Abstract
Seven ligands bind to and activate the mammalian epidermal growth factor (EGF) receptor (EGFR/ERBB1/HER1): EGF, transforming growth factor-alpha (TGFA), heparin-binding EGF-like growth factor (HBEGF), betacellulin (BTC), amphiregulin (AREG), epiregulin (EREG), and epigen (EPGN). Of these, EGF, TGFA, HBEGF, and BTC are thought to be high-affinity ligands, whereas AREG, EREG, and EPGN constitute low-affinity ligands. This focused review is meant to highlight recent studies related to actions of the individual EGFR ligands, the interesting biology that has been uncovered, and relevant advances related to ligand interactions with the EGFR.
Keywords: epidermal growth factor, EGFR, Amphireguin, Transforming growth factor-alpha, Epiregulin, Betacellulin, HBEGF, epigen
Introduction
Although the role of the epidermal growth factor receptor (EGFR) in the generation of biological responses has been reviewed extensively, an analysis of EGFR ligands, crucial initiators of these responses, has not been reviewed recently 1– 8. All EGFR ligands are synthesized as type 1 transmembrane precursors that undergo extracellular domain cleavage to release soluble ligands, which then bind to and activate the EGFR. This cleavage event is usually mediated by members of the a disintegrin and metalloprotease (ADAM) family. Understanding how these ligands are trafficked within the cell and released at the cell surface has the potential to produce significant new insights in cell biology. For example, the role of a trafficking adaptor for the transforming growth factor-alpha (TGFA) precursor (see ‘Transforming growth factor-alpha’ section) and the role of exosomal ligands in mediating receptor activation (see ‘Amphiregulin’ section) have been identified during these studies.
Aspects of individual ligands
In the following paragraphs, recent advances for each of the seven EGFR ligands are discussed. Although in this section we discuss the ligands individually, we point out that these ligands do not act in isolation but rather affect the behavior of each other to accomplish a diverse repertoire of biological responses through EGFR signaling. From a focused view in this section, the next section highlights the differences, similarities, and cross-talk among the ligands.
Epidermal growth factor
EGF is the prototypic and founding member of the EGFR ligand family, first identified from submaxillary gland extracts during nerve growth factor studies 9. The EGF-EGFR ligand-receptor system has greatly enhanced our understanding of receptor tyrosine kinase signaling, as evidenced by more than 70,000 publications for EGF alone. A recent review has distilled our current understanding of EGF and its actions 3. More recently, a study uncovered that EGF-induced EGFR signaling enhances production of intracellular reactive oxygen species (ROS) by dual oxidase 1 (DUOX1) 10. This nicely complements earlier studies in which ROS were shown to enhance EGFR signaling by modulating both positive and negative regulators of EGFR signaling (ADAMs and protein tyrosine phosphatases) 11– 14. In another recent study, urinary EGF has been shown to be an independent risk factor for progression of chronic kidney disease, substantiating earlier findings by Harris and colleagues 15– 18.
Transforming growth factor-alpha
A historical perspective of key advances for TGFA, including TGFA regulation at the level of expression, trafficking, and processing, has been provided in a recent review 7. Studies with transmembrane TGFA precursor (pro-TGFA) uncovered a novel interaction with Naked2 (NKD2) and showed that NKD2 acts as a cargo recognition and targeting (CaRT) protein for pro-TGFA 19, 20. In polarized epithelial cells, NKD2 envelops pro-TGFA-containing exocytic vesicles and directs them to the basolateral surface where the vesicles dock and fuse in an NKD2 myristoylation-dependent manner 19. In Madin-Darby canine kidney (MDCK) cells expressing myristoylation-deficient NKD2 (glycine at the second position is replaced by an alanine, G2A-NKD2), the vesicles accumulate at the basolateral “corner” and pro-TGFA is trapped in the cytoplasm 20. Basolateral delivery of pro-AREG and pro-EREG, unlike pro-TGFA, is unaffected in G2A-NKD2-expressing MDCK cells, suggesting utilization of alternate trafficking machinery 21, 22. More recently, the Schekman lab has demonstrated that cornichon-1 (CNIH) acts as a cargo receptor for pro-TGFA in the early secretory pathway 23. These findings indicate that each ligand has distinct nuances as to its biosynthetic trafficking, cell surface delivery, and ectodomain cleavage. Upstream of regulation at the protein trafficking level, TGFA can be regulated at the level of translation by microRNAs (miRs) directly or indirectly (for example, by miR-374a 24 and miR-505 25 directly and by miR-124 through Slug 26, in addition to other miRs reported earlier 7, 27). miR-203 has been noted to be a broad EGF family regulator that binds to 3′ untranslated regions of AREG, EREG, and TGFA mRNA and regulates their stability 28.
