Abstract
Pseudouridylation of messenger RNA emerges as an abundant modification involved in gene expression regulation. Pseudouridylation of stop codons in eukaryotic and bacterial cells results in stop-codon read through. The structural mechanism of this phenomenon is not known. Here we present a 3.1-Å crystal structure of E. coli release factor 1 (RF1) bound to the 70S ribosome in response to the ΨAA codon. The structure reveals that recognition of a modified stop codon does not differ from that of a canonical stop codon. Our in vitro biochemical results support this finding by yielding nearly identical rates for peptide release from E. coli ribosomes programmed with pseudouridylated and canonical stop codons. The crystal structure also brings insight into E. coli RF1-specific interactions and suggests involvement of L27 in bacterial translation termination. Our results are consistent with a mechanism in which read through of a pseudouridylated stop codon in bacteria results from increased decoding by near-cognate tRNAs (miscoding) rather than from decreased efficiency of termination.
Keywords: pseudouridine, post-transcriptional modification of mRNA, translation regulation, stop-codon read through, translation termination
Graphical abstract
INTRODUCTION
Recent studies show that pseudouridylation (isomerization of uridine to pseudouridine) of messenger RNA is more abundant than has been previously thought [1–4]. Pseudouridylation may play roles in translation regulation, as suggested by the observation of increased mRNA modification under stress conditions [1]. Such roles must be mediated by chemical differences between pseudouridine and uridine. Pseudouridine differs from uridine in that the aromatic ring contains an additional hydrogen-bond donor N1 and connects with the ribose via a C-glycosidic bond (Fig. 1a). Pseudouridylation of sense codons or untranslated regions may contribute to translation regulation by affecting RNA structure—e.g., by increasing secondary structure stability [5, 6]. Pseudouridylation of stop codons strongly suppresses translation termination—i.e., promotes stop-codon read through—both in yeast [7] and E. coli [8]. Read through could result from increased efficiency of recognition of a pseudouridylated stop codon by a near-cognate tRNA, decreased efficiency of translation termination, or both. Structural analysis of a 30S ribosome complex formed with a modified ΨAG stop codon and a near-cognate anticodon stem loop tRNASer fragment revealed non-canonical base-pair formation, similar to that formed by an UAG stop codon [8]. Thus, whether pseudouridylation of a stop codon affects tRNA selection remains unclear.
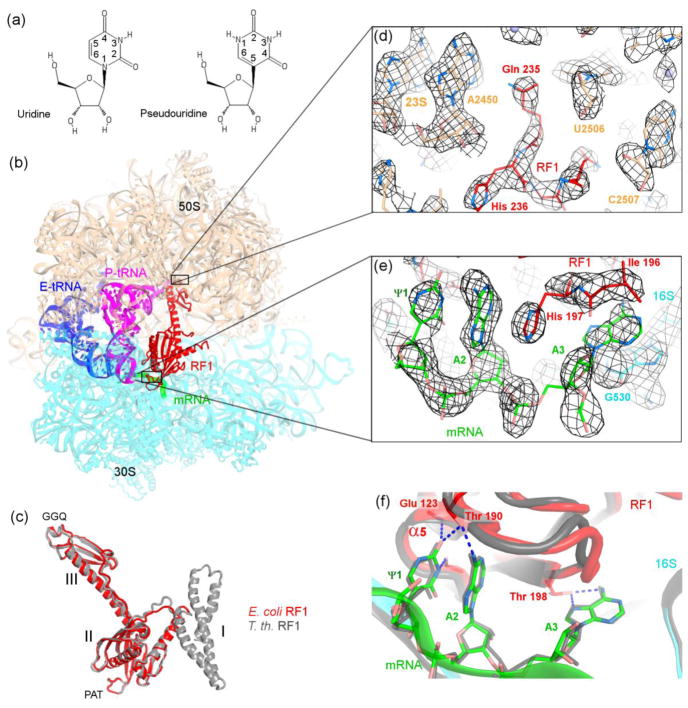
Figure 1.
