), suggesting that patulin-induced p62-TRAF6 complexes might be degraded by the ubiquitin-proteasome pathway. The increase of p62-TRAF6 interaction by patulin treatment was also observed in RAW264.7 cells after LPS stimulation (Fig. 6C).
FIGURE 6.
Patulin enhanced Nrf2 accumulation and p62-TRAF6 interaction. A, after patulin and LPS (100 ng/ml) treatment (16 h), RAW264.7 cells were lysed and separated into soluble and insoluble fractions. Nrf2 expression was examined by immunoblotting. B, 293T cells were transfected with plasmids to introduce p62-GFP and Myc-tagged TRAF6 expression for 48 h and further treated with patulin (1 μm) for 8 h. MG132 (10 μm) was added at 4 h before cell harvest. p62-TRAF6 interaction was evaluated by co-immunoprecipitation (IP). ←, exogenous p62-GFP; ◀, endogenous p62. C, patulin-pretreated RAW264.7 cells were stimulated with LPS for 6 h and with MG132 for 4 h. p62-TRAF6 interaction was evaluated by co-immunoprecipitation. D, RAW264.7 cells were pretreated with PMI (2–50 μm) for 2 h and then stimulated with various LPS (100 ng/ml) for 16 h in the presence of PMI (n = 6). IL-6 concentration, mitochondrial metabolic activity, and cytotoxicity were measured by an ELISA, an MTT assay, and a lactate dehydrogenase-releasing assay. Ub, ubiquitin. Error bars, S.E.
Patulin treatment induced mitochondrial dysfunction and p62-dependent mitophagy activation, and the presence of an autophagy blocker partially restored the immunosuppression activity of patulin. Therefore, a newly developed compound, p62-mediated mitophagy inducer (PMI) (45), was used to evaluate whether it is possible to modulate TLR-dependent cytokine production through direct activation of mitophagy. The p62 induction at the protein level was examined by immunoblotting, but p62 was not obviously induced in PMI-treated RAW267.4 cells from 2 to 50 μm (data not shown). Upon LPS treatment, PMI did not disturb cytokine production, but the MTT activity was significantly reduced (Fig. 6D). It is known that MTT activity is mainly attributable to mitochondrial enzymes. The reduction of MTT activity in PMI-treated cells could be caused by cytotoxicity or mitophagy activation-dependent mitochondria reduction. Therefore, lactate dehydrogenase release activity, a cell membrane integrity-based cytotoxicity, was used to examine the cytotoxicity of PMI. Even at 50 μm, PMI treatment did not cause too much toxicity in RAW267.4 cells (Fig. 6D, left), suggesting that PMI can effectively activate mitophagy but not influence cytokine production in RAW267.4 cells.
Patulin Inhibited RIG-I/MAVS-dependent IL-6 Production
We previously reported that hepatic expression of HCV RNA-dependent RNA polymerase (RdRp) activated the RIG-I/MAVS pathway, resulting in IL-6 and type I interferon production (13). Because our results suggested that patulin blocked TLR-dependent cytokine production, we considered whether patulin could also inhibit RIG-I/MAVS-dependent cytokine induction. Hepatocyte NeHepLxHT cells were infected with recombinant adenovirus Ad-del to drive expression of the HCV RdRp active mutant (FLAG-tagged Del mutant), resulting in IL-6 production. Patulin treatment did not disturb FLAG-tagged RdRp Del protein expression but reduced RIG-I/MAVS-dependent IL-6 production (Fig. 7A). Patulin-dependent immunosuppression on RIG-I/MAVS-dependent IL-6 production was restored by high GSH concentrations (1 mm) (Fig. 7B). p62 induction following patulin treatment was also observed in NeHepLxHT cells (Fig. 7C), and the immunosuppressive effects of patulin were reversed by 3-MA (Fig. 7D). Overall, these results suggested that p62-dependent mitophagy was involved in patulin-mediated immunosuppression of RIG-I/MAVS-dependent cytokine production.
FIGURE 7.
