Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
PTPRD lesions are among the most recurrent alterations in NMZL and appear to be enriched in this lymphoma type across mature B-cell tumors.
NMZL and SMZL genetics overlap with the exceptions of PTPRD lesions, supporting their distinction as independent entities.
Abstract
Nodal marginal zone lymphoma (NMZL) is a rare, indolent B-cell tumor that is distinguished from splenic marginal zone lymphoma (SMZL) by the different pattern of dissemination. NMZL still lacks distinct markers and remains orphan of specific cancer gene lesions. By combining whole-exome sequencing, targeted sequencing of tumor-related genes, whole-transcriptome sequencing, and high-resolution single nucleotide polymorphism array analysis, we aimed at disclosing the pathways that are molecularly deregulated in NMZL and we compare the molecular profile of NMZL with that of SMZL. These analyses identified a distinctive pattern of nonsilent somatic lesions in NMZL. In 35 NMZL patients, 41 genes were found recurrently affected in ≥3 (9%) cases, including highly prevalent molecular lesions of MLL2 (also known as KMT2D; 34%), PTPRD (20%), NOTCH2 (20%), and KLF2 (17%). Mutations of PTPRD, a receptor-type protein tyrosine phosphatase regulating cell growth, were enriched in NMZL across mature B-cell tumors, functionally caused the loss of the phosphatase activity of PTPRD, and were associated with cell-cycle transcriptional program deregulation and increased proliferation index in NMZL. Although NMZL shared with SMZL a common mutation profile, NMZL harbored PTPRD lesions that were otherwise absent in SMZL. Collectively, these findings provide new insights into the genetics of NMZL, identify PTPRD lesions as a novel marker for this lymphoma across mature B-cell tumors, and support the distinction of NMZL as an independent clinicopathologic entity within the current lymphoma classification.
Introduction
The World Health Organization (WHO) Classification of Tumors of Hematopoietic and Lymphoid Tissues recognizes 3 types of marginal zone lymphoma (MZL), namely extranodal MZL (EMZL) of the mucosa associated lymphoid tissue (MALT), splenic MZL (SMZL), and nodal MZL (NMZL).1 Among MZL, only EMZL of the MALT represents a well-defined entity in terms of clinicopathologic presentation, genetics, and treatment. Conversely, the lack of typical markers, with the exception of IRTA1 expression in a fraction of NMZL,2,3 and the absence of clear consensus criteria for diagnosis, make the pathologic classification of NMZL and SMZL difficult, laborious, and not easily reproducible.4
NMZL and SMZL share similar morphology and phenotype and are distinguished solely by the different pattern of dissemination. SMZL involves the spleen without concomitant lymphoadenopathy, whereas NMZL is primarily a nodal B-cell tumor without clinical evidence of extranodal or splenic disease.1,4 In this context, understanding the biological bases of NMZL and SMZL may be helpful to: (1) generate disease-specific markers for differential diagnosis and for their correct identification, and (2) understand whether they represent a continuum of the same disease entity that home in different anatomical sites, or, alternatively, they should be kept as separate disorders.
The genetics of SMZL has been clarified to a certain detail and is characterized by mutations of genes involved in the physiologic differentiation and homeostasis of MZ B cells.5-11 First, the zinc finger transcription factor KLF2 is mutated in 10% to 40% of SMZL, thus representing the most frequently mutated individual gene in this lymphoma. Second, the NOTCH pathway is affected in ∼30% to 40% of SMZL, including mutations of NOTCH2, the master regulator of MZ B-cell differentiation.5-10 Finally, the noncanonical NF-κB pathway is mutated in ∼25% of cases.11
Conversely, the current understanding of the biology of NMZL is elusive and, among B-cell tumors, this is one of the few disease entities still remaining orphan of specific genetic lesions. By combining whole-exome sequencing (WES), targeted sequencing of tumor-related genes, high-resolution SNP arrays, and RNA sequencing (RNA-seq), here we show that: (1) PTPRD lesions are among the most recurrent alterations in NMZL, where they appear to be enriched compared with other mature B-cell tumors; and (2) the genetics of NMZL and SMZL overlap, with the exception of PTPRD mutations, supporting their distinction as independent clinicopathologic entities.
Material and methods
Samples
The study included 35 NMZL (18 discovery, 17 screening cases; supplemental Table 1, available on the Blood Web site) diagnosed according to the World Health Organization classification,1 and confirmed by: (1) pathologic revision of lymph node histology; and (2) lack of clinical and imaging evidence of MALT or splenic disease either at diagnosis or during follow-up by medical and radiologic charts revision. Consistent with NMZL, cases: (1) lacked CD5, CD10, and cyclin D1 expression, 7q deletion, t(11;14), t(14;18), t(11;18), and t(1;14) translocations; (2) recurrently harbored +3 (17% of cases), +12 (14% of cases), and +18 (11% of cases); and (3) preferentially used the IGHV4-34 immunoglobulin heavy variable gene (17% of cases). In all cases, large cells were <20%, without clustering in distinct aggregates. A third independent cohort of NMZL (n = 8) provided with the same inclusion criteria as the discovery and screening cases was investigated to validate PTPRD mutations recurrence. For comparative purposes, 39 SMZL cases (see supplemental Appendix) and 3 putative SMZL cell lines (VL51, SSK-41, and KARPAS-1718) were also analyzed. Among primary NMZL and SMZL, tumor DNA and RNA have been derived from fresh or frozen lymph node (NMZL) or spleen (SMZL) sections obtained at diagnosis and containing a tumor representation >70%, as documented by immunohistochemistry and confirmed by single nucleotide polymorphism (SNP) array genomic profiling of the immunoglobulin gene loci. Matched normal DNA was obtained from saliva or peripheral blood granulocytes in 27 NMZL (18 discovery, 8 screening, and 1 validation cases) and 14 SMZL patients, and confirmed to be tumor-free by polymerase chain reaction (PCR) of tumor-specific IGHV-D-J rearrangements. Patients provided informed consent in accordance with local institutional review board requirements and the Declaration of Helsinki. The study was approved by the Ethical Committee of the Ospedale Maggiore della Carità di Novara affiliated with the Amedeo Avogadro University of Eastern Piedmont (Protocol Code CE 116/12).
Whole-exome sequencing
Tumor and germline genomic DNA from 18 discovery NMZL cases was enriched in protein coding sequences using SureSelectXT HumanAllExon V5+UTRs (Agilent Technologies). The captured targets were sequenced using the HiSequation 2500 analyzer (Illumina) with the paired-end 2 × 100 bp read option. Exome capture and sequencing were performed at the HiSeq Service of Fasteris SA (Plan-les-Ouates, Switzerland). Details are in the supplemental Appendix.