Structural basis for recognition of the ΨAA codon by release factor 1. (a) Chemical structures of pseudouridine and uridine. (b) Crystal structure of the 70S•ΨAA•RF1 termination complex (this work). The 50S subunit is shown in wheat and 30S subunit in cyan. (c) Similar conformations of E. coli (this work) and T. thermophilus release factors in 70S termination complexes. (d–e) Unbiased feature-enhanced density map [37] shows well-resolved features of RF1 in the peptidyl-transferase center in (d) and ΨAA and RF1 in the decoding center in (e). (mRNA and RF1 were not included in the simulated-annealing-refined 70S model used for map calculation.) (f) Comparison of the conformations of the ΨAA (this work) and UAA (gray) codons in the 70S•RF1 termination complexes. In panels (c) and (f), the structure of T. thermophilus 70S•RF1 complex (PDB ID: 4V63, [11]) is shown in gray.
It is not known whether translation termination on a pseudouridylated stop codon contributes to stop codon read through. Termination occurs when a stop codon (UAA, UAG or UGA in all three domains of life) at the A (aminoacyl-tRNA) site of the ribosome is recognized by release factors (RF1 and RF2 in bacteria; and eRF1 in eukaryotes) that catalyze peptidyl-tRNA hydrolysis. High-resolution structures of bacterial [9–14] and eukaryotic [15, 16] termination complexes demonstrate that conserved elements of release factors recognize each nucleotide of a stop codon. Notably, the uridine at the first position of a stop codon constitutes a stringent requirement for release factor specificity. Substitution of the uridine to another nucleotide strongly reduces binding of release factors [17]. In line with this observation, recent computational analyses suggest that recognition of pseudouridine instead of uridine would require a conformational rearrangement of a stop codon [18]. The rearrangement was proposed to be necessary to align pseudouridine’s dipole moment (which is distinct from that of uridine) with the dipole moment of a release factor’s recognition helix α5 [18].
In this work, we test the hypothesis that modification of the uridine at the first position of a stop codon to pseudouridine affects the efficiency of translation termination. To visualize the mechanism of recognition of a pseudouridylated stop codon, we have determined a 3.1-Å resolution structure of E. coli RF1 bound to the T. thermophilus 70S ribosome programmed with the ΨAA codon in the A site (Fig. 1b). The structure of an E. coli RF1 bound to the 70S ribosome brings insights into the termination mechanism in E. coli, which has been the primary bacterial model system. Furthermore, this work allows us to address structural differences between the mesophilic and previously studied thermophilic release factors. The structure reveals that the conformations of the pseudouridylated stop codon, the decoding residues of RF1 and the ribosomal A site are nearly identical to those in the canonical UAA-bound termination complexes [9, 11], suggesting identical structural mechanisms for recognition of the ΨAA and UAA codons. Our structural findings are complemented by in vitro studies, in which we tested whether the efficiency of peptide release by RF1 or RF2 is affected when a uridine in a stop codon is substituted by pseudouridine.
RESULTS AND DISCUSSION
Crystal structure reveals similar mechanisms of recognition of the ΨAA and UAA stop codons
We have determined a crystal structure of the bacterial translation termination complex formed with the ΨAA stop codon, using Thermus thermophilus 70S ribosomes bound with tRNAfMet in the P (peptidyl-tRNA) site and E. coli release factor RF1 in the A site (Fig. 1b–1e). RF1 and mRNA were resolved in the initial unbiased electron density maps, which allowed unambiguous interpretation of protein and RNA conformations (Fig. 1d, 1e). The ribosomal 50S and 30S subunits adopt the non-rotated (classical) conformation, similar to that observed in structures of termination complexes formed with the canonical (non-modified) stop codons [9–12]. The conformation of the release factor (Fig. 1c) is similar to that in the previous structures [10, 11], but domain 1 of the release factor is disordered. We note that domain 1 was the least resolved part of RF1 in previous termination complexes formed with T. thermophilus 70S ribosomes and T. thermophilus RF1 [11].
Previous structures demonstrated that release factors employ domain 2 to recognize the stop codon and domain 3 to catalyze the hydrolysis of peptidyl-tRNA (reviewed in [19, 20]). The “recognition head” of domain 2 of RF1 contains the conserved PxT motif critical for codon recognition. The tip of domain 3 contains the universally conserved GGQ motif, required for catalysis. As described below, the structural recognition of the ΨAA stop codon and positioning of the catalytic GGQ motif in the peptidyl-transferase center are nearly identical to those observed in the canonical 70S termination complexes [10, 11].