Patulin blocked RLR-dependent IL-6 production. A, human hepatocyte HeHepLxHT cells were infected with recombinant adenovirus Ad-del to drive hepatitis C virus RdRp active mutant expression with Ad-GFP as control. FLAG-tagged RdRp expression in cells infected for 16 h was detected by immunoblotting with anti-FLAG antibody (left). RdRp-dependent IL-6 production in the presence or absence of patulin treatment (1 μm) was measured by ELISA (n = 6). B, RdRp-dependent IL-6 induction in NeHepLxHT cells was evaluated in the presence of patulin (1 μm) and in combination with GSH (1 mm) (n = 6). C, the expression of the autophagy marker p62 in recombinant adenovirus-infected NeHepLxHT was evaluated by immunoblotting with specific antibodies. D, IL-6 induction of Ad-del-infected NeHepLxHT cells in the presence of patulin (1 μm) and 3-MA (10, 50, or 100 μm) was measured by ELISA (n = 3). p62, FLAG-tagged RdRp, GFP, and tubulin were detected by immunoblotting. E, HeHepLxHT cells were infected with Ad-del and treated with patulin (1 μm) or PMI (2, 10, or 50 μm). IL-6 production in the culture medium was measured by ELISA (n = 6). p62, FLAG-tagged RdRp, GFP, and tubulin expression in the infected cells at 16 h after infection were detected by immunoblotting (left). Error bars, S.E.
Whether PMI can modulate RLR-dependent cytokine production through direct activation of mitophagy was also examined. As shown in Fig. 7E, Ad-del infection-dependent IL-6 production was blocked in the presence of 10–50 μm PMI treatment in a dose-dependent manner. The p62 expression was enhanced by PMI treatment at 2 and 10 μm but not significantly at 50 μm (Fig. 6E). We suspect that protein synthesis might be affected by mitochondrial clearance after long term culture, or another feedback mechanism may exist.
Discussion
The present study showed that the mycotoxin patulin had immunosuppressive effects on TLR- and RLR-dependent cytokine production by decreasing GSH levels and causing mitochondrial dysfunction that resulted in p62 expression (Fig. 8). p62 accumulation enhanced mitophagy to clear damaged mitochondria and signaling molecules involved in TLR and RLR. Data presented in this report demonstrated how patulin modulated innate immune responses in addition to describing a critical role for mitochondria in TLR and RLR activation.
FIGURE 8.

Patulin disturbed TLR- and RLR-dependent cytokine production.
Mitochondria are an important organelle requisite for cell respiration and various metabolic processes. Furthermore, healthy mitochondria are essential for cytokine production that is suppressed during mitophagy. Therefore, maintaining mitochondrial homeostasis is critical for physiological cell functions. Damaged or depolarized mitochondria are removed by selective autophagy, namely mitophagy, through a parkin-ubiquitin-p62-mediated pathway. Patulin-depleted intracellular GSH and disturbed mitochondrial function resulted in the activation of p62-dependent mitophagy in both macrophages and non-hematopoietic cells (hepatocyte), and in the presence of high patulin concentrations (>50 μm), mitochondrial activity was significantly impaired in RAW264.7 cells. Because patulin induced GSH depletion and mitochondrial dysfunction, some consequential events, such as oxidative stress, endoplasmic reticulum stress, and apoptosis (18–22), may occur in the presence of a high concentration of patulin treatment (>5 μm). Therefore, all of the experiments in the study were limited at low concentration (1 μm) to prevent nonspecific effects caused by disturbed cell physiology.
Patulin disturbed LPS-dependent NF-κB activation and suppressed IL-6 mRNA expression levels by up to 50%. In contrast, IL-6 expression was completely blocked by patulin. We therefore speculated that patulin treatment might also affect cytokine production at the post-transcriptional level, such as protein translation. Patulin was shown to block protein synthesis in liver cells (46, 47). Previous work demonstrated that CCCP and oligomycin treatment reduced ATP levels and protein synthesis (48, 49). When mitochondrial dysfunction was induced, TLR-dependent cytokine production was partially decreased by CCCP and olivomycin and completely blocked by patulin. CCCP and oligomycin did not influence TLR-dependent NF-κB activity and did not induce significant p62 expression, suggesting that the CCCP and oligomycin might have affected cytokine protein synthesis levels.