Sequence mapping and identification of tumor-specific variants
Paired-end reads (∼109 million per case) obtained by high-throughput sequencing were aligned to the human genome reference hg19/NCBI GRCh37 using the BWA alignment tool version v.0.6.2, and provided a mean depth of ∼60× with at least 70% of the target exome covered at 30× (supplemental Table 2). The SAVI (Statistical Algorithm for Variant Identification) algorithm was used for variant calling.12-14 Details are in the supplemental Appendix.
Targeted sequencing
Genes discovered by WES in the discovery panel were investigated by targeted next-generation sequencing (NGS) in the screening panel of 17 NMZL (paired tumor and normal DNA in 8 cases and solely tumor DNA in 9 cases). Genes discovered by WES in NMZL were also investigated in SMZL (32 primary cases and 3 cell lines; paired tumor and normal DNA in 6 cases and solely tumor DNA in 26 cases), whereas genes previously discovered by WES in SMZL5 were studied in NMZL. Candidate gene-coding exons and splice sites were enriched through a custom-designed SeqCap EZ Choice Libraries (NimbleGen) system. The obtained libraries were sequenced using the MiSeq analyzer (Illumina) with the paired-end 2 × 250 bp read option. The mean depth of coverage of the targeted resequencing was 369×. Details of the bioinformatics pipeline for variant calling are in the supplemental Appendix.
Sanger sequencing
Mutation analysis of cytokine signaling genes that did not emerge from WES in NMZL, but are known to be mutated in lymphomas, namely PTPN1, PTPN2, SOCS1, STAT3, STAT6, and JAK1,15-20 as well as mutation analysis of PTPRD in EMZL and in the validation cohort of NMZL, were performed by Sanger sequencing. Details are in the supplemental Appendix.
High-density SNP array analysis
Genome-wide DNA profiles of the 35 NMZL cases (paired-tumor and normal DNA in 23 cases and solely tumor DNA in 12 cases) from the discovery and screening panels were obtained by CytoScan HD Array (Affymetrix, Santa Clara, CA). The identification of regions of abnormal copy number was performed by 2 approaches: (1) Chromosome Analysis Suite (ChAS v.2.0.1.2, Affymetrix) software, genome build hg19; and (2) circular binary segmentation as detailed in previously published papers,21,22 and adapted to the CytoScan HD array. MCRs of aberration were identified as previously described.22 Details are in the supplemental Appendix.
RNA-seq, gene fusion, and gene-expression analyses
Whole-transcriptome sequencing of 11 NMZL discovery cases provided with high-quality tumor RNA was performed at the HiSeq Service of Fasteris SA (Plan-les-Ouates, Switzerland) using paired-end 2 × 100 bp read option (average 73M reads per case). Gene fusions were predicted using the Pegasus pipeline.23 RNA sequencing was also used to derive the transcriptome profile of NMZL.24-29 Details are in the supplemental Appendix.
IGHV-IGHD-IGHJ rearrangement analysis
IGHV-IGHD-IGHJ rearrangement analysis was performed as previously described.30 Details are in the supplemental Appendix.
Fluorescence in situ hybridization
Translocations of candidate genes were assessed by fluorescence in situ hybridization (FISH), using the following probes: (1) LSI IGH break apart, LSI BIRC3-MALT1 translocation dual fusion, LSI IGH-MALT1 dual fusion; (2) XL t(11;14) dual-fusion probe, XL t(14;18) dual-fusion probe (Metasystem); and (3) BCL10 FISH DNA probe break apart (BCL10 translocation) (Dako). Details are in the supplemental Appendix.
Clonal composition and mutational signature analysis
Tumor purity was estimated by using the Absolute algorithm.31 PyClone algorithm,32 with default parameters was used to identify clonal population structures. Wellcome Trust Sanger Institute’s framework was used for deciphering mutational processes.33 Both synonymous and nonsynonymous variants were used to identify mutational signatures. We assessed signature stability and computed average Frobenius reconstruction error for k = 1 to k = 10 number of signatures. Insignificant reduction in reconstruction error after k = 1, as well as stable reproducibility only at k = 1, indicated only a single recognized signature by the algorithm.
Cell sorting
Normal spleen samples were obtained through the Servizio di Immunologia dei Trapianti, Città della Salute e della Scienza Hospital (Turin, Italy). Cells were isolated by Ficoll density gradient centrifugation (GE Healthcare) and stained using anti-CD19, -CD21, -CD27, -CD38, -IgM, and -IgD. All antibodies were purchased from Miltenyi Biotec, except from CD21 (e-Bioscience). B cells were identified as CD19+/CD21+ and sorted on a FACS ARIA III (BD Biosciences) as follows: CD38+/IgDlow (“germinal center, GC”), IgMhigh/IgDlow (“marginal zone, MZ”), IgDhigh/CD27– (“naïve”).34
RNA and protein expression
Expression of PTPRD mRNA was monitored by quantitative reverse-transcription PCR. Standard immunoblotting protocols8 were used to assess the expression of PTPRD (C-18; #sc-10867; 1/600; Santa Cruz Biotechnology), STAT3 (124H6; #9139S; 1/1000; Cell Signaling), Y705-phospho STAT3 (Y705; #9145S; 1/1000; Cell Signaling), α-tubulin (#T6074; 1/5000; Sigma Aldrich), and α-actin (#A2066; 1/5000; Sigma Aldrich). Formalin-fixed tumor sections were processed for immunohistochemical analyses on a semiautomated stainer.35 The following antibodies were used: anti-Ki-67 (MB-1, Dako, dilution 1:50), anti-phospho-STAT3 (Y705) (Rabbit mAb#9145, Cell Signaling Technology; dilution 1:100). Details are in the supplemental Appendix.
Plasmid constructs and mutagenesis
PTPRD (NM_002839) Human ORF clone tagged at the C-terminal with green fluorescent protein (GFP) (RG 214115, Origene Technologies) was subjected to mutagenesis using QuickChange II Site-Directed Mutagenesis kit (Stratagene). Empty vector was generated by SgfI, MluI, and EcoRV restriction enzyme digestion of the PTPRD (NM_002839) human cDNA ORF clone followed by ligation. Each plasmid was transfected into the HEK 293 Blue IL-6 cells (Invivogen) using Lipofectamine 2000 (Invitrogen). Transfected cells were cultured for 24 hours. GFP expression by flow cytometry (FACSCalibur, CellQuest software; BD) was used to determine transfection efficiency (>90%). After 24 hours from transfection, the HEK 293 Blue IL-6 cells, both nontransfected, transfected with the empty, wild-type, and mutant PTPRD constructs were treated with or without 5 or 10 ng/mL of IL-6, or IL-4 as control (Invivogen), for 14 hours and then total cell lysates were prepared.
