Abstract
The members of the genus Thermoactinomyces are known for their protein degradative capacities. Thermoactinomyces sp. strain AS95 is a Gram-positive filamentous bacterium, isolated from moderately saline water in the Thamelaht region of Algeria. This isolate is a thermophilic aerobic bacterium with the capacity to produce extracellular proteolytic enzymes. This strain exhibits up to 99 % similarity with members of the genus Thermoactinomyces, based on 16S rRNA gene sequence similarity. Here we report on the phenotypic features of Thermoactinomyces sp. strain AS95 together with the draft genome sequence and its annotation. The genome of this strain is 2,558,690 bp in length (one chromosome, but no plasmid) with an average G + C content of 47.95 %, and contains 2550 protein-coding and 60 RNA genes together with 64 ORFs annotated as proteases.
Keywords: Thermoactinomyces sp. strain AS95, Genome, Thermophilic, Proteolytic activity, Taxonomo-genomics
Introduction
Modern metagenomic approaches have provided insights on the evolution and functional capacity of microbial communities resistant to classical culture-based methods [1]. However, these classical techniques remain crucial for understanding the molecular adaptations of microbial guilds, especially those with potential biotechnological applications [2, 3]. Consequently, efforts to isolate novel taxa, particularly from environmentally extreme habitats remain widespread [4, 5].
The genus Thermoactinomyces is a member of the family Thermoactinomycetaceae. The first known representative from this genus (Thermoactinomyces vulgaris) was isolated from decaying straw and manure [6]. Since then, a number of isolates, from a wide array of extreme habitats [7–10] have been validly described. Currently, this genus comprises ten validly published species, and a few of these are; Thermoactinomyces vulgaris [6], Thermoactinomyces intermedius [11], Thermoactinomyces daqus [7] and Thermoactinomyces guangxiensis [8]. These species are all Gram-positive, aerobic, non-acid-fast, chemoorganotrophic, filamentous and thermophilic bacteria.
Here, we report the draft genome sequence of Thermoactinomyces sp. strain AS95, which was isolated from a sebkha (endorheic salt pan) in the Thamelaht region ofAlgeria. We present a summary of the classification and set of phenotypic features for Thermoactinomyces sp. strain AS95 together with the description of the non-contiguous genome sequence and its annotation with particular reference to ORFs encoding proteolytic enzymes.
Organism information
Classification and features
Thermoactinomyces strain AS95 was isolated from a sebkha water sample collected in June 2013 from the Thamelaht region ofAlgeria (Table 1). This isolate is a Gram-positive, aerobic, thermophilic, filamentous bacterium (Fig. 1) belonging to the order Bacillales. Based on the 16S rRNA gene sequence similarity searches by BLASTN against the NCBI-NT database, strain AS95 showed 97–99 % sequence similarity to members of the genus Thermoactinomyces. A 16S rRNA gene-based phylogenetic tree of Thermoactinomyces sp. strain AS95 was constructed (Fig. 2), based on neighbor-joining and maximum composite likelihood models with 1000 bootstrap replications using MEGA 7 [12]. The Thermoactinomyces sp. strain AS95 (KU942442) 16S rRNA gene sequence exhibited high identity (99 %) with Thermoactinomyces vulgaris RVH210302 (AY114167), the closest validly published Thermoactinomyces species.
Table 1.
Classification and general features of Thermoactinomyces sp. strain AS95
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain: Bacteria | TAS [20] | |
| Phylum: Firmicutes | TAS [21–23] | ||
| Class: Bacilli | TAS [24, 25] | ||
| Order: Bacillales | TAS [26, 27] | ||
| Family: Thermoactinomycetaceae | TAS [25, 28] | ||
| Genus: Thermoactinomyces | TAS [6] | ||
| Species: Thermoactinomyces sp. | IDA | ||
| Strain: AS95 | IDA | ||
| Gram stain | Positive | IDA | |
| Cell shape | Filamentous | IDA | |
| Motility | Non-motile | IDA | |
| Sporulation | Endospores on unbranched sporophores | IDA | |
| Temperature range | 40–65 °C (Thermophilic) | IDA | |
| Optimum temperature | 55 °C | IDA | |
| pH range; Optimum | 5.6–8.6; 7.2 | IDA | |
| Carbon source | Peptides | IDA | |
| GS-6 | Habitat | Saline water | IDA |
| MIGS-6.3 | Salinity | 5.0 % total salt (w/v) | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Non-pathogen | IDA |
| MIGS-4 | Geographic location | Thamelaht,, Algeria | IDA |
| MIGS-5 | Sample collection time | 20 June 2013 | IDA |
| MIGS-4.1 | Latitude | 36°32'18.29"N | IDA |
| MIGS-4.2 | Longitude | 5°11'48.89"E | IDA |
| MIGS-4.4 | Altitude | 890 m above sea level | IDA |
aEvidence codes – IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e. a direct report exists in the literature). These evidence codes are from the Gene Ontology Project [29]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements
Fig. 1.