Amphiregulin
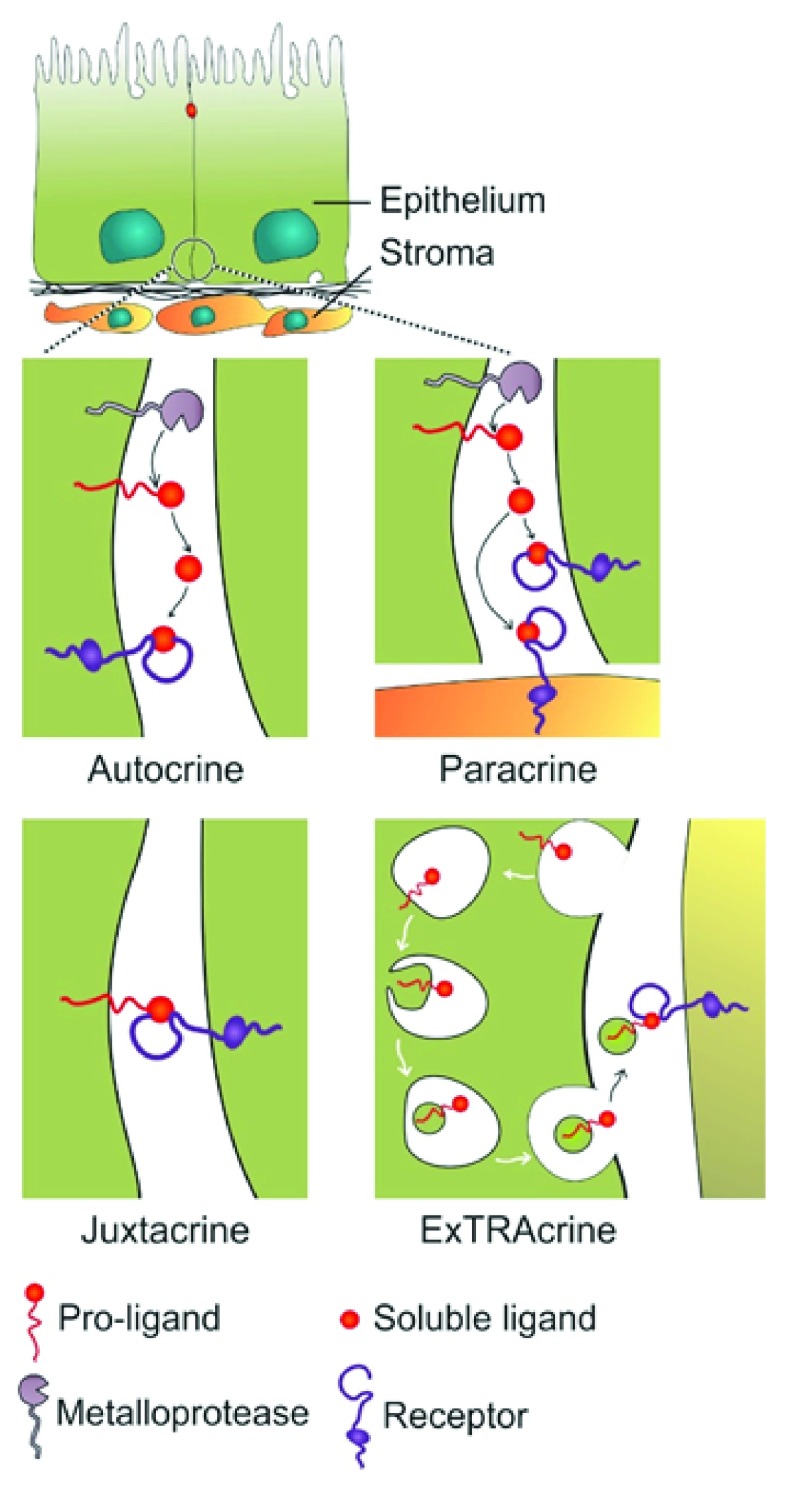
A recent review has highlighted our current understanding of AREG 2. During pro-AREG studies, a new mode of EGFR ligand signaling via exosomes was discovered 29. pro-AREG is packaged into exosomes, and pro-AREG-containing exosomes increase the invasiveness of recipient breast cancer cells. Exosomal uptake is partially dependent on ligand-receptor interaction as treatment of recipient cells with EGFR blocking monoclonal antibody attenuated the uptake of pro-AREG-containing exosomes. We have termed this new mode of EGFR activation by exosomal ligands as ExTRAcrine (exosomal targeted receptor activation) signaling. As noted in Figure 1, ExTRAcrine signaling has features of autocrine, paracrine, and juxtacrine signaling. It is possibly involved in endocrine signaling as well, since EGFR and pro-AREG can be detected in human plasma exosomes 30.
Figure 1. Modes of signaling via epidermal growth factor receptor (EGFR) ligands.
Autocrine signaling occurs when a ligand is released from a cell and binds to EGFR on that same cell. Paracrine signaling refers to the released ligand acting on a nearby cell, usually a different cell type. Juxtacrine signaling occurs when a non-cleaved, transmembrane ligand binds to EGFR on an adjacent cell; this is best documented for heparin-binding epidermal growth factor-like growth factor (HBEGF). Amphiregulin (AREG), transforming growth factor-alpha (TGFA), and HBEGF, as well as EGFR, can be packaged into signaling competent exosomes. Uptake of exosomal AREG by recipient cells is, at least in part, dependent on EGFR, leading to the term exosomal targeted receptor activation (ExTRAcrine). ExTRAcrine signaling has features of autocrine, paracrine, and juxtacrine signaling as well as possibly endocrine signaling since EGFR and AREG can be detected in human plasma exosomes 30. Adapted from Singh and Coffey 36.
There is accumulating evidence that AREG is produced in a number of cells other than epithelial cells and fibroblasts. Artis and colleagues have identified roles for AREG in immune surveillance 31. Damaged epithelial cells release interleukin-33 (IL-33), IL-25, and thymic stromal lymphopoietin (TSLP), which can activate group 2 innate lymphoid cells to release AREG, as well as IL-5, IL-9, and IL-13. It is thought that AREG ameliorates injury by binding to epithelial EGFR and stimulating proliferation and repair. This does not exclude an effect of AREG on EGFR-expressing, non-epithelial cells, including fibroblasts, polymorphonuclear cells, and Fox3 + regulatory T cells; the latter two have been reported to express EGFR by flow cytometry 31, 32. A difficulty in studying the actions of the ligands in the mouse is the lack of robust antibodies to examine mouse EGFR by immunofluorescence or immunohistochemistry, or both.
A variety of stresses, such as inflammation (with lipopolysaccharides), ischemia, and hypoxia, induce AREG and EREG expression in the brain (cortex, striatum, and hippocampus). Under these stresses, glial cells show upregulation of EREG and AREG, which, when released, may protect against neuronal cell death. In Neuro2a cells, administration of EREG or AREG inhibits tunicamycin-induced endoplasmic reticulum (ER) stress and cell death 33. Recent work by Elder’s group continues to implicate a role for the C-terminal domain of AREG in promoting keratinocyte proliferation 34. In addition, Yarden’s group has shown preclinical efficacy for an AREG neutralizing antibody in ovarian cancer 35.
