Abstract
Glucocorticoid receptor (GR) signaling has recently been shown to play a direct role in the regulation of cardiomyocyte function. In this study, we investigated the potential role of KLF13 as a downstream effector of GR action utilizing both in vivo and in vitro approaches. Our data show that KLF13 mRNA and protein levels are significantly diminished in the hearts of mice lacking GR in cardiomyocytes. Glucocorticoid administration up-regulated Klf13 mRNA in the mouse heart, in isolated primary cardiomyocytes, and in immortal cardiomyocyte cell lines. Glucocorticoid Klf13 gene expression was abolished by treatment with a GR antagonist (RU486) or by knockdown of GR in cardiomyocytes. Moreover, glucocorticoid induction of Klf13 mRNA was resistant to de novo protein synthesis inhibition, demonstrating that Klf13 is a direct glucocorticoid receptor gene target. A glucocorticoid responsive element (GRE) was identified in the Klf13 gene and its function was verified by chromatin immunoprecipitation in HL-1 cells and mouse hearts. Functional studies showed that GR regulation of Klf13 is critical to protect cardiomyocytes from DNA damage and cell death induced by cobalt(II) chloride hexahydrate (CoCl2·6H2O) and the antineoplastic drug doxorubicin. These results established a novel role for GR and KLF13 signaling in adult cardiomyocytes with potential clinical implications for the prevention of cardiotoxicity induced heart failure.
Keywords: cardiomyocyte, gene regulation, glucocorticoid receptor, hypoxia, Kruppel-like factor (KLF)
Introduction
Glucocorticoids are essential steroid hormones synthesized and secreted by the adrenal glands in a diurnal fashion and in response to stress (1, 2). They act by binding to the glucocorticoid receptor (GR) 3 (NR3C1), a member of the nuclear receptor superfamily of transcription factors (3). Upon ligand binding, glucocorticoid receptor translocates to the nucleus to regulate the expression of thousands of genes through direct DNA binding, by interacting with other transcription factors, and by regulation of RNA turnover (4–6). Glucocorticoid receptors are highly expressed in heart cardiomyocytes, fibroblasts, and endothelial cells (7–12), and several direct actions of glucocorticoids in the heart have been described in vivo (13, 14). For example, Rog-Zielinska et al. (13) recently demonstrated that glucocorticoid signaling in cardiomyocytes is necessary for heart maturation during development. Mice lacking glucocorticoid receptors in cardiomyocytes develop spontaneous cardiac hypertrophy, left ventricular systolic dysfunction, heart failure, and death (14). Global gene expression analysis of the hearts of these mice revealed significant dysregulation of several gene networks associated with cardiomyocyte homeostasis and function. Among these genes, we observed two members of the Krüppel-like factor (KLF) family of transcriptional regulators, Klf15 and Klf13 (14).
Krüppel-like factors are zinc finger DNA-binding transcription factors homologous to the Drosophila gap gene Krüppel (15). They have been implicated in the regulation of an array of cellular processes (16), including cardiac biology (14, 17, 18). KLF13 and KLF15, in particular, have both been suggested to play roles in the heart (14, 17, 19), and KLF15 expression is directly induced by glucocorticoids (18, 20). KLF15 is known to be a transcriptional repressor of pathological cardiac hypertrophy (14, 17), and as a regulator of cardiac lipid metabolism (18). KLF13 has been reported to modulate cardiomyocyte growth and differentiation during heart development and morphogenesis in Xenopus (19). Mice with global deficiency of KLF13 have been reported to have defects in erythropoiesis and splenomegaly; in addition these mice have been reported to have enlarged hearts, however, this component of their phenotype has apparently not been studied (21).
In this study, we utilized cardiomyocyte-specific GR knock-out mice (cardioGRKO), wild-type mice, murine primary adult cardiomyocytes, HL-1 mouse, and H9C2 rat cardiomyocytes to understand the role of glucocorticoid regulation of KLF13 in the heart. Our results show that glucocorticoids regulate Klf13 expression in the mouse heart, primary cardiomyocytes, and immortal cells. Glucocorticoids directly regulate Klf13 gene transcription through binding of glucocorticoid receptors to a newly identified intragenic glucocorticoid response element (GRE). Moreover, KLF13 regulation by glucocorticoid receptor has profound effects on the expression of genes associated with cell death and survival, GR regulation of KLF13 appears to be critical to protect cardiomyocytes from DNA damage and cell death induced by cobalt(II) chloride hexahydrate (CoCl2· 6H2O) and the antineoplastic drug doxorubicin.
Materials and Methods
Reagents and Antibodies
Dexamethasone and mifepristone (RU486) were purchased from Steraloids (Newport, RI). Cycloheximide, cobalt(II) chloride hexahydrate (CoCl2·6H2O), and doxorubicin were purchased from Sigma. The anti-KLF13 antibody was purchased from Proteintech Group (Chicago, IL), the anti-troponin I antibody was purchased from Millipore (Billerica, MA). Generation and use of the rabbit polyclonal anti-GR 57 antibody has been previously described (22).
RNA Extraction and Real-time Quantitative PCR (RT-PCR)
Total RNA was isolated from tissues and cells using the RNeasy Mini Kit and RNase-Free DNase Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions (14). mRNA levels were determined using the TaqMan one-step RT-PCR procedure on a 7900HT sequence detection system (Applied Biosystems, Rockville, MD).
Western Blotting Analysis
Heart tissue and cells were homogenized and lysed in Tris glycine SDS sample buffer (Invitrogen) supplemented with 2.5% β-mercaptoethanol (14). Membranes with equivalent amounts of protein were incubated with primary antibodies and developed using the LI-COR Odyssey imaging system (LI-COR Biotechnology). GR and KLF13 protein levels were quantified by densitometry using NIH ImageJ analysis software.
Animal Experiments
Hearts from 1–2-month-old control (Floxed GR mice, GRloxP/loxP) and cardioGRKO (GRloxP/loxPαMHCCre/+) male mice were harvested for RNA isolation and Western blotting. CardioGRKO mice were generated by crossing floxed GR mice (GRloxP/loxP) with mice expressing Cre recombinase under the control of the α-myosin heavy chain promoter (αMHCCre/+) to specifically delete GR in cardiomyocytes as previously described by Oakley RH et al. (14). Adrenalectomized 8-week-old C57BL/6 male mice were purchased from Charles River Laboratories (Wilmington, MA). For glucocorticoid treatment, mice were administered 1 mg of dexamethasone/kg of body weight or vehicle control (PBS) intraperitoneally (i.p.). Six and 24 h after the treatment hearts were harvested for RNA isolation, immunofluorescence, and in vivo ChIP assays. All mice were euthanized by cervical dislocation. Animals were maintained in accordance with the National Institutes of Health directives for the care and use of laboratory animals. These studies were approved by the National Institute of Environmental Health Sciences' Animal Care and Use Committee.
Immunofluorescence
Hearts from vehicle and dexamethasone-treated C57BL/6 mice were perfused with PBS and fixed with freshly prepared 4% paraformaldehyde. Slides were prepared and processed for immunofluorescence for KLF13 and troponin I. Images were obtained on a Zeiss LSM 710 inverted confocal microscope equipped with a ×40 (oil) objective.
Cardiomyocyte Isolation
Adult cardiomyocytes from 8-week-old C57BL/6 male mice were isolated using a commercially available kit (Perfusion Adumyts Cardiomyocyte Isolation Kit, Cellutron, Baltimore, MD). Cells were then treated with 100 nm dexamethasone or vehicle control for 6 h. Cells were blocked and incubated overnight with a primary antibody to KLF13 and troponin I. Images were obtained on a Zeiss LSM 710 inverted confocal microscope equipped with a ×40 (oil) objective.
Cell Culture
The cardiac mouse cell line HL-1 was kindly provided by Dr. W. C. Claycomb (Louisiana State University, New Orleans, LA) and cultured as described (23). The rat embryonic cardiomyocyte H9C2 cells (American Type Culture Collection, Manassas, VA) were cultured in high-glucose (4500 mg/liter) DMEM supplemented with 10% heat-inactivated fetal bovine serum, 1 mm sodium pyruvate, 100 μg/ml of streptomycin, and 100 units/ml of penicillin (24). All treatments of cells were performed in media containing 10% charcoal/dextran-stripped fetal calf serum.
Cells ChIP Assays
ChIP assays were performed in HL-1 cells following the described protocol (25). Quantitative RT-PCR was performed on both input and immunoprecipitated DNA.
Heart Tissue ChIP Assays
Heart tissue was used for ChIP assays according to the protocol of the ChIP assay kit (Millipore, 17–295), with minor modifications. The immunoprecipitated DNA was then quantified using real-time quantitative PCR employing the same primers and conditions used for in vitro ChIP assays.
Luciferase Assays
The Klf13 gene was analyzed for GREs using Sequence Motif Search and rVista 2.0. Fragments from the KLF13 promoter and intron-1 were amplified from HL-1 cell genomic DNA using a FastStart High-fidelity PCR System (Roche Applied Science). The Klf13 intron-1 GRE sequence GAGAACATGCTGTTCC was mutated to GAGAACAAGCAGTTCC (366Thr → Ala) using the QuikChange site-directed mutagenesis kit (Stratagene) to generate Klf13 intron1-mut. The cells were transiently transfected using FuGENE® 6 transfection reagent (Promega, Madison, WI) with either the control pGL3 luciferase plasmid or the pGL3 luciferase construct, using the Renilla luciferase reporter as a control for transfection. Luciferase activity was measured using the Dual Glo Luciferase Reporter Assay (Promega).
RNA Interference
Non-targeting control (NTC), GR, and KLF13 siRNAs were purchased from SMARTpool siRNA (Thermo Scientific). HL-1 cells were transfected with 50 nm of each siRNA using Dharmafect1 transfection reagent (Thermo Scientific). Forty-eight hours after transfection, cells were replated and treated with 100 nm dexamethasone or vehicle control. RNA was isolated from these cells 24 h following dexamethasone treatment.
