Abstract
Outside of well characterized model eukaryotes, relatively little is known about the translocons that transport proteins across the two membranes that surround the mitochondrion. Apicomplexans are a phylum of intracellular parasites that cause major diseases in humans and animals and are evolutionarily distant from model eukaryotes such as yeast. Apicomplexans harbor a mitochondrion that is essential for parasite survival and is a validated drug target. Here, we demonstrate that the apicomplexan Toxoplasma gondii harbors homologues of proteins from all the major mitochondrial protein translocons present in yeast, suggesting these arose early in eukaryotic evolution. We demonstrate that a T. gondii homologue of Tom22 (TgTom22), a central component of the translocon of the outer mitochondrial membrane (TOM) complex, is essential for parasite survival, mitochondrial protein import, and assembly of the TOM complex. We also identify and characterize a T. gondii homologue of Tom7 (TgTom7) that is important for parasite survival and mitochondrial protein import. Contrary to the role of Tom7 in yeast, TgTom7 is important for TOM complex stability, suggesting the role of this protein has diverged during eukaryotic evolution. Together, our study identifies conserved and modified features of mitochondrial protein import in apicomplexan parasites.
Keywords: intracellular trafficking, mitochondria, parasite, protein import, Toxoplasma gondii
Introduction
Mitochondria arose through the endosymbiotic acquisition of an α-proteobacterium, likely in a single event that occurred early in eukaryotic evolution (1, 2). Most known extant eukaryotes retain a mitochondrion or homologous organelle (3). Similar to their bacterial antecedents, most mitochondria retain a genome that encodes for some proteins. However, most proteins that function in the mitochondrion are nucleus-encoded and must post-translationally target across the two membranes that surround the mitochondrion. Proteins targeted to the mitochondrion contain either N-terminal presequences or internal targeting signals. These targeting signals interact with various protein translocons in the inner and outer membranes to ensure protein translocation across the mitochondrial membranes and into the appropriate compartment of the organelle.
Mitochondrial protein import is a core organellar function that was a critical early step in the evolution of mitochondria from α-proteobacteria (4). The last common ancestor of extant eukaryotes harbored a mitochondrion and also bore a functional mitochondrial protein import apparatus. However, it is becoming increasingly apparent that mitochondrial protein import machinery has diversified through the course of eukaryotic evolution.
Mitochondrial import has been most extensively studied in the yeast Saccharomyces cerevisiae (5). Translocation across the outer membrane is mediated by the translocon of the outer mitochondrial membrane (TOM) 3 complex. The TOM complex represents the entry point into the organelle for most mitochondrial proteins. In yeast, the TOM complex includes the β-barrel protein Tom40, which forms the central pore of the complex (6). The TOM complex also includes the receptor proteins Tom20 and Tom70, three small Tom proteins (Tom5, Tom6, and Tom7) that function in regulating TOM complex assembly and function, and Tom22, a single-pass transmembrane protein that has several functions. The cytosolic N-terminal region of yeast Tom22 functions as a receptor domain that interacts with proteins as they enter from the cytosol (7). The transmembrane domain is critical for assembling the TOM complex into a higher order structure (8). The intermembrane space-localized C-terminal domain of Tom22 interacts with presequence-containing proteins as they pass through the TOM complex, an interaction that is critical for translocation of these proteins to the translocase of the inner membrane (9).
The outer mitochondrial membrane contains several β-barrel proteins. The targeting of these proteins involves translocation into the intermembrane space through the TOM complex, and subsequent insertion into the outer membrane by an outer membrane insertase called the sorting and assembly machinery (SAM) complex, the central component of which is known as Sam50 (10).
The presequence translocase, also known as the translocon of the inner mitochondrial membrane 23 (TIM23) complex, translocates presequence-containing proteins across the inner membrane. Tim23 forms the pore through this membrane, whereas Tim50 functions as a receptor for proteins as they translocate from the TOM complex (11, 12). The presequence translocase can recruit a motor complex called the presequence-translocase associated motor (PAM) to drive ATP-dependent translocation of proteins into the mitochondrial matrix. The central component of the PAM complex is a mitochondrial Hsp70 that associates with J-domain proteins such as Pam18 (13). Upon translocation into the matrix, the presequence is proteolytically cleaved by a mitochondrial processing peptidase (MPP) to yield the mature protein (14), which can then fold and carry out its function.
Many mitochondrial proteins lack N-terminal presequences and instead harbor internal signals to direct them to the mitochondrion. Mitochondrial solute carrier proteins are inner membrane proteins that fall into this category. Mitochondrial carrier proteins enter the mitochondrion through the TOM complex and then interact with small Tim proteins such as Tim9 and Tim10 in the intermembrane space. These function to deliver carrier proteins to the TIM22 complex, which inserts carrier proteins into the inner membrane. The core component of this insertase is the protein Tim22 (15).
Although the mechanisms of mitochondrial import are well characterized in yeast and related organisms such as animals, this is less true of other eukaryotic lineages. The core components of the mitochondrial import machinery, including the TOM, TIM23, TIM22, PAM, and SAM complexes, are present in plants (16). There are, however, several major differences between plants and yeast, most notably features of the TOM complex. Plants lack homologues of the receptor proteins Tom70 and Tom20 and have evolved alternative receptor proteins (17, 18). Additionally, the plant Tom22 homologue is truncated at the N terminus and may not function as a presequence receptor (18, 19).
Another phylum where the molecular mechanisms of mitochondrial import have been functionally examined is the trypanosomatids, a group of parasites that include Trypanasoma brucei, the causative agent of African sleeping sickness. The outer membrane translocon of trypanosomatids is termed the archaic translocase of the outer mitochondrial membrane (ATOM) complex and appears different from the TOM complex in yeast. The central component of the ATOM complex is a β-barrel protein called Atom40 (20). There is debate as to whether Atom40 is homologous to Tom40 (21, 22). Studies have identified other ATOM protein components, including Atom69, Atom46, Atom36, Atom14, Atom12, and Atom11 (23, 24). These components lack clear homologues in yeast, although hidden Markov model analysis suggests that Atom14 is homologous to Tom22 (21). Trypanosomatid genomes harbor a single TIM translocase protein homologue, which is essential for import of presequence-containing proteins (25, 26).
Apicomplexans are a phylum of intracellular parasites. They include important human parasites such as the Plasmodium species, the causative agents of malaria, and Toxoplasma gondii, a parasite that causes congenital disease in unborn children and encephalitis in immunocompromised people. Most apicomplexans harbor a mitochondrion, which is the target of major and emerging anti-apicomplexan drugs such as atovaquone, endochin-like quinolones, and triazolopyrimidines (27–29). Studies of the mitochondrial protein import machinery of apicomplexans have been limited to comparative genomic approaches (30–32). These have identified putative homologues of the core components of mitochondrial import, including members of the TOM, TIM23, TIM22, PAM, and SAM complexes.
The TOM complex of apicomplexans appears divergent from that found in yeast. Apicomplexan genomes lack identifiable homologues to TOM complex receptors such as Tom70 and Tom20. Like plants, the N terminus of Plasmodium Tom22 appears truncated (19). Additionally, these comparative approaches have not identified Tom7 homologues in apicomplexan genomes, making apicomplexans one of the few lineages where a TOM complex is present that appears to lack Tom7 (19, 21).
The last common ancestor of apicomplexans and yeast contained a mitochondrion and must have had a means of targeting nucleus-encoded proteins into this organelle. In this paper, we ask the following. What features of this mitochondrial import apparatus have been conserved from the last common ancestor? What new features have arisen? We use T. gondii as our model system to perform the first broad functional analysis of the mitochondrial protein import machinery in Apicomplexa. In particular, we focus on the TOM complex and assay the role of two TOM complex components in mitochondrial import and TOM complex biogenesis. These studies provide insights into conserved and novel functions of mitochondrial import machinery across their evolution, and they provide a platform for future studies of mitochondrial import in an evolutionarily interesting and medically important group of organisms.
Results
Identity and Localization of Putative Mitochondrial Import Proteins in T. gondii
To identify candidate mitochondrial import proteins, we performed Basic Local Alignment Search Tool (BLAST) searches, querying the T. gondii genome with characterized mitochondrial import proteins from the yeast S. cerevisiae, the plant Arabidopsis thaliana, and the trypanosomatid T. brucei. Matches were confirmed through reciprocal BLAST searches against the T. gondii genome. The results of these searches are summarized in supplemental Table S1. We identified homologues of major subunits from each of the translocases and insertases involved in protein import into the mitochondrion. These include homologues of Tom40, Sam50, Tim23, Tim17, Tim50, Hsp70, Mge1, Tim44, Pam18, Tim22, and a host of small Tims. We also identified a T. gondii homologue of Oxa1, a protein that functions in the insertion of inner membrane proteins encoded on the mitochondrial genome (supplemental Table S1) (33).
Some notable proteins lacked detectable homologues in T. gondii. This was particularly the case for the TOM complex, for which we were unable to identify candidates for receptor proteins (e.g. Tom70 and Tom20) or small Tom proteins (e.g. Tom5 and Tom7). Additionally, we were unable to identify components of the SAM and carrier insertase complexes beyond the central Sam50 and Tim22 subunits. We were also unable to identify a homologue of MIA40, a protein with a central role in the import of small Tim proteins into the intermembrane space (34).
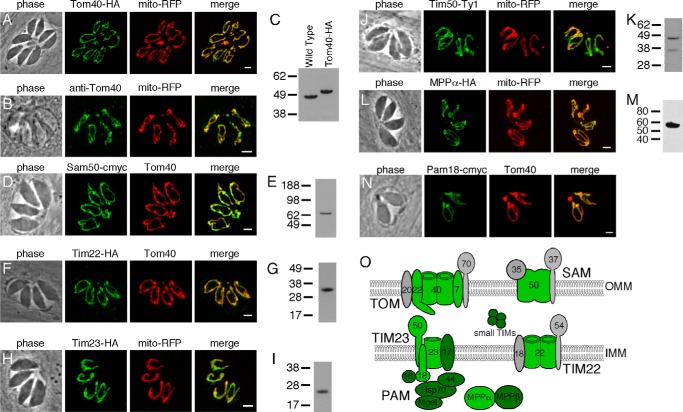
To determine whether these candidate mitochondrial import proteins localized to the mitochondrion, we epitope-tagged select members of each protein complex. We first tagged TgTom40, the putative import pore in the outer membrane, and demonstrated co-localization with mitochondrially targeted RFP (Fig. 1A). Additionally, we generated polyclonal rabbit antibodies against TgTom40. Immunofluorescence assays using this antibody supported the mitochondrial localization of TgTom40 (Fig. 1B). Analysis by Western blotting revealed that TgTom40 is a protein with a mass of 48 kDa (Fig. 1C). Through co-localization with either Tom40 or mitochondrial RFP, and associated Western blottings, we demonstrated mitochondrial localization and protein expression for the following proteins: TgSam50 (Fig. 1, D and E), consistent with the mitochondrial localization of this protein reported in a recent study (35); TgTim22 (Fig. 1, F and G); TgTim23 (Fig. 1, H and I); TgTim50 (Fig. 1, J and K); and TgMPPα (Fig. 1, L and M). We were unable to generate a parasite strain that stably expressed epitope-tagged TgPam18. Instead, we transiently overexpressed c-Myc-tagged TgPam18 in T. gondii parasites and performed an immunofluorescence assay. This revealed mitochondrial localization of TgPam18 (Fig. 1N). Taken together, these data indicate that T. gondii homologues of each of the major mitochondrial protein translocases and insertases localize to the mitochondrion (Fig. 1O).