Mitochondria play a role in innate immune responses elicited by either viruses or bacteria (50). Some signaling molecules associated with NLR, TLR, and RLR pathways, respectively, reside within mitochondria or translocate within mitochondria following ligand activation. For example, mitochondrial cardiolipin required for Nlrp3 inflammasome activation and mitochondrial dysfunction is a prerequisite for Nlrp3 inflammasome activation (51). TRAF6 translocation into the mitochondrial membrane following activation via the LPS/TLR4 pathway is critical for bacterial clearance (52). MAVS resides on the mitochondrial outer membrane and is the key adaptor protein for RLR receptors (53). These observations suggest that mitochondria are a key signaling platform required for the induction of innate immune responses and, as a consequence, represent a target for immunomodulatory interventions, as evidenced by the effects of patulin on TLR- and RLR-dependent cytokine expression.
In contrast to CCCP and oligomycin, patulin treatment induced GSH depletion and induced robust p62 expression, leading to inefficient TLR-dependent NF-κB activation and complete inhibition of cytokine production. We therefore speculated that activation of p62-dependent mitophagy might be a potential immunomodulatory target. Interestingly, p62 expression was also induced in RAW264.7 7 h after LPS stimulation, and 3-MA treatment also enhanced IL-6 production in control cells, suggesting that p62-dependent mitophagy provided negative feedback to TLR activation. The increase of TRAF6-p62 interaction and the enhancement of ubiquitinated protein aggregate formation for degradation after patulin treatment may be an important mechanism for the suppression of cytokine expression.
Considering that RLR/MAVS activate downstream signaling in mitochondria, mitophagy might directly reduce RLR signaling molecules and therefore affect innate immune responses, further supported by the observation that RLR-dependent cytokine production was sensitive to PMI treatment. Because TRAF6 is also involved in MAVS-dependent antiviral response (54), TRAF6-p62 interaction might also contribute to the inactivation of RLR/MAVS signaling. Hypothetically, PMI should be able to block cytokine production if p62 was induced by PMI to a level as high as it was induced by patulin in RAW264.7 cells. However, p62 expression was not significantly elevated in PMI-treated RAW264.7 cells even up to 50 μm. This may explain why PMI did not show an immunosuppressive activity in RAW264.7 cells even with a very low dose of LPS (1 ng/ml).
The immunomodulatory properties of patulin have demonstrated how mitochondria can be targeted to modulate the magnitude of innate immune responses. Through GSH depletion, mitochondrial dysfunction, and p62-mitophagy activation, patulin blocked both TLR- and RLR-dependent cytokine production. Because mitochondria are important for various metabolic processes, mitochondrial dysfunction may affect normal cellular physiology and may therefore not be an ideal approach for immunomodulation; however, activation of mitophagy is a natural negative feedback mechanism and may be considered as an approach to reducing excessive inflammation resulting from a cytokine storm that can be elicited following infection. More research will be required to test whether inducing mitophagy represents a viable immunomodulatory approach for the treatment of inflammation-associated diseases.
Experimental Procedures
Cell Culture and Virus Infection
Mouse macrophage RAW264.7 cells were maintained in RPMI medium (Gibco) with 10% FBS (Gibco). J774A.1 cells were cultured in DMEM high glucose medium (Thermo) supplemented with 10% FBS. Mouse peritoneal macrophages were collected from C57B/6 mice 4 days after mice were injected intraperitoneally with 3% Brewer thioglycollate (Creative Media Products, Chapel Hill, NC). All of the mouse-related experiments were conducted in compliance with the guidelines of the Laboratory Animal Center of the National Health Research Institutes and an approved animal protocol. Macrophages were maintained in DMEM/F-12 medium (Gibco) with 10% FBS (55) and allowed to attach for 6–8 h before they were used in TLR-related experiments. HEK293T cells were cultured in DMEM (Gibco) with 10% FBS. Human hepatocyte NeHepLxHT cells were maintained in DMEM/F-12 with 10% FBS supplemented with dexamethasone (40 ng/ml), insulin (2 μg/ml), and EGF (20 ng/ml) on collagen-coated plates. For RLR/MAVS signaling activation, NeHepLxHT cells were infected with Ad-GFP and Ad-Del (13) (multiplicity of infection = 10) in serum-free medium for 2 h and then maintained in normal growth medium.