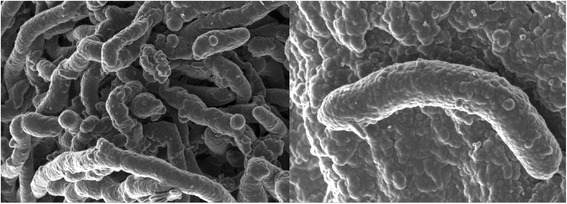
Scanning electron microscopy of Thermoactinomyces sp. strain AS95 using a Cryo-SEM (JEOL)
Fig. 2.

Phylogenetic tree based on 16S rRNA gene sequences showing the relationship between strain AS95 (1435 bp) and strains of related genera of the family Thermoactinomycetaceae. The strains and their corresponding Genbank accession numbers are shown following the organism name and indicated in parentheses. The phylogenetic tree was made using the neighbor-joining method with maximum composite likelihood model implemented in MEGA 7. The tree includes the 16S rRNA gene sequence of Sulfobacillus acidophilus DSM 10332T as outgroup. Bootstrap consensus trees were inferred from 1000 replicates, only bootstrap values >50 % are indicated. The scale bar represents 0.02 nucleotide changes per position. (♦) indicates the isolate assessed in the current study, Thermoactinomyces sp. strain AS95
The strain was cultivated on Thermus medium agar containing 2.0 g NaCl, 4.0 g yeast extract, 8.0 g peptone and 30.0 g agar per liter of distilled water. The bacterium grew optimally at 55 °C, with a broad temperature growth range of between 40 and 65 °C (Table 1). The strain grew in liquid media at pH values from 5.6 to 8.6, but optimal growth occurred at a pH of 7.2. Morphologically, the isolate forms white colonies and abundant aerial mycelia with the appearance of well-developed, branched and septate substrate mycelia. The micromorphology of the cells was examined using scanning electron microscopy (Fig. 1). The predominant menaquinone was MK-7. Major fatty acids included iso-C15:0, and significant amounts of iso-C17:0 were also present.
Genome sequencing information
Genome project history
A high-quality draft genome sequence is deposited at DDBJ/EMBL/GenBank under the accession LSVF00000000 and consists of 11 scaffolds of 11 contigs. A summary of the project information and its association with MIGS version 2.0 compliance are shown in Table 2 [13].
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | One paired-end 300 bp library |
| MIGS-29 | Sequencing platforms | MiSeq-Illumina |
| MIGS-31.2 | Fold coverage | 40.0× |
| MIGS-30 | Assemblers | SPAdes 3.5.0 |
| MIGS-32 | Gene calling method | NCBI Prokaryotic Genome, Annotation Pipeline |
| Genbank ID | LSVF00000000 | |
| Genbank Date of Release | April 04, 2016 | |
| BIOPROJECT | PRJNA312744 | |
| GOLD ID | Gs0118400 | |
| MIGS-13 | Project relevance | Biotechnological, Environmental |
Growth conditions and genomic DNA preparation
Thermoactinomyces sp. strain AS95 was grown aerobically on Thermus medium agar (pH 7.2) at 55 °C for 24 h. Genomic DNA was extracted using a modification of a previously described protocol [14]. The quantity and quality of the genomic DNA was measured using a NanoDrop Spectrophotometer and a Qubit™ Fluorometer (Thermo Fisher Scientific Inc.).
Genome sequencing and assembly
Genomic DNA samples of Thermoactinomyces sp. strain AS95 were sequenced at MR DNA (Shallowater, TX, USA). Genome sequencing was performed on a MiSeq (Illumina, Inc.) generating 2 x 300 bp paired-end libraries. The sequencing run produced a total of 5,085,250 reads, with a mean length of 265.58 bp. The raw paired-end sequences were subjected to the fastxtools software [15] for quality trimming using a phred quality score ≥ 20. After trimming, a total of 3,013,639 reads with a mean length of 171.11 bp were assembled using SPAdes, version 3.5.0 [16]. The final assembly resulted in a total of 11 scaffolds, which generated a genome size of 2.56 Mb.
Genome annotation
Genome annotation was carried out on the RAST server [17] and using the NCBI Prokaryotic Genome Annotation Pipeline tools [18]. This Whole Genome Shotgun sequence project has been deposited at DDBJ/EMBL/GenBank under accession LSVF00000000. The version described in this paper is version LSVF00000000.