FIGURE 1.
Localization of putative mitochondrial import proteins in T. gondii. A, B, D, F, H, J, L, and N, target proteins are labeled in green, and in all cases they co-localize with a mitochondrial marker (either RFP fused to the mitochondrial matrix-targeting leader sequence of TgHsp60 or anti-Tom40 antibodies; red). Scale bars, 2 μm. C, E, G, I, K, and M, Western blottings of each protein are shown to the right of each panel. For the TgTom40 Western blotting, protein extracts from wild-type (WT) and the HA-tagged TgTom40 strain were probed with anti-TgTom40 antibodies. Our transfection experiments did not produce tagged TgPam18 in sufficient quantity to detect by Western blotting. O, schematic depicting the putative composition of the TOM, SAM, TIM23, PAM, and TIM22 complexes in T. gondii. Proteins localized in this study are depicted in light green, and proteins identified bioinformatically are depicted in dark green, and notable proteins without homologues in T. gondii are depicted in gray. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane.
Localization of the Tom22 Homologue of T. gondii
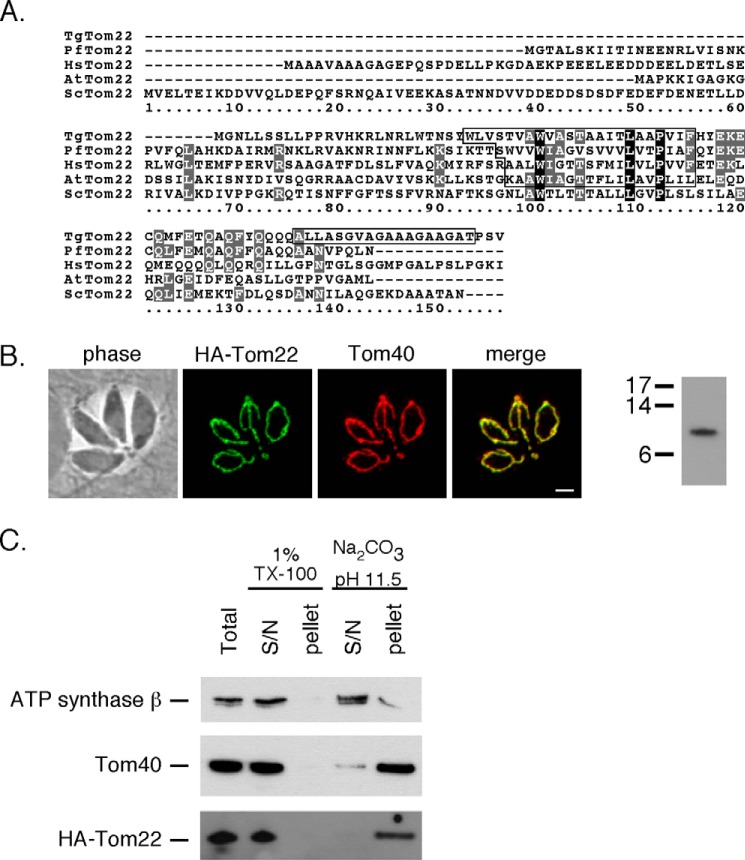
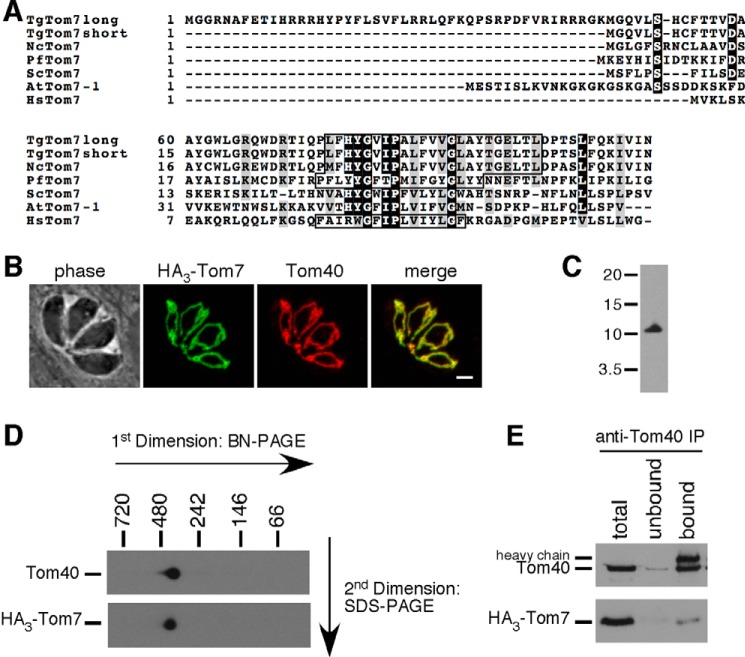
Our bioinformatics survey (supplemental Table S1) identified a T. gondii homologue of Tom22 that we called TgTom22. We performed an alignment of TgTom22 with Tom22 homologues from Plasmodium falciparum, yeast, humans, and plants (Fig. 2A). This alignment revealed that the N terminus of TgTom22 is truncated compared with yeast and human Tom22, as noted previously for Tom22 homologues in plants and Plasmodium (19). We identified two predicted transmembrane domains in TgTom22, one in an equivalent position to the transmembrane domain predicted for other Tom22 proteins and the second at the very C terminus of the protein (Fig. 2A). Sequence conservation among putative Tom22 homologues is generally poor, with only three identical residues found in all the proteins presented in the alignment, all three of which are found in the predicted transmembrane domain.
FIGURE 2.
TgTom22 is a mitochondrial membrane protein with homology to Tom22 proteins from other eukaryotes. A, multiple protein sequence alignment of TgTom22 with homologues from P. falciparum (PfTom22), humans (HsTom22), A. thaliana (AtTom22), and S. cerevisiae (ScTom22). Transmembrane domains of each protein (predicted using TMHMM) are highlighted by a box. B, immunofluorescence assay demonstrating co-localization of HA-Tom22 protein (green) with the mitochondrion marker Tom40 (red). Scale bar, 2 μm. A Western blotting analysis of HA-Tom22, probed with anti-HA antibodies, reveals a protein with a mass of 10 kDa. C, Western blotting analysis of HA-Tom22-expressing parasites, where proteins were extracted in 1% Triton X-100 (TX-100) or in sodium carbonate (Na2CO3) at pH 11.5. HA-Tom22 partitions into the pellet phase of the Na2CO3 extraction, much like the integral mitochondrial membrane protein Tom40 and unlike the soluble mitochondrial matrix protein ATP synthase β subunit, which partitions predominantly into the supernatant (S/N) phase.
To localize TgTom22, and to facilitate subsequent functional analysis of this protein, we introduced an N-terminal hemagglutinin (HA) epitope-tagged TgTom22 into the TATi strain of T. gondii. This exogenous version of TgTom22 was placed under the control of an anhydrotetracycline (ATc)-regulated promoter. We termed the resultant strain rTom22/eTom22, to indicate the presence of both regulatable and endogenous copies of TgTom22. We then knocked out the native Tom22 locus, verifying successful knock-out by PCR analysis (Fig. 3, A and B). The resultant strain was termed rTom22/Δtom22 and expressed only the ATc-regulatable HA-tagged TgTom22. Immunofluorescence analysis of this strain revealed that HA-Tom22 co-localizes with TgTom40, indicating that TgTom22 is a mitochondrial protein (Fig. 2B). Western blotting revealed that HA-Tom22 is 10 kDa in mass, close to the predicted mass of 12 kDa for the HA-tagged protein. To test whether TgTom22 is a transmembrane protein, we extracted T. gondii proteins in the rTom22/Δtom22 cell line with either the detergent Triton X-100 or sodium carbonate (Na2CO3) at pH 11.5. We determined that the HA-Tom22 protein localizes to the soluble fraction in Triton X-100 extraction but to the membrane fraction upon Na2CO3 extraction (Fig. 2C). This is similar to the predicted integral membrane protein TgTom40 and distinct from the β-subunit of ATP synthase, a predicted soluble protein. We conclude that TgTom22 is a mitochondrial integral membrane protein.
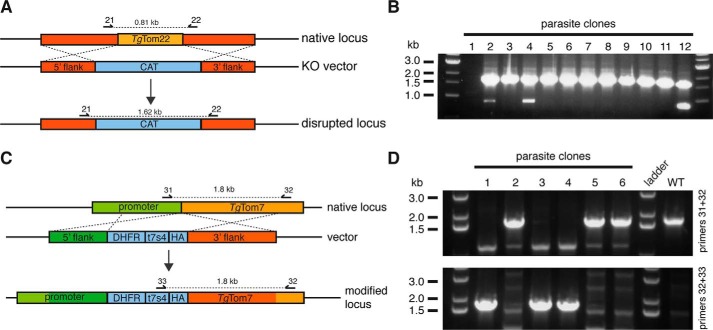
FIGURE 3.
Generation of mutant parasite strains. A, schematic describing knock-out of the native TgTom22 gene. The native TgTom22 locus (orange) was replaced with a chloramphenicol acetyltransferase (CAT)-selectable marker (blue), in a background strain expressing an ATc-regulatable copy of TgTom22 (data not shown). B, PCR screen of 12 parasite clones transfected with the TgTom22 knock-out construct with primers 21 and 22. The primer pair will produce a band of 0.8 kb if the native gene is present and a band of 1.6 kb when the regulatable copy of TgTom22 is present. Clones 3 and 5–11 lack the native gene, indicating successful knock-out of the TgTom22 native locus in these parasites. C, schematic describing replacement of the native TgTom7 promoter with an ATc-regulated promoter (t7s4) and 5′ HA tag. The TATi/Δku80 parasite strain was transfected with a vector containing 5′ and 3′ flanks of the TgTom7 gene, the t7s4 ATc-regulated promoter, a 5′ HA tag, and a pyrimethamine-resistant T. gondii DHFR-selectable marker. D, PCR screen of six parasite clones transfected with the promoter replacement vector and screened with primer pairs 31 and 32 to detect the native TgTom7 locus (top) or primer pairs 32 and 33 to detect the modified locus (bottom). DNA from wild-type (WT) TATi/Δku80 parasites was used as a control. Clones 1, 3, and 4 lack the band corresponding to the presence of the native gene but harbor the band corresponding to the modified locus, indicating successful promoter replacement at the TgTom7 locus in these parasites.
