Abstract
The DUF156 family of DNA-binding transcriptional regulators includes metal sensors that respond to cobalt and/or nickel (RcnR, InrS) or copper (CsoR) plus CstR, which responds to persulfide, and formaldehyde-responsive FrmR. Unexpectedly, the allosteric mechanism of FrmR from Salmonella enterica serovar Typhimurium is triggered by metals in vitro, and variant FrmRE64H gains responsiveness to Zn(II) and cobalt in vivo. Here we establish that the allosteric mechanism of FrmR is triggered directly by formaldehyde in vitro. Sensitivity to formaldehyde requires a cysteine (Cys35 in FrmR) conserved in all DUF156 proteins. A crystal structure of metal- and formaldehyde-sensing FrmRE64H reveals that an FrmR-specific amino-terminal Pro2 is proximal to Cys35, and these residues form the deduced formaldehyde-sensing site. Evidence is presented that implies that residues spatially close to the conserved cysteine tune the sensitivities of DUF156 proteins above or below critical thresholds for different effectors, generating the semblance of specificity within cells. Relative to FrmR, RcnR is less responsive to formaldehyde in vitro, and RcnR does not sense formaldehyde in vivo, but reciprocal mutations FrmRP2S and RcnRS2P, respectively, impair and enhance formaldehyde reactivity in vitro. Formaldehyde detoxification by FrmA requires S-(hydroxymethyl)glutathione, yet glutathione inhibits formaldehyde detection by FrmR in vivo and in vitro. Quantifying the number of FrmR molecules per cell and modeling formaldehyde modification as a function of [formaldehyde] demonstrates that FrmR reactivity is optimized such that FrmR is modified and frmRA is derepressed at lower [formaldehyde] than required to generate S-(hydroxymethyl)glutathione. Expression of FrmA is thereby coordinated with the accumulation of its substrate.
Keywords: allosteric regulation, Escherichia coli (E. coli), metal, Salmonella enterica, zinc, RcnR/CsoR, cobalt, formaldehyde, glutathione
Introduction
Formaldehyde (H2C=O), as a strong electrophile, is capable of alkylating and cross-linking the reactive groups (such as thiols and amines) of proteins and DNA (1–5). This reactivity and subsequent damage to biological macromolecules make formaldehyde a highly cytotoxic compound. In addition to environmental sources, formaldehyde is generated intracellularly by a number of cellular processes. In methylotrophic and methanotrophic bacteria, it is well known that formaldehyde is generated as a by-product of methanol and methane oxidation (6–9), consistent with the presence of genetically encoded formaldehyde detoxification systems in these organisms (2, 8, 10–12). Intracellular formaldehyde generation in bacteria that do not use these C1 substrates as a carbon source has been less well studied. Formaldehyde is produced by the alternative heme degradation pathway (IsdG and IsdI) in Staphylococcus aureus to acquire iron (13, 14). The recent detection of trimethylamine N-oxide (TMAO) 3 demethylase activity in cell extracts suggests that this activity may be an endogenous source of formaldehyde in Escherichia coli (15). Demethylation of nucleic acids and production of methylglyoxal from glyceraldehyde 3-phosphate and dihydroxyacetone phosphate during glycolysis represent more widespread physiological sources of formaldehyde (16–18). In addition, several mechanisms for the generation of formaldehyde at the host-pathogen interface have recently been proposed (2).
Inducible formaldehyde detoxification mechanisms have now been recognized in most bacteria (2, 3, 19). A glutathione-dependent pathway represents the most widespread formaldehyde detoxification system, although the functional proteins and/or genomic arrangement may vary (19–26). In E. coli, this pathway is encoded by the frmRAB operon, which includes frmA, encoding a Zn(II)-binding glutathione-dependent formaldehyde dehydrogenase, and frmB, encoding S-formylglutathione hydrolase (Figs. 1 and 2A) (26–29). Regulation of the frmRAB operon upon formaldehyde accumulation is mediated by the first gene product, FrmR, a DNA-binding transcriptional regulator (26).
FIGURE 1.
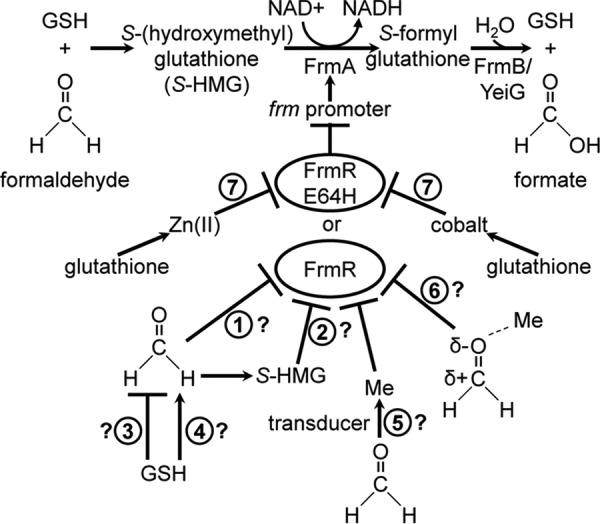
Formaldehyde detoxification and sensing. Spontaneous reaction of formaldehyde with GSH generates S-(hydroxymethyl)glutathione (S-HMG), the substrate oxidized by FrmA to S-formylglutathione (28). Following hydrolysis of S-formylglutathione by FrmB, formate is produced as the final product, and GSH is regenerated (22, 29). An additional enzyme, YeiG, is also implicated in formaldehyde detoxification because it demonstrates hydrolytic activity against S-formylglutathione, although yeiG is not FrmR-regulated (27). Notably, FrmB is present in the E. coli but not the Salmonella frm operon. Salmonella FrmR (or FrmRE64H) represses expression from the frm promoter, which is alleviated by exogenous formaldehyde. The intracellular effector of (any) FrmR is unknown, and possibilities include formaldehyde alone (1) or S-(hydroxymethyl)glutathione (2), in which case GSH could act negatively (3) or positively (4) on FrmR-mediated derepression. Alternatively, FrmR derepression may be transduced by a metal intermediate (5) or require activation of formaldehyde by metal (6). FrmRE64H additionally responds to Zn(II) and cobalt; however, the response to metals is lost in cells lacking glutathione (7) (50).
FIGURE 2.
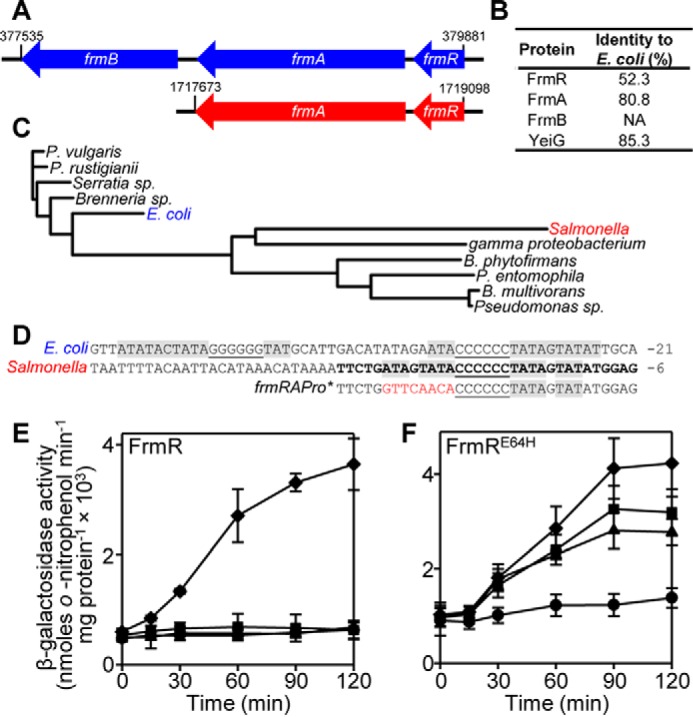
E. coli and Salmonella FrmRs have distinct origins but Salmonella FrmR and FrmRE64H retain their effector sensitivities in E. coli. A, schematic representation of the frm operon (to scale) from E. coli K12 (blue) and Salmonella (strain SL1344; red), with the nucleotide position at the start and end of each gene cluster indicated. B, percentage identity of Salmonella proteins required for formaldehyde detoxification compared with their E. coli orthologue. C, rooted phylogenetic tree of 11 sequences from the DUF156 FrmR subgroup identified previously (45). Organism details and UniProtKB identifiers are outlined under “Experimental Procedures.” D, alignment of the frm promoter from E. coli and Salmonella. The position relative to the translational start site is labeled. G/C tracts are underlined. T/A-rich inverted repeats are highlighted in gray. The sequence corresponding to one strand of frmRAPro, used for fluorescence anisotropy, is in boldface type. Mutations to generate frmRAPro* are highlighted in red. E and F, β-galactosidase activity as a function of time in E. coli BW25113ΔfrmR containing PfrmRA-frmR (E) or PfrmRA-frmRE64H (F), fused to lacZ following exposure of logarithmic cells to MNIC formaldehyde (50 μm; diamonds), Zn(II) (50 μm; triangles), Co(II) (5 μm; squares), or untreated control (circles). Values are means of at least three biological replicates (each performed in triplicate) with S.D. (error bars).
FrmR is a member of the RcnR/CsoR family (DUF156) of (predominantly) metal-sensing transcriptional repressors (30–32). This family can be divided into subgroups that have evolved to detect distinct and specific effectors in a cellular context by modification of a relatively conserved protein scaffold, in a manner similar to ArsR, MerR, and Fur family regulators (33–36). In addition to FrmR, characterized DUF156 subgroups to date include the metal sensors RcnR and DmeR, which respond to Ni(II)/Co(II); CsoR and RicR, which respond to Cu(I); InrS, which responds to Ni(II); and the non-metal sensor CstR, which undergoes cysteine modification by sodium sulfite, selenite, and tellurite (31, 32, 37–41). Upon binding of an allosteric effector (e.g. metal ion), affinity for DNA is weakened, alleviating repression from the target operator-promoter (30). At the time of writing, CsoR represents the only member of this family for which a structure has been reported (31, 42–44). CsoR forms a three-helix bundle that adopts a tetrameric assembly made up of a dimer of dimers. The known effector sensory sites of metal-sensing DUF156 proteins exploit side chains of conserved residues at a dimer interface, denoted the WXYZ fingerprint, characteristic of each subgroup but all involving a conserved Cys-thiolate (position X) located at the amino-terminal end of helix α2 (31, 45, 46). Analogous information is not yet available for the sensory sites of FrmR.
E. coli FrmR-mediated transcriptional repression is alleviated following exposure of cells to exogenous formaldehyde, CO-releasing molecules, and chloride treatment and during anaerobic respiration using TMAO as the terminal electron acceptor (15, 26, 47–49). However, the effector directly detected by FrmR in each case remains unexplored. We recently identified an FrmR homologue in Salmonella enterica serovar Typhimurium strain SL1344 (hereafter referred to as Salmonella), which, as observed for E. coli, responds to exogenous formaldehyde in vivo (Fig. 1) (50). Unlike E. coli FrmR (containing two), Salmonella FrmR possesses three (four including Glu64) putative metal ligands at positions WXY, within the metal-binding fingerprint of metal-sensing DUF156 members (50). Moreover, Salmonella FrmR can bind Co(II), Cu(I), and Zn(II). Unexpectedly, Cu(I) and Zn(II) are capable of triggering an allosteric response that weakens FrmR DNA affinity in vitro (50). Metal responsiveness is not observed in vivo because FrmR is less sensitive than the endogenous Salmonella sensors for these metals. However, generation of a variant FrmR, responsive to cobalt and Zn(II) in addition to formaldehyde in vivo, is achieved by single amino acid substitution at the putative metal-binding site (Glu64 → His) (Fig. 1). The combined effect of tighter metal affinity and weaker DNA affinity of the apo-form, relative to wild type FrmR, confers metal-sensing gain of function to FrmRE64H (50). Evidence that Salmonella FrmR is competent to respond to metals raises the possibility that formaldehyde sensing could be indirect and mediated by an effect on metal availability to FrmR by formaldehyde (Fig. 1). Notably, FrmA also requires Zn(II) for catalytic activity (47). The extent to which Zn(II) might be required to act as a signal transducer of formaldehyde accumulation in a cell now needs to be addressed.
In addition to FrmR, transcriptional regulators that respond following exposure to exogenous formaldehyde include HxlR (MarR family) from Bacillus subtilis and NmlR/AdhR (MerR family) identified in Neisseria sp. and other Gram-positive pathogens (51–56). However, the effector(s) detected by any formaldehyde-responsive transcriptional regulator has yet to be biochemically identified. Despite the requirement of glutathione for formaldehyde detoxification by FrmA, the extent to which glutathione plays a role in the regulation of expression of glutathione-dependent formaldehyde dehydrogenase in any organism is unknown (Fig. 1).