Epiregulin
EREG binds to and activates EGFR and ERBB4. EREG is weakly expressed in normal adult tissues but is overexpressed in diseases like cancer. Multiple roles for EREG in normal physiology and disease states have been reviewed recently 4. However, the basic biological processes, such as the polarized distribution of pro-EREG within cells and the subsequent spatial control of EGFR activity, have been understudied. We have identified a new mode of epithelial transformation by apical mistrafficking of pro-EREG 22. In polarized MDCK cells, pro-EREG is delivered to the basolateral membrane and its delivery is dependent on a tyrosine-based (YXXΦ) sorting motif. Disruption of this motif leads to complete apical delivery; however, pro-EREG basolateral sorting is independent of the usual YXXΦ-recognizing clathrin adaptor, AP1B 22. This apical rerouting of pro-EREG leads to activation of apical EGFR signaling, which has a different activation profile compared with basolateral EGFR signaling. MDCK cells expressing an apical mistrafficking mutant of EREG (Y156A) are more transforming than their wild-type, basolateral EREG-expressing counterparts when injected subcutaneously in nude mice. Additionally, there are mutations in human cancers that would disrupt the sorting motifs of the majority of EGFR ligands, including pro-EREG 36. In a recent study with breast epithelial MCF10A cells, EREG expression was shown to contribute to tumor progression during early stages of cancer 37. A comprehensive understanding of pathways that govern spatial compartmentalization of the EGFR ligands might reveal alternate approaches to treat cancer.
Recently, additional functions of EREG have been uncovered. In circulating monocytes, EREG is upregulated acutely during short bursts of exercise and intermittent hypoxia in rat aortic smooth muscle cells. These facts may have implications for atherosclerosis 38. Additionally, EREG is shown to assist in the proliferation, repair, or regeneration (or a combination of these) in liver, colon, and salivary gland acinar cells 39– 42. EREG also plays a role in odontogenesis by enhancing proliferation of dental apical papilla stem cells and inducing oral epithelial cell differentiation by dental papilla cells 43.
Wrana and colleagues have identified EGFR-dependent Yap signaling in the intestine which contributes to regeneration and tumorigenesis 44. They reported that EREG, but not EGF or AREG, is able to maintain growth of organoids generated from Yap null mice; however, it should be noted that the concentration of recombinant mouse EREG given was very high and the source of the other ligands was unclear.
Betacellulin
Betacellulin (BTC) is a dual-specificity ligand that binds to and activates EGFR and ERBB4. In a recent review, structural details and key functions of BTC gleaned from knockouts, transgenic animals, patient samples, and in vitro studies have been reviewed 8. Recently, it has also been shown that pro-BTC localizes to the basolateral membrane 45. pro-BTC basolateral sorting is dependent on a cytoplasmic EEXXXL motif, and disruption of this motif, or introduction of a human cancer mutation (E156K) within this motif, leads to pro-BTC mistrafficking. An analogous EEXXXL motif is also responsible for basolateral sorting of pro-AREG 21. This report also demonstrated that pro-BTC mistrafficking induces an EGFR-dependent hepatic polarity phenotype (apical surfaces on the side, between two cells) in otherwise columnarly polarized MDCK cells, a finding not observed with any of the other EGFR ligands.
Recent publications have uncovered additional functions of BTC. BTC transgenic mice display high cortical bone mass 46. Additionally, in bone metastases associated with castration-resistant prostate cancer, BTC is upregulated in osteoblasts and contributes to osteoblastic activity 47. BTC transgenic mice also develop urothelial hyperplasia and show sex-dependent reduction in urinary protein content, which appears to be independent of EGFR signaling, suggesting a role for ERBB4 48. BTC has also been identified as a novel modulator of interferon (IFN) response and enhances the anti-viral action of IFN 49. In a large cytokine and chemokine screen to modulate IFN responses, BTC was identified as one of the most potent modulators of the IFN response. Moreover, miR-200 has been shown to control BTC (and AREG) translation 50.
Heparin-binding epidermal growth factor-like growth factor
Roles for heparin-binding EGF-like growth factor (HBEGF) in multiple cellular processes in normal and disease states have been recently reviewed 5. Among the EGFR ligands, pro-HBEGF has the longest residency time at the cell surface, perhaps explaining why pro-HBEGF functions as a receptor for the B fragment of Diphtheria toxin. Since the toxin binds only to human and monkey pro-HBEGF, mouse modelers have exploited this selective binding by introducing human pro-HBEGF into the ATG start site of mouse genes expressed in selected cell types, which can then be eliminated by administration of the toxin 38, 39. However, when repurposing pro-HBEGF in this way, possible “side effects” of human HBEGF (that can bind to mouse EGFR and ERBB4) need to be considered 51.
Local administration of HBEGF helps mice recover from chronic suppurative otitis media, a chronic inflammation of the middle ear 52. Delivery of EGF or FGF2 was not effective 53. It is unclear whether auto- or cross-induction (or both) of other ligands play a role in this process 54, 55. HBEGF mRNA is also a target for miR-132, both of which play a major role in wound healing 56. During the transition from inflammation to proliferation in wound healing, miR-132 expression was upregulated together with a concomitant decrease in HBEGF levels. Surprisingly, HBEGF downregulation coincided with the proliferative phase during wound healing and increased receptor activity. Other miRs (for example, miR-96, miR-212, and miR1192) have also been shown to target HBEGF 57– 59.