Characterization of the TOM Complex of T. gondii
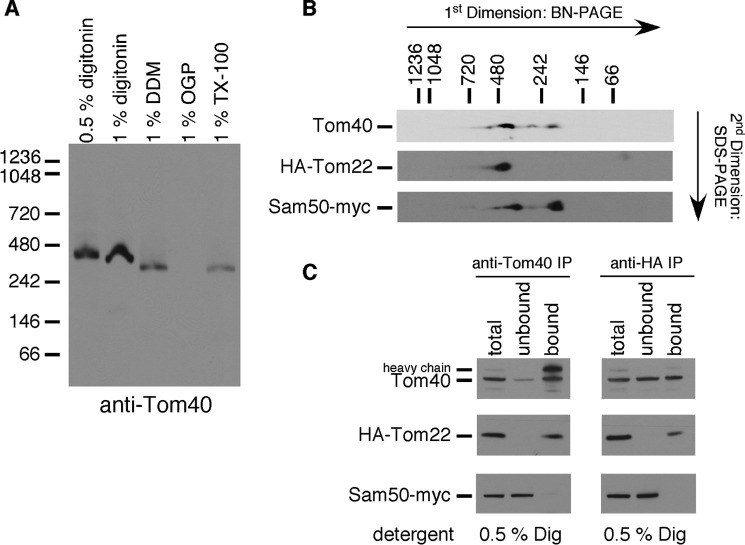
As a first step to characterizing the TOM complex of T. gondii, we performed blue native-PAGE (BN-PAGE) on rTom22/Δtom22 strain T. gondii parasites, probing Western blottings with the TgTom40 antibody. When solubilized in either 0.5 or 1% digitonin, TgTom40 localizes to a protein complex of around 400 kDa (Fig. 4A). When solubilized in 1% dodecyl maltoside or 1% Triton X-100, TgTom40 localizes to a slightly smaller complex of around 325 kDa. The TgTom40 complex appeared insoluble in octyl β-d-glucopyranoside. We conclude that the TgTom40 protein is part of an ∼400-kDa protein complex in the mitochondrion of T. gondii, which is similar in mass to characterized TOM complexes measured by BN-PAGE in yeast (36). We refer to this complex as the T. gondii TOM complex.
FIGURE 4.
T. gondii harbors a TOM complex of ∼400 kDa that contains TgTom40 and TgTom22. A, Western blotting probing TgTom40 proteins separated by BN-PAGE extracted in 0.5% (w/v) digitonin, 1% (w/v) digitonin, 1% (w/v) dodecyl maltoside (DDM), 1% (w/v) octyl β-d-glucopyranoside (OGP), and 1% (w/v) Triton X-100 (TX-100). B, Western blotting probing TgTom40, HA-Tom22, and Sam50-cMyc proteins separated by BN-PAGE in a first dimension and SDS-PAGE in a second dimension. C, Western blottings of immunoprecipitated parasite proteins extracted in 0.5% digitonin and pulled down with anti-Tom40 antibodies (left) or anti-HA antibodies (right). Western blottings were probed for TgTom40 (top), HA-Tom22 (middle), and Sam50-cMyc (bottom). Total proteins, unbound proteins, and bound proteins were loaded for each experiment, and each lane contains proteins extracted from an equivalent number of parasites. The heavy chain of anti-Tom40 antibodies was detectable in the anti-Tom40 Western blotting.
We have identified one other putative outer membrane protein involved in import, namely TgSam50, which is predicted to function in the outer membrane insertase complex. We hypothesized that TgTom22 localizes to the TOM complex, whereas TgSam50 localizes to a separate complex. To test this, we expressed a c-Myc epitope-tagged TgSam50 in the rTom22/Δtom22 cell line. We initially performed one-dimensional BN-PAGE and were unable to identify either HA-Tom22 or TgSam50-Myc protein, likely because the epitope tags on these proteins were inaccessible to antibodies when in the complete complex. 4 We therefore performed two-dimensional BN-PAGE, separating protein complexes in the second dimension using SDS-PAGE. These studies revealed that TgTom40 is predominantly found in a 400-kDa complex that corresponds in mass to the TOM complex we observed in one-dimensional BN-PAGE (Fig. 4B). Lesser amounts of TgTom40 were observed in smaller complexes of around 300 and 200 kDa (Fig. 4B). HA-Tom22 localizes exclusively in a 400-kDa complex that corresponds in mass to the 400-kDa complex containing TgTom40. TgSam50-c-Myc localizes in complexes of 200 and 350 kDa (Fig. 4B).
To further test the composition of the T. gondii TOM complex, we performed co-immunoprecipitation experiments on the TgSam50-c-Myc/rTom22/Δtom22 cell line. We solubilized T. gondii proteins in 0.5% digitonin and immunoprecipitated TgTom40 and all interacting proteins using anti-TgTom40 antibodies. HA-Tom22, but not TgSam50, co-immunoprecipitated with TgTom40 antibodies (Fig. 4C).
Next, we performed pulldowns of HA-Tom22-interacting proteins using anti-HA antibodies. These studies revealed that a large fraction of TgTom40, but not TgSam50, associated with HA-Tom22 (Fig. 4C). Together, these results indicate that TgTom40 and TgTom22 form part of a core 400-kDa TOM complex that does not include TgSam50.
TgTom22 Is Essential for Parasite Growth
Having generated an ATc-regulatable TgTom22 cell line, we sought to determine whether TgTom22 was essential for parasite survival. We first measured knockdown of HA-Tom22 by growing parasites for 0–2 days in ATc. This revealed that HA-Tom22 protein levels are much reduced 1 day after the addition of ATc and are undetectable after 2 days (Fig. 5A).
FIGURE 5.
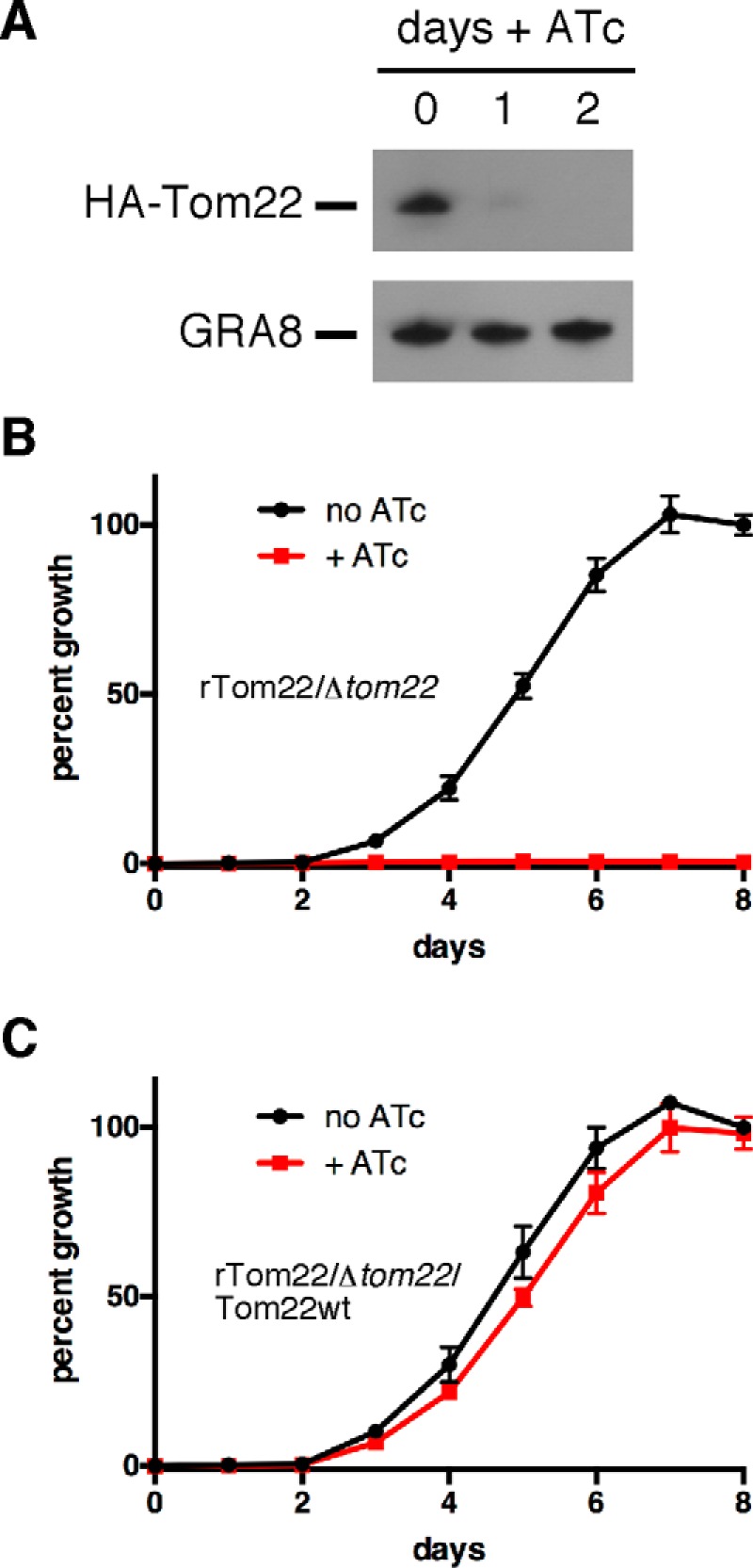
TgTom22 is essential for parasite growth. A, Western blotting of rTom22/Δtom22 parasites grown for 0–2 days on ATc and probed with anti-HA antibodies to detect HA-Tom22 protein or anti-GRA8 antibodies as a loading control. B and C, fluorescence growth assays of TgTom22 knockdown (rTom22/Δtom22) (B) and TgTom22-complemented (rTom22/Δtom22/Tom22WT) (C) strains. Parasites were grown in the absence (black) or presence (red) of ATc for 0–8 days. Error bars represent the standard deviation of three technical replicates.
To determine whether TgTom22 is required for growth of T. gondii parasites, we introduced a tandem tomato red fluorescent protein into the rTom22/Δtom22 cell line. This allows daily quantification of parasite growth as a function of well fluorescence in a 96-well plate (37, 38). rTom22/Δtom22 parasites cultured in the presence of ATc showed no detectable growth across the 8-day experiment (Fig. 5B). These data indicate that TgTom22 is critical for parasite growth. To determine whether the observed defect in growth in the presence of ATc is dependent on TgTom22, we complemented the rTom22/Δtom22 cell line with constitutively expressed TgTom22, producing the strain we termed rTom22/Δtom22/Tom22WT. Parasite growth in this strain was equivalent with or without ATc (Fig. 5C), consistent with the observed growth effect in mutant parasites resulting solely from loss of TgTom22.
