Abstract
Currently available methods for interrogating DNA-protein interactions at individual genomic loci have significant limitations, and make it difficult to work with unmodified cells or examine single-copy regions without specific antibodies. In this study, we describe a physiological application of the Hybridization Capture of Chromatin-Associated Proteins for Proteomics (HyCCAPP) methodology we have developed. Both novel and known locus-specific DNA-protein interactions were identified at the ENO2 and GAL1 promoter regions of S. cerevisiae, and revealed subgroups of proteins present in significantly different levels at the loci in cells grown on glucose versus galactose as the carbon source. Results were validated using chromatin immunoprecipitation. Overall, our analysis demonstrates that HyCCAPP is an effective and flexible technology that does not require specific antibodies nor prior knowledge of locally occurring DNA-protein interactions and can now be used to identify changes in protein interactions at target regions in the genome in response to physiological challenges.
1. Introduction
Genome control and function in every organism is tightly regulated and modulated by complex interactions of the DNA molecule with a large number of proteins. Technologies like chromatin immunoprecipitation (ChIP) and DNase footprinting have revealed a number of those interactions [1,2]. While ChIP is able to look at a histone, transcription factor, or any other DNA-binding protein, and analyze all the regions in the genome that bind to this one particular protein at a given time, approaches like DNase footprinting indicate what regions in the genome are more likely to be occupied by DNA-interacting proteins without precise knowledge of the individual proteins [3]. Clearly, the obvious limitation is that ChIP only allows studying one protein at a time, and the protein to be analyzed is targeted using a specific antibody, without any information on other proteins or co-factors binding in the same genomic region. DNase footprinting, in contrast, reveals protein occupancy at any given locus in the genome, but without an efficient way to identify and characterize bound proteins.
A number of emerging technologies have recognized these challenges and approach them in different ways [4]. Some follow ChIP-like procedures to capture individual proteins of interest and then identify additional proteins bound to the enriched chromatin fragments, instead of retrieving the DNA sequences [5-7], while others target specific DNA sequences to be enriched for proteomic analysis [8-10]. These approaches have been used to target multi-copy regions, or exploited the insertion of specialized plasmids.
Here, we present a novel technology we recently developed [11] called Hybridization Capture of Chromatins-Associated Proteins for Proteomics (HyCCAPP). The method uses hybridization to enrich specific cross-linked genomic regions for proteomic analysis. We previously used HyCCAPP in Saccharomyces cerevisiae to study high copy regions within the rDNA locus, the telomere adjacent X-element and a single copy region at the upstream activator sequence for the GAL10 and GAL1 genes (UASGAL). Our previous efforts aimed to obtain comprehensive lists of all associated proteins in a single state, including proteins of unknown relevant function crosslinked to the target locus. Our current efforts were aimed at adapting the approach to identify DNA-binding proteins likely to mediate changes in transcriptional activity in response to a physiological stimulus. As we show, this application minimizes false positive identifications, and uncovers biologically relevant protein binding differences at individual single copy regions in cells grown under different conditions. Specifically, we demonstrated the feasibility of the HyCCAPP approach using yeast cells grown with either glucose or galactose as the carbon source. The method modifications we have introduced here highlight the flexibility of HyCCAPP to directly address biologically relevant changes in DNA-protein interactions. Our approach allows the study of single copy loci in unaltered cells and identifies proteins that are enriched at a particular locus under a given condition without any prior knowledge of putative binding proteins or antibody reagents.
2. Materials and methods
2.1. Cell growth and chromatin extraction
Saccharomyces cerevisiae Y1788 cells were grown in yeast extract peptone media with either dextrose or galactose as the carbon source (Sigma-Aldrich). Cell were grown at 30 °C to an average cell density of 3×107 cells per ml. Cells were crosslinked in 3% formaldehyde (Sigma-Aldrich) for 15 minutes at 30 °C and quenched in 250mM Tris HCl pH 8. Crosslinked cells were pelleted at 4 °C and washed twice in ice cold PBS pH 7.4 (Life Technologies).
Cell pellets equivalent to 1 liter of cell culture were lysed in 15 ml of lysis buffer (75mM Tris HCl pH 8, 75mM NaCl, protease inhibitors) using a French Pressure Cell Press at 1200 psig. After lysis, 1 ml of RNase A/T1 mix (Thermo Scientific) was added and incubated for 2 hours at 30 °C. SDS (Sigma-Aldrich) was added to a final concentration of 4 %. Cell lysates were layered over 5-8 M urea gradients (5 ml of lysate per 30 ml of gradient) and centrifuged at 4 °C at 100,000g for 16 hours [12]. Pellets were rinsed in TE buffer and resuspended in 3 ml of buffer S (50mM Tris HCl pH 8, 10mM EDTA, 1% SDS, and protease inhibitors).
A sample amount equivalent to 2×1011 cells was placed in a rosette on ice and sonicated 15 times using a 30 seconds on 40 seconds off cycle at level 5 using a High Intensity Ultrasonic Processor (Sonics Materials) and a tapped step horn with tip. Resulting samples were centrifuged at 4 °C at 14,000g for 10 minutes. Supernatants were collected and concentrated using 100K filter spin columns (Amicon). Stabilization buffer (50mM Potassium acetate, 20mM Tris acetate, 10mM Magnesium acetate, 1mM DTT, protease inhibitors) was added (1/3 of the final volume). Chromatin was immediately used for HyCCAPP experiments or stored at −80 °C.