Genome properties
The genome is composed of 2,558,690 nucleotides with 47.95 % G + C content (Table 3) and comprised 11 scaffolds of 11 contigs. The genome contains a total of 2649 genes, 2550 of which were protein coding, 39 pseudogenes and 60 RNA coding genes. The majority of protein-coding genes (75.45 %) were assigned a putative function while the remaining genes were annotated as hypothetical. The distribution of genes in COGs functional categories is presented in Table 4.
Table 3.
Genome statistics of the Thermoactinomyces sp. strain AS95
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 2,558,690 | 100.00 |
| DNA coding region (bp) | 2,214,681 | 86.56 |
| DNA G + C (bp) | 1,226,817 | 47.95 |
| DNA scaffolds | 11 | |
| Total genes | 2,649 | 100.00 |
| Protein coding genes | 2,550 | 96.26 |
| RNA genes | 60 | 2.26 |
| Pseudo genes | 39 | 1.47 |
| Genes in internal clusters | ND | ND |
| Genes with function prediction | 1,296 | 50.82 |
| Genes with Pfam domains | 2,001 | 78.47 |
| Genes assigned to COGs | 1,924 | 75.45 |
| Genes with signal peptides | 164 | 6.43 |
| Genes with transmembrane helices | 655 | 25.69 |
| CRISPR repeats | 2 | ND |
aThe total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome. ND: Not determined
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 154 | 9.96 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 145 | 5.68 | Transcription |
| L | 100 | 3.92 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 27 | 1.05 | Cell cycle control, mitosis and meiosis |
| V | 32 | 1.25 | Defense mechanisms |
| T | 71 | 2.78 | Signal transduction mechanisms |
| M | 99 | 3.88 | Cell wall/membrane biogenesis |
| N | 8 | 0.31 | Cell motility |
| Z | 0 | 0.03 | Cytoskeleton |
| U | 33 | 1.29 | Intracellular trafficking and secretion |
| O | 85 | 3.33 | Posttranslational modification, protein turnover, chaperones |
| C | 135 | 5.29 | Energy production and conversion |
| G | 122 | 4.78 | Carbohydrate transport and metabolism |
| E | 213 | 8.35 | Amino acid transport and metabolism |
| F | 70 | 2.74 | Nucleotide transport and metabolism |
| H | 108 | 4.23 | Coenzyme transport and metabolism |
| I | 109 | 4.27 | Lipid transport and metabolism |
| P | 101 | 3.96 | Inorganic ion transport and metabolism |
| Q | 53 | 2.07 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 249 | 9.76 | General function prediction only |
| S | 196 | 7.68 | Function unknown |
| - | 626 | 24.54 | Not in COGs |
aThe total is based on the total number of protein coding genes in the annotated genome
A blastp comparison was conducted against the MEROPS database. A total of 64 protein-coding genes (2.4 %) were predicted to share homology with various categories of proteases (Table 5). Of these predictions indicated that 36 were putatively secreted in a classical pathway (SignalP), whereas the other 28 were secreted in a non-classical pathway (SecretomeP). Only 2 of the 64 protein-coding genes share sequence similarities with proteases of the Thermoactinomyces vulgaris and sp. E79 families of peptidases in the MEROPS database.
Table 5.
The four major types of proteases predicted in Thermoactinomyces sp. strain AS95
| Type | Classical (SignalP) | Non-classical (SecretomeP) |
|---|---|---|
| Cysteine | 6 | 3 |
| Metallo | 18 | 12 |
| Serine | 11 | 10 |
| Threonine | 0 | 2 |
Conclusions
This study describes the draft genome sequence of Thermoactinomyces sp. strain AS95, which is associated with a high level of extracellular proteolytic activities. To date, only a few metabolic pathways involved in protein degradation have been characterized for the genus Thermoactinomyces [19]. The genome sequence and characteristics of strain AS95 will provide new insights into the mechanisms of protein degradation in the genus Thermoactinomycetes, and towards establishing a comprehensive genomic catalog of the metabolic diversity of the genus Thermoactinomyces.
Acknowledgements
We wish to acknowledge the following organizations for providing financial support for this project: The Genomics Research Institute and the University of Pretoria (OKIB, DAC, TPM), the National Research Foundation (MWVG, DAC, TPM). The Algerian Ministry of Higher Education and Scientific Research is also acknowledged for funding (MAG and KK).