We present the first in vitro evidence that formaldehyde is a direct allosteric effector of Salmonella FrmR. The FrmR sensory site is particularly reactive to formaldehyde such that the related Salmonella RcnR sensor is less responsive to formaldehyde in vitro and in vivo. We determine the crystal structure of FrmRE64H to define the effectors and sensory site(s) of this formaldehyde- and metal-sensing variant. Residues required for Zn(II)/Co(II) and formaldehyde sensing are determined and support a mechanism involving an FrmR-specific formaldehyde cross-link between Pro2 and Cys35. An RcnR variant with enhanced sensitivity for formaldehyde in vitro was generated based on the deduced FrmR sensory site and mechanism. Implications for the basis of effector specificity within DUF156 family proteins and the chemical species detected by FrmR in vivo are discussed.
Results
Salmonella FrmR and FrmRE64H Retain Responsiveness to Formaldehyde and Metals When Expressed in E. coli
Despite E. coli and Salmonella being co-linear for most genes (57), the Salmonella frm operon occurs at a distinct genomic location compared with E. coli and lacks the frmB gene for S-formylglutathione hydrolase (Fig. 2A). E. coli and Salmonella FrmRs share only 52.3% sequence identity (Fig. 2B) compared with an average ∼85% identity for orthologous gene products between these organisms (58, 59), and analysis of the DUF156 FrmR subgroup demonstrates that they are polyphyletic (Fig. 2C). This is also reflected by the distinct operator-promoter sequences upstream of each frm locus (Fig. 2D). The significance of these differences in relation to formaldehyde detoxification remains unknown but may reflect specific requirements for formaldehyde detoxification in the respective cellular backgrounds. To investigate the response of Salmonella FrmR in an E. coli cytosol, PfrmRA-frmR reporter constructs comprising the Salmonella frmRA promoter (PfrmRA) and frmR coding sequence fused to lacZ were expressed in E. coli cells that lacked the endogenous E. coli frmR gene (ΔfrmR) (Fig. 2E). As observed in Salmonella (50), expression from PfrmRA-frmR was derepressed in the heterologous E. coli host following exposure of cells to maximum non-inhibitory concentrations (MNICs) of formaldehyde, whereas exposure to MNIC CoCl2 and ZnCl2 did not alleviate repression (Fig. 2E). FrmR variant, FrmRE64H, which responds to CoCl2, ZnCl2, and formaldehyde in Salmonella cells (50), retains the same effector responsiveness when PfrmRA-frmRE64H is expressed in E. coli ΔfrmR (Fig. 2F). This demonstrates that the ability of FrmRE64H to respond to metals (and formaldehyde) in vivo is not exclusive to Salmonella cells.
FrmR Senses Formaldehyde Directly
Repression by FrmR (and FrmRE64H) is alleviated by exogenous formaldehyde in vivo (Fig. 2, E and F), but DNA binding to the target frmRA operator-promoter (frmRAPro) (Fig. 2D) is weakened by Zn(II) (and Cu(I)) in vitro (50). To explore whether the in vivo response might be transduced by metals during formaldehyde stress or whether formaldehyde is able to act directly on FrmR, fluorescence anisotropy was used to monitor the interaction of FrmR with fluorescently labeled frmRAPro in the presence of formaldehyde (Fig. 3A). FrmR has previously been shown to bind frmRAPro with a stoichiometry of two tetramers per DNA molecule and a KDNA of 9.9 ± 0.3 × 10−8 m for each tetramer, in the absence of effector (50) (also confirmed here in Fig. 4C). Consequently, a limiting concentration (10 nm) of frmRAPro was used for titration with FrmR in the presence of 10 or 20 μm formaldehyde, concentrations chosen to minimize nonspecific formaldehyde cross-linking, which is likely at higher formaldehyde concentrations (60). EDTA was included as a metal chelator to eliminate any effect that may arise due to the presence of (allosterically effective) trace metals. The anisotropy data were fit to a model describing the binding of two non-dissociable FrmR tetramers per DNA molecule and revealed that DNA binding of FrmR to frmRAPro was weakened by ∼6.5-fold and ≥70-fold (compared with the published value (50); Table 1) in the presence of 10 and 20 μm formaldehyde, respectively (Fig. 3A). This identifies formaldehyde as a direct allosteric effector of FrmR.
FIGURE 3.
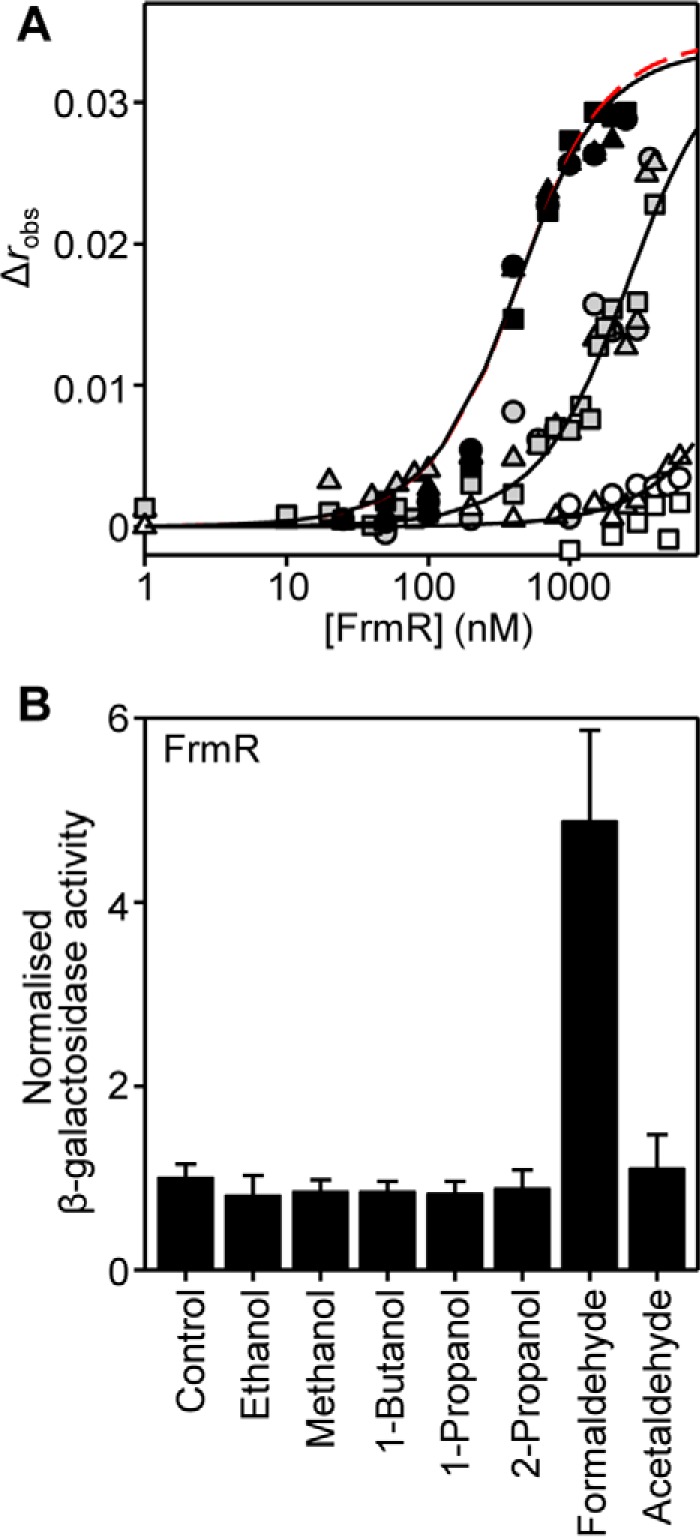
FrmR responds specifically to formaldehyde in vitro and in vivo. A, anisotropy change upon titration of a limiting concentration of frmRAPro (10 nm) with FrmR in the presence of 5 mm EDTA and either 20 μm acetaldehyde (black symbols), 10 μm formaldehyde (gray symbols), or 20 μm formaldehyde (open symbols). Symbol shapes represent individual experiments. Data were fit to a model describing a 2:1 protein tetramer (nondissociable)/DNA stoichiometry (binding with equal affinity) (50, 86), and lines represent simulated curves produced from the average (apparent) KDNA determined across the experimental replicates shown. The dashed red line (largely obscured) is a simulated curve based on the published KDNA of apo-FrmR (50), presented here for comparative purposes. B, β-galactosidase activity in SL1344 containing PfrmRA-frmR fused to lacZ grown to mid-exponential phase in M9 minimal medium in the absence (control) or presence of MNIC indicated alcohol or aldehyde (see “Experimental Procedures” for concentrations). Values are means of three biological replicates (each performed in triplicate) with S.D. (error bars).
FIGURE 4.
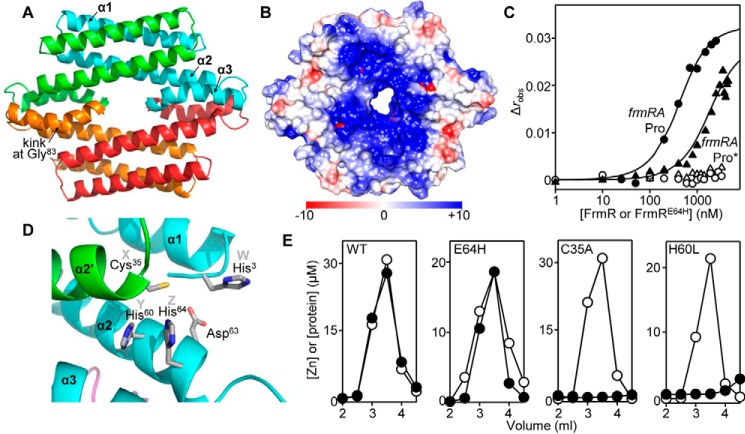
Structure of FrmRE64H and inferred Zn(II)/Co(II)-sensing site. A, ribbon representation of the 2.19 Å resolution crystal structure of FrmRE64H tetramer (Protein Data Bank code 5LCY; see Table 2 for a summary of the crystallographic data). Each monomer is colored differently, and secondary structural units are labeled on the cyan monomer. B, electrostatic surface potential of FrmRE64H tetramer using Chimera (103). The color scale is from −10 (negative potential; red) to +10 (positive potential; blue) kcal/mol·e. C, anisotropy change upon titration of a limiting concentration (10 nm) of frmRAPro (solid symbols) or frmRAPro* (half-site defined in Fig. 2D; open symbols) with FrmR (circles) or FrmRE64H (triangles) in the presence of 5 mm EDTA. The lines are fits of the data to a model describing a 2:1 protein tetramer (nondissociable)/DNA stoichiometry (binding with equal affinity) (50, 86). D, expansion of the dimeric interface with backbone helices from two different monomers shaded green and cyan (the same colors as used in A). The inferred Zn(II)/Co(II)-binding site comprises Cys35 from α2′, and His60 and His64 from α2 (belonging to the XYZ motif required for metal binding in DUF156 members CsoR, RcnR, and InrS (39, 46, 68), with His3 from α1 (position W (46, 61)) and Asp63 presenting candidate fourth ligands. E, analysis of fractions (0.5 ml) for protein by Bradford assay (open circles) and metal by inductively coupled plasma MS (filled circles) following size exclusion chromatography of FrmR, FrmRE64H, FrmRC35A (50 μm, monomer), or FrmRH60L (in this case, [monomer] = 32.5 μm), preincubated with 150 μm ZnCl2.
TABLE 1.
DNA binding affinities and allosteric coupling free energies for FrmR and RcnR
Values were determined from fluorescence anisotropy experiments. The conditions used were as follows: 25 °C, 10 mm HEPES, pH 7.0, 60 mm NaCl, 240 mm KCl with the addition of 5 mm EDTA for titrations with apoprotein or 5 μm NiCl2 or CoCl2 for metal-loaded titrations. RcnR was incubated with 1.2 molar eq of NiCl2 or CoCl2 per monomer, as indicated.
| Protein | Effector | KDNAa |
|---|---|---|
| m | ||
| FrmR | Apob | 9.9 ± 0.3 × 10−8 |
| FrmRC35A | Apo | 1.6 ± 0.2 × 10−7 |
| FrmRP2S | Apo | 1.5 ± 0.2 × 10−7 |
| RcnR | Apo | 1.5 ± 0.8 × 10−7 |
| RcnR | Ni(II) | ≥5.9 ± 1.3 × 10−6 |
| RcnR | Co(II) | ≥1.5 ± 0.2 × 10−5 |
| RcnRS2P | Apo | 1.6 ± 0.1 × 10−7 |
a Data were fit to a model describing two nondissociable tetramers of FrmR or RcnR binding with equal affinity to frmRAPro or rcnRAPro, respectively.
b Determined previously under the same conditions (50).
