Summary
Photosynthetic microbes exhibit light‐dependent electron export across the cell membrane, which can generate electricity in biological photovoltaic (BPV) devices. How electrons are exported remains to be determined; the identification of mechanisms would help selection or generation of photosynthetic microbes capable of enhanced electrical output. We show that plasma membrane NADPH oxidase activity is a significant component of light‐dependent generation of electricity by the unicellular green alga Chlamydomonas reinhardtii. NADPH oxidases export electrons across the plasma membrane to form superoxide anion from oxygen. The C. reinhardtii mutant lacking the NADPH oxidase encoded by RBO1 is impaired in both extracellular superoxide anion production and current generation in a BPV device. Complementation with the wild‐type gene restores both capacities, demonstrating the role of the enzyme in electron export. Monitoring light‐dependent extracellular superoxide production with a colorimetric assay is shown to be an effective way of screening for electrogenic potential of candidate algal strains. The results show that algal NADPH oxidases are important for superoxide anion production and open avenues for optimizing the biological component of these devices.
Keywords: alga, biophotovoltaic, Chlamydomonas, energy, NADPH oxidase
Introduction
Renewable power sources such as wind farms, solar panels or hydro turbines are now in relatively widespread use, yet the feasibility of these to replace fossil fuels is uncertain due to their higher cost and lower energy density (Cho, 2010). Therefore, in the past decade, there has been a considerable shift in research focus into harvesting energy from high‐biomass biological materials. However, considerable areas of arable land are required to sustain biofuel production, which is at odds with food production (Jones and Mayfield, 2012). As a result, the use of algae as novel biofuel sources has rapidly gained popularity in recent years. Individually, algae are simple photosynthetic organisms, yet together constitute over 300 000 species with great evolutionary diversity. Their rapid growth, high‐biomass yields and ability to thrive in freshwater, salt water and wastewater could be highly advantageous for biofuel production. They do not compete for space with commercial crop species (Scott et al., 2010), and their photosynthetic capacity is excellent, with energy efficiency levels on a par with land plants (Larkum, 2010). At the level of the primary light reaction, when the energy of light is used to do photosynthetic work in terms of water photolysis, up to 37% efficiency has been estimated (Zhu et al., 2010). Total worldwide energy consumption is around 15 terawatts per year, so with over 85 000 terawatts of solar energy reaching the Earth annually, this represents a lucrative source of renewable power even if only some of it could be harnessed (Jones and Mayfield, 2012).
Algae also have the potential for direct production of electricity, in addition to providing combustible biomass for its generation. This is increasingly relevant as the quest for renewable energy has included the development of microbial fuel cells that utilize biological electron export processes to generate electricity (Rosenbaum et al., 2010). Using photosynthetic microbes such as microalgae in these fuel cells reduces the need for a supply of fixed carbon and effectively creates a biological solar panel, often termed biological photovoltaic (BPV) devices (Bombelli et al., 2011; Samsonoff et al., 2014; Figure 1a and b). Light energy is captured by the photosynthetic apparatus, leading to electron export across the cell membrane (Bradley et al., 2012). Electrons can then be captured by a soluble electron carrier in the aqueous medium for delivery to the anode and current generation (Bombelli et al., 2011; Figure 1c). Alternatively, electrons can be delivered directly to the anode by biofilms growing on its surface (McCormick et al., 2011). A limiting factor is cellular electron export and so identifying the electron transport proteins that contribute power output in the devices remains a fundamental goal in the development of these biological solar cells.
Figure 1.
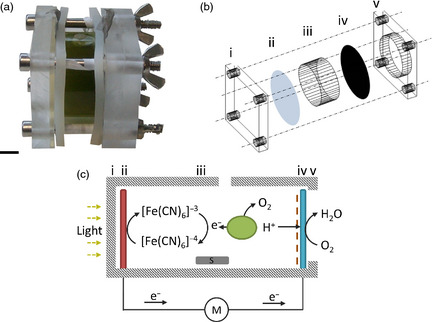
BPV design and function. (a) Image of assembled BPV device containing Chlamydomonas reinhardtii cells in the anodic chamber. Scale bar, 1 cm. (b) Exploded schematic of the anodic chamber. The front clamp (i) permits light to enter via the anode (ii) into the algal chamber (iii) and the back clamp (v) has a cut‐out to allow oxygen contact with the cathode (iv). (c) Principles of operation of a BPV device. Upon illumination, cells (green ovoid) within the algal chamber release electrons (e−) which reduce extracellular ferricyanide ([Fe(CN)6]4−) to ferrocyanide ([Fe(CN)6]3−). Ferrocyanide shuttles electrons to the anode (red rectangle, ii) into the external circuit via an external resister and multimeter (V) through to the cathode (blue rectangle, iv). Simultaneously, protons (H+) diffuse from the chamber via a dialysis and Nafion membrane (dotted line) to the cathode combining with electrons and oxygen to form water. Cells are kept in suspension by a magnetic stirrer (S) and evaporation limited by sealing chamber (B). Wires are connected via crocodile clips to two stainless steel strips. Numbers above selected components in (b) correspond to those shown in (c).