Antibodies and Reagents
The mouse TLR1–8 agonist kit was purchased from InvivoGen (San Diego, CA). LPS (Escherichia coli serotype O110:B4 or 055:B5), patulin (P1639), GSH (G4251), CCCP (C2759), oligomycin (O4876), 3-MA (M9281), actinomycin D (A1410), and MG132 (C2211) were purchased from Sigma. The anti-IκBα (sc-371, Santa Cruz Biotechnology, Inc.) and IκBβ (sc-969, Santa Cruz Biotechnology), tubulin (Sigma), FLAG (Sigma), SQSTM1 (p62) (Genetex or Abcam), LC3I/II (Genetex), Nrf2 (sc-721, Santa Cruz Biotechnology), ubiquitin (sc-8017, Santa Cruz Biotechnology), Myc (sc-40, Santa Cruz Biotechnology), NLRP3 (AG-25B-0006, Adipogen), IL-1β (AF-401-NA, R&D Systems), TNF (AB2148P, Chemicon), caspase-1 (sc-514, Santa Cruz Biotechnology), and GAPDH (GTX627408, GeneTex) were used for immunoblotting. PMI (45) was synthesized and provided by Dr. Jinq-Chyi Lee.
ELISA, Nitric Oxide (NO) Production, MTT Assays, Lactate Dehydrogenase Assays, and GSH Measurement
To evaluate the immunosuppressive activity of patulin, RAW267.4 cells were stimulated with LPS (O110:B4, 1 or 0.1 μg/ml) in the presence of natural product for 16 h. Cell culture supernatants were collected for the determination of IL-6 and NO production. IL-6 and TNFα ELISA (eBioscience, San Diego, CA) and IFNβ (PBL, Piscataway, NJ) ELISA kits were used to measure cytokine as described by the manufacturers. NO was estimated from the accumulation of nitrite (NO2−), a stable end product of NO metabolism in the medium using the Griess reagent (Sigma). Equal amounts of culture supernatants and Griess reagent (1% sulfanilamide in 5% phosphoric acid and 0.1% α-naphthylethylenediamine dihydrochloride in distilled water) were mixed and incubated for 15 min at room temperature. The amount of violet-colored reaction product was measured at A540nm using an ELISA reader, and the nitrite concentration was calculated from a nitrite standard curve. To examine cell viability after the respective treatments, MTT (Sigma) stock solution (5 mg/ml) was added to the cells at a volume equal to 0.1× the original culture volume and incubated for 4 h. The resulting purple MTT formazan was solubilized with acidic isopropyl alcohol (0.1 n HCl in isopropyl alcohol), and the optical density was measured at an A570 nm using a spectrophotometer. A lactate dehydrogenase-releasing assay was performed with a cytotoxicity detection kit (Roche Applied Science) by following the manufacturer's instructions. For intracellular GSH measurement, cells were lysed to a concentration of 1 × 105/ml, and total GSH concentration was measured with a GSH/GSSG ratio detection kit (Abcam, Cambridge, UK).
IL-1β and Caspase-1 Activation Detection
J774A.1 cells pretreated with or without LPS (055:B5; 500 ng/ml) for 4 h were stimulated with 5 mm ATP for 30 min. The cells were then washed with PBS and incubated in low serum medium for an additional 4 h. Culture supernatants were harvested and mixed with one-tenth volume of 100% (w/v) trichloroacetic acid and were incubated for at least 30 min at −20 °C to precipitate proteins. The precipitated pellets were collected by centrifugation, washed once with 400 μl of cold acetone, and subjected to immunoblotting using antibodies against IL-1β and caspase-1 (p20).