The ability of FrmR to respond directly to formaldehyde opens up the possibility that other aldehydes or related alcohols may also act as allosteric effectors. To test this hypothesis, DNA binding was monitored in the presence of acetaldehyde (CH3HC=O), which differs from formaldehyde by replacement of a carbonyl-bonded proton with a methyl group. Inclusion of 20 μm acetaldehyde did not weaken the DNA binding affinity of FrmR (Fig. 3A), and expression from PfrmRA-frmR was not derepressed when Salmonella cells were exposed to MNIC acetaldehyde (Fig. 3B). Similarly, FrmR did not respond to ethanol, methanol, 1-butanol, 1-propanol, and 2-propanol in vivo (Fig. 3B). Together, these data show that the response of FrmR demonstrates specificity for formaldehyde over other organic molecules and suggest that metal ions are not required to transduce the formaldehyde signal to FrmR in vivo.
Structure of FrmRE64H and Visualization of Its Inferred Metal (Zn(II)/Co(II))-sensing Site
To identify the sensory site(s) of FrmRE64H for metal and formaldehyde, diffraction quality crystals were generated, and an x-ray crystal structure was determined to 2.19 Å resolution (Fig. 4A). FrmRE64H exists as a homotetrameric assembly composed of a dimer of dimers as observed for the structurally characterized metal-sensing regulator CsoR (31, 42–44) (Fig. 4A). FrmRE64H has a kink (∼45º) in helix α3, not observed in (any) CsoR structure, which is enabled by Gly83, a residue specific to Salmonella FrmR (distinct from Ile83 in E. coli FrmR). The electrostatic surface potential highlights a region of positive potential composed of positively charged residues from helices α1 and α2 within a single monomer subunit (Fig. 4B). This region (as suggested for metal sensor CsoR (31, 43, 45)) is anticipated to enable binding of FrmRE64H to the frmRA operator-promoter, although the precise nature of the protein-DNA interactions for any DUF156 member are unknown. The Salmonella frmRA operator-promoter comprises a C6 tract flanked by a T/A-rich inverted repeat (Fig. 2D). The requirement of these inverted repeats for FrmR and FrmRE64H binding was investigated by fluorescence anisotropy using a modified frmRAPro dsDNA fragment in which one flanking repeat had been mutated (frmRAPro*) (Figs. 2D and 4C). No binding of FrmR or FrmRE64H to frmRAPro* (10 nm) was detected, indicating a considerably weaker DNA binding affinity (>10−5 m) than determined for frmRAPro. This demonstrates that the T/A-rich inverted repeat is required for tight affinity (physiologically relevant) DNA binding to the frmRA operator-promoter. The frmRA operator-promoter supports binding of two FrmR (or FrmRE64H) tetramers (50), and these data are consistent with obligatory tetramer interaction with the frmRAPro inverted repeat.
A candidate metal-binding site of FrmRE64H is formed by the side chains of His60 and His64 from one subunit and Cys35 from the second subunit within the dimeric assembly (Fig. 4D). These residues match the XYZ motif required for metal binding in related metal sensors RcnR, CsoR, and InrS, and this was the rationale behind the Glu64 → His substitution (31, 39, 45, 46, 50). To investigate the role of Cys35 and His60 in metal binding, site-directed mutants FrmRC35A and FrmRH60L were generated and assayed for their ability to bind Zn(II). Following preincubation with excess ZnCl2, neither variant retained Zn(II) during size exclusion chromatography in contrast to wild type FrmR and FrmRE64H, which each co-migrate with 1 molar eq of Zn(II) (Fig. 4E) (50). This indicates that the affinities of FrmRC35A and FrmRH60L for Zn(II) are considerably weaker than wild type FrmR and implicates these residues in Zn(II) (and by inference Co(II)) coordination. Candidate residues for a fourth ligand required for the tetrahedral coordination geometry observed for Co(II) and inferred for Zn(II) (50) include His3 (denoted position W in RcnR (46, 61)), Asp63, the amino terminus, or solvent (Fig. 4D).
Proposed Formaldehyde Sensory Site and Reaction Mechanism
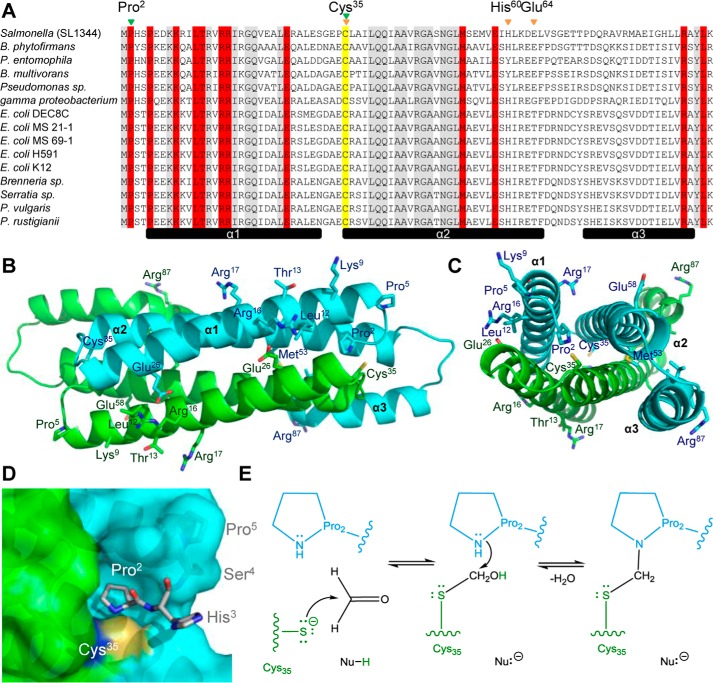
To define the functional formaldehyde sensory site, residues specifically conserved within the FrmR subgroup of the DUF156 family of transcriptional regulators were identified. Protein sequences previously ascribed to the FrmR subgroup (45) were used to generate a multiple-sequence alignment with Salmonella FrmR (Fig. 5A). Twelve residues are conserved within the FrmR subgroup but absent from the closely related Ni(II)/Co(II)-sensing RcnR subgroup. Two-thirds of the conserved residues are clustered in helix α1 based on the structure of FrmRE64H (Fig. 5, A–C). Sensing of formaldehyde may proceed via reaction with Cys35, also implicated in the FrmR metal site (Fig. 4, D and E) due to its conservation in all characterized DUF156 members. Formation of an S-formyl adduct at this Cys-thiol followed by reaction with a primary amine has been suggested as a possible reaction mechanism of FrmR with formaldehyde (30). The pyrrolidine side chain of proline residue 2 (α1) is in close proximity (3.0–3.2 Å in the four independent locations within the tetrameric structure) to Cys35 from α2′ (Fig. 5, B and C, and supplemental Fig. S1A). A second FrmR-specific proline (Pro5) acts to terminate helix α1 and positions the amino terminus of FrmRE64H adjacent to Cys35 (Fig. 5B). Pro2 is the first residue identified in the FrmRE64H structure and is positioned in a pocket at the dimer interface, leaving no space (and no observed electron density) for the amino-terminal methionine predicted by the primary sequence (Fig. 5D and supplemental Fig. S1A). The amino-terminal region has been implicated in the coordination of Ni(II)/Co(II) by RcnR and of Ni(II) by InrS (61, 62). In the absence of Met1, the terminal secondary amine of Pro2 and a Cys35-thiolate are both ideal candidates for nucleophilic addition to formaldehyde (Fig. 5, D and E) (63, 64). Either reaction with Pro2 followed by Cys35 via an N-methylol intermediate or reciprocally via an S-hydroxymethyl intermediate is plausible (Fig. 5E). In both cases, the end product would be a methylene bridge between the two residues, requiring a 1:1 formaldehyde/FrmR monomer (4 possible sites/tetramer) reaction stoichiometry.
FIGURE 5.
Conservation of residues in the DUF156 FrmR subgroup and proposed formaldehyde-sensing site. A, alignment of Salmonella FrmR with nonredundant UniProtKB DUF156 sequences previously attributed to the FrmR subgroup (45). Organism details and UniProtKB identifiers are outlined under “Experimental Procedures.” Highlighted in gray are residues conserved in both FrmR and RcnR subgroups. Highlighted in red are residues conserved in the FrmR but not RcnR subgroup. Highlighted in yellow is the invariant cysteine present in all DUF156 proteins. The secondary structure elements of the FrmRE64H crystal structure are shown below (black bars). The inferred Zn(II)/Co(II)-sensing site is identified by orange arrows. The proposed formaldehyde sensing site is identified by green arrows. B and C, dimeric representation of FrmRE64H with the side chains for Cys35 and FrmR subgroup-specific residues labeled. Each monomer is colored differently (using the same colors as in Fig. 3A) with secondary structure units labeled on the cyan subunit. D, solvent-accessible surface representation of the proposed formaldehyde-binding site, which comprises Pro2 (subunit 1, cyan) and Cys35 (subunit 2, green). E, proposed reaction of formaldehyde with FrmR Cys35 (green) followed by Pro2 (cyan) (both deprotonated ultimately to water) via an S-hydroxymethyl intermediate. The reciprocal reaction with Pro2 followed by Cys35 via an N-methylol intermediate is also possible. In both cases, a methylene bridge (black) between the two residues is the final product. The nucleophile(s) responsible for deprotonation of Cys35 and Pro2 remain unknown.
The cleavage of FrmR Met1 was examined by multiple-reaction monitoring MS using purified FrmR. Amino-terminal peptide PHSPEDK was detected, confirming that FrmR is a substrate for methionine aminopeptidase (Fig. 6A). To investigate the requirement of Cys35 and Pro2 for formaldehyde sensing, transcriptional fusions of PfrmRA-frmRC35A and PfrmRA-frmRP2S with lacZ were generated, introduced into Salmonella, and compared with wild type (PfrmRA-frmR). Expression from PfrmRA-frmR is derepressed by exposure of cells to formaldehyde in a concentration-dependent manner (Fig. 6B). Repression of PfrmRA is retained following mutation of either Cys35 or Pro2, but derepression in response to formaldehyde is completely abolished (Fig. 6B). To confirm that formaldehyde was unable to act as an allosteric effector of these FrmR variants, the DNA binding properties of FrmRC35A and FrmRP2S were characterized using fluorescence anisotropy (Fig. 6, C and D). Titration of frmRAPro with apo-FrmRC35A or apo-FrmRP2S in the presence of excess EDTA revealed KDNA values comparable with wild type FrmR and consistent with the observed repression by both mutants in vivo (KDNAapo-FrmRC35A = 1.6 ± 0.2 × 10−7 m and KDNAapo-FrmRP2S = 1.5 ± 0.2 × 10−7 m) (Fig. 6, C and D, and Table 1). However, in contrast to wild type FrmR (Fig. 3A), DNA binding by FrmRC35A was unaffected by the presence of 20 μm formaldehyde, indicating a loss of formaldehyde reactivity (Fig. 6C). The reactivity of FrmRP2S to formaldehyde was significantly decreased compared with wild type FrmR with apparent DNA binding affinity weaker than apo-FrmRP2S by only ∼4-fold in the presence of 20 μm formaldehyde (compared with ≥70-fold for wild type FrmR (Fig. 3A)) (Fig. 6D). Consequently, the FrmRC35A substitution impairs formaldehyde reactivity more severely than FrmRP2S (Fig. 6E). The proposed mechanism (1:1 formaldehyde/FrmR stoichiometry (Fig. 5E)) and observed DNA binding by FrmR implies an affinity at the allosteric site(s) of FrmR for formaldehyde in the 10–20 μm range, whereas formaldehyde affinities of FrmRC35A and FrmRP2S variants are inferred to be ≫100 μm and >50 μm, respectively (Fig. 6E). These data implicate Cys35 and Pro2 in formaldehyde-mediated derepression in vivo and impaired DNA binding in vitro (Fig. 6). The Zn(II)/Co(II) site also requires Cys35 (Fig. 4, D and E), implying overlap between the two effector sensory sites.
FIGURE 6.
