Abstract
The fungi Aspergillus niger and A. welwitschiae are morphologically indistinguishable species used for industrial fermentation and for food and beverage production. The fungi also occur widely on food crops. Concerns about their safety have arisen with the discovery that some isolates of both species produce fumonisin (FB) and ochratoxin A (OTA) mycotoxins. Here, we examined FB and OTA production as well as the presence of genes responsible for synthesis of the mycotoxins in a collection of 92 A. niger/A. welwitschiae isolates from multiple crop and geographic origins. The results indicate that (i) isolates of both species differed in ability to produce the mycotoxins; (ii) FB-nonproducing isolates of A. niger had an intact fumonisin biosynthetic gene (fum) cluster; (iii) FB-nonproducing isolates of A. welwitschiae exhibited multiple patterns of fum gene deletion; and (iv) OTA-nonproducing isolates of both species lacked the ochratoxin A biosynthetic gene (ota) cluster. Analysis of genome sequence data revealed a single pattern of ota gene deletion in the two species. Phylogenetic analysis suggest that the simplest explanation for this is that ota cluster deletion occurred in a common ancestor of A. niger and A. welwitschiae, and subsequently both the intact and deleted cluster were retained as alternate alleles during divergence of the ancestor into descendent species. Finally, comparison of results from this and previous studies indicate that a majority of A. niger isolates and a minority of A. welwitschiae isolates can produce FBs, whereas, a minority of isolates of both species produce OTA. The comparison also suggested that the relative abundance of each species and frequency of FB/OTA-producing isolates can vary with crop and/or geographic origin.
Keywords: fum cluster, ota cluster, fumonisin, ochratoxin, biosynthetic gene cluster, Aspergillus niger, Aspergillus welwitschiae
Introduction
Aspergillus niger is one of the most important filamentous fungi used for biotechnological purposes and has been labeled by the US Food and Drug Administration as “generally recognized as safe” (GRAS). It is widely used in industrial fermentation of organic acids and enzymes, particularly citric acid and extracellular enzymes, and in fermentation of some oriental foods and beverages. The fungus is also a common member of microbial communities on food crops (Varga et al., 2011; Susca et al., 2013, 2014a,b). A. niger is part of a taxonomic grouping of at least 11 species known as the A. niger “aggregate,” which in turn is part of formal taxon Aspergillus section Nigri, known less formally as the black aspergilli (Varga et al., 2011). Species of black aspergilli are morphologically similar, and in some cases indistinguishable, but can be reliably distinguished by DNA sequence analysis of housekeeping genes, such as the calmodulin and beta tubulin genes (Perrone et al., 2011; Hong et al., 2013). Among species of the black aspergilli, only A. niger and its sister species A. welwitschiae have been reported to produce both fumonisin (FB) (Frisvad et al., 2007, 2011; Mogensen et al., 2010) and ochratoxin A (OTA) (Medina et al., 2005; Battilani et al., 2006). Due to the toxicity of these mycotoxins, there is a need to assess the potential risk that FB and OTA-producing black aspergilli pose to human health. Such assessments will be aided by information on production of the mycotoxins by Aspergillus strains used in food and beverage fermentation, mycotoxin production during fermentation processes, frequencies of producing and nonproducing isolates in microbial communities on crops, and the genetic bases for production vs. nonproduction.
The genome sequence of A. niger revealed the presence of homologs of the fumonisin (fum) and ochratoxin (ota) biosynthetic gene clusters that had been described in other fungi (Geisen et al., 2006; Alexander et al., 2009; Gallo et al., 2009). The A. niger fum cluster consists of homologs of 10 genes that had been previously characterized in the Fusarium fum cluster and an additional gene, sdr1, that is not present in Fusarium (Figure 1; Baker, 2006; Susca et al., 2014a). The functions of ota genes in ochratoxin biosynthesis have not been characterized as extensively as fum genes in fumonisin biosynthesis. Only the polyketide synthase gene (for convenience in this study we will refer to it as ota1), and the non-ribosomal peptide synthetase (NRPS) gene (ota2) have been subjected to functional analysis and demonstrated to be required for ochratoxin production (Geisen et al., 2006; Gallo et al., 2012, 2014). The three other genes, ota3–ota5, located upstream of ota1 and ota2 are hypothesized to be part of the cluster because of their proximity to ota1 and ota2, and because they exhibit expression patterns similar to ota1 and ota2 (Ferracin et al., 2012; Gil-Serna et al., 2015). In addition, the presence of a chlorine atom in ochratoxin A is consistent with the predicted halogenase function of the ota5-encoded protein (Geisen et al., 2006; Gallo et al., 2012). Likewise, the presence of oxygen atoms in the structures of multiple ochratoxins is consistent with the predicted cytochrome P450 monooxygenase function of the ota3-encoded protein. Fungal gene clusters responsible for synthesis of secondary metabolites, including mycotoxins, often include a transcription factor that controls expression of the cluster genes (Keller et al., 2005; Brakhage, 2013). Thus, the predicted bZIP transcription factor function of the ota4-encoded protein is also consistent with it being part of the ochratoxin cluster.
Figure 1.
The fumonisin biosynthetic (fum) and ochratoxin biosynthetic (ota) gene clusters in A. niger, as well as structures of an analog of each of the corresponding mycotoxins. Arrows represent genes and point in the direction of transcription. Due to space limitations, only numbers are given for some fum and ota genes (i.e., in the fum cluster, 19, 15, and 21 indicate fum19, fum15, and fum21 respectively). The predicted functions of the ota genes based on sequence homology to genes of known function are indicated below each gene: PKS for polyketide synthase; NRPS for non-ribosomal peptide synthetase; P450 for cytochrome P450 monooxygenase; TF for bZIP transcription factor, and HAL for halogenase. ota1, ota2, ota3, ota4, and ota5 correspond to gene models An15g07880, An15g07890, An15g07900, An15g07910, and An15g07920, respectively, in A. niger strain CBS 513.88 (Pel et al., 2007; Ferracin et al., 2012) and gene models 151162, 402289, 398584, 407763, and 51750, respectively, in the genome database for A. carbonarius strain ITEM 5010 at the Joint Genome Institute (http://jgi.doe.gov/carbonarius/) (Nordberg et al., 2014). In the fum cluster, the gene labeled as s is the dehydrogenase gene sdr1 (Susca et al., 2014a).
Although fumonisin and ochratoxin production have been reported in both A. niger and A. welwitschiae, not all strains of these species produce the mycotoxins in laboratory culture (Susca et al., 2010, 2014a; Varga et al., 2010; Frisvad et al., 2011; Storari et al., 2012; Palumbo et al., 2013; Gherbawy et al., 2015; Massi et al., 2016). PCR, Southern and genome sequence data indicate that FB-nonproducing strains of A. welwitschiae have a partially deleted fum cluster. An analysis of a collection of FB-nonproducing isolates of A. welwitschiae from grapes grown in the Mediterranean Basin indicated that all the isolates exhibited the same eight-gene deletion within the fum cluster (Susca et al., 2014a), whereas, A. welwitschiae isolates from raisins produced in California exhibited two different, although similar, patterns of gene deletion within the fum cluster (Palumbo et al., 2013). In contrast, the same types of analyses indicated that FB-nonproducing isolates of A. niger from Mediterranean grapes and Californian raisins had an intact fum cluster (Palumbo et al., 2013; Susca et al., 2014a). Sequence analysis indicated that in the clusters of FB-nonproducing isolates of A. niger, the fum gene homologs were functional; i.e., the genes did not have insertions, deletions, transitions or transversions predicted to render the genes nonfunctional (Susca et al., 2014a). Together, these findings suggested that the genetic basis for FB nonproduction differs in A. niger and A. welwitschiae.
Differences in homologs of the ota cluster in A. niger and A. welwitschiae have received less attention than the fum cluster. However, the genome sequence of OTA-producing A. niger strain CBS 513.88 has an intact ota cluster, whereas the genome sequence of OTA-nonproducing strain ATCC 1015 has a 22-kb deletion that includes ota1–ota5 (Andersen et al., 2011; Ferracin et al., 2012). Furthermore, in a recent study of a collection of 89 A. niger isolates and 86 A. welwitschiae isolates recovered from multiple crops in Brazil, analysis of the presence of ota1 and ota5 by PCR indicated that most OTA-nonproducing isolates of both species lacked the two genes.
The objective of the current study was to gain further insight into the biodiversity of A. niger and A. welwitschiae with respect to variation in OTA and FB production as well as variation in the gene content of the fum and ota clusters. To meet this objective, we examined a collection of 92 isolates from multiple crops covering a wider range of geographic locations than previously included in one study. The isolates were examined for FB and OTA production as well as for the presence and absence of 10 genes within the fum cluster and 4 genes within the ota cluster. The findings indicate that mycotoxin production varies among the isolates, provide evidence for novel patterns of gene deletion within the fum cluster in FB-nonproducing isolates of A. welwitschiae, and provide evidence for deletion of the entire ota cluster in OTA-nonproducing isolates of both A. niger and A. welwitschiae.