TgTom22 Is Essential for Mitochondrial Protein Import
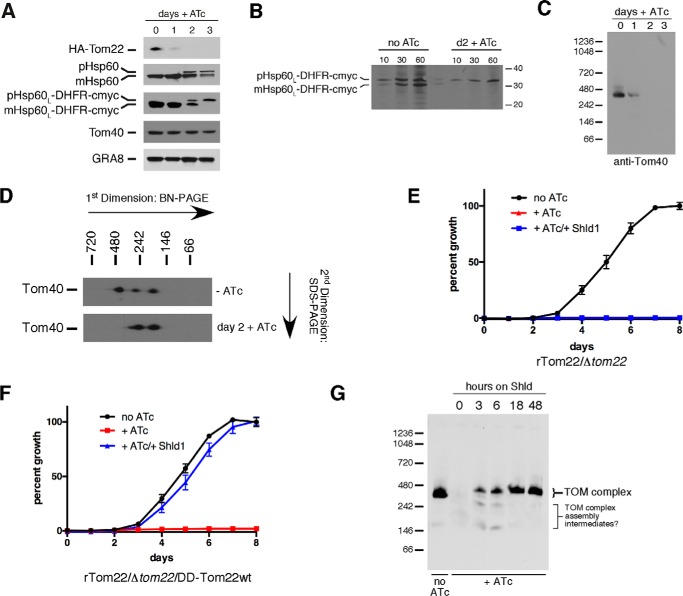
We next wanted to determine the function(s) of TgTom22. We hypothesized that TgTom22 plays a role in mitochondrial protein import. Proteins targeted to the matrix of mitochondria harbor an N-terminal presequence that is removed upon entry into the matrix. Presequence cleavage therefore serves as a robust measure for mitochondrial protein import. To monitor this, we fused the presequence leader of the mitochondrial protein TgHsp60 (Hsp60L) to a c-Myc-tagged mouse DHFR (mDHFR) reporter protein (mDHFR has been used extensively in studies of mitochondrial import in other systems) and introduced this Hsp60L-mDHFR-cMyc protein construct into rTom22/Δtom22 parasites. We grew parasites for 0–3 days on ATc and measured the abundance of a range of proteins by Western blotting. As shown previously, HA-Tom22 was undetectable after 2 days on ATc (Fig. 6A). In the presence of HA-Tom22, we observed a single Hsp60L-mDHFR-cMyc band that corresponds in mass to the mature (processed) form of this protein, where the presequence has been removed (Fig. 6A). Upon loss of HA-Tom22, we noted an accumulation of the precursor form of Hsp60L-mDHFR-cMyc, as well as the precursor form of the native TgHsp60 protein (Fig. 6A). Notably, Tom40 levels remain unchanged with the loss of HA-Tom22.
FIGURE 6.
TgTom22 is essential for mitochondrial protein import and functions in assembly of the TOM complex. A, Western blottings of HA-Tom22 parasites expressing c-Myc-tagged mouse DHFR fused to the mitochondrion-targeting leader sequence of TgHsp60 (Hsp60L-DHFR-cMyc). Parasites were grown for 0–3 days in ATc, and protein extracts were probed with anti-HA, anti-Hsp60, anti-cMyc, anti-TgTom40, and anti-GRA8 antibodies as a loading control. Precursor and mature forms of the Hsp60 and Hsp60L-DHFR-cMyc protein are indicated. This experiment was performed in three biological replicates, and a representative experiment is shown. B, import of radiolabeled Hsp60L-DHFR-cMyc protein into the mitochondrion of rTom22/Δtom22 parasites grown for 0 (no ATc) or 2 (d2 +ATc) days on ATc. Intracellular parasites were incubated for 10, 30, and 60 min in growth medium containing [35S]cysteine and [35S]methionine. Proteins were extracted in detergent, and Hsp60L-DHFR-cMyc protein was purified by immunoprecipitation before separation by SDS-PAGE. Precursor (pHsp60L-DHFR-cMyc) and mature (mHsp60L-DHFR-cMyc) isoforms of the Hsp60L-DHFR-cMyc protein were detected. This experiment was performed in two biological replicates, and a representative experiment is shown. C, anti-Tom40 Western blotting of proteins extracted from rTom22/Δtom22 parasites grown for 0–3 days in ATc and separated by BN-PAGE. D, Western blotting of proteins extracted from rTom22/Δtom22 parasites grown in the absence (top) or presence of ATc for 2 days (bottom). Proteins were separated by BN-PAGE in a first dimension and SDS-PAGE in a second dimension and then probed with anti-Tom40 antibodies. E and F, fluorescence growth assays of rTom22/Δtom22 parasites (E) and rTom22/Δtom22 (F) complemented with DD-tagged wild-type TgTom22 (rTom22/Δtom22/DD-Tom22WT) and grown in the absence (black) or presence (red) of ATc or in the presence of ATc and Shld1 (blue). Note that the red +ATc line in E is not visible behind the blue +ATc/+Shld1 line. Error bars represent the standard deviation of three technical replicates. G, anti-Tom40 Western blotting of proteins extracted from rTom22/Δtom22/DD-Tom22WT parasites separated by BN-PAGE. Parasites were grown in the absence of ATc or in the presence of ATc for 2 days. Parasites grown in ATc were also grown for 0–48 h in Shld1.
As a more sensitive measure of mitochondrial protein import, we performed a radioactive labeling experiment. We incubated rTom22/Δtom22 parasites expressing Hsp60L-mDHFR-cMyc for 10, 30, and 60 min in medium containing [35S]methionine and [35S]cysteine. We then performed immunoprecipitations to purify Hsp60L-mDHFR-cMyc protein and separated proteins by SDS-PAGE. In the absence of ATc, we observed two protein bands, corresponding in mass to the precursor and mature forms of Hsp60L-mDHFR-cMyc (Fig. 6B). Only a small amount of mature Hsp60L-mDHFR-cMyc was present at the 10-min time point in the absence of ATc, but this increased upon longer incubation in the radioactive medium, consistent with protein synthesis followed by precursor processing upon mitochondrial import. In contrast, we observed only the precursor form of Hsp60L-mDHFR-cMyc in parasites grown for 2 days on ATc, indicative of an absence of mitochondrial protein import in these parasites. We conclude that TgTom22 is critical for mitochondrial protein import.
TgTom22 Is Critical for TOM Complex Assembly
We next sought to determine the functional roles of TgTom22. We first asked whether TgTom22 has a role in TOM complex assembly. We performed BN-PAGE on proteins extracted from rTom22/Δtom22 parasites grown for 0–3 days on ATc. We observed a marked decrease in the 400-kDa TOM complex after 1 day on ATc and complete loss of this complex after 2 days (Fig. 6C), concomitant with loss of the HA-Tom22 protein.
The apparent absence of the TOM complex in this experiment was puzzling, because TgTom40 protein levels remain unchanged upon loss of HA-Tom22 (Fig. 6A). We wondered whether loss of HA-Tom22 upon the addition of ATc resulted in a complex that masked the TgTom40 protein to antibodies in this one-dimensional BN-PAGE approach. We performed two-dimensional BN-PAGE on rTom22/Δtom22 parasites grown in the absence of ATc or in the presence of ATc for 2 days. In the absence of ATc, most Tom40 was present in the 400-kDa TOM complex, whereas some was also present in ∼200- and ∼300-kDa complexes (Fig. 6D), as observed previously (Fig. 4B). Upon loss of HA-Tom22, we could detect Tom40, but this was exclusively in the 200- and 300-kDa protein complexes.
To further test the role of TgTom22 in TOM complex assembly, we fused an FKBP-based destabilization domain (DD) (39) to the N terminus of wild-type TgTom22 and introduced this into rTom22/Δtom22 parasites to derive the parasite strain rTom22/Δtom22/DD-Tom22WT. The stability of DD-tagged proteins can be controlled through the addition of the small molecule Shld1, allowing for rapid control of protein levels (39). We monitored the growth of both rTom22/Δtom22 and rTom22/Δtom22/DD-Tom22WT parasites using the previously described fluorescence growth assay, incubating parasites either in the absence of ATc and Shld1, in the presence of ATc and absence of Shld1, or in the presence of both ATc and Shld1 (to turn off expression of regulatable HA-Tom22 and to stabilize DD-Tom22WT). Growth of rTom22/Δtom22 parasites was negligible in the presence of ATc, regardless of the presence of Shld1 (Fig. 6E). In the rTom22/Δtom22/DD-Tom22WT parasite strain, however, parasite growth in the presence of ATc was restored to near wild-type levels upon the addition of Shld1 (Fig. 6F). This indicates that DD-Tom22WT can complement the rTom22/Δtom22 mutant.
We grew rTom22/Δtom22/DD-Tom22WT parasites for 0 or 48 h in ATc. For the parasites grown for 48 h in ATc, we added Shld1 0, 3, 6, 18, or 48 h before harvesting. We then performed BN-PAGE on proteins extracted from these cell lines. In the absence of ATc, TgTom40 is found in the ∼400-kDa TOM complex, whereas the addition of ATc disrupts the 400-kDa complex in the absence of Shld1 (0 h; Fig. 6G). Three hours after the addition of Shld1, we see re-formation of the 400-kDa TOM complex, and the abundance of the TOM complex increases with increased time on Shld1 (Fig. 6G). Three and 6 h after the addition of Shld1, we observed some TgTom40 in smaller protein complexes of ∼150 and ∼300 kDa, which may represent intermediates in TOM complex assembly. We conclude that TgTom22 has a critical, and likely direct, role in TOM complex assembly.
Apicomplexan Parasites Have a Tom7 Homologue That Is Important for TOM Complex Assembly and Parasite Growth
T. gondii harbors a TOM complex of around 400 kDa that contains TgTom40 and TgTom22. We reasoned that other proteins must be part of this complex as well. Previous bioinformatics approaches have been unable to identify homologues of small TOM complex proteins such as Tom7 in apicomplexans, even though these are present in the genomes of many other eukaryotes (19, 21). Given the limited primary sequence conservation, we reasoned that homology searches might be insufficient to identify Tom7 homologues in these parasites. From an alignment of Tom7s from multiple organisms, we noted that the conserved feature of all of these is a G(X)2P(X)5G motif in the single transmembrane domain. We searched the genome of the apicomplexan parasite P. falciparum for predicted proteins that harbor this motif. We then screened these candidates further for proteins of less than 15 kDa that harbored a single transmembrane domain. This produced one candidate, a hypothetical protein annotated as PF3D7_0823700. Using this as a query sequence, we identified a homologous protein encoded on the T. gondii genome (TGME49_210255) and termed this TgTom7. We performed 5′-rapid amplification of cDNA ends (5′-RACE) to identify the open reading frame and establish its sequence.
We constructed an alignment of the putative TgTom7 protein with Tom7 homologues in other organisms, revealing the presence of the G(X)2P(X)5G in the transmembrane domain, but little further conservation (Fig. 7A). The TgTom7 open reading frame contains two potential start codons. Alignments of TgTom7 sequence with the homologous protein in Neospora caninum, a close relative of T. gondii, revealed no conservation upstream of the second start codon (Fig. 7A). Furthermore, subsequent analysis revealed that the longer form of TgTom7 exhibits similar localization and mutant phenotypes to the short form, suggesting that the region upstream of the second start codon is dispensable for function.4 Together, these data suggest that the second codon may represent the true start codon in this gene.
FIGURE 7.