2.2. RNA sequencing
RNA was isolated using the RNeasy Mini Kit (Qiagen). Indexed cDNA libraries were generated using the ScriptSeq Complete Gold Kit (Epicentre). Libraries were quantified by qPCR and sequenced on the MiSeq (Illumina) platform using a 150 cycle flow cell. Output paired-read sequences were analyzed using Partek Flow software v 3.0.14.0910. Raw read pre-alignment quality was assessed with FastQC v1.0 and quality trimming to a minimum Phred score of 20 and minimum read length of 25 bp was performed with Cutadapt v 1.2.1. Processed reads were then aligned and mapped to the sacCer3 genome assembly with STAR aligner v 2.3.1j. Mapped reads were quantified by the Partek E/M method, and gene-specific analyses were performed to assess differential expression.
2.3. HyCCAPP
Streptavidin-coated magnetic Sera-Mag SpeedBeads (Thermo Scientific) were washed in Hybridization buffer (100mM MES, 1M NaCl, 20mM EDTA, 0.01% Tween-20). Samples were pre-cleared for 1 hour at 42 °C in 250 μl of beads per 100 femtomoles of DNA. After removal of beads, biotinylated capture oligonucleotides (IDT) (Supporting Information Tables S1 and S2) were added at a 4,000:1 oligo:DNA ratio. Samples were incubated at 42 °C for 2 hours with end over end rotation at 10 rpm. Additional beads were washed in fresh Hybridization buffer, and 2 μl of beads were used for every 7 picomoles of capture oligonucleotides (twice the saturation volume). Samples were incubated 30 minutes at room temperature with rotation. Samples were placed on a magnet and the supernatant removed. The remaining beads were washed in wash buffer (200mM NaCl, 0.2% SDS, 50mM Tris pH 8). All washes were performed at room temperature (unless otherwise noted) with end over end rotation. Beads were washed in ½ the hybridization volume three times for 5 minutes and one time for 1 hour. Beads were then resuspended in twice the original bead volume for the remaining washes. One time for 30 minutes, one time for 15 minutes at 37 °C and one time for 5 minutes. For mass spectrometry (MS) analysis, beads were reconstituted in 1× the original volume in DNase buffer (10mM Tris-HCl, 2.5mM MgCl2, 0.5mM CaCl2, pH 7.6) and DNase I was added to a final concentration of 12 U/ml (New England Biolabs) and incubated at 37 °C for 30 minutes. Samples were quickly vortexed and placed on a magnet. The supernatant was transferred to a new tube and incubated at 94 °C for 10 minutes. Samples were stored at −80 °C for mass spectrometry analysis. For DNA analysis, an aliquot of washed beads was diluted and incubated at 94 °C for 5 minutes to remove DNA from the beads. A minimum of three independent replicates were used per sample.
2.4. Real-time polymerase chain reaction analysis
Primers and probes for real-time RT-PCR and qPCR were designed using PrimerQuest (IDT). All probes were designed with a 5′-FAM label and a double quencher system (internal ZEN and 3′-IBFQ) (See Supporting Information Table S3 for complete sequences). Reactions were carried out in triplicate. RT-PCR reactions were run using the TaqMan One-Step RT-PCR Master Mix Reagents Kit (Applied Biosystems). Actin2 was used as reference gene, and samples analyzed using the ΔΔCT method [13]. qPCR reactions were run using the TaqMan Universal PCR Master Mix (Applied Biosystems). PCR amplicons of the target regions were quantified to generate standard curves.
2.5. DNA sequencing
Eluted samples from the HyCCAPP procedure were processed using the TrueSeq ChIP Sample Preparation Kit (Illumina). The libraries were quantified by qPCR and sequenced on the MiSeq (Illumina) platform using a paired-end 2×150 run. Output paired-read sequences were analyzed using Partek Flow software v 3.0.14.0910. Raw read pre-alignment quality was assessed with FastQC v1.0 and quality trimming was performed with Cutadapt v 1.2.1. Processed reads were then aligned and mapped to the sacCer3 genome masked for simple repeats and abundant rRNA, tRNA, and mtRNA sequences. The Bowtie 2 aligner v2.1.0 with “very sensitive” settings, an ambiguous character penalty of 100, and exclusion of mixed or discordant alignments to avoid single stranded mapping was used. Aligned reads were filtered to a quality threshold of Phred 30 and PCR duplicates were removed with Picard v1.44. BAM alignment files were analyzed with the Partek Genomics Suite ChIP-Seq workflow. Peak detection was performed over 1 kb windows with a 0.001 FDR cut off.
2.6. Mass spectrometry
6M urea was added to each sample and incubated at 30 °C for 30 min. 10mM DTT was added and incubated at 30 °C for 20 min. 55mM iodoacetamide was added and incubated for 30 min in the dark. The sample was diluted in 100mM ammonium bicarbonate to reduce the final urea concentration to 1M. Resulting samples (~4 μg) were digested overnight at 37 °C in 0.25 μg of trypsin. The solution was acidified using 0.5% TFA and desalted using both C18 and C4 tips (Millipore). The eluted samples were taken to near dryness by rotary evaporation and reconstituted in 23 μl of 0.1% formic acid. The solutions were sonicated for 5 min in a bath sonicator. Samples were analyzed on an Orbitrap Elite tandem mass spectrometer (Thermo Scientific) using a 2 hour gradient elution method with top 15 MS/MS scans.