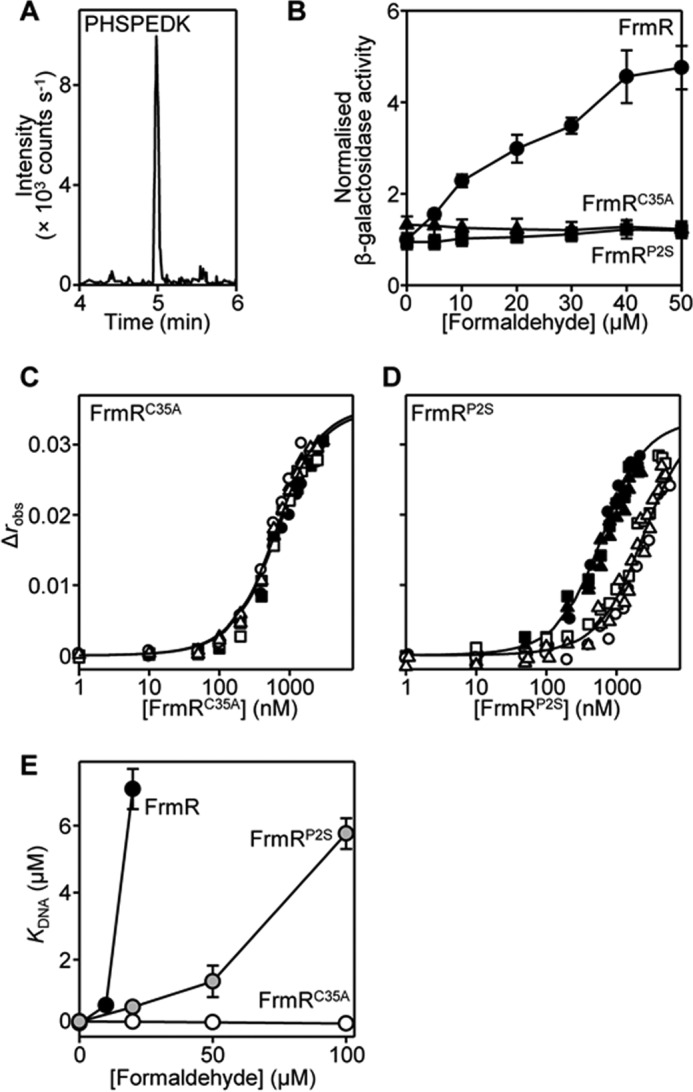
Pro2 and Cys35 are required for formaldehyde sensing by FrmR. A, LC-MS chromatogram following multiple-reaction monitoring of purified FrmR. Ion transition 405.19/488.24 is for analyte PHSPEDK. B, β-galactosidase activity in SL1344 containing PfrmRA-frmR (circles), PfrmRA-frmRC35A (triangles), or PfrmRA-frmRP2S (squares) fused to lacZ grown to mid-exponential phase in M9 minimal medium in the presence of formaldehyde (MNIC = 50 μm). Values are means of three biological replicates (each performed in triplicate) with S.D. C and D, anisotropy change upon titration of a limiting concentration of frmRAPro (10 nm) with FrmRC35A (C) or FrmRP2S (D) in the presence of 5 mm EDTA (closed symbols) and with the addition of 20 μm formaldehyde (open symbols). Data were fit to a model describing a 2:1 protein tetramer (nondissociable)/DNA stoichiometry (binding with equal affinity) (50, 86), and lines represent simulated curves produced from the average (apparent) KDNA determined across the experimental replicates shown. Symbol shapes represent individual experiments. E, apparent KDNA values of FrmR (black symbols), FrmRP2S (gray symbols), and FrmRC35A (open symbols) with increasing formaldehyde concentration. Values are means of three replicates with S.D. (error bars).
FrmR Is More Sensitive to Formaldehyde than RcnR
Although FrmR shares sequence similarities with Ni(II)/Co(II)-sensing RcnR (30, 32, 45, 46), expression from Salmonella rcnR-PrcnRA fused to lacZ is not derepressed by formaldehyde in vivo (Fig. 7A). Analysis of the rcnR-rcnA intergenic region identified two putative RcnR DNA-binding sequences in the target RcnR operator-promoter (supplemental Table S1). The interaction of RcnR with a fluorescently labeled double-stranded DNA fragment containing these sequences, rcnRAPro, was monitored by fluorescence anisotropy. The stoichiometry of Salmonella RcnR binding to rcnRAPro was first confirmed by titration of RcnR into a relatively high concentration of DNA (2.5 μm) with saturation observed at 8 molar eq of RcnR (monomer) consistent with binding of two tetramers (one per site) (Fig. 7B) as observed for E. coli RcnR (65). A limiting concentration of DNA (10 nm) and a model describing the binding of two non-dissociable RcnR tetramers per DNA molecule were subsequently used to determine the KDNA of RcnR and rcnRAPro as 1.5 ± 0.8 × 10−7 m for apo-RcnR (Fig. 7C and Table 1). As predicted, titration of rcnRAPro with either Ni(II)-RcnR or Co(II)-RcnR dramatically weakened DNA binding (Fig. 7C and Table 1); KDNANi(II)-RcnR ≥ 5.9 ± 1.3 × 10−6 m and KDNACo(II)-RcnR ≥ 1.5 ± 0.2 × 10−5 m. The allosteric coupling free energy (ΔGC), which couples effector binding to DNA binding (66–68), is calculated to be ≥+2.2 ± 0.2 and ≥+2.7 ± 0.2 kcal mol−1 for Ni(II)- and Co(II)-RcnR, respectively. Conversely, DNA binding by RcnR is unaffected by the inclusion of 20 μm formaldehyde (Fig. 7D), a concentration that weakens FrmR DNA binding by ≥70-fold (Fig. 6E). Importantly, these data establish a degree of specificity of FrmR over RcnR for formaldehyde.
FIGURE 7.
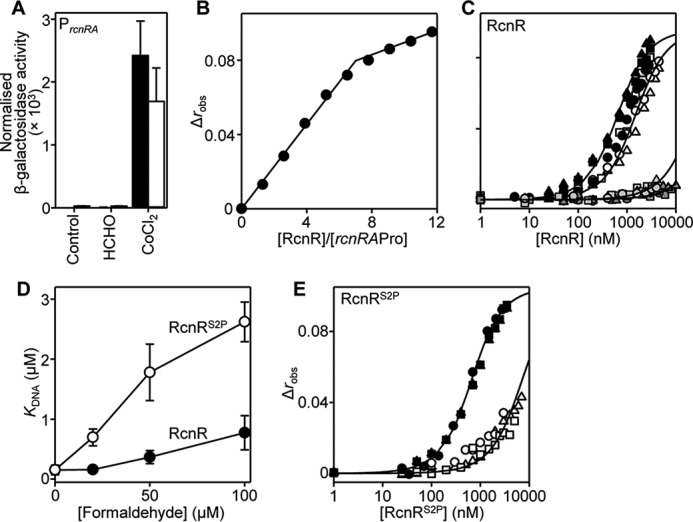
RcnR is less formaldehyde-responsive but RcnRS2P gains reactivity. A, β-galactosidase activity in SL1344 containing rcnR-PrcnRA (solid bars) or rcnRS2P-PrcnRA (open bars) fused to lacZ grown to mid-exponential phase in M9 minimal medium in the absence or presence of MNIC formaldehyde (50 μm) or CoCl2 (1 μm). Values are means of three biological replicates (each performed in triplicate) with S.D. (error bars). B and C, anisotropy change upon titration of a high concentration of rcnRAPro (2.5 μm) with RcnR in the presence of 5 mm EDTA (B) or a limiting concentration of rcnRAPro (10 nm) with RcnR in the presence of 5 mm EDTA (black symbols) and with the addition of 50 μm formaldehyde (open symbols) or Ni(II)-RcnR (light gray symbols) and Co(II)-RcnR (dark gray symbols) in the presence of 5 μm NiCl2 or 5 μm CoCl2, respectively (C). Symbol shapes represent individual experiments. Data were fit to a model describing a 2:1 protein tetramer (nondissociable)/DNA stoichiometry (binding with equal affinity) (50, 86), and lines represent simulated curves produced from the average (apparent) KDNA determined across the experimental replicates shown. D, apparent KDNA values of RcnR (black symbols) and RcnRS2P (open symbols) with increasing formaldehyde concentration. Values are means of three replicates with S.D. E, as described in C except with RcnRS2P in the presence of 5 mm EDTA (black symbols) and with the addition of 50 μm formaldehyde (open symbols).
Generation of an RcnRS2P Variant with Enhanced Response to Formaldehyde in Vitro
Increasing the concentration of formaldehyde during fluorescence anisotropy to 50 and 100 μm does impair binding of RcnR to rcnRAPro by ∼2.5- and ∼5-fold (relative to apo-RcnR), respectively (Fig. 7, C and D), providing an assay to monitor changes in RcnR formaldehyde reactivity. Introduction of the proposed formaldehyde sensing site of FrmR into Salmonella RcnR was achieved by mutation of Ser2 to Pro2 (Cys35 is conserved in both proteins). Titration of RcnRS2P into rcnRAPro (10 nm) confirms that this variant binds rcnRAPro with equal affinity to wild type RcnR (KDNAapo-RcnRS2P =1.6 ± 0.1 × 10−7 m) (Fig. 7E and Table 1). DNA binding by RcnRS2P was subsequently assessed in the presence of formaldehyde; inclusion of 50 and 100 μm formaldehyde weakened the apparent DNA affinity of RcnRS2P by ∼11- and ∼17-fold, respectively (Fig. 7, D and E). Thus, the single Ser2 → Pro point mutation generates an RcnR variant with increased reactivity to formaldehyde compared with wild type RcnR (Fig. 7D). However, this increase was not sufficient to gain formaldehyde sensing by rcnRS2P-PrcnRA in vivo (Fig. 7A). Repression from PrcnRA under control conditions (without inducer), combined with an observed cobalt responsiveness, confirmed that the expressed RcnRS2P was functional (Fig. 7A). It is inferred that the threshold for formaldehyde detection is not met by RcnRS2P in vivo.
Glutathione Inhibits Formaldehyde Sensing
The substrates of the FrmR-regulated alcohol dehydrogenase from Salmonella (FrmA) are predicted to be the formaldehyde and nitrosylated adducts of GSH, S-(hydroxymethyl)glutathione and S-nitrosoglutathione, respectively, by analogy to E. coli (Fig. 2B) (47, 69). Despite the evidence that DNA binding by FrmR is directly weakened by formaldehyde in vitro (Fig. 3A), glutathione adducts of formaldehyde might represent the predominant available species during formaldehyde stress conditions. Notably, glutathione has been shown to act positively on metal detection by FrmRE64H in vivo, suggesting that the protein may interact with glutathione adducts (Fig. 1) (50). Deletion of gshA, encoding γ-glutamate-cysteine ligase (70), required for the first step in glutathione biosynthesis, renders Salmonella more sensitive to exogenous formaldehyde compared with the wild type strain (Fig. 8A), as expected if (as in E. coli) glutathione is required for formaldehyde detoxification in Salmonella by formation of S-(hydroxymethyl)glutathione. However, formaldehyde-mediated derepression of PfrmRA-frmR was not impaired in ΔgshA cells (Fig. 8B), indicating that formation of formaldehyde-glutathione adducts is not an absolute requirement for FrmR responsiveness to formaldehyde in vivo. Indeed, expression levels from PfrmRA-frmR were higher in ΔgshA than in wild type, at equivalent exogenous formaldehyde concentrations (Fig. 8B), consistent with FrmR detecting increased formaldehyde accumulation in the cytosol of ΔgshA cells, due to reduced FrmA activity and/or due to glutathione acting negatively on the modification of FrmR by formaldehyde.
FIGURE 8.