Plasma membrane NADPH oxidases (NOX) are found in animals, plants and algae. They are encoded in animals by NOX genes and in plants by respiratory burst oxidase homologue (RBOH) genes (named from homology with the animal respiratory burst NOX; Figure 2). They catalyse the transfer of electrons from cytosolic NADPH to extracellular oxygen, so generating extracellular superoxide anion (; Sagi et al., 2004). The ability of NOX to generate an electrical current was demonstrated by Schrenzel et al. (1998) by whole‐cell patch clamping of human phagocytes (in which NOX are the main transmembrane redox component). Current was dependent on the presence of intracellular NADPH and was diminished by the widely used NOX inhibitor diphenylene iodonium chloride (DPI). Additionally, mutated cells lacking a functional NOX showed no detectable current, demonstrating the importance of NOX as an electrogenic component (Schrenzel et al., 1998). Subsequently, power output of a fuel cell containing human neutrophils was shown to be dependent on NOX activity (Sakai et al., 2009). The presence of DPI or inhibitors of NOX protein assembly inhibited power output. These enzymes in photosynthetic microbes are therefore targets for development in the improvement of BPV output.
Figure 2.
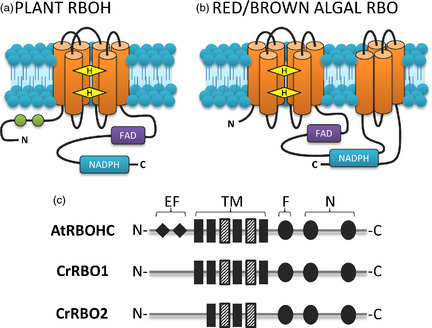
Comparison of plant and algal NADPH oxidase structures. (a) Plant NADPH oxidase (RBOH) structure based on sequence data from Arabidopsis thaliana and other plant NOX described in Torres et al. (1998) and Kawahara et al. (2007). (b) Algal RBO structure based on sequence data from Chondrus crispus, Cyanidioschyzon merolae, Porphyra Yezoensis and Phaeodactylum tricornutum described in Hervé et al. (2005). NOX in these algae have an additional four transmembrane domains in between the NADPH‐binding domain. These are absent from the Chlamydomonas RBO. (c) Schematic of RBO proteins CrRBO1 and CrRBO2 compared to Arabidopsis thaliana AtRBOHC. Abbreviations: EF, EF hand; F, FAD‐binding domain; N, NADPH‐binding domains; TM, transmembrane domain. Dashed TM domains indicate the presence of conserved haem‐binding histidine residues. Proteins are orientated with the cytosolic side at bottom of image. Orange cylinders represent transmembrane domains, green circles represent EF hands, and yellow diamonds represent haem molecules. Binding regions for FAD and NADPH are highlighted.
In plants, mutant studies have shown that NOX are involved in immunity, development and stress responses (Foreman et al., 2003; Kadota et al., 2014; Lee et al., 2013). In contrast to the research on animal and plant NOX, the research on algal NOX (encoded by RBO genes; Figure 2) is much less advanced. Extracellular superoxide production has been recorded in many species of red, green and brown algae and diatoms (Marshall et al., 2005), suggesting that algal NOX may be widespread. They have been proposed to be involved in growth, stress and immune responses (reviewed by Anderson et al., 2011). Production can be inhibited by DPI, suggesting that this is a useful tool in delineating NOX activity (Bouarab, 1999; Küpper et al., 2001; Ross et al., 2005; Coelho et al., 2008; Pérez‐Pérez et al., 2012). In the unicellular raphidophyte Chattonella marina, superoxide production is at the plasma membrane and stimulated by light (Kim et al., 2004, 2007) implying that reductants for superoxide generation are sourced from photosynthesis. Molecular data for the Chondrus crispus NOX showed that typical human NOX2 features are highly conserved, including the substrate‐binding sites, haem‐binding histidine residues and transmembrane domains. There is substantial evidence for NOX in algae, and therefore, they may function as plasma membrane electron transporters for use in BPV devices (Bombelli et al., 2011).