Microarray Studies
HL-1 cells were transfected with NTC, GR siRNA, or KLF13 siRNA. Forty-eight hours following transfection, cells were replated as described above and treated with 100 nm dexamethasone or vehicle control for 24 h. Genome-wide microarray analysis was performed using Agilent Whole Mouse Genome 4 × 44 multiplex format oligonucleotide arrays (product number 014868; Agilent Technologies) according to the manufacturer's instructions (25, 26). The data were processed using package Agi4 × 44PreProcess and limma in Bioconductor for the R software environment. Expression measures for each probe set were log-transformed, followed by background correction, and normalization between samples. The significance of the log ratio for each probe was determined by calculating one modified t statistic per probe using an empirical Bayesian approach. Multiple test correction was used to reduce the number of false positives. Probes with Benjamini-Hochberg multiple test corrected p value <0.05 were considered to be differentially expressed. Heat map was generated using BioConductor package HeatPlus. The lists of probe sets generated were visually sorted by using a Venn diagram generator and further analyzed with Pathway Analysis version 6.5 (Ingenuity Systems).
Hypoxia
To induce hypoxia, HL-1 cells were incubated 48 h in a controlled hypoxic incubator (Thermo Scientific, Forma, Series II Water Jacketed, model 3130 incubator). All experiments were performed in serum-free medium. The time control group consisted of cells without the hypoxic stimulus kept in complete medium in a normoxic incubator at 37 °C. Cell viability was measured by assessing plasma membrane integrity by flow cytometry analysis of 10 μg/ml of propidium iodide (PI; Invitrogen) exclusion. Ten thousand cells were analyzed using a BD LSRII flow cytometer (San Jose, CA) equipped with FACSDiVa software. Cells were excited with a 561-nm laser and PI fluorescence was detected at 585 nm. A gate was drawn on a PI histogram for the control sample to determine the percent of viable and dead experimental cells.
Induction of Cell Death and DNA Damage
Cell death and DNA damage were induced by treating non-targeting control (NTC), (−)GR, and (−)KLF13 cells with 500 μm CoCl2·6H2O (27, 28) or 1 μm doxorubicin (29). For all treatments cells were cultured in complete medium supplemented with 0.1–1 μm dexamethasone and maintained in a humidified 5% CO2 atmosphere at 37 °C for 48 h. Following this treatment CoCl2·6H2O or doxorubicin was added for 24–48 h. Plasma membrane integrity of HL-1 cells was then analyzed by flow cytometry analysis of 10 μg/ml of PI (Invitrogen) exclusion. DNA degradation was analyzed by flow cytometry. Briefly, cells were fixed in 70% ethanol overnight at 4 °C. Cells were then pelleted and washed once in PBS. The cells were then stained with a PI solution (20 μg/ml of PI, 10 units/ml RNase One, Promega in PBS) and incubated in the dark for 20 min. Cells were examined on a forward-scatter versus side-scatter dot plot, a forward-scatter histogram, and a PI histogram to analyze the cell cycle. At least 7,500 cells in a LO flow rate (the rate of cells should be less than 400 cells/second) were analyzed using a BD LSRII flow cytometer (San Jose, CA) equipped with FACSDiVa software.
KLF13 Overexpression
Lipofectamine 2000 (Invitrogen) was used to transfect Klf13 plasmid (Origene, MR224297) into HL-1 cells. Twenty-four hours after transfection, 2× Laemmli sample buffer was used to make cell lysates of transfectants for Western blot analysis. After confirming KLF13 overexpression, cells were treated with doxorubicin for 48 h and analyzed by flow as described above.
Statistical Analysis
Data were presented as a mean ± S.E. Statistical significance was determined by analysis of variance with Tukey's post hoc analysis. All analyses were performed using GraphPad Prism 6 software.
Results
Klf13 Expression Is Significantly Reduced in the Hearts of Mice Lacking GR in Cardiomyocytes
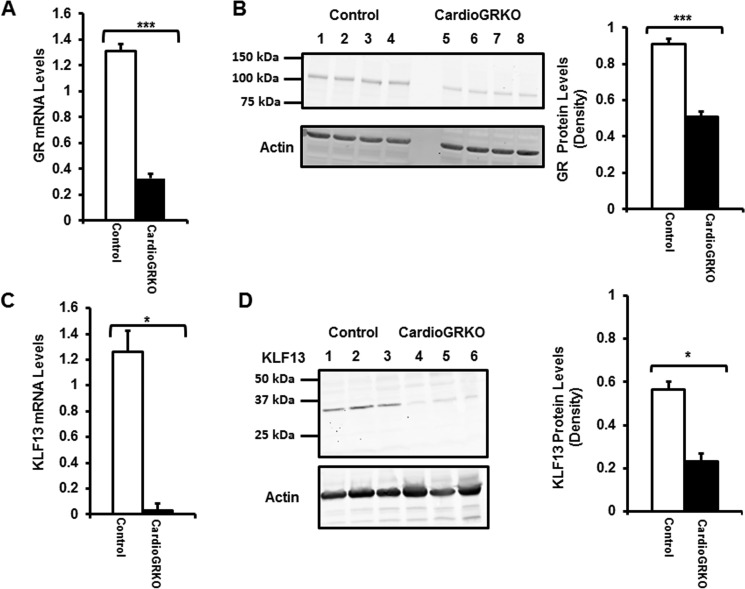
Global gene expression analysis of the hearts of mice lacking glucocorticoid receptors in cardiomyocytes (cardioGRKO mice) identified Klf13 as expressed in the mouse heart, and its transcription is significantly reduced in the hearts of the cardioGRKO mice (14). To assess these data at both the RNA and protein levels, we evaluated these parameters in hearts of cardioGRKO mice and their littermate controls. GR levels were significantly reduced at both mRNA and protein levels in cardioGRKO hearts as previously described (14) (Fig. 1, A and B). Klf13 mRNA levels were dramatically diminished in hearts from cardioGRKO mice (Fig. 1C). Consistent with this reduction in mRNA, KLF13 protein levels were also significantly decreased in hearts from cardioGRKO mice (Fig. 1D). These results confirm that Klf13 expression is profoundly compromised in the hearts of mice lacking GR in cardiomyocytes.
FIGURE 1.
GR and KLF13 mRNA and protein levels are diminished in the heart of the cardioGRKO mouse model. A, RT-PCR of GR mRNA from hearts of 2-month-old control and cardioGRKO mice. B, representative Western blotting of GR protein from hearts of control and cardioGRKO mice. C, RT-PCR of KLF13 mRNA from hearts of 2-month-old control and cardioGRKO mice. D, representative Western blotting of KLF13 protein from hearts of control and cardioGRKO mice. Data represent mean ± S.E. (n = 3–4 mice per group). *, p < 0.05; ***, p < 0.001.
Cardiac Klf13 Expression Is Regulated by Glucocorticoids in Vivo and in Primary Cardiomyocytes
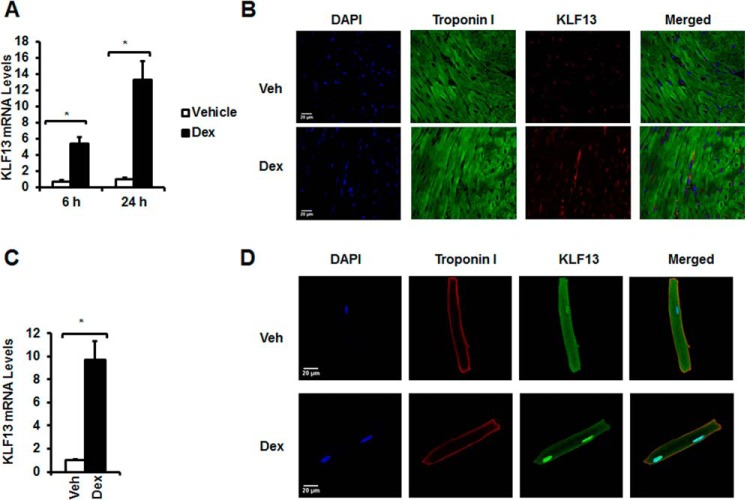
To investigate whether glucocorticoids control KLF13 expression in vivo, 8-week-old C57BL/6 mice were adrenalectomized and treated with vehicle (PBS) or glucocorticoids (dexamethasone, Dex). Hearts were harvested at 6 and 24 h following this treatment. Six hours after glucocorticoid injection, Klf13 mRNA levels were increased 5.42 ± 0.79-fold (p < 0.05) over basal levels, and were increased 13.28 ± 2.34-fold (p < 0.05) over basal levels at 24 h (Fig. 2A). KLF13 was also detected under basal conditions in the mouse heart (Fig. 2B) and its levels were increased upon glucocorticoid treatment (Fig. 2B). These data indicate that glucocorticoids regulate KLF13 expression in the mouse heart.
FIGURE 2.
KLF13 mRNA and protein levels are regulated by glucocorticoids in the heart and primary adult cardiomyocytes. Adrenalectomized (ADX) C57BL/6 male mice were injected with vehicle (Veh) or 1 mg/kg of Dex. RNA was isolated at 6 and 24 h after the treatment. Hearts were harvested for immunofluorescence at 24 h following Dex treatment. A, KLF13 mRNA levels in Veh (white bars)- and Dex (black bars)-treated mice. B, representative immunofluorescence staining of heart sections from control and cardioGRKO mice with anti-troponin I (green) and anti-KLF13 (red) antibodies. DAPI is shown in blue. Primary cardiomyocytes were treated with vehicle or Dex for 6 h. C, KLF13 gene expression levels in vehicle (white bars)- and Dex (black bars)-treated cardiomyocytes. D, representative immunofluorescence staining of Veh- and Dex-treated cardiomyocytes. Troponin I (red), KLF13 (green), and DAPI (blue nuclear staining) are indicated. Data represent mean ± S.E. (n = 3–5 mice per group). *, p < 0.05.