The ΨAA codon is bound in the A site (decoding center) of the 30S subunit and adopts a conformation similar to that of canonical stop codons (Fig. 1f) in the presence of RF1 [10, 11] or RF2 [9, 13, 14]. Specifically, the first two nucleotides, Ψ1 and A2, are stacked, whereas the base plane of A3 is nearly orthogonal to that of A2. A3 is sandwiched between I196 of RF1 and the universally conserved G530 of 16S ribosomal RNA (Fig. 1e). The bases of the codon interact with the recognition head of domain 2. The Watson-Crick face of Ψ1 forms hydrogen bonds with the backbone atoms of G120 and Q123 at the N-terminus of α5. The O2 atom of pseudouridine forms a hydrogen bond with the hydroxyl group from threonine of the PxT motif (residues 189–191), which also hydrogen bonds with the N6 atom on the second stop-codon nucleotide (Fig. 1f). The Hoogsteen face of A3 forms a hydrogen bond with T198 of domain 2 (Fig. 1f). Domain 3 connects domain 2 and the large ribosomal subunit (Fig. 1b). The catalytic GGQ motif of domain 3 is docked into the peptidyl-transferase center, ~80 Å away from the PxT motif of domain 2. The glutamine of the GGQ motif abuts the ribose of the 3′-terminal nucleotide of the P-site tRNA, consistent with its catalytic function, as described previously [11].
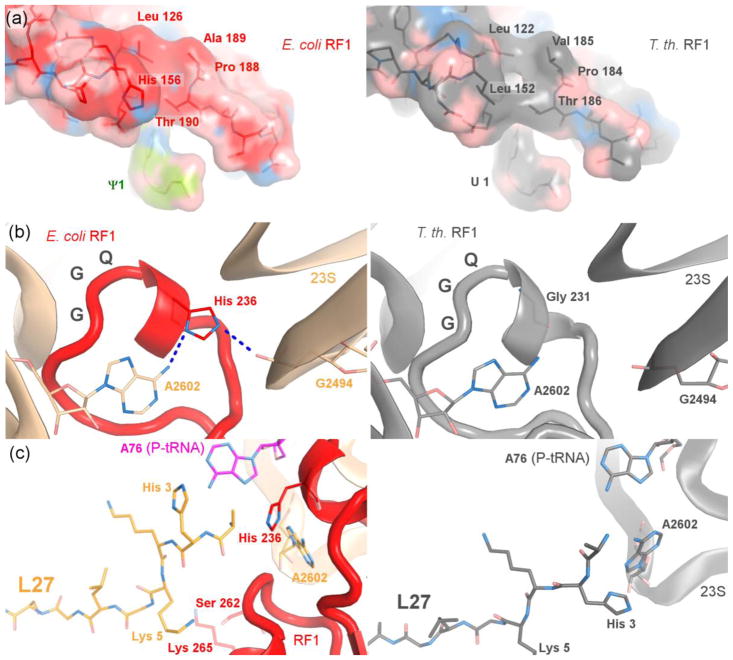
Our structure provides insight into the differences between the mesophilic and extremophilic release factors and reveals interaction of protein L27 with the release factor, which we discuss in a subsequent section (Fig. 2). Previous structural analyses of bacterial termination were based on 70S complexes obtained with T. thermophilus release factors [9–11, 13, 14]. The functional centers of the mesophilic release factors (e.g., E. coli RF1 in this work) are distinct from those of the thermophilic release factors. The PxT codon-recognition motif contains alanine (i.e., PAT) in E. coli and valine (i.e., PVT) in T. thermophilus. The alanine of the PAT motif of E. coli RF1 packs on L126 (Fig. 2a). In T. thermophilus RF1, the valine of PVT is positioned similarly and interacts with L122, which in turn packs on L152 (H156 in E. coli). The more hydrophobic environment of the codon-recognition head in the thermophilic release factor likely contributes to conformational stability of this key functional center [21].
Fig. 2.