Materials and methods
Fungal strains
In this study, we examined a collection of 92 fungal isolates that were previously identified by colony morphology as A. niger-A. welwitschiae. The isolates were selected to represent strains recovered from a diversity of crop species and geographic origins (Table 2). Thirty-four isolates originated from raisins, 32 from grapes, 8 from cashew nuts, 7 from maize, 5 from pistachio, 3 from almonds, and 3 from walnuts. Furthermore, 33 of the isolates were from Turkey, 19 from Italy, 10 from USA, 7 from Brazil, 8 from Argentina, 6 from Greece, 4 from Portugal, 2 from Chile, and one isolate each from China, India, Iran, and Spain. For comparison purposes, the study also included the A. niger type strain (ITEM 4501) as well as a second A. niger reference strain (ITEM 9568, syn CBS513.88) and a A. welwitschiae reference strain (ITEM 4552) (Perrone et al., 2011; Hong et al., 2013). Thirty-six of the isolates from grape were previously identified to species, as well as previously analyzed for fumonisin production and the presence of fum genes (Susca et al., 2014a). All isolates were obtained from the Agri-Food Toxigenic Fungi Culture (ITEM) collection at the Institute of Sciences of Food Production CNR-ISPA. Detailed information about the strain features (year of isolation, depositor, toxin production capability, etc.) are available from the ITEM database (http://www.ispa.cnr.it/Collection).
Growth conditions and DNA extraction
For each strain, conidia were scraped from the surface of PDA cultures and inoculated in 100 ml of Wikerham's medium (40 g of glucose, 5 g of peptone, 3 g of yeast extract, and 3 g of malt extract, distilled water up to 1 L). The resulting liquid cultures were then incubated with shaking (150 rpm) at 25°C for 2 days. Mycelia were filtered, lyophilized and ground using iron beads in a Mill MM 301 mixer (Retsch, Germany). DNA isolation was done with “Wizard® Magnetic Purification System for Food” kit (Promega, USA) according to manufacturer's instructions and starting from 10 mg of ground dried mycelium. Quality and yield of resulting DNA were evaluated by agarose gel electrophoresis, and final quantification was performed with NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA).
Fungal species identification
Species identification was performed through a DNA sequence-based approach by comparing a 650-nt, PCR-amplified fragment of the calmodulin gene (CaM) from each isolate against the corresponding sequence from reference strains of A. niger (ITEM 4501, AY585536) and A. welwitschiae (ITEM 4509, syn CBS 557.65, AJ964874) (Perrone et al., 2011; Hong et al., 2013). PCR products obtained using CL1/CL2A primers (O'Donnell et al., 2000) were purified with the enzymatic mixture EXO/FastAP (Exonuclease I and Thermosensitive Alkaline Phosphatase, Thermo Fisher Scientific, USA) and submitted to bidirectional sequencing with the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc., USA). Labelled amplicons were purified by gel filtration through Sephadex G-50 (GE Healthcare, UK) and analyzed with the ABI PRISM 3730 Genetic Analyzer (Applied Biosystems Inc., USA). Multiple alignment of CaM sequences, and similarity score against reference sequences were performed by Bionumerics 5.1 software (Applied Maths, Belgium), determining the best-scoring reference sequence of the similarity output (≥99% against references sequences).
Mycotoxins analysis
Fumonisin B2 (FB2) and OTA production capabilities were assessed according to Frisvad et al. (2007) for each strain, on CY20S and YES agar media, respectively, as described below. In vitro production of OTA and FB2 was examined on a subset of 88 and 95 isolates, respectively, including species-reference strains. For FB2 analysis, each CY20S agar cultures (1 g) was extracted with 5 ml of methanol/water (70:30, v/v) on orbital shaker for 60 min. One hundred microliter were diluted with 900 μl of acetonitrile/water (30:70, v/v) and then filtered using RC 0.2 μm filters (Phenomenex, USA). Fifty microliter of the extract were derivatized with 50 μL of o-phtaldialdehyde (OPA) and mixed for 50 s using an Agilent 1100 HPLC autosampler (Agilent Technologies, USA). One hundred microliter of the resulting derivatized samples were injected by full loop at 3 min after adding the OPA reagent for fumonisin analysis. The analytical column was a SymmetryShield RP18 (15 cm × 4.6 mm i.d., 5 μm) with a guard column inlet filter (0.5 μm × 3 mm i.d.) (Rheodyne, Idex Corporation, USA). The mobile phase consisted of 57% of solvent A (water/acetic acid, 99:1, v/v) and 43% of solvent B (acetonitrile/acetic acid, 99:1, v/v). The initial composition was kept constant for 5 min, then solvent B was linearly increased to 54% over 21 min, then up to 58% at 25 min and kept constant for 5 min. The column temperature was 30°C. The flow rate of the mobile phase was 0.8 ml/min. The fluorometric detector was set at wavelengths 335 nm for excitation and 440 nm for emission. FB2 was quantified by measuring peak areas, and comparing them to a calibration curve obtained with standard solutions. The detection limit for FB2 was 0.1 μg/g based on a signal-to-noise ratio of 3:1.
For extraction and detection of OTA, YES agar (1 g) cultures were extracted with 5 ml of methanol/water (70:30, v/v) on an orbital shaker for 60 min. One hundred microliter were diluted with 900 μl of acetonitrile/water (30:70, v/v) and then filtered using RC 0.2 μm filters (Phenomenex, USA). Fifty microliter of the extract were injected into an Agilent 1100 HPLC autosampler (Agilent Technologies, USA) with a full loop injection system. The analytical column was a Zorbax SB-C18 (4.6 mm × 150 mm i.d., 5 μm) with a guard column inlet filter (0.5 μm × 3 mm i.d.) (Rheodyne, Idex Corporation, USA). The mobile phase consisted of a mixture of acetonitrile/water/glacial acetic acid (99:99:2, v/v/v) at a flow rate of 1 ml/min. The fluorometric detector was set at 340 nm for excitation and 460 nm for emission. Ochratoxin A was measured by comparing peak areas with a calibration curve obtained with OTA standard solutions (Sigma-Aldrich, USA). The detection limit for OTA was 5 μg/kg based on a signal-to-noise ratio of 3:1.
PCR primers and amplification conditions
The presence/absence of 10 genes and 3 intergenic regions within the putative fum cluster was assessed by PCR-based approach using primers previously described by Susca et al. (2014a). The amplicons represented 13 regions along almost the entire length of the FUM cluster described in A. niger strain ATCC 1015 (GenBank accession ACJE00000000) and strain CBS513.88 (GenBank accessions AM269948; AM270415). Eleven of the amplicons corresponded to the following 10 fum genes (listed in their order along the chromosome): fum1, fum15, fum21 (two amplicons), fum14, fum13, fum8, fum3, fum7, fum10, and fum6. In addition, one amplicon corresponded to a region downstream of fum6 (fum6 ds), and another amplicon corresponded to the fum19-fum15 intergenic region (fum19-15 IGR). The presence and absence of four genes within the ota cluster was also assessed by a PCR-based approach using four primer sets. One primer set (pks15ksF/R) targeted the polyketide synthase gene ota1 and was previously described (Ferracin et al., 2012). The three other primer sets were designed during the course of the current study and are based on genomic DNA sequence of A. niger strain CBS 513.88, which was available from the GenBank database of the National Center for Biotechnology Information (NCBI). Each of the three latter primer sets were designed to amplify a fragment of a different ota gene: ota2, ota3, and ota5. The primer pairs were designed using Primer Express® 2.0 software (Applied Biosystems Inc., USA). Table 1 summarizes the target genes, accession numbers, predicted functions, sequences of and amplicon sizes for each of the primer sets designed to amplify fum and ota gene fragments.
Table 1.