Epigen
Epigen (EPGN), the most recently discovered EGFR ligand, seems to be a low-affinity EGFR ligand. The localization of pro-EPGN in polarized epithelial cells is not known and its cytoplasmic domain lacks any recognized basolateral sorting motifs 36. Schneider and Yarden have recently reviewed EPGN structure and function 6. EPGN knockout mice do not display an obvious phenotype 60; however, transgenic overexpression of EPGN during embryonic development induces sebaceous gland hyperplasia 61. Interestingly, activation of the transcription factor Nrf2, a master regulator of cellular anti-oxidant defense, causes sebaceous gland enlargement in an EPGN-dependent manner 62. Pharmacologic activation of Nrf2 has been employed as a cancer prevention strategy and this is due in part to its role in ROS detoxification 63. However, Nrf2 activation-induced EPGN upregulation and subsequent EGFR activation might actually be pro-tumorigenic and act counter to its anti-cancer effects. EGFR signaling also regulates Nrf2 activity; in cortical neurons, astragaloside IV (extracted from Astragalus membranaceus) induces HBEGF-dependent EGFR transactivation that leads to Ser40 phosphorylation of Nrf2 and its nuclear translocation 64. EPGN transgenic mice also show a peripheral demyelinating neuropathy, leading to late-onset muscular dystrophy 65.
Different ligands, differing functions
EGFR can be activated by seven related, but distinct, ligands. Multiple publications have noted the capacity of these different ligands to act in a “functionally selective” manner (that is, to produce quantitatively and, to a lesser extent, qualitatively distinct cellular responses 66– 68). It is important to note that EGFR is one of four members of a family of related receptors. The others are designated as ERBB2/HER2, ERBB3/HER3, and ERBB4/HER4. Of the ligands that bind to the EGFR, three (EREG, HBEGF, and BTC) are known to bind to and activate ERBB4. None of the EGFR ligands is known to interact with ERBB2 or ERBB3. A family of EGF-related ligands, termed neuregulins, binds to ERBB3 and ERBB4. There is no known ligand for ERBB2.
In some cases, the ligand functional selectivity could be due to the capacity of some of the ligands to activate ERBB4; however, differences also exist in cells that do not express detectable levels of ERBB4. As noted above, four ligands (EGF, TGFA, AREG, and EPGN) interact solely through EGFR, yet they do not produce identical biological responses. In particular, AREG is often regarded as a low-affinity ligand for the EGFR. Crystallographic studies have described high-resolution ligand:ectodomain structures for EGF:EGFR, TGFA:EGFR, and NRG-1β:ERBB4 69. However, these studies do not provide an explanation for downstream differences in biological activity.
Recently, a comparative study 70 of EGF, TGFA, AREG, and BTC binding to the EGFR and subsequent dimer formation has made clear that EGF and TGFA have a distinct preference to produce EGFR:ERBB2 heterodimers compared with EGFR:EGFR homodimers, but that BTC and AREG produced dimers of both types equally. In addition, AREG produced significantly (50%) fewer dimers of either type compared with the other ligands. These initial data point to dimerization-competent, conformation-based receptor differences provoked by different ligands as a potential basis for heterogeneity in signaling and biological outcomes. These data and the related issues of receptor methylation and receptor antagonists (discussed below) will need to be evaluated.
Another fact, sometimes forgotten or ignored, is that EGFR ligands can auto- and cross-induce one another, adding a layer of complexity to studies of an individual ligand 54, 55. In addition, the often observed co-expression of EPGN, EREG, AREG, and BTC, which may be due to their chromosomal clustering on human 4q13-21 and 5E1 in the mouse, further complicates our understanding of the role of individual ligands in biological processes 55. ExTRAcrine signaling and auto- and cross-induction of EGFR ligands are variables that merit consideration in systems biology approaches to the merging fields of autocrine signaling and quorum sensing, as reviewed elsewhere 71.
Interestingly, exogenous administration of ligands has recently been used as a treatment strategy for certain disease conditions in mice. Table 1 lists distinct actions recently observed for individual EGFR ligands in vivo. Once again, this is a highly selective compilation and we apologize for any omissions. We have chosen to highlight studies that identify new functions of the individual ligands.
Table 1. In vivo administration of epidermal growth factor receptor ligands as treatment strategies.
| Ligand | Mode of administration | Effect | Reference |
|---|---|---|---|
| EGF | One-week perfusion with
ciliary neurotrophic factor via mini-osmotic pump |
Acinar to beta cell transdifferentiation for
up to 248 days in adult mice with chronic hyperglycemia |
88 |
| HBEGF | Intranasal | Reduced oligodendrocytic death when
given immediately after injury in a mouse model of pre-term brain injury |
89 |
| HBEGF | HBEGF-containing hydrogel
injected through the external auditory canal |
Regeneration of chronic tympanic
membrane perforations in mice |
52 |
| HBEGF | Topical application | Accelerated wound healing in a diabetic
mouse model |
90 |
Exogenous administration of the soluble epidermal growth factor receptor ligands for the treatment of various disease states in animal disease models. EGF, epidermal growth factor; HBEGF, heparin-binding epidermal growth factor-like growth factor.
Ligand processing and delivery to the cell surface
As noted above, all seven mammalian EGFR ligands are synthesized as a type 1 transmembrane precursor and are processed through biosynthetic compartments common to other secretory and cell surface proteins. We have mentioned earlier that mammalian ligands are usually cleaved by ADAMs. Owing to the focus on EGFR ligands in this review, we have not elaborated on ADAM activity in cleavage of EGFR itself. ADAM-mediated EGFR cleavage acts as a negative feedback for EGFR signaling and acts in concert with the positive feedback through ligand shedding 72. Drosophila genetics has identified the involvement of particular gene products (Rhomboid, Star) for intracellular trafficking of the fly EGFR ligands. Recent work in mammalian cells has found at least two novel roles for inactive rhomboids (iRhoms) in the processing of EGFR ligand precursors 73.