Authors’ contributions
OKIB performed the analysis, and led the drafting of the manuscript. MAG isolated the strain and conducted confirmatory analysis using 16S rRNA gene sequencing. RP performed the assembly and annotation. MWVG performed the SEM and helped draft the manuscript. KK supervised the isolation of the strain. DAC provided support in drafting the manuscript. TPM conceived the study and provided support in drafting the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cowan DA, Ramond J-B, Makhalanyane TP, De Maayer P. Metagenomics of extreme environments. Curr Opin Microbiol. 2015;25:97–102. doi: 10.1016/j.mib.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27(7):398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Hahn MW, Lünsdorf H, Wu Q, Schauer M, Höfle MG, Boenigk J, Stadler P. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol. 2003;69(3):1442–1451. doi: 10.1128/AEM.69.3.1442-1451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison JP, Gheeraert N, Tsigelnitskiy D, Cockell CS. The limits for life under multiple extremes. Trends Microbiol. 2013;21(4):204–212. doi: 10.1016/j.tim.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Dash HR, Mangwani N, Chakraborty J, Kumari S, Das S. Marine bacteria: potential candidates for enhanced bioremediation. Appl Microbiol Biotechnol. 2013;97(2):561–571. doi: 10.1007/s00253-012-4584-0. [DOI] [PubMed] [Google Scholar]
- 6.Tsilinsky P. On the thermophilic moulds. Ann Inst Pasteur. 1899;13:500–505. [Google Scholar]
- 7.Yao S, Liu Y, Zhang M, Zhang X, Li H, Zhao T, Xin C, Xu L, Zhang B, Cheng C. Thermoactinomyces daqus sp. nov., a thermophilic bacterium isolated from high-temperature Daqu. Int J Syst Evol Microbiol. 2014;64(1):206–210. doi: 10.1099/ijs.0.055509-0. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Liu B, Pan S. Thermoactinomyces guangxiensis sp. nov., a thermophilic actinomycete isolated from mushroom compost. Int J Syst Evol Microbiol. 2015;65(9):2859–2864. doi: 10.1099/ijs.0.000342. [DOI] [PubMed] [Google Scholar]
- 9.Mokrane S, Bouras N, Meklat A, Lahoum A, Zitouni A, Verheecke C, Klenk HP. Thermoactinomyces khenchelensis sp. nov., a filamentous bacterium isolated from soil sediment of a terrestrial hot spring. Antonie van Leeuwenhoek. 2016;109(2):311–317. [DOI] [PubMed]
- 10.Yao S, Xu Y, Xin C, Xu L, Liu Y, Li H, Li J, Zhao J, Cheng C. Genome sequence of Thermoactinomyces daqus H-18, a novel thermophilic species isolated from high-temperature Daqu. Genome announcements. 2015;3(1):e01394–01314. doi: 10.1128/genomeA.01394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurup V, Hollick G, Pagan E. Thermoactinomyces intermedius, a new species of amylase negative thermophilic actinomycetes. Science-Ciencia Bol Cien Sur. 1980;7:104–108. [Google Scholar]
- 12.Kumar S, Stecher G, Tamura K. Kumar S, Stecher G, Tamura K. MEGA7. Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–1874. [DOI] [PMC free article] [PubMed]
- 13.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller D, Bryant J, Madsen E, Ghiorse W. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65(11):4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.FASTX-Toolkit T: http://hannonlab.cshl.edu/fastx_toolkit/. Accessed Mar 2016.
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Ciufo S, Li W. Prokaryotic genome annotation pipeline. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Białkowska A, Gromek E, Florczak T, Krysiak J, Szulczewska K, Turkiewicz M. Extremophilic Proteases: Developments of Their Special Functions, Potential Resources and Biotechnological Applications. In: Biotechnology of Extremophiles. Switzerland: Springer; 2016: 399–444.
- 20.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol. 1978;28(1):1–6. doi: 10.1099/00207713-28-1-1. [DOI] [Google Scholar]
- 22.Garrity GM, Holt JG. The Road Map to the Manual. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual® of Systematic Bacteriology: Volume One: The Archaea and the Deeply Branching and Phototrophic Bacteria. New York, NY: Springer New York; 2001. pp. 119–166. [Google Scholar]
- 23.Murray R. Bergey’s Manual of Systematic Bacteriology. 1984. The higher taxa, or, a place for everything. [Google Scholar]
- 24.Ludwig WW, Whitman WB. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2009. pp. 19–20. [Google Scholar]
- 25.Euzeby J. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2006;56(5):925–927. doi: 10.1099/ijs.0.64380-0. [DOI] [PubMed] [Google Scholar]
- 26.Skerman V, McGowan V, Sneath PHA. Approval lists of bacterial names. Int J Syst Bacteriol. 1980;30:255–420. [Google Scholar]
- 27.Hauduroy P, Ehringer G. Dictionnaire des bactéries pathogènes. Paris: Masson; 1953. [Google Scholar]
- 28.Goodfellow M, Jones AL. “Thermoactinomycetaceae”. Bergey's Manual of Systematics of Archaea and Bacteria. New York: John Wiley & Sons, Ltd; 2015. p. 1–18.
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