TgTom7 is a mitochondrial protein and a constituent of the TOM complex. A, multiple protein sequence alignment of the long and short isoforms of TgTom7 with homologues from N. caninum (NcTom7), P. falciparum (PfTom7), S. cerevisiae (ScTom7), A. thaliana (AtTom7-1), and humans (HsTom7). Transmembrane domains of each protein (predicted using TMHMM) are highlight by a box. B, immunofluorescence assay demonstrating co-localization of HA-tagged version of the short isoform of TgTom7 (HA3-Tom7; green) with the mitochondrial marker Tom40 (red). Scale bar, 2 μm. C, Western blotting analysis of HA3-Tom7, probed with anti-HA antibodies, reveals a protein of 11 kDa. D, Western blotting of proteins extracted from the TATi/Δku80/rTom7 parasite strain and separated by BN-PAGE in a first dimension and SDS-PAGE in a second dimension. Blots were probed with anti-Tom40 antibodies (top) and anti-HA antibodies to detect HA3-Tom7 protein (bottom). E, Western blottings of parasite proteins extracted from TATi/Δku80/rTom7 strain parasites in 0.5% digitonin, immunoprecipitated with anti-Tom40 antibodies, and separated by SDS-PAGE. Blots were probed with anti-Tom40 antibodies (top) and anti-HA antibodies (bottom). Total proteins, unbound proteins, and bound proteins were loaded for each experiment, and each lane contains proteins extracted from an equivalent number of parasites. The heavy chain of anti-Tom40 antibodies was detectable in the anti-Tom40 Western blotting.
To localize TgTom7, we fused a region encoding a 3×HA tag to the 5′ end of the open reading frame to enable detection of the resultant protein (which we termed HA3-Tom7). To facilitate subsequent functional analysis, we also replaced the native promoter of this gene with an ATc-regulated promoter in the TATi/Δku80 parasite strain (41) to generate the parasite strain we termed TATi/Δku80/rTom7. We confirmed successful integration of the 5′-HA tag and ATc-regulated promoter through PCR analysis (Fig. 3, C and D). We performed an immunofluorescence assay, which demonstrated co-localization of HA3-Tom7 with TgTom40 (Fig. 7B), indicating that TgTom7 is a mitochondrial protein. Western blotting analysis indicated that HA3-Tom7 is 11 kDa in mass (Fig. 7C), correlating to the predicted molecular mass of 11 kDa for HA3-Tom7.
Two-dimensional BN-PAGE analysis revealed that TgTom7 exists in a protein complex of around 400 kDa, corresponding in mass to the TOM complex (Fig. 7D). Curiously, and in contrast to our previous analyses performed on the rTom22/Δtom22 strain of parasites, we did not observe the ∼300- and ∼200-kDa protein complexes in the TgTom40 Western blotting. This may suggest that the rTom22/Δtom22 mutant has some defects in TOM complex assembly even in the absence of ATc, possibly due to the presence of the N-terminal HA tag or resulting from slight modification of native expression levels in using the ATc-regulated promoter.
We performed a co-immunoprecipitation with antibodies against TgTom40. Most of the HA3-Tom7 protein appeared in the bound fraction (Fig. 7E), indicating that HA3-Tom7 is part of a complex with TgTom40. We attempted the reciprocal experiment of co-immunoprecipitating with anti-HA-coupled beads. We were unable to purify either HA3-Tom7 or TgTom40,4 suggesting that the HA tag on TgTom7 may be hidden from antibodies within the complex.
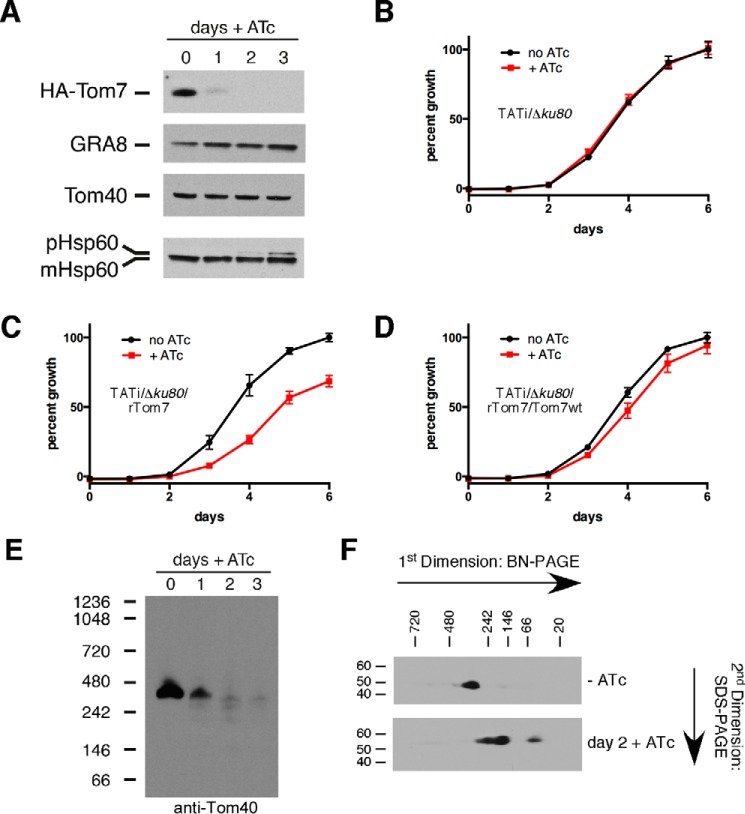
We next asked whether TgTom7 was important for parasite growth. First, we determined whether the addition of ATc resulted in knockdown of the HA3-Tom7 protein. We cultured parasites in ATc for 0–3 days and measured protein abundance. HA3-Tom7 protein was undetectable after 2 days growth on ATc (Fig. 8A). To test whether TgTom7 is important for parasite growth, we introduced a tandem tomato red fluorescent protein into the rTom22/Δtom22 cell line. We performed fluorescence growth assays on tomato-expressing parental (TATiΔku80) and TATi/Δku80/rTom7 parasites. These revealed that TATi/Δku80/rTom7 strain parasites alone exhibited growth impairment in the presence of ATc (Fig. 8, B and C).
FIGURE 8.
TgTom7 is important for parasite growth, mitochondrial protein import, and TOM complex assembly. A, Western blotting of proteins extracted from TATi/Δku80/rTom7 grown for 0–3 days in ATc. Blots were probed with antibodies against HA3-Tom7, TgGRA8 (loading control), TgTom40, and TgHsp60. Precursor and mature forms of TgHsp60 are indicated. This experiment was performed in three biological replicates, and a representative experiment is shown. B–D, fluorescence growth assays of TATi/Δku80 parental control (B). C, TgTom7 knockdown (TATi/Δku80/rTom7) and D, TgTom7 complemented (TATi/Δku80/rTom7/Tom7wt) strains. Parasites were grown in the absence (black) or presence (red) of ATc for 0 to 6 days. Error bars represent the standard deviation of three technical replicates. E, anti-Tom40 Western blotting of proteins extracted from TATi/Δku80/rTom7 parasites grown for 0–3 days in ATc and separated by BN-PAGE. F, Western blotting of proteins extracted from TATi/Δku80/rTom7 parasites grown in the absence (top) or presence of ATc for 2 days (bottom). Proteins were separated by BN-PAGE in a first dimension and SDS-PAGE in a second dimension, and probed with anti-Tom40 antibodies.
To ascertain whether this growth phenotype resulted solely from down-regulation of TgTom7 expression, we complemented the TATi/Δku80/rTom7 strain with TgTom7 expressed from a constitutive promoter to produce a cell line termed TATi/Δku80/rTom7/Tom7WT. We then introduced a tandem tomato red fluorescent protein and performed fluorescent growth assays. This revealed that this complemented strain was fully restored in growth (Fig. 8D), suggesting that the growth defect we observed results entirely from the decrease in TgTom7 expression.
We next wanted to determine whether TgTom7 has a role in mitochondrial protein import. Attempts to generate a TATi/Δku80/rTom7 strain expressing the mitochondrial Hsp60L-mDHFR-cMyc construct were unsuccessful. Instead, we performed Western blotting on TATi/Δku80/rTom7 parasites grown for 0–3 days in ATc and probed with antibodies against mitochondrial TgHsp60. When Tom7 was present (day 0), we observe a single TgHsp60 protein species that corresponds to the mature mitochondrially localized TgHsp60 protein (Fig. 8A). Two days after the addition of ATc and concomitant with TgTom7 knockdown, we observed the appearance of a higher molecular mass species, corresponding to presequence-containing TgHsp60. This band increased in abundance after 3 days on ATc (Fig. 8A). Notably, the abundance of TgTom40 remains unchanged upon the addition of ATc. These data are consistent with a role for TgTom7 in mitochondrial protein import that does not depend on regulating the stability or turnover of TgTom40.
Given the role of TgTom22 in TOM complex assembly, we wanted to determine whether TgTom7 was also important for this process. We performed BN-PAGE to measure TOM complex assembly upon HA3-Tom7 knockdown. These studies revealed loss of the TOM complex concomitant with HA3-Tom7 knockdown (Fig. 8E). We performed two-dimensional BN-PAGE on TATi/Δku80/rTom7 parasites grown in the absence of ATc or in the presence of ATc for 2 days. In the absence of ATc, Tom40 was present in the ∼400-kDa TOM complex. Upon HA3-Tom7, knockdown, Tom40 was present in smaller complexes of ∼240, 150, and 50 kDa. These observations are consistent with a role for TgTom7 in TOM complex formation and/or assembly.
Discussion
The means to import proteins into mitochondria must have arisen early in mitochondrial and therefore eukaryotic evolution (42). Less clear are the origins of the proteins and protein complexes involved in this process and how these have changed over time. Recent studies of mitochondrial import in plants and trypanosomes have revealed that canonical yeast features such as an outer membrane TOM complex and separate inner membrane TIM23 and TIM22 complexes are either extensively modified or lacking from these organisms (17, 20, 21, 23, 26). We demonstrate here that T. gondii harbors mitochondrially localized candidate proteins from all the major mitochondrial translocons and insertases (Fig. 1O; supplemental Table S1). This implies that the last common ancestor of apicomplexans and yeast had homologues of these mitochondrial import components and that these likely functioned in the mitochondrion. The fact that homologues of these complexes are found in other eukaryotes, such as plants and the amoeba Dictyostelium (43), suggests that the common ancestor of all these major eukaryotic lineages harbored homologues of these proteins.
Comparative genomics implies but cannot truly assess conservation of protein function. We therefore set about characterizing select aspects of mitochondrial protein import in T. gondii, with a particular view to understanding whether the function of candidate translocon components has changed across evolution. We focused our attention on the putative T. gondii TOM complex, which our comparative genomic approach indicated had interesting differences from the TOM complex of yeast.
We demonstrate that the TOM complex of T. gondii is ∼400 kDa in mass (Fig. 4), corresponding to the mass of the core TOM complex in yeast but is considerably larger than the ∼230-kDa complex found in plants (44, 45). The T. gondii TOM complex contains TgTom40, TgTom22, and TgTom7 and excludes TgSam50. Instead, TgSam50 exists predominantly in a protein complex of ∼200 kDa, similar in mass to the SAM complex of yeast (46, 47), although a larger complex of ∼350 kDa is also apparent.