2.7. Data analysis
Precursor MS and MS/MS spectra were searched against the S. cerevisiae fasta protein database (Uniprot database containing 6,812 sequences and combined with cRAP contaminant database from GPM) using Sequest HT via Proteome Discoverer (Thermo Scientific). Oxidized methionine (+15.995 Da) and carbamidomethylated cysteines (+57.021 Da) were allowed as dynamic modifications. Up to 3 missed trypsin cleavages were permitted. The precursor match tolerance was set at 10 ppm and the CID fragment match tolerance was set at 0.8 Da. The search results were validated using the Percolator algorithm using a decoy database search FDR of 5% based on q-values. The generated data consisting of individual protein peptides counts in each sample, were first filtered based on a minimum peptide spectral match (PSMs) of 2 reducing the multiple testing burden. Counts over multiple runs of the baseline samples and HyCCAPP runs were summed, yielding separate total counts for each protein pre and post HyCCAPP. A one-sided Fisher’s exact test was used to examine whether the count for a given protein was increased comparing lysate and HyCCAPP samples, conditional on the total number of proteins counts in all samples. A one-sided test was used to solely focus on those proteins enriched in HyCCAPP runs. Fisher’s exact test was also used to identify differentially enriched proteins between samples obtained under different growth conditions.
2.8. Chromatin immunoprecipitation (ChIP) analysis
ChIP followed a previously described protocol [14] with a few modifications. Briefly, TAP-tagged cells were grown for each protein to be validated under the same conditions as for HyCCAPP experiments. Cells lysed in nuclear lysis buffer (75mM NaCl, 75mM Tris pH 8, 1% SDS, protease inhibitors) were kept cold and sonicated to an average size of 500 bp. Samples were centrifuged at 14,000 rpm for 10 minutes at 4 °C. The chromatin and protein content in the supernatant was measured by a Qubit assay (Life technologies). For each IP, 4 μg of chromatin (approx. 700 μg of protein) was diluted in 5 volumes of IP dilution buffer (0.92 % Triton X-100, 0.008 % SDS, 1mM EDTA, 13.9mM Tris–HCl pH 8, 13.9mM NaCl, 12.5mM sodium butyrate) and pre-cleared with A/G-sepharose beads for 30 min at 4 °C. Samples were immunoprecipitated overnight at 4 °C with a TAP-Tag antibody (Thermo Scientific), followed by 90 min incubation at 4 °C with protein A/G-sepharose beads. Beads were washed, eluted and formaldehyde crosslinks reversed in 300mM NaCl. The qPCR analysis was run as described above. Three different qPCR assays (Supporting Information Table S3) accounted for the potential difference in size between the HyCCAPP targets and the ChIP fragments.
3. Results
3.1. RNA expression analysis reveals ENO2 as a candidate for HYCCAPP analysis
For this study, we initially sought to identify genes involved in carbohydrate metabolism that would exhibit high transcriptional activity, but with clear differences in expression in cells grown with either glucose or galactose as the carbon source. A gene actively transcribed under both conditions should have a relatively open chromatin configuration, facilitating hybridization capture and allow HyCCAPP to identify differences in protein binding that potentially mediate any differential expression observed between the growth conditions.
To identify suitable gene targets, RNA sequencing analysis was performed on Saccharomyces cerevisiae grown under the two growth conditions. Differential expression between conditions was seen in 2,502 genes, while 3,825 genes did not show any substantial difference. As expected, cells grown under galactose showed very strong upregulation of transcription for genes involved in galactose metabolism, including GAL10, GAL 1, GAL7, HXK1 and GAL2, (Supporting Information Table S4). Several glycolysis-related genes also showed increased expression, including TDH3, GPM1, ENO1, GPD1 and PGK1. Of these glycolysis genes, only Fructose 1,6 bisphosphate aldolase (FBA1) and Enolase II (ENO2) were among the top 1% of genes expressed under both conditions. Of those two genes, ENO2 showed the largest difference in expression between cells grown under glucose and galactose, showing a 2-fold upregulation when using galactose instead of glucose as the carbon source (Supporting Information Table S5). This result was validated by qPCR. Based on these findings, the ENO2 promoter region was selected as an initial HyCCAPP target.
3.2. Enrichment of DNA-protein complexes from single copy regions
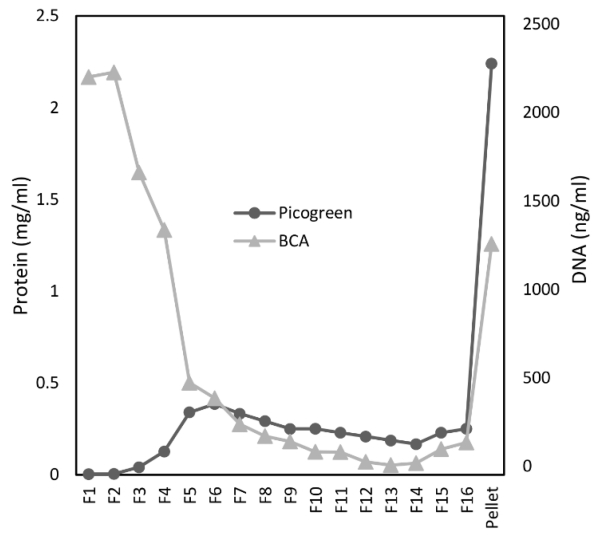
The HyCCAPP procedure follows the workflow illustrated in Figure 1. In order to isolate DNA-protein complexes, cells were crosslinked in 3% formaldehyde prior to sample processing. Cell lysates were ultracentrifuged in urea gradients (5-8M) based on previous methods described for enrichment of crosslinked DNA-protein complexes [12]. Protein and DNA content profiles resembled previous reports, clearly showing enrichment of crosslinked DNA-protein complexes in the pellet and separated from non DNA-bound proteins, retained in the upper fractions of the gradient (Figure 2). This approach generates a starting material for HyCCAPP capture experiments that primarily consists of DNA-protein complexes, and that has been separated from unbound DNA and proteins, resulting in a decrease in background capture of DNA molecules not bound to proteins.