The relationship between glutathione and formaldehyde sensing by FrmR. A, survival of wild type Salmonella SL1344 (solid circles) or ΔgshA (open circles) grown to mid-exponential phase in M9 minimal medium in the presence of formaldehyde. Values are means of three biological replicates (each performed in triplicate) with S.D. (error bars). B, β-galactosidase activity of SL1344 (solid symbols) or ΔgshA (open symbols) containing PfrmRA-frmR fused to lacZ grown to mid-exponential phase in M9 minimal medium in the presence of formaldehyde (MNIC = 50 and 20 μm for wild type and ΔgshA, respectively; see supplemental Fig. S2 for corresponding growth data). Values are means of at least three biological replicates (each performed in triplicate) with S.D. C, anisotropy change upon titration of a limiting concentration of frmRAPro (10 nm) with FrmR in the presence of 5 mm EDTA and 800 μm GSH in the absence (gray symbols) or presence (open symbols) of 20 μm formaldehyde. Symbol shapes represent individual experiments. Data were fit to a model describing a 2:1 protein tetramer (nondissociable)/DNA stoichiometry (binding with equal affinity) (50, 86), and lines represent simulated curves produced from the average (apparent) KDNA determined across the experimental replicates shown. D, intracellular glutathione concentration in Salmonella cells following growth to exponential phase in M9 minimal medium aerobically (O2) or anaerobically with TMAO as an alternative electron acceptor. Values are means of three biological replicates with S.D. E, representative (n = 3) LC-MS chromatograms of ion transitions detected in mid-logarithmic Salmonella SL1344 cells under aerobic growth conditions. Transitions are for analyte GQVEALER (solid black line) or labeled internal standard (IS) (GQVEALER[13C6,15N4], where R[13C6,15N4] represents 13C,15N-labeled arginine) (dashed gray line). F, fractional modification by formaldehyde of FrmR (solid black line), GSH (solid gray line), or FrmR (dashed red line) and RcnR (dashed blue line; tighter limit as indicated by the blue arrow) in the presence of GSH in Salmonella cells grown anaerobically with TMAO. Formaldehyde affinities of 10−5, 10−4 (tighter limit), and 1.77 × 10−3 m (73) were used for FrmR, RcnR, and GSH, respectively. Intracellular abundance was determined for FrmR (16.1 ± 0.2 nm) and GSH (1.2 ± 0.4 mm) and estimated for RcnR, as described under “Experimental Procedures.” G, the role of glutathione in formaldehyde detoxification and sensing in Salmonella. In the absence of effector, Salmonella FrmR represses the frm promoter. Formaldehyde directly modifies FrmR (reaction 1 in Fig. 1) via a deduced intersubunit methylene bridge between Pro2 and Cys35 (Fig. 5, up to four per tetramer) derepressing frm expression. GSH inhibits formaldehyde detection (reaction 3 in Fig. 1), and despite the high [glutathione], the affinity of FrmR for formaldehyde is sufficiently tight relative to GSH to enable expression of FrmA to coincide with the appearance of its substrate. The Salmonella frm operon lacks frmB, and YeiG may catalyze the final detoxification step. S-HMG, S-(hydroxymethyl)glutathione.
Binding of FrmR to frmRAPro was monitored by fluorescence anisotropy in the presence of 800 μm GSH. GSH alone has a minimal but detectable effect on DNA binding by apo-FrmR (∼2.5-fold tighter) (Fig. 8C). The ability of FrmR to respond to formaldehyde in the presence of GSH was assessed by subsequent titration of FrmR into frmRAPro in the presence of both formaldehyde (20 μm) and excess GSH (800 μm). The apparent DNA affinity of FrmR was weaker (relative to FrmR and GSH alone, without formaldehyde) by ∼9-fold, but critically, the magnitude of the response by FrmR to formaldehyde is diminished by GSH (compare open symbols in Figs. 3A and 8C). These data show that GSH competes with FrmR for formaldehyde rather than contributing toward its reactivity. How then can FrmR detect free formaldehyde in vivo, since glutathione is expected to be in a large molar excess? Under aerobic conditions, the intracellular glutathione concentration in Salmonella cells was determined to be 4.2 ± 0.5 mm (Fig. 8D), whereas the abundance of FrmR was 9.7 ± 2.6 tetramers/cell (16.1 ± 0.2 nm), as determined by quantitative mass spectrometry (Fig. 8E and supplemental Table S2). Repression by E. coli FrmR is alleviated during TMAO-mediated anaerobic respiration, probably due to TMAO demethylase activity and intracellular formaldehyde generation (15). The concentration of glutathione in Salmonella drops to 1.2 ± 0.4 mm when cells are grown anaerobically using TMAO as an alternative electron acceptor (Fig. 8D). These data have been used to model formation of S-(hydroxymethyl)glutathione and the modification of FrmR as a function of [formaldehyde], with implications for the species detected by FrmR in vivo discussed below (Fig. 8F).
Discussion
Detection of metals and formaldehyde by Salmonella FrmRE64H is retained when the sensor is expressed in a heterologous E. coli host (Fig. 2). Zn(II) is not required to transduce the formaldehyde signal in vivo because formaldehyde directly allosterically activates wild type Salmonella FrmR in vitro (Fig. 3). The allosteric response to organic molecules is specific to formaldehyde and not acetaldehyde in vitro and in vivo (Fig. 3). Deduced sensory sites for Zn(II)/Co(II) and for formaldehyde overlap with both effectors requiring Cys35 (Figs. 4 and 5). Substitution of either Cys35 or Pro2 decreases the reactivity of FrmR to formaldehyde in vitro and abolishes sensing in vivo (Fig. 6). The sensory site of FrmR is more reactive to formaldehyde than the related Ni(II)/Co(II) sensor RcnR in vitro, and RcnR does not respond to formaldehyde in vivo (Fig. 7). Introduction of the deduced formaldehyde sensory site to generate RcnRS2P confers increased reactivity to formaldehyde in vitro (Fig. 7). Although S-(hydroxymethyl)glutathione is a substrate for FrmA, free formaldehyde is the allosteric effector of FrmR, and glutathione competes with FrmR for formaldehyde both in vitro and in vivo (Fig. 8).
The unexpected ability of Zn(II) and Cu(I) to weaken Salmonella FrmR KDNA in vitro (50) raised the possibility that metals might act as signal transducers of intracellular formaldehyde accumulation (Fig. 1). Moreover, there is precedence for a Zn(II)-dependent alcohol dehydrogenase being regulated in response to Zn(II) by Zap1 (zinc-responsive activator protein) transcription factor as a Zn(II)-sparing mechanism in yeast (71, 72). FrmR-regulated glutathione-dependent formaldehyde dehydrogenase, FrmA, similarly requires Zn(II) for activity (47). However, here we eliminate the requirement of Zn(II) during FrmR-mediated derepression of frmRA because formaldehyde is shown by fluorescence anisotropy to be a direct allosteric effector of FrmR (Figs. 2 (E and F) and 3). The related metal sensor RcnR (which shares 40% identity with FrmR) is less reactive to formaldehyde by at least an order of magnitude (Figs. 3A, 6E, and 7 (C and D)). Candidate effector sensory sites for formaldehyde and Zn(II)/Co(II) were identified by structural characterization of FrmRE64H (Figs. 4 and 5) and shown by site-directed mutagenesis to each require Cys35 (Figs. 4E and 6 (B, C, and E)). We show that an FrmR-specific amino terminus, Pro2, is also required to react with formaldehyde and propose formation of an interdimer methylene bridge between the two residues (Figs. 5 and 6 (D and E) and supplemental Fig. S1A). Introduction of such a cross-link would only alter the distance between Cys35 and Pro2 by ∼+0.5 Å relative to the crystal structure. Future studies should aim to visualize the proposed methylene bridge and the nature of allosteric coupling between formaldehyde modification and DNA binding. Indeed, such coupling is yet to be characterized for any DUF156 family member. The unique (to date) Gly83 in Salmonella FrmR and the resulting kink in α3 may make this protein distinct.
Glutathione is not required for FrmR to respond to formaldehyde in vivo (Fig. 8B). Rather than aid detection, glutathione competes with FrmR for formaldehyde in vitro and inhibits the response in vivo (Fig. 8, B and C). In contrast, although glutathione acts positively toward cobalt detection by FrmRE64H in vivo (50), the present data argue against a suggestion that FrmRE64H preferentially detects cobalt due to its interaction with glutathione conjugates (Fig. 8). Because glutathione is such an abundant biomolecule (Fig. 8D), S-(hydroxymethyl)glutathione (the substrate for FrmA) might be expected to predominate over formaldehyde in a cell. However, the affinity of FrmR for formaldehyde is inferred to be ∼10−5 m from measured DNA affinities (Fig. 6E), substantially tighter than the affinity for formation of S-(hydroxymethyl)glutathione from formaldehyde and GSH (1.77 × 10−3 m) (73) (Fig. 8G). Thus, although GSH is at least 5 orders of magnitude more abundant than FrmR (determined to be 16.1 ± 0.2 nm; Fig. 8E and supplemental Table S2), FrmR will nonetheless be >85% modified by formaldehyde at cellular [formaldehyde], where only 4% of the GSH pool is in the S-(hydroxymethyl)glutathione form (Fig. 8 (F and G), dashed red line and gray line). Crucially, this means that expression of frmA will be derepressed as cellular S-(hydroxymethyl)glutathione begins to accumulate (Fig. 8F). Importantly, the ability of FrmR to respond to formaldehyde directly prevents [formaldehyde] from rising to levels where cross-linking of other cellular proteins (e.g. RcnR) (Fig. 8F, blue dashed line) or significant depletion of the GSH pool would occur.
The Salmonella and E. coli frm operons are distinct (Fig. 2), which could reflect requirements specific to pathogenicity with a suggestion that formaldehyde generation may arise following the macrophage respiratory burst (2). Consistent with this, the frmRA locus is known to be up-regulated during intracellular survival of Salmonella within macrophages (74, 75). Unlike E. coli, the Salmonella frm locus does not present a complete formaldehyde detoxification pathway (or recycling of GSH) due to the lack of frmB. YeiG, capable of catalyzing the formation of formate and GSH from S-formylglutathione in E. coli (27), is also present in Salmonella (Fig. 2B) and is an obvious candidate to function in the absence of FrmB (Fig. 8G). Furthermore, yeiG, which is not FrmR-regulated, is co-expressed with genes encoded by the Salmonella pathogenicity island-2 (SPI-2), notable for being up-regulated and absolutely required during replication within eukaryotic cells (74–76). It is formally possible that differences may emerge between the effectors and sensory sites of Salmonella versus E. coli FrmR (e.g. residues surrounding sensory sites, specificities to organic molecules, metals as allosteric effectors, nature of allostery) that reflect the demands for survival in the distinctive niches inhabited by each organism.
The FrmR sensory site is more reactive to formaldehyde than, for example, RcnR. The amino terminus of mature FrmR becomes a pyrrolidine secondary amine from Pro2, in contrast to the primary amino group of RcnR (from Ser2). Consequently, the nucleophilic reactivity of the FrmR amino terminus is predicted to be greater than RcnR (63, 64) and more able to undergo nucleophilic addition to the formaldehyde carbonyl group (Fig. 5). A Cys35-thiolate in both proteins also presents a particularly nucleophilic group capable of this reaction (Fig. 5) (63, 64). We propose that the presence of this reactive pair in FrmR would allow formation of an interdimer cross-link (Fig. 5). Consistent with this, Pro2 and Cys35 are required for formaldehyde detection by FrmR (Fig. 6), and creation of the proposed formaldehyde site in RcnRS2P increases sensitivity to formaldehyde in vitro (Fig. 7). However, mutation of the RcnR amino terminus alone is not sufficient to confer the same degree of reactivity exhibited by FrmR; therefore, it is likely that additional residues optimize formation of a cross-link. Most notable is Pro5, another FrmR-specific residue that terminates helix α1 and may confer a degree of rigidity to the amino terminus, positioning Pro2 into the sensory site adjacent to Cys35.
Effector selectivity of DUF156 family transcriptional repressors can be changed by relatively modest sequence alterations. Conservation of a cysteine at the effector site is now confirmed to be common to a formaldehyde-sensing family member (Figs. 5 and 6) as well as the metal sensors. Changing single residues proximal to this active cysteine has 1) increased the ability of RcnR to sense formaldehyde in vitro in RcnRS2P (Fig. 7, D and E), 2) enabled metal sensing in vivo by FrmRE64H (Fig. 2F) (50), and 3) switched the metal specificity of RcnRH3E in vivo (61). Notably, although RcnRS2P is more reactive to formaldehyde than RcnR in vitro, it still cannot respond in vivo (Fig. 7). Furthermore, FrmR can respond to metal in vitro but not in vivo (Fig. 2E) (50), the latter being achieved by the FrmRE64H variant. In the case of FrmRE64H, the threshold for Zn(II) responsiveness in vivo was met by a tighter Zn(II) affinity and weaker apo-DNA affinity (relative to wild type FrmR), rendering FrmRE64H competitive relative to cognate Zn(II) sensors, ZntR and Zur (50). FrmR responds to formaldehyde in a cell, placing it above some threshold of reactivity for this effector (Figs. 2E, 3B, and 8F). FrmRC35A and FrmRP2S variants, along with RcnR and RcnRS2P, must be below the threshold for formaldehyde sensing (Figs. 6 (B–E) and 7 (C–E)). Presumably, cells do not survive at [effector] sufficient to trigger such sensors. Among the FrmR-RcnR DUF156 proteins (and yet to be tested for CstR and CsoR), subtle quantitative changes to effector responses tune these sensors above or below different cellular thresholds, and this is sufficient to confer the necessary level of specificity in vivo.