We were unable to identify homologs to known TOM receptor proteins in apicomplexans. One striking feature of TOM complex receptors is that no two major eukaryotic lineages yet examined have identifiable homologous receptor proteins. Tom20 and Tom70 are unique to opisthokonts; plant Tom20 and mtToc64 are unique to plants; ATOM69 and ATOM46 are unique to trypanosomatids; and no homologues of these proteins are apparent in apicomplexans. This suggests that TOM receptor proteins were either absent from the last common ancestor of these eukaryotes or that novel receptors have replaced the original ones. One theory of mitochondrial evolution posits that the evolution of receptor proteins on the TOM complex will have created a powerful selective advantage (42). An intriguing possibility is that the evolution of TOM complex receptors occurred multiple times in separate lineages and that this contributed to the radiation, and subsequent success, of the extant eukaryotic lineages. Identifying and characterizing the TOM complex receptors in apicomplexans are now the major priorities in this area.
We demonstrate that loss of TgTom22 leads to defects in maturation of mitochondrial matrix proteins, consistent with a role for TgTom22 in mitochondrial protein import. Studies from yeast have identified a key role for Tom22 in TOM complex assembly (8). Our data suggest TgTom22 has a direct role in TOM complex assembly, mirroring the role of Tom22 in yeast. In yeast, loss of Tom22 results in disassembly of the TOM complex to form an ∼100-kDa complex. In T. gondii, TgTom22 knockdown leads to the formation of two major protein complexes of 200 and 300 kDa (Fig. 6D). It is unclear whether these are simply disassembled TOM complexes or precursors of the TOM complex, although it is interesting to note that SAM50 forms complexes of ∼200 kDa (Fig. 4B), suggesting the smaller of the complexes could be a SAM-TOM complex intermediate. Also unclear is whether these depleted TgTom40-containing complexes can function in import. Regardless, the conserved role of Tom22 in the TOM complex assembly between yeast and apicomplexans suggests that one role of Tom22 in the last common ancestor of these lineages was assembly of the TOM complex. This points to an ancient and conserved role for Tom22 in TOM complex assembly.
In yeast, Tom7 is a non-essential protein that negatively regulates TOM complex assembly (48, 49). Loss of Tom7 in yeast therefore promotes TOM complex assembly. Previously, apicomplexans were thought to be one of only a few TOM complex-containing eukaryotic lineages that lacked a Tom7 homologue (19). We revisited this and were able to identify a candidate Tom7 homologue in both T. gondii and P. falciparum. We could demonstrate that TgTom7 was indeed a component of the core TOM complex in these parasites and important for mitochondrial protein import.
In contrast to yeast, loss of TgTom7 leads to a growth defect in T. gondii parasites and impairment of TOM complex assembly. We observe TOM complex dissociation concomitantly with TgTom7 knockdown, suggesting the two processes are directly linked. Nevertheless, we were unable to generate a tightly regulated DD-tagged TgTom7 cell line, which precluded a more robust test of this hypothesis. Notably, although knockdown of both TgTom7 and TgTom22 results in dissociation of the TOM complex, loss of TgTom22 leads to a considerably stronger growth defect (cf. Figs. 5B and 8C), and a qualitatively more severe defect in mitochondrial protein import (cf. Figs. 6A and 8A). This suggests that protein import is not entirely ablated upon TOM complex disassembly. It also suggests that, in addition to its role in TOM complex assembly, TgTom22 has other roles in mitochondrial protein import. The C-terminal intermembrane space domain of Tom22 in other organisms is critical for translocating proteins from the TOM complex to the presequence translocase (9), and it is conceivable that TgTom22 has a role in this process.
Apicomplexan Tom7 is the first non-opisthokont Tom7 that has been functionally characterized. The contrasting role of Tom7 between apicomplexans and opisthokonts suggests that, even though it is conserved as part of the “core” TOM complex, the functions of Tom7 have diverged substantially during eukaryotic evolution. This is in contrast to our findings with Tom22, where the role of Tom22 in TOM complex assembly appears conserved. From our functional analyses, we can conclude that the TOM complex of apicomplexan parasites contains both conserved and unique features when compared with the TOM complex of yeast.
Experimental Procedures
Parasite Culture and Growth Assays
Parasites were grown in human foreskin fibroblasts using Dulbecco's modified Eagle's medium supplemented with 1% fetal calf serum and antibiotics. Strains used included RH, TATi, Δku80, and TATi/Δku80 (41, 50, 51). All parasite strains described in this work were cloned by limiting dilution. Where required, we added ATc at a final concentration of 0.5 μg/ml and Shld1 at a final concentration of 0.75 μm. Fluorescence growth assays were performed in optical bottom 96-well plates, as described previously (37, 38), and read using a FLUOstar Optima fluorescence plate reader (BMG Labtech).
Plasmid Construction
To determine the localization of the candidate mitochondrial import-related proteins, we introduced epitope tags at the 3′ end of either the endogenous locus of the genes encoding these proteins or of the open reading frame of candidate genes expressed from constitutive promoters. To localize TgTom40, we amplified the 3′ region with primers 1 and 2 (Table 1) and inserted this into the vector pHA3.LIC.DHFR (a kind gift from Michael White, University of South Florida) by ligation-independent cloning (LIC), as described previously (50), before linearization with EcoRV, transfection into Δku80 strain parasites, and selection on pyrimethamine as described (52). To localize TgSam50, we amplified the open reading frame with primers 3 and 4, digested the resulting product with BglII and AvrII, ligated into the vector pBTM3 (53), transfected into parasites, and selected on phleomycin as described (54). To localize TgTim22, we amplified the 3′ region with primers 5 and 6, digested the resulting product with BglII and AvrII, ligated into pHH vector, a modified version of the pgCM3 vector described previously (55) that has a selectable marker for mycophenolic acid selection. The vector was linearized with AflII, transfected into parasites, and selected on mycophenolic acid as described (52). To localize TgTim23, we amplified the 3′ region with primers 7 and 8, digested the resulting product with BglII and AvrII, and ligated into the pBH vector a modified version of the pHH vector that contains a phleomycin resistance marker. This vector was linearized with NcoI, transfected into parasites, and selected on phleomycin. To localize TgTim50, we amplified the 3′ region with primers 9 and 10, inserted the product into the vector pTy.LIC.DHFR, a modified version of pHA3.LIC.DHFR where the HA tag has been replaced by a 1xTy1 tag, through LIC. The resulting vector was digested with NsiI, transfected into parasites, and selected on pyrimethamine. To localize TgPam18, we amplified the open reading frame with primers 11 and 12, digested the resulting product with BglII and AvrII, and ligated into the vector pBTM3. We transfected the resulting vector into parasites and observed transient expression after 1 day. To localize TgMPPα, we amplified the 3′ region with primers 13 and 14, inserted this into pHA3.LIC.DHFR by LIC, linearized with NcoI, transfected into parasites, and selected on pyrimethamine.
TABLE 1.
Primers used for generating vectors and screening parasite strains
The following abbreviations are used: fwd, forward; rvs, reverse; Ab, antibody; 3′rep, 3′-replacement; comp, complementation.
| No. | Primer name | Primer sequence |
|---|---|---|
| 1 | TgTom40 3′rep fwd | 5′-TACTTCCAATCCAATTTAGCTAAGCTGCAATTGGGAGGCG |
| 2 | TgTom40 3′rep rvs | 5′-TCCTCCACTTCCAATTTTAGCCATCGGAGGAGGCTGTTCC |
| 3 | TgSam50 ORF fwd | 5′-GATCAGATCTACTAGTAAAATGGCGGGGTCAGCTCCTACC |
| 4 | TgSam50 ORF rvs | 5′-CAGTCCTAGGCTCGGGGAGTCTTCCAGAAGAAAGAC |
| 5 | TgTim22 3′rep fwd | 5′-GATCAGATCTACTAGTCAGATGCGGCAGCAATATCGGAAC |
| 6 | TgTim22 3′rep rvs | 5′-CAGTCCTAGGCGCTCCCATGTTTCCAAAAGGCTG |
| 7 | TgTim23 3′rep fwd | 5′-GTCAAGATCTACTAGTAAGGCAGGCGCTCAGCTTGGC |
| 8 | TgTim23 3′rep rvs | 5′-CAGTCCTAGGGACGTACTTGCGAAGGTACTGG |
| 9 | TgTim50 3′rep fwd | 5′-TACTTCCAATCCAATTTAGCAAATCGTGCTTCCTTGTGGTTG |
| 10 | TgTim50 3′rep rvs | 5′-TCCTCCACTTCCAATTTTAGCGGCCTTTTCCCACTTTTGTCTCTG |
| 11 | TgPam18 ORF fwd | 5′-AGATCTACTAGTAAAATGTGGGCACTGGCCTGCTTCCTG |
| 12 | TgPam18 ORF rvs | 5′-CCTAGGCCGCCGGCTGTCCTTCAGCAGCTTC |
| 13 | TgMPPβ 3′rep fwd | 5′-TACTTCCAATCCAATTTAGCGAGTTTCAAATCAGTTCCCTCTTCAG |
| 14 | TgMPPβ 3′rep rvs | 5′-TCCTCCACTTCCAATTTTAGCCTTGCCGACGCCCGCGGC |
| 15 | TgTom22 ORF fwd | 5′-TCCCCCCGGGGCTAGCATGGGGAACCTCCTGTCCTCGC |
| 16 | TgTom 22 ORF rvs | 5′-ATCGGATATCGACGTCTCACACCGAAGGAGTCGCC |
| 17 | TgTom22 5′flank fwd | 5′-ACTAGTCTTAGGGGTGTGCAGATTGGTGCCTT |
| 18 | TgTom22 5′flank rvs | 5′-AGATCTTTTGGAGGAAACAGGAGAGCGAGTGAC |
| 19 | TgTom22 3′flank fwd | 5′-GTCGACGGGGACCTCGAATGCCGTTTGAGGTAT |
| 20 | TgTom22 3′flank rvs | 5′-GGGCCCGGCACGCCTCATTTTCCGGTCTTG |
| 21 | TgTom22 screen fwd | 5′-TTTCTCGGCTGGTTTCTTAG |
| 22 | TgTom22 screen fwd | 5′-GGGCGAGATTGACGAAGC |
| 23 | TgTom22 comp fwd | 5′-GATCCCCGGGCTTAAGCCATGGCATATGGGGAACCTCCTGTCCTCG |
| 24 | TgTom22 comp rvs | 5′-GATCGACGTCTCACACCGAAGGAGTCGCCC |
| 25 | TgTom7 5′-RACE | 5′-CTGGAATAAGCTCGTCGGATCCAGAGTC |
| 26 | TgTom7 5′-RACE nested | 5′-CTCTCCCGTGTAGGCCAGGCCAAC |
| 27 | TgTom7 3′flank fwd | 5′-GATCCCCGGGATGGGCCAGGTTTTGAGTCATTG |
| 28 | TgTom7 3′flank rvs | 5′-CGATGCGGCCGCCAATCACCGAGTGGCGTTAATACC |
| 29 | TgTom7 5′flank fwd | 5′-CGATGGGCCCCTATCTGTCGGACCCAACTACTCAAAC |
| 30 | TgTom7 5′flank rvs | 5′-CGTAGGCGCGCCTAGTGCTTCCACAAATTGCTGTTGG |
| 31 | TgTom7 screen fwd | 5′-CGTCGGTGGAATCTGCTG |
| 32 | TgTom7 screen fwd | 5′-TACGTGCACCAGCAATGTC |
| 33 | t7s4 screen fwd | 5′-AAGGGGACGCAGTTCTCGGA |
| 34 | TgTom7 comp fwd | 5′-GATCCCATGGCCCGGGATGGGCCAGGTTTTGAGTCATTG |
| 35 | TgTom7 comp rvs | 5′-GATCCAATTGTCAGTTGATGACAATTTTCTGGAA |
| 36 | TgTom40 Ab fwd | 5′-GGGTCCTGGTTCGATGAAAACGTCTTCTCGCCATGAAGAGC |
| 37 | TgTom40 Ab rvs | 5′-CTTGTTCGTGCTGTTTATTAGCCTGCCACGGCCATCCATTC |
To generate an ATc-regulated knockdown strain of TgTom22, we amplified the TgTom22 open reading frame with primers 15 and 16, digested the resulting product with XmaI and AatII, and ligated into the equivalent sites of the pDt7s4H vector (56). This vector was transfected into TATi strain parasites and selected on pyrimethamine. To knock-out the native TgTom22 gene, we amplified a region upstream of the TgTom22 start codon with primers 17 and 18, digested the resulting product with SpeI and BglII, and ligated into the equivalent sites on the vector pTCY (38). We then amplified a region downstream of the TgTom22 stop codon using primers 19 and 20, digested the resulting product with SalI and ApaI, and ligated into equivalent sites of the pTCY vector containing the TgTom22 5′ flank. We transfected this into parasites expressing the ATc-regulated copy of TgTom22, selected parasites on chloramphenicol as described (52), and subjected parasites to negative YFP selection as described (57). We screened clones for TgTom22 knock-out using primers 21 and 22.