Information on target genes/regions and primers used for PCR in this study.
| Gene/Intergenic region name | Gene locus tag | Accession number | Predicted gene function | Primer sequences (5g′-3′) | Amplicon size | References | |
|---|---|---|---|---|---|---|---|
| fum cluster | downstream fum6 | – | NT_166518.1 | Intergenic region | f: CAAAAGACACCGCCCGTCT r: TTGACGCCCTGTACAAGGC |
667 bp | Susca et al., 2014a |
| fum6 | ANI_1_2654014 | NT_166518.1 | NADPH-cyt P450 reductase | f: CTGTGAGGCCCTGGCACTT r: TCTGCCGGAGCTCAACGTA |
849 bp | Susca et al., 2014a | |
| fum10 | ANI_1_2658014 | NT_166518.1 | Peroxisomal-coenzyme A synthetase | f: GTCATTATTCCTCCGGCCCT r: TGGGATTCGAAAGCATACCG |
651 bp | Susca et al., 2014a | |
| fum7 | ANI_1_2660014 | NT_166518.1 | Fe-containing alcohol dehydrogenase | f: CAACAGCCCGAATCCCAGTA r: GCTCAGTCTTGCCCATCGTG |
681 bp | Susca et al., 2014a | |
| fum3 | ANI_1_892014 | NT_166518.1 | Dioxygenase | f: TACCATGGACCACTTTCCCG r: AAGTTCCTCAAGCGGCAGTC |
651 bp | Susca et al., 2014a | |
| fum8 | ANI_1_894014 | NT_166518.1 | a-oxoamine synthase | f: TTCGTTTGAGTGGTGGCA r: CAACTCCATASTTCWWGRRAGCCT |
651 bp | Susca et al., 2014a | |
| fum13 | ANI_1_2662014 | NT_166518.1 | short chain dehydrogenase | f: ATGCTCTTCACCTCCTCCGG r: CACTCAACGAGGAGCCTTCG |
651 bp | Susca et al., 2014a | |
| fum14 | ANI_1_2664014 | NT_166518.1 | NRPS-like condensation domain | f: TTGGGCTGATGTGCTCTGTC r: CCTCGTAGACGTAATTGAGTAGTCCT |
730 bp | Susca et al., 2014a | |
| fum21 region I | – | NT_166518.1 | Zn(II)2Cys6 DNA-binding protein | f: CATTTCATGGGACCTCAGCC r: AAGCACAGGTTCCGAATTTGA |
703 bp | Susca et al., 2014a | |
| fum21 region II | – | NT_166518.1 | Zn(II)2Cys6 DNA-binding protein | f: GGGTCCCATTGCCTCAATT r: CAATGGAGTCGACGGTGTCAC |
705 bp | Susca et al., 2014a | |
| fum15 | ANI_1_2668014 | NT_166518.1 | Cytochrome P450 monoxygenase | f: CGATTGGTAGCCCGAGGAA r: CTTGATATTGCGGAGTGGTCC |
701 bp | Susca et al., 2014a | |
| fum 19-15 IGR | – | NT_166518.1 | Intergenic region | f: ACACCGCGAGAATTCCATG r: GCAGGCTGGTAGTAGCGACAT |
868 bp | Susca et al., 2014a | |
| fum1 | ANI_1_2672014 | NT_166518.1 | Polyketide synthase | f: GGGTTCCAGGCAGAATCGTAC r: GAACTCACATCCTTTTGGGCC |
701 bp | Susca et al., 2014a | |
| ota cluster | ota1 | ANI_1_1836134 | NT_166530.1 | Polyketide synthase | f: CAATGCCGTCCAACCGTATG r: CCTTCGCCTCGCCCGTAG |
776 bp | Ferracin et al., 2012 |
| ota2 | ANI_1_1832134 | NT_166530.1 | Nonribosomal peptide-synthetase | f: GGGAAYRCTGAYGTCGTGTTT r: TCCCACGAGCAWACAGCCTC |
644 bp | This study | |
| ota3 | ANI_1_1830134 | NT_166530.1 | Cytochrome P450 | f: TTAGACAAACTGCGCGAGGA r: GCGTCGCTATGCCCAGATA |
613 bp | This study | |
| ota5 | ANI_1_1826134 | NT_166530.1 | radH flavin-dependent halogenase | f: TCCCTCGGTAAGAGTATCCTCGT r: GCGAGTTCTTGGTTCATGACG |
845 bp | This study | |
| CaM | ANI_1_1116184 | NT_166539.1 | Calmodulin | f: GARTWCAAGGAGGCCTTCTC r: TTTTTGCATCATGAGTTGGAC |
650 bp | O'Donnell et al., 2000 |
The locus tags and accession numbers are GenBank/NCBI gene and contig designations, respectively, for A. niger strain CBS 513.88. Predicted gene functions are based on sequence homology to genes with known functions present in the GenBank/NCBI database.
PCR reactions were carried out in a 10 μl volume containing: 0.25 U of HotMaster™ Taq DNA Polymerase (5 Prime, Germany), 1x HotMaster™ Taq DNA Polymerase buffer, 300 nM each primer, 200 μM deoxynucleotide mix (5 Prime, Germany) and approximately 20 ng of fungal DNA template. The PCR amplification program for fum genes was the same for the 13 primer sets, and was as follows: denaturation at 95°C for 2 min; 35 cycles of denaturation at 94°C for 50 s, annealing at 58°C for 50 s, extension at 72°C for 50 s; final extension at 72°C for 7 min, followed by cooling at 4°C until samples recovery. The PCR amplification program for genes in the ota cluster was the same for the 4 primer sets, and was as follows: denaturation at 95°C for 2 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 30 s; final extension at 72°C for 7 min, followed by cooling at 4°C until samples recovery. Primer annealing temperature (60°C) for 3 OTA primer sets, designed in the present study, was empirically determined through gradient analysis.
Amplification of the approximately 650-bp fragment of the calmodulin gene, using CL1/CL2A primer (O'Donnell et al., 2000), was used as internal control for PCR assays. A. niger CBS 513.88 (ITEM 9568) was used as reference strain for all PCR experiments, because its genome sequence includes all fum and ota genes (Pel et al., 2007; Andersen et al., 2011). PCR products were electrophoresed in a 1.0% w/v agarose gel, stained with GelRed™ (Biotium Inc., USA) and visualized under UV transillumination.
Examination of fum and ota cluster regions in genome sequences
fum and ota cluster regions in strains of A. niger and A. welwitschiae were retrieved from newly and previously generated genome sequences of these fungi. The previously generated sequences were from four strains of A. niger (ATCC 1015, ATCC 13496, CBS 513.88, and ITEM 10355) and three strains of A. welwitschiae (ATCC 13157, ITEM 7468, ITEM 11945). The ATCC 1015, ATCC 13157, and ATCC 13496 sequences were examined using the Joint Genome Institute (JGI) Genome Portal website (http://genome.jgi.doe.gov/), and the CBS 513.88 sequence was examined using the NCBI website (http://www.ncbi.nlm.nih.gov/). The genome sequences for strains ITEM 7468, ITEM 10355, and ITEM 11945 were previously generated by Susca et al. (2014a).
In order to examine the fum and ota cluster regions in selected isolates of A. welwitschiae, whole genome sequences were generated for the following isolates: ITEM 4552, ITEM 6142, ITEM 6144, ITEM 7097, ITEM 10353, ITEM 10929, ITEM 10932, ITEM 11209, ITEM 11980, ITEM 11984, ITEM 14309, ITEM 15179, ITEM 15309. Each isolate was grown in YEDP medium (0.1 yeast extract, 0.1 peptone, 2% dextrose) for 2 days at room temperature with shaking at 200 rpm. Mycelia were harvested by filtration, lyophilized, ground to a powder, and genomic DNA was extracted using the method described by Raeder and Broda (1985). The resulting DNA was further purified with the UltraClean DNA purification kit according to the specifications of the manufacturer (MoBio Laboratories, Inc.). DNA libraries were prepared using a NExtera XT DNA library Preparation Kit, and sequence data were generated with an Illumina MiSeq sequencing platform as specified by the manufacturer (Illumina, San Diego, California). CLC Genomics Workbench (CLC bio, Qiagen, Aarhus, Denmark) was used to process the resulting sequence reads and obtain a de novo assembly of each genome.
For ITEM isolates, sequences of the fum and ota cluster regions were retrieved from the genome sequences via the BLASTn (Altschul et al., 1997) function in CLC Genomics Workbench and using A. niger homologs of fum, ota, and cluster flanking genes, which have been previously reported (Pel et al., 2007; Gallo et al., 2012, 2014; Susca et al., 2014a). Sequences corresponding to the fum and ota cluster regions from strains have been deposited into the GenBank database at NCBI as accessions KX267735–KX267737. Phylogenetic analysis of homologous sequences from the ota cluster region was done using the phylogenetic analysis software MEGA 5 (Tamura et al., 2011). The sequences were aligned using Muscle, and the resulting alignment was subjected to a model selection analysis to determine the best nucleotide substitution model and to maximum likelihood analysis.
Results
Fungal identification
The species identity of the 92 black aspergilli isolates, which had A. niger morphology in culture based on characteristic conidial morphology (Kozakiewicz, 1989), was determined by comparison of the DNA sequence of a PCR-amplified caM fragment from each isolate to the caM sequences for reference strains of A. niger and A. welwitschiae. Previous analyses have shown that the caM sequence can be used to distinguish between A. niger and A. welwitschiae (Perrone et al., 2011). The comparisons revealed that sequences from 67 isolates were 99.75–100.00% identical to the reference sequence from A. welwitschiae strains ITEM 4509 and ITEM 4552, and sequences from 25 isolates was 99.75–100.00% identical to the reference sequence from A. niger strain ITEM 4501. Previous phylogenetic analyses of some of the isolates employed in this study indicate that such differences in sequence identity are reliable indicators of the species identity of A. niger and A. welwitschiae (Susca et al., 2014a,b). Based on this, we concluded that 67 isolates in the collection were A. welwitschiae, and 25 were A. niger.