Interactions of E. coli RF1 with the ribosome and rearrangement of L27 upon RF1 binding. (a) Comparison of the PxT codon-recognition domains of E. coli (PAT, left; current work) and T. thermophilus RF1 (PVT, right; PDB ID: 4V63, [11]). (b) Comparison of the GGQ regions of RF1 from E. coli (left, current work) and T. thermophilus (right; PDB ID: 4V63, [11]) in the peptidyl-transferase center. Interactions of H236 are shown. (c) Rearrangement of L27 upon binding of RF1. L27 in the presence of RF1 (left; this work); L27 from the 70S ribosome with a vacant A site (right; PDB ID: 4W2H, [33]) in the absence of RF1.
In domain 3, the GGQ catalytic motif is followed by a histidine (H236) in both E. coli RF1 and RF2 and by glycine in T. thermophilus RF1 and RF2 (Fig. 2b). The GGQH motif is highly conserved in mesophilic bacteria, including pathogens, and in the human mitochondrial release factor mtRF1a. The histidine forms hydrogen bonds with the backbone of nucleotide G2494 and with the N6 atom in the nucleobase of the universally conserved A2602 of 23S rRNA (Fig. 2b). A2602 is buried in a pocket of domain 3 of RF1 and is critical for translation termination [22]. H236 therefore likely contributes to stabilization of the GGQ motif and peptidyl-transferase center conformations for catalysis in mesophilic release factors.
Pseudouridylation does not affect kinetics of peptide release
We next asked whether the structural similarity of the 70S•RF1 structures formed in the presence of ΨAA and UAA codons reflects the similarity of stop codon recognition mechanisms. To this end, we tested the efficiency of peptide release by both E. coli release factors, RF1 and RF2, on canonical and pseudouridylated stop codons (Table 1). We employed a standard in vitro assay to measure the rates of release of [35S]-labeled N-formyl-methionine from E. coli 70S ribosomes bound with fMet-tRNAfMet [23].
Table 1.
Rates of [35S]-fMet release mediated by E. coli RF1 and RF2 from E. coli 70S ribosomes programmed with canonical or pseudouridylated stop codons.
| Stop codon | ||||||
|---|---|---|---|---|---|---|
| UAA | UAG | ΨAA | ΨAG | AAA | no mRNA | |
|
| ||||||
| kobs, s−1 | ||||||
| 10 nM RF1 | 0.031 ± 0.004 | 0.039 ± 0.005 | 0.032 ± 0.005 | 0.028 ± 0.004 | (1.7 ± 0.2) ·10−5 | (9.1 ± 1.0)·10−6 |
| 30 nM RF1 | 0.045 ± 0.004 | not measured | 0.044 ± 0.005 | not measured | (2.8 ± 0.3)·10−5 | (1.7 ± 0.3)·10−5 |
| 100 nM RF1 | 0.058 ± 0.008 | 0.046 ± 0.005 | 0.052 ± 0.006 | 0.048 ± 0.005 | (7.9 ± 0.6)·10−5 | (2.5 ± 0.2)·10−5 |
| 500 nM RF1 | 0.055 ± 0.006 | 0.059 ± 0.006 | 0.066 ± 0.008 | 0.059 ± 0.006 | (1.8 ± 0.1)·10−4 | (4.2 ± 0.1)·10−5 |
| 10 nM RF2 | (5.5 ± 0.8)·10−4 | not measured | (5.7 ± 1.0)·10−4 | not measured | not measured | not measured |
| 25 nM RF2 | (4.9 ± 1.0)·10−4 | not measured | (5.9 ± 0.7)·10−4 | not measured | not measured | not measured |
| 2 μM RF2 | (4.3 ± 0.3)·10−4 | not measured | (5.4 ± 0.4)·10−4 | not measured | (1.2 ± 0.2)·10−5 | (6.0 ± 1.9)·10−6 |
|
| ||||||
| T. th. 70S + 500 nM E. coli RF1 | 0.023 ± 0.002 | not measured | 0.020 ± 0.003 | not measured | not measured | not measured |
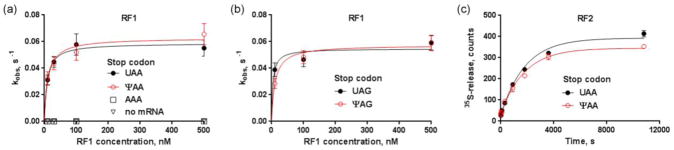
The efficiencies of peptide release catalyzed by RF1 and RF2 were similar for E. coli 70S complexes formed on UAA, ΨAA, UAG or ΨAG codons (Table 1). Apparent KM values for RF1-catalyzed reactions were in the range of 5 to 10 nM (Fig. 3a, 3b), consistent with the KM of 8.3 nM determined by Ehrenberg and colleagues for the canonical stop codons [17].