PTPRD methylation analysis
Bisulfite modification of genomic DNA was carried out using the MethylEdge Bisulfite Convertion System Kit (Promega). Primer sequences for the methylation-specific PCR (MSP) were as follows: PTPRD_MSP3_UM_forward 5′-TGGTGGGGTTTGTTTAGGTTGTG-3′, PTPRD_MSP3_UM_reverse 5′-ATACTCCAAACACCCACTAAAAAAAAAAACAACA-3′, PTPRD_MSP3_M_forward 5′-GGGGTTCGTTTAGGTCGC-3′, PTPRD_MSP3_M_reverse 5′-CGCCCGCTAAAAAAAAAAACGACG-3′.
Estimation of PTPRD mutation and deletion prevalence in mature B-cell tumors
Mutation of PTPRD in mature B-cell tumors was experimentally assessed by Sanger sequencing in EMZL or aggregated from our own or published reports.5-7,9,15,22,36-49 Copy number (CNA) alteration frequency of PTPRD in SMZL, EMZL, diffuse large B-cell lymphoma (DLBCL), chronic lymphocytic leukemia, and mantle cell leukemia was derived from the following datasets: GSE50252, GSE24881, GSE15127, GSE8123, and GSE4176.
Statistical analysis
Categorical variables were compared by χ2 test or Fisher exact test. Continuous variables were compared by Student t test or Mann-Whitney U test. All statistical tests were 2-sided. Statistical significance was defined as P < .05. The analysis was performed with SPSS v.21.0
Accession codes
WES data are deposited in SRA (http://www.ncbi.nlm.nih.gov/sra; accession number SRP059441). Copy number data are deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE68078).
Results
The coding genome of NMZL
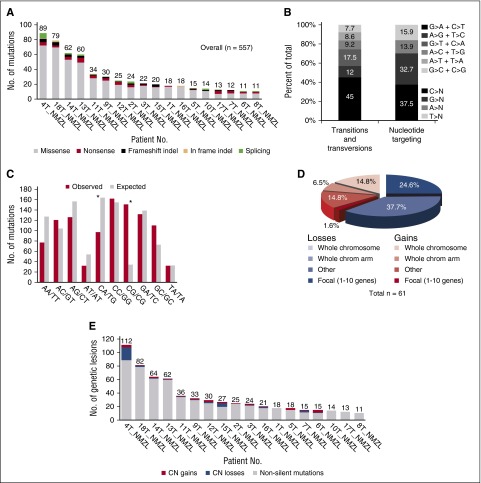
To discover somatic, nonsilent mutations and CNAs that are clonally represented in the NMZL coding genome, we performed WES and high-density SNP array analysis of paired tumor and normal DNA from 18 untreated NMZL patients. We identified a total of 557 nonsilent somatic mutations involving 504 genes, including 453 missense, 48 nonsense, 24 splice-site, and 32 indel mutations (Figure 1A; supplemental Table 3), and a total of 61 CNAs, including 23 gains (1 focal) and 38 losses (15 focal) (Figure 1D; supplemental Table 4). As in other B-cell lymphoproliferative disorders,5,48,49 NMZL was characterized by a predominance of the age-related mutational signature involving C>T substitutions at NpCpG trinucleotides (Figure 1B-C; supplemental Figure 1).50 When combining point mutations and CNAs, the overall load of tumor-acquired lesions was heterogeneous across the 18 NMZL cases investigated, ranging from 11 to 112 lesions per case (mean load, 34.4 lesions/case) (Figure 1E), which is consistent with the load of somatic lesions observed also in SMZL.5-9 The clonal composition of NMZL was assessed by integrating CNAs and somatic mutations to estimate average ploidy of cancer cells and the mutant cancer cell fraction. By this approach, most NMZL (83%, 15/18) showed a simple clonal architecture composed by only one dominant clone (supplemental Figure 2). Transcriptome analysis, available for 11 NMZL cases, showed that coding variants affecting recurrently mutated genes were expressed at the RNA level (supplemental Table 5). At variance with EMZL, but similar to SMZL,5,51 NMZL did not harbor any highly recurrent gene fusions, as documented by transcriptome sequencing (data not shown).
Figure 1.
NMZL coding genome complexity. (A) Number and type of nonsilent somatic mutations identified in the 18 discovery genomes. (B) The pattern of nucleotide substitutions in the discovery genomes revealed a predominance of transitions over transversions (296:224, ratio of 1.3) and a preferential targeting of G and C nucleotides (70.2% affecting G/C compared with 29.8% affecting A/T nucleotides). (C) Mutation frequency at specific dinucleotides (red bars). The expected frequencies (gray bars) correspond to the dinucleotide sequence composition of the Consensus CDS. Asterisks denote statistically significant differences in overrepresented changes. (D) Frequency and type of somatically acquired copy number abnormalities (CNAs). (E) Combined load of somatically acquired genetic lesions in the discovery genomes, including nonsilent mutations and CNAs.
Recurrent targets of genetic alterations in NMZL
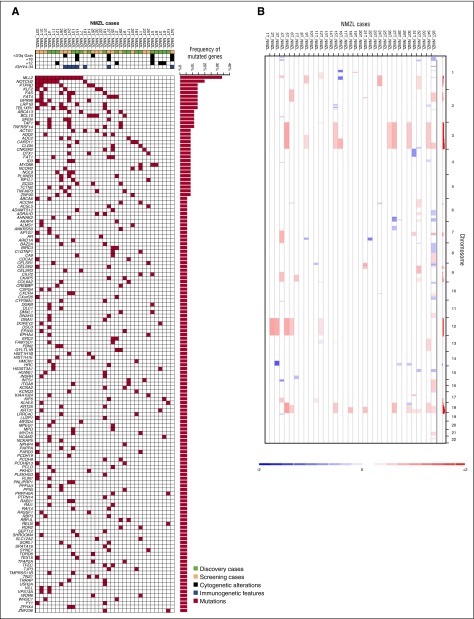
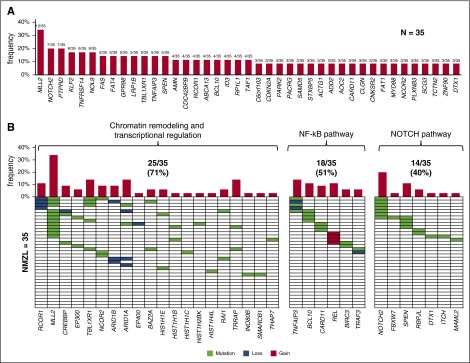
Genes (n = 504) discovered through exome sequencing in the 18 discovery NMZL were further assessed for their mutation recurrence by targeted NGS in an independent screening panel of 17 NMZL cases meeting the same inclusion criteria as the discovery panel (supplemental Table 3). The 17 screening NMZL were also characterized for CNAs by high-density SNP array analysis (supplemental Table 4). By compiling the results of WES, targeted resequencing (Figure 2A), and high-resolution SNP array analysis (Figure 2B), 41 genes were recurrently affected in ≥3 of 35 (9%) of NMZL by mutations (n = 32 genes) or focal CNA (n = 9 genes), including MLL2 (also known as KMT2D, 34%), PTPRD (20%), NOTCH2 (20%), and KLF2 (17%) (Figure 3A; supplemental Tables 3 and 4). Affected genes pointed to the molecular deregulation of specific programs in NMZL, namely chromatin remodeling/transcriptional regulation (71% of cases), NOTCH (40% of cases), and NF-κB (51% of cases) (Figure 3B).