The sample sizes of isolates from almonds, cashews, maize, pistachios and walnuts ranged from two to eight, making it difficult to evaluate whether one species occurred more frequently than the other on these crops. However, the sample sizes from grapes and raisins were 33 and 34 respectively, and A. welwitschiae occurred more frequently on both; 82% of the isolates from these two crops were A. welwitschiae, and 18% were A. niger.
Mycotoxin production
The levels of FB2 and OTA produced by the collection of A. niger and A. welwitschiae isolates varied markedly. Among producing isolates, the levels of OTA produced (30–590 μg/kg) were typically higher than the levels of FB2 (0.1–44.4 μg/g). The frequency of FB2-producing and nonproducing isolates was markedly different in the two species. That is, a majority of A. niger isolates (74%) produced FB2, but a minority of A. welwitschiae isolates (16%) produced FB2 (Table 2). In contrast, a minority of isolates of both species produced OTA: 4% of A. niger isolates and 25% of A. welwitschiae isolates. The frequency of isolates that produced both mycotoxins was similar in the two species: 4% of A. niger isolates and 8% of A. welwitschiae isolates (Table 2). In contrast, the frequency of isolates that produced neither mycotoxin differed markedly: 13% of A. niger isolates and 68% of A. welwitschiae isolates (Table 2).
Table 2.
Strain designations, crop and geographic origins, FB2 and OTA production, and mycotoxin biosynthetic gene amplification patterns for A. niger and A. welwitschiae isolates examined in this study.
| ITEM | Species | Substrate | Origin | FB2 (μg/g) | fum amplicon pattern | OTA (μg/kg) | ota amplicon pattern |
|---|---|---|---|---|---|---|---|
| 4501 | A. niger | 0.4 | f-1 | − | o-1 | ||
| 9568 | A. niger | + | f-1 | + | o-1 | ||
| 11461 | A. niger | Almonds | USA | 6.4 | nd | − | o-2 |
| 11432 | A. niger | Cashew nuts | Brazil | 18.7 | nd | − | o-2 |
| 11433 | A. niger | Cashew nuts | Brazil | 36.7 | nd | − | o-2 |
| 11446 | A. niger | Cashew nuts | Brazil | 41.7 | nd | − | o-2 |
| 11449 | A. niger | Cashew nuts | Brazil | 40.4 | nd | − | o-2 |
| 11451 | A. niger | Cashew nuts | Brazil | 29.1 | nd | − | o-2 |
| 11558 | A. niger | Cashew nuts | India | 3.2 | nd | − | o-2 |
| 5218 | A. niger | Grape | Greece | − | f-1 | − | o-2 |
| 5219 | A. niger | Grape | Greece | − | f-1 | − | o-2 |
| 5276 | A. niger | Grape | Greece | 0.6 | f-1 | − | o-2 |
| 7090 | A. niger | Grape | Italy | − | f-1 | − | o-2 |
| 7091 | A. niger | Grape | Italy | 3.3 | f-1 | − | o-2 |
| 10355 | A. niger | Grape | Italy | − | f-1 | − | o-2 |
| 15310 | A. niger | Maize | USA | − | f-1 | n.t. | nd |
| 15342 | A. niger | Maize | USA | − | f-1 | n.t. | nd |
| 15369 | A. niger | Maize | USA | − | f-1 | n.t. | nd |
| 15374 | A. niger | Maize | USA | 0.1 | f-1 | n.t. | nd |
| 12736 | A. niger | Pistachio | Italia | 9.5 | nd | − | o-2 |
| 10939 | A. niger | Pistachio | USA | 4.3 | nd | − | o-2 |
| 10924 | A. niger | Raisins | Turkey | 2.8 | nd | − | o-2 |
| 10927 | A. niger | Raisins | Turkey | 10.7 | nd | − | o-2 |
| 11930 | A. niger | Raisins | Turkey | 7.8 | nd | − | o-2 |
| 11941 | A. niger | Raisins | Turkey | 1.1 | nd | − | o-2 |
| 14275 | A. niger | Raisins | Turkey | 0.6 | f-1 | − | o-2 |
| 14281 | A. niger | Raisins | Turkey | 0.5 | f-1 | − | o-2 |
| 11196 | A. welwitschiae | Almonds | Italy | − | f-13 | − | o-2 |
| 11197 | A. welwitschiae | Almonds | Italy | − | f-13 | − | o-2 |
| 11435 | A. welwitschiae | Cashew nuts | Brazil | 19.2 | f-1 | − | o-2 |
| 11447 | A. welwitschiae | Cashew nuts | Brazil | 44.4 | f-1 | − | o-2 |
| 11774 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 11778 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 11786 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 11828 | A. welwitschiae | Grape | Argentina | − | f-13 | 40.0 | nd |
| 11836 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 11844 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 11854 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 11867 | A. welwitschiae | Grape | Argentina | − | f-13 | − | o-2 |
| 5253 | A. welwitschiae | Grape | Greece | − | f-13 | − | o-2 |
| 5267 | A. welwitschiae | Grape | Greece | − | f-13 | − | o-2 |
| 5277 | A. welwitschiae | Grape | Greece | 10.9 | f-1 | − | o-2 |
| 4717 | A. welwitschiae | Grape | Italy | − | f-13 | − | o-2 |
| 4853 | A. welwitschiae | Grape | Italy | − | f-13 | − | o-2 |
| 4858 | A. welwitschiae | Grape | Italy | − | f-13 | − | o-2 |
| 4863 | A. welwitschiae | Grape | Italy | − | f-13 | − | o-2 |
| 6122 | A. welwitschiae | Grape | Italy | − | f-13 | − | nd |
| 6126 | A. welwitschiae | Grape | Italy | − | f-13 | 280.0 | nd |
| 6127 | A. welwitschiae | Grape | Italy | − | f-12 | − | nd |
| 6128 | A. welwitschiae | Grape | Italy | − | f-12 | − | o-2 |
| 7097 | A. welwitschiae | Grape | Italy | 7.5 | f-1 | 590.0 | o-1 |
| 7468 | A. welwitschiae | Grape | Italy | − | f-13 | − | o-2 |
| 10353 | A. welwitschiae | Grape | Italy | − | f-9 | − | o-2 |
| 4552 | A. welwitschiae | grape | Portugal | − | f-13 | 45.0 | o-1 |
| 6140 | A. welwitschiae | Grape | Portugal | − | f-7 | − | o-2 |
| 6142 | A. welwitschiae | Grape | Portugal | − | f-7 | 545.0 | o-1 |
| 6144 | A. welwitschiae | Grape | Portugal | − | f-9 | 380.0 | nd |
| 4947 | A. welwitschiae | Grape | Spain | − | f-13 | − | o-2 |
| 15095 | A. welwitschiae | Maize | Italy | 0.1 | f-1 | n.t. | nd |
| 15179 | A. welwitschiae | Maize | Italy | − | f-3 | n.t. | nd |
| 15309 | A. welwitschiae | Maize | USA | − | f-2 | n.t. | nd |
| 13288 | A. welwitschiae | Pistachio | Iran | − | f-13 | − | o-2 |
| 11734 | A. welwitschiae | Pistachio | USA | − | f-13 | − | o-2 |
| 12780 | A. welwitschiae | Pistachio | USA | − | f-12 | − | o-2 |
| 10635 | A. welwitschiae | Raisins | Turkey | − | f-12 | − | o-2 |
| 10636 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | o-2 |
| 10637 | A. welwitschiae | Raisins | Turkey | − | f-12 | − | o-2 |
| 10929 | A. welwitschiae | Raisins | Turkey | − | f-4 | − | nd |
| 10932 | A. welwitschiae | Raisins | Turkey | − | f-11 | − | o-2 |
| 10935 | A. welwitschiae | Raisins | Turkey | 2.9 | f-1 | − | o-2 |
| 11925 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | o-2 |
| 11931 | A. welwitschiae | Raisins | Turkey | − | f-11 | − | o-2 |
| 11943 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | o-2 |
| 11945 | A. welwitschiae | Raisins | Turkey | 25.3 | f-1 | − | o-2 |
| 11948 | A. welwitschiae | Raisins | Turkey | − | f-12 | 30.0 | nd |
| 11980 | A. welwitschiae | Raisins | Turkey | − | f-10 | 50.0 | nd |
| 11982 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | o-2 |
| 11984 | A. welwitschiae | Raisins | Turkey | − | f-12 | 55.0 | nd |
| 11990 | A. welwitschiae | Raisins | Turkey | − | f-12 | − | o-2 |
| 12113 | A. welwitschiae | Raisins | Turkey | − | f-12 | − | o-2 |
| 12115 | A. welwitschiae | Raisins | Turkey | − | f-13 | 40.0 | nd |
| 12246 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | nd |
| 12591 | A. welwitschiae | Raisins | Turkey | − | f-5 | 110.0 | nd |
| 12594 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | o-2 |
| 14297 | A. welwitschiae | Raisins | Turkey | 9.5 | f-1 | 35.0 | nd |
| 14303 | A. welwitschiae | Raisins | Turkey | 3.5 | f-1 | 45.0 | nd |
| 14305 | A. welwitschiae | Raisins | Turkey | 3.6 | f-1 | 45.0 | nd |
| 14307 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | nd |
| 14308 | A. welwitschiae | Raisins | Turkey | 1.0 | f-1 | 40.0 | nd |
| 14309 | A. welwitschiae | Raisins | Turkey | − | f-8 | 45.0 | nd |
| 14310 | A. welwitschiae | Raisins | Turkey | − | f-13 | − | nd |
| 12120 | A. welwitschiae | Raisins | USA | − | f-13 | − | o-2 |
| 10908 | A. welwitschiae | Walnuts | Chile | − | f-13 | − | o-2 |
| 10910 | A. welwitschiae | Walnuts | Chile | − | f-13 | − | o-2 |
| 11209 | A. welwitschiae | Walnuts | China | − | f-6 | − | o-2 |
−: Not detected with the limit of detection (LOD): 5 μg/kg (OTA), 0.1 μg/g (FB2). nt: not tested; nd: not determined. bold type: species reference strain. +: strain reported in literature as mycotoxin producer (Andersen et al., 2011).