Experimental Procedures
Bacterial Strains and DNA Manipulations
S. enterica sv. Typhimurium strain SL1344 was used as wild type, and strain LB5010a was used as a restriction-deficient modification-proficient host for DNA manipulations (50). Deletion derivatives of SL1344 lacking frmR and gshA were generated previously (50). E. coli strains BW25113ΔfrmR, in which the frmR coding sequence is disrupted by a kanR cassette (77), was used for β-galactosidase assays. This was a gift from D. Weinkove (Durham University). E. coli strain DH5α was used for routine cloning, and strain BL21(DE3) was used for recombinant protein overexpression. Bacteria were cultured aerobically (with shaking) at 37 °C in LB medium or M9 minimal medium (78), supplemented with thiamine (0.001%, w/v) and either l-histidine (20 μg ml−1) for Salmonella or 1 μm C6H5FeO7 for E. coli. Carbenicillin (100 μg ml−1), kanamycin (25 μg ml−1), and TMAO (40 mm) were added where appropriate. Cells were transformed to antibiotic resistance as described (78, 79). For glutathione quantification under aerobic and anaerobic conditions, glucose was replaced with glycerol as a non-fermentable carbon source. For survival assays, overnight cultures were grown in M9 minimal medium, diluted 1:50 into fresh medium in 14-ml culture tubes containing the indicated concentrations of formaldehyde, and grown to mid-logarithmic phase. Growth was assessed by measuring absorbance at 600 nm and calculating the percentage survival compared with the control condition for each strain. Experiments were performed in triplicate on at least three separate occasions. Generated plasmid constructs were checked by sequence analysis. Primers are listed in supplemental Table S1.
Bioinformatic Analysis
Fourteen FrmR and nine RcnR non-redundant primary amino acid sequences identified in (45) and still present in UniProtKB (80) were aligned with the S. enterica serovar Typhimurium SL1344 FrmR sequence (UniProtKB identifier: A0A0H3NLH8) using the PRALINE multiple-sequence alignment tool (81). UniProtKB identifiers for the FrmR sequences were as follows: B2SZZ0, Burkholderia phytofirmans (strain DSM 17436/PsJN); Q1IAA5, Pseudomonas entomophila (strain L48); B9BFA7, Burkholderia multivorans CGD1; F0DZ53, Pseudomonas sp. (strain TJI-51); B5JUQ3, Gammaproteobacterium HTCC5015; H4ZQC4, E. coli DEC8C; D8A2B2, E. coli (strain MS 21–1); D7ZJL9, E. coli MS 69–1; F4VAD6, E. coli H591; P0AAP3 (blue), E. coli (K12); G7LSK1, Brenneria sp. EniD312; I0QLA2, Serratia sp. M24T3; Q8KKB0, Proteus vulgaris; D1P3L2, Providencia rustigianii DSM 4541. Residues present in FrmR but not RcnR sequences were identified as FrmR-specific. For three sequences (D8A2B2, D7ZJL9, and F4VAD6) amino-terminal residues annotated by UniprotKB were not predicted to be coding residues using the NCBI (National Center for Biotechnology Information) database and were removed. Phylogenetic analysis was performed using ClustalW2 phylogeny (82); E. coli FrmR sequences except for E. coli K12 were removed. Distance values relate to the number of substitutions as a proportion of the length of the alignment (excluding gaps). Amino acid sequence identities were determined using Clustal Omega (83).
Generation of Promoter-lacZ Fusion Constructs and β-Galactosidase Assays
Promoter-lacZ fusions PfrmRA-frmR, PfrmRA-frmRE64H, and rcnR-PrcnRA have been described previously (50). Subcloning vector pGEM-T containing either the PfrmRA-frmR or rcnR-PrcnRA DNA fragment (50) was used as a template for site-directed mutagenesis via the QuikChange® protocol (Stratagene) using primers 1 and 2 to generate PfrmRA-frmRP2S, primers 3 and 4 to generate PfrmRA-frmRC35A, or primers 5 and 6 to generate rcnRS2P-PrcnRA (primers listed in supplemental Table S1). Digested fragments were cloned into the SmaI/BamHI site of pRS415 (84). Constructs were introduced into E. coli strain BW25113ΔfrmR as appropriate or Salmonella strain LB5010a before SL1344 (and derivatives). β-Galactosidase assays were performed as described (50, 85), in triplicate, and on at least three separate occasions. Briefly, overnight cultures were grown in M9 minimal medium; diluted 1:50 in fresh medium supplemented with up to MNIC (defined as the maximum concentration that inhibited growth by ∼10%) of metal, alcohol, or aldehyde; and grown to mid-logarithmic phase before assays. MNICs under these growth conditions were 5 μm CoCl2, 50 μm ZnCl2, 300 mm ethanol, 600 mm methanol, 5 mm 1-butanol, 50 mm 1-propanol, 200 mm 2-propanol, 50 μm formaldehyde, and 3 mm acetaldehyde, with the exception that 1 μm CoCl2 was the MNIC for cells expressing rcnR-PrcnRA or rcnRS2P-PrcnRA. Time course experiments were performed as described (50) by exposing logarithmic cells to MNIC metal or formaldehyde for 2 h at 25 °C. Where stated, β-galactosidase activity (nmol of o-nitrophenol min−1 mg of protein−1) was normalized to the control data for cells expressing the wild type protein conducted in parallel.
Protein Expression and Purification
Vectors for overexpression of FrmR, FrmRE64H, and RcnR have been described previously (50). Site-directed mutagenesis was conducted as described above using template pETfrmR and primers 7–12 to generate pETfrmRP2S, pETfrmRC35A, and pETfrmRH60L or using template pETrcnR and primers 13 and 14 to generate pETrcnRS2P. Proteins were expressed and purified as described previously (50). Mutant variants were purified exactly as described for the respective wild type protein. Protein purity was assessed by SDS-PAGE. Anerobic protein stocks (maintained in an anaerobic chamber) were prepared as described and confirmed to be ≥90% reduced and ≥95% metal-free (50). FrmR and variants were stored in 100 mm NaCl, 400 mm KCl, 10 mm HEPES, pH 7.0. RcnR and variants were stored in 200 mm NaCl, 800 mm KCl, 10 mm HEPES, pH 7.0. All in vitro experiments were carried out under anaerobic conditions using Chelex-treated and N2-purged buffers as described previously (50). Due to the absence of any thiol groups, experiments with FrmRC35A were carried out under aerobic conditions.
Inductively Coupled Plasma MS
Anaerobic protein stocks (10–20 μm) or size exclusion chromatography fractions were diluted 10-fold in 2.5% Suprapur HNO3 (Merck Millipore). Quantitative analysis of metal content was determined using an XSERIES-2 inductively coupled plasma mass spectrometer (Thermo Fisher Scientific) following calibration with elemental standards that were matrix-matched to the sample by inclusion of an appropriate buffer system.
Fluorescence Anisotropy
Fluorescently labeled double-stranded DNA probe, frmRAPro, containing the identified FrmR-binding site has been described previously (50). Complementary single-stranded oligonucleotides 15 (hexachlorofluorescein-labeled) and 16 (containing two identified RcnR-binding sites (32, 65) and flanking oligonucleotides) or 17 (hexachlorofluorescein-labeled) and 18 (frmRAPro but with mutation of one T/A-rich inverted repeat) (supplemental Table S1) were annealed by heating a 10 or 200 μm concentration of each strand in 150 mm NaCl, 10 mm HEPES, pH 7.0, to 95 °C and cooled slowly overnight, to generate rcnRAPro (35 bp) or frmRAPro* (33 bp). Fluorescently labeled annealed probes were analyzed by native PAGE (12% (w/v)), and RcnR/rcnRAPro stoichiometry experiments were performed as described (50) by titration of RcnR (prepared in 100 mm NaCl, 400 mm KCl, 10 mm HEPES, pH 7.0, and 5 mm EDTA) into 2.5 μm rcnRAPro in 60 mm NaCl, 240 mm KCl, 10 mm HEPES, pH 7.0, and 5 mm EDTA. For KDNA determination in the absence of effector, frmRAPro or rcnRAPro was diluted to 10 nm in the same buffer. Formaldehyde, acetaldehyde, and GSH were included as outlined in the figure legends. For metal-loaded experiments, EDTA was replaced with 5 μm NiCl2 or CoCl2. FrmR (and variants) and RcnR (and variants) were prepared as described previously (50) and described above or by replacing EDTA with 1.2 molar eq/protein monomer of NiCl2 or CoCl2 as appropriate. Formaldehyde was prepared daily from single-use sealed ampules of methanol-free 16% (v/v) formaldehyde (Pierce) and stored under anaerobic conditions for the course of the experiment to prevent oxidation. A concentrated GSH stock (5 mm) was prepared in Chelex-treated, N2-purged 100 mm NaCl, 400 mm KCl, 10 mm HEPES, pH 7.0, under anaerobic conditions. This stock was confirmed to be ≥90% reduced by reaction with DTNB, stored anaerobically to prevent oxidation, and used within 3 days. Changes in anisotropy (Δrobs) were measured using a modified Cary Eclipse fluorescence spectrophotometer (Agilent Technologies) as described (50). Control titrations of apo-FrmR and apo-FrmRE64H into frmRAPro (Fig. 4C) are new unpublished data sets and are presented here to demonstrate reproducibility and for comparative purposes. Data (for both FrmR and RcnR) were fit to a model describing binding of two non-dissociable tetramers (Ktet fixed at 10−20 m) to a target DNA probe with equal affinity (50, 86), using Dynafit (87) (see Figs. 3, 4, 6, and 7 legends and Table 1 for details; sample Dynafit script shown in the supplemental material). For experiments where DNA binding did not saturate, the average fitted Δrobs maximum value from apoprotein experiments was used as a fixed parameter. Coupling free energies (ΔGC) linking DNA binding to effector binding (62, 66, 67) were determined as described previously (50), calculated from the full set of (equally weighted) possible pairwise permutations of KC.
Crystallization of FrmRE64H and Data Collection
Concentrated FrmRE64H (∼1 mm) was diluted to 0.5 mm in 400 mm NaCl, 1 mm EDTA, 1 mm DTT, and 10 mm HEPES, pH 7.0, and stored aerobically at 4 °C for up to 2 weeks. Initial crystallization trials were conducted using the Screenmaker 96 + 8TM Xtal (Innovadyne Technologies) and commercially available screening kits (Molecular Dimensions). Subsequent FrmRE64H crystals were obtained in 20 mm NaCl, 23% (w/v) poly(ethylene glycol) 4000, and 10 mm BisTris, pH 6.5, by hanging drop vapor diffusion at 20 °C. Crystals were physically fragile and disintegrated rapidly when cryoprotectants were added. Multiple crystals using a wide range of cryoconditions were frozen and tested. Results were obtained from a crystal soaked in 25% (v/v) glycerol mounted in cryoloops (88). Overall data quality was compromised by residual ice rings and anisotropic diffraction, potentially giving rise to higher than expected R-factors of the final model. FrmRE64H diffraction data were collected at the Diamond Light Source on beamline I03 at 77 K with a Pilatus pixel detector (89). Diffraction data were initially processed using Mosfilm (90) to a resolution of 2.1 Å to enable ab initio solution and reprocessed (to 2.19 Å) with Xia2 (91) for structure refinement. Initial molecular replacement trials using MolRep (92) and Phaser (93) using Protein Data Bank entry 2HH7 (Cu(I)-CsoR from Mycobacterium tuberculosis) (31) were unsuccessful, presumably due to differences in the orientation of the three helices of the monomer and significant differences in monomer-monomer as well as dimer-dimer orientations in the homotetramer. The structure was solved using Arcimboldo installed on a Condor grid computer (94, 95). The initial model was completed by iterative cycles of model building and refinement using COOT (96) and REFMAC (97). The final model contained one homotetramer in the asymmetric unit with each chain containing residues 2–89 and residue 90 in chains B and C and 103 water molecules. The structure was refined against intensities with local non-crystallographic symmetry restraints (98), using Phenix (99). Applying local non-crystallographic symmetry restraints enabled the tracing of all four chains despite relatively weak density, particularly in α3. This confers higher than expected overall real-space R value Z-score and R-factors. A number of polar surface residues where no electron density was observed for the side chains were refined as alanines (Chain A: His3, Lys8, Lys9, Glu69, Ile82; Chain B: Lys62, Glu69, Ile82, Leu90; Chain C: Glu30, Glu69, Leu90; Chain D: His3, Lys8, Lys9, Glu55, Lys62, Glu69, Ile82). Ramachandran plot analysis using RAMPAGE Ramachandran plot assessment (100) of FrmRE64H demonstrates that 98.8% of residues are in the favored region (supplemental Fig. S1B). The final data and refinement statistics are provided in Table 2 with the structure deposited in the Protein Data Bank under accession code 5LCY.