Rhomboid gene products are known to be seven membrane-spanning molecules, which function as intramembrane serine proteases that cleave various transmembrane molecules within the cell or at the cell surface 73. The rhomboid family also includes catalytically inactive proteins termed iRhoms. Although rhomboid cleavage of the fly EGFR ligand precursor is required for processing to the cell surface, the presence of iRhoms in the biosynthetic pathway prevents this cleavage and leads to intracellular degradation of EGFR ligand precursors 74. iRhoms localize to the ER. In this way, iRhoms block the cell surface expression of multiple EGFR ligands, several of which are not substrates for the catalytic rhomboids. Hence, the iRhoms may not just compete with rhomboids, but rather regulate EGFR ligand levels by an independent mechanism, suggesting that the iRhom gene may have evolved a distinct regulatory function. Consistent with this idea, during evolution the iRhoms have acquired large segments of sequence not shared with the catalytic rhomboids. In flies, the independent mechanism that regulates intracellular ligand levels is identified as the ER-associated protein degradation pathway that leads to proteosomal degradation 73.
A second, less direct, mechanism by which iRhoms control the production of bioactive secreted EGFR ligands is through control of the ultimate step in processing: the metalloprotease-mediated, cell-surface cleavage of the ligand precursor. This step is executed by members of the ADAM protease family of which ADAM17 is probably the most significant member for EGFR ligands. ADAMs are single-pass transmembrane proteins that are trafficked through the ER and Golgi before reaching the cell surface, where the ectodomain proteolytically cleaves the ectodomain of substrates, such as EGFR ligand precursors. During intracellular trafficking, ADAMs are converted by furin-dependent proteolytic processing in the Golgi from an inactive form to a mature active species, and this requires iRhoms 75– 78. When mammalian iRhoms are deleted or knocked down, no mature cell surface ADAM17 molecules are produced and in turn ADAM17 substrates, such as EGFR ligand precursors are not cleaved. In mammals, there are two iRhoms (RHBDF1/iRhom1 and RHBDF2/iRhom2), which seem to have overlapping effects on ADAM17 maturation, depending on the cell type. iRhom effects, however, are selective toward ADAM17. Since ADAM17 also cleaves the inflammatory tumor necrosis factor precursor, iRhoms may influence both cell proliferation and inflammatory pathways. Interestingly, dominant iRhom2 mutations have been detected in an inherited syndrome, tylosis (thickening of the palms and soles), in patients with esophageal cancer 79. The disorder appears to be due to mutations in the N-terminal cytosolic domain of iRhom2 that stabilize the protein. These mutations have been linked to increased release of AREG (and HBEGF) and increased EGFR activity. The impact of these mutations on ADAM17 activity is unsettled 79– 81. A spontaneous recessive mutation in this domain of iRhom2 has been identified in a mouse with a hairless phenotype called curly bare (cub) 82. Of interest, there is a suppressor of Cub, Mcub, in which there is a loss-of-function mutation in mouse AREG 82.
Ligand interactions with receptors
Although the complexities presented by multiple ligands binding to multiple ErbB receptors are described above and in more detail elsewhere 68, 70, a few recent publications give additional parameters to consider. First, an antagonist has now been described for the mammalian receptors 83, which adds to the negative control of ligand receptor interaction described some time ago for the Drosophila EGFR system. In flies, the secreted molecule Argos is able to associate with fly EGF and prevent ligand binding to the Drosophila EGF receptor (DER) 84, 85. Although an Argos equivalent has not been detected in mammals, Zheng and colleagues 83 report that migration inhibitory factor (MIF), which is O-glycosylated and secreted, binds to EGFR and blocks EGF binding. This antagonist is, therefore, mechanistically distinct from Argos. The report does show that MIF blocks EGF binding to its receptor in cell culture, preventing activation of the receptor and downstream signaling pathways, and that recombinant MIF interacts with the recombinant EGFR ectodomain in a purified system, but there are a few missing pieces to this provocative and potentially significant report. Does MIF interact with the EGFR at biologically significant concentrations? What is the receptor interaction site for MIF and does it overlap with the known ligand binding sites? Is there specificity to the MIF interaction within the ERBB system or unrelated receptors? Importantly, this study also shows that EGF activation of its receptor induces the secretion of a metalloprotease (MMP 13) that degrades MIF and thus provides a feedback loop to this EGFR antagonist.
The EGFR is subject to a variety of co- and post-translational modifications that have significant roles in the capacity of the receptor to transduce second messenger systems following ligand binding. Recently, the role of methylation in mediating high-affinity ligand binding has been described 86. Methylation at R198 and R200 within the EGFR ectodomain is reported to mediate high-affinity EGF binding. When receptor methylation is prevented by mutagenesis of the two Arg residues or by knockdown of the relevant methyltransferase (PRMT1), dissociation constant (Kd) values are altered approximately threefold to reflect a loss of higher affinity binding compared with the wild-type receptor. Decreased ligand-dependent receptor dimerization, activation, and downstream signaling, including tumorigenesis, are observed in the absence of receptor methylation, compared with wild-type receptor. Similarly, exogenous expression of PRMT1 increased high-affinity ligand binding to the EGFR, as well as receptor-mediated downstream signaling events. The study also provides evidence that a pool of PRMT1 is localized within the ER.
The EGFR ectodomain is often subdivided into four regions (D1, D2, D3, and D4) with EGF binding requiring molecular contacts with D1 and D3. In untreated cells, over 90% of the receptor exists in an inactive or “tethered” state involving interaction of residues in D2 and D4 that sterically prevent ligand binding to the D1 and D3 regions. Methylation of R198 and R200, located in D2, is proposed on the basis of molecular modeling to destabilize the tethered conformation and thereby increase receptor in the extended conformation, which allows high-affinity binding to D1 and D3. In the past, the basis of high-affinity binding has been attributed to receptor heterogeneity or negative cooperativity 87. Recent studies that have supported these mechanisms may have to be adjusted to include receptor methylation status. The level of methylated receptor is estimated to be approximately 10% of the EGFR population, which is about the same as the high-affinity binding receptor pool. Since methylation of the EGFR ectodomain is reported to occur within the lumen of the ER/Golgi during biosynthesis, the authors concluded that the level of PRMT1 activity in that compartment may determine the size of the high-affinity receptor pool. If the above studies are confirmed, methylation may also contribute to high-affinity binding.