Figure 2.
Mutated genes and copy number abnormalities in NMZL. (A) Heatmap showing the distribution of mutations in the 142 recurrently mutated genes (>5% of cases) among NMZL of the discovery plus screening cohorts (n = 35). Each row represents a gene and each column represents a primary tumor. Mutations are color coded in red. The horizontal bar graph shows the gene mutation frequency. The plot below the case label indicates sample characteristics (green, discovery cases; yellow, screening cases; blue, immunogenetic features; black, cytogenetic aberrations). (B) Copy number analysis of the NMZL cases. Curated segmentation data for the 35 NMZL samples. In the red-blue scale, white corresponds to a normal (diploid) copy number log ratio, blue is a deletion, and red is a gain.
Figure 3.
Genes and pathways that are recurrently affected by genetic lesions in NMZL. (A) Genes (n = 41) that were recurrently affected by mutations and/or focal copy number aberrations in ≥3 of 35 of NMZL. The bar graph represents the frequency of molecular lesions in each gene. (B) Molecularly deregulated pathways in NMZL. In the heatmap, rows correspond to genes and columns represent individual patients. Color coding is based on gene alteration status (white, wild-type; green, mutated; blue, loss; red, gain). The bar graph represents the frequency of molecular lesions in each gene. The overall frequency of genetic lesions in each pathway is indicated.
The mutational signature of NMZL largely overlaps with that of SMZL with the exception of MLL2 and PTPRD mutations
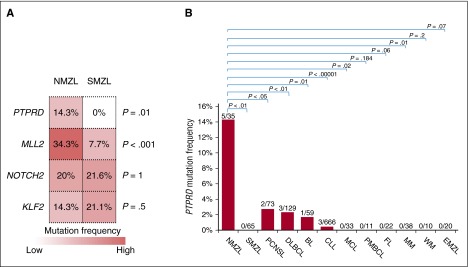
To assess whether the genetic signatures of SMZL and NMZL overlap, we analyzed a series of SMZL (n = 20) by targeted NGS of the 504 genes discovered by WES in NMZL, and our cohort of NMZL (n = 35; discovery and screening panels) by targeted NGS of the 191 genes we previously identified by WES in SMZL.5 MLL2, PTPRD, NOTCH2, and KLF2 mutation analysis was also extended to additional SMZL cases to further validate their recurrence in this lymphoma (MLL2, NOTCH2, and KLF2 mutations were previously reported5,8) (supplemental Table 3). This approach revealed that, though NMZL and SMZL largely shared a common mutation signature characterized by variants of NOTCH2 and KLF2, NMZL was significantly enriched with MLL2 mutations (NMZL 34.3% vs SMZL 7.7%, P < .001) and harbored mutations of the PTPRD gene that were otherwise absent in SMZL (Figure 4A).52-55
Figure 4.
PTPRD mutations are enriched in NMZL among mature B-cell tumors. (A) The heatmap represents the frequency of mutations in the top genes that are affected in at least 15% cases of NMZL and/or SMZL. P values from the comparison of the mutation frequency in NMZL vs SMZL by Bonferroni corrected Fisher exact test. (B). Prevalence of nonsynonymous PTPRD mutations among mature B-cell tumors (NMZL, nodal marginal zone lymphoma, data from this study; SMZL, splenic marginal zone lymphoma, data from this study and others5,6,7,9; DLBCL, diffuse large B-cell lymphoma, data from various studies22,37,45,47; BL, Burkitt lymphoma, data from various studies40,41; CLL, chronic lymphocytic leukemia, data from various studies38,39,44,49; MCL, mantle cell lymphoma, data from Beà et al43; PMBCL, primary mediastinal large B-cell lymphoma, data from Gunawardana et al15; FL, follicular lymphoma, data from various studies37,48; MM, multiple myeloma, data from Chapman et al36; WM, Waldenström macroglobulinemia, data from various studies42,46; EMZL, extranodal marginal zone lymphoma, data from this study; PCNSL primary central nervous system lymphoma, data from various studies52-55).
PTPRD alterations in NMZL
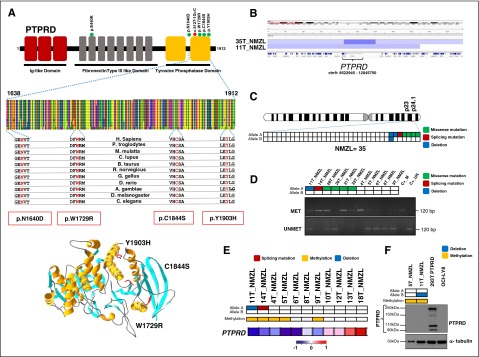
PTPRD encodes the receptor-type-protein-tyrosine-phosphatase-δ, a tumor suppressor gene involved in cell growth regulation and affected by somatic mutations in solid cancer.56-59 Among NMZL, PTPRD mutations occurred in 14.3% (5/35) of the compiled discovery and screening cases, including (Figure 5A): (1) one splice-site variant that, by cDNA analysis, caused aberrant transcripts, leading to the retention of intronic sequences, loss of its coding potential, and truncation of the tyrosine phosphatase domain (supplemental Figure 3); (2) three missense substitutions that clustered within the tyrosine phosphatase domain, affected conserved amino acids, and were predicted to be deleterious according to PolyPhen-2, which is suggestive of a potential mutational hotspot within this region; and (3) one missense mutation affecting the second fibronectine-type domain predicted to be deleterious according to PolyPhen-2 (supplemental Table 3). PTPRD mutations were monoallelic and clonally represented in the tumor (supplemental Table 3). Whenever possible, cDNA sequencing confirmed that PTPRD mutations (not shown) were expressed at the transcript level. The somatic origin of missense mutations was confirmed in all tested cases by analysis of paired normal DNA, which was available for 3 variants (supplemental Figure 3). Analysis of a third independent cohort of NMZL (n = 8) further validated the occurrence of PTPRD mutations in NMZL by disclosing a missense substitution affecting the PTPRD tyrosine phosphatase domains in 1 of 8 (12.5%) cases (Figure 5A).60
Figure 5.