The unicellular green alga Chlamydomonas reinhardtii generates extracellular superoxide (Hema et al., 2007; Hemschemeier et al., 2013; Pérez‐Pérez et al., 2012). Its two RBO genes are predicted to encode NOX with the conserved NADPH‐binding domains and the transmembrane domains with haem‐liganding histidine residues that are essential for electron transport (Hervé et al., 2005; Anderson et al., 2011; Figure 2). However, at present, the contribution of the two RBO proteins to electron export and extracellular production is unknown. We therefore tested the hypothesis that an algal NOX could contribute to power output in a BPV device by comparing C. reinhardtii strain cw15 (a standard laboratory strain in which the cell wall is greatly reduced) and its sta6rbo1 mutant (Li et al., 2010), in which STA6 is mutated and RBO1 is deleted (Blaby et al., 2013; Table 1). The results show that RBO1 is indeed responsible for extracellular superoxide anion production and that this generates a significant part of the current measured in the BPV device. RBO expression in C. reinhardtii can be maintained even under nutrient limitation (Allen et al., 2007) and RBOL2 expression increases under anaerobiosis (Hemschemeier et al., 2013). As algae produce even under adverse conditions including as a response to predation or infection (Hervé et al., 2005; Hema et al., 2007; Kim et al., 2007; Küpper et al., 2001; Anderson et al., 2011), these photosynthetic microbes would be a robust, exploitable and renewable resource for application in biological solar cells.
Table 1.
Summary of algal strains used in this study
| Strain | Description |
|---|---|
| cw92 | Cell wall‐deficient, WT for STA6 and RBO1 |
| cw15 | Cell wall‐deficient, WT for STA6 and RBO1 |
| sta6rbo1 | cw15 carrying mutated STArchless6 and with a deleted RBO1 |
| sta6rbo1(RBO1) | cw15 carrying mutated STArchless6 and with a deleted RBO1 but complemented for RBO1 |
| sta6rbo1(STA6) | cw15 carrying mutated STArchless6 and with a deleted RBO1 but complemented for STA6 |
Results
Production of extracellular superoxide anion is light‐dependent and requires RBO1
We first confirmed that extracellular superoxide anion production could be detected from C. reinhardtii by assaying the reduction of cell‐impermeable XTT (2,3‐bis‐(2‐methoxy‐4‐nitro‐5‐sulfophenyl)‐2H‐tetrazolium‐5‐carboxanilide (Sutherland and Learmonth, 1997). The cell wall‐deficient strain cw92, which is wild type for both RBO1 and STA6 (STArchless6 encoding the small subunit of ADP‐Glc pyrophosphorylase; Table 1), was grown to mid‐logarithmic phase. These cells supported light‐dependent production of extracellular superoxide anion that was significantly inhibited by DPI, indicating NOX activity (Figure 3). Extracellular superoxide anion production by cw92 was indistinguishable from that of mid‐logarithmic cw15, the cell wall‐deficient STA6RBO1 strain from which the sta6rbo1 mutant is derived (Figure 4; Table 1). Both cw15 and cw92 were derived from mutagenesis of the WT 137c (Davies and Plaskitt, 1971; Pröschold et al., 2005). The genetic basis for the cw mutation remains unknown, but cw92 contains a glycoprotein in its residual cell wall that is absent from cw15 (Voigt et al., 1996). Light‐dependent extracellular production by cw15 was significantly inhibited by DPI, indicating production by NOX. Critically, light‐dependent production by sta6rbo1 was significantly impaired even without DPI addition, which suggests that RBO1 was responsible for the majority of production (Figure 4). The residual production by sta6rbo1 that was DPI‐sensitive could be due to the continued low expression of RBO2 (Blaby et al., 2013). The DPI‐insensitive production shown by all three strains suggests that alternative cell membrane redox enzymes are still in operation.
Figure 3.
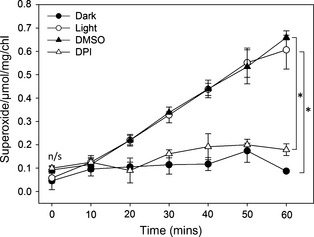
Chlamydomonas reinhardtii extracellular superoxide anion production is light‐dependent and DPI sensitive. Time course of production by mid‐logarithmic cells of the Chlamydomonas reinhardtii strain cw92, determined using XTT reduction. Mean ± SEM production was prevented by dark incubation and inhibited in the light by DPI (20 μm) as a NOX inhibitor. The equivalent dimethylsulphoxide (DMSO) concentration was used as the DPI solvent control (0.0075% v/v). Asterisks denote significant difference (P < 0.05) from control (Student's t‐test; n/s, no significance; n = 3).
Figure 4.