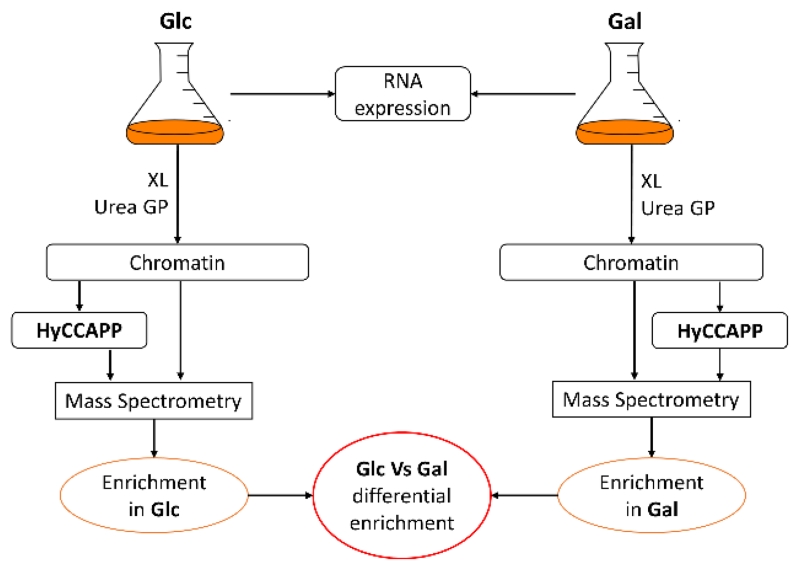
Figure 1.
HyCCAPP workflow diagram. Gene expression for yeast cells grown under glucose or galactose is measured to help identify relevant regions for HyCCAPP experiments. Cells are crosslinked, harvested and the chromatin is purified using gradient ultracentrifugation. MS is used to identify proteins in the chromatin and in samples resulting from the HyCCAPP process. Both general chromatin-associated proteins and HyCCAPP-captured proteins that are enriched under one of the two growth conditions are identified. Glc, glucose; Gal, galactose; XL, crosslinked; GP, gradient purification.
Figure 2.
Urea gradient profile. DNA and protein contents are shown for each individual fraction and the remaining pellet after ultracentrifugation in a 5-8M urea gradient.
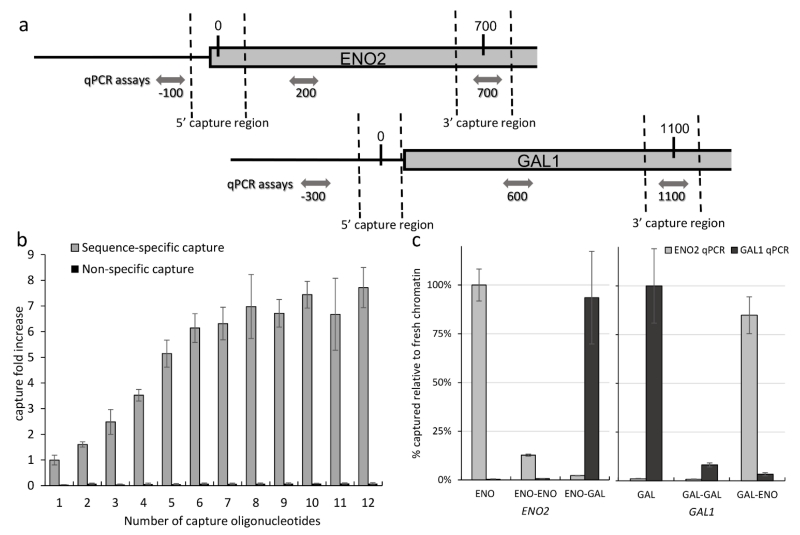
Capture oligonucleotides were targeted to the promoter region of the ENO2 gene, located on yeast chromosome VIII (Figure 3a). These oligonucleotides were specific for both strands and both ends of the 700bp promoter target. Different capture oligonucleotides and combinations were tested, but as shown in Figure 3b, optimization of capture yield using combinations of capture oligonucleotides revealed a plateau after 6-7 oligonucleotides, resulting in a > 6 fold increase in target capture when compared to any single oligonucleotide, but no further increase was seen with additional oligonucleotides. Despite the increase in capture oligonucleotides targeting different sequences, non-specific capture of other genomic regions was not increased, demonstrating the high specificity of the hybridization capture. Based on these results and sequence constraints due to strong homology to the ENO1 region in chromosome VII, a cocktail of 7 oligonucleotides (Supporting Information Table S1) was used for all ENO2 HyCCAPP experiments. Repeat captures of the ENO2 region from the same chromatin material (Figure 3c) with the same oligonucleotides resulted in negligible yields (<10% of original capture), demonstrating that most of the capture-amenable chromatin material is extracted during a single hybridization capture. In contrast, other chromatin regions can be subsequently captured with only minimally reduced efficiencies, demonstrating that the lack of capture of ENO2 in the second hybridization is not due to degradation of the chromatin.
Figure 3.