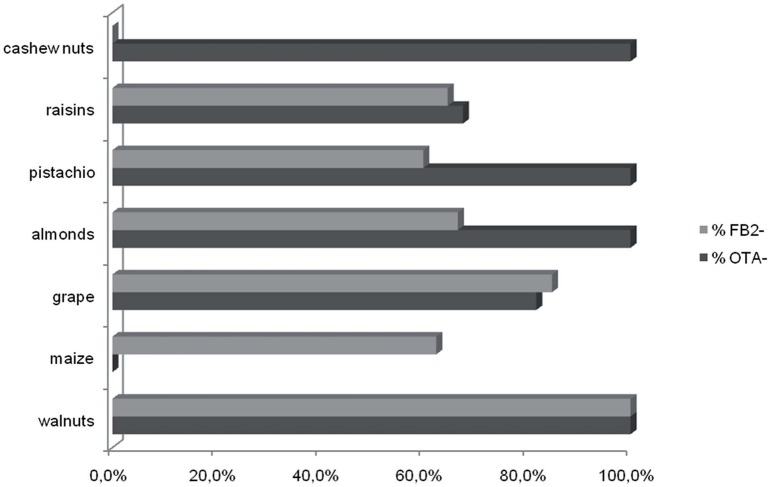
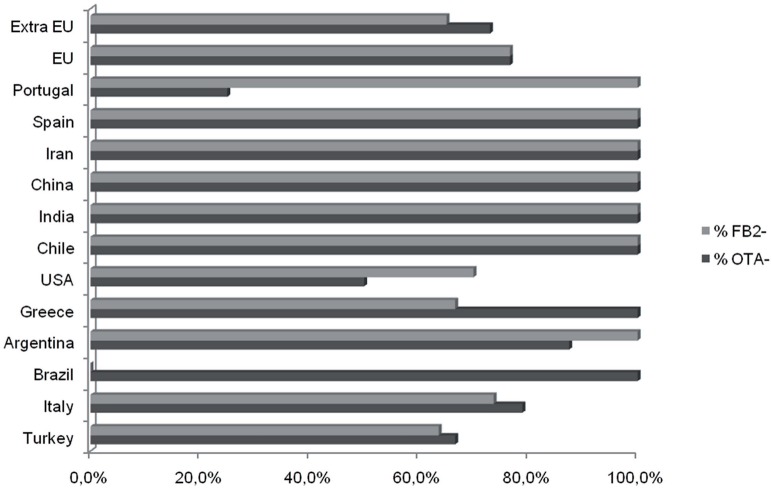
Although the sample sizes for sets of isolates recovered from a given crop were small, the data indicate some possible differences among isolates from different crops (Figure 2). As noted above, the majority of A. niger isolates produced FB2. However, all A. niger isolates from cashews and raisins produced FB2, whereas most isolates from grape and maize did not (Table 2). Although most isolates of A. welwitschiae did not produce FB2 or OTA, there was a relatively high proportion of FB2 and OTA-producing isolates from raisins: 6 of 27 isolates produced FB2, and 10 of 27 isolates produced OTA. In addition, 4 of the 5 A. welwitschiae isolates that produced both mycotoxins originated on raisins. OTA-nonproducing isolates occurred with almost equal frequencies in European (77%) and non-European (73%) regions, while FB2-nonproducing isolates occurred slightly more frequently in European (77%) than in non-European (65%) locations (Figure 3).
Figure 2.
Occurrence of FB2- and OTA-nonproducing isolates on different crops. Values are percentages for each crop.
Figure 3.
Occurrence FB2- and OTA-nonproducing isolates from different geographic origins. Values are percentages for each Country.
Variation in mycotoxin biosynthetic gene content
fum cluster genes
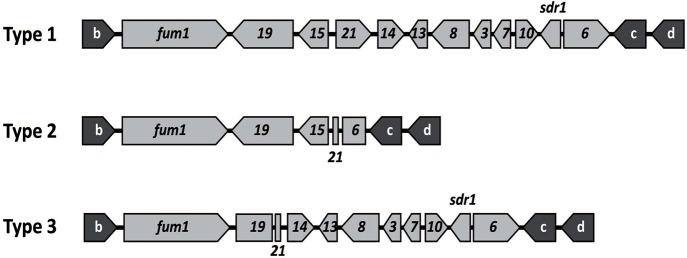
The presence and absence of the 10 genes in the fum cluster were assessed initially by the PCR assay described previously (Susca et al., 2014a) using the primers shown in Table 1. The seven FB2-producing and 7 FB2-nonproducing isolates of A. niger examined yielded amplicons for all fum genes and intergenic regions examined. These PCR results indicated that there were no apparent differences in the gene content of the fum cluster of FB-producing and nonproducing isolates of A. niger, a finding that is consistent with previous analyses of the cluster in this species (Palumbo et al., 2013; Susca et al., 2014a). A larger set of 8 FB2-producing and 57 nonproducing isolates of A. welwitschiae was also subjected to the fum-gene PCR assay. The analysis yielded 13 amplicon patterns: the two patterns previously described for FB-producing and nonproducing isolates of the species (Susca et al., 2014a) as well as 11 novel patterns. In Table 2, the previously described amplification patterns for FB-producing and nonproducing isolates of A. welwitschiae were designated as Patterns f-1 and f-2, respectively, while the 11 novel amplification patterns were designated as Patterns f-3 through f-13. To determine whether the novel amplification patterns correspond to undescribed deletions within the fum cluster, we generated whole-genome sequence data for isolates of A. welwitschiae representative of the novel patterns, except f-5. Retrieval and analysis of sequences of the fum cluster region from each genome sequence revealed that, in all isolates examined except ITEM 10929, the gene content of the fum cluster in isolates with the novel PCR amplification patterns was identical to one of the two fum cluster types previously described for A. welwitschiae (Susca et al., 2014a), and designated as cluster Types 1 and 2 in Figure 4. In Type 1, there were full-length homologs of all 11 fum genes and the sdr1 gene. In Type 2, six fum genes and sdr1 were absent, and fum6 and fum21 were truncated, whereas, fum1, fum15, and fum19 were intact. Thus, the novel fum gene amplification patterns observed in the PCR assay were not consistent with the genomic sequences.
Figure 4.
Three fum cluster types observed in newly generated genome sequences of isolates of A. welwitschiae. Type 1, an intact cluster, was observed in isolates ITEM 7097, 11209, and 15309. Type 2, a partially deleted cluster, was observed in isolates ITEM 4552, 6142, 10353, 10932, 11209, 11980, 11984, 14309, and 15309. Type 3, also a partially deleted cluster, was observed in isolate ITEM 10929 only. The gene content and arrangement in Type 1 is identical to the fum cluster previously described in the FB-producing A. welwitschiae isolate ITEM 11945 as well as all A. niger isolates examined; and the gene content and arrangement in Type 2 is identical to the fum cluster previously described in A. welwitschiae isolate ITEM 7468 (Susca et al., 2014a). As far as we are aware, Type 3 has not been described previously. Gene designations are as in Figure 1. Genes shown as arrows have full-length coding regions and are most likely functional, whereas, genes shown as rectangles/squares are truncated and therefore nonfunctional. Genes labeled b, c, and d are previously described fum cluster flanking genes (Susca et al., 2014a).
The analysis of the genome sequences of A. welwitschiae isolates ITEM 11209 and ITEM 15309 revealed that the fum cluster homologs in these two strains contained all 11 fum genes as well as sdr1. As far as we are aware, this is the first report of a full-length fum clusters in FB-nonproducing isolates of A. welwitschiae. In ITEM 15309, all fum genes appeared to be functional in that there were no insertions or deletions that would result in frameshifts or premature stop codons in the gene coding regions of any of the genes. Thus, the sequence of the fum cluster in this strain did not provide clues as to the genetic basis for the FB-nonproduction phenotype of the isolate. In ITEM 11209, all the fum genes appeared to be functional except for fum1, which had a C-to-T transition that changed codon 2163 from CGA to TGA, a stop codon. This premature stop codon would most likely render the fum1-encoded polyketide synthase nonfunctional, because it would block translation of the mRNA before synthesis of keto reductase and phosphopantetheine domains, two domains that are essential for function of the polyketide synthase. As noted above, isolate ITEM 10929 was an exception; it had a fum cluster in which fum19 and fum21 were truncated, fum15 was absent, but the other eight fum genes and sdr1 were intact and apparently functional (Figure 4). Thus, the fum cluster in ITEM 10929 differed from the two previously described cluster types in A. welwitschiae.