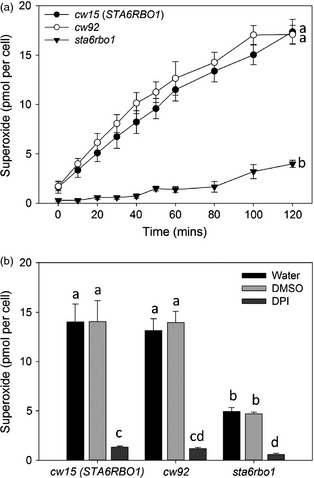
Chlamydomonas reinhardtii extracellular superoxide anion production requires RBO1. (a) Time course of production by mid‐logarithmic cells of the Chlamydomonas reinhardtii cell wall‐deficient strains cw15, cw92 (both STA6RBO1) and the RBO1‐deficient sta6rbo1 mutant, determined using XTT reduction. Levels of production were indistinguishable between both cell wall‐less strains cw15 and cw92. Different letters denote significant differences (one‐way ANOVA, P < 0.05). Data are mean ± SEM (n = 3). (b) DPI (20 μm) inhibited light‐dependent production by all strains. Values are for 120 min. (one‐way ANOVA, P < 0.05; mean ± SEM, n = 3). There was no effect of equivalent concentration (v/v) of DMSO as the solvent for DPI. Different letters denote significant differences (one‐way ANOVA, P < 0.05). Data are mean ± SEM (n = 3).
Complementation with RBO1 restores production
If RBO1 were the source of extracellular production then it follows that complementing the sta6rbo1 mutant with RBO1 would restore production. To test this, the sta6rbo1 mutant was complemented with a synthetic gene construct containing the complete RBO1 gene model (Table 1). Three independently transformed strains were selected by antibiotic resistance and confirmed for RBO1 by PCR (Figure 5a). These were compared for light‐dependent production against two sta6rbo1 strains that had previously been transformed to express only STA6 and so retained loss of RBO1 function (Blaby et al., 2013; Table 1). Only complementation with RBO1 restored production to levels comparable to (and not significantly different from) that of the cw15 STA6RBO1 parental strain (Figure 5b). Complementation with STA6 did not increase production above that of the sta6rbo1 mutant (Figure 5b). These data show that production is indeed RBO1 dependent.
Figure 5.
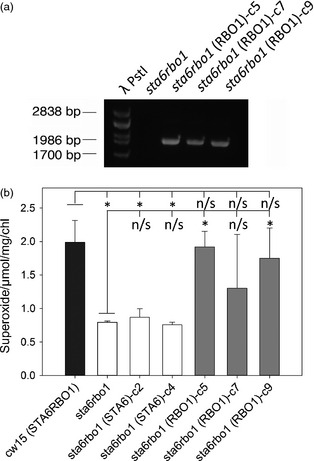
Superoxide anion production is restored by RBO1. (a) Validation of pSL18_RBO1 insertion into sta6rbo1 (CC‐4348) genome. To verify integration of the RBO1‐containing construct, PCRs were performed using primers specific to the plasmid. The predicted amplicon size is 1979 bp. (b) Mean ± SEM production in light (120 min) from cw15, RBO1‐deficient sta6, two lines of sta6rbo1(STA6) (Blaby et al., 2013) and the three RBO1‐complemented lines shown in (a), determined using XTT (n ≤ 5). Only the presence of RBO1 was effective in restoring production. Asterisks denote significant difference (P < 0.05) from indicated control (Student's t‐test; n/s, no significance).
RBO1 contributes to current output in the BPV device
Having shown that RBO1 expression results in extracellular production, which is in turn the result of electron extrusion across the plasma membrane, we tested for a relationship between this activity and an ability to generate current in the BPV device (Figure 1). A representative current output from mid‐logarithmic cw15 (wild type for STA6 and RBO1) is shown in Figure 6a. A first application of light caused current output in the BPV device for over 12 h (Light Effect 1, LE1; Figure 6a). The return to darkness saw a decline in current, and a second application of light (LE2) was less effective in promoting current generation (Figure 6a). The reason(s) for this lower output in LE2 is not yet known. The addition of DPI to the BPV device impaired electrode function, so it was not possible to demonstrate that cw15 treated with DPI phenocopied sta6rbo1 as an additional line of evidence for NOX‐dependent current generation.
Figure 6.
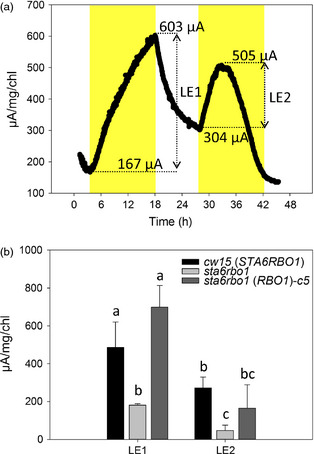
RBO1‐dependent current from Chlamydomonas reinhardtii. (a) Example current from the cw15 parental strain in the BPV device. Yellow indicates light application; LE1 and LE2 denote the first and second light effect (LE) current. (b) Mean ± SEM of LE1 and LE2 currents from Chlamydomonas reinhardtii cultures (n = 3–4), standardized to chlorophyll (Chl) content. sta6rbo1(RBO1)‐c5 is one of the three strains complemented with RBO1 and exhibiting normal production (Figure 5b). Letters denote significant differences as determined by one‐way ANOVA (P < 0.05).