Hybridization strategy. (a) Diagram depicting the target regions for ENO2 and GAL1 promoter regions. Target oligonucleotides are designed targeting both strands and both ends of the target regions. For each HyCCAPP target region, three qPCR assays were designed accounting for the size differences between the HyCCAPP process and the ChIP validations. Distances in base pairs from the middle of the 5′ capture region are shown. (b) Hybridization capture experiments were carried out with an increasing number of capture oligonucleotides without altering the total final concentration of capture oligonucleotides. The efficiency was measured through qPCR after reversing the crosslinking of the captured material. Fold increases were calculated relative to hybridization with one oligonucleotide. Depicted error bars represent the standard deviation. No significant change was observed in non-specific capture across all samples. (c) Subsequent captures using oligonucleotides targeting the ENO2 and GAL1 regions. Capture efficiency was measured through qPCR after reversing the crosslinking of the captured material. ENO and GAL refer to captures using fresh chromatin, while ENO-ENO/GAL and GAL-ENO/GAL, refer to captures using chromatin previously used for captures targeting the ENO2 and GAL1 regions, respectively.
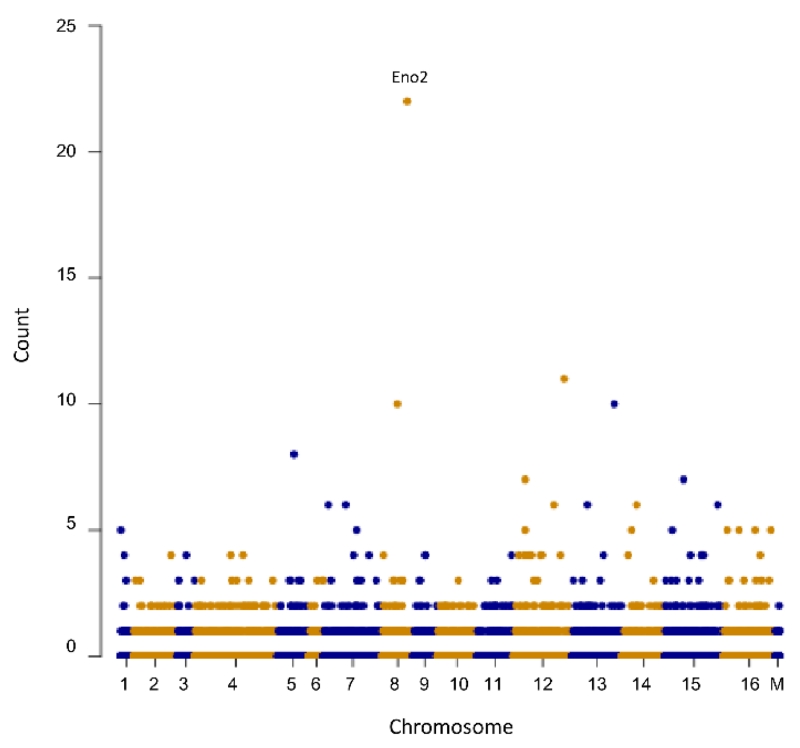
Our described protocol for capture resulted in a final average capture efficiency of 3.8% ± 1.0% across all samples and an average enrichment of 175 ± 23 fold when compared to the enrichment of independent control regions of the yeast genome. The overall unbiased capture specificity of the process was assessed by sequencing the DNA of the captured material. Reads were aligned to the reference genome, and sequence coverage was assessed in 1 kb windows. The ENO2 promoter region targeted by HyCCAPP was clearly the most abundantly enriched fragment (Figure 4). The aligned reads in the contig covered 693 bp out of the 1 kb window, consistent with the target region of 700 bp. The next most abundant region was a genomic interval near UTP21, with the contig covering only 111bp in length. Less than 2.7% of all 1 kb windows had more than 1 read, and only 0.39% had 4 or more reads. The ENO2 promoter region was the only contig in the alignment with more than 400 bp in length. 98.6% of the genome did not have any reads aligned to them.
Figure 4.
Sequencing of captured material. DNA extracted from HyCCAPP experiments was sequenced and aligned to the yeast genome. The plot depicts read counts in 1 kb windows throughout the genome. The target region (ENO2 promoter in chromosome VIII) had twice as many counts as the next most abundant region (UTP21 in chromosome XII). A total of 86.94% of all 1 kb windows had a count of 0 reads. Detected reads represent less than 1.4% of the whole genome.
3.3. Mass spectrometry analysis identifies enriched and differentially bound proteins
DNase I digestion was used to elute proteins from captured chromatin to minimize release of non-specific proteins bound to the streptavidin-coated beads. This digestion approach selectively elutes proteins bound to dsDNA, but not proteins directly bound to the beads. Mass spectrometry analyses were performed on samples before (lysate) and after the HyCCAPP procedure (Figure 1). Only proteins found to be significantly enriched when compared to the pre-capture chromatin material were included in subsequent analyses. At the ENO2 promoter region, 62 and 56 proteins were found to be significantly enriched when yeast were grown under galactose and glucose, respectively, of which 15 proteins were shared between captured samples from both growth conditions (Supporting Information Table S6). Using gene ontology (GO) enrichment analysis, proteins involved in chromatin remodeling were enriched among proteins identified at the ENO2 promoter region under galactose growth conditions. In contrast, under glucose growth conditions, GO enrichment analysis identified proteins involved in glycolytic processes. Little overlap was observed between GO enrichments between the two growth conditions with just a few proteins (Lcd1 for chromatin organization and Tdh3 and Tdh2 for glycolytic process) identified in both samples.