Fig. 3.
Kinetics of peptide release from 70S ribosomes programmed with pseudouridylated stop codons are similar to those with non-modified stop codons. (a) Michaelis-Menten curves for RF1-mediated release of 35S-fMet from E. coli ribosomes programmed with the stop codons UAA (filled circles) or ΨAA (clear circles, red curve), sense codon AAA (clear squares), or not bound with an mRNA (clear triangles). (b) Michaelis-Menten curves for RF1-mediated release of [35S]-fMet from E. coli ribosomes programmed with the UAG (filled circles) or ΨAG (clear circles, red curve) codon. (c) Time progress curves for RF2-mediated release of [35S]-fMet from E. coli ribosomes programmed with the UAA (filled circles) or ΨAA (clear circles, red curve) codon at 10 nM RF2.
We also tested whether our heterologous release complex (E. coli RF1 and T. thermophilus ribosome) used for crystallography behaves similarly to E. coli 70S complexes. Although E. coli RF1 on the T. thermophilus ribosome is almost three times slower than on E. coli ribosomes, the rates of release on the ΨAA and UAA codons are nearly identical (0.020 ± 0.003 s−1 and 0.023 ± 0.002 s−1 respectively). Similarly, at concentrations between 10 nM and 2 μM, E. coli RF2 (K12 strain) on ΨAA and UAA codons catalyzes peptide release with nearly identical rates (Table 1; Fig. 3c). Since the rates of peptide release are sensitive to pH [24], we have measured reaction rates at pH 6.5 (Table 1) and pH 7.5. Consistent with previous work [24], the release is faster at higher pH, however the rates remain similar for ΨAA and UAA (0.21 ± 0.04 and 0.18 ± 0.03 for 100 nM RF1; (9.0 ± 1.0)·10−4 and (9.5 ± 0.9)·10−4 for 100 nM RF2). As expected, negative control 70S complexes formed with a sense codon AAA or in the absence of mRNA are inefficient in N-formyl-methionine release (Table 1 and Fig. 3a).
Our data demonstrate that RF1 does not distinguish between the ΨAA and UAA codons, or between the ΨAG and UAG codons on E. coli and T. thermophilus ribosomes. Furthermore, RF2 does not distinguish between the ΨAA and UAA stop codons.
Interaction with L27
Bacterial ribosomes contain a conserved ribosomal protein L27, whose N-terminal tail reaches the 3′-terminal nucleotide of the P-site tRNA in an elongation complex [25]. The function of this interaction is not fully understood. Deletion of three N-terminal amino acids of L27 inhibits cell growth and impairs peptidyl-transferase activity [26]. Recent work, however, argues against the involvement of L27 in the peptidyl-transferase activity, as ΔL27 ribosomes were found to catalyze peptidyl transfer similarly to wild-type ribosomes in vitro [27]. Because the peptidyl-transferase centers of archaeal and eukaryotic ribosomes do not contain an L27 homolog and the mechanism of peptidyl transfer is universally conserved, bacterial L27 is unlikely to be critical for peptidyl transfer. L27 might, however, contribute to bacterial translation termination, which is mechanistically distinct from that in eukaryotes, as indicated by sequence and structure divergence between bacterial and eukaryotic release factors RF1/2 and eRF1, different mechanisms of stop codon recognition [9–12, 15, 16], and requirement for a translational GTPase eRF3 in eukaryotes [28–32].
Since the N-terminal tail of L27 was not modeled into previous structures of termination complexes, we examined the corresponding region in our crystal structure. Electron density in the peptidyl-transferase center reveals the N-terminal amino acids of L27 near the GGQ region of RF1 (Fig. 2c). The N-terminus of L27 packs in a cavity formed by RF1, P-site tRNA, 23S rRNA (near G2253) and L16 (near R82). N-terminal residue A2 of L27 reaches H235 of RF1 and A76 of the P-site tRNA. K5 of L27 is positioned to interact with RF1 in the vicinity of S262 and K265. This conformation of the N-terminus is different from that in the 70S ribosome with a vacant A site [33]. In our structure, the N-terminal tail is displaced by ~3 Å (measured at the backbone nitrogen of H3 of L27) to avoid steric clash with RF1 (Fig. 2c).