OTA cluster genes
The presence and absence of four ota cluster genes (ota1-ota3 and ota5) was also assessed in a subset of 69 isolates using a PCR-based assay. There were only two patterns of amplicons observed. The first pattern (Pattern o-1) consisted of amplicons for the four ota cluster genes examined. All OTA-producing isolates of both species (A. niger ITEM 9568 and A. welwitschiae ITEM 7097, ITEM 6142, ITEM 4552) exhibited Pattern o-1. The amplicon sizes were consistent with the expected sizes based on the design of PCR primers: 776 bp for ota1, 645 bp for ota2, 614 bp for ota3, and 846 bp for ota5. In the second ota PCR pattern (Pattern o-2) none of the amplicons was present. All OTA-nonproducing isolates of both A. niger (21 isolates) and A. welwitschiae (44 isolates) exhibited Pattern o-2 (Table 2). The absence of amplicons for the 4 putative ota cluster genes is consistent with the previously described 21-Kb deletion in the ota cluster region of the A. niger OTA-nonproducing strain ATCC 1015 (Andersen et al., 2011).
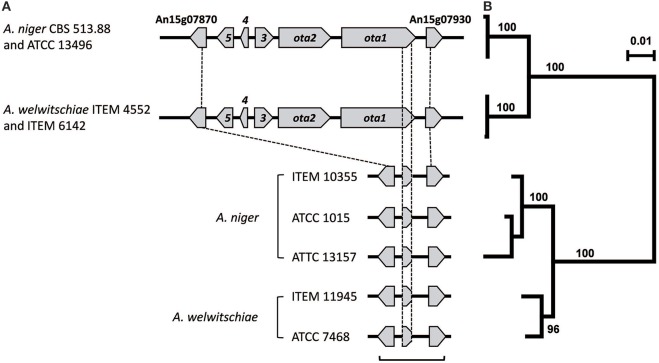
To further evaluate whether the amplification Patterns o-1 and o-2 were consistent with the presence and absence of ota genes, we examined the genome sequences from a subset of one A. niger and seven A. welwitschiae isolates for which amplification-pattern data were determined. This analysis revealed that the three A. welwitschiae isolates (ITEM 4552, ITEM 6142, and ITEM 7097) that exhibited Pattern o-1 had an intact ota cluster; i.e., they had all five ota genes, and the genes were apparently functional. Further, the sequence analysis indicated that the one A. niger isolate (ITEM 10355) and the four A. welwitschiae isolates (ITEM 7468, ITEM 10353, ITEM 10932, and ITEM 11209) with Pattern o-2 did not have any of the ota genes, except for an approximately 1100-nucleotide region near the 3′ end of ota1. These results indicate that the ota amplification patterns were consistent with the presence and absence of ota genes. Furthermore, the results of both the PCR and genome sequence data provide evidence that the OTA-nonproduction phenotype in the majority of isolates of both A. niger and A. welwitschiae results from the absence of ota genes in the isolates. Examination of previously generated genome sequences for the ota cluster confirmed that the cluster was intact in A. niger strain CBS 513.88 and absent in A. niger strain ATCC 1015 as previously reported (Andersen et al., 2011). This analysis also revealed that an intact ota cluster was present in A. niger strains ATCC 13157 and ATCC 13496, but not in A. niger strain ITEM 10355 as well as A. welwitschiae strains ITEM 7468 and ITEM 11945 (Figure 5A). Further examination of both previously generated genome sequence data and sequence data generated during this study (20 genome sequences in total) revealed that all strains examined have homologs of the ota cluster flanking genes An15g07870 and An15g07930 regardless of species and whether the strains have an intact cluster (Figure 5A). In both A. niger and A. welwitschiae sequences that lack the ota cluster, the An15g07870-An15g07930 intergenic region ranges in length from 2508 to 2800 nucleotides. 94% of this aligned intergenic sequence was homologous to three sequence elements in A. niger and A. welwitschiae strains with an intact ota cluster: 18% of the region was homologous to the sequence immediately 5′ to the An15g07870 start codon; 41% was homologous to sequence near the 3′ end of ota1; and 35% was homologous to sequence immediately 5′ to the An15g07930 start codon (Figure 5A). The identity of homologs of the three sequence elements ranged from 79 to 92% in strains with an intact vs. a deleted ota cluster. In strains that had a deleted cluster, the sequence corresponding to ota1 was 1127–1178 nucleotides in length and was homologous to nucleotides 7012–8126 of the intact ota1 coding region (with introns included). Variation in the length of the ota1-fragment homologs resulted from deletions and insertions within the homologs. Regardless of species, the ota1-fragment homologs began and ended at the same positions, nucleotides 7012 and 8126 respectively, of the intact ota1 coding region. The homology of the three sequence elements within the An15g07870-An15g07930 intergenic region facilitated alignment and phylogenetic analysis of the sequences. The tree inferred from the analysis indicated that, regardless of species, sequences from strains with the ota cluster deletion are more closely related to one another than to sequences from strains with the intact ota cluster (Figure 5B).
Figure 5.
(A) Comparison of the intact and deleted ota cluster region in selected isolates of A. niger and A. welwitschiae. Arrows and gene designations are as in Figure 1. An15g07870 and An15g07930 are genes flanking each side of the ota cluster; and the An15g07870 and An15g07930 designations are the original gene model designations for GenBank accessions for the genome sequence of A. niger strain CBS 513.88. (B) Maximum likelihood tree inferred from the An15g07870-An15g07930 region: i.e., the region spanned by the bracket at the bottom of (A) in strains that lack an ota cluster as well as the homologous sequences from strains that have an intact cluster. The scale at the upper right indicates the number of substitutions per site. On the JGI website, ATCC 13157 is classified as A. phoenicis. Although A. phoenicis is considered to be synonymous with A. welwitschiae (Hong et al., 2013), our analysis indicates that ATCC 13157 is a strain of A. niger sensu stricto (Figure S1).
Discussion
The black aspergilli A. niger and A. welwitschiae are used in fermentation of food and beverages. However, concerns about the safety of these fungi have been raised with the discovery that some isolates can produce the mycotoxins FBs and OTA (Frisvad et al., 2007, 2011; Susca et al., 2014b). As a result, multiple studies have been initiated to investigate the occurrence and distribution of FB and OTA production as well as the genetic basis for nonproduction in both industrial and field strains of these fungi. The studies indicate that both species exist as mixed populations of FB-producing or FB-nonproducing individuals as well as OTA-producing or OTA-nonproducing individuals. In addition, all possible combinations of FB/OTA production and nonproduction have been reported. That is, isolates that are FB-producing but OTA-nonproducing, FB-nonproducing but OTA-producing, FB-nonproducing and OTA-nonproducing, or FB-producing and OTA-producing have been reported. The FB-producing and OTA-producing phenotype has been reported less frequently than the other phenotypes: 15 of 175 isolates examined for both mycotoxins in a study by Massi et al. (2016), and six of 88 isolates examined for both mycotoxins in the current study (Table 2).
Published surveys of FB and OTA production in A. niger and A. welwitschiae generally do not indicate whether the collections of isolates examined represent random samples or biased selections of isolates. As a result, caution must be exercised when drawing conclusions about trends in production and nonproduction of isolates from multiple crops and geographic regions that appear to be evident from comparisons of the surveys. Nevertheless, a comparison of results of the current study and five previously reported studies indicate some trends. First, the results from all studies indicate that a majority of A. niger isolates examined can produce FBs (Table 3). In contrast, most of the studies indicate that only a minority of A. welwitschiae isolates can produce FBs (Table 3). The one exception to this is from a collection of A. welwitschiae isolates recovered from onion grown in Saudi Arabia (Gherbawy et al., 2015), where approximately half (48%) of the isolates produced FBs (Table 3). The studies also indicate that only a minority of both A. niger and A. welwitschiae isolates can produce OTA (Table 3). In previous studies, 0–1% of A. welwitschiae isolates produced OTA. In current study, by contrast, a substantially higher percentage (25%) of A. welwitschiae isolates produced OTA. Thus, it is possible that environmental factors such plant species, location, climate and/or agricultural practices can alter the frequency of FB- and OTA-producing isolates.
Table 3.