To determine whether glucocorticoids regulate Klf13 expression in mammalian cardiomyocytes, primary adult cardiomyocytes were isolated from 8-week-old C57BL/6 male mice. Glucocorticoid treatment of primary cardiomyocytes led to significant up-regulation of Klf13 mRNA levels at 6 h (9.72 ± 1.58-fold; p < 0.05) (Fig. 2C). KLF13 protein levels were also significantly increased in cardiomyocytes following glucocorticoid treatment (Fig. 2D). These results demonstrate that Klf13 is expressed in primary adult cardiomyocytes and its gene expression and protein levels are regulated by glucocorticoids.
Glucocorticoids Induce Klf13 Gene Expression in Immortal Rat and Mouse Cardiomyocytes
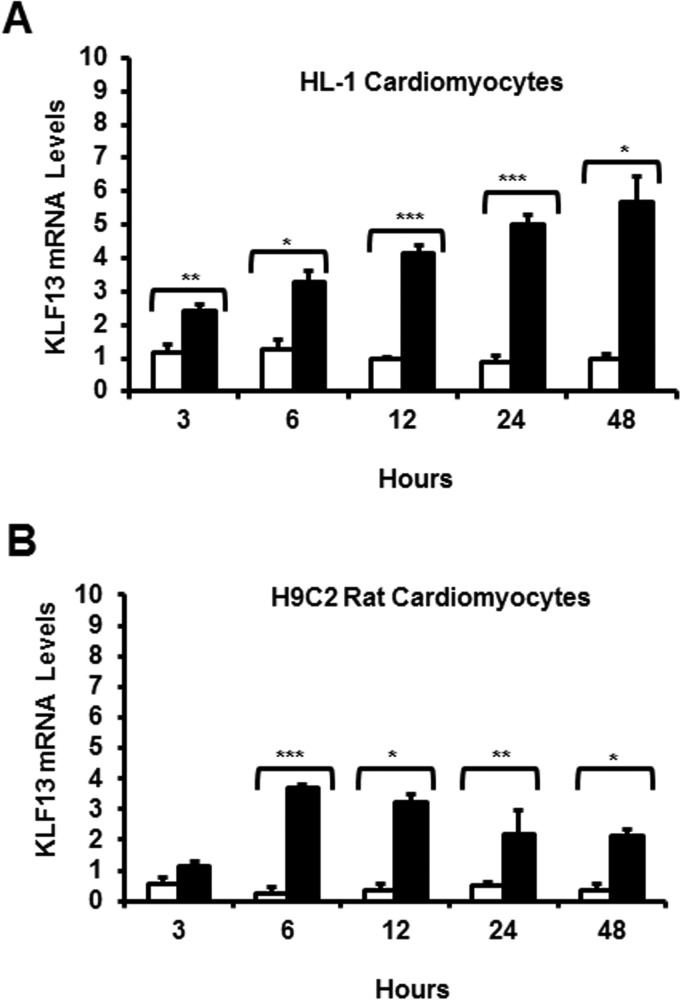
To further define the mechanisms of glucocorticoid regulation of KLF13; we employed two different cell lines: H9C2 rat immortal cardiomyocytes (24) and HL-1 mouse immortal cardiomyocytes (23). Both H9C2 and HL-1 cardiomyocytes were treated with glucocorticoids, and then RNA was isolated at different time points (0–48 h). Increases in Klf13 gene expression were observed in both cell lines in response to glucocorticoids (Fig. 3, A and B). These data support our studies in primary cardiomyocytes, and revealed that KLF13 is indeed a glucocorticoid-regulated gene in adult cardiomyocytes.
FIGURE 3.
Glucocorticoids regulate KLF13 gene expression in H9C2 rat cardiomyocytes and HL-1 mouse cardiomyocytes. A and B, H9C2 and HL-1 cells were treated with Veh (white bars) or 100 nm Dex (black bars) for the indicated times (3–48 h) and KLF13 levels were analyzed by RT-PCR. C, HL-1 KLF13 protein levels at 48 h following Dex treatment. KLF13 levels were normalized to β-actin. A representative Western blotting is shown. Data represent mean ± S.E. from three or four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The Glucocorticoid Receptor Directly Regulates Klf13 Transcription in HL-1 Mouse Adult Cardiomyocytes
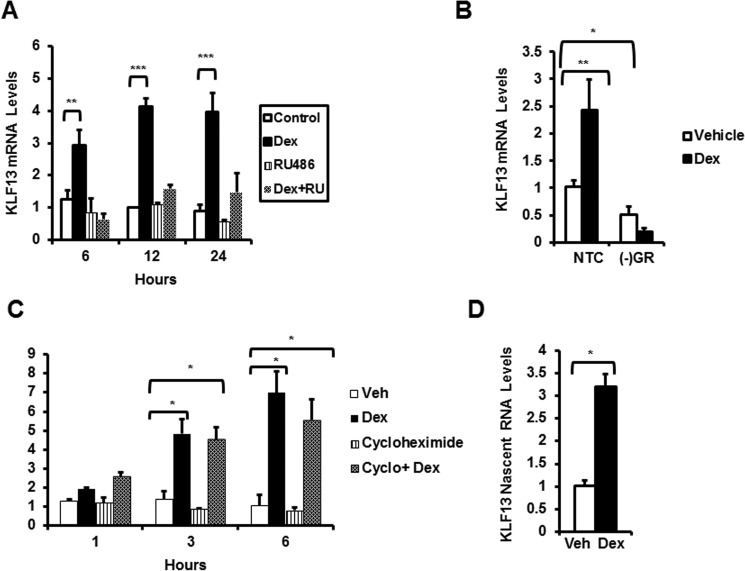
To elucidate the mechanism(s) underlying glucocorticoid regulation of KLF13, we first determined if glucocorticoid receptors are required for the observed changes in Klf13 gene expression. HL-1 cardiomyocytes were treated with the glucocorticoid receptor-antagonist RU486. Exposure of HL-1 cells to RU486 prevented glucocorticoid-mediated induction of Klf13 mRNA at the times examined (Fig. 4A). Silencing of the glucocorticoid receptor gene by siRNA significantly decrease KLF13 basal levels and abolished glucocorticoid-induced up-regulation of Klf13 mRNA in HL-1 cardiomyocytes (Fig. 4B). These findings indicate that activated glucocorticoid receptors are necessary for glucocorticoid regulatory effects on Klf13 expression in cardiomyocytes. We determined if KLF13 is a primary or secondary target for the glucocorticoid receptor. For these studies, HL-1 cells were treated with the protein synthesis inhibitor cycloheximide for 1 to 6 h. Glucocorticoid treatment significantly increased Klf13 mRNA levels in the presence and absence of cycloheximide (Fig. 4C), leading us to conclude that KLF13 is a primary target of glucocorticoid regulation. Finally, to determine whether glucocorticoids regulate transcription of the Klf13 gene, nascent RNA was measured using primers targeting intronic sequences of the gene. KLF13 nascent RNA (3.19 ± 0.28-fold over basal levels) was significantly increased in response to glucocorticoids (Fig. 4D). We conclude that glucocorticoids regulate Klf13 cardiomyocyte gene expression at the transcriptional level through activated glucocorticoid receptors.
FIGURE 4.