Light‐dependent current output was significantly impaired in the sta6rbo1 mutant for both LE1 and LE2. Output in LE1 was fully restored by complementation with RBO1 (Figure 6b). This was shown using the sta6rbo1(RBO1)‐c5 strain in which extracellular production (XTT assay) was equal to that of the cw15 parental strain (Figure 5b). As in the parental strain, the current generation was lower on a second light exposure (LE2). Overall, significant components of both extracellular production and light‐dependent current output require RBO1.
Discussion
It is clear that RBO1 makes a significant contribution to light‐dependent current generation in the BPV device, and the overall current output from C. reinhardtii is promising. From Figure 6b, up to approximately 800 μA/mgChl can be generated, which equals 800 μC/mgChl/s. An AAA zinc–carbon battery could have a nominal capacity of 500–700 mAh. A battery with a 650 mAh capacity holds 2340 C. A litre of Chlamydomonas culture with a chlorophyll concentration of 15 μg/mL would contain 15 mg of chlorophyll. It could generate 12 mC/s (800 μC/mgChl/s × 15) and would therefore take 2.25 days to generate the charge held by the battery (2340 C/0.012 C/s = 195 000 s = 2.25 days). Whether the contribution to output by RBO1 can be increased now needs to be examined. Additionally, the electron export mechanisms that were evident in the absence of RBO1 should now be identified.
Selecting algae for both biofilm formation and high NOX activity is a realistic goal. Extracellular superoxide anion production is widespread and has already been recorded from other green algae such as Dasycladus vermicularis (Ross et al., 2005), Ulva pertusa (Goh et al., 2010), Ulva compressa (Gonzalez et al., 2010), the brown alga Laminaria digitata (Küpper et al., 2001), the red alga Chondrus crispus (Bouarab, 1999) and red intertidal algae (Liu and Pang, 2010). In the unicellular raphidophyte Chattonella marina, superoxide production is essential for growth. The level of extracellular superoxide anion peaks during the exponential growth phase, and growth is inhibited in the presence of superoxide dismutase (Oda et al., 1995). Additionally, superoxide levels were stimulated by light (Kim et al., 2004) implying reductants for superoxide generation by C. marina are sourced from photosynthesis (Marshall et al., 2005). Although it has been speculated that NOX could make a contribution to production, there has been no genetic evidence up until now and the findings here on Chlamydomonas RBO1 will enable future studies on the physiological roles and regulation of algal NOX that could be exploited for power output. Undoubtedly however, there will be other limiting factors besides electron export to overcome to ensure growth and productivity of algal cultures.
Assaying production with XTT affords a more rapid assessment of NOX activity than measuring current and could be used for screening. The widespread presence of NOX genes in freshwater and marine algae makes these organisms an attractive prospect for deployment in BPV devices. We envisage niche, off‐grid applications in which small‐scale algal production could power, for example, mobile phones or LED lights.
Experimental procedures
Algal strains and growth conditions
Chlamydomonas reinhardtii strains cw15 (CC‐1883) and cw92 (CC‐503) were gifts from Dr Saul Purton (UCL, London, UK). sta6rbo1 (CC‐4348) and complemented strains sta6rbo1(STA6)‐c2 (CC‐4565) and sta6rbo1(STA6)‐c4 (CC‐4566) were from Dr. Ursula Goodenough (Washington University, St. Louis, MO). Strains are described in Table 1. They were grown axenically in Tris‐acetate phosphate (TAP) medium prepared as described (http://www.chlamy.org/methods.html). Cultures were inoculated using cells from stationary phase stock cultures (inoculum density 20 000 cells/mL) and grown under continuous light (~40 μE/m2/s, 21 °C, 140 rpm; INFORS HT, Reigate, Surrey, UK). Chlorophyll was extracted according to Inskeep and Bloom (1985).
Complementation of RBO1
To restore rbo1 function to sta6rbo1 (CC‐4348), a synthetic gene construct containing the complete RBO1 gene model (Genscript, Piscataway, New Jersey, USA) designed based on the v5.3 assembly of the C. reinhardtii genome (http://www.phytozome.net/chlamy.php) was excised from pUC57 by NdeI SpeI digest and inserted into the NdeI SpeI sites of pSL18 (Fischer and Rochaix, 2001) to generate pSL18_RBO1. pSL18_RBO1 was linearized by XhoI digestion and transformed into CC‐4348. Transformants were selected for by paromomycin resistance (5 μg/mL) and candidate colonies were PCR‐validated using the primers specific to the plasmid (5′ to 3′: forward primer ATTATGTATCAATATTGTTGCGTTCG, reverse primer GTCACCACAAACACAACGGCGAG). PCR amplicons were sequenced to confirm product.