The conformational rearrangement of L27 and specific interactions with RF1 suggest that L27 may help to position the catalytic GGQ motif in the peptidyl-transferase center. A recent study reports that a saturating concentration (1.5 μM) of RF2 catalyzes peptide release at nearly identical rates from wild-type and ΔL27 ribosomes [27], suggesting that L27 does not modulate the catalytic activity of release factors. Instead, L27 may regulate termination efficiency by modulating the affinity of release factors.
Concluding remarks
Our structural and biochemical studies argue against the hypothesis that pseudouridylation promotes stop-codon read through by reducing the efficiency of translation termination. Interactions of the ΨAA stop codon with the ribosome and RF1 are similar to those of the UAA codon. Bacterial release factors catalyze peptide release on pseudouridylated and non-modified stop codons with similar rates. Rather than decreasing the efficiency of termination, pseudouridylation may exert its effect through increased miscoding. Previous crystallographic analyses suggest that the structural mechanism of miscoding of a pseudouridylated stop codon by tRNASer is the same as that of a canonical stop codon [8], which echoes our findings for the termination mechanism. Biochemical studies are required to test whether the efficiency of miscoding is affected by pseudouridylation. Furthermore, structural and biochemical analyses of eukaryotic termination complexes will reveal whether pseudouridylation modulates translation termination in eukaryotes.
Materials and Methods
Purification of ribosomes and ligands
70S T. thermophilus ribosomes were purified from the HB27 strain as described [11]. E. coli 70S ribosomes were purified from MRE600 cells [35], and E. coli RF1 (C-terminally His-tagged) and E. coli (K12) RF2 (N-terminally His-tagged) were purified essentially as described [9, 11]. Messenger RNAs GGCAAGGAGGUAAAAAUGXYZAAAAAA, where XYZ stands for a stop codon used in this study, were purchased from IDT (non-modified) and Dharmacon (pseudouridylated).
Crystallization and structure determination
Crystallization of the 70S•ΨAA•RF1 complex was performed essentially as described [36]. 4 μM (all concentrations are given for the final crystallization solution) T. thermophilus ribosome was incubated with 12 μM ΨAA-containing mRNA, 10 μM tRNAfMet, and 16 μM E. coli RF1 in buffer containing 25 mM Tris acetate (pH 7.0), 50 mM potassium acetate, 10 mM ammonium acetate, 10 mM magnesium acetate, and 2.8 mM Deoxy Big CHAPS (Soltec Ventures). Crystallization drops were obtained by mixing 3 μl of the 70S•ΨAA•RF1 complex and 3 μl of crystallization solution containing 0.1 M Tris·HCl (pH 7.5), 4% (v/v) PEG 20000 (Hampton Research), 8% (v/v) 2-Methyl-2,4-pentanediol (Hampton Research), and 0.2 M KSCN. Crystallization was carried out via hanging-drop vapor diffusion over 300 μl of 0.4–0.6 M NaCl. Crystals were cryoprotected by gradually (in 4 overnight steps) changing the mother liquor to 0.1 M Tris·HCl (pH 7.5), 4.5% (v/v) PEG 20000, 10% (v/v) PEG 200 (Hampton Research), 30% (v/v) 2-methyl-2,4-pentanediol, and 0.2 M KSCN. Crystals were frozen by plunging into liquid nitrogen.