Comparison of results from this and five previous studies on production of FBs and OTA in field isolates of A. niger and A. welwitschiae.
| Fumonisin analysis | Ochratoxin analysis | |||
|---|---|---|---|---|
| No. isolatesa | Producersb(%) | No. isolatesa | Producersb (%) | |
| A. NIGER | ||||
| This Study | 27 | 74 | 24 | 4 |
| Massi et al., 2016 | 89 | 74 | 89 | 31 |
| Qi et al., 2016 | 10 | 100 | 10 | 0 |
| Susca et al., 2014a | 19 | 63 | nd | nd |
| Storari et al., 2012 | 41 | 76 | 41 | 7 |
| A. WELWITSCHIAE | ||||
| This Study | 68 | 16 | 65 | 25 |
| Massi et al., 2016 | 86 | 34 | 86 | 1 |
| Qi et al., 2016 | 109 | 26 | 109 | 0 |
| Gherbawy et al., 2015 | 37 | 48 | 37 | 0 |
| Susca et al., 2014a | 35 | 29 | nd | nd |
| Storari et al., 2012 | 27 | 37 | 27 | 0 |
Number of isolates of A. niger or A. welwitschiae examined in each study for fumonisin or ochratoxin production. nd indicated not determined.
Percentage of isolates examined that produced fumonisin or ochratoxin. nd indicated not determined.
In addition to the frequency of mycotoxin production phenotypes, the comparison of studies suggests that A. niger and A. welwitschiae can vary in their frequencies of occurrence. A. welwitschiae appears to occur more frequently than A. niger in multiple host/location combinations. This was the case for grape and raisin isolates in the current study, for grape isolates in Canada (Qi et al., 2016), and for onion isolates from Brazil (Massi et al., 2016) and Saudi Arabia (Gherbawy et al., 2015; Table 3). However, this trend does not appear to exist for all crops and locations. For example, the numbers of A. niger and A. welwitschiae isolates from Brazil nuts grown in Brazil were similar, and the number of A. niger isolates (28) from grapes in Brazil was approximately two times greater than the number of A. welwitschiae isolates.
Several previous studies on A. niger and A. welwitschiae have also provided evidence that the genetic basis for the FB-nonproduction phenotype differs in the two species: the fum cluster is partially deleted in FB-nonproducing isolates of A. welwitschiae, whereas, the fum cluster is intact in FB-nonproducing isolates of A. niger. In the current study, PCR results for all A. niger isolates were consistent with previous results, whereas, results for some FB-nonproducing isolates of A. welwitschiae were not. However, sequence analysis of selected nonproducing isolates of A. welwitschiae indicated that, in most cases, the isolates had either one of the two fum cluster types that were previously described (Figure 4; Susca et al., 2014a). Therefore, the PCR results did not always accurately reflect the gene content of the fum cluster in A. welwitschiae, a phenomenon that has been noted previously (Palumbo et al., 2013). In addition, we identified an isolate (ITEM 15309) that had an intact fum cluster, but did not produce FBs, a situation that has been reported previously for A. niger but not A. welwitschiae (Susca et al., 2014a). Because isolate ITEM 15309 has apparently functional fum genes, it is not clear why this isolate does not produce FBs. In contrast, FB-nonproducing isolate ITEM 11209 had a fum cluster with a point mutation in fum1 but apparently functional homologs of other fum genes. Because the point mutation likely renders fum1 nonfunctional, and because fum1 is required for the first committed biochemical reaction in fumonisin biosynthesis (Proctor et al., 1999), the mutation would almost certainly block FB production in ITEM 11209. Thus, the nonfunctional fum1 in ITEM 11209 provides a possible explanation for why the isolate does not produce FBs. Likewise, the truncation of fum21 in the novel fum cluster type in A. welwitschiae isolate ITEM 10929 could explain the lack of FB production in this isolate, because analysis of the fum21 homolog in Fusarium has demonstrated that it is required for expression of other fum cluster genes and, therefore, FB production (Brown et al., 2007).
The PCR and sequence analyses of ota genes were consistent and indicated that in OTA-nonproducing isolates of both A. niger and A. welwitschiae the ota cluster is almost completely deleted. These results contrast those of Massi et al. (2016), who used PCR to examine 146 OTA-nonproducing isolates of these species for the presence of ota1 and ota5. Although Massi et al. did not detect either gene in the majority of nonproducing isolates examined, they did detect the two genes in one nonproducing isolate of A. niger and six nonproducing isolates of A. welwitschiae. These findings could be an indication that factors other than the absence of ota genes contribute to the lack of OTA production. Alternatively, the findings could be an indication that multiple patterns of ota gene deletion exist among OTA-nonproducing isolates of A. niger and A. welwitschiae.
Regardless of the difference in the ota-based PCR results reported in the current study and the study by Massi et al. (2016), genome sequence data indicate deletion of DNA within the ota cluster region is almost identical in OTA-nonproducing strains of A. niger and A. welwitschiae. We propose two alternative scenarios to explain this observation. In the first scenario, deletion of the ota cluster resulted from independent events in the two species: one event (or series of events) in A. niger and another independent event (or series of events) in A. welwitschiae. In this scenario, however, the deletion events were nonrandom and left almost identical sequence elements, including an ota1 fragment, in the An15g07870-An15g07930 intergenic region. In the second scenario, the intact and deleted ota clusters in the two species are descendants of ancestral alleles. That is, deletion of the ota cluster occurred in a common ancestor of A. niger and A. welwitschiae, and resulted in the formation of two alleles: an intact ota-cluster allele and a deleted ota-cluster allele. Subsequently, as A. niger and A. welwitschiae diverged from the ancestor, the two alleles were retained by both species. We propose that the second scenario is the most parsimonious, because it requires only one deletion of the cluster and provides a relatively simple explanation for the high level of similarity of An15g07870, An15g07930, and the intergenic region between them in OTA-producing versus nonproducing isolates of A. niger and A. welwitschiae.
The results of the current study and comparisons with previously published studies provide further insights into the distribution of FB and OTA production among field isolates of the black aspergilli A. niger and A. welwitschiae. The results also provide evidence for the first time of FB-nonproducing isolates of A. welwitschiae with an intact or almost intact fum cluster, like the intact fum cluster in FB-nonproducing isolates of A. niger. The presence of intact fum clusters in isolates of A. niger and A. welwitschiae suggest that such isolates might produce FBs under conditions other than those employed in the current study and other studies (Frisvad et al., 2007, 2011; Palumbo et al., 2013). The relatively high frequency of occurrence of strains that lack the genetic potential to produce either FBs or OTA points to the potential to reduce Aspergillus-induced FB/OT contamination of crops by preemptive application of mycotoxin-nonproducing strains of A. niger and/or A. welwitschiae. That such an approach would be possible is supported by successful control of aflatoxin contamination in multiple crops by preemptive application of aflatoxin-nonproducing isolates of A. flavus and A. parasiticus (Abbas et al., 2006; Dorner and Horn, 2007). Furthermore, efforts to control Aspergillus-induced mycotoxin contamination could be supported by research aimed at understanding the ecological advantage(s) for Aspergillus species to exist as mixed populations of mycotoxin-producing and nonproducing individuals.