Superoxide measurements
Algal cells in TAP medium were collected by centrifugation (4000 g, 5 min), and cells were washed in buffer (Tris‐HCl, 20 mm, pH 7.0) and resuspended in buffer plus 100 μm XTT (Sigma‐Aldrich, Haverhill, Suffolk, UK). Cells were transferred to a transparent Perspex chamber (except 5b for which cells were assayed in microfuge tubes), continually mixed to ensure cell suspension and illuminated with white LED lights (OSRAM Parathom Par 16, Stockport, UK; 100 μE/m2/s). Samples (250 μL) were removed, cells pelleted, and the absorbance at 470 nm of the supernatant measured with a plate reader (Fluostar Optima, BMG Labtech, Aylesbury, Bucks, UK) in 96‐well flat‐bottom plates (Greiner Bio‐One, Stonehouse, South Lanarkshire, UK). Superoxide was quantified according to the extinction coefficients of Sutherland and Learmonth (1997). All experiments were conducted in the light (100 μE/m/s2) unless stated otherwise. In inhibitor experiments, cells were exposed to DPI or DMSO for 1 h prior to experiment and then resuspended in buffer (Tris‐HCl, 20 mm pH 7.0) with DPI/DMSO present at the same concentration used in the pretreatment.
BPV device construction and operation
BPV devices were formed from 10‐mm‐thick transparent Perspex (anodic chamber external diameter 50 mm, internal diameter 44 mm, depth 20 mm with nominal internal volume 30.5 mL and 15.2 cm2 illuminated area), sealed with polydimethylsiloxane, similar to the device described in Bombelli et al. (2011). An 80 × 80 mm carbon–platinum cathode assembly impregnated on one side (50 × 50 mm coated area) with Nafion (Ion Power, New Castle, Delaware, USA) formed the back wall of the anodic chamber. A single layer of dialysis membrane (SnakeSkin Dialysis Tubing 10 K MCWO, Sigma‐Aldrich) was layered on to the cathode assembly. Transparent indium tin oxide (ITO, 60 Ω/sq) on plastic polyethylene terephthalate (Sigma‐Aldrich) formed the front of the anodic chamber. Electrodes were attached to the external circuit via a contact formed from a strip of stainless steel. Current was monitored via a multimeter (Model UT70B, Uni‐Trend, Hong Kong) and recorded using UT70B interface software v3.04 (Uni‐Trend, Kowloon, Hong Kong, China). A bias potential (500 mV) was applied between the anode and cathode by means of an external power box made in‐house. The device was illuminated through the ITO side by a panel of red LED bulbs (maximum emission peak 630 nm) controlled by a PSU130 power unit (LASCAR, Salisbury, UK) set at 100 μE/m2/s. The apparatus was completely covered by black velvet fabric to ensure complete darkness. Mid‐logarithmic algal cultures in TAP medium were loaded directly into the anodic chamber. Potassium ferricyanide (FeCN) was added to a final concentration of 1 mm, and cultures were continually mixed via a magnetic stirrer (Multistirrer 6, VELP Scientifica, Usmate (MB), Italy; Setting 4). Devices were operated at room temperature (~21 °C). Dialysis membrane was replaced for each experiment, and the other components washed with deionized water before reuse.
Acknowledgements
We acknowledge the UK EPSRC, EnAlgae (European Regional Development Fund: INTERREG IVB NEW programme) and the US Department of Energy for funding. I.K.B. was supported by a training grant from the National Institutes of Health (T32ES015457). Work in the Merchant laboratory was supported by the U.S. Department of Energy (grant no. DE–FC02–02ER63421).
Accession numbers: Rbo1: XM_001691803.1, Rbo2: XM_001691804.1, Sta6: XM_001691802.1
References
- Allen, M.D. , del Campo, J.A. , Kropat, J. and Merchant, S.S. (2007) FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron‐deficient Chlamydomonas reinhardtii . Eukaryot. Cell 6, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, A. , Bothwell, J.H. , Laohavisit, A. , Smith, A.G. and Davies, J.M. (2011) NOX or not? Evidence for algal NADPH oxidases. Trends Plant Sci. 16, 579–581. [DOI] [PubMed] [Google Scholar]
- Blaby, I.K. , Glaesner, A.G. , Mettler, T. , Fitz‐Gibbon, S.T. , Gallaher, S.D. , Liu, B.S. , Boyle, N.R. , Kropat, J. , Stitt, M. , Johnson, S. , Benning, C. , Pellegrini, M. , Casero, D. and Merchant, S.S. (2013) Systems‐level analysis of nitrogen starvation‐induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell, 25, 4305–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombelli, P. , Bradley, R.W. , Scott, A.M. , Philips, A.J. , McCormick, A.J. , Cruz, S.M. , Anderson, A.A. , Yunus, K. , Bendall, D.S. , Cameron, P. , Davies, J.M. , Smith, A.G. , Howe, C.J. and Fisher, A.C. (2011) Quantitative analysis of the factors limiting solar power transduction by Synechocystis sp. PCC 6803 in biological photovoltaic devices. Energy Environ. Sci. 4, 4690–4698. [Google Scholar]