Generally, more proteins were found with annotations related to DNA binding (17 Vs 7 proteins), nuclear localization (21 Vs 10 proteins) and gene regulation (11 Vs 8 proteins) in yeast samples grown under galactose compared to under glucose (Supporting Information Table S6). Of the enriched proteins, 7 proteins showed significant differences in abundance between the growth conditions (Table 1). Nst1, Ark1 and Rsm24 were found to be significantly enriched under glucose growth conditions at the ENO2 locus, while Htb2, Pab1, Rim1 and Sec28 were found to be significantly enriched under galactose growth conditions. Of these last four proteins, Pab1 and Rim1 were also identified under glucose growth conditions but at significantly lower levels than in galactose samples. Htb2 is a core histone protein and would be expected to be bound to DNA, while Sec28 is a coatomer protein and has been shown to bind to DNA at other loci in yeast [11].
Table 1. Differentially enriched proteins at the ENO2 and GAL1 promoter regions.
| Protein id | Name | Gene | Fold enrichment in Gal (p value) |
Fold enrichment in Glc (p value) |
Fold enrichment between conditions (p value) |
|
|---|---|---|---|---|---|---|
| ENO2 | P02294 | Histone H2B | HTB2 | 13.3 (1×10−07) | Gal only (4×10−02) | |
| P04147 | Poly(A) binding protein | PAB1 | 347.6 (5×10−228) | 66.5 (2×10−32) | Gal 3.1 (7×10−08) | |
| P32445 | Replication in mitochondria | RIM1 | 301.9 (5×10−67) | 59.1 (1×10−03) | Gal 11.4 (1×10−05) | |
| P40509 | SECretory | SEC28 | 172.5 (2×10−14) | Gal only (4×10−02) | ||
| P53935 | Negatively affects salt tolerance | NST1 | NDC (2×10−10) | Glc only (8×10−03) | ||
| P53974 | Actin regulating kinase | ARK1 | NDC (2×10−10) | Glc only (8×10−03) | ||
| Q03976 | Ribosomal small subunit | RSM24 | NDC (2×10−10) | Glc only (8×10−03) | ||
|
| ||||||
| GAL1 | P00924 | Phosphopyruvate hydratase enolase | ENO1 | 3.5 (6×10−04) | Glc only (1×10−02) | |
| P04147 | Poly(A) binding protein | PAB1 | 148.2 (6×10−31) | 32.3 (1×10−05) | Gal 2.8 (4×10−02) | |
Gal, galactose; Glc, glucose; NDC, not detected in control (lysate)
3.4. ChIP validates identified proteins
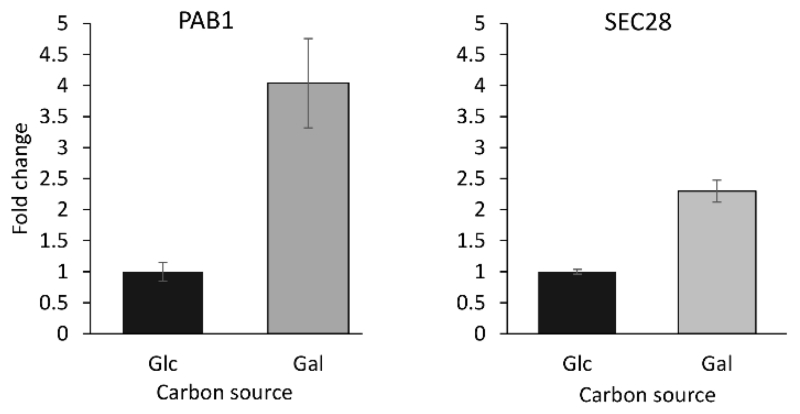
Of the proteins identified in our analysis, five were selected for chromatin immunoprecipitation (ChIP) validation: three proteins that showed differences between growth conditions (Pab1, Rim1 and Sec28), and two proteins that were enriched under both conditions but did not show differences in abundance between glucose and galactose growth conditions (Tdh2 and Rpa190). ChIP data for the analysis of Rim1 were not reproducible and highly variable, and could not be used for confirmation of the HyCCAPP results. ChIP assays for Pab1 and Sec28 revealed significantly higher enrichment in galactose treated cells compared to glucose treated cells (Figure 5), consistent with the HyCCAPP results. Both Tdh2 and Rpa190 showed enrichment compared to the negative control, but no significant difference between the two growth conditions, also in accordance with the HyCCAPP results.
Figure 5.
ChIP validation. TAP-Tag strains for PAB1 and SEC28 were used for ChIP-qPCR validations. Fold changes for galactose relative to glucose grown cells are shown for the ENO2 promoter region.
3.5. HyCCAPP analysis of the promoter region of the GAL1 gene
We previously reported an analysis of the upstream activator sequence located between the GAL10 and GAL1 genes (UASGAL) in cells grown under glucose as carbon source [11]. Similarly, Byrum et al. [8,9] applied their approach, Chromatin Affinity Purification with Mass Spectrometry (ChAP-MS), to study the promoter of the GAL1 gene, a region adjacent to the UASGAL. In order to more accurately evaluate our technology, and compare it to findings using other technologies, we targeted the GAL1 promoter region (Figure 3a) in cells grown under glucose and galactose as carbon source. Since GAL1 is a gene required for galactose metabolism, this region of the genome is highly transcriptionally active when galactose is present, but repressed when glucose is used as a carbon source, which was clearly confirmed in our RNA-Seq results, where GAL1 and GAL10 were the 5th and 2nd highest overall expressed genes under galactose growth conditions, but fell to the bottom 20% when grown under glucose (Supporting Information Table S4). As noted before, capture of repressed regions is more challenging, especially in hybridization-based approaches, due to the condensation of inactive chromatin.