Immunofluorescence Staining
For p62 and Mitotracker staining, cells were exposed to Mitotracker Red FM (Invitrogen) at 300 nm for 15 min and fixed in methanol for 10 min at −20 °C. Fixed cells were then stained with rabbit anti-p62 (Genetex), Alexa Fluor® 488 goat anti-rabbit IgG followed by DAPI staining (Invitrogen). Fluorescent images were captured using a Leica TCS SP5 confocal microscope. Colocalization of mitochondria and p62-containing aggregates was examined by double immunofluorescence staining with mouse anti-p62 (Abcam) and Alexa Fluor® 488 goat anti-mouse IgG followed by an incubation with rabbit anti-AIF (mitochondrial marker; Cell Signaling Technology, Danvers, MA) and Alexa Fluor® 594 goat anti-mouse IgG staining. Nuclei were stained using DAPI. Images captured using a Leica TCS SP5 confocal microscope were used to perform line plot quantification. To prepare GFP- and p62-GFP-expressing cells, RAW264.7 cells were transfected with pEGFP-N2 (Clontech) or SQSTM1/pCMV6-AC-GFP (Origene RG203214) by Lipofectamine 2000 (Invitrogen) and selected by Geneticin (Invitrogen). Images of GFP and p62-GFP expression patterns were captured by a Leica DM2500 fluorescence microscope.
Quantitative PCR
Total RNA was extracted from cultured RAW264.7 cells using TRIzol (Invitrogen) and used for cDNA synthesis with the SuperScript first strand synthesis system (Invitrogen). cDNAs were then used as templates for RT-PCR using the ΔΔCt method using cyclophilin A cDNA as an internal control. Primer sequences were as follows: cyclophilin A, 5′-atggtcaaccccaccgtgt-3′ (forward) and 5′-ttcttgctgtctttggaactttgtc-3′ (reverse); IL-6, 5′-gaggataccactcccaacagacc-3′ (forward) and 5′-aagtgcatcatcgttgttcataca-3′ (reverse) or 5′-cgctatgaagttcctctctgc-3′ (forward) and 5′-ttgggagtggtatcctctgtg-3′ (reverse); IFN-β, 5′-agctccaagaaaggacgaacat-3′ (forward) and 5′-gccctgtaggtgaggttgatct-3′ (reverse); TNF-α, 5′-gaccaggctgtcgctacatca-3′ (forward) and 5′-cgtaggcgattacagtcacgg-3′ (reverse); IL-1β, 5′-caaccaacaagtgatattctccatg-3′ (forward) and 5′-gatccacactctccagctgca-3′ (reverse).
Subcellular Fractionation
Mitochondria isolation was performed with the mitochondria isolation kit for cultured cells (Thermo Scientific, Rockford, IL) by following the instructions from the manufacturer. RAW264.7 cell lysate was also separated into detergent-soluble and -insoluble fractions by following the method described previously (41). In brief, soluble cell extracts were prepared in extraction buffer containing 50 mm Tris (pH 8.0), 150 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Triton X-100, and 1× protease inhibitor mixture (Roche Applied Science). After separated by centrifugation at 10,000 × g at 4 °C for 10 min, the detergent-insoluble fractions were solubilized in the buffer with 1% SDS with brief sonication. The detergent-soluble and -insoluble fractions (20 μg) were subjected to immunoblotting.
Co-immunoprecipitation
p62-TRAF6 interaction in RAW264.7 or 293T cells was examined by a co-immunoprecipitation approach, as described previously (40). 293T cells were transfected with pRK5-Myc-TRAF6 and SQSTM1/pCMV6-AC-GFP DNA with Lipofectamine 2000 (Invitrogen) to introduce Myc-TRAF6 and p62-GFP expression for 48 h. After patulin and MG132 treatment, RAW264.7 or 293T cells were lysed with lysis buffer (20 mm HEPES, 150 mm NaCl, 20% glycerol, 0.1% Nonidet P-40, 1 mm PMSF, 10 mm NaF, 1 mm Na3VO4, 10 mm N-ethylmaleimide, 1× protease inhibitor mixture) at 4 °C for 30 min. After brief sonication to shear chromosomal DNA, the lysate (0.5–1 mg) precleared with protein A and G magnetic beads (Invitrogen) was incubated with 1 μg of anti-GFP (sc-8334, Santa Cruz Biotechnology) or anti-TRAF6 (sc-7221, Santa Cruz Biotechnology) antibody at 4 °C overnight and further incubated with protein A and G beads for 6 h at 4 °C. After washing three times in lysis buffer, precipitates were eluted in 1× SDS sample buffer and subjected to immunoblotting.