- Bouarab, K. (1999) Sulfated oligosaccharides mediate the interaction between a marine red alga and its green algal pathogenic endophyte. Plant Cell, 11, 1635–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, R.W. , Bombelli, P. , Rowden, S.J.L. and Howe, C.J. (2012) Biological photovoltaics: intra‐ and extra‐cellular electron transport by cyanobacteria. Biochem. Soc. Trans. 40, 1302–1307. [DOI] [PubMed] [Google Scholar]
- Cho, A. (2010) Energy's tricky tradeoffs. Science, 329, 786–787. [DOI] [PubMed] [Google Scholar]
- Coelho, S.M.B. , Brownlee, C. and Bothwell, J.H.F. (2008) A tip‐high Ca2+‐interdependent, reactive oxygen species gradient is associated with polarized growth in Fucus serratus zygotes. Planta, 227, 1037–1046. [DOI] [PubMed] [Google Scholar]
- Davies, D.R. and Plaskitt, A. (1971) Genetic and structural analyses of cell‐wall formation in Chlamydomonas reinhardtii . Genet. Res. 17, 33–43. [Google Scholar]
- Fischer, N. and Rochaix, J.D. (2001) The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii . Mol. Genet. Genomics, 265, 888–894. [DOI] [PubMed] [Google Scholar]
- Foreman, J. , Demidchik, V. , Bothwell, J.H.F. , Mylona, P. , Miedema, H. , Torres, M.A. , Linstead, P. , Costa, S. , Brownlee, C. , Jones, J.D.G. , Davies, J.M. and Dolan, L. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature, 422, 442–446. [DOI] [PubMed] [Google Scholar]
- Goh, C.‐H. , Oh, S.‐J. , Jun, S.‐S. and Han, T. (2010) External K+ deficiency inhibits photosynthetic activity through superoxide anion production in protoplasts isolated from the thallus of Ulva pertusa . J. Plant Biol. 53, 155–164. [Google Scholar]
- Gonzalez, A. , Vera, J. , Castro, J. , Dennett, G. , Mellado, M. , Morales, B. , Correa, J.A. and Moenne, A. (2010) Co‐occurring increases of calcium and organellar reactive oxygen species determine differential activation of antioxidant and defense enzymes in Ulva compressa (Chlorophyta) exposed to copper excess. Plant, Cell Environ. 33, 1627–1640. [DOI] [PubMed] [Google Scholar]
- Hema, R. , Senthil‐Kumar, M. , Shivakumar, S. , Chandrasekhara Reddy, P. , Udayakumar, M. , Shivakumar, R.H.M.S.S. and Udayakumar, P.C.R.M. (2007) Chlamydomonas reinhardtii, a model system for functional validation of abiotic stress responsive genes. Cultures, 226, 655–670. [DOI] [PubMed] [Google Scholar]
- Hemschemeier, A. , Casero, D. , Liu, B. , Benning, C. , Pellegrini, M. , Happe, T. and Merchant, S.S. (2013) COPPER RESPONSE REGULATOR1‐dependent and ‐independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell, 25, 3186–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé, C. , Tonon, T. , Collén, J. , Corre, E. and Boyen, C. (2005) NADPH oxidases in Eukaryotes: red algae provide new hints!. Curr. Genet. 49, 190–204. [DOI] [PubMed] [Google Scholar]
- Inskeep, W.P. and Bloom, P.R. (1985) Extinction coefficients of chlorophyll a and b in n, n‐dimethylformamide and 80% acetone. Plant Physiol. 77, 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C.S. and Mayfield, S.P. (2012) Algae biofuels: versatility for the future of bioenergy. Curr. Opin. Biotechnol. 23, 346–351. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, J.D.G. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kawahara, B. , Quinn, M.T. and Lambeth, J.D. (2007) Molecular evolution of the reactive oxygen‐generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol. Biol. 7, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Watanabe, M. , Nakayasu, Y. and Kohata, K. (2004) Production of superoxide anion and hydrogen peroxide associated with cell growth of Chattonella antiqua . Aquat. Microb. Ecol. 35, 57–64. [Google Scholar]
- Kim, D. , Nakashima, T. , Matsuyama, Y. , Niwano, Y. , Yamaguchi, K. and Oda, T. (2007) Presence of the distinct systems responsible for superoxide anion and hydrogen peroxide generation in red tide phytoplankton Chattonella marina and Chattonella ovata . J. Plankton Res. 29, 241–247. [Google Scholar]