PTPRD mutations in NMZL. (A) Schematic diagram of the PTPRD protein with its key functional domains (top). Color-coded symbols indicate the type and position of the mutations on the PTPRD protein (green, missense mutations; red, splicing site mutation). Multiple alignment of the PTPRD phosphatase domain amino acid sequences with the 11 orthologous PTPRD proteins. Conserved amino acids affected by mutations are color coded in red. Representation of the 3D structure of the PTPRD phosphatase domain mutations (bottom). The structural view of the PTPRD phosphatase domain was generated in DeepView-Swiss-PdbViewer (http://spdbv.vital-it.ch/) using the coordinates of the crystal structure of the PTPRS phosphatase domain (99% identity with PTPRD) (PDB 2fh7). Residues targeted by somatic mutations in NMZL are highlighted. (B) Graphic representation of segmentation data from 2 NMZL cases harboring PTPRD copy number losses visualized with Integrative Genomics Viewer (IGV) software. Each track represents one sample, where blue indicates region of a copy number loss. Individual genes in the region are aligned in the bottom panel. (C) Allelic (A or B) distribution of PTPRD genetic lesions in individual NMZL cases (green, missense mutation; red, splicing mutation; blue, deletion). (D) Methylation-specific PCR documenting aberrant methylation of the PTPRD-promoter CpG sites in NMZL cases (MET, methylated sample; UNMET, unmethylated sample; C+_M, positive control of methylated reaction; C+_UN, positive control of unmethylated reaction; bp, base pair). In the heatmap, rows correspond to the allelic status of PTPRD gene in NMZL (white, wild-type; green, mutated; blue, deleted). (E) Transcriptome analysis of PTPRD expression in primary NMZL. In the heatmaps, the first 2 rows correspond to the allelic status of the PTPRD gene in NMZL (white, wild-type; green, mutated; blue, deleted), the third row shows the methylation status of PTPRD (white, unmethylated; yellow, methylated), the last row shows the expression of PTPRD in NMZL (red, high expression; blue, low expression). (F) Western blot analysis of PTPRD protein expression in primary NMZL samples. The HEK 293T cells tranfected with a vector containing the wild-type PTPRD tagged with GFP at the C-terminal were used as positive control. The OCI-LY8 cells harboring a biallelic deletion of PTPRD were used as negative control. In the positive control, western blot analysis shows the GFP-tagged full-length PTPRD protein (250 kDa), as well as 2 N-terminal PTPRD protein products (150 kDa and 90k Da), and a GFP-tagged C-terminal PTPRD product (110 kDa) after cleavage process by a physiologic posttranslational modification called ectodomain shedding60 α-tubulin was used as a loading control. In the heatmaps, the first 2 rows correspond to the allelic status of PTPRD gene in NMZL (white, wild-type; blue, deleted), and the third row shows the methylation status of PTPRD (white, unmethylated; yellow, methylated).
PTPRD mutations appeared to be enriched in NMZL, being absent in other MZL entities, and rare in other mature B-cell tumors (Figure 4B). The PTPRD locus was also affected by deletions in 5.7% (2/35) NMZL, including one biallelic loss. The focal PTPRD loss identified in one case (case 35T) defined a minimal common region of deletion and supported PTPRD as the specific target of the deletions (Figure 5B). By combining mutations and deletions, the PTPRD gene was affected in 20% (7/35) NMZL (Figure 5C).
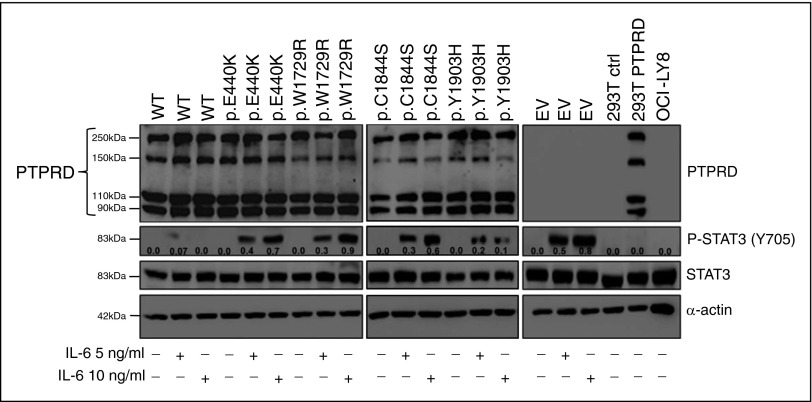
Upon transient transfection of HEK 293 cells, mutant PTPRD (n = 4; including the p.C1844S, p.Y1903H, p.E440K, and p.W1729R substitutions) were expressed at similar levels compared with the wild-type PTPRD, as assessed by western blot analysis with anti-PTPRD antibody, suggesting that mutations do not have major effects on destabilizing the PTPRD protein (Figure 6). Phospho-STAT3 (Y705) is an experimentally validated substrate of PTPRD used as functional readout of its phosphatase activity.56,61,62 Therefore, to evaluate how phosphatase activity was affected by the missense PTPRD mutations identified in NMZL, we expressed wild-type PTPRD or mutant PTPRD with p.C1844S, p.Y1903H, p.E440K, and p.W1729R substitutions in HEK 293 cells engineered to express the human interleukin 6 (IL-6) receptor (HEK 293-IL6) (Figure 6). In this system, stimulation with increasing doses of IL-6 led to dose-dependent phosphorylation of STAT6 Y705, which was markedly reduced in the presence of the exogenous wild-type PTPRD phosphatase (Figure 6). In contrast, we observed sustained Y705 phosphorylation of STAT3 upon expression of all PTPRD mutants (Figure 6), confirming these alterations as deleterious, and indicating that, as previously reported in other cancers, mutations of both the tyrosine phosphatase and the extracellular domains affect the catalytic activity of PTPRD.56,62
Figure 6.
Mutations impair the tyrosine phosphatase activity of PTPRD. Expression of the STAT3 protein and of the phosphorylated STAT3 protein (pSTAT3, Y705) in HEK 293 cells transfected with constructs expressing wild-type PTPRD-GFP (WT), mutant PTPRD-GFP, or empty vector (EV). Nontransfected HEK 293T cells (293T ctrl) and OCI-LY8 were used as negative control for PTPRD expression. The HEK 293T cells transfected with a vector containing the wild-type PTPRD tagged with GFP at the C-terminal were used as positive control for PTPRD expression (western blot shows the GFP-tagged full-length PTPRD protein [250 kDa], as well as 2 N-terminal PTPRD protein products [150 kDa and 90 kDa], and a GFP-tagged C-terminal PTPRD product [110 kDa] after cleavage process by a physiologic posttranslational modification called ectodomain shedding60). Protein lysates were prepared from cell cultures treated with or without 5 or 10 ng/mL human interleukin-6 (IL-6). Relative densitometric values of the phosphorylated STAT3 protein (pSTAT3, Y705) (shown for each band) were normalized against the levels of the internal loading control (α-actin).