KLF13 is a direct target of the glucocorticoid receptor in HL-1 mouse adult cardiomyocytes. A, HL-1 cells were treated with vehicle (control, white bars), 100 nm Dex (black bars), 1 μm RU486 (striped bars), or both 1 μm RU486 and 100 nm Dex (cross-hatched bars). KLF13 mRNA were measured by RT-PCR at the indicated times. B, HL-1 cells transfected with NTC siRNA or GR siRNA (−GR) were treated with 100 nm Dex for 3 h. mRNA levels were measured by RT-PCR. C, HL-1 cells were pretreated for 1–6 h with vehicle (control) or 10 μg/ml of cycloheximide (cyclo) then cells were exposed to 100 nm Dex for 3 h. KLF13 mRNA levels were assessed by RT-PCR. D, nascent RNA was measured by RT-PCR after treating the cells for 3 h with 100 nm Dex or vehicle (control). Data represent mean ± S.E. from three or four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
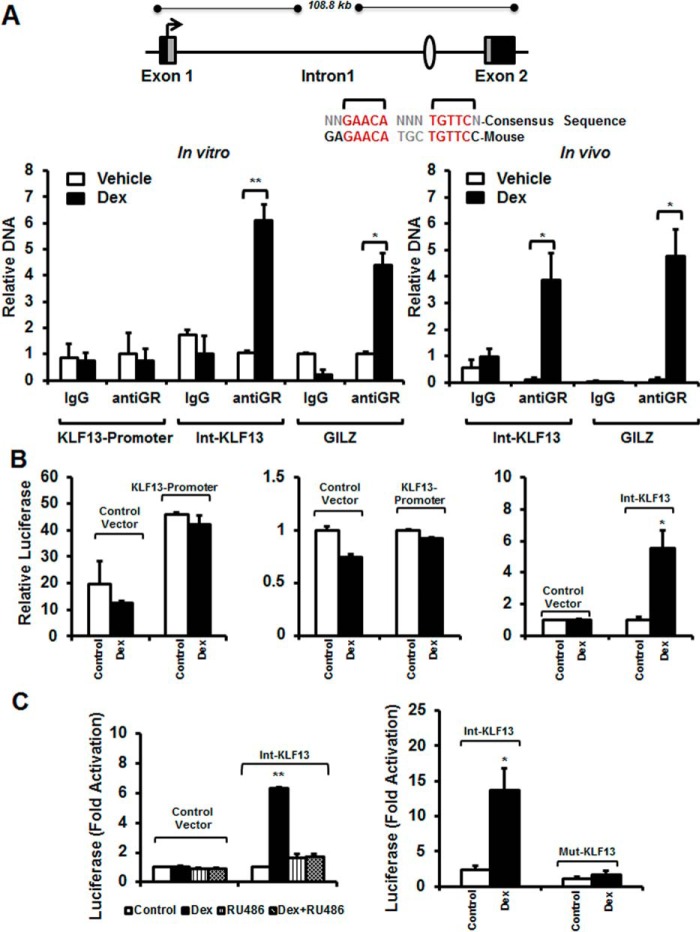
The Glucocorticoid Receptor Regulates Klf13 Expression by Binding to an Intragenic GRE
One mechanism by which glucocorticoid receptors modulate gene transcription is through direct binding to DNA sequences known as GREs (5, 30). In silico analysis of the Klf13 gene identified one putative GRE, with all critical nucleotides conserved as compared with the consensus GRE sequence, located in the promoter region (5 kb upstream of the transcription start site) and one putative GRE in intron-1 (Fig. 5A). ChIP assays were performed on vehicle- and glucocorticoid-treated HL-1 cells to determine whether glucocorticoid receptors are recruited to the identified GREs in its native chromatin context. We observed that glucocorticoid receptors were strongly recruited to the Klf13 intron-1 GRE when HL-1 cells were exposed to glucocorticoids for 90 min (Fig. 5A, left panel). A similar robust enrichment (4.37 ± 0.47-fold, p < 0.05) of activated glucocorticoid receptors was observed at a functional GRE in the promoter of the classical glucocorticoid-regulated gene Gilz (Fig. 5A, left panel). In contrast, glucocorticoid receptors were not significantly recruited to the putative GRE in the Klf13 promoter (Fig. 5A, left panel). In vivo ChIP assays using whole heart samples from glucocorticoid-treated mice yield very similar results further validating the data obtained in HL-1 cardiomyocytes. Glucocorticoid receptors were strongly recruited in the Klf13 intron-1 GRE in hearts from glucocorticoid-treated mice (Fig. 5A, right panel). These data show that activated glucocorticoid receptors bind to the intron-1 GRE to regulate Klf13 transcription in vitro (HL-1 cardiomyocytes) and in vivo (heart tissue). To determine whether the Klf13 intron-1 GRE functions as a glucocorticoid-dependent enhancer in a heterologous system, a 500-bp fragment of the Klf13 intron containing the putative GRE and a 1.7-kb fragment of the Klf13 promoter were cloned into luciferase reporter plasmids (31). HL-1 cells were transfected with the reporter plasmids, and luciferase activity was measured after an overnight treatment with either vehicle or glucocorticoid. The Klf13 promoter resulted in constitutive luciferase activity (45.95 ± 3.34-fold greater than the control vector) (Fig. 5B, left panel); however, in the absence of the intragenic GRE construct, luciferase activity was not regulated by glucocorticoid treatment (Fig. 5B, left panel). In contrast, glucocorticoid treatment induced a 5.55 ± 1.09-fold increase (p < 0.05) in luciferase activity in the construct containing the intragenic GRE (Fig. 5B, right panel). To define if the increase in luciferase expression exhibited by the construct with the intragenic GRE (intron-KLF13) requires glucocorticoid receptor binding to an agonist, transfected HL-1 cells were co-treated with dexamethasone and the glucocorticoid receptor-antagonist RU486. The glucocorticoid-mediated increase in luciferase activity was significantly inhibited in response to RU486 (Fig. 5C, left panel), indicating that the GRE in intron-1 of the Klf13 gene is functional. Finally, to analyze if glucocorticoid receptor binding to the intronic GRE confers glucocorticoid receptor-dependent induction of the target gene, the Klf13 intron-1 GRE sequence GAGAACATGCTGTTCC was mutated to GAGAACAAGCAGTTCC (366Thr → Ala). Mutation of the Klf13-intron-1 GRE significantly inhibited glucocorticoid induction of luciferase expression (Fig. 5C, right panel). These data reveal that the GRE in intron-1 of the Klf13 gene is functional, and it is activated by ligand occupied glucocorticoid receptors in cardiomyocytes.
FIGURE 5.
Glucocorticoid receptor is recruited to a functional GRE located in the intron of the KLF13 gene in vitro and in vivo. A, schematic representation of the KLF13 gene. Sequence alignment of the consensus GRE and GRE identified within the KLF13 intron of the mouse and human genes. HL-1 cells were treated with vehicle-control (white bars) or 100 nm Dex (black bars) for 3 h, and ChIP assays were performed with equivalent amounts of rabbit IgG or rabbit anti-GR antibody. Coimmunoprecipitated DNA was analyzed by RT-PCR using primers to the KLF13 promoter, the KLF13 intron (Int-KLF13) and a functional GRE previously identified in the promoter of the glucocorticoid-regulated gene leucine zipper (GILZ) (left panel). Adrenalectomized C57BL/6 male mice were injected with vehicle (Veh, white bars) or 1 mg/kg of Dex (black bars). Six h after this treatment hearts were harvested. Coimmunoprecipitated DNA was analyzed by RT-PCR using primers to the KLF13 intron (Int-KLF13) and GILZ promoter GRE (right panel). Results are plotted as a function of input DNA. B and C, HL-1 cells were transfected with the following luciferase reporter plasmids: control vector (empty vector), KLF13 promoter, intron-KLF13 (Int-KLF13), and mutant-KLF13 (Mut-KLF13). Twenty-four h after transfection cells were treated for 18 h with vehicle control (white bars), 100 nm Dex (black bars), 1 μm RU486 (striped bars) or both 1 μm RU486 and 100 nm Dex (cross-hatched bars). Cells were harvested and luciferase activity was measured. Data represent ± S.E. for four independent experiments and n = 5–10 mice per group. *, p < 0.05; **, p < 0.01.
Glucocorticoid Receptor Regulation of Klf13 Is Critical to Glucocorticoid Regulation of Cardiomyocyte Gene Expression
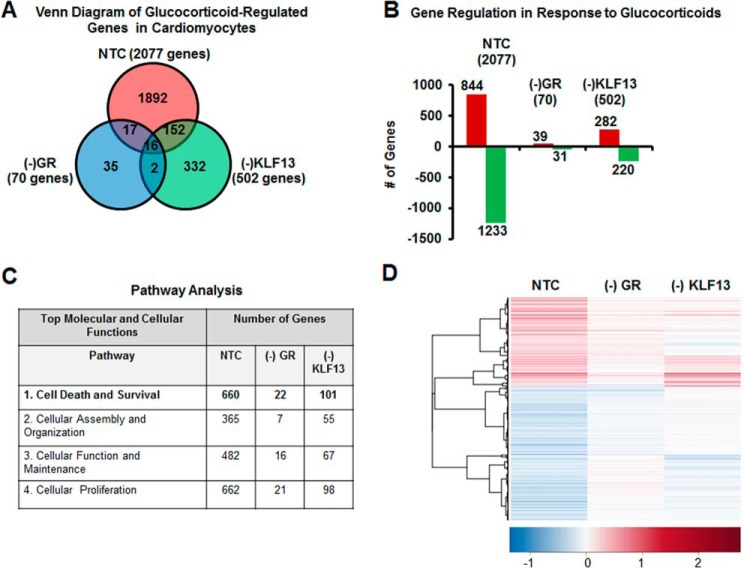
Our goal was to define the physiological significance of glucocorticoid regulation of Klf13 in adult cardiomyocytes. To address this issue, we examined the effects of silencing GR and KLF13 on the glucocorticoid-regulated transcriptome of HL-1 cardiomyocytes. For these studies, HL-1 cells transfected with NTC siRNA, GR siRNA (−GR), or KLF13 siRNA (−KLF13) and then treated with glucocorticoids or vehicle control for 24 h. Cells were then subjected to genome-wide microarray analysis. Gene lists for each condition were generated and are reported in supplemental Tables S1–S3. To identify unique and commonly regulated genes among the three treatments, gene lists were sorted by Venn diagram (Fig. 6A). In control cells (NTC), 2077 genes were regulated in response to glucocorticoids (Fig. 6A), whereas only 70 genes were regulated by hormone treatment in the (−)GR group (Fig. 6A). Only 502 genes were observed in (−)KLF13 cells (Fig. 5A). Venn diagram analysis showed that 1892 genes were uniquely regulated by glucocorticoids in control cells (Fig. 6A, red circle). In contrast, only 35 unique genes were observed in the (−)GR group (Fig. 6A, blue circle). Venn analysis also showed that 332 genes were unique to the (−)KLF13 cells (Fig. 6A, green circle). Only 16 genes were commonly regulated by glucocorticoids among control, (−)GR and (−)KLF13 groups (Fig. 6A, circle intersection). Analysis of the hormone-regulated genes showed that an equal proportion of up- and down-regulated genes were observed in the control cells (Fig. 6B). Similar results were found in the absence of GR and KLF13 (Fig. 6B) suggesting that GR or KLF13 knockdown did not alter the proportion of induced versus repressed genes in response to glucocorticoids, but rather the overall number of regulated genes (Fig. 6, A and B). These results suggest that the majority of the glucocorticoid-regulated genes (1892 of a total 2077 genes) in HL-1 cardiomyocytes depend on both hormone binding to their cognate receptor (GR) and on the modulation of KLF13 gene expression by glucocorticoid receptors.
FIGURE 6.
KLF13 regulation by glucocorticoids is important to maintain cardiomyocytes global gene expression. HL-1 cells were transfected with non-target siRNA (NTC), GR siRNA ((−)GR), or KLF13 siRNA ((−)KLF13) and treated with vehicle-control (Veh) or 100 nm Dex. mRNA was isolated and analyzed using whole mouse genome 4 × 44 multiplex format oligo array (Agilent) for gene expression. A, the glucocorticoid-regulated genes within each group were sorted by a Venn diagram. B, the number of probes that were regulated by glucocorticoids is organized as either induced (red) or repressed (green) according to treatment group in NTC, (−)GR and (−)KLF13. C, results from microarray analysis of the glucocorticoid-regulated genes found in the three treatments were loaded into Ingenuity Pathway Analysis software. Glucocorticoid treatment significantly regulated an important number of genes associated to different cellular and molecular biological pathways. Silencing of GR and KLF13 by siRNA inhibited glucocorticoid regulation of the majority of these genes. Cell death and survival (bold letters) was ranked as the top 1 molecular and cellular function. D, glucocorticoid treatment regulated 660 genes associated to cell death and survival in NTC cells. Only 22 and 101 of these same genes were regulated in the absence of GR and KLF13, respectively. A heat map was generated with Heatplus software (BioConductor) to visualize the differences in the gene expression of cell death and survival genes between treatment groups.