Crystals were screened at the Argonne National Laboratory (beam lines 23 ID-B, 23 ID-D), Stanford Synchrotron Radiation Lightsource (beam line 12–2), and Brookhaven National Laboratory (NSLS-I, beam line X25). The data sets that resulted in the crystal structure were collected at the Argonne National Lab (beam line 23 ID-D, 1.033 Å wavelength, detector Dectris PILATUS3 6M) and SSRL (beam line 12–2, 0.9795 Å wavelength, detector Dectris PILATUS 6M) using the 0.2° oscillation range. The final data set was obtained by merging seven data sets, which were integrated and scaled using XDS. Two percent of reflections were used as a test set (Rfree set). The starting model for molecular replacement was created using two ribosome structures, which do not contain release factors (PDB ID: 5D8B [36] and PDB ID: 1VY5 [34]), from which the structural model of mRNA was omitted. Density for mRNA and RF1 was present in unbiased feature-enhanced maps (FEM) calculated as implemented [37] using PHENIX [38]. The starting model for E. coli RF1 was obtained by homology modeling from T. thermophilus RF1 (PDB ID: 4V63 [11]) using SWISSPROT [39]. Well-resolved densities for ions or solvent coordinated by the ribosome were modeled as magnesium ions, using CNS [40, 41]. Refinement was carried out using PHENIX [38]. Refinement yielded the structure with Rwork / Rfree 0.228/0.261 (Table 2). Figures were rendered using PyMOL [42].
Table 2.
Data collection and structure refinement statistics.
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 211.54, 454.40, 619.47 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 70–3.1 (3.2–3.1) |
| Rp.i.m.# | 0.237 (1.99) |
| CC(1/2)## | 99.4 (25.9) |
| I / σI | 8.28 (1.01) |
| Completeness (%) | 100 (100) |
| Redundancy | 37.4 (36.6) |
| Structure Refinement | |
| Resolution (Å) | 50–3.1 |
| No. reflections | 1,174,254 |
| Rwork / Rfree | 0.228 / 0.261 |
| Total No. atoms | 301,065 |
| Ions/water (modeled as Mg2+) | 2893 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.081 |
Values in parentheses are for the high-resolution shell.
Rp.i.m. (precision-indicating merging R factor [44]) was calculated using SCALA, which is part of the CCP4 [45] package.
CC(1/2) is the percentage of correlation between intensities from random half-datasets as defined by Karplus and Diederichs [46].
Peptide release assay
[35S]-methionine (Perkin Elmer) was used to aminoacylate E. coli tRNAfMet (Chemical Block), as previously described [43]. Termination assays were carried out essentially as described [23]. Pre-termination complex was formed by incubating [35S]-fMet-tRNAfMet with 1.5-fold molar excess of E. coli 70S ribosome for 15 minutes at 37°C in buffer containing 20 mM Tris acetate (pH 6.5, Table 1) or 20 mM Tris chloride (pH 7.5, as described in Results and Discussion), 100 mM ammonium acetate, and 20 mM magnesium acetate. The solution was diluted 22-fold with the same buffer containing 0.05% Triton X-100 (OmniPur), resulting in 10 nM pre-termination complex (10 nM [35S]-fMet-tRNAfMet and 15 nM ribosome). 1 μM mRNA (final) was added to the pre-termination complex. The complex was incubated for 5 minutes at 37°C, cooled to room temperature, and 4.5 μl of the mixture was quenched in 30 μl of 0.1 M HCl to represent the zero-time point. 45 μl of the pre-termination complex was mixed with 5 μl of a 10× solution of release factor in the buffer used to prepare the pre-termination complex, and 5 μl aliquots collected at different time points were quenched in 30 μl of 0.1 M HCl. [35S]-labeled N-formyl-methionine was extracted with 700 μl of ethylacetate, and 600 μl of extract was mixed with 3.5 ml of scintillation cocktail (Econo-Safe). Samples were counted using a scintillation counter (Beckman).
Rate constants (kobs) were obtained by single exponential fitting of the time progress curves. Values for kobs were plotted as a function of release factor concentration and fit to a hyperbola, yielding apparent KM values (Fig. 3a–b.). All experimental data were obtained from experiments performed at least twice. Fitting and visualization of kinetic data were carried out using GraphPad Prism 6.
Highlights.
Structural recognition of the ΨAA stop codon by RF1 is identical to that of the UAA codon
Biochemical experiments show that RF1 and RF2 do not distinguish between pseudouridylated and canonical stop codons
The tail of L27 in the peptidyl-transferase center rearranges upon RF1 binding
Acknowledgments
We thank the staff members of beam lines 23ID-B, 23ID-D of Advanced Photon Source (Argonne National Laboratory), X25 of National Synchrotron Light Source I (Brookhaven National Laboratory) and 12-2 of Stanford Synchrotron Radiation Lightsource (SLAC National Accelerator Laboratory) for assistance with crystal screening and data collection. We thank Daryl Conte Jr. and members of the Korostelev Lab for helpful comments on the manuscript. The study was supported by US NIH grant GM107465.