We assessed the expression of PTPRD in normal mature B-cell subpopulations (ie, naïve, germinal center, and marginal zone B-cells) isolated from normal human spleen by cell sorting. PTPRD mRNA was expressed in normal germinal center B cells and, though at lower levels, also in naïve and marginal zone B cells (supplemental Figure 4). Beside molecular lesions, the PTPRD gene may be epigenetically affected in tumors by aberrant methylation.56 In NMZL, cases harboring monoallelic PTPRD mutation or deletion (n = 6) were also characterized by aberrant methylation of PTPRD-promoter CpG sites that have been previously shown to associate with reduced PTPRD expression,56 suggesting epigenetic downregulation of the retained wild-type allele (Figure 5D). Transcriptome analysis revealed low PTPRD expression in primary NMZL cases harboring a monoallelic deletion or a disrupting mutation coupled with promoter methylation, as well as in cases harboring promoter methylation in the absence of structural abnormalities (Figure 5E). Nonetheless, additional mechanisms appeared to contribute to PTPRD inactivation in NMZL, because a fraction of primary NMZL showed low levels of PTPRD transcript even in the absence of PTPRD structural alterations or promoter methylation (Figure 5D). Western blot analysis of primary NMZL samples confirmed the lack of PTPRD protein expression in one case harboring monoallelic deletion coupled with promoter methylation and in one case lacking genetic/epigenetic lesions (Figure 5F).
Though PTPRD and CDKN2A genetic lesions are known to co-occur in solid cancers,56 in NMZL, only 1 of 35 cases harbored concurrent deletions of PTPRD and CDKN2A, whereas the remaining 6 NMZL harboring PTPRD lesions showed neither CDKN2A loss nor CDKN2A mutations (supplemental Table 6). In glioblastoma and head and neck squamous cell carcinoma, PTPRD mutations result in phospho-STAT3 (Y705) expression.56-59,61,62 Conversely, primary NMZL samples showed no or low levels of phospho-STAT3 (Y705), independent of PTPRD genetic status (supplemental Figure 5A-B). Also, although performed on a limited number of cases, gene set enrichment analysis of transcripts that were over- and underexpressed in PTPRD-disrupted NMZL did not show any clue of deregulated STAT3-mediated programs (supplemental Figure 5C). These observations suggest that, at variance with solid cancer, PTPRD does not cooperate with CDKN2A in promoting lymphomagenesis, and has functional consequences that do not involve STAT3 in NMZL. Consistent with the lack of cytokine signaling deregulation in NMZL, mutations in lymphoma genes associated with this pathway15-20 were rare or absent in NMZL.
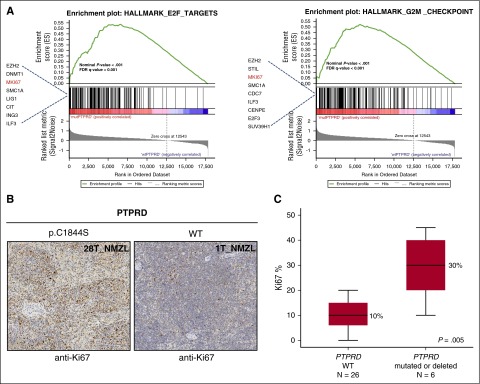
To gain insights into the molecular pathways associated with PTPRD lesions, we compared the transcriptome profile of PTPRD mutated or deleted NMZL vs PTPRD wild-type cases. Though underpowered to appreciate the full gene-expression program associated with PTPRD lesions, transcriptome analysis showed a significant enrichment of cell-cycle genes in NMZL harboring PTPRD lesions compared with PTPRD wild-type cases (Figure 7A), including higher expression of the MKI67 gene that encodes for the Ki-67 proliferation marker (supplemental Figure 6). To validate this observation, we assessed Ki-67 expression by immunohistochemical analysis of the lymph node biopsy from NMZL cases of the study cohort (n = 32; Figure 7B). Consistent with the results of transcriptome analysis, PTPRD mutated or deleted cases showed a larger fraction of Ki-67+ cells compared with PTPRD wild-type cases (Figure 7C).
Figure 7.
Primary NMZL cases harboring PTPRD lesions show a cell-cycle signature and an increased proliferation index. (A) GSEA plot illustrating the enrichment and upregulation of different biologically relevant gene sets (E2F pathway and G2M cell-cycle checkpoints) in PTPRD mutated/deleted vs PTPRD wild-type. Genes with an enrichment score <0.1 are shown. GSEA, Gene Set Enrichment Analysis; FDR, false discovery rate. (B) Ki-67 expression by immunohistochemical analysis of the lymph node biopsy from one exemplificative PTPRD-mutated NMZL case and one exemplificative PTPRD wild-type case (original magnification ×10). (C) Comparison of the Ki-67 proliferation index assessed by immunohistochemistry between PTPRD-mutated/deleted cases vs PTPRD wild-type NMZL (median, black line inside the bar; quartiles, margin of the box; range, whiskers). P value by Mann-Whitney U test.
Discussion
The main results of this study have both pathogenetic and clinical implications for NMZL. We identified somatic coding-sequence mutations and deletions of the receptor-type tyrosine phosphatase gene PTPRD as a molecular feature of NMZL among indolent B-cell tumors. PTPRD encodes a receptor-type protein tyrosine phosphatase that has not been previously described as recurrently mutated in indolent and aggressive lymphomas, with the exception of a few cases of primary central nervous system DLBCL.52 In NMZL, PTPRD lesions either remove the entire PTPRD gene or damage the tyrosine phosphatase function of the protein, suggesting that they have been selected to interfere with PTPRD regulation of cellular mechanisms that depend upon tyrosine phosphorylation. PTPRD is a tumor-suppressor gene disrupted in many solid tumors by mutations or deletions.56,61,62 Y705-phospho STAT3 is one of the few validated substrates of PTPRD,56,61,62 and, consistently, PTPRD disruption activate Y705-phospho STAT3 in solid cancers to promote tumorigenesis.61 However, NMZL, including PTPRD-affected cases, carry neither Y705-phospho STAT3 nor any other phenotypic or genetic clue of cytokine-signaling deregulation, thus indicating that PTPRD genetic lesions do not act through STAT3 constitutive activation in NMZL. The observation that PTPRD missense substitutions interfere with Y705-phospho STAT3 dephosphorylation by PTPRD in a biochemical assay confirms the deleterious effect of these variants and supports the notion that they are not passenger events. However, given the lack of detectable Y705-phospho STAT3 in primary NMZL cases under steady-state conditions, further investigations will be necessary to elucidate the mechanism by which PTPRD mutations promote NMZL pathogenesis.