Analyses of colorectal tumor tissue showed an increased level of methylated EGFR compared with control tissue. Methylated receptor was correlated with a worse overall patient survival and higher recurrence rate. Cetuximab, an EGFR neutralizing monoclonal antibody, is used to treat certain cancers, particularly colon cancer. This antibody binds to the D3 region of the EGFR, thereby blocking ligand binding. Methylation not only increased high-affinity EGF binding but also decreased the capacity of cetuximab to interfere with ligand binding. In tissues of patients with cancer, the presence of methylated receptor was a predictor of poor patient response to this therapeutic agent.
In summary, study of the EGFR and its ligands, since their discovery more than fifty years ago, continues to yield important insights into multiple biological processes. From oocyte maturation, blastocyst implantation, and embryonic development to organ development and maintenance and diseases like cancer, this line of investigation continues to be broadly relevant and clinically important.
Abbreviations
ADAM, a disintegrin and metalloprotease; AREG, amphiregulin; BTC, betacellulin; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; EPGN, epigen; ER, endoplasmic reticulum; EREG, epiregulin; ExTRAcrine, exosomal targeted receptor activation; HBEGF, heparin-binding epidermal growth factor-like growth factor; IFN, interferon; IL, interleukin; iRhom, inactive rhomboid; MDCK, Madin-Darby canine kidney; MIF, migration inhibitory factor; miR, microRNA; NKD2, Naked2; TGFA, transforming growth factor-alpha.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Edward Leof, Division of Pulmonary and Critical Care Medicine, Department of Medicine, Mayo Clinic, Rochester, MN, USA; Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN, USA
Mark A. Lemmon, Department of Pharmacology, Yale University School of Medicine, New Haven, CT, USA; Cancer Biology Institute, Yale University, West Haven, CT, USA
Douglas A. Lauffenburger, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA
Funding Statement
The work is supported by National Cancer Institute grants R01-CA46413, R01-CA163563, and P50-CA95103 to Robert J. Coffey.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
References
- 1. Harris R, Chung E, Coffey RJ: EGF receptor ligands. Exp Cell Res. 2003;284:2–13. 10.1016/S0014-4827(02)00105-2 [DOI] [PubMed] [Google Scholar]
- 2. Berasain C, Avila MA: Amphiregulin. Semin Cell Dev Biol. 2014;28:31–41. 10.1016/j.semcdb.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 3. Zeng F, Harris RC: Epidermal growth factor, from gene organization to bedside. Semin Cell Dev Biol. 2014;28:2–11. 10.1016/j.semcdb.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riese DJ 2nd, Cullum RL: Epiregulin: roles in normal physiology and cancer. Semin Cell Dev Biol. 2014;28:49–56. 10.1016/j.semcdb.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor SR, Markesbery MG, Harding PA: Heparin-binding epidermal growth factor-like growth factor (HB-EGF) and proteolytic processing by a disintegrin and metalloproteinases (ADAM): a regulator of several pathways. Semin Cell Dev Biol. 2014;28:22–30. 10.1016/j.semcdb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- 6. Schneider MR, Yarden Y: Structure and function of epigen, the last EGFR ligand. Semin Cell Dev Biol. 2014;28:57–61. 10.1016/j.semcdb.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh B, Coffey RJ: From wavy hair to naked proteins: the role of transforming growth factor alpha in health and disease. Semin Cell Dev Biol. 2014;28:12–21. 10.1016/j.semcdb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dahlhoff M, Wolf E, Schneider MR: The ABC of BTC: structural properties and biological roles of betacellulin. Semin Cell Dev Biol. 2014;28:42–8. 10.1016/j.semcdb.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 9. Cohen S: Purification of a nerve-growth promoting protein from the mouse salivary gland and its neuro-cytotoxic antiserum. Proc Natl Acad Sci U S A. 1960;46(3):302–11. 10.1073/pnas.46.3.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sirokmány G, Pató A, Zana M, et al. : Epidermal growth factor-induced hydrogen peroxide production is mediated by dual oxidase 1. Free Radic Biol Med. 2016;97:204–11. 10.1016/j.freeradbiomed.2016.05.028 [DOI] [PubMed] [Google Scholar]
- 11. Singh B, Schneider M, Knyazev P, et al. : UV-induced EGFR signal transactivation is dependent on proligand shedding by activated metalloproteases in skin cancer cell lines. Int J Cancer. 2009;124(3):531–9. 10.1002/ijc.23974 [DOI] [PubMed] [Google Scholar]
- 12. Knebel A, Rahmsdorf HJ, Ullrich A, et al. : Dephosphorylation of receptor tyrosine kinases as target of regulation by radiation, oxidants or alkylating agents. EMBO J. 1996;15(19):5314–25. [PMC free article] [PubMed] [Google Scholar]
- 13. Tomic S, Greiser U, Lammers R, et al. : Association of SH2 domain protein tyrosine phosphatases with the epidermal growth factor receptor in human tumor cells. Phosphatidic acid activates receptor dephosphorylation by PTP1C. J Biol Chem. 1995;270(36):21277–84. 10.1074/jbc.270.36.21277 [DOI] [PubMed] [Google Scholar]
- 14. Heppner DE, van der Vliet A: Redox-dependent regulation of epidermal growth factor receptor signaling. Redox Biol. 2016;8:24–7. 10.1016/j.redox.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Safirstein R, Price PM, Saggi SJ, et al. : Changes in gene expression after temporary renal ischemia. Kidney Int. 1990;37:1515–21. 10.1038/ki.1990.143 [DOI] [PubMed] [Google Scholar]
- 16. Storch S, Saggi S, Megyesi J, et al. : Ureteral obstruction decreases renal prepro-epidermal growth factor and Tamm-Horsfall expression. Kidney Int. 1992;42(1):89–94. 10.1038/ki.1992.265 [DOI] [PubMed] [Google Scholar]