To generate a strain expressing the mitochondrial targeting sequence of TgHsp60 fused to c-Myc-tagged mouse DHFR, we digested the TgHsp60 leader sequence from the vector Hsp60-RFP in pBTR (58) with BglII and AvrII, ligated this into the equivalent sites of the vector mDHFR in pBTM3 (53), transfected into the TgTom22 knockdown cell line, and selected on phleomycin.
To generate a parasite strain expressing TgTom22 fused to an N-terminal DD tag, we amplified the open reading frame of TgTom22 with the primers 23 and 24, digested with XmaI and AatII, and ligated this into the equivalent sites of the vector pCTDDnM, a modified version of the vector pCTDDnH (58), which contains a c-Myc tag in place of an HA tag. We then replaced the chloramphenicol resistance marker in this vector with a phleomycin resistance marker (to generate the vector pBTDDnM-TgTom22), transfected into TgTom22 knockdown parasites, and selected on phleomycin.
To determine the correct 5′ region of the TgTom7 cDNA, we performed 5′-RACE using the SMARTer RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions. We used primer 25 for the first reaction and primer 26 for the nested reaction.
To determine the localization and function of TgTom7, we replaced the native promoter of TgTom7 with an ATc-regulated promoter, introducing an N-terminal 3×HA tag onto TgTom7 in the process. We amplified a 3′ flank of TgTom7 with primers 27 and 28, digested the product with XmaI and NotI, and ligated into the equivalent sites of the vector pPR2-HA3 (55). We next amplified the 5′ flank of the TgTom7 gene with primers 29 and 30, digested the product with ApaI and AscI, and ligated into the equivalent sites of the pPR2-HA3 vector harboring the TgTom7 3′ flank. We linearized this vector with NotI, transfected into TATi/Δku80 strain parasites (41), and selected on pyrimethamine. Clones were screened for successful promoter replacement by PCR, using primers 31 and 32, which only gives a band if the native locus is present, and primers 32 and 33, which only give a band if the 5′ region of TgTom7 is replaced by the regulated promoter.
To complement the TgTom7 knockdown mutant, we amplified the entire open reading frame of TgTom7 with primers 34 and 35 using T. gondii cDNA as template. We digested the product with NcoI and MfeI and ligated this into the equivalent sites of the pBTDDnTy vector, a modified version of the pBTDDnM vector that has a 5′-Ty1 epitope tag instead of a c-Myc tag. We digested the resulting vector with AvrII and NotI, which excises the Ty1-TgTom7 open reading frame, and we ligated this into equivalent sites of pUgCTH3, 5 which places Ty1-tagged TgTom7 downstream of the constitutive α-tubulin promoter. We linearized this vector with AatII, transfected into the TgTom7 knockdown cell line, and selected parasites on chloramphenicol.
Anti-TgTom40 Antibody Generation
To generate an antibody against TgTom40, we PCR-amplified a region of the TgTom40 open reading frame (encoding residues 2–173 of the TgTom40 protein) with primers 36 and 37 and integrated the product into the vector pAVA0421 by LIC (59). The plasmid was transformed into the BL21 strain Escherichia coli, and TgTom40 expression was induced through the addition of isopropyl 1-thio-β-d-galactopyranoside. His-tagged TgTom40 was purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen) according to the manufacturer's instructions, and purified protein was used to immunize rabbits (IVMS Vet Services, Adelaide, Australia).
BN-PAGE
For preparation of proteins for BN-PAGE, parasites were filtered through a 3-μm filter and pelleted by centrifugation at 1500 × g, 10 min, 4 °C. Parasites were washed once in ice-cold phosphate-buffered saline (PBS), then solubilized to a final concentration of 2.5 × 105 parasites/μl in Native-PAGE buffer (Life Technologies, Inc.), and supplemented with EDTA-free Complete protease inhibitor mixture (Roche Applied Sciences), 2 mm EDTA, and an appropriate detergent. Samples were incubated for 30 min at 4 °C and then centrifuged at 20,000 × g for 30 min at 4 °C to remove insoluble material. Samples were separated on a 4–16% Native-PAGE BisTris gel (Life Technologies, Inc.) according to the manufacturer's instructions, then transferred to PVDF membrane, or subjected to a second dimension SDS-polyacrylamide gel (Life Technologies, Inc.) according to the manufacturer's instructions, before transferring to nitrocellulose membrane and subsequent Western blotting.
Sodium Carbonate Extractions, Immunoprecipitations, and 35S Radiolabeling
Sodium carbonate extractions and immunoprecipitations were performed as described previously (38). Antibodies used for immunoprecipitations were rat anti-HA conjugated to agarose beads (Roche Applied Sciences), mouse anti-c-Myc conjugated to agarose beads (Thermo Scientific), and anti-TgTom40 antibodies conjugated to protein A-Sepharose CL-4B beads (Pierce). Co-immunoprecipitations were performed on parasite extracts solubilized in 0.5% digitonin in a buffer consisting of 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, and Complete protease inhibitors. 35S radiolabeling was performed as described previously (38), except that parasites were incubated in Expre35S35S protein labeling mix (PerkinElmer Life Sciences) for 10–60 min before harvesting.
Western Blotting and Immunofluorescence Assays
Western blotting and immunofluorescence assays were performed as described previously (38). Samples were probed with the following antibodies: rat anti-HA (1:100 to 1:500, Roche Applied Science); mouse anti-c-Myc (1:100 to 1:500, Thermo Scientific); mouse anti-Ty1 (1:200 to 1:1000 (60)); mouse anti-GRA8 (1:100,000, a kind gift from Gary Ward, University of Vermont (61)); rabbit anti-TgTom40 (1:2000, this study); rabbit anti-E. coli GroEL (1:1000; which we have previously shown also binds the mitochondrial Hsp60 of apicomplexan parasites (62)); rabbit anti-ATP synthase β-subunit (1:3000, Agrisera); AlexaFluor 488-conjugated goat anti-rat IgG (1:200, Life Technologies, Inc.); AlexaFluor 488-conjugated goat anti-mouse IgG (1:200 to 1:500, Life Technologies, Inc.); AlexaFluor 546-conjugated goat anti-rabbit IgG (1:500, Life Technologies, Inc.); horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:5000 to 1:10,000, Thermo Scientific); HRP-conjugated goat anti-mouse IgG (1:5000 to 1:10,000, Thermo Scientific); HRP-conjugated goat anti-rat IgG (1:5000; Thermo Scientific); TrueBlot HRP-conjugated anti-mouse IgG (1:1000, Rockland Immunochemicals); and TrueBlot HRP-conjugated anti-rabbit IgG (1:1000, Rockland Immunochemicals). Fluorescence images were acquired on a Leica TCS SP2 inverted laser scanning confocal microscope or a DeltaVision Elite system (GE Healthcare) using an inverted Olympus IX71 microscope. DeltaVision images were deconvolved using SoftWoRx Suite 2.0 software. All images were adjusted for contrast and brightness.
Author Contributions
G. v. D., B. S., and G. I. M. conceived the study. G. v. D. and L. M. Y. designed and performed the experiments. G. v. D., L. M. Y., B. S., and G. I. M. analyzed the data. G. v. D. wrote the manuscript with contributions from all the authors.
Supplementary Material
Acknowledgments
We thank Ming Kalanon for helpful discussions, Gary Ward and Michael White for sharing reagents, and Harpreet Vohra and Julie Nelson for assistance with flow cytometry.
This work was supported by Discovery Grant DP110103144 from the Australian Research Council (to G. v. D.), a National Health and Medical Research Program Grant (to G. I. M.), and Grant RO1AI 64671 from the National Institutes of Health (to B. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
G. van Dooren, unpublished observations.
E. Rajendran and G. van Dooren, manuscript in preparation.
- TOM
- translocon of the outer mitochondrial membrane
- SAM
- sorting and assembly machinery
- TIM
- translocon of the inner mitochondrial membrane
- PAM
- presequence translocase-associated motor
- MPP
- mitochondrial processing peptidase
- ATOM
- archaic translocase of the outer mitochondrial membrane
- ATc
- anhydrotetracycline
- Shld1
- Shield-1
- LIC
- ligation-independent cloning
- BN-PAGE
- blue native-PAGE
- DDM
- dodecyl maltoside
- mDHFR
- mouse dihydrofolate reductase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DD
- destabilization domain
- RFP
- red fluorescent protein
- 5′-RACE
- 5′-rapid amplification of cDNA ends.