Consistent with a tumor-suppressor function of PTPRD, its knockout promotes tumorigenesis in mice, including lymphoma development.61,63 At variance with solid cancers arising in ptprd knockout mice, lymphomas arising in these animal models do not carry any clue of Y705-phospho STAT3 activation,63 which is consistent with our observation in human NMZL patients and indicates that other yet unknown PTPRD substrates and oncogenic signaling are affected by its deletion or mutations in lymphomas. The observation that PTPRD lesions associate with proliferation in NMZL prompts investigations aimed at characterizing the biochemical mechanisms linking PTPRD mutations to the cell-cycle program in this lymphoma and its clinical implications. Indeed, though NMZL is generally characterized by an indolent course, its prognosis is heterogeneous and considered less favorable than that of SMZL and EMZL.4
The notion that NMZL share with SMZL mutations of NOTCH2 and KLF2, which are physiologically involved in the commitment of mature B cells to the marginal zone (MZ),64,65 points to MZ differentiation as one of the major programs deregulated in MZL arising outside MALT. Although the genetic signatures of NMZL and SMZL largely overlap, the association of PTPRD mutations with NMZL, alongside the lack of 7q deletion in this lymphoma,66 support the distinction of NMZL and SMZL as different entities rather than different clinical presentations of the same disease, and is consistent with their separation in the WHO Classification of hematopoietic tumors.1
The absence of a clear consensus for diagnosis makes the classification of NMZL difficult, laborious, and not easily reproducible, often resulting in a diagnosis of exclusion.67-69 From a diagnostic standpoint, PTPRD lesions are enriched in NMZL among lookalike mature B-cell tumors, and thus may represent a genetic biomarker that, though not highly sensitive, is provided with a positive predictive value for NMZL specification. The increasing identification of tumor-associated genetic lesions among indolent B-cell lymphoproliferative disorders otherwise lacking a specific immunophenotype (ie, NMZL in this study and lymphoplasmacytic lymphoma42) prompts diagnostic accuracy studies aimed at verifying whether molecular profiling may assist the differentiation of pathologically challenging cases.
Acknowledgments
This study was supported by Fondazione Cariplo grant 2012-0689; Special Program Molecular Clinical Oncology 5 × 1000 grant 10007; My First AIRC grant 13470, and iCARE 17860, Associazione Italiana per la Ricerca sul Cancro Foundation Milan, Italy; Progetto Ricerca Finalizzata grants RF-2010-2307262 and RF-2011-02349712, Ministero della Salute, Rome, Italy; Futuro in Ricerca 2012 grant RBFR12D1CB, Ministero dell'Istruzione, dell'Università e della Ricerca, Rome, Italy; Ateneo-San Paolo Program, Torino, Italy; National Cancer Institute, National Institutes of Health grant 1 U54-CA193313-01; and grant KFS-3746-08-2015, Swiss Cancer League, Bern, Switzerland.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.R. and G.G. designed the study, interpreted data, and wrote the manuscript; V.S. contributed to data interpretation and manuscript preparation; V.S., M.M., S.M., A.B., F.D., M.C., and S.C. performed molecular studies; V.S. and E.S. performed functional experiments; H.K., L.C., A.B.H., and S.Z. analyzed exome sequencing and RNA-seq data; L.A., M.L., F.T., R.M., S.D., M. Ponzoni, A.R., and E.T. provided study material and contributed to manuscript revision; and M. Paulli, B.F., L.P., G.I., F.B., R.F., and R.R. contributed to data interpretation and manuscript revision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.K. is Center for Systems and Computational Biology, Rutgers Cancer Institute, Rutgers University, New Brunswick, NJ.
Correspondence: Davide Rossi, Division of Hematology, Department of Translational Medicine, Amedeo Avogadro University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy; e-mail: rossidav@med.unipmn.it; and Gianluca Gaidano, Division of Hematology, Department of Translational Medicine, Amedeo Avogadro University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy; e-mail: gianluca.gaidano@med.uniupo.it.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. Lyon, France: IARC; 2008. World Health Organization Classification of Tumours, Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 2.Falini B, Agostinelli C, Bigerna B, et al. IRTA1 is selectively expressed in nodal and extranodal marginal zone lymphomas. Histopathology. 2012;61(5):930–941. doi: 10.1111/j.1365-2559.2012.04289.x. [DOI] [PubMed] [Google Scholar]
- 3.Falini B, Tiacci E, Pucciarini A, et al. Expression of the IRTA1 receptor identifies intraepithelial and subepithelial marginal zone B cells of the mucosa-associated lymphoid tissue (MALT). Blood. 2003;102(10):3684–3692. doi: 10.1182/blood-2003-03-0750. [DOI] [PubMed] [Google Scholar]
- 4.Thieblemont C, Davi F, Noguera ME, Brière J. Non-MALT marginal zone lymphoma. Curr Opin Hematol. 2011;18(4):273–279. doi: 10.1097/MOH.0b013e3283477815. [DOI] [PubMed] [Google Scholar]
- 5.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiel MJ, Velusamy T, Betz BL, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209(9):1553–1565. doi: 10.1084/jem.20120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez N, Almaraz C, Vaqué JP, et al. Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation. Leukemia. 2014;28(6):1334–1340. doi: 10.1038/leu.2013.365. [DOI] [PubMed] [Google Scholar]
- 8.Piva R, Deaglio S, Famà R, et al. The Krüppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia. 2015;29(2):503–507. doi: 10.1038/leu.2014.294. [DOI] [PubMed] [Google Scholar]
- 9.Clipson A, Wang M, de Leval L, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia. 2015;29(5):1177–1185. doi: 10.1038/leu.2014.330. [DOI] [PubMed] [Google Scholar]
- 10.Parry M, Rose-Zerilli MJ, Ljungström V, et al. Genetics and Prognostication in Splenic Marginal Zone Lymphoma: Revelations from Deep Sequencing. Clin Cancer Res. 2015;21(18):4174–4183. doi: 10.1158/1078-0432.CCR-14-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi D, Deaglio S, Dominguez-Sola D, et al. Alteration of BIRC3 and multiple other NF-κB pathway genes in splenic marginal zone lymphoma. Blood. 2011;118(18):4930–4934. doi: 10.1182/blood-2011-06-359166. [DOI] [PubMed] [Google Scholar]
- 12.Trifonov V, Pasqualucci L, Tiacci E, Falini B, Rabadan R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst Biol. 2013;7(Suppl 2):S2. doi: 10.1186/1752-0509-7-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Silva Almeida AC, Abate F, Khiabanian H, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet. 2015;47(12):1465–1470. doi: 10.1038/ng.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Reports. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunawardana J, Chan FC, Telenius A, et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat Genet. 2014;46(4):329–335. doi: 10.1038/ng.2900. [DOI] [PubMed] [Google Scholar]