- 17. Safirstein R, Zelent AZ, Price PM: Reduced renal prepro-epidermal growth factor mRNA and decreased EGF excretion in ARF. Kidney Int. 1989;36(5):810–5. 10.1038/ki.1989.266 [DOI] [PubMed] [Google Scholar]
- 18. Ju W, Nair V, Smith S, et al. : Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med. 2015;7(316):316ra193. 10.1126/scitranslmed.aac7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li C, Franklin JL, Graves-Deal R, et al. : Myristoylated Naked2 escorts transforming growth factor alpha to the basolateral plasma membrane of polarized epithelial cells. Proc Natl Acad Sci U S A. 2004;101(15):5571–6. 10.1073/pnas.0401294101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li C, Hao M, Cao Z, et al. : Naked2 acts as a cargo recognition and targeting protein to ensure proper delivery and fusion of TGF-alpha containing exocytic vesicles at the lower lateral membrane of polarized MDCK cells. Mol Biol Cell. 2007;18(8):3081–93. 10.1091/mbc.E07-02-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gephart JD, Singh B, Higginbotham JN, et al. : Identification of a novel mono-leucine basolateral sorting motif within the cytoplasmic domain of amphiregulin. Traffic. 2011;12(12):1793–804. 10.1111/j.1600-0854.2011.01282.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh B, Bogatcheva G, Washington MK, et al. : Transformation of polarized epithelial cells by apical mistrafficking of epiregulin. Proc Natl Acad Sci U S A. 2013;110(22):8960–5. 10.1073/pnas.1305508110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang P, Schekman R: Distinct stages in the recognition, sorting, and packaging of proTGFα into COPII-coated transport vesicles. Mol Biol Cell. 2016;27(12):1938–47. 10.1091/mbc.E16-02-0090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu H, Liu Y, Shu XO, et al. : MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis. 2016;37(6):567–75. 10.1093/carcin/bgw038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen S, Sun KX, Liu BL, et al. : MicroRNA-505 functions as a tumor suppressor in endometrial cancer by targeting TGF-α. Mol Cancer. 2016;15:11. 10.1186/s12943-016-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin W, Pan Y, Zheng X, et al. : MicroRNA-124 regulates TGF-α-induced epithelial-mesenchymal transition in human prostate cancer cells. Int J Oncol. 2014;45(3):1225–31. 10.3892/ijo.2014.2506 [DOI] [PubMed] [Google Scholar]
- 27. Kalinowski FC, Giles KM, Candy PA, et al. : Regulation of epidermal growth factor receptor signaling and erlotinib sensitivity in head and neck cancer cells by miR-7. PLoS One. 2012;7(10):e47067. 10.1371/journal.pone.0047067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siu MK, Abou-Kheir W, Yin JJ, et al. : Loss of EGFR signaling regulated miR-203 promotes prostate cancer bone metastasis and tyrosine kinase inhibitors resistance. Oncotarget. 2014;5(11):3770–84. 10.18632/oncotarget.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Higginbotham JN, Demory Beckler M, Gephart JD, et al. : Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21(9):779–86. 10.1016/j.cub.2011.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Higginbotham JN, Zhang Q, Jeppesen DK, et al. : Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles. 2016;5:29254. 10.3402/jev.v5.29254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaiss DM, Gause WC, Osborne LC, et al. : Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42(2):216–26. 10.1016/j.immuni.2015.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lewkowicz N, Lewkowicz P, Banasik M, et al. : Predominance of Type 1 cytokines and decreased number of CD4 +CD25 +high T regulatory cells in peripheral blood of patients with recurrent aphthous ulcerations. Immunol Lett. 2005;99(1):57–62. 10.1016/j.imlet.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 33. Zhan L, Zheng L, Hosoi T, et al. : Stress-induced neuroprotective effects of epiregulin and amphiregulin. PLoS One. 2015;10(2):e0118280. 10.1371/journal.pone.0118280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stoll SW, Stuart PE, Lambert S, et al. : Membrane-Tethered Intracellular Domain of Amphiregulin Promotes Keratinocyte Proliferation. J Invest Dermatol. 2016;136(2):444–52. 10.1016/j.jid.2015.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carvalho S, Lindzen M, Lauriola M, et al. : An antibody to amphiregulin, an abundant growth factor in patients' fluids, inhibits ovarian tumors. Oncogene. 2016;35(4):438–47. 10.1038/onc.2015.93 [DOI] [PubMed] [Google Scholar]
- 36. Singh B, Coffey RJ: Trafficking of epidermal growth factor receptor ligands in polarized epithelial cells. Annu Rev Physiol. 2014;76:275–300. 10.1146/annurev-physiol-021113-170406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farooqui M, Bohrer LR, Brady NJ, et al. : Epiregulin contributes to breast tumorigenesis through regulating matrix metalloproteinase 1 and promoting cell survival. Mol Cancer. 2015;14:138. 10.1186/s12943-015-0408-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radom-Aizik S, Zaldivar FP Jr, Haddad F, et al. : Impact of brief exercise on circulating monocyte gene and microRNA expression: implications for atherosclerotic vascular disease. Brain Behav Immun. 2014;39:121–9. 10.1016/j.bbi.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tomita K, Haga H, Mizuno K, et al. : Epiregulin promotes the emergence and proliferation of adult liver progenitor cells. Am J Physiol Gastrointest Liver Physiol. 2014;307(1):G50–7. 10.1152/ajpgi.00434.2013 [DOI] [PubMed] [Google Scholar]