Footnotes
Accession numbers
Coordinates and structure factors were deposited in Protein Data Bank (PDB ID: 5J4D).
Author Contributions
E.S. and A.A.K. designed the project. R.M. purified proteins, ribosome and optimized the release assay. E.S. performed crystallization and peptide release experiments. E.S. and A.A.K. processed the crystallographic data and refined the structure. E.S. and A.A.K. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–6. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu G, Huang C, Yu YT. Pseudouridine in mRNA: Incorporation, Detection, and Recoding. Methods Enzymol. 2015;560:187–217. doi: 10.1016/bs.mie.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–6. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CH, Tinoco I., Jr Conformation studies of 13 trinucleoside diphosphates by 360 MHz PMR spectroscopy. A bulged base conformation. I. Base protons and H1′ protons. Biophys Chem. 1980;11:283–94. doi: 10.1016/0301-4622(80)80031-7. [DOI] [PubMed] [Google Scholar]
- 7.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395-+. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–10. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–9. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–85. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–7. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 12.Santos N, Zhu J, Donohue JP, Korostelev AA, Noller HF. Crystal structure of the 70S ribosome bound with the Q253P mutant form of release factor RF2. Structure. 2013;21:1258–63. doi: 10.1016/j.str.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci U S A. 2010;107:8593–8. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–6. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–6. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matheisl S, Berninghausen O, Becker T, Beckmann R. Structure of a human translation termination complex. Nucleic Acids Res. 2015;43:8615–26. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci U S A. 2000;97:2046–51. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parisien M, Yi C, Pan T. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA. 2012;18:355–67. doi: 10.1261/rna.031351.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–21. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura Y, Ito K. tRNA mimicry in translation termination and beyond. Wiley Interdiscip Rev RNA. 2011;2:647–68. doi: 10.1002/wrna.81. [DOI] [PubMed] [Google Scholar]
- 21.Gromiha MM, Pathak MC, Saraboji K, Ortlund EA, Gaucher EA. Hydrophobic environment is a key factor for the stability of thermophilic proteins. Proteins. 2013;81:715–21. doi: 10.1002/prot.24232. [DOI] [PubMed] [Google Scholar]
- 22.Polacek N, Gomez MJ, Ito K, Xiong L, Nakamura Y, Mankin A. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell. 2003;11:103–12. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 23.Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci U S A. 2013;110:12283–8. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Indrisiunaite G, Pavlov MY, Heurgue-Hamard V, Ehrenberg M. On the pH dependence of class-1 RF-dependent termination of mRNA translation. J Mol Biol. 2015;427:1848–60. doi: 10.1016/j.jmb.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 26.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol Cell. 2005;20:427–35. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Maracci C, Wohlgemuth I, Rodnina MV. Activities of the peptidyl transferase center of ribosomes lacking protein L27. RNA. 2015;21:2047–52. doi: 10.1261/rna.053330.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–41. [PMC free article] [PubMed] [Google Scholar]
- 29.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–78. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–73. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–72. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1 center dot eRF3 center dot GTP center dot Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Research. 2006;34:3947–54. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polikanov YS, Osterman IA, Szal T, Tashlitsky VN, Serebryakova MV, Kusochek P, et al. Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome. Mol Cell. 2014;56:531–40. doi: 10.1016/j.molcel.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21:787–93. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–94. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 36.Svidritskiy E, Korostelev AA. Ribosome Structure Reveals Preservation of Active Sites in the Presence of a P-Site Wobble Mismatch. Structure. 2015;23:2155–61. doi: 10.1016/j.str.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Afonine PV, Moriarty NW, Mustyakimov M, Sobolev OV, Terwilliger TC, Turk D, et al. FEM: feature-enhanced map. Acta Crystallogr D Biol Crystallogr. 2015;71:646–66. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 39.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Rice LM. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5:325–36. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- 43.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell. 2005;20:623–32. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Weiss MS. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130–5. [Google Scholar]
- 45.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–3. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]