Author contributions
AS, AL, AM, RP: Substantial contributions to the conception or design of the work; MM, MH, AG: the acquisition, analysis, or interpretation of data for the work; AS, AL, AM, RP, MM, MH, AG: Drafting the work or revising it critically for important intellectual content; AS, AL, AM, RP, MM, MH, AG: Final approval of the version to be published; AS, AL, AM, RP, MM, MH, AG: Agree for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The present work was funded by the European Union's Horizon 2020 research and innovation programme under Grant Agreement No. 678781 (MycoKey). We are grateful for the excellent technical assistance of Alessandra Villani (from ISPA) Gaetano Stea, Filomena Epifani from ISPA-CNR, and Amy McGovern, Nathane Orwig ran the MiSeq sequencher. Stephanie Folmar, Marcie Moore and Crystal Probyn from USDA-ARS.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01412
References
- Abbas H. K., Zablotowicz R. M., Bruns H. A., Abel C. A. (2006). Biocontrol of aflatoxin in corn by inoculation with non-aflatoxigenic Aspergillus flavus isolates. Biocontrol. Sci. Technol. 16, 437–449. 10.1080/09583150500532477 [DOI] [Google Scholar]
- Alexander N. J., Proctor R. H., McCormick S. P. (2009). Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 28, 198–215. 10.1080/15569540903092142 [DOI] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Millwer W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. R., Salazar M. P., Schaap P. J., van de Vondervoort P. J. I., Culley D., Thykaer J, et al. (2011). Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 21, 885–897. 10.1101/gr.112169.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. E. (2006). Aspergillus niger genomics: past, present and into the future. Med. Mycol. 44, S17–S21. 10.1080/13693780600921037 [DOI] [PubMed] [Google Scholar]
- Battilani P., Barbano C., Marin S., Sanchis V., Kozakiewicz Z., Magan N. (2006). Mapping of Aspergillus section Nigri in Southern Europe and Israel based on geostatistical analysis. Int. J. Food Microbiol. 111, S72–S82. 10.1016/j.ijfoodmicro.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Brakhage A. A. (2013). Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11, 21–32. 10.1038/nrmicro2916 [DOI] [PubMed] [Google Scholar]
- Brown D. W., Butchko R. A. E., Busman M., Proctor R. H. (2007). The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryot. Cell 6, 1210–1218. 10.1128/EC.00400-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner J. W., Horn B. W. (2007). Separate and combined applications of nontoxigenic Aspergillus flavus and A. parasiticus for biocontrol of aflatoxin in peanuts. Mycopathologia 163, 215–223. 10.1007/s11046-007-9004-0 [DOI] [PubMed] [Google Scholar]
- Ferracin L. M., Fier C. B., Vieira M. L. C., Monteiro-Vitorello C. B., Varani Ade M., Rossi M. M., et al. (2012). Strain-specific polyketide synthase genes of Aspergillus niger. Int. J. Food Microbiol. 155, 137–145. 10.1016/j.ijfoodmicro.2012.01.020 [DOI] [PubMed] [Google Scholar]
- Frisvad J. C., Larsen T. O., Thrane U., Meijer M., Varga J., Samson R. A., et al. (2011). Fumonisin and ochratoxin production in industrial Aspergillus niger strains. PLoS ONE 6:e23496. 10.1371/journal.pone.0023496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J. C., Smedsgaard J., Samson R. A., Larsen T. O., Thrane U. (2007). Fumonisin B2 production by Aspergillus niger. J. Agric. Food Chem. 55, 9727–9732. 10.1021/jf0718906 [DOI] [PubMed] [Google Scholar]
- Gallo A., Bruno K. S., Solfrizzo M., Perrone G., Mulè G., Visconti A., et al. (2012). New insight into the ochratoxin A biosynthetic pathway through deletion of a nonribosomal peptide synthetase gene in Aspergillus carbonarius. Appl. Environ. Microbiol. 78, 8208–8218. 10.1128/AEM.02508-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A., Knox B. P., Bruno K. S., Solfrizzo M., Baker S. E., Perrone G. (2014). Identification and characterization of the polyketide synthase involved in ochratoxin A biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 179, 10–17. 10.1016/j.ijfoodmicro.2014.03.013 [DOI] [PubMed] [Google Scholar]
- Gallo A., Perrone G., Solfrizzo M., Epifani F., Abbas A., Dobson A. D., et al. (2009). Characterisation of a pks gene which is expressed during ochratoxin A production by Aspergillus carbonarius. Int. J. Food Microbiol. 129, 8–15. 10.1016/j.ijfoodmicro.2008.10.022 [DOI] [PubMed] [Google Scholar]
- Geisen R., Schmidt-Heydt M., Karolewiez A. (2006). A gene cluster of the ochratoxin A biosynthetic genes in Penicillium. Mycotoxin Res. 22, 134–141. 10.1007/BF02956777 [DOI] [PubMed] [Google Scholar]
- Gherbawy Y., Elhariry H., Kocsube S., Bahobial A., Deeb B. E., Altalhi A., et al. (2015). Molecular characterization of black Aspergillus species from onion and their potential for ochratoxin A and fumonisin B2 production. Foodborne Pathog. Dis. 12, 414–423. 10.1089/fpd.2014.1870 [DOI] [PubMed] [Google Scholar]
- Gil-Serna J, Vázquez, C, González-Jaén, M. T., Patiño B. (2015). Clustered array of ochratoxin A biosynthetic genes in Aspergillus steynii and their expression patterns in permissive conditions. Int. J. Food Microbiol. 214, 102–108. 10.1016/j.ijfoodmicro.2015.07.020 [DOI] [PubMed] [Google Scholar]
- Hong S. B., Lee M., Kim D. H., Varga J., Frisvad J. C., Perrone G., et al. (2013). Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. PLoS ONE 8:e63769. 10.1371/journal.pone.0063769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P., Turner G., Bennett J. W. (2005). Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. 10.1038/nrmicro1286 [DOI] [PubMed] [Google Scholar]
- Kozakiewicz Z. (1989). Aspergillus Species on Stored Products. Mycological Papers, Vol. 161, Wallingford, CT: CABI Publishing. [Google Scholar]
- Massi F. P., Sartori D, de Souza Ferranti, L., Iamanaka B. T., Taniwaki M. H., Vieira M. L., et al. (2016). Prospecting for the incidence of genes involved in ochratoxin and fumonisin biosynthesis in Brazilian strains of Aspergillus niger and Aspergillus welwitschiae. Int. J. Food Microbiol. 221, 19–28. 10.1016/j.ijfoodmicro.2016.01.010 [DOI] [PubMed] [Google Scholar]
- Medina C., Mateo R., López-Ocaña L., Valle-Algarra F. M., Jiménez M. (2005). Study on Spanish grape mycobiota and ochratoxin A production by isolates of Aspergillus tubingensis and other members of Aspergillus section Nigri. Appl. Environ. Microbiol. 71, 4696–4702. 10.1128/AEM.71.8.4696-4702.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen J. M., Frisvad J. C., Thrane U., Nielsen K. F. (2010). Production of Fumonisin B2 and B4 by Aspergillus niger on grapes and raisins. J. Agric. Food. Chem. 58, 954–958. 10.1021/jf903116q [DOI] [PubMed] [Google Scholar]
- Nordberg H., Cantor M., Dusheyko S., Hua S., Poliakov A., Shabalov I., et al. (2014). The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 42, D26–D31. 10.1093/nar/gkt1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Nirenberg H. I., Aoki T., Cigelnik E. (2000). A Multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41, 61–78. 10.1007/BF02464387 [DOI] [Google Scholar]
- Palumbo J. D., O'Keeffe T. L., Gorski L. (2013). Multiplex PCR analysis of fumonisin biosynthetic genes in fumonisin-nonproducing Aspergillus niger and A. awamori strains. Mycologia 105, 277–284. 10.3852/11-418 [DOI] [PubMed] [Google Scholar]
- Pel H. J., de Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25, 221–231. 10.1038/nbt1282 [DOI] [PubMed] [Google Scholar]
- Perrone G., Stea G., Epifani F., Varga J., Frisvad J. C., Samson R. A. (2011). Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biol. 115, 1138–1150. 10.1016/j.funbio.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Proctor R. H., Desjardins A. E., Plattner R. D., Hohn T. M. (1999). A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27, 100–112. 10.1006/fgbi.1999.1141 [DOI] [PubMed] [Google Scholar]
- Qi T. F., Renaud J. B., McDowell T., Seifert K. A., Yeung K. K., Sumarah M. W. (2016). Diversity of mycotoxin-producing black aspergilli in Canadian vineyards. J. Agric. Food Chem. 64, 1583–1589. 10.1021/acs.jafc.5b05584 [DOI] [PubMed] [Google Scholar]
- Raeder U., Broda P. (1985). Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–20. 10.1111/j.1472-765X.1985.tb01479.x [DOI] [Google Scholar]
- Storari M., Dennert F. G., Bigler L., Gessler C., Broggini G. A. L. (2012). Isolation of mycotoxins producing black aspergilli in herbal teas available on the Swiss market. Food Control 26, 157–161. 10.1016/j.foodcont.2012.01.026 [DOI] [Google Scholar]
- Susca A., Moretti A., Stea G., Villani A., Haidukowski M., Logrieco A., et al. (2014b). Comparison of species composition and fumonisin production in Aspergillus section Nigri populations in maize kernels from USA and Italy. Int. J. Food Microbiol. 188, 75–82. 10.1016/j.ijfoodmicro.2014.06.031 [DOI] [PubMed] [Google Scholar]
- Susca A., Perrone G., Cozzi G., Stea G., Logrieco A. F., Mulè G. (2013). Multilocus sequence analysis of Aspergillus Sect. Nigri in dried vine fruits of worldwide origin. Int. J. Food Microbiol. 165, 163–168. 10.1016/j.ijfoodmicro.2013.04.027 [DOI] [PubMed] [Google Scholar]
- Susca A., Proctor R. H., Butchko R. A, Haidukowski M., Stea G., Logrieco A. F., et al. (2014a). Variation in the fumonisin biosynthetic gene cluster in fumonisin-producing and nonproducing black aspergilli. Fungal Genet. Biol. 73, 39–52. 10.1016/j.fgb.2014.09.009 [DOI] [PubMed] [Google Scholar]
- Susca A., Proctor R. H., Mulè G., Stea G., Ritieni A., Logrieco A., et al. (2010). Correlation of mycotoxin fumonisin B2 production and presence of the fumonisin biosynthetic gene fum8 in Aspergillus niger from Grape. J. Agric. Food Chem. 58, 9266–9272. 10.1021/jf101591x [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J. C., Kocsubé S., Brankovics B., Tóth B., Szigeti G., et al. (2011). New and revisited species in Aspergillus section Nigri. Stud. Mycol. 69, 1–17. 10.3114/sim.2011.69.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Kocsubé S., Suri K., Szigeti G., Szekeres A., Varga M., et al. (2010). Fumonisin contamination and fumonisin producing black Aspergilli in dried vine fruits of different origin. Int. J. Food Microbiol. 143, 143–149. 10.1016/j.ijfoodmicro.2010.08.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.