- 40. Lee D, Pearsall RS, Das S, et al. : Epiregulin is not essential for development of intestinal tumors but is required for protection from intestinal damage. Mol Cell Biol. 2004;24(20):8907–16. 10.1128/MCB.24.20.8907-8916.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagai K, Arai H, Okudera M, et al. : Epiregulin is critical for the acinar cell regeneration of the submandibular gland in a mouse duct ligation model. J Oral Pathol Med. 2014;43(5):378–87. 10.1111/jop.12145 [DOI] [PubMed] [Google Scholar]
- 42. Cao Y, Xia DS, Qi SR, et al. : Epiregulin can promote proliferation of stem cells from the dental apical papilla via MEK/Erk and JNK signalling pathways. Cell Prolif. 2013;46(4):447–56. 10.1111/cpr.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao B, Zhou X, Zhou X, et al. : BMP7 and EREG Contribute to the Inductive Potential of Dental Mesenchyme. Sci Rep. 2015;5: 9903. 10.1038/srep09903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gregorieff A, Liu Y, Inanlou MR, et al. : Yap-dependent reprogramming of Lgr5 + stem cells drives intestinal regeneration and cancer. Nature. 2015;526(7575):715–8. 10.1038/nature15382 [DOI] [PubMed] [Google Scholar]
- 45. Singh B, Bogatcheva G, Starchenko A, et al. : Induction of lateral lumens through disruption of a monoleucine-based basolateral-sorting motif in betacellulin. J Cell Sci. 2015;128(18):3444–55. 10.1242/jcs.170852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider MR, Mayer-Roenne B, Dahlhoff M, et al. : High cortical bone mass phenotype in betacellulin transgenic mice is EGFR dependent. J Bone Miner Res. 2009;24(3):455–67. 10.1359/jbmr.081202 [DOI] [PubMed] [Google Scholar]
- 47. Larson SR, Chin J, Zhang X, et al. : Prostate cancer derived prostatic acid phosphatase promotes an osteoblastic response in the bone microenvironment. Clin Exp Metastasis. 2014;31(2):247–56. 10.1007/s10585-013-9625-2 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Schulz H, Dahlhoff M, Glogowska A, et al. : Betacellulin transgenic mice develop urothelial hyperplasia and show sex-dependent reduction in urinary major urinary protein content. Exp Mol Pathol. 2015;99(1):33–8. 10.1016/j.yexmp.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 49. Al-Yahya S, Mahmoud L, Al-Zoghaibi F, et al. : Human Cytokinome Analysis for Interferon Response. J Virol. 2015;89(14):7108–19. 10.1128/JVI.03729-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang Y, Ahn YH, Chen Y, et al. : ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J Clin Invest. 2014;124(6):2696–708. 10.1172/JCI72171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Blijswijk J, Schraml BU, Rogers NC, et al. : Altered lymph node composition in diphtheria toxin receptor-based mouse models to ablate dendritic cells. J Immunol. 2015;194(1):307–15. 10.4049/jimmunol.1401999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Santa Maria PL, Kim S, Varsak YK, et al. : Heparin binding-epidermal growth factor-like growth factor for the regeneration of chronic tympanic membrane perforations in mice. Tissue Eng Part A. 2015;21(9–10):1483–94. 10.1089/ten.tea.2014.0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Santa Maria PL, Weierich K, Kim S, et al. : Heparin Binding Epidermal Growth Factor-Like Growth Factor Heals Chronic Tympanic Membrane Perforations With Advantage Over Fibroblast Growth Factor 2 and Epidermal Growth Factor in an Animal Model. Otol Neurotol. 2015;36(7):1279–83. 10.1097/MAO.0000000000000795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coffey RJ Jr, Derynck R, Wilcox JN, et al. : Production and auto-induction of transforming growth factor-alpha in human keratinocytes. Nature. 1987;328(6133):817–20. 10.1038/328817a0 [DOI] [PubMed] [Google Scholar]
- 55. Barnard JA, Graves-Deal R, Pittelkow MR, et al. : Auto- and cross-induction within the mammalian epidermal growth factor-related peptide family. J Biol Chem. 1994;269(36):22817–22. [PubMed] [Google Scholar]
- 56. Li D, Wang A, Liu X, et al. : MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. J Clin Invest. 2015;125(8):3008–26. 10.1172/JCI79052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang M, Pan Y, Zhou Y: miR-96 promotes osteogenic differentiation by suppressing HBEGF-EGFR signaling in osteoblastic cells. FEBS Lett. 2014;588(24):4761–8. 10.1016/j.febslet.2014.11.008 [DOI] [PubMed] [Google Scholar]
- 58. Wei LQ, Liang HT, Qin DC, et al. : MiR-212 exerts suppressive effect on SKOV3 ovarian cancer cells through targeting HBEGF. Tumour Biol. 2014;35(12):12427–34. 10.1007/s13277-014-2560-2 [DOI] [PubMed] [Google Scholar]
- 59. Yu S, Geng Q, Ma J, et al. : Heparin-binding EGF-like growth factor and miR-1192 exert opposite effect on Runx2-induced osteogenic differentiation. Cell Death Dis. 2013;4:e868. 10.1038/cddis.2013.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dahlhoff M, Schäfer M, Wolf E, et al. : Genetic deletion of the EGFR ligand epigen does not affect mouse embryonic development and tissue homeostasis. Exp Cell Res. 2013;319(4):529–35. 10.1016/j.yexcr.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 61. Dahlhoff M, Frances D, Kloepper JE, et al. : Overexpression of epigen during embryonic development induces reversible, epidermal growth factor receptor-dependent sebaceous gland hyperplasia. Mol Cell Biol. 2014;34(16):3086–95. 10.1128/MCB.00302-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schäfer M, Willrodt AH, Kurinna S, et al. : Activation of Nrf2 in keratinocytes causes chloracne (MADISH)-like skin disease in mice. EMBO Mol Med. 2014;6(4):442–57. 10.1002/emmm.201303281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lieder F, Reisen F, Geppert T, et al. : Identification of UV-protective activators of nuclear factor erythroid-derived 2-related factor 2 (Nrf2) by combining a chemical library screen with computer-based virtual screening. J Biol Chem. 2012;