Abstract
There has been a recent interest in the broader physiological importance of multispecific “drug” transporters of the SLC and ABC transporter families. Here, a novel multi-tiered systems biology approach was used to predict metabolites and signaling molecules potentially affected by the in vivo deletion of organic anion transporter 1 (Oat1, Slc22a6, originally NKT), a major kidney-expressed drug transporter. Validation of some predictions in wet-lab assays, together with re-evaluation of existing transport and knock-out metabolomics data, generated an experimentally validated, confidence ranked set of OAT1-interacting endogenous compounds enabling construction of an “OAT1-centered metabolic interaction network.” Pathway and enrichment analysis indicated an important role for OAT1 in metabolism involving: the TCA cycle, tryptophan and other amino acids, fatty acids, prostaglandins, cyclic nucleotides, odorants, polyamines, and vitamins. The partly validated reconstructed network is also consistent with a major role for OAT1 in modulating metabolic and signaling pathways involving uric acid, gut microbiome products, and so-called uremic toxins accumulating in chronic kidney disease. Together, the findings are compatible with the hypothesized role of drug transporters in remote inter-organ and inter-organismal communication: The Remote Sensing and Signaling Hypothesis (Nigam, S. K. (2015) Nat. Rev. Drug Disc. 14, 29). The fact that OAT1 can affect many systemic biological pathways suggests that drug-metabolite interactions need to be considered beyond simple competition for the drug transporter itself and may explain aspects of drug-induced metabolic syndrome. Our approach should provide novel mechanistic insights into the role of OAT1 and other drug transporters implicated in metabolic diseases like gout, diabetes, and chronic kidney disease.
Keywords: ABC transporter, diabetes, kidney, microbiome, systems biology, Recon, SLC transporter, chronic kidney disease, genome-scale metabolic reconstruction, pharmacophore
Introduction
A great deal of recent evidence suggests that solute carriers (SLC) 2 play a much broader role in physiology, including signaling and metabolism, than has been previously appreciated (1, 2). Indeed, there has been a recent call for more systematic analysis of the roles of SLCs in metabolism and signaling (1). Perhaps due to heavy emphasis on their key role in pharmacokinetics, members of the SLC (and ABC as well) “drug” transporter families are not generally depicted in biochemical pathways involving the endogenous metabolites they transport (2). Such an omission could have clinical consequences, because drugs directly or indirectly affect pathways normally involved in the movement of key metabolites, pathway intermediates, and signaling molecules, thereby fundamentally affecting cell and organ physiology in normal, pathophysiological, and developmental situations (1, 2).
According to the remote sensing and signaling hypothesis, SLC and ABC drug transporters are important in regulating the movement of small endogenous molecules such as key metabolites (e.g. α-ketoglutarate, tryptophan metabolites), signaling molecules (e.g. cAMP, prostaglandins, polyamines), vitamins, antioxidants (e.g. uric acid), and certain hormones (e.g. thyroxine) between tissues, organs, and even organisms (2–11). According to this theory, the SLC and ABC transporters form an integrated network allowing remote communication between different tissues via small endogenous molecules. This integrated network functions in a manner similar to the neuroendocrine system and is, in fact, interlinked with it. The ability of SLC and ABC drug and other transporters to regulate or modulate broad aspects of systemic physiology suggests that drug-metabolite interactions might extend well beyond simple competition for transport at the binding site(s) and provide an explanation for aspects of certain drug-induced metabolic syndromes (e.g. those seen with diuretic use or chronic HIV antiviral treatment (12, 13)). Furthermore, elucidation of their physiological role is likely to be useful for further defining the roles of drug transporters in modulating common metabolic diseases, such as diabetes, gout, and chronic kidney disease (2, 4, 14, 15).
Among the aforementioned transporters, organic anion transporter 1 (OAT1/SLC22A6, originally identified as NKT (16, 17)), likely the main basolateral probenecid-sensitive organic anion drug transporter of the kidney, mediates rate-limiting steps in the renal elimination of organic anionic drugs and a few cationic drugs (7, 18–22). This drug transporter has also long been suggested to play a role in key endogenous functions (21, 23–25), whereas analysis of Oat1 knock-out mice have provided critical information about its potential role in basal physiology (23, 26, 27). For example, targeted and untargeted metabolomics analyses have revealed significant changes in the concentrations of a number of endogenous metabolites in the knock-out animal, including key biochemical pathway intermediates and signaling molecules (23, 27). Moreover, OAT1 (as well as other SLC22 transporters) has also been associated with metabolic abnormalities and disease (2, 3, 7, 11, 25, 26, 28, 29). Taken together, this suggests an important, if underappreciated, role for SLC drug transporters in metabolic processes and signaling.
Taking a cue from our previous systems biology efforts to analyze the physiological role of OAT1 (11), we sought to build a detailed map of metabolic and signaling pathways modulated by “drug” transporters such as OAT1. A systems biology approach involving integration of OAT1 knock-out and wild type gene expression data into a genome-scale metabolic reconstruction (GEM) together with constraint-based modeling (i.e. flux variability analysis (FVA)) was used to predict metabolites affected by the absence of OAT1. Pharmacophore-based virtual screening, re-evaluation of existing transport, and knock-out metabolomics data, as well as wet-lab validation were then used to constrain and rank the predicted compounds based on their potential to directly interact with OAT1. Input of this data into the Cytoscape plug-in, MetScape, enabled the generation of a largely experimentally validated, confidence-ranked OAT1-centered metabolic interaction network. Pathway and enrichment analysis supported an important role for OAT1 in several key metabolic and signaling pathways. Moreover, the results indicate the feasibility of this novel hierarchical, integrative approach in the context of generally understanding drug transporter-related metabolism, as well as other biological processes involving SLC and ABC transporters. The results appear consistent with the Remote Sensing and Signaling Hypothesis (2–11).
Results
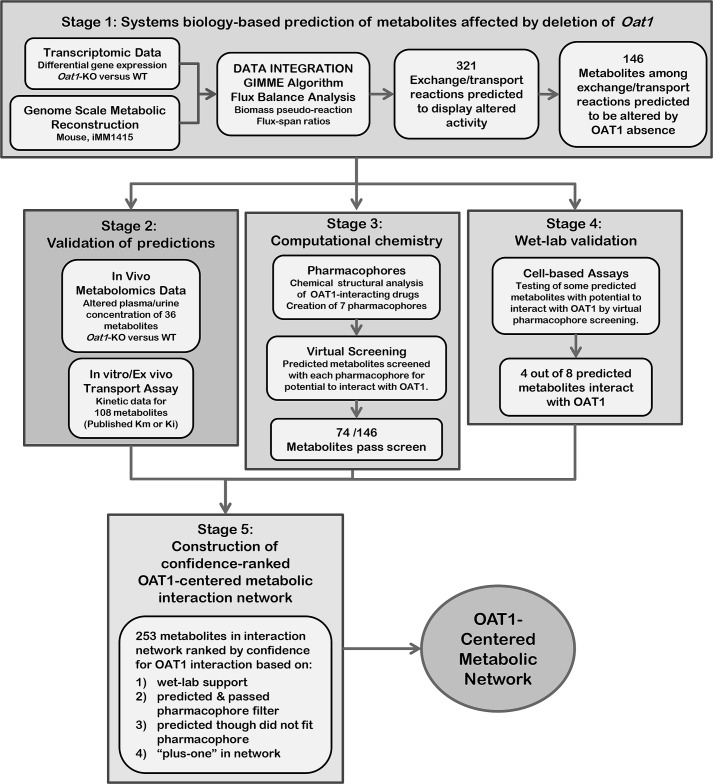
We have previously used GEMs to identify non-obvious, novel OAT1 substrates that were experimentally validated (3, 11). However, it was also noted that the GEMs failed to detect some of the known OAT1 substrates. Thus we hypothesized that by using constraint-based modeling (to capture systems level interactions) in conjunction with pharmacophore modeling (to capture molecular substrate-receptor (transporter) binding interactions), the strengths of each methodology could be leveraged to overcome their respective weakness. We performed a loosely constrained, context-specific analysis of transcriptomic data of a global metabolic model to catch all possible metabolites potentially interacting with OAT1 (either directly or indirectly) followed by pharmacophore-based virtual screening, along with comparison to in vivo/in vitro databases and wet-lab assays to provide the specificity filters to differentiate those metabolites that would directly interact with OAT1 from those likely to indirectly be part of the pathway (but not an OAT1 substrate). Ultimately, this multi-tiered systems level analysis, together with metabolomics (from knock-out and wild type animals) and transport data from OAT1-expressing cells was used to build a detailed confidence-ranked OAT1-centered metabolic interaction network that includes many small molecules with well established functions in metabolic and signaling pathways (Fig. 1). We arrived at the final network by working through 5 stages:
FIGURE 1.