- Küpper, F.C. , Kloareg, B. , Guern, J. and Potin, P. (2001) Oligoguluronates elicit an oxidative burst in the brown algal kelp Laminaria digitata . Plant Physiol. 125, 278–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum, A.W.D. (2010) Limitations and prospects of natural photosynthesis for bioenergy production. Curr. Opin. Biotechnol. 21, 271–276. [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Rubio, M.C. , Alassimone, J. and Geldner, N. (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Han, D.X. , Hu, G.R. , Dauvillee, D. , Sommerfeld, M. , Ball, S. and Hua, Q. (2010) Chlamydomonas starchless mutant defective in ADP‐glucose pyrophosphorylase hyper‐accumulates triacylglycerol. Metab. Eng. 12, 387–391. [DOI] [PubMed] [Google Scholar]
- Liu, F. and Pang, S.J. (2010) Stress tolerance and antioxidant enzymatic activities in the metabolisms of the reactive oxygen species in two intertidal red algae Grateloupia turuturu and Palmaria palmata . J. Exp. Mar. Bio. Ecol. 382, 82–87. [Google Scholar]
- Marshall, J.‐A. , Ross, T. , Pyecroft, S. , Hallegraeff, G. , Salas, M. and Oda, T. (2005) Superoxide production by marine microalgae. Mar. Biol. 147, 533–540. [Google Scholar]
- McCormick, A.J. , Bombelli, P. , Scott, A.M. , Philips, A.J. , Smith, A.G. , Fisher, A.C. and Howe, C.J. (2011) Photosynthetic biofilms in pure culture harness solar energy in a mediatorless bio‐photovoltaic cell (BPV) system. Energy Environ. Sci. 4, 4699. [Google Scholar]
- Oda, T. , Moritomi, J. , Kawano, I. , Hamaguchi, S. , Ishimatsu, A. and Muramatsu, T. (1995) Catalase‐ and superoxide dismutase‐induced morphological changes and growth inhibition in the red tide phytoplankton Chattonella marina . Biosci. Biotechnol. Biochem. 59, 2044–2048. [Google Scholar]
- Pérez‐Pérez, M.E. , Couso, I. and Crespo, J.L. (2012) Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii . Autophagy, 8, 376–388. [DOI] [PubMed] [Google Scholar]
- Pröschold, T. , Harris, E.H. and Coleman, A.W. (2005) Portrait of a species: Chlamydomonas reinhardtii . Genetics, 170, 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, M. , He, Z. and Angenent, L.T. (2010) Light energy to bioelectricity: photosynthetic microbial fuel cells. Curr. Opin. Biotechnol. 21, 259–264. [DOI] [PubMed] [Google Scholar]
- Ross, C. , Küpper, F.C. , Vreeland, V. , Herbert Waite, J. and Jacobs, R.S. (2005) Evidence of a latent oxidative burst in relation to wound repair in the giant unicellular chlorophytes Dasycladus vermicularis . J. Phycol. 41, 531–541. [Google Scholar]
- Sagi, M. , Davydov, O. , Orazova, S. , Yesbergenova, Z. , Ophir, R. , Stratmann, J.W. and Fluhr, R. (2004) Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum . Society, 16, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, M. , Vonderheit, A. , Wei, X. , Kuttel, C. and Stemmer, A. (2009) A novel biofuel cell harvesting energy from activated human macrophages. Biosens. Bioelectron. 25, 68–75. [DOI] [PubMed] [Google Scholar]
- Samsonoff, N. , Ooms, M. and Sinton, D. (2014) A photosynthetic‐plasmonic voltaic cell: excitation of photosynthetic bacteria and current collection through a plasmonic substrate. App. Phys. Lett. 104, 043704. [Google Scholar]
- Schrenzel, J. , Serrander, L. , Bánfi, B. , Nüsse, O. , Fouyouzi, R. , Lew, D.P. , Demaurex, N. and Krause, K.H. (1998) Electron currents generated by the human phagocyte NADPH oxidase. Nature, 392, 734–737. [DOI] [PubMed] [Google Scholar]
- Scott, S.A. , Davey, M.P. , Dennis, J.S. , Horst, I. , Howe, C.J. , Lea‐Smith, D. and Smith, A.G. (2010) Biodiesel from algae: challenges and prospects. Curr. Opin. Biotechnol. 21, 277–286. [DOI] [PubMed] [Google Scholar]
- Sutherland, M.W. and Learmonth, B.A. (1997) The tetrazolium dyes MTS and XTT provide new quantitative assays for superoxide and superoxide dismutase. Free Radic. Res. 27, 283–289. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Onouchi, H. , Hamada, S. , Machida, C. , Hammond‐kosack, K.E. and Jones, J.D.G. (1998) Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91 phox). Plant J. 14, 365–370. [DOI] [PubMed] [Google Scholar]
- Voigt, J. , Liebich, I. , Hinkelmann, B. and Kiees, M. (1996) Immunological identification of a putative precursor of the insoluble glycoprotein framework of the Chlamydomonas cell wall. Plant Cell Physiol. 37, 91–101. [DOI] [PubMed] [Google Scholar]
- Zhu, X.G. , Long, S.P. and Ort, D.R. (2010) Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. [DOI] [PubMed] [Google Scholar]