Glucocorticoid Regulation of KLF13 Activity Has a Marked Influence on the Expression of Genes Involved in Cell Death and Survival
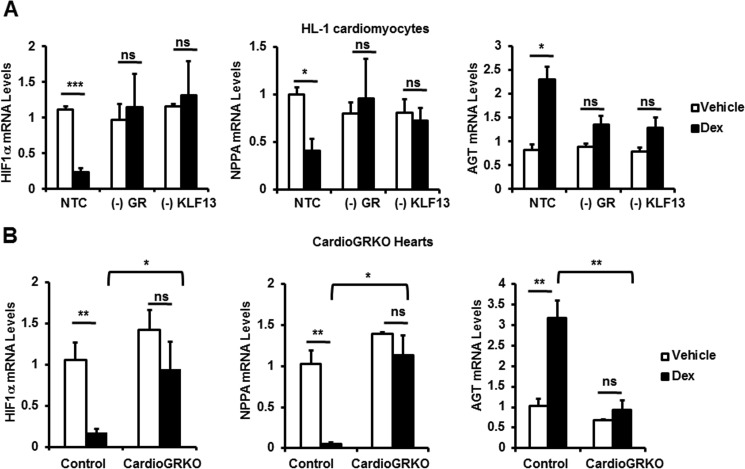
To elucidate the significance of the glucocorticoid-mediated regulation of the cardiomyocyte transcriptome, gene lists were analyzed by Ingenuity Pathway Analysis software (IPA). The analysis showed that genes associated with cell death and survival were highly regulated in NTC cells in response to glucocorticoids (Fig. 6C and supplemental Tables S4–S6). The regulation of these same genes differed significantly when GR was silenced by siRNA. 660 genes were associated with cell death and survival in control cells, and only 22 of these genes were regulated in these cells when GR was silenced (Fig. 6C). When KLF13 was knocked down in HL-1 cardiomyocytes, only 101 of the 660 cell death and survival genes were regulated by glucocorticoids (Fig. 6C). A heat map of averaged sample replicates within treatment groups and hierarchical clustering is shown in Fig. 6D to visualize cardiomyocyte cell death and survival gene expression profiles in response to glucocorticoids in our three group treatments. These results suggest that GR regulation of KLF13 activity has a marked influence in the expression of genes involved in pathways important for cardiomyocyte survival. Further bioinformatic analysis of the 660 genes regulated by glucocorticoids in control cells revealed that 39 of these genes have been reported to have a role in myocardial damage and ischemic injury (supplemental Table S7). Among these 39 genes, we found 3 genes involved in myocardial injury due to oxygen deprivation (supplemental Table S7): hypoxia inducible factor 1, α subunit (Hif1α), angiotensinogen (Agt), and atrial natriuretic peptide (Nppa). To validate the microarray results, control cells, (−)GR, and (−)KLF13 HL-1 cells were treated with vehicle or glucocorticoids for 24 h and the mRNA levels of this subset of genes were measured by RT-PCR. In agreement with our microarray data, we observed that changes in the expression of Hif1α, Nppa, and Agt in response to glucocorticoids depend on both GR and KLF13 (Fig. 7A). To further validate the microarray data, 8-week-old cardioGRKO mice and littermate controls were adrenalectomized and treated with vehicle (PBS) or glucocorticoids (Dex). Hearts were harvested at 6 h following this treatment. Six hours after glucocorticoid administration we observed significant changes in the mRNA levels of Hif1α, Agt, and Nnpa in control hearts; in contrast, no changes in the expression of these genes were found in glucocorticoid-treated cardioGRKO hearts (Fig. 7B). These data correlate with the results obtained in HL-1 cardiomyocytes and suggest that an intact GR-KLF13 axis is important for the regulation of these target genes. Thus, we explore the role of GR-KLF13 in hypoxia-induced cell death.
FIGURE 7.
Glucocorticoids regulate genes involved in cardiomyocyte death and survival in vitro and in vivo. A, microarray validation. HL-1 cells were transfected with NTC siRNA, GR siRNA (−GR), or KLF13 siRNA (−KLF13). Cells were then treated with vehicle (control, white bars) or 100 nm Dex (black bars) for 24 h. mRNA levels were determined for HIF1α, AGT, and NPPA by RT-PCR. B, adrenalectomized cardioGRKO male mice were injected with vehicle (Veh) or 1 mg/kg of Dex. RNA was isolated 6 h after the treatment. HIF1α, AGT, and NPPA mRNA levels in Veh (white bars)- and Dex (black bars)-treated mice. Values are presented as fold-change relative to the gene expression detected in cells treated with vehicle control. Data represent mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, no significant.
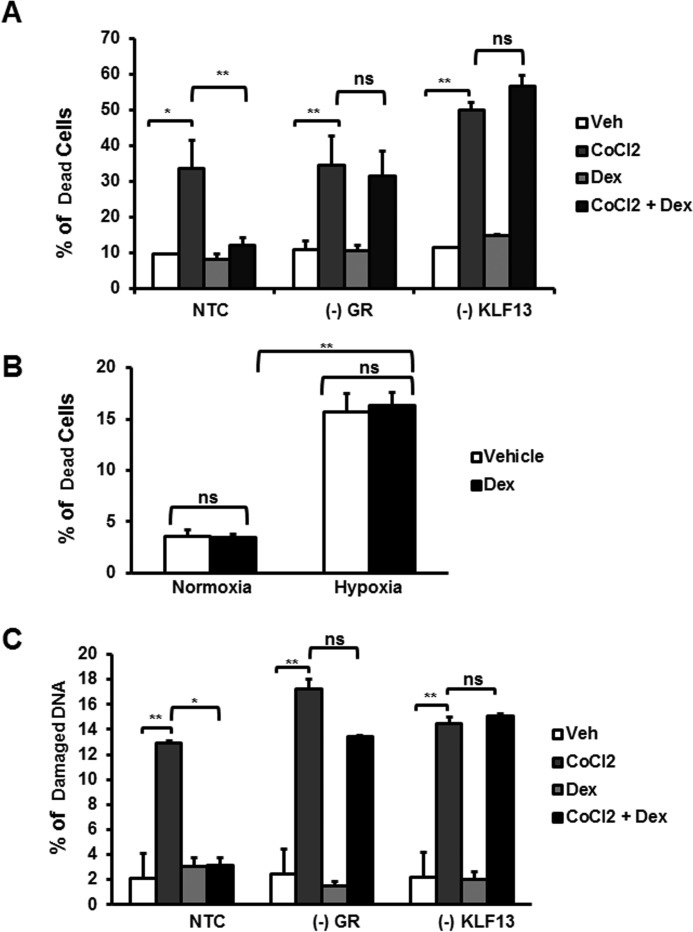
Glucocorticoids Protect Cardiomyocytes from Cell Death Triggered by CoCl2 Independently of Hypoxia Signaling Pathways
Hypoxia is an important factor involved in apoptotic cell death associated with ischemic heart disease (32). Studies have shown that glucocorticoids influence hypoxia-inducible gene expression in vitro (33). In addition, administration of glucocorticoids to mice significantly reduces cardiac damage after ischemia reperfusion injury (7). However, the protective effects afforded by glucocorticoids have not been explored, particularly at the cellular level. CoCl2 has been reported to induced hypoxia like gene expression in mammalian cell cultures (27), and prolonged exposure to this chemical can result in cell death (34). Therefore, to explore if GR and KLF13 play a role in CoCl2-induced cell death, NTC, (−)GR, and (−)KLF13 HL-1 cells were treated with CoCl2 for 24 h in the presence and absence of glucocorticoids. Following these treatments, cell death was measured by membrane integrity by flow cytometry (propidium iodide exclusion), which is characteristic of apoptosis. In response to CoCl2, we observed a 33.6% increase in cell death in NTC cells compared with vehicle-treated cells (Fig. 8A). Similar increases in cell death were observed with GR- and KLF13-deficient cells in the absence of glucocorticoids (Fig. 8A). Glucocorticoid treatment protected control cells from CoCl2-induced cell death (Fig. 8A), whereas glucocorticoid treatment did not protect HL-1 cardiomyocytes from cell death in the absence of GR or KLF13 (Fig. 8A). Together, these data indicate that one possible mechanism by which glucocorticoids protects HL-1 cardiomyocytes from CoCl2-induced death is through KLF13 regulation by GR. To further investigate if glucocorticoids protect cardiomyocytes from cell death under hypoxic conditions, HL-1 cells were subjected to hypoxia for 48 h in the presence/absence of glucocorticoids. Analysis of cell viability showed that glucocorticoids failed to protect HL-1 cardiomyocytes from hypoxia-induced cell death (Fig. 8B). Therefore, the protecting effects exerted by glucocorticoids in response to CoCl2 are likely independent of hypoxia signaling pathways.
FIGURE 8.