We also observed more efficient capture of the GAL1 promoter region when the gene was active under galactose growth conditions. Nevertheless, we were also able to capture the GAL1 promoter region in its repressed form under glucose growth conditions (capture oligonucleotides are listed in Supporting Information Table S2), in contrast to other previously reported approaches [9]. Following the same mass spectrometry and statistical procedures described above, we were able to identify 20 proteins enriched under glucose growth conditions, and 20 proteins enriched with galactose as the carbon source (Supporting Information Table S7). Of those, 6 proteins were shared between both conditions while each group had 14 unique proteins. Eno1 was found to be significantly enriched under glucose treatment, when compared to galactose growth, and poly (A) binding protein (Pab1) was significantly enriched under galactose (Table 1). Both of these proteins were also identified by Byrum et al. [8] using the ChAP-MS approach. A total of 6 (Adh1, Cdc19, Eno1, Pab1, Npl3 and Tdh1) and 4 (Tdh3, Tdh1, Pab1 and Ror1) proteins were identified by both HyCCAPP and ChAP-MS under glucose and galactose treatment, respectively.
As described above for the ENO2 locus, we used ChIP to validate the HyCCAPP findings. Both Pab1 and Tdh2, targeted as part of the ENO2 HyCCAPP validation, were analyzed, and showed enrichment at the GAL1 locus. Furthermore, Pab1 showed increased binding to GAL1 under galactose growth conditions, consistent with HyCCAPP results.
4. Discussion
The present study describes HyCCAPP as a method capable not only to identify DNA-bound proteins at specific single-copy genomic loci, but to also identify changes in protein-DNA interactions under different physiological conditions, including those that lead to changes in transcriptional activity of target regions. HyCCAPP allowed the identification of a number of proteins that are differentially bound to the ENO2 promoter region when comparing yeast cells grown under different carbon sources, highlighting the potential of this approach. Selected proteins analyzed by ChIP showed the same quantitative changes between different growth conditions as revealed by HyCCAPP, suggesting that the careful and stringent processing and analysis of the captured chromatin described here reveals predominantly true DNA-protein interactions.
The few technologies currently available that are capable to identify novel proteins binding to DNA at specific genomic loci without the use of specific antibodies require either genetic engineering [8], plasmid insertions [9] or use of locked nucleic acid probes on multi-copy regions [10]. We have previously shown that HyCCAPP can be used on multi-copy and single-copy genomic regions without the need to alter the cell [11]. However, our analyses identified a large number of bound proteins, and it is unclear how many of the DNA-protein interactions are biologically relevant, or just a by-product of DNA-protein crosslinking. We used urea gradient ultracentrifugation prior to capture hybridization, and DNase-mediated elution, to optimize the HyCCAPP protocol, and ensure that the analysis exclusively focuses on proteins bound to chromatin as this approach removes any unbound proteins from the starting material for HyCCAPP. The urea gradient centrifugation step proved to be very effective in enriching protein-bound chromatin with enough sample integrity for hybridization and downstream mass spectral analyses. Furthermore, we clearly showed that the modest yields we obtained are a direct result of the scarcity of hybridization-amenable regions in the cross-linked chromatin and not due to a deficient hybridization step. Additionally, the mass spectral profile comparison between pre- and after-capture, and between conditions, further enables to discriminate proteins that were crosslinked unspecifically to the target DNA. By running multiple independent replicates, we were able to increase the confidence of the identified proteins. Intrinsic features of any method aimed at sequence-specifically enriching cross-linked DNA-protein complexes from the genome include modest enrichment yields and potentially significant background levels. By sequencing the DNA of the captured samples, we showed that the capture process had good specificity, and the target region was enriched with high selectivity. The ENO2 promoter region was clearly the most abundant contig. And even though some other contigs were present in significant levels and are not precluded from potentially contributing to the proteomic identifications, their lower levels and far shorter sizes makes them less likely of substantially altering the results. In our application where we compared samples grown under different physiological conditions, HyCCAPP allowed the identification of proteins enriched under each condition. Furthermore, based on the sequencing results, we estimate that we capture >6000 copies of the ENO2 target region in a full scale HyCCAPP experiment. Regions captured at even lower numbers are unlikely to provide sufficient amounts of bound proteins, and therefore are unlikely to result in reproducible mass spectral protein identifications. However, larger amounts of captured material may be necessary to identify additional low abundance proteins that are known to bind the target regions we investigated, but we were not able to detect in our analyses.
Despite the high confidence in the identified proteins for both the ENO2 and the GAL1 promoter loci, the biological relevance of the identified proteins remains unclear at this point. We were able to identify a number of nuclear proteins involved in gene transcription. As an example, we identified Asf1 at the ENO2 promoter region under galactose growth, a well-known derepressor protein [15], Ino80, a protein involved in chromatin rearrangement and histone mobilization [16], and Rpa190 and Rpa43, two RNA polymerase subunits. Under glucose growth we identified Gsm1, a transcription factor with binding sites proximal to our target region [17], and Spt7 a member of the SAGA protein complex [18]. We did identify many proteins, however, with unknown function in the regulation of these loci. This is also true for the well-studied GAL1 promoter region [19]. We did not detect any of the previously described interacting proteins Gal4, Gal80 [20] or Mig1 [21]. As in our current study, the efforts using the ChAP-MS [8,9] technologies, and our previous efforts targeting the adjacent UASGal [11], also failed to identify these proteins. As we mentioned before, we did identify a number of proteins that were also previously identified by the ChAP-MS approach. A highly intriguing one is Pab1, a mRNA binding protein that to our knowledge has not been implicated in chromatin interactions or DNA binding. We have been able to identify it by HyCCAPP and validate the interaction with ChIP, and the protein was also reported to bind to the GAL1 promoter in the previous study using the ChAP-MS approach [8]. Additionally we identified Swt1, a protein that interacts with the TREX complex during transcription [22], and Nhp6a, a high-mobility group protein involved in nucleosome remodeling [23]. While the previously reported functions of these proteins could potentially explain their binding to the ENO2 and GAL1 promoter regions, they have not been reported previously, highlighting the potential of the HyCCAPP approach to uncover novel DNA-binding proteins.