- 16.Kleppe M, Tousseyn T, Geissinger E, et al. Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin’s lymphoma and T-cell non-Hodgkin’s lymphoma. Haematologica. 2011;96(11):1723–1727. doi: 10.3324/haematol.2011.041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mottok A, Renné C, Seifert M, et al. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood. 2009;114(20):4503–4506. doi: 10.1182/blood-2009-06-225839. [DOI] [PubMed] [Google Scholar]
- 18.Odejide O, Weigert O, Lane AA, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yildiz M, Li H, Bernard D, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125(4):668–679. doi: 10.1182/blood-2014-06-582650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crescenzo R, Abate F, Lasorsa E, et al. European T-Cell Lymphoma Study Group, T-Cell Project: Prospective Collection of Data in Patients with Peripheral T-Cell Lymphoma and the AIRC 5xMille Consortium “Genetics-Driven Targeted Management of Lymphoid Malignancies”. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516–532. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pounds S, Cheng C, Mullighan C, Raimondi SC, Shurtleff S, Downing JR. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics. 2009;25(3):315–321. doi: 10.1093/bioinformatics/btn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abate F, Zairis S, Ficarra E, et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;8:97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer AL, Wright G, Yang L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 30.Rossi D, Spina V, Cerri M, et al. Stereotyped B-cell receptor is an independent risk factor of chronic lymphocytic leukemia transformation to Richter syndrome. Clin Cancer Res. 2009;15(13):4415–4422. doi: 10.1158/1078-0432.CCR-08-3266. [DOI] [PubMed] [Google Scholar]
- 31.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Reports. 2013;3(1):246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seifert M, Sellmann L, Bloehdorn J, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med. 2012;209(12):2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piva R, Agnelli L, Pellegrino E, et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol. 2010;28(9):1583–1590. doi: 10.1200/JCO.2008.20.9759. [DOI] [PubMed] [Google Scholar]
- 36.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter J, Schlesner M, Hoffmann S, et al. ICGC MMML-Seq Project. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treon SP, Xu L, Yang G, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 43.Beà S, Valdés-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110(45):18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter ZR, Xu L, Yang G, et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood. 2014;123(11):1637–1646. doi: 10.1182/blood-2013-09-525808. [DOI] [PubMed] [Google Scholar]
- 47.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okosun J, Bödör C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puente XS, Beà S, Valdés-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 50.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farinha P, Gascoyne RD. Molecular pathogenesis of mucosa-associated lymphoid tissue lymphoma. J Clin Oncol. 2005;23(26):6370–6378. doi: 10.1200/JCO.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 52.Vater I, Montesinos-Rongen M, Schlesner M, et al. The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia. 2015;29(3):677–685. doi: 10.1038/leu.2014.264. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Aguilar A, Idbaih A, Boisselier B, et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res. 2012;18(19):5203–5211. doi: 10.1158/1078-0432.CCR-12-0845. [DOI] [PubMed] [Google Scholar]
- 54.Braggio E, Van Wier S, Ojha J, et al. Genome-Wide Analysis Uncovers Novel Recurrent Alterations in Primary Central Nervous System Lymphomas. Clin Cancer Res. 2015;21(17):3986–3994. doi: 10.1158/1078-0432.CCR-14-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukumura K, Kawazu M, Kojima S, et al. Genomic characterization of primary central nervous system lymphoma. Acta Neuropathol. 2016;131(6):865–875. doi: 10.1007/s00401-016-1536-2. [DOI] [PubMed] [Google Scholar]
- 56.Veeriah S, Brennan C, Meng S, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci USA. 2009;106(23):9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walia V, Prickett TD, Kim JS, et al. Mutational and functional analysis of the tumor-suppressor PTPRD in human melanoma. Hum Mutat. 2014;35(11):1301–1310. doi: 10.1002/humu.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304(5674):1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 60.Pulido R, Krueger NX, Serra-Pagès C, Saito H, Streuli M. Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J Biol Chem. 1995;270(12):6722–6728. doi: 10.1074/jbc.270.12.6722. [DOI] [PubMed] [Google Scholar]
- 61.Ortiz B, Fabius AW, Wu WH, et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc Natl Acad Sci USA. 2014;111(22):8149–8154. doi: 10.1073/pnas.1401952111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peyser ND, Du Y, Li H, et al. Loss-of-Function PTPRD Mutations Lead to Increased STAT3 Activation and Sensitivity to STAT3 Inhibition in Head and Neck Cancer. PLoS One. 2015;10(8):e0135750. doi: 10.1371/journal.pone.0135750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ortiz B, White JR, Wu WH, Chan TA. Deletion of Ptprd and Cdkn2a cooperate to accelerate tumorigenesis. Oncotarget. 2014;5(16):6976–6982. doi: 10.18632/oncotarget.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hart GT, Wang X, Hogquist KA, Jameson SC. Krüppel-like factor 2 (KLF2) regulates B-cell reactivity, subset differentiation, and trafficking molecule expression. Proc Natl Acad Sci USA. 2011;108(2):716–721. doi: 10.1073/pnas.1013168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol. 2009;9(11):767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- 66.Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood. 2011;117(5):1595–1604. doi: 10.1182/blood-2010-01-264275. [DOI] [PubMed] [Google Scholar]
- 67.Campo E, Miquel R, Krenacs L, Sorbara L, Raffeld M, Jaffe ES. Primary nodal marginal zone lymphomas of splenic and MALT type. Am J Surg Pathol. 1999;23(1):59–68. doi: 10.1097/00000478-199901000-00006. [DOI] [PubMed] [Google Scholar]
- 68.Camacho FI, Algara P, Mollejo M, et al. Nodal marginal zone lymphoma: a heterogeneous tumor: a comprehensive analysis of a series of 27 cases. Am J Surg Pathol. 2003;27(6):762–771. doi: 10.1097/00000478-200306000-00006. [DOI] [PubMed] [Google Scholar]
- 69.van den Brand M, van Krieken JH. Recognizing nodal marginal zone lymphoma: recent advances and pitfalls. A systematic review. Haematologica. 2013;98(7):1003–1013. doi: 10.3324/haematol.2012.083386. [DOI] [PMC free article] [PubMed] [Google Scholar]