Statistics
Statistical significance was determined using Student's t test, and p values <0.05 were considered significant.
Author Contributions
W. T. T., Y. C. L., and M. S. W. conducted most of the experiments. C. Y. L. initiated the compound screening platform. Y. P. K., Y. H. L., Y. T., and K. C. C. performed experiments for candidate compound validation. C. H. Y. and J. C. L. conducted PMI synthesis. T. H. C., L. C. H., and J. T. H. participated in experimental design, data interpretation, and manuscript preparation with G. Y. Y.
Acknowledgment
We thank the Core Instrument Center at the National Health Research Institutes, which provided confocal microscopy assistance.
This study was supported by National Health Research Institutes (NHRI) Grant IV-103-pp-21 and National Science Council Grant NSC 100-2320-B-400-006-MY3 from Taiwan (to G. Y. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
- TLR
- Toll-like receptor(s)
- RLR
- RIG-I-like receptor(s)
- NLR
- Nod-like receptor(s)
- HCV
- hepatitis C virus
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- 3-MA
- 3-methyladenine
- PMI
- p62-mediated mitophagy inducer
- RdRp
- RNA-dependent RNA polymerase.
References
- 1. Iwasaki A., and Medzhitov R. (2015) Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 3. Fukata M., and Abreu M. T. (2008) Role of Toll-like receptors in gastrointestinal malignancies. Oncogene 27, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castello G., Scala S., Palmieri G., Curley S. A., and Izzo F. (2010) HCV-related hepatocellular carcinoma: from chronic inflammation to cancer. Clin. Immunol. 134, 237–250 [DOI] [PubMed] [Google Scholar]
- 5. Horner S. M., and Gale M. Jr. (2013) Regulation of hepatic innate immunity by hepatitis C virus. Nat. Med. 19, 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J. Q., Jeelall Y. S., Ferguson L. L., and Horikawa K. (2014) Toll-like receptors and cancer: MYD88 mutation and inflammation. Front. Immunol. 5, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castello G., Costantini S., and Scala S. (2010) Targeting the inflammation in HCV-associated hepatocellular carcinoma: a role in the prevention and treatment. J. Transl. Med. 8, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. Y., Zhao L., and Hwang D. H. (2010) Modulation of pattern recognition receptor-mediated inflammation and risk of chronic diseases by dietary fatty acids. Nutr. Rev. 68, 38–61 [DOI] [PubMed] [Google Scholar]
- 9. He G., Yu G. Y., Temkin V., Ogata H., Kuntzen C., Sakurai T., Sieghart W., Peck-Radosavljevic M., Leffert H. L., and Karin M. (2010) Hepatocyte IKKβ/NF-κB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell 17, 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naugler W. E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A. M., and Karin M. (2007) Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317, 121–124 [DOI] [PubMed] [Google Scholar]
- 11. Rebouissou S., Amessou M., Couchy G., Poussin K., Imbeaud S., Pilati C., Izard T., Balabaud C., Bioulac-Sage P., and Zucman-Rossi J. (2009) Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature 457, 200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Capone F., Costantini S., Guerriero E., Calemma R., Napolitano M., Scala S., Izzo F., and Castello G. (2010) Serum cytokine levels in patients with hepatocellular carcinoma. Eur. Cytokine Netw. 21, 99–104 [DOI] [PubMed] [Google Scholar]
- 13. Yu G. Y., He G., Li C. Y., Tang M., Grivennikov S., Tsai W. T., Wu M. S., Hsu C. W., Tsai Y., Wang L. H., and Karin M. (2012) Hepatic expression of HCV RNA-dependent RNA polymerase triggers innate immune signaling and cytokine production. Mol. Cell 48, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyd E. M. (1944) Patulin. Can. Med. Assoc. J. 50, 159. [PMC free article] [PubMed] [Google Scholar]