TABLE 2.
Crystallographic data collection and refinement statistics for FrmRE64H
| Parameter | Value |
|---|---|
| Data collection | |
| Beam line | I03 |
| Wavelength (Å) | 0.9762 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 68.79, 25.68, 100.50 |
| α, β, γ (degrees) | 90, 103.1, 90 |
| Resolution (Å) | 29.7–2.19 |
| Rmerge | 0.081 (0.495)a |
| I/σI | 12.1 (3.2)a |
| Multiplicity | 6.3 (6.4)a |
| Completeness (%) | 99.3 |
| Wilson B-factorb | 33 |
| Refinement | |
| Resolution (Å) | 29.7–2.19 |
| No. reflections | 17990 |
| Rwork/Rfree | 0.24/0.31 |
| No. of atoms | |
| Protein | 2767 |
| Water | 103 |
| B-factors (Å2) | |
| Chain A | 51 |
| Chain B | 47 |
| Chain C | 47 |
| Chain D | 53 |
| Protein | 30 |
| Water | 48 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (degrees) | 1.1 |
a The values in parentheses refer to the highest resolution shell (2.25–2.19 Å).
b Calculated using phenix.xtriage (99).
Protein Metal Migration by Size Exclusion Chromatography
Experiments were carried out as described previously (50). FrmR, FrmRE64H, FrmRC35A, or FrmRH60L was incubated (120 min) with an excess of ZnCl2 in 100 mm NaCl, 400 mm KCl, 10 mm HEPES, pH 7.0, and an aliquot (0.5 ml) was resolved by size exclusion chromatography (PD10 Sephadex G25, GE Healthcare) in the same buffer conditions. Fractions were analyzed for zinc by inductively coupled plasma MS and for protein by a Bradford assay as described (50). The control experiments with FrmR and FrmRE64H (Fig. 4E) are new unpublished data sets and are presented here to demonstrate reproducibility and for comparative purposes.
FrmR in Vivo Quantification and Detection of Met1 Cleavage by Liquid Chromatography-Tandem Mass Spectrometry
Quantification of FrmR in cellular lysates of SL1344 was performed exactly as described previously using aerobically grown logarithmic cells (50). To detect FrmR Met1 cleavage, a tryptic digest was performed with 5 μg of FrmR and 14 μg of trypsin in 50 mm NH4HCO3 with shaking (1000 rpm) at 37 °C for 16 h and stopped by the addition of 15% (v/v) formic acid (5 μl). The digested samples were separated by gradient elution at 0.3 ml min−1 using a Zorbax Eclipse Plus C18 column (2.1 × 150 mm, 3.5-μm particles, Agilent Technologies) at room temperature. Mobile phase A and B consisted of 0.1% (v/v) formic acid in water and 0.1% (v/v) formic acid in acetonitrile, respectively. Detection of FrmR amino-terminal peptide PHSPEDK was achieved by applying an aliquot (10 μl) to a 6500 triple quadrupole mass spectrometer (AB Sciex) operating in positive ionization mode. Acquisition methods used the following parameters: 5500 V ion spray voltage, 25 p.s.i. curtain gas, 60 p.s.i. source gas, 550 °C interface heating temperature, 40 V declustering potential, 26 V collision energy, and 27 V collision cell exit potential. Scheduled multiple-reaction monitoring of ion transition 405.19/488.24 was performed with a 90-s multiple-reaction monitoring detection window and 1.00-s target scan time.
Quantification of Intracellular Glutathione
Intracellular glutathione was measured as described (50). Lysates from logarithmically growing cells were prepared from overnight cultures grown in M9 minimal medium with glycerol as a carbon source, diluted 1:50 in fresh medium, and grown at 37 °C either in round bottom flasks with shaking to maintain aerobic conditions or with the addition of TMAO (40 mm) and static incubation of completely filled Parafilm-sealed 1.5-ml tubes to maintain anaerobic conditions. No growth was observed under anaerobic conditions when TMAO was not included as an electron acceptor. Viable cells were enumerated on LB agar, and cell volume was estimated as 1 fl.
Fractional Occupancy Model to Describe Formaldehyde Modification in Vivo
Fractional modification of FrmR, RcnR, and GSH with formaldehyde as a function of formaldehyde concentration was determined using Dynafit (87) with the following values as fixed parameters. Affinities of FrmR and RcnR for formaldehyde were estimated following fluorescence anisotropy to be 10−5 and 10−4 m, respectively; the dissociation constant for GSH and S-(hydroxymethyl)glutathione was 1.77 × 10−3 m (73); the total intracellular concentration of glutathione during anaerobic growth with TMAO as an electron acceptor was 1.2 × 10−3 m and was assumed to be in the reduced form (90–99% of the glutathione pool is GSH in resting Salmonella cells (101, 102)); the intracellular concentration of FrmR tetramer under aerobic conditions was calculated to be 1.61 × 10−8 m and was used as an estimate for the intracellular concentrations of FrmR and RcnR tetramers under anaerobic growth conditions. Cell volume was 1 fl. A sample Dynafit script is shown in the supplemental material.
Author Contributions
D. O. carried out the in vivo survival and gene expression experiments and bioinformatic analysis and developed the fractional occupancy model for formaldehyde modification. C. P. did the in vitro metal-binding and fluorescence anisotropy experiments and generated FrmRE64H crystals. E. P. and I. U. solved the x-ray crystal structure of FrmRE64H. J. C. and T. G. H performed the LC-MS/MS. L. N. S. and C. P. determined the intracellular glutathione concentration. D. O. and C. P. made equivalent contributions to data preparation and, with N. J. R., interpreted the significance of the data. D. O. and N. J. R. drafted the manuscript. N. J. R. had overall responsibility for the design and coordination of the program. All authors reviewed the results and edited and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Diamond Light Source for access to beamline I03. We thank DBS Genomics (Durham University) for sequencing of plasmid constructs, David Weinkove (Durham University) for the E. coli frmR deletion strain, and Jeff Green (University of Sheffield) and Peter T. Chivers and Andrew W. Foster (both from Durham University) for insightful discussions. The support and advice of Elena Lurie-Luke and Charlie Saunders (both from Procter and Gamble) is gratefully acknowledged.
This work was supported by a Biotechnology and Biological Sciences Research Council Industrial Partnership Award BB/J017787/1. This work received support from Procter and Gamble.

This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 5LCY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TMAO
- trimethylamine N-oxide
- MNIC
- maximum non-inhibitory concentrations
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Bolt H. M. (1987) Experimental toxicology of formaldehyde. J. Cancer Res. Clin. Oncol. 113, 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen N. H., Djoko K. Y., Veyrier F. J., and McEwan A. G. (2016) Formaldehyde stress responses in bacterial pathogens. Front. Microbiol. 7, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conaway C. C., Whysner J., Verna L. K., and Williams G. M. (1996) Formaldehyde mechanistic data and risk assessment: endogenous protection from DNA adduct formation. Pharmacol. Ther. 71, 29–55 [DOI] [PubMed] [Google Scholar]
- 4. Feldman M. Y. (1973) Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog. Nucleic Acid Res. Mol. Biol. 13, 1–49 [DOI] [PubMed] [Google Scholar]
- 5. Ma T. H., and Harris M. M. (1988) Review of the genotoxicity of formaldehyde. Mutat. Res. 196, 37–59 [DOI] [PubMed] [Google Scholar]
- 6. Chistoserdova L., Kalyuzhnaya M. G., and Lidstrom M. E. (2009) The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63, 477–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanson R. S., and Hanson T. E. (1996) Methanotrophic bacteria. Microbiol. Rev. 60, 439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vorholt J. A. (2002) Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178, 239–249 [DOI] [PubMed] [Google Scholar]
- 9. Yurimoto H., Kato N., and Sakai Y. (2005) Assimilation, dissimilation, and detoxification of formaldehyde, a central metabolic intermediate of methylotrophic metabolism. Chem. Rec. 5, 367–375 [DOI] [PubMed] [Google Scholar]
- 10. Ferenci T., Strom T., and Quayle J. R. (1974) Purification and properties of 3-hexulose phosphate synthase and phospho-3-hexuloisomerase from Methylococcus capsulatus. Biochem. J. 144, 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marx C. J., Chistoserdova L., and Lidstrom M. E. (2003) Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185, 7160–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marx C. J., Miller J. A., Chistoserdova L., and Lidstrom M. E. (2004) Multiple formaldehyde oxidation/detoxification pathways in Burkholderia fungorum LB400. J. Bacteriol. 186, 2173–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsui T., Nambu S., Ono Y., Goulding C. W., Tsumoto K., and Ikeda-Saito M. (2013) Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry 52, 3025–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Streit B. R., Kant R., Tokmina-Lukaszewska M., Celis A. I., Machovina M. M., Skaar E. P., Bothner B., and DuBois J. L. (2016) Time-resolved studies of IsdG protein identify molecular signposts along the non-canonical heme oxygenase pathway. J. Biol. Chem. 291, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Denby K. J., Rolfe M. D., Crick E., Sanguinetti G., Poole R. K., and Green J. (2015) Adaptation of anaerobic cultures of Escherichia coli K-12 in response to environmental trimethylamine-N-oxide. Environ. Microbiol. 17, 2477–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thornalley P., Wolff S., Crabbe J., and Stern A. (1984) The autoxidation of glyceraldehyde and other simple monosaccharides under physiological conditions catalysed by buffer ions. Biochim. Biophys. Acta 797, 276–287 [DOI] [PubMed] [Google Scholar]
- 17. Trewick S. C., Henshaw T. F., Hausinger R. P., Lindahl T., and Sedgwick B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 [DOI] [PubMed] [Google Scholar]
- 18. Thornalley P. J. (1993) The glyoxalase system in health and disease. Mol. Aspects Med. 14, 287–371 [DOI] [PubMed] [Google Scholar]
- 19. Harms N., Ras J., Reijnders W. N., van Spanning R. J., and Stouthamer A. H. (1996) S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178, 6296–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goenrich M., Bartoschek S., Hagemeier C. H., Griesinger C., and Vorholt J. A. (2002) A glutathione-dependent formaldehyde-activating enzyme (Gfa) from Paracoccus denitrificans detected and purified via two-dimensional proton exchange NMR spectroscopy. J. Biol. Chem. 277, 3069–3072 [DOI] [PubMed] [Google Scholar]
- 21. Wilson S. M., Gleisten M. P., and Donohue T. J. (2008) Identification of proteins involved in formaldehyde metabolism by Rhodobacter sphaeroides. Microbiology 154, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uotila L., and Koivusalo M. (1974) Purification and properties of S-formylglutathione hydrolase from human liver. J. Biol. Chem. 249, 7664–7672 [PubMed] [Google Scholar]
- 23. Kaulfers P. M., and Marquardt A. (1991) Demonstration of formaldehyde dehydrogenase activity in formaldehyde-resistant Enterobacteriaceae. FEMS Microbiol. Lett. 63, 335–338 [DOI] [PubMed] [Google Scholar]
- 24. Rosario Fernández M., Jörnvall H., Moreno A., Kaiser R., and Parés X. (1993) Cephalopod alcohol dehydrogenase: purification and enzymatic characterization. FEBS Lett. 328, 235–238 [DOI] [PubMed] [Google Scholar]
- 25. Gutheil W. G., Kasimoglu E., and Nicholson P. C. (1997) Induction of glutathione-dependent formaldehyde dehydrogenase activity in Escherichia coli and Hemophilus influenza. Biochem. Biophys. Res. Commun. 238, 693–696 [DOI] [PubMed] [Google Scholar]
- 26. Herring C. D., and Blattner F. R. (2004) Global transcriptional effects of a suppressor tRNA and the inactivation of the regulator frmR. J. Bacteriol. 186, 6714–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gonzalez C. F., Proudfoot M., Brown G., Korniyenko Y., Mori H., Savchenko A. V., and Yakunin A. F. (2006) Molecular basis of formaldehyde detoxification: characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 281, 14514–14522 [DOI] [PubMed] [Google Scholar]
- 28. Mason R. P., Sanders J. K. M., Crawford A., and Hunter B. K. (1986) Formaldehyde metabolism by Escherichia coli: detection by in vivo carbon-13 NMR spectroscopy of S-(hydroxymethyl)glutathione as a transient intracellular intermediate. Biochemistry 25, 4504–4507 [DOI] [PubMed] [Google Scholar]
- 29. Uotila L., and Koivusalo M. (1974) Formaldehyde dehydrogenase from human liver: purification, properties, and evidence for the formation of glutathione thiol esters by the enzyme. J. Biol. Chem. 249, 7653–7663 [PubMed] [Google Scholar]