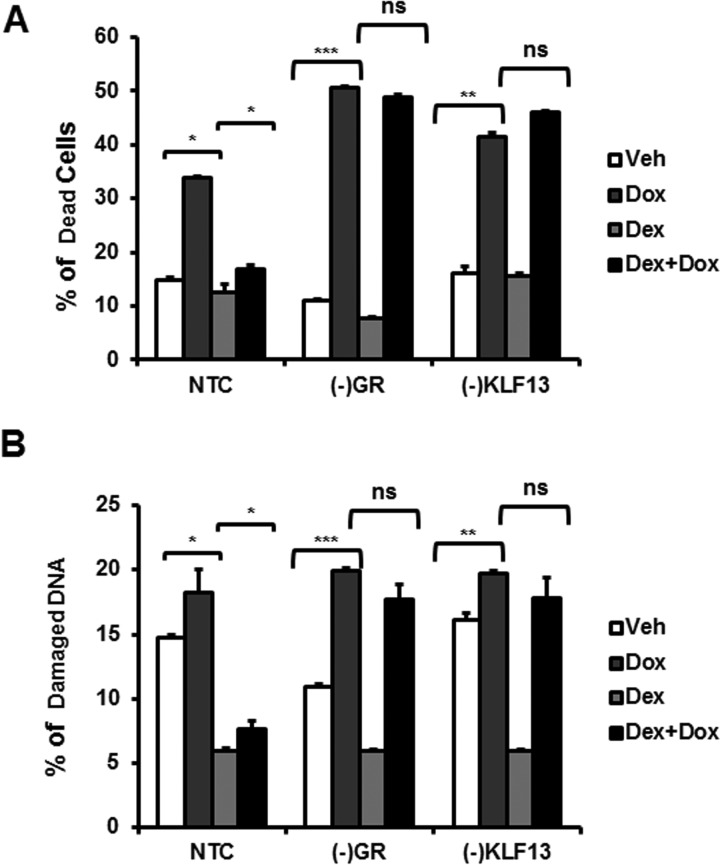
Glucocorticoid receptor and KLF13 expression is essential to protect cardiomyocytes against cell death and DNA damage triggered by CoCl2 independently of hypoxia pathways. A, HL-1 cardiomyocytes treated with non-target (NTC) siRNA, GR siRNA ((−)GR), or KLF13 siRNA ((−)KLF13) were co-treated with vehicle (Veh) or Dex and CoCl2 for 24 h. Cell viability and was then analyzed by flow cytometry for PI staining. The plot represents the percentage (%) of dead cells in each group treatment. B, HL-1 cells were subjected to hypoxia for 48 h in the presence/absence of glucocorticoids. Cell viability was then analyzed by flow cytometry for PI staining. The plot represents percentage (%) of dead cells in normoxia and hypoxia. C, HL-1 cardiomyocytes treated with non-target (NTC) siRNA, GR siRNA ((−)GR), or KLF13 siRNA ((−)KLF13) were co-treated with vehicle (Veh) or Dex and CoCl2 for 24 h. DNA degradation was analyzed by flow cytometry. The plot represents percentage (%) of degraded DNA in each group treatment. Data represented as the mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ns, no significant.
Glucocorticoids Protect Cardiomyocytes from DNA Degradation Triggered by CoCl2 and Doxorubicin
CoCl2 is known to enhance the production of reactive oxygen species in mammalian cell cultures (35), which in turn leads to DNA damage and cell death (36, 37). Glucocorticoids have been reported to protect cells from DNA damage and cell death by inducing the expression of anti-apoptotic genes and repressing pro-apoptotic genes (38). To investigate if glucocorticoids protect cardiomyocytes from DNA damage induced by CoCl2, DNA degradation was analyzed by flow cytometry. Control cells (NTC) treated with glucocorticoids were protected for DNA degradation in response to CoCl2 (Fig. 8C). In contrast, glucocorticoids failed to prevent DNA damage in the absence of GR or KLF13. These results suggest that glucocorticoids inhibit cell death induced by CoCl2 by preventing DNA damage through GR-KLF13 signaling activation.
To investigate the potential clinical implication of these results, HL-1 cardiomyocytes were treated with doxorubicin (Dox), an antineoplastic drug known to induce cardiomyopathy by damaging DNA (39, 40), NTC, (−)GR, and (−)KLF13 cells were cultured with dexamethasone for 48 h, and then treated with 1 μm Dox for 48 h. Glucocorticoid-treated NTC cells were protected from Dox-induced cell death and DNA degradation (Fig. 9, A and B). In contrast, glucocorticoid failed to protect cells in the absence of GR and KLF13 expression (Fig. 9, A and B). These results suggest that glucocorticoids protect cardiomyocytes from DNA damage and death through the regulation of KLF13.
FIGURE 9.
Glucocorticoids protect cardiomyocytes from DNA degradation triggered by doxorubicin. NTC, (−)GR, and (−)KLF13 cells were culture with dexamethasone for 48 h, and then treated with 1 μm Dox for 48 h. Cell viability and DNA damage were analyzed by flow cytometry. A, percentage (%) of dead cells in each group treatment. B, the plot represents the percentage (%) of degraded DNA in each group treatment. Data are represented as the mean ± S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, no significant.
KLF13 Protects Cardiomyocytes from Cell Death and DNA Degradation
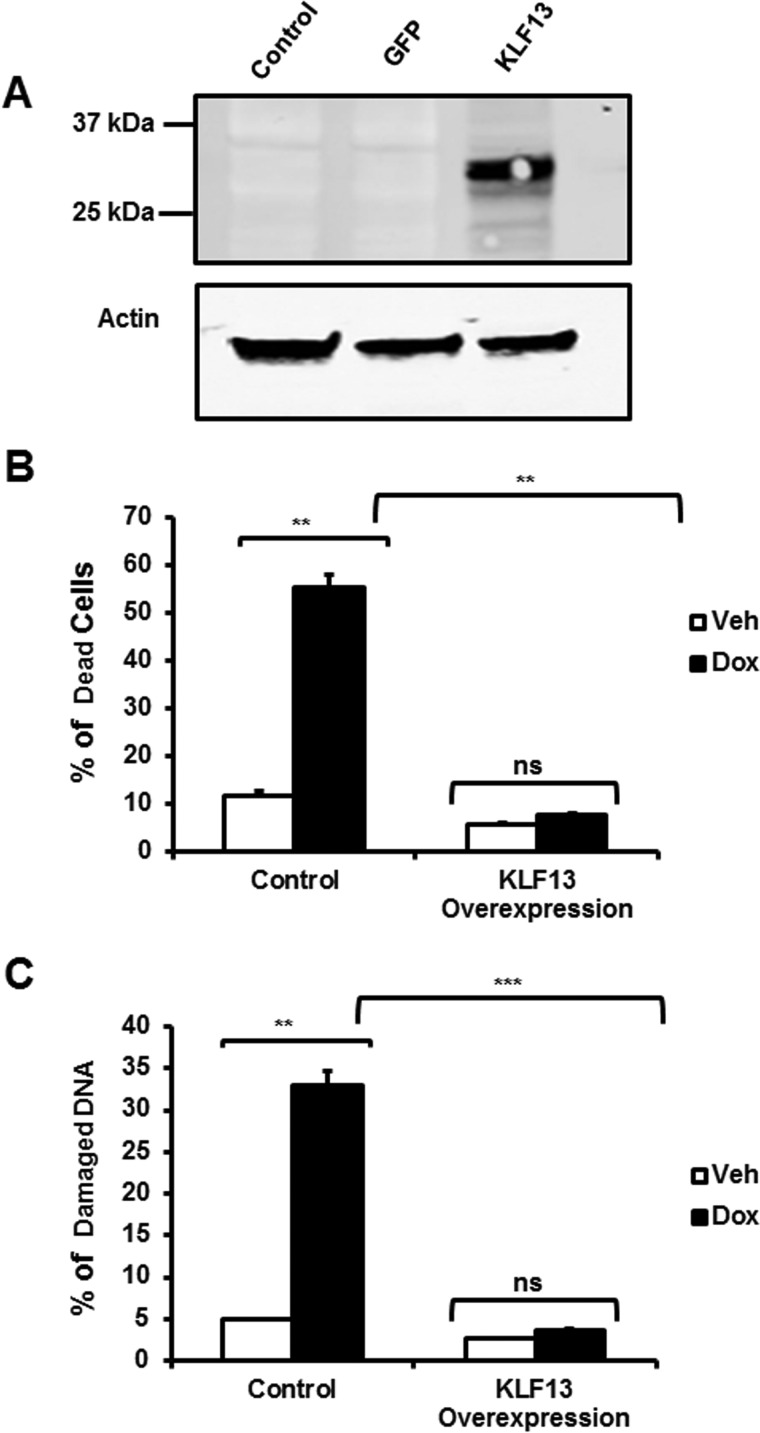
To investigate the protective role of KLF13 in cardiomyocytes, KLF13 was overexpressed in HL-1 cardiomyocytes and cells were then treated with 1 μm Dox. KLF13 overexpression was confirmed by Western blot analysis (Fig. 10A). Cells overexpressing KLF13 were protected from those Dox deleterious effects. No significant differences in cell survival were found between Dox-treated KLF13 overexpressing cells and vehicle-treated cells (Fig. 10B). In agreement with these results, KLF13 overexpressing cells were resistant to DNA damage induced by Dox (Fig. 10C). These data suggest that manipulation of KLF13 signaling protects cardiomyocytes against Dox-induced toxicity.
FIGURE 10.
KLF13 overexpression protects cardiomyocytes from cell death and DNA degradation triggered by doxorubicin. HL-1 cells were transfected with a KLF13 plasmid and treated with Dox for 48 h. A, representative Western blotting showing KLF13 protein levels in HL-1-transfected cells. Control (empty vector), GFP (control for transfection efficiency), and KLF13 plasmid are shown. B, percentage (%) of dead cells in control and KLF13 overexpressing cells. C, percentage (%) of degraded DNA in each group treatment. Data represented as the mean ± S.E. from three independent experiments. **, p < 0.01; ***, p < 0.001; ns, no significant.