- 15. Fliege R., and Metzler M. (2000) Electrophilic properties of patulin: N-acetylcysteine and glutathione adducts. Chem. Res. Toxicol. 13, 373–381 [DOI] [PubMed] [Google Scholar]
- 16. Burghardt R. C., Barhoumi R., Lewis E. H., Bailey R. H., Pyle K. A., Clement B. A., and Phillips T. D. (1992) Patulin-induced cellular toxicity: a vital fluorescence study. Toxicol. Appl. Pharmacol. 112, 235–244 [DOI] [PubMed] [Google Scholar]
- 17. Barhoumi R., and Burghardt R. C. (1996) Kinetic analysis of the chronology of patulin- and gossypol-induced cytotoxicity in vitro. Fundam. Appl. Toxicol. 30, 290–297 [DOI] [PubMed] [Google Scholar]
- 18. Zhang B., Peng X., Li G., Xu Y., Xia X., and Wang Q. (2015) Oxidative stress is involved in Patulin induced apoptosis in HEK293 cells. Toxicon 94, 1–7 [DOI] [PubMed] [Google Scholar]
- 19. Wu T. S., Liao Y. C., Yu F. Y., Chang C. H., and Liu B. H. (2008) Mechanism of patulin-induced apoptosis in human leukemia cells (HL-60). Toxicol. Lett. 183, 105–111 [DOI] [PubMed] [Google Scholar]
- 20. Kwon O., Soung N. K., Thimmegowda N. R., Jeong S. J., Jang J. H., Moon D. O., Chung J. K., Lee K. S., Kwon Y. T., Erikson R. L., Ahn J. S., and Kim B. Y. (2012) Patulin induces colorectal cancer cells apoptosis through EGR-1 dependent ATF3 up-regulation. Cell. Signal. 24, 943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fribley A. M., Cruz P. G., Miller J. R., Callaghan M. U., Cai P., Narula N., Neubig R. R., Showalter H. D., Larsen S. D., Kirchhoff P. D., Larsen M. J., Burr D. A., Schultz P. J., Jacobs R. R., Tamayo-Castillo G., et al. (2011) Complementary cell-based high-throughput screens identify novel modulators of the unfolded protein response. J. Biomol. Screen. 16, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boussabbeh M., Ben Salem I., Prola A., Guilbert A., Bacha H., Abid-Essefi S., and Lemaire C. (2015) Patulin induces apoptosis through ROS-mediated endoplasmic reticulum stress pathway. Toxicol. Sci. 144, 328–337 [DOI] [PubMed] [Google Scholar]
- 23. Doi K., and Uetsuka K. (2014) Mechanisms of mycotoxin-induced dermal toxicity and tumorigenesis through oxidative stress-related pathways. J. Toxicol. Pathol. 27, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marin M. L., Murtha J., Dong W., and Pestka J. J. (1996) Effects of mycotoxins on cytokine production and proliferation in EL-4 thymoma cells. J. Toxicol. Environ. Health 48, 379–396 [DOI] [PubMed] [Google Scholar]
- 25. Luft P., Oostingh G. J., Gruijthuijsen Y., Horejs-Hoeck J., Lehmann I., and Duschl A. (2008) Patulin influences the expression of Th1/Th2 cytokines by activated peripheral blood mononuclear cells and T cells through depletion of intracellular glutathione. Environ. Toxicol. 23, 84–95 [DOI] [PubMed] [Google Scholar]
- 26. Yu H. B., and Finlay B. B. (2008) The caspase-1 inflammasome: a pilot of innate immune responses. Cell Host Microbe 4, 198–208 [DOI] [PubMed] [Google Scholar]
- 27. Pelegrin P., Barroso-Gutierrez C., and Surprenant A. (2008) P2X7 receptor differentially couples to distinct release pathways for IL-1β in mouse macrophage. J. Immunol. 180, 7147–7157 [DOI] [PubMed] [Google Scholar]
- 28. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., and Lee J. O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 29. Kang J. Y., Nan X., Jin M. S., Youn S. J., Ryu Y. H., Mah S., Han S. H., Lee H., Paik S. G., and Lee J. O. (2009) Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31, 873–884 [DOI] [PubMed] [Google Scholar]
- 30. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., and Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 31. Chuang T. H., Lee J., Kline L., Mathison J. C., and Ulevitch R. J. (2002) Toll-like receptor 9 mediates CpG-DNA signaling. J. Leukoc. Biol. 71, 538–544 [PubMed] [Google Scholar]