References
- 1. Embley T. M., and Martin W. (2006) Eukaryotic evolution, changes and challenges. Nature 440, 623–630 [DOI] [PubMed] [Google Scholar]
- 2. Sagan L. (1967) On the origin of mitosing cells. J. Theor. Biol. 14, 255–274 [DOI] [PubMed] [Google Scholar]
- 3. van der Giezen M., and Tovar J. (2005) Degenerate mitochondria. EMBO Rep. 6, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dolezal P., Likic V., Tachezy J., and Lithgow T. (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 [DOI] [PubMed] [Google Scholar]
- 5. Dudek J., Rehling P., and van der Laan M. (2013) Mitochondrial protein import: common principles and physiological networks. Biochim. Biophys. Acta 1833, 274–285 [DOI] [PubMed] [Google Scholar]
- 6. Hill K., Model K., Ryan M. T., Dietmeier K., Martin F., Wagner R., and Pfanner N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395, 516–521 [DOI] [PubMed] [Google Scholar]
- 7. Brix J., Dietmeier K., and Pfanner N. (1997) Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 272, 20730–20735 [DOI] [PubMed] [Google Scholar]
- 8. van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J., Moczko M., Wagner R., Meijer M., Guiard B., Hönlinger A., and Pfanner N. (1999) Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401, 485–489 [DOI] [PubMed] [Google Scholar]
- 9. Moczko M., Bömer U., Kübrich M., Zufall N., Hönlinger A., and Pfanner N. (1997) The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 17, 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gentle I., Gabriel K., Beech P., Waller R., and Lithgow T. (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geissler A., Chacinska A., Truscott K. N., Wiedemann N., Brandner K., Sickmann A., Meyer H. E., Meisinger C., Pfanner N., and Rehling P. (2002) The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111, 507–518 [DOI] [PubMed] [Google Scholar]
- 12. Truscott K. N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A. J., Rassow J., Pfanner N., and Wagner R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 13. Truscott K. N., Voos W., Frazier A. E., Lind M., Li Y., Geissler A., Dudek J., Müller H., Sickmann A., Meyer H. E., Meisinger C., Guiard B., Rehling P., and Pfanner N. (2003) A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F. U., and Neupert W. (1988) Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell 53, 795–806 [DOI] [PubMed] [Google Scholar]
- 15. Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H. E., Kühlbrandt W., Wagner R., Truscott K. N., and Pfanner N. (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299, 1747–1751 [DOI] [PubMed] [Google Scholar]
- 16. Duncan O., Murcha M. W., and Whelan J. (2013) Unique components of the plant mitochondrial protein import apparatus. Biochim. Biophys. Acta 1833, 304–313 [DOI] [PubMed] [Google Scholar]
- 17. Lister R., Carrie C., Duncan O., Ho L. H., Howell K. A., Murcha M. W., and Whelan J. (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19, 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rimmer K. A., Foo J. H., Ng A., Petrie E. J., Shilling P. J., Perry A. J., Mertens H. D., Lithgow T., Mulhern T. D., and Gooley P. R. (2011) Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J. Mol. Biol. 405, 804–818 [DOI] [PubMed] [Google Scholar]
- 19. Maćasev D., Whelan J., Newbigin E., Silva-Filho M. C., Mulhern T. D., and Lithgow T. (2004) Tom22′, an 8-kDa trans-site receptor in plants and protozoans, is a conserved feature of the TOM complex that appeared early in the evolution of eukaryotes. Mol. Biol. Evol. 21, 1557–1564 [DOI] [PubMed] [Google Scholar]
- 20. Pusnik M., Schmidt O., Perry A. J., Oeljeklaus S., Niemann M., Warscheid B., Lithgow T., Meisinger C., and Schneider A. (2011) Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 [DOI] [PubMed] [Google Scholar]
- 21. Mani J., Meisinger C., and Schneider A. (2016) Peeping at TOMs-diverse entry gates to mitochondria provide insights into the evolution of eukaryotes. Mol. Biol. Evol. 33, 337–351 [DOI] [PubMed] [Google Scholar]
- 22. Zarsky V., Tachezy J., and Dolezal P. (2012) Tom40 is likely common to all mitochondria. Curr. Biol. 22, R479–R481 [DOI] [PubMed] [Google Scholar]
- 23. Mani J., Desy S., Niemann M., Chanfon A., Oeljeklaus S., Pusnik M., Schmidt O., Gerbeth C., Meisinger C., Warscheid B., and Schneider A. (2015) Mitochondrial protein import receptors in kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat. Commun. 6, 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pusnik M., Mani J., Schmidt O., Niemann M., Oeljeklaus S., Schnarwiler F., Warscheid B., Lithgow T., Meisinger C., and Schneider A. (2012) An essential novel component of the noncanonical mitochondrial outer membrane protein import system of trypanosomatids. Mol. Biol. Cell 23, 3420–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gentle I. E., Perry A. J., Alcock F. H., Likić V. A., Dolezal P., Ng E. T., Purcell A. W., McConnville M., Naderer T., Chanez A. L., Charrière F., Aschinger C., Schneider A., Tokatlidis K., and Lithgow T. (2007) Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol. Biol. Evol. 24, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 26. Singha U. K., Peprah E., Williams S., Walker R., Saha L., and Chaudhuri M. (2008) Characterization of the mitochondrial inner membrane protein translocator Tim17 from Trypanosoma brucei. Mol. Biochem. Parasitol. 159, 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doggett J. S., Nilsen A., Forquer I., Wegmann K. W., Jones-Brando L., Yolken R. H., Bordón C., Charman S. A., Katneni K., Schultz T., Burrows J. N., Hinrichs D. J., Meunier B., Carruthers V. B., and Riscoe M. K. (2012) Endochin-like quinolones are highly efficacious against acute and latent experimental toxoplasmosis. Proc. Natl. Acad. Sci. U.S.A. 109, 15936–15941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phillips M. A., Gujjar R., Malmquist N. A., White J., El Mazouni F., Baldwin J., and Rathod P. K. (2008) Triazolopyrimidine-based dihydroorotate dehydrogenase inhibitors with potent and selective activity against the malaria parasite Plasmodium falciparum. J. Med. Chem. 51, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Srivastava I. K., Rottenberg H., and Vaidya A. B. (1997) Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J. Biol. Chem. 272, 3961–3966 [DOI] [PubMed] [Google Scholar]
- 30. Alcock F., Webb C. T., Dolezal P., Hewitt V., Shingu-Vasquez M., Likić V. A., Traven A., and Lithgow T. (2012) A small Tim homohexamer in the relict mitochondrion of Cryptosporidium. Mol. Biol. Evol. 29, 113–122 [DOI] [PubMed] [Google Scholar]
- 31. Deponte M., Hoppe H. C., Lee M. C., Maier A. G., Richard D., Rug M., Spielmann T., and Przyborski J. M. (2012) Wherever I may roam: protein and membrane trafficking in P. falciparum-infected red blood cells. Mol. Biochem. Parasitol. 186, 95–116 [DOI] [PubMed] [Google Scholar]
- 32. van Dooren G. G., Stimmler L. M., and McFadden G. I. (2006) Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 30, 596–630 [DOI] [PubMed] [Google Scholar]
- 33. Hell K., Neupert W., and Stuart R. A. (2001) Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20, 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., and Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sheiner L., Fellows J. D., Ovciarikova J., Brooks C. F., Agrawal S., Holmes Z. C., Bietz I., Flinner N., Heiny S., Mirus O., Przyborski J. M., and Striepen B. (2015) Toxoplasma gondii Toc75 functions in import of stromal but not peripheral apicoplast proteins. Traffic 16, 1254–1269 [DOI] [PubMed] [Google Scholar]
- 36. Ahting U., Thun C., Hegerl R., Typke D., Nargang F. E., Neupert W., and Nussberger S. (1999) The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gubbels M. J., Li C., and Striepen B. (2003) High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob. Agents Chemother. 47, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Dooren G. G., Tomova C., Agrawal S., Humbel B. M., and Striepen B. (2008) Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc. Natl. Acad. Sci. U.S.A. 105, 13574–13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G., and Wandless T. J. (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deleted in proof
- 41. Sheiner L., Demerly J. L., Poulsen N., Beatty W. L., Lucas O., Behnke M. S., White M. W., and Striepen B. (2011) A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 7, e1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cavalier-Smith T. (2006) Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc. Biol. Sci. 273, 1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dolezal P., Dagley M. J., Kono M., Wolynec P., Likić V. A., Foo J. H., Sedinová M., Tachezy J., Bachmann A., Bruchhaus I., and Lithgow T. (2010) The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog. 6, e1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dekker P. J., Ryan M. T., Brix J., Müller H., Hönlinger A., and Pfanner N. (1998) Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18, 6515–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Werhahn W., Niemeyer A., Jänsch L., Kruft V., Schmitz U. K., and Braun H. (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol. 125, 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., and Neupert W. (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426, 862–866 [DOI] [PubMed] [Google Scholar]
- 47. Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M. T., Pfanner N., and Meisinger C. (2003) Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424, 565–571 [DOI] [PubMed] [Google Scholar]
- 48. Becker T., Wenz L. S., Thornton N., Stroud D., Meisinger C., Wiedemann N., and Pfanner N. (2011) Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J. Mol. Biol. 405, 113–124 [DOI] [PubMed] [Google Scholar]
- 49. Yamano K., Tanaka-Yamano S., and Endo T. (2010) Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J. Biol. Chem. 285, 41222–41231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huynh M. H., and Carruthers V. B. (2009) Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meissner M., Schlüter D., and Soldati D. (2002) Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298, 837–840 [DOI] [PubMed] [Google Scholar]
- 52. Striepen B., and Soldati D. (2007) in Toxoplasma gondii. The Model Apicomplexan–Perspectives and Methods (Weiss L. D., and Kim K., eds) pp. 391–415, Elsevier, London [Google Scholar]
- 53. Glaser S., van Dooren G. G., Agrawal S., Brooks C. F., McFadden G. I., Striepen B., and Higgins M. K. (2012) Tic22 is an essential chaperone required for protein import into the apicoplast. J. Biol. Chem. 287, 39505–39512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Messina M., Niesman I., Mercier C., and Sibley L. D. (1995) Stable DNA transformation of Toxoplasma gondii using phleomycin selection. Gene 165, 213–217 [DOI] [PubMed] [Google Scholar]
- 55. Katris N. J., van Dooren G. G., McMillan P. J., Hanssen E., Tilley L., and Waller R. F. (2014) The apical complex provides a regulated gateway for secretion of invasion factors in Toxoplasma. PLoS Pathog. 10, e1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Agrawal S., van Dooren G. G., Beatty W. L., and Striepen B. (2009) Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. J. Biol. Chem. 284, 33683–33691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mazumdar J., H Wilson E., Masek K., A Hunter C., and Striepen B. (2006) Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 103, 13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Dooren G. G., Reiff S. B., Tomova C., Meissner M., Humbel B. M., and Striepen B. (2009) A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr. Biol. 19, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alexandrov A., Vignali M., LaCount D. J., Quartley E., de Vries C., De Rosa D., Babulski J., Mitchell S. F., Schoenfeld L. W., Fields S., Hol W. G., Dumont M. E., Phizicky E. M., and Grayhack E. J. (2004) A facile method for high-throughput co-expression of protein pairs. Mol. Cell. Proteomics 3, 934–938 [DOI] [PubMed] [Google Scholar]
- 60. Bastin P., Bagherzadeh Z., Matthews K. R., and Gull K. (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235–239 [DOI] [PubMed] [Google Scholar]
- 61. Carey K. L., Donahue C. G., and Ward G. E. (2000) Identification and molecular characterization of GRA8, a novel, proline-rich, dense granule protein of Toxoplasma gondii. Mol. Biochem. Parasitol. 105, 25–37 [DOI] [PubMed] [Google Scholar]
- 62. Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., and McFadden G. I. (2004) Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.