- 30. Higgins K. A., and Giedroc D. (2014) Insights into protein allostery in the CsoR/RcnR family of transcriptional repressors. Chem. Lett. 43, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., and Giedroc D. P. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 [DOI] [PubMed] [Google Scholar]
- 32. Iwig J. S., Rowe J. L., and Chivers P. T. (2006) Nickel homeostasis in Escherichia coli: the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 62, 252–262 [DOI] [PubMed] [Google Scholar]
- 33. Brown N. L., Stoyanov J. V., Kidd S. P., and Hobman J. L. (2003) The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27, 145–163 [DOI] [PubMed] [Google Scholar]
- 34. Lee J. W., and Helmann J. D. (2007) Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499 [DOI] [PubMed] [Google Scholar]
- 35. Osman D., and Cavet J. S. (2010) Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 27, 668–680 [DOI] [PubMed] [Google Scholar]
- 36. Pennella M. A., and Giedroc D. P. (2005) Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals 18, 413–428 [DOI] [PubMed] [Google Scholar]
- 37. Rubio-Sanz L., Prieto R. I., Imperial J., Palacios J. M., and Brito B. (2013) Functional and expression analysis of the metal-inducible dmeRF system from Rhizobium leguminosarum bv. viciae. Appl. Environ. Microbiol. 79, 6414–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Festa R. A., Jones M. B., Butler-Wu S., Sinsimer D., Gerads R., Bishai W. R., Peterson S. N., and Darwin K. H. (2011) A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foster A. W., Patterson C. J., Pernil R., Hess C. R., and Robinson N. J. (2012) Cytosolic Ni(II) sensor in cyanobacterium: nickel detection follows nickel affinity across four families of metal sensors. J. Biol. Chem. 287, 12142–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grossoehme N., Kehl-Fie T. E., Ma Z., Adams K. W., Cowart D. M., Scott R. A., Skaar E. P., and Giedroc D. P. (2011) Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luebke J. L., Arnold R. J., and Giedroc D. P. (2013) Selenite and tellurite form mixed seleno- and telluro-trisulfides with CstR from Staphylococcus aureus. Metallomics 5, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coyne H. J. 3rd, and Giedroc D. P. (2013) Backbone resonance assignments of the homotetrameric (48 kD) copper sensor CsoR from Geobacillus thermodenitrificans in the apo- and Cu(I)-bound states: insights into copper-mediated allostery. Biomol. NMR Assign. 7, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dwarakanath S., Chaplin A. K., Hough M. A., Rigali S., Vijgenboom E., and Worrall J. A. (2012) Response to copper stress in Streptomyces lividans extends beyond genes under direct control of a copper-sensitive operon repressor protein (CsoR). J. Biol. Chem. 287, 17833–17847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakamoto K., Agari Y., Agari K., Kuramitsu S., and Shinkai A. (2010) Structural and functional characterization of the transcriptional repressor CsoR from Thermus thermophilus HB8. Microbiology 156, 1993–2005 [DOI] [PubMed] [Google Scholar]
- 45. Chang F. M., Coyne H. J., Cubillas C., Vinuesa P., Fang X., Ma Z., Ma D., Helmann J. D., García-de los Santos A., Wang Y. X., Dann C. E. 3rd, and Giedroc D. P. (2014) Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J. Biol. Chem. 289, 19204–19217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iwig J. S., Leitch S., Herbst R. W., Maroney M. J., and Chivers P. T. (2008) Ni(II) and Co(II) sensing by Escherichia coli RcnR. J. Am. Chem. Soc. 130, 7592–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gutheil W. G., Holmquist B., and Vallee B. L. (1992) Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry 31, 475–481 [DOI] [PubMed] [Google Scholar]
- 48. Nobre L. S., Al-Shahrour F., Dopazo J., and Saraiva L. M. (2009) Exploring the antimicrobial action of a carbon monoxide-releasing compound through whole-genome transcription profiling of Escherichia coli. Microbiology 155, 813–824 [DOI] [PubMed] [Google Scholar]
- 49. Wang S., Deng K., Zaremba S., Deng X., Lin C., Wang Q., Tortorello M. L., and Zhang W. (2009) Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl. Environ. Microbiol. 75, 6110–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Osman D., Piergentili C., Chen J., Chakrabarti B., Foster A. W., Lurie-Luke E., Huggins T. G., and Robinson N. J. (2015) Generating a metal-responsive transcriptional regulator to test what confers metal sensing in cells. J. Biol. Chem. 290, 19806–19822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yurimoto H., Hirai R., Matsuno N., Yasueda H., Kato N., and Sakai Y. (2005) HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis. Mol. Microbiol. 57, 511–519 [DOI] [PubMed] [Google Scholar]
- 52. Kidd S. P., Potter A. J., Apicella M. A., Jennings M. P., and McEwan A. G. (2005) NmlR of Neisseria gonorrhoeae: a novel redox responsive transcription factor from the MerR family. Mol. Microbiol. 57, 1676–1689 [DOI] [PubMed] [Google Scholar]
- 53. Kidd S. P., Jiang D., Tikhomirova A., Jennings M. P., and McEwan A. G. (2012) A glutathione-based system for defense against carbonyl stress in Haemophilus influenzae. BMC Microbiol. 12, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stroeher U. H., Kidd S. P., Stafford S. L., Jennings M. P., Paton J. C., and McEwan A. G. (2007) A pneumococcal MerR-like regulator and S-nitrosoglutathione reductase are required for systemic virulence. J. Infect. Dis. 196, 1820–1826 [DOI] [PubMed] [Google Scholar]
- 55. McEwan A. G., Djoko K. Y., Chen N. H., Couñago R. L., Kidd S. P., Potter A. J., and Jennings M. P. (2011) Novel bacterial MerR-like regulators their role in the response to carbonyl and nitrosative stress. Adv. Microb. Physiol. 58, 1–22 [DOI] [PubMed] [Google Scholar]
- 56. Chen N. H., Couñago R. M., Djoko K. Y., Jennings M. P., Apicella M. A., Kobe B., and McEwan A. G. (2013) A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of Neisseria meningitidis in biofilms. Antioxid. Redox. Signal. 18, 743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanderson K. E. (1976) Genetic relatedness in the family Enterobacteriaceae. Annu. Rev. Microbiol. 30, 327–349 [DOI] [PubMed] [Google Scholar]
- 58. McClelland M., Florea L., Sanderson K., Clifton S. W., Parkhill J., Churcher C., Dougan G., Wilson R. K., and Miller W. (2000) Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 28, 4974–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sharp P. M. (1991) Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J. Mol. Evol. 33, 23–33 [DOI] [PubMed] [Google Scholar]
- 60. Travers A. A., and Buckle M. (eds) (2000) DNA-protein Interactions: A Practical Approach, pp. 141–150, Oxford University Press, NY [Google Scholar]
- 61. Higgins K. A., Chivers P. T., and Maroney M. J. (2012) Role of the N-terminus in determining metal-specific responses in the Escherichia coli Ni- and Co-responsive metalloregulator, RcnR. J. Am. Chem. Soc. 134, 7081–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Foster A. W., Pernil R., Patterson C. J., and Robinson N. J. (2014) Metal specificity of cyanobacterial nickel-responsive repressor InrS: cells maintain zinc and copper below the detection threshold for InrS. Mol. Microbiol. 92, 797–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brotzel F., Chu Y. C., and Mayr H. (2007) Nucleophilicities of primary and secondary amines in water. J. Org. Chem. 72, 3679–3688 [DOI] [PubMed] [Google Scholar]
- 64. Brotzel F., and Mayr H. (2007) Nucleophilicities of amino acids and peptides. Org. Biomol. Chem. 5, 3814–3820 [DOI] [PubMed] [Google Scholar]
- 65. Iwig J. S., and Chivers P. T. (2009) DNA recognition and wrapping by Escherichia coli RcnR. J. Mol. Biol. 393, 514–526 [DOI] [PubMed] [Google Scholar]
- 66. Grossoehme N. E., and Giedroc D. P. (2012) Illuminating allostery in metal sensing transcriptional regulators. Methods Mol. Biol. 875, 165–192 [DOI] [PubMed] [Google Scholar]
- 67. Guerra A. J., and Giedroc D. P. (2012) Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch. Biochem. Biophys. 519, 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ma Z., Jacobsen F. E., and Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu L., Hausladen A., Zeng M., Que L., Heitman J., and Stamler J. S. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 [DOI] [PubMed] [Google Scholar]
- 70. Hibi T., Nii H., Nakatsu T., Kimura A., Kato H., Hiratake J., and Oda J. (2004) Crystal structure of γ-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101, 15052–15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bird A. J., Gordon M., Eide D. J., and Winge D. R. (2006) Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 25, 5726–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., and Eide D. J. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sanghani P. C., Stone C. L., Ray B. D., Pindel E. V., Hurley T. D., and Bosron W. F. (2000) Kinetic mechanism of human glutathione-dependent formaldehyde dehydrogenase. Biochemistry 39, 10720–10729 [DOI] [PubMed] [Google Scholar]
- 74. Eriksson S., Lucchini S., Thompson A., Rhen M., and Hinton J. C. D. (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47, 103–118 [DOI] [PubMed] [Google Scholar]
- 75. Srikumar S., Kröger C., Hébrard M., Colgan A., Owen S. V., Sivasankaran S. K., Cameron A. D. S., Hokamp K., and Hinton J. C. D. (2015) RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 11, e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shea J. E., Hensel M., Gleeson C., and Holden D. W. (1996) Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 93, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 79. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Simossis V. A., and Heringa J. (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 33, W289–W294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 83. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Simons R. W., Houman F., and Kleckner N. (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96 [DOI] [PubMed] [Google Scholar]
- 85. Osman D., Waldron K. J., Denton H., Taylor C. M., Grant A. J., Mastroeni P., Robinson N. J., and Cavet J. S. (2010) Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 285, 25259–25268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu T., Chen X., Ma Z., Shokes J., Hemmingsen L., Scott R. A., and Giedroc D. P. (2008) A Cu(I)-sensing ArsR family metal sensor protein with a relaxed metal selectivity profile. Biochemistry 47, 10564–10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 88. Teng T-Y. (1990) Mounting of crystals for macromolecular crystallography in a free-standing thin film. J. Appl. Cryst. 23, 387–391 [Google Scholar]
- 89. Broennimann C., Eikenberry E. F., Henrich B., Horisberger R., Huelsen G., Pohl E., Schmitt B., Schulze-Briese C., Suzuki M., Tomizaki T., Toyokawa H., and Wagner A. (2006) The PILATUS 1M detector. J. Synchrotron Radiat. 13, 120–130 [DOI] [PubMed] [Google Scholar]
- 90. Powell H. R., Johnson O., and Leslie A. G. W. (2013) Autoindexing diffraction images with iMosflm. Acta Crystallogr. D Biol. Crystallogr. 69, 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Winter G. (2010) xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 [Google Scholar]
- 92. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 93. Bunkóczi G., Echols N., McCoy A. J., Oeffner R. D., Adams P. D., and Read R. J. (2013) Phaser.MRage: automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 69, 2276–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Millán C., Sammito M., and Usón I. (2015) Macromolecular ab initio phasing enforcing secondary and tertiary structure. IUCrJ 2, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pröpper K., Meindl K., Sammito M., Dittrich B., Sheldrick G. M., Pohl E., and Usón I. (2014) Structure solution of DNA-binding proteins and complexes with ARCIMBOLDO libraries. Acta Crystallogr. D. Biol. Crystallogr. 70, 1743–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 97. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Usón I., Pohl E., Schneider T. R., Dauter Z., Schmidt A., Fritz H.-J., and Sheldrick G. M. (1999) 1.7 Å structure of the stabilized REIv mutant T39K: application of local NCS restraints. Acta Crystallogr. D Biol. Crystallogr. 55, 1158–1167 [DOI] [PubMed] [Google Scholar]
- 99. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 101. Henard C. A., Bourret T. J., Song M., and Vázquez-Torres A. (2010) Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J. Biol. Chem. 285, 36785–36793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Song M., Husain M., Jones-Carson J., Liu L., Henard C. A., and Vázquez-Torres A. (2013) Low-molecular-weight thiol-dependent antioxidant and antinitrosative defences in Salmonella pathogenesis. Mol. Microbiol. 87, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.