Discussion
The nature of glucocorticoid-mediated actions in the heart remains controversial. Systemic high levels of glucocorticoids are linked with adverse cardiovascular events, but glucocorticoid administration has also been shown to be beneficial in the treatment of atherosclerosis and in the prevention of cardiomyocyte death triggered by ischemia reperfusion (7). Recent studies of mice with cardiomyocyte-specific deletion of normal glucocorticoid receptor revealed that endogenous glucocorticoid receptor signaling is essential for the maintenance of cardiac function (14). Deletion of glucocorticoid receptor in vivo leads to the dysregulation of genes associated with cardiovascular disease, including several members of the Krüppel-like factor family, among those Klf13 (Fig. 1) (14). Our data reveal that Klf13 is a direct glucocorticoid receptor-regulated gene, and its expression is induced by glucocorticoids in vivo (mouse heart) and in vitro (primary and immortal cell lines) (Figs. 2 and 3). These findings are supported by studies that have demonstrated that glucocorticoid receptor directly regulates several members of the KLF family (41, 42). We also found that KLF13 is a direct target of GR and its regulation involves binding of activated glucocorticoid receptors to a conserved and functional GRE within intron 1 of the Klf13 gene (Figs. 4 and 5). These results are consistent with the mechanism proposed for the regulation of KLF9 and KLF15 by glucocorticoids, which also involves binding of activated glucocorticoid receptor to two intronic consensus GREs (20, 42).
To elucidate the molecular and functional consequences of glucocorticoid regulation of Klf13 in adult cardiomyocytes, gene expression studies were performed. Microarray analysis suggests that one mechanism by which glucocorticoids exert effects on the adult cardiomyocyte transcriptional program is by the induction of KLF13. Silencing of GR or KLF13 by siRNA significantly altered the number of glucocorticoid-regulated gene probes in HL-1 cardiomyocytes (Fig. 6). Analysis of this cohort of genes by Ingenuity Pathway Analysis software showed that genes involved in cell death and survival were regulated by glucocorticoids in control HL-1 cardiomyocytes and the expression of these same genes was modified in the absence of Klf13 gene expression (Fig. 6). Consistent with these results, our previous studies on rat H9C2 cardiomyocytes have shown that glucocorticoid treatment leads to significant changes in gene expression profiles that are characterized by dysregulation of genes associated with cardiac hypertrophy and cell death (12); however, the molecular targets responsible for these actions remain unknown. Our microarray data presented here clearly suggest that some of the effects exerted by glucocorticoids in cardiomyocytes are mediated by GR signaling through KLF13.
To understand the precise role of GR secondary signaling in cardiomyocytes, a detailed analysis of the cell death and survival genes regulated by glucocorticoids in control cells was performed. This analysis showed that 39 of these genes have been reported to play a role in myocardial damage and ischemic injury (supplemental Table S7). Among these genes, we found HIF1α, AGT, and NPPA. HIF1 is a transcription factor that regulates the expression of pro- and anti-apoptotic genes that are affected by oxygen deprivation (43–45). HIF1α is a subunit of HIF1 and changes in its expression by hypoxia induction leads to cardiomyocyte apoptosis (45, 46). AGT is a component of the renin-angiotensin system, which is involved in the regulation of blood pressure and has been shown to play a role in cardiac function (47). NPPA is a vasodilator that exerts beneficial effects when used clinically for the treatment of heart failure (48) and protects cardiomyocytes from apoptosis in vitro (49). Changes in the expression of these genes were validated in HL-1 cells (Fig. 7A) and mouse hearts (Fig. 7B). In vivo data showed that glucocorticoid administration led to significant changes in the mRNA levels of Hif1α, Agt, and Nnpa in control hearts (expressing GR). In contrast, no changes in the expression of these genes were found in the absence of GR in cardiomyocytes (Fig. 7B). These data correlate with the results obtained in vitro and suggest GR and KLF13 play an important role in the regulation of signaling pathways involved in cardiomyocyte damage.
Numerous studies employing in vivo models of heart disease have shown that apoptosis plays a pivotal role during heart failure (50), we wanted to further evaluate the role of GR-KLF13 signaling (27). Our results show that glucocorticoids protect cardiomyocytes from CoCl2-induced cell death in a GR-KLF13-dependent manner (Fig. 8). These data are consistent with reports that glucocorticoid receptor signaling in cardiomyocytes inhibits cells death triggered by ischemia reperfusion, mechanical stress, serum starvation, or TNFα (7, 12, 33). However, our results showed that the cardioprotective effects exerted by glucocorticoids in response to CoCl2 were likely independent of hypoxia inducible factor pathways (Fig. 8). Glucocorticoids failed to protect HL-1 cardiomyocytes from exposure to low oxygen concentration (Fig. 8). CoCl2 has also been shown to induce the production of reactive oxygen species in mammalian cell cultures (35). Increase in reactive oxygen species is an important factor in cell death and DNA damage (36, 37). In agreement with these published data, we found that glucocorticoid prevented DNA damage induced by CoCl2 in HL-1 cells (Fig. 8), and this protection is dependent upon GR and KLF13 expression (Fig. 8).
Prevention of cardiomyocyte DNA damage has important clinical implications, the use of many effective chemotherapy agents, including anthracyclines, is limited by their cardiotoxicity (51). Oxidative stress is the primary mechanism by which anthracyclines induce cardiomyocyte injury and death (52). Doxorubicin is an anthracycline used for the treatment of breast cancer and leukemia (29). In addition to induction of oxidative stress, doxorubicin interacts with DNA topoisomerase causing cardiomyocyte damage and death (40). For example, Chen et al. (29) has also shown that glucocorticoids protect cardiomyocytes from cell death induced by doxorubicin. Consistent with the literature, our results showed that glucocorticoids protected HL-1 cardiomyocytes from cell death and DNA damage induced by Dox (Fig. 9). We found that one potential mechanism by which glucocorticoids attenuate cardiomyocyte toxicity in response to Dox is by GR-dependent up-regulation of KLF13, which in turn leads to the regulation of both anti- and pro-cell death genes (Fig. 9). These results were further confirmed by overexpressing KLF13 in HL-1 cardiomyocytes. Cells overexpressing KLF13 were protected from Dox-induced cell death and DNA damage (Fig. 10). Therefore, our findings establish a function for KLF13 in adult cardiomyocytes and provide evidence of the dynamic role of glucocorticoids in the modulation of cardiomyocyte function and identify a novel signaling pathway regulated by these hormones in cardiomyocytes.
Author Contributions
D. C. T. and B. H. conducted most of the experiments, analyzed the results, and wrote the paper. X. X. analyzed microarray data and pathways. J. A. C. directed the project and wrote the paper with D. C. T.
Supplementary Material
Acknowledgments
We thank Dr. W. C. Claycomb (Louisiana State University, New Orleans, LA) for kindly providing the HL-1 cell line used for our studies. We also acknowledge Dr. Carl Bortner from the Flow Cytometry Center and Dr. Kevin Gerrish from the Molecular Genomics Core for their technical assistance. Finally, we acknowledge Lois Wyrick from NIEHS arts and photography.
This work was supported by the Intramural Research Program of the National Institutes of Health, NIEHS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S7.
- GR
- glucocorticoid receptor
- KLF
- Kruppel-like factor
- GRE
- glucocorticoid response element
- PI
- propidium iodide
- NTC
- non-targeting control
- Dex
- dexamethasone
- HIF1α
- hypoxia inducible factor 1, α subunit
- AGT
- angiotensinogen, NPPA, atrial natriuretic peptide
- Dox
- doxorubicin.
References
- 1. Sapolsky R. M., Romero L. M., and Munck A. U. (2000) How do glucocorticoids influence stress responses? integrating permissive, suppressive, stimulatory, and preparative actions. Endocr. Rev. 21, 55–89 [DOI] [PubMed] [Google Scholar]
- 2. Rhen T., and Cidlowski J. A. (2005) Antiinflammatory action of glucocorticoids: new mechanisms for old drugs. N. Engl. J. Med. 353, 1711–1723 [DOI] [PubMed] [Google Scholar]
- 3. Evans R. M. (1988) The steroid and thyroid hormone receptor superfamily. Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kadmiel M., and Cidlowski J. A. (2013) Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 34, 518–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oakley R. H., and Cidlowski J. A. (2013) The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J. Allergy Clin. Immunol. 132, 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smoak K., and Cidlowski J. A. (2006) Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor α inflammatory signaling. Mol. Cell. Biol. 26, 9126–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tokudome S., Sano M., Shinmura K., Matsuhashi T., Morizane S., Moriyama H., Tamaki K., Hayashida K., Nakanishi H., Yoshikawa N., Shimizu N., Endo J., Katayama T., Murata M., Yuasa S., et al. (2009) Glucocorticoid protects rodent hearts from ischemia/reperfusion injury by activating lipocalin-type prostaglandin D synthase-derived PGD2 biosynthesis. J. Clin. Invest. 119, 1477–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Czerwinski-Helms S. M., and Hickson R. C. (1987) Specificity of activated glucocorticoid receptor expression in heart and skeletal muscle types. Biochem. Biophys. Res. Commun. 142, 322–328 [DOI] [PubMed] [Google Scholar]
- 9. Katz S. E., Penefsky Z. J., and McGinnis M. Y. (1988) Cytosolic glucocorticoid receptors in the developing rat heart. J. Mol. Cell Cardiol. 20, 323–328 [DOI] [PubMed] [Google Scholar]
- 10. Kayes-Wandover K. M., and White P. C. (2000) Steroidogenic enzyme gene expression in the human heart. J. Clin. Endocrinol. Metab. 85, 2519–2525 [DOI] [PubMed] [Google Scholar]
- 11. Sheppard K. E., and Autelitano D. J. (2002) 11β-Hydroxysteroid dehydrogenase 1 transforms 11-dehydrocorticosterone into transcriptionally active glucocorticoid in neonatal rat heart. Endocrinology 143, 198–204 [DOI] [PubMed] [Google Scholar]
- 12. Ren R., Oakley R. H., Cruz-Topete D., and Cidlowski J. A. (2012) Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology 153, 5346–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rog-Zielinska E. A., Thomson A., Kenyon C. J., Brownstein D. G., Moran C. M., Szumska D., Michailidou Z., Richardson J., Owen E., Watt A., Morrison H., Forrester L. M., Bhattacharya S., Holmes M. C., and Chapman K. E. (2013) Glucocorticoid receptor is required for foetal heart maturation. Hum. Mol. Genet. 22, 3269–3282 [DOI] [PubMed] [Google Scholar]
- 14. Oakley R. H., Ren R., Cruz-Topete D., Bird G. S., Myers P. H., Boyle M. C., Schneider M. D., Willis M. S., and Cidlowski J. A. (2013) Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proc. Natl. Acad. Sci. U.S.A. 110, 17035–17040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zuo P., Stanojević D., Colgan J., Han K., Levine M., and Manley J. L. (1991) Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev. 5, 254–264 [DOI] [PubMed] [Google Scholar]
- 16. McConnell B. B., and Yang V. W. (2010) Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 90, 1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fisch S., Gray S., Heymans S., Haldar S. M., Wang B., Pfister O., Cui L., Kumar A., Lin Z., Sen-Banerjee S., Das H., Petersen C. A., Mende U., Burleigh B. A., Zhu Y., et al. (2007) Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 104, 7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prosdocimo D. A., Anand P., Liao X., Zhu H., Shelkay S., Artero-Calderon P., Zhang L., Kirsh J., Moore D., Rosca M. G., Vazquez E., Kerner J., Akat K. M., Williams Z., Zhao J., et al. (2014) Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. J. Biol. Chem. 289, 5914–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lavallée G., Andelfinger G., Nadeau M., Lefebvre C., Nemer G., Horb M. E., and Nemer M. (2006) The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 25, 5201–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasse S. K., Mailloux C. M., Barczak A. J., Wang Q., Altonsy M. O., Jain M. K., Haldar S. M., and Gerber A. N. (2013) The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol. Cell. Biol. 33, 2104–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon A. R., Outram S. V., Keramatipour M., Goddard C. A., Colledge W. H., Metcalfe J. C., Hager-Theodorides A. L., Crompton T., and Kemp P. R. (2008) Splenomegaly and modified erythropoiesis in KLF13−/− mice. J. Biol. Chem. 283, 11897–11904 [DOI] [PubMed] [Google Scholar]
- 22. Cidlowski J. A., Bellingham D. L., Powell-Oliver F. E., Lubahn D. B., and Sar M. (1990) Novel antipeptide antibodies to the human glucocorticoid receptor: recognition of multiple receptor forms in vitro and distinct localization of cytoplasmic and nuclear receptors. Mol. Endocrinol. 4, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 23. Claycomb W. C., Lanson N. A. Jr, Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., and Izzo N. J. Jr. (1998) HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hescheler J., Meyer R., Plant S., Krautwurst D., Rosenthal W., and Schultz G. (1991) Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9c2) line from rat heart. Circ. Res. 69, 1476–1486 [DOI] [PubMed] [Google Scholar]
- 25. Ramamoorthy S., and Cidlowski J. A. (2013) Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol. Cell. Biol. 33, 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whirledge S., Xu X., and Cidlowski J. A. (2013) Global gene expression analysis in human uterine epithelial cells defines new targets of glucocorticoid and estradiol antagonism. Biol. Reprod. 89, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu D., and Yotnda P. (2011) Induction and testing of hypoxia in cell culture. J. Vis. Exp. 54, e2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vassilopoulos A., and Papazafiri P. (2005) Attenuation of oxidative stress in HL-1 cardiomyocytes improves mitochondrial function and stabilizes Hif-1α. Free Radic. Res. 39, 1273–1284 [DOI] [PubMed] [Google Scholar]
- 29. Chen Q. M., Alexander D., Sun H., Xie L., Lin Y., Terrand J., Morrissy S., and Purdom S. (2005) Corticosteroids inhibit cell death induced by doxorubicin in cardiomyocytes: induction of antiapoptosis, antioxidant, and detoxification genes. Mol. Pharmacol. 67, 1861–1873 [DOI] [PubMed] [Google Scholar]
- 30. So A. Y., Chaivorapol C., Bolton E. C., Li H., and Yamamoto K. R. (2007) Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oakley R. H., Revollo J., and Cidlowski J. A. (2012) Glucocorticoids regulate arrestin gene expression and redirect the signaling profile of G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 17591–17596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim N. H., and Kang P. M. (2010) Apoptosis in cardiovascular diseases: mechanism and clinical implications. Korean Circ. J. 40, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kodama T., Shimizu N., Yoshikawa N., Makino Y., Ouchida R., Okamoto K., Hisada T., Nakamura H., Morimoto C., and Tanaka H. (2003) Role of the glucocorticoid receptor for regulation of hypoxia-dependent gene expression. J. Biol. Chem. 278, 33384–33391 [DOI] [PubMed] [Google Scholar]
- 34. Chachami G., Simos G., Hatziefthimiou A., Bonanou S., Molyvdas P. A., and Paraskeva E. (2004) Cobalt induces hypoxia-inducible factor-1α expression in airway smooth muscle cells by a reactive oxygen species- and PI3K-dependent mechanism. Am. J. Respir. Cell Mol. Biol. 31, 544–551 [DOI] [PubMed] [Google Scholar]
- 35. Lison D., Carbonnelle P., Mollo L., Lauwerys R., and Fubini B. (1995) Physicochemical mechanism of the interaction between cobalt metal and carbide particles to generate toxic activated oxygen species. Chem. Res. Toxicol. 8, 600–606 [DOI] [PubMed] [Google Scholar]
- 36. Patlolla A., Patlolla B., and Tchounwou P. (2010) Evaluation of cell viability, DNA damage, and cell death in normal human dermal fibroblast cells induced by functionalized multiwalled carbon nanotube. Mol. Cell. Biochem. 338, 225–232 [DOI] [PubMed] [Google Scholar]
- 37. Wang G., Hazra T. K., Mitra S., Lee H. M., and Englander E. W. (2000) Mitochondrial DNA damage and a hypoxic response are induced by CoCl2 in rat neuronal PC12 cells. Nucleic Acids Res. 28, 2135–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stojadinovic O., Lee B., Vouthounis C., Vukelic S., Pastar I., Blumenberg M., Brem H., and Tomic-Canic M. (2007) Novel genomic effects of glucocorticoids in epidermal keratinocytes: inhibition of apoptosis, interferon-γ pathway, and wound healing along with promotion of terminal differentiation. J. Biol. Chem. 282, 4021–4034 [DOI] [PubMed] [Google Scholar]
- 39. Keizer H. G., Pinedo H. M., Schuurhuis G. J., and Joenje H. (1990) Doxorubicin (adriamycin): a critical review of free radical-dependent mechanisms of cytotoxicity. Pharmacol. Ther. 47, 219–231 [DOI] [PubMed] [Google Scholar]
- 40. Singal P. K., Li T., Kumar D., Danelisen I., and Iliskovic N. (2000) Adriamycin-induced heart failure: mechanism and modulation. Mol. Cell. Biochem. 207, 77–86 [DOI] [PubMed] [Google Scholar]
- 41. Bagamasbad P., Ziera T., Borden S. A., Bonett R. M., Rozeboom A. M., Seasholtz A., and Denver R. J. (2012) Molecular basis for glucocorticoid induction of the Kruppel-like factor 9 gene in hippocampal neurons. Endocrinology 153, 5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Asada M., Rauch A., Shimizu H., Maruyama H., Miyaki S., Shibamori M., Kawasome H., Ishiyama H., Tuckermann J., and Asahara H. (2011) DNA binding-dependent glucocorticoid receptor activity promotes adipogenesis via Kruppel-like factor 15 gene expression. Lab. Invest. 91, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wenger R. H., and Gassmann M. (1997) Oxygen(es) and the hypoxia-inducible factor-1. Biol. Chem. 378, 609–616 [PubMed] [Google Scholar]
- 44. Jiang B. H., Zheng J. Z., Leung S. W., Roe R., and Semenza G. L. (1997) Transactivation and inhibitory domains of hypoxia-inducible factor 1α: modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272, 19253–19260 [DOI] [PubMed] [Google Scholar]
- 45. Carmeliet P., Dor Y., Herbert J. M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., Koch C. J., Ratcliffe P., Moons L., Jain R. K., Collen D., Keshert E., and Keshet E. (1998) Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394, 485–490 [DOI] [PubMed] [Google Scholar]
- 46. Wang X., Ma S., and Qi G. (2012) Effect of hypoxia-inducible factor 1-α on hypoxia/reoxygenation-induced apoptosis in primary neonatal rat cardiomyocytes. Biochem. Biophys. Res. Commun. 417, 1227–1234 [DOI] [PubMed] [Google Scholar]
- 47. De Mello W. C., and Danser A. H. (2000) Angiotensin II and the heart: on the intracrine renin-angiotensin system. Hypertension 35, 1183–1188 [DOI] [PubMed] [Google Scholar]
- 48. Colucci W. S., Sawyer D. B., Singh K., and Communal C. (2000) Adrenergic overload and apoptosis in heart failure: implications for therapy. J. Card. Fail. 6, 1–7 [PubMed] [Google Scholar]
- 49. Kato T., Muraski J., Chen Y., Tsujita Y., Wall J., Glembotski C. C., Schaefer E., Beckerle M., and Sussman M. A. (2005) Atrial natriuretic peptide promotes cardiomyocyte survival by cGMP-dependent nuclear accumulation of zyxin and Akt. J. Clin. Invest. 115, 2716–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kang P. M., and Izumo S. (2000) Apoptosis and heart failure: a critical review of the literature. Circ. Res. 86, 1107–1113 [DOI] [PubMed] [Google Scholar]
- 51. Grenier M. A., and Lipshultz S. E. (1998) Epidemiology of anthracycline cardiotoxicity in children and adults. Semin. Oncol. 25, 72–85 [PubMed] [Google Scholar]
- 52. Zhou S., Palmeira C. M., and Wallace K. B. (2001) Doxorubicin-induced persistent oxidative stress to cardiac myocytes. Toxicol. Lett. 121, 151–157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.