Unlike most genomic analyses where comprehensive readouts can be expected, mass spectrometry will only identify a fraction of the proteins present. Which proteins are detected will obviously depend on the abundance of such proteins, but also on the binding affinity and occupancy, as well as technical aspects of protein mass spectrometry such as ionization efficiency or ion interference in complex samples. The field of chromatin-protein analysis using mass spectrometry is rapidly advancing [24], and it is allowing HyCCAPP to contribute in the understanding of dynamic DNA-protein interactions and their role in chromatin regulation.
In summary, we present here a useful novel methodology to study locus-specific DNA-protein interactions, and an initial demonstration that HyCCAPP can elucidate how these interactions change under different physiological conditions. The tools developed here significantly reduce the number of false positives, and allow HyCCAPP to be adapted as a flexible new technology to investigate other genomic regions and physiological conditions.
Supplementary Material
Highlights.
An unbiased method to study DNA-protein interactions in vivo is proposed.
Sequence-specific hybridization of crosslinked chromatin fragments is described.
Whole genome sequencing validates the specificity of the process.
Novel DNA-protein interactions at single-copy regions are identified.
ACKNOWLEDGMENT
This work was supported by NIH/NHGRI grant P50HG004952.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
The manuscript was written with contributions from all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Elnitski L, Jin VX, Farnham PJ, Jones SJM. Locating mammalian transcription factor binding sites: A survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- 2.Walhout AJM. Unraveling transcription regulatory networks by protein–DNA and protein–protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- 3.Boyle AP, Song L, Lee B-K, London D, Keefe D, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillen-Ahlers H, Shortreed MR, Smith LM, Olivier M. Advanced methods for the analysis of chromatin-associated proteins. Physiol Genomics. 2014;46:441–447. doi: 10.1152/physiolgenomics.00041.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert J-P, Fillingham J, Siahbazi M, Greenblatt J, Baetz K, et al. Defining the budding yeast chromatin-associated interactome. Mol Syst Biol. 2010;6:448. doi: 10.1038/msb.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soldi M, Bonaldi T. The Proteomic Investigation of Chromatin Functional Domains Reveals Novel Synergisms among Distinct Heterochromatin Components. Mol Cell Proteomics. 2013;12 doi: 10.1074/mcp.M112.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, et al. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol. 2013;20:202–209. doi: 10.1038/nsmb.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrum SD, Raman A, Taverna SD, Tackett AJ. ChAP-MS: A Method for Identification of Proteins and Histone Posttranslational Modifications at a Single Genomic Locus. Cell Rep. 2012;2:198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrum SD, Taverna SD, Tackett AJ. Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Déjardin J, Kingston RE. Purification of Proteins Associated with Specific Genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy-Darling J, Guillen-Ahlers H, Shortreed MR, Scalf M, Frey BL, et al. Discovery of Chromatin-Associated Proteins via Sequence-Specific Capture and Mass Spectrometric Protein Identification in Saccharomyces cerevisiae. J Proteome Res. 2014 doi: 10.1021/pr5004938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Belle I, Cai S, Kohwi-Shigematsu T. The Genomic Sequences Bound to Special AT-rich Sequence-binding Protein 1 (SATB1) In Vivo in Jurkat T Cells Are Tightly Associated with the Nuclear Matrix at the Bases of the Chromatin Loops. J Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Oduro AK, Fritsch MK, Murdoch FE. Chromatin context dominates estrogen regulation of pS2 gene expression. Exp Cell Res. 2008;314:2796–2810. doi: 10.1016/j.yexcr.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 16.Udugama M, Sabri A, Bartholomew B. The INO80 ATP-Dependent Chromatin Remodeling Complex Is a Nucleosome Spacing Factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, et al. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 2008;36:e21. doi: 10.1093/nar/gkm1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, et al. Functional Organization of the Yeast SAGA Complex: Distinct Components Involved in Structural Integrity, Nucleosome Acetylation, and TATA-Binding Protein Interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lue NF, Chasman DI, Buchman AR, Kornberg RD. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987;7:3446–3451. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston M, Flick JS, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14 doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Röther S, Clausing E, Kieser A, Strässer K. Swt1, a Novel Yeast Protein, Functions in Transcription. Journal of Biological Chemistry. 2006;281:36518–36525. doi: 10.1074/jbc.M607510200. [DOI] [PubMed] [Google Scholar]
- 23.Rhoades AR, Ruone S, Formosa T. Structural Features of Nucleosomes Reorganized by Yeast FACT and Its HMG Box Component, Nhp6. Mol Cell Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soldi M, Cuomo A, Bremang M, Bonaldi T. Mass spectrometry-based proteomics for the analysis of chromatin structure and dynamics. Int J Mol Sci. 2013;14:5402–5431. doi: 10.3390/ijms14035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.