- 32. Akira S., Uematsu S., and Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 33. Perry S. W., Norman J. P., Barbieri J., Brown E. B., and Gelbard H. A. (2011) Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. BioTechniques 50, 98–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., and Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 35. Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y. S., Kimura M., Sato S., Hattori N., Komatsu M., Tanaka K., and Matsuda N. (2010) p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15, 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Randow F., and Youle R. J. (2014) Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seglen P. O., and Gordon P. B. (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allen G. F., Toth R., James J., and Ganley I. G. (2013) Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 14, 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jain A., Lamark T., Sjøttem E., Larsen K. B., Awuh J. A., Øvervatn A., McMahon M., Hayes J. D., and Johansen T. (2010) p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285, 22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J. Y., and Ozato K. (2009) The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-κB activity. J. Immunol. 182, 2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fujita K., Maeda D., Xiao Q., and Srinivasula S. M. (2011) Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 1427–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moscat J., Diaz-Meco M. T., and Wooten M. W. (2007) Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 43. Häcker H., Vabulas R. M., Takeuchi O., Hoshino K., Akira S., and Wagner H. (2000) Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J. Exp. Med. 192, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanz L., Diaz-Meco M. T., Nakano H., and Moscat J. (2000) The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. East D. A., Fagiani F., Crosby J., Georgakopoulos N. D., Bertrand H., Schaap M., Fowkes A., Wells G., and Campanella M. (2014) PMI: a DeltaPsim independent pharmacological regulator of mitophagy. Chem. Biol. 21, 1585–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hatey F., and Moulé Y. (1979) Protein synthesis inhibition in rat liver by the mycotoxin patulin. Toxicology 13, 223–231 [PubMed] [Google Scholar]
- 47. Arafat W., and Musa M. N. (1995) Patulin-induced inhibition of protein synthesis in hepatoma tissue culture. Res. Commun. Mol. Pathol. Pharmacol. 87, 177–186 [PubMed] [Google Scholar]
- 48. Breitbart H. (1981) Effect of ionophores and metabolic inhibitors on protein synthesis in rabbit reticulocytes. Biochim. Biophys. Acta 656, 160–166 [DOI] [PubMed] [Google Scholar]
- 49. Otero M. J., and Carrasco L. (1984) Action of oligomycin on cultured mammalian cells: permeabilization to translation inhibitors. Mol. Cell. Biochem. 61, 183–191 [DOI] [PubMed] [Google Scholar]
- 50. West A. P., Shadel G. S., and Ghosh S. (2011) Mitochondria in innate immune responses. Nat. Rev. Immunol. 11, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iyer S. S., He Q., Janczy J. R., Elliott E. I., Zhong Z., Olivier A. K., Sadler J. J., Knepper-Adrian V., Han R., Qiao L., Eisenbarth S. C., Nauseef W. M., Cassel S. L., and Sutterwala F. S. (2013) Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. West A. P., Brodsky I. E., Rahner C., Woo D. K., Erdjument-Bromage H., Tempst P., Walsh M. C., Choi Y., Shadel G. S., and Ghosh S. (2011) TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seth R. B., Sun L., Ea C. K., and Chen Z. J. (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 54. Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y. T., Sun L., and Chen Z. J. (2013) MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2, e00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang X., Goncalves R., and Mosser D. M. (2008) The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]