Overall strategy employed. A diagram of the overall multi-tiered hierarchal approach employed in this study. As described under “Results,” the approach is comprised of 5 stages: 1) systems biology analysis of transcriptomic data for the prediction of metabolites altered by the absence of OAT1 (supplemental Fig. S1 and Table S1); 2) validation of predictions using metabolomics and kinetic data (supplemental Fig. S1); 3) computational chemistry analysis, generation of pharmacophore hypotheses, and the application of pharmacophore filtering (supplemental Fig. S2 and Table S2); 4) wet-lab validation and identification of new OAT1 metabolites/signaling molecules (Fig. 3); and 5) construction of a substantially validated OAT1-centered metabolic network (Fig. 4, supplemental Fig. S3).
Stage 1: Prediction of Metabolites Affected by the Deletion of OAT1
The first phase involved a systems biology approach in which transcriptomic data derived from the Oat1-KO mouse was integrated into the mouse GEM, iMM1415 (30), to predict metabolites potentially affected by the absence of this transporter (supplemental Fig. S1 and Table S1). Context-specific wild type (WT) and knock-out (KO) models were constructed from kidney gene expression data using the Gene Inactivity Moderated by Metabolism and Expression (GIMME) algorithm (31) with iMM1415. This systems biology approach simulates the metabolic state of a tissue/organ based on transcriptomic data and the execution of specific biological processes by converting the metabolic network into mathematical equations followed by the application of linear programming to calculate a solution of fluxes (i.e. measurement of rate of production or depletion of metabolites) for each of the reactions. We confirmed that the generated models could produce many of the small molecules requisite for the normal physiologic function of kidneys.
FVA (32), which calculates the boundary points of the steady state solution space, is one of most common constraint-based approaches used in investigations of the effects of gene deletion on a system (33); however, it requires an appropriate objective function (e.g. biomass production, ATP production, substrate uptake) that yields realistic flux distributions (34). However, the current metabolic models do not adequately encapsulate the in vivo metabolic alterations likely resulting from the loss of the drug transporter (OAT1) in a complex organ, and we were forced to apply approximations to the data to gain some understanding of the in vivo role of OAT1 in metabolism. Thus, whereas several objective functions were considered, including the biomass objective function, the ATP objective function and the renal objective function (35), we ultimately decided to use the biomass objective function because it is: 1) comprised of many metabolites involved in core cellular processes including those involved in regulating/modulating cell maintenance; and 2) applicable to non-proliferative cells, which either produce or consume these metabolites. Furthermore, many of the essential metabolites involved in regulating/modulating cell maintenance (e.g. Krebs cycle intermediates, prostaglandins, vitamins, uric acid, and polyamines) are transported by OAT1 and/or other closely related SLC22 family members (2, 36, 37). (Note that, in the simulations and comparative analyses between the wild type and knock-out models, we are not optimizing for growth, thus we are not assuming that biomass is being maximized, but rather only that the biomass components can be produced by the cells, which we know to be the case, biologically.)
To increase the sensitivity of model predictions, we used broad constraints at this initial stage of investigation allowing the inclusion of all possible metabolites that were then ranked in confidence based on the application of pharmacophore screens, as well as available data on their ability to directly interact with OAT1 (from either in vivo metabolomics data or pharmacokinetic data). The resulting WT and KO models consisted of 2233 and 2143 reactions, respectively (supplemental Fig. S1). FVA was used to compare metabolic differences and to find the maximum and minimum flux values for each reaction in the network, enabling the calculation of flux span ratios of the KO over WT for each reaction. Reactions with flux span ratios equal to 1 indicated no change in reaction activities due to the deletion of Oat1; flux span ratios less than 1 implied decreased reaction activities, whereas ratios greater than 1 indicated increased reaction activities. It was assumed that reactions that require transport of metabolites would change in the OAT1 KO, and 1026 reactions with altered activities were identified, including 321 exchange/transport reactions, which were responsible for the handling of 177 metabolites. After excluding water and other uninformative molecules (see “Experimental Procedures”), 146 metabolites remained (supplemental Table S1), which were predicted to be linked to OAT1-mediated transport.
Stage 2: Validation of the Models Using in Vivo Data from the Oat1-KO, as Well as with in Vitro/ex Vivo Transport Data
Multiple approaches were used to validate the metabolic reconstruction, as well as to provide a measure of confidence in the ability of the predicted metabolites to interact with OAT1. Initially, metabolomics data from the Oat1-KO were interrogated to determine whether experimental observations corresponded with computational predictions. Previous metabolomics profiles identified 36 metabolites (some of which are also signaling molecules), with significantly altered plasma and/or urine concentrations between the WT and Oat1-KO (Fig. 1, supplemental Fig. S1 and Table S1) (23, 27). Among these 36 metabolites, only 19 were actually included in the iMM1415 GEM (supplemental Table S1), and direct comparison with this metabolomics data revealed that 10 of these 19 metabolites were predicted by the GEM analysis (supplemental Table S1). Applying a hypergeometric test to this sample clearly indicates statistically significant enrichment in such metabolites in our population of 146 predicted metabolites (p < 0.01), which suggests that the systems biology approach integrating transcriptomics data together with loosely constrained FVA modeling makes reasonable predictions of the in vivo metabolic differences between WT and KO.
In addition, because metabolites that interact with OAT1 would be expected to be affected by the absence of this transporter, a search of the literature was performed and a list of 108 metabolites for which kinetic data exists (i.e. Km and/or Ki) indicating interaction with OAT1 was identified. Among these 108 metabolites, 56 are present in the iMM1415 GEM, of which 21 were found within the 146 GEM-predicted metabolites (supplemental Table S1). Once again, applying a hypergeometric test to this sample reveals statistically significant enrichment in OAT1-interacting metabolites in our population of 146 predicted metabolites (p < 0.01). Moreover, combining the metabolomics results with the kinetic data generated a list of 65 non-overlapping metabolites with wet-lab support (either in vivo metabolomics or in vitro/ex vivo kinetic data) for interaction with OAT1 in the iMM1415 GEM and out of these 65 metabolites 24 were predicted by the systems biology analysis and applying the hypergeometric test indicates significant enrichment in OAT1-interacting metabolites among the 146 GEM-predicted metabolites (p < 0.01). Taken together, the significant enrichment of the 146 GEM-predicted metabolites in OAT1-interacting metabolites indicates the utility of the loosely constrained systems biology approach utilized in our study.
Stage 3: Pharmacophore Analysis of Metabolites' Potential to Interact with OAT1
As described above, the broad constraints were applied in Stage 1 to identify all possible reactions and maximize the prediction of potential endogenous metabolites and signaling molecules likely affected by the absence of this drug transporter. To generate an OAT1-centered interaction network, this broad list of metabolites was then filtered and ranked by their potential to interact with OAT1. QSAR and pharmacophore modeling have been used to analyze limited sets of OAT drugs/substrates (6, 9, 18, 38, 39). Although many drugs appear to be related to metabolites and signaling molecules (40, 41), the availability of chemical libraries and computational tools have led to more systematic comparisons of metabolites, natural compounds, and drugs (42–45). Indeed, pharmacophores based on OAT1 metabolites have previously been used to virtually screen chemical libraries and identify potential inhibitors that have been experimentally validated (27, 46). We thus reasoned that the chemical features of known OAT1-transported drugs might be used to rank the predicted metabolites for their potential to interact with OAT1. In addition, some of these metabolites could then be prioritized for later wet-lab validation to assess direct interaction with OAT1. Therefore, OAT1 pharmacophore models based on a large set of well established drug ligands were built.
To construct pharmacophore models for OAT1, 61 drugs having a published Km or Ki less than 100 μm for OAT1 were selected as “actives” for model building and model validation; two-thirds of the drugs in this group (41 drugs) were used to build the pharmacophore models (training set), and one-third (20 drugs) of the actives was used as a validating set (supplemental Fig. S2A and Table S2). Because the drugs possess diverse chemical structures (consistent with the known multispecific nature of OAT1), they were first clustered into groups using their atomic property fields (APF) (e.g. hydrogen bond donors, hydrogen bond acceptors, SP2 hybridization, lipophilicity, sizes of large atoms, positive and negative charges, etc.) as discriminators (47–48). Thus, the training actives were grouped into 7 distinct clusters (Fig. 2A), and pharmacophore models were then built for each cluster based upon the alignment of its members (Fig. 2, B and C). Fig. 2B demonstrates how members of cluster 1 were first aligned, and “pharmacophore model 1” was built to represent the three-dimensional atomic properties shared among the members of that cluster. Then, the pharmacophore models were validated based on the validating set (known positives) and drugs from Drugbank database (serving as true negatives), and a ROC curve was generated (supplemental Fig. S2B). The calculated area under curve was 80.58, which supports the utility of these pharmacophores to identify OAT1-interacting compounds.
FIGURE 2.
Pharmacophore generation for characterization of metabolites. A, clustering of 41 OAT1 drugs based on APF (supplemental Table S2 and Fig. S2). Seven clusters of 3 or more drugs were created and each cluster was used for the generation of a single pharmacophore hypothesis. B, the alignment of drugs from Cluster 1 and the creation of the pharmacophore model: (a) the chemical structures of the drugs in the cluster were aligned; (b) chemical determinants are superimposed on the cluster alignment as three-dimensional pharmacophore features; (c) three-dimensional pharmacophore model of the aligned drugs created and used for virtual screening. C, pharmacophore models that were generated from each of the 7 drug clusters; blue, hydrogen bond donor; red, hydrogen bond acceptor; white, aromaticity; yellow, hydrophobicity; light red, negative charges; light blue, positive charges.
The 7 pharmacophores (Fig. 2C) were then used as three-dimensional chemical space constraints for virtual screening of the predicted molecules that revealed that 74 of the 146 predicted metabolites satisfied the constraints (supplemental Table S1) and were therefore predicted to have direct interaction with OAT1. Of these 74 metabolites, 18 are known to have direct interaction with OAT1 based on previous experimental in vitro observations (supplemental Table S1). Thus, compared with the original list of 146 metabolites that had 21 metabolites with kinetic data indicating interaction with OAT1 (prior to pharmacophore filtering), the percentage of metabolites known to have direct interactions was enriched about 2-fold (from 14.4% (21 of 146) to 24.3% (18 of 74) after filtering) and this enrichment in OAT1-interacting metabolites was found to be statistically significant by the hypergeometric test (p < 0.01).
Stage 4: Wet-lab Validation and Identification of Novel OAT1 Ligands
Based on their ability to fit the pharmacophore models, a subset of 8 commercially available metabolites out of the remaining 56 metabolites predicted to directly interact with OAT1 in the pharmacophore screen, but for which there is no wet-lab kinetic data indicating actual interaction with this transporter, were then randomly selected for validation in wet-lab transport assays. Of these 8 metabolites/signaling molecules, four were found to interact with OAT1 in transfected cells (Fig. 3, Supplemental Table S1). These metabolites/signaling molecules were dihydrofolic acid, palmitoleic acid, 16-hydroxy-hexadecanoic acid, and prostaglandin E1 with calculated Ki values of 93, 200, 13, and 12 μm, respectively (Fig. 3); values that are well within the documented range for many compounds shown to interact with OAT1 (supplemental Table S2). These metabolites are important in whole-body physiology, signaling, and cellular metabolism. For example, prostaglandin E1, an endogenous vasodilator, serves to increase peripheral blood flow (48), whereas dihydrofolic acid is required to synthesize both purines and pyrimidines. Palmitoleic acid, a long-chained fatty acid serving as a potential lipokine, is important in the regulation of lipid metabolism (49).
FIGURE 3.
Wet-lab validation of predicted metabolites and the identification of novel OAT1 metabolites. The IC50 curves for the metabolites characterized in the in vitro uptake inhibition assay using OAT1-expressing CHO cells. The assay was done by testing for inhibition of uptake of 10 μm 6-carboxyfluorescein, a fluorescent tracer that is also an OAT1 substrate (35, 36) (Km = 3.9 μm).
Taken together with the metabolomics and existing kinetic data described above, the number of wet-lab supported metabolites was increased and the application of a hypergeometric statistical test to the overall method for prediction and identification of novel OAT1 metabolites (the combined in silico and in vitro approach), indicates significant enrichment in the number of OAT1-interacting metabolites (p < 0.01).
Stage 5: Construction of an OAT1-centered Metabolic Interaction Network
The next phase of this study involved construction and analysis of a substantially validated, confidence ranked OAT1-centered metabolite interaction network (Figs. 4, 5, supplemental Fig. S3 and Table S3). To link OAT1 to multiple metabolic pathways, an interaction network was built based on the results of the aforementioned systems biology/pharmacophore approach and the wet-lab data (validated here or published previously), using Metscape, a Cytoscape plug-in used to construct and visualize metabolic networks based on the KEGG database (50). The resulting broader OAT1-centered metabolic network (see “Experimental Procedures”) consisted of a total of 253 metabolites, including 176 experimentally validated and/or computationally predicted and 77 “plus-one” (directly connected) metabolites (Fig. 4). Of these 176 metabolites, 73 had wet-lab support for interactions with OAT1 either by in vivo metabolomics from the knock-out or in vitro assays that were performed in this study or published. Moreover, ∼3% of the 2272 (i.e. 78/2272) metabolites comprising the MetScape database at the time of analysis were among those for which OAT1-interaction data were available; however, within the 253 metabolites comprising the OAT1-interaction network more than 28% (i.e. 73/253) have been shown to interact with OAT1. This represented a significant enrichment for OAT1-interacting metabolites in the OAT1-centered interaction network (p < 0.01). These metabolites were thus placed in the group termed “wet-lab” support (supplemental Table S3) and had the highest level of confidence for being part of an OAT1-centered metabolic network based on their ability to interact with OAT1.
FIGURE 4.
An OAT1-centered metabolic interaction network contains several essential biochemical pathways. Interaction network consisting of metabolites classified according to the level of confidence for OAT1 interaction: 1) wet-lab supported; 2) predicted to interact with high confidence; 3) predicted to be affected indirectly; and 4) first neighbor (plus-one) (supplemental Table S3). The level of confidence of metabolites is reflected by the size and color of nodes (larger and darker nodes have a higher ranking). In the network, some well represented metabolic pathways are shown. For example, purine metabolism is shown as 25 nodes: 5 wet-lab supported (urate, GMP, hypoxanthine, inosine, 3′,5′-cyclic GMP); 7 predicted to interact with high confidence (guanosine, deoxyguanosine, deoxyinosine, dGDP, deoxyadenosine, dADP, L-glutamine); 2 predicted to be affected indirectly (3′, 5′-cyclic AMP, L-aspartate); and 10 first neighbors (guanine, dGMP, reduced glutaredoxin, dAMP, adenine, AMP, IMP, xanthosine 5′-phosphate, GMP, 5-phospho-alpha-D-ribose 1-diphosphate).
FIGURE 5.
Pathway enrichment analysis of the OAT1-centered metabolic interaction network identifies several essential biochemical pathways affected by the absence of OAT1. A, graph of pathway enrichment analysis comparing the -log(p) to the impact on the various pathways for the network metabolites. Some of the affected pathways are indicated with circles colored based on the p value (darker red indicates a lower p value, whereas yellow indicates less significance) and sized based on the impact on the pathway (larger circles have a greater impact). Pathway impact accounts for both the number of affected nodes and its importance with the maximum importance of each pathway being 1. B, the TCA cycle pathway is shown as an example of an affected pathway as it has the highest percentage of hits with wet-lab support (see Table 1). Affected pathway metabolites are highlighted in red and those with wet-lab support for OAT1 interaction have a green border. The number within the rectangle is the KEGG ID for each metabolite, the seven metabolites shown in red are: C00022-pyruvate, C00149-malate, C00042-succinate, C00122-fumarate, C00158-citrate, C00311-isocitrate, C00026-2-oxoglutarate.
Three other groups of metabolites were included, in order of level of confidence (supplemental Table S3). The metabolites with the next level of confidence were those first predicted by GEM and which also passed pharmacophore filters; these were termed “metabolites with high confidence of interacting with OAT1” (supplemental Table S3). Metabolites only predicted by GEM (but having structures such that they did not pass the drug-based pharmacophore filters with high confidence) were classified as “metabolites likely to be affected indirectly.” Finally, the remaining plus-one metabolites were termed “OAT1-first neighbor compounds” (Fig. 4). The network revealed that, in the revised OAT1-centered metabolite pathways, the majority of metabolites were interconnected to constitute a main component, and there were also a number of small self-connected components (Fig. 4); network parameters were measured and some are shown in supplemental Fig. S4. The metabolites within the network participated in more than 20 different canonical metabolic pathways, suggesting the broad importance of OAT1 in metabolism. Pathway enrichment analysis of these 253 metabolites was performed using the online bioinformatics resource, Metaboanalyst (51). This over-representation analysis provides statistical information on the impact of the metabolites on various pathways (Table 1 and Fig. 5). The affected metabolite/signaling pathways included carbohydrate (e.g. Kreb's cycle, galactose metabolism, etc.), lipid (glycosphingolipid metabolism, bile acid biosynthesis, etc.), amino acid (alanine, aspartate and glutamate, etc.), nucleotide (purine and pyrimidine), and cofactor and vitamin (vitamin A, B2, B3, B5, B6, and B9) metabolism (Table 1; Fig. 5).
TABLE 1.
Top pathways affected by OAT1 deficiency as determined by pathway analysis and ranked by the number of hits (%) with wet-lab support
The represented metabolic pathways are ranked according to their percentage of hits with wet-lab support. TCA cycle, tyrosine, and tryptophan metabolism are some of the most well validated pathways represented in the network.
| Top pathways | Total | Hits | p Value | FDR | Hits with wet-lab support | Hits with wet-lab support |
|---|---|---|---|---|---|---|
| % | ||||||
| TCA cycle | 20 | 7 | 0.001972 | 0.011266 | 5 | 71.4 |
| Tyrosine metabolism | 76 | 17 | 0.000728 | 0.004852 | 10 | 58.8 |
| Alanine, aspartate, and glutamate metabolism | 24 | 12 | 5.34E-07 | 1.42E-05 | 7 | 58.3 |
| Butanoate metabolism | 40 | 12 | 0.000264 | 0.002344 | 7 | 58.3 |
| Arginine and proline metabolism | 77 | 13 | 0.032714 | 0.084423 | 7 | 53.8 |
| Tryptophan metabolism | 79 | 15 | 0.007898 | 0.033253 | 8 | 53.3 |
| Nicotinate and nicotinamide metabolism | 44 | 12 | 0.000697 | 0.004852 | 5 | 41.7 |
| Valine, leucine, and isoleucine degradation | 40 | 10 | 0.003935 | 0.018519 | 4 | 40.0 |
| Nitrogen metabolism | 39 | 13 | 4.24E-05 | 0.000565 | 5 | 38.5 |
| Glyoxylate and dicarboxylate metabolism | 50 | 12 | 0.002356 | 0.012563 | 4 | 33.3 |
| Propanoate metabolism | 35 | 10 | 0.001322 | 0.008134 | 3 | 30.0 |
| Glycine, serine, and threonine metabolism | 48 | 13 | 0.00045 | 0.003598 | 3 | 23.1 |
| Purine metabolism | 92 | 27 | 5.52E-08 | 2.21E-06 | 5 | 18.5 |
| Pyrimidine metabolism | 60 | 22 | 1.11E-08 | 8.85E-07 | 3 | 13.6 |
The most highly represented metabolic pathways having at least 7 metabolites are shown and ranked according to the validation percentage in Table 1 (validation percentage is equal to the number of “wet-lab supported” metabolites of the pathway divided by the total number of metabolites in the pathway). Among these pathways, the two with the highest “hits with wet-lab support” were tyrosine metabolism and TCA cycle (58.8 and 71.4%, respectively). The TCA cycle is noteworthy, because it includes metabolites known to be classical substrates of OAT1, such as α-ketoglutarate, citrate, fumarate, and succinate (2, 9, 22, 52). Also included among these top-ranked pathways was tryptophan metabolism, which had considerable wet-lab support for 8 of 15 metabolites (53.3%), including anthranilate, xanthurenic acid, kynurenine, and indole-acetic acid, which are also putative uremic toxins associated with chronic kidney disease (CKD) (27, 53, 54).
Discussion
As previously described, systemic deletion of OAT1 was accomplished using a standard homologous recombination approach and thus resulted in the generalized loss of the transporter (23). OAT1 is largely responsible for the uptake of anionic substrates from the blood, and from the viewpoint of organ physiology, any change in the function of OAT1 (e.g. Oat1 knock-out) would concomitantly alter the concentration of metabolites cleared by this transporter not only in the bloodstream, but also in the cells of the tissue in which it is expressed.
In the mouse, the kidney is the predominant site of OAT1 expression and Northern blot and immunohistochemical analyses of kidneys from OAT1 knock-out animals revealed undetectable levels of gene products (i.e. RNA and protein) (23). OAT1 is also found, albeit at much lower levels of expression, in some other mouse tissues, including the choroid plexus, inner ear, eye, brain, and spleen. Thus, whereas we cannot completely eliminate the possibility that these other tissues contribute to the observed metabolic alterations seen in OAT1-KO animals, based on the relatively minor contributions expected from these other tissues, it would seem likely that the observed metabolic alterations in the blood are driven mainly by the lack of kidney-specific expression of the transporter. Moreover, despite the generalized deletion of this important drug transporter, mutant animals are born at expected sex ratios and both male and female mice are viable, appear healthy, and have a normal life expectancy (23).
The OAT1 drug transporter is a focus of regulatory agencies concerned about side effects of drugs due to interaction at the level of the transporter (55, 56). For some time, it has been clear that OAT1 and other drug transporters play key roles in regulating levels of endogenous metabolites and signaling molecules (11, 21, 27, 57). For example, the high but shifting embryonic expression of OAT1 and other SLC22 transporters (OAT3, OCT1, URAT1) in the developing nervous system and other developing tissues led to the hypothesis in 2000 that these drug transporters transported endogenous small molecules that could affect morphogenesis (21). There has recently been a call for more systematic analyses of the roles of these transporters, particularly the SLCs, in metabolism and signaling (1). There is also growing evidence showing relevance of OAT1 to metabolic disease, including chronic kidney disease (15, 25, 58). Taken together with the finding that these highly conserved drug transporters are differentially and highly expressed in various epithelial tissues lining body fluid compartments, it has been hypothesized that they potentially participate in remote communication (“remote sensing and signaling”) between organs and organisms (2–4, 7, 9, 10, 27). This may apply to other SLC families as well (1).
The remote sensing and signaling hypothesis for SLC and ABC drug transporters, which are mainly found on the apical and basolateral surfaces of epithelial cells lining body fluid compartments (e.g. blood, CSF, bile, amniotic fluid), theorizes that, together, these transporters function analogously to the endocrine system (2, 4, 7, 10). As we show here, they play a key role in regulating or modulating biochemical pathways involving a wide variety of small endogenous molecules with high informational content from the perspective of systemic or local metabolism and signaling. Our analysis thus builds upon prior work (7, 11, 23, 27) implicating OAT1 in the in vivo regulation of pathways involving essential metabolites (e.g. TCA cycle intermediates, tryptophan metabolites derived from the gut microbiome), key signaling molecules (e.g. prostaglandins, polyamines, cyclic nucleotides), molecules with antioxidant activity (e.g. ascorbic acid, urate), hormones (e.g. thyroxine), vitamins, and cofactors (e.g. panthothenic acid) (Fig. 5, Table 1). Crucially, from our analyses we are able to construct a partly validated OAT1-centered metabolic network (Figs. 4, 5; Table 1).
The molecules in the network are central to classical metabolism and signaling pathways, as well as organ and systemic physiology. Moreover, they are important in pathophysiological states like hyperuricemia, metabolic syndrome, diabetes, and chronic kidney disease. For example, some metabolites known to be transported by OAT1 are potential classical uremic toxins (e.g. indoxyl sulfate, kynurenate, polyamines, and uric acid), which accumulate as renal function declines in CKD (53, 54). Moreover, many of these pathophysiological states have been linked to SNPs or altered expression and/or function of members of the OAT subfamily of SLC22 (2, 4, 7, 10, 14, 15, 25, 58–61).
An important corollary has to do with the role of OATs in drug-metabolite interactions due to competition for transport. This may also be important in chronic kidney disease, where uremic toxins such as those mentioned above are transported by OAT1. For example, polyamines can inhibit transport of the drug methotrexate by OAT1 (11). If the competition is more substantial, as might occur with the drug probenecid (which binds OAT1 with high affinity), our data suggests that the metabolic changes could be quite significant, potentially resulting in many metabolic alterations that substantially overlap with the OAT1 knock-out noted here. In this regard, it is worth noting that there are a number of drug-induced metabolic syndromes that are associated with OAT1-transported drugs (12, 13).
Furthermore, the complexity of the OAT1-centered network points to the possibility of unexpected metabolic changes that could go well beyond the relatively straightforward concept of transporter-level competition for the ligand binding site. For example, in this study, we were able to separate likely direct versus indirect interactions of metabolites with OAT1; thus, a drug that tightly binds OAT1 may not only alter metabolites that directly compete for transport but also others in the OAT1-centered network that are not directly transported by OAT1. Because thiazide diuretics and HIV antivirals are transported by OAT1 (6, 62, 63), the OAT1-centered network may help in understanding the drug-induced metabolic syndromes associated with chronic treatment with these drugs (64–67).
In summary, we have described and validated a novel approach for systems driven discovery through integration of distinct and complementary systems biology, computational and wet-lab approaches to gain further insight into the substrate specificity and function of the OAT1 transporter in basal physiology (Fig. 1). As more wet lab transport data accumulates, and with further improvement of pathway analysis and other computational tools–including metabolic reconstruction approaches (particularly with respect to mammalian transporters), it should be possible to further refine and validate the OAT1-centered network proposed here. Furthermore, based on our results, we suggest that the types of multifaceted analyses described here for OAT1, enabling the construction of a “drug transporter”-centered metabolic network (Figs. 4, 5; Table 1), can be applied to other SLC and ABC drug transporters to generate a more comprehensive picture of the role that these transporters play in metabolism. Eventually, this approach could potentially connect cellular metabolism in different organs via molecules transported by multispecific drug transporters (as well as other transport systems such as those involving other types of transporters or channels) as proposed in the remote sensing and signaling hypothesis (2, 4, 7, 9, 10).
Importantly, once drug-transporter metabolic networks are created for other SLC and ABC multispecific transporters, the systems biology approach employed here may be useful for explicitly predicting the metabolic alterations expected for new drugs in healthy or diseased populations with globally altered metabolism (e.g. CKD, liver disease, metabolic syndrome, diabetes, multiorgan injury).
Experimental Procedures
The overall approach taken in the study is depicted in schematic flow charts (Fig. 1), which consists of the following stages: 1) systems biology analysis to predict metabolites affected by the deletion of OAT1 (supplemental Fig. S1); 2) validation of models using in vivo and in vitro data (Fig. 1); 3) computational chemistry analysis (Fig. 2, supplemental Fig. S2); 4) wet-lab validation (Fig. 3); and 5) construction of an OAT1-centered metabolic interaction network (Fig. 4, supplemental Fig. S3).
Materials
Water-soluble probenecid was purchased from Molecular Probes. The fluorescent tracer (6-carboxyfluorescein), tested metabolites, and signaling molecules (prostaglandin E1, pristanic acid, elaidic acid, trans-vaccenic acid, dihydrofolic acid, palmitoleic acid, β-nicotinamide mononucleotide, and 16-hydroxy-hexadecanoic acid) were purchased from Sigma.
Analysis of Transcriptomic Data with Mouse GEM Network
The transcriptomic and metabolomics data from previous studies (23, 27, 63, 68–70) were used for the context-specific analysis. The mouse GEM, iMM1415 (30), which contains the biochemical transformations for numerous tissues and cells in mice, was utilized (see supplemental Fig. S1 for more details). To create context-specific models, gene expression data from wild type (WT) and Oat1 knock-out (KO) mice were analyzed by Microarray Suite version 5.0 to assign present/absent calls. A minimum of 3 sets of microarray data for each condition (WT and KO) was analyzed separately, and for a gene to be considered present, it had to be present in at least 2 of 3 sets of data. In this way, the gene expression data were converted into a “binary” classification (i.e. either absent or present). This binary data without regard to actual expression values was then integrated into the genome-scale metabolic reconstruction model using the GIMME algorithm using the default setting of 90% for the percentage of the objective function needed to be met to generate the model (31).
GEM reconstructions and the application of constraint-based analysis of metabolic networks are widely used in systems biology approaches. The initial planned approach for this study described here was to apply statistical analysis or sampling of the networks with FVA. Unfortunately, the use of strictly constrained FVA models not only resulted in the prediction of a limited number of metabolites with which to work, but they also failed to capture some important interactions known to involve the transporter. Although this is not an uncommon occurrence, it is particularly problematic for this study when one takes into account the fact that our current understanding and the scope of the current metabolic models do not adequately encapsulate the metabolic interactions/alterations resulting from the deletion of SLC drug transporter. In other words, the initial approach was limited by a metabolic model that was not specifically designed to address the questions that were being asked and thus required a modified approach described below.
Sensitivity Filter Through Comparison of the Metabolic Differences between KO and WT with FVA and Model Validation
FVA was used to compare functional differences between the WT and KO mice based on maximal achievable reaction flux ranges (32). A reduced model was created by removing those reactions classified as absent based on gene expression data from the WT and KO conditions. When establishing the lower and upper bound for exchange reactions, uptake constraints for WT and KO models were set to be the same (10 μmol/h) for most metabolites, and to make the model more renal-specific, a few metabolites that were listed for the previously published renal objective function, which was used to analyze blood pressure regulation (35), were set to be either secreted or absorbed accordingly. In our analysis, a biomass maintenance function, which was defined to represent the metabolic composition necessary for the maintenance of mouse tissues, was used as the constraint to generate the context-specific models using the GIMME algorithm. We also considered the ATP objective function, as well as the so-called “renal objective function” (35). As with the biomass objective function, application of these other objective functions to the data requires some degree of approximation. Although the ATP objective function produced similar results to the biomass objective function, the application of the later produced a broader range of results. Interestingly, despite its name, the application of the renal objective function resulted in a more limited set of metabolites for subsequent analysis. This is likely due to the fact that it was designed for the analysis of the role of the kidney in regulating/modulating blood pressure.
The biomass pseudoreaction was ultimately selected to ensure that the cells could produce the complement of small molecules requisite for its function and cellular maintenance as well as metabolites/signaling molecules known to be transported by OAT1 have roles in cell maintenance and growth. Flux spans were then calculated as the differences between the maximum and minimum of reaction fluxes and the pairwise ratio between the KO and WT flux spans were calculated (i.e. [(max flux WT) − (min flux KO)] ÷ [(max flux WT) − (min flux WT)]), resulting in a set of flux span ratios for the set of reactions shared between the two models. Flux span ratios with a value of 1 were considered to be unchanged; in this loosely constrained model, any variation from 1 was considered to be affected by the absence of OAT1 and a successful model was expected to generate a list of “GEM-predicted OAT1 metabolites” that could be further evaluated (supplemental Table S4).
Specificity Filter through Pharmacophore Model Building and Validation Based on Drugs Known to Interact with OAT1
Computational chemistry analysis was performed with ICM software developed by Molsoft L.L.C (San Diego, CA). The software was used to perform clustering, alignment, and pharmacophore building based on the APF (47) of OAT1-interacting drugs or tracers (pharmaceuticals) with known Km (substrate affinity) or Ki (inhibitory affinity) of less than 100 μm were selected. A total of 61 pharmaceuticals were selected; among which two-thirds (41 drugs) were selected for a training set for the model generation, and one-third (20 drugs) were placed in a validating set. The structure-data files (pubchem.ncbi.nlm.nih.gov) for the OAT1-interacting drugs were input into ICM, which superimposed and aligned the drugs based on using the APF superimposition method. This method takes into consideration a number of three-dimensional structural parameters, hydrogen bond donors, hydrogen bond acceptors, SP2 hybridization, lipophilicity, size of large atoms, and positive and negative charges (71, 72). Based upon these alignments, 7 pharmacophore hypotheses were generated. After the model generation, the validating set (serving as true positives) and all the drugs from the Drugbank database (serving as true negatives) were screened against the pharmacophores. Using these screening results, a ROC curve was generated (supplemental Fig. S2).
Screening the List of Metabolites Predicted by the Metabolic Networks with Pharmacophore Models
The GEM-predicted OAT1 metabolites were compared with each of the 7 pharmacophore models and ranked by how well they fit with the three-dimensional molecular space defined by the drugs known to interact with OAT1. The 30 metabolites that best fit each of the 7 pharmacophores were selected as having potential to directly interact with OAT1. Many metabolites fit more than one pharmacophore model. After eliminating overlaps, 74 metabolites remained, and these were termed “top-ranked GEM-predicted OAT1 metabolites” (passing OAT1 pharmacophore filter).
Uptake Inhibition Assay
Oat1-transfected CHO cells cultured on 96-well plates were incubated in the presence of 10 μm 6-carboxyfluorescein (6, 9, 62) with or without individual metabolites (with controls treated with the OAT1 inhibitor probenecid). The IC50 curves for novel ligands were plotted in Prism Software (GraphPad Inc., San Diego, CA), and the IC50 values were converted to Ki (inhibition affinity) using the Cheng-Prusoff equation.
 |
Construction of an OAT1-centered Metabolic Interaction Network
The OAT1-centered metabolic interaction network was generated based on the categorization and ranking of metabolites according to the confidence for their interaction with OAT1. Those metabolites for which kinetic data exists indicating the ability to interact with OAT1 were merged together and labeled as “wet-lab supported” metabolites. Metabolites predicted by the GEM analysis, which also fit the pharmacophore filtering were designated as “metabolites predicted to interact with high confidence.” Metabolites that were predicted by the GEM analysis but which did not fit the pharmacophore screen were designated as “metabolites predicted to be affected indirectly.” The KEGG IDs for all of these metabolites were input then into Metscape, a Cytoscape plug-in (50). Metabolites present in Metscape were used as “input metabolites” to build a metabolic network. The construction of the network also introduced many “plus-one” or “first-neighbor” metabolites, and because there is currently no evidence supporting their interactions with OAT1, they were deemed to have the lowest confidence for interacting with OAT1. The network was then trimmed to eliminate uninformative nodes using the following criteria: small molecules (such as water, carbon dioxide, etc), energy-related molecules (NADH, ATP, etc.), and large peptides not known to interact with OAT1 or related transporters (somatostatin, kinetensin, etc.) were removed; unnecessary “dead-ended” and “inter-connecting” plus-ones were removed to create a more concise network (in other words, dead-ended plus-one nodes, which connected only to one node, were removed, as were inter-connecting plus-ones that did not affect connections between wet-lab validated or predicted nodes). The final network thus consisted of metabolites that fell into four categories, which in the order of level of confidence of their potential to interact with OAT1, the categories were: 1) wet-lab supported; 2) predicted to interact with high confidence; 3) predicted to be affected indirectly; and 4) plus-one after trimming.
Metabolic Pathway Analysis
Pathway and enrichment analyses were performed using Metaboanalyst 3.0 for pathway analysis and visualization (51). Lists of the KEGG IDs for the metabolites were input into this online bioinformatics resource to either the Pathway Analysis or Enrichment Analysis functionalities on the MetaboAnalyst 3.0 website. For the pathway analysis, the Homo sapiens (human) pathway library was selected and all compounds in the selected pathway were used. The algorithms specified were the hypergeometric test for the over-representation analysis and the relative betweeness centrality for the pathway topology analysis. For enrichment analysis, the pathway-associated metabolite set was selected as the library and all compounds in the metabolite set library were used.
Statistics
To determine whether the overall in silico approach results in significant enrichment of metabolites known to have direct interaction with OAT1, a hypergeometric-based test was performed to calculate the various p values. The hypergeometric calculation, which is based on certain assumptions, has been used in systems biological analyses for determining the probability of a result occurring just by chance (73–75). The hypergeometric test can be used as a measure of over-representation and takes into account the overall population size, the number of successes within this population, the sample size, and the number of successes within the sample population.
Author Contributions
H. C. L., N. J., and Y. C. performed the system biology analysis of the transcriptomic data; H. C. L. and S. Y. C. performed the pharmacophore analysis and virtual screening; H. C. L. and S. Y. C. performed the wet-lab transport assays; S. A. E., V. B., W. W., and K. T. B. provided methodological assistance; V. B. and K. T. B. assisted with the statistical analysis; R. A. and B. O. P. contributed analytical tools and provided assistance with their use; S. K. N. conceived the hypothesis and supervised the experiments; H. C. L., K. T. B., W. W., and S. K. N. wrote or contributed to the writing of the manuscript.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM098449, RO1GM104098, and U54-HD07160. R. Abagyan has a significant financial interest in Molsoft or its products. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. S1–S4 and Tables S1–S4.
- SLC
- solute carrier
- OAT1
- organic anion transporter 1
- ABC
- ATP-binding cassette
- FVA
- flux variability analysis
- GEM
- genome-scale metabolic reconstruction
- APF
- atomic property field
- GIMME
- gene inactivity moderated by metabolism and expression
- CKD
- chronic kidney disease
- ROC
- receiving operating characteristic.
References
- 1. César-Razquin A., Snijder B., Frappier-Brinton T., Isserlin R., Gyimesi G., Bai X., Reithmeier R. A., Hepworth D., Hediger M. A., Edwards A. M., and Superti-Furga G. (2015) A call for systematic research on solute carriers. Cell 162, 478–487 [DOI] [PubMed] [Google Scholar]
- 2. Nigam S. K. (2015) What do drug transporters really do? Nat. Rev. Drug Discov. 14, 29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu W., Jamshidi N., Eraly S. A., Liu H. C., Bush K. T., Palsson B. O., and Nigam S. K. (2013) Multispecific drug transporter Slc22a8 (Oat3) regulates multiple metabolic and signaling pathways. Drug Metab. Dispos. 41, 1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu W., Dnyanmote A. V., and Nigam S. K. (2011) Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol. Pharmacol. 79, 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu W., Bush K. T., Liu H. C., Zhu C., Abagyan R., and Nigam S. K. (2015) Shared ligands between organic anion transporters (OAT1 and OAT6) and odorant receptors. Drug Metab. Dispos. 43, 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Truong D. M., Kaler G., Khandelwal A., Swaan P. W., and Nigam S. K. (2008) Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J. Biol. Chem. 283, 8654–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nigam S. K., Bush K. T., Martovetsky G., Ahn S. Y., Liu H. C., Richard E., Bhatnagar V., and Wu W. (2015) The organic anion transporter (OAT) family: a systems biology perspective. Physiol. Rev. 95, 83–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaler G., Truong D. M., Sweeney D. E., Logan D. W., Nagle M., Wu W., Eraly S. A., and Nigam S. K. (2006) Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem. Biophys. Res. Commun. 351, 872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaler G., Truong D. M., Khandelwal A., Nagle M., Eraly S. A., Swaan P. W., and Nigam S. K. (2007) Structural variation governs substrate specificity for organic anion transporter (OAT) homologs: potential remote sensing by OAT family members. J. Biol. Chem. 282, 23841–23853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahn S. Y., and Nigam S. K. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol. Pharmacol. 76, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahn S. Y., Jamshidi N., Mo M. L., Wu W., Eraly S. A., Dnyanmote A., Bush K. T., Gallegos T. F., Sweet D. H., Palsson B. Ø., and Nigam S. K. (2011) Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J. Biol. Chem. 286, 31522–31531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hester E. K. (2012) HIV medications: an update and review of metabolic complications. Nutr. Clin. Pract. 27, 51–64 [DOI] [PubMed] [Google Scholar]
- 13. Buscemi S., Nicolucci A., Lucisano G., Galvano F., Grosso G., Massenti F. M., Amodio E., Bonura A., Sprini D., and Rini G. B. (2013) Impact of chronic diuretic treatment on glucose homeostasis. Diabetol. Metab. Syndr. 5, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prentice K. J., Luu L., Allister E. M., Liu Y., Jun L. S., Sloop K. W., Hardy A. B., Wei L., Jia W., Fantus I. G., Sweet D. H., Sweeney G., Retnakaran R., Dai F. F., and Wheeler M. B. (2014) The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 19, 653–666 [DOI] [PubMed] [Google Scholar]
- 15. Sharma K., Karl B., Mathew A. V., Gangoiti J. A., Wassel C. L., Saito R., Pu M., Sharma S., You Y. H., Wang L., Diamond-Stanic M., Lindenmeyer M. T., Forsblom C., Wu W., Ix J. H., et al. (2013) Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J. Am. Soc. Nephrol. 24, 1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lopez-Nieto C. E., You G., Barros E. J. G., Beier D. R., and Nigam S. K. (1996) Molecular cloning and characterization of a novel transport protein with very high expression in the kidney. J. Am. Soc. Nephrol. 7, 1301 [Google Scholar]
- 17. Lopez-Nieto C. E., You G., Bush K. T., Barros E. J., Beier D. R., and Nigam S. K. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J. Biol. Chem. 272, 6471–6478 [DOI] [PubMed] [Google Scholar]
- 18. Ahn S. Y., Eraly S. A., Tsigelny I., and Nigam S. K. (2009) Interaction of organic cations with organic anion transporters. J. Biol. Chem. 284, 31422–31430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burckhardt G., and Burckhardt B. C. (2011) In vitro and in vivo evidence of the importance of organic anion transporters (OATs) in drug therapy. Handb. Exp. Pharmacol. 201, 29–104 [DOI] [PubMed] [Google Scholar]
- 20. Emami Riedmaier A., Nies A. T., Schaeffeler E., and Schwab M. (2012) Organic anion transporters and their implications in pharmacotherapy. Pharmacol. Rev. 64, 421–449 [DOI] [PubMed] [Google Scholar]
- 21. Pavlova A., Sakurai H., Leclercq B., Beier D. R., Yu A. S., and Nigam S. K. (2000) Developmentally regulated expression of organic ion transporters NKT (OAT1), OCT1, NLT (OAT2), and Roct. Am. J. Physiol. Renal Physiol. 278, F635–643 [DOI] [PubMed] [Google Scholar]
- 22. VanWert A. L., Gionfriddo M. R., and Sweet D. H. (2010) Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm. Drug Dispos. 31, 1–71 [DOI] [PubMed] [Google Scholar]
- 23. Eraly S. A., Vallon V., Vaughn D. A., Gangoiti J. A., Richter K., Nagle M., Monte J. C., Rieg T., Truong D. M., Long J. M., Barshop B. A., Kaler G., and Nigam S. K. (2006) Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem. 281, 5072–5083 [DOI] [PubMed] [Google Scholar]
- 24. Møller J. V., and Sheikh M. I. (1982) Renal organic anion transport system: pharmacological, physiological, and biochemical aspects. Pharmacol Rev 34, 315–358 [PubMed] [Google Scholar]
- 25. Saito H. (2010) Pathophysiological regulation of renal SLC22A organic ion transporters in acute kidney injury: pharmacological and toxicological implications. Pharmacol. Ther. 125, 79–91 [DOI] [PubMed] [Google Scholar]
- 26. Eraly S. A., Vallon V., Rieg T., Gangoiti J. A., Wikoff W. R., Siuzdak G., Barshop B. A., and Nigam S. K. (2008) Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol. Genomics 33, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wikoff W. R., Nagle M. A., Kouznetsova V. L., Tsigelny I. F., and Nigam S. K. (2011) Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J. Proteome Res. 10, 2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vallon V., Eraly S. A., Wikoff W. R., Rieg T., Kaler G., Truong D. M., Ahn S. Y., Mahapatra N. R., Mahata S. K., Gangoiti J. A., Wu W., Barshop B. A., Siuzdak G., and Nigam S. K. (2008) Organic anion transporter 3 contributes to the regulation of blood pressure. J. Am. Soc. Nephrol. 19, 1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vallon V., Eraly S. A., Rao S. R., Gerasimova M., Rose M., Nagle M., Anzai N., Smith T., Sharma K., Nigam S. K., and Rieg T. (2012) A role for the organic anion transporter OAT3 in renal creatinine secretion in mice. Am. J. Physiol. Renal Physiol. 302, F1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sigurdsson M. I., Jamshidi N., Steingrimsson E., Thiele I., and Palsson B. Ø. (2010) A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst Biol 4, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becker S. A., and Palsson B. O. (2008) Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 4, e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahadevan R., and Schilling C. H. (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 [DOI] [PubMed] [Google Scholar]
- 33. Raman K., and Chandra N. (2009) Flux balance analysis of biological systems: applications and challenges. Brief Bioinform. 10, 435–449 [DOI] [PubMed] [Google Scholar]
- 34. Knies D., Wittmüß P., Appel S., Sawodny O., Ederer M., and Feuer R. (2015) Modeling and simulation of optimal resource management during the diurnal cycle in Emiliania huxleyi by genome-scale reconstruction and an extended flux balance analysis approach. Metabolites 5, 659–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang R. L., Xie L., Xie L., Bourne P. E., and Palsson B. Ø. (2010) Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput. Biol. 6, e1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Förster J., Famili I., Fu P., Palsson B. Ø., and Nielsen J. (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bordbar A., and Palsson B. O. (2012) Using the reconstructed genome-scale human metabolic network to study physiology and pathology. J. Intern. Med. 271, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duan P., Li S., Ai N., Hu L., Welsh W. J., and You G. (2012) Potent inhibitors of human organic anion transporters 1 and 3 from clinical drug libraries: discovery and molecular characterization. Mol. Pharm. 9, 3340–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wolman A. T., Gionfriddo M. R., Heindel G. A., Mukhija P., Witkowski S., Bommareddy A., and Vanwert A. L. (2013) Organic anion transporter 3 interacts selectively with lipophilic β-lactam antibiotics. Drug Metab. Dispos. 41, 791–800 [DOI] [PubMed] [Google Scholar]
- 40. Elion G. B. (1989) Nobel lecture: the purine path to chemotherapy. Biosci. Rep. 9, 509–529 [DOI] [PubMed] [Google Scholar]
- 41. Hitchings G. H., Jr.(1989) Nobel lecture in physiology or medicine–1988: selective inhibitors of dihydrofolate reductase. In Vitro Cell Dev. Biol. 25, 303–310 [DOI] [PubMed] [Google Scholar]
- 42. Gupta S., and Aires-de-Sousa J. (2007) Comparing the chemical spaces of metabolites and available chemicals: models of metabolite-likeness. Mol. Divers 11, 23–36 [DOI] [PubMed] [Google Scholar]
- 43. Karakoc E., Sahinalp S. C., and Cherkasov A. (2006) Comparative QSAR- and fragments distribution analysis of drugs, druglikes, metabolic substances, and antimicrobial compounds. J. Chem. Inf. Model 46, 2167–2182 [DOI] [PubMed] [Google Scholar]
- 44. Khanna V., and Ranganathan S. (2009) Physiochemical property space distribution among human metabolites, drugs and toxins. BMC Bioinform. 10, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Hagan S., and Kell D. B. (2015) Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front Pharmacol 6, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kouznetsova V. L., Tsigelny I. F., Nagle M. A., and Nigam S. K. (2011) Elucidation of common pharmacophores from analysis of targeted metabolites transported by the multispecific drug transporter-Organic anion transporter1 (Oat1). Bioorg. Med. Chem. 19, 3320–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Totrov M., and Abagyan R. (2008) Flexible ligand docking to multiple receptor conformations: a practical alternative. Curr. Opin. Struct. Biol. 18, 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu H. C., Goldenberg A., Chen Y., Lun C., Wu W., Bush K. T., Balac N., Rodriguez P., Abagyan R., and Nigam S. K. (2016) Analysis of molecular properties of drugs interacting with SLC22 transporters, OAT1, OAT3, OCT1, OCT2: A machine-learning approach. J. Pharmacol. Exp. Ther. pii: jpet.116.232660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hodson L., and Karpe F. (2013) Is there something special about palmitoleate? Curr. Opin. Clin. Nutr. Metab. Care 16, 225–231 [DOI] [PubMed] [Google Scholar]
- 50. Karnovsky A., Weymouth T., Hull T., Tarcea V. G., Scardoni G., Laudanna C., Sartor M. A., Stringer K. A., Jagadish H. V., Burant C., Athey B., and Omenn G. S. (2012) Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 28, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xia J., Sinelnikov I. V., Han B., and Wishart D. S. (2015) MetaboAnalyst 3.0: making metabolomics more meaningful. Nucleic Acids Res. 43, W251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pritchard J. B. (1995) Intracellular α-ketoglutarate controls the efficacy of renal organic anion transport. J. Pharmacol. Exp. Ther. 274, 1278–1284 [PubMed] [Google Scholar]
- 53. Vanholder R., De Smet R., Glorieux G., Argiles A., Baurmeister U., Brunet P., Clark W., Cohen G., De Deyn P. P., Deppisch R., Descamps-Latscha B., Henle T., Jorres A., Lemke H. D., et al. (2003) Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 63, 1934–1943 [DOI] [PubMed] [Google Scholar]
- 54. Duranton F., Cohen G., De Smet R., Rodriguez M., Jankowski J., Vanholder R., Argiles A., and European Uremic Toxin Work Group (2012) Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 23, 1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bush K. T., Nagle M., Truong D. M., Bhatnagar V., Kaler G., Eraly S. A., Wu W., and Nigam S. K. (2014) Organic anion transporters. in Drug transporters: Molecular characterization and role in drug disposition (You G., and Morris M. E., eds) 2nd Ed., pp. 25–42, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 56. Lee S. C., Zhang L., and Huang S. M. (2014) Regulatory science perspectives on transporter studies in drug development. in Drug Transporters: Molecular Characterization and Role in Drug Disposition (You G., and Morris M. E., eds) 2nd Ed., pp. 473–487, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 57. Eraly S. A., Blantz R. C., Bhatnagar V., and Nigam S. K. (2003) Novel aspects of renal organic anion transporters. Curr. Opin. Nephrol. Hypertens. 12, 551–558 [DOI] [PubMed] [Google Scholar]
- 58. Nigam S. K., Wu W., Bush K. T., Hoenig M. P., Blantz R. C., and Bhatnagar V. (2015) Handling of drugs, metabolites, and uremic toxins by kidney proximal tubule drug transporters. Clin. J. Am. Soc. Nephrol. 10, 2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bhatnagar V., Richard E. L., Wu W., Nievergelt C. M., Lipkowitz M. S., Jeff J., Maihofer A. X., and Nigam S. K. (2016) Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin. Kidney J. 9, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schuetz J. D., Swaan P. W., and Tweedie D. J. (2014) The role of transporters in toxicity and disease. Drug Metab. Dispos. 42, 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu C., Nigam K. B., Date R. C., Bush K. T., Springer S. A., Saier M. H. Jr, Wu W., and Nigam S. K. (2015) Evolutionary analysis and classification of OATs, OCTs, OCTNs, and other SLC22 transporters: structure-function implications and analysis of sequence motifs. PLoS ONE 10, e0140569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nagle M. A., Truong D. M., Dnyanmote A. V., Ahn S. Y., Eraly S. A., Wu W., and Nigam S. K. (2011) Analysis of three-dimensional systems for developing and mature kidneys clarifies the role of OAT1 and OAT3 in antiviral handling. J. Biol. Chem. 286, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nagle M. A., Wu W., Eraly S. A., and Nigam S. K. (2013) Organic anion transport pathways in antiviral handling in choroid plexus in Oat1 (Slc22a6) and Oat3 (Slc22a8) deficient tissue. Neurosci. Lett. 534, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sinxadi P. Z., Dave J. A., Samuels D. C., Heckmann J. M., Maartens G., Levitt N. S., Wester C. W., Haas D. W., and Hulgan T. (2013) Mitochondrial genomics and antiretroviral therapy-associated metabolic complications in HIV-infected black South Africans: a pilot study. AIDS Res. Hum. Retroviruses 29, 1031–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Monier P. L., and Wilcox R. (2004) Metabolic complications associated with the use of highly active antiretroviral therapy in HIV-1-infected adults. Am. J. Med. Sci. 328, 48–56 [DOI] [PubMed] [Google Scholar]
- 66. Anderson P. L., Kakuda T. N., and Lichtenstein K. A. (2004) The cellular pharmacology of nucleoside- and nucleotide-analogue reverse-transcriptase inhibitors and its relationship to clinical toxicities. Clin. Infect. Dis. 38, 743–753 [DOI] [PubMed] [Google Scholar]
- 67. Lewis W. (2003) Mitochondrial DNA replication, nucleoside reverse-transcriptase inhibitors, and AIDS cardiomyopathy. Prog. Cardiovasc. Dis. 45, 305–318 [DOI] [PubMed] [Google Scholar]
- 68. Vallon V., Rieg T., Ahn S. Y., Wu W., Eraly S. A., and Nigam S. K. (2008) Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am. J. Physiol. Renal Physiol. 294, F867–873 [DOI] [PubMed] [Google Scholar]
- 69. Sweeney D. E., Vallon V., Rieg T., Wu W., Gallegos T. F., and Nigam S. K. (2011) Functional maturation of drug transporters in the developing, neonatal, and postnatal kidney. Mol. Pharmacol. 80, 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Torres A. M., Dnyanmote A. V., Bush K. T., Wu W., and Nigam S. K. (2011) Deletion of multispecific organic anion transporter Oat1/Slc22a6 protects against mercury-induced kidney injury. J. Biol. Chem. 286, 26391–26395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen Y. C., Totrov M., and Abagyan R. (2014) Docking to multiple pockets or ligand fields for screening, activity prediction and scaffold hopping. Future Med. Chem. 6, 1741–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Totrov M. (2008) Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem. Biol. Drug Des. 71, 15–27 [DOI] [PubMed] [Google Scholar]
- 73. Mitrea C., Taghavi Z., Bokanizad B., Hanoudi S., Tagett R., Donato M., Voichit̨a C., and Drăghici S. (2013) Methods and approaches in the topology-based analysis of biological pathways. Front. Physiol. 4, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang R. S., Oldham W. M., and Loscalzo J. (2014) Network-based association of hypoxia-responsive genes with cardiovascular diseases. New J. Phys. 16, 105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Y., Fan H., Xu J., Xiao Y., Xu Y., Li Y., and Li X. (2013) Network analysis reveals functional cross-links between disease and inflammation genes. Sci. Rep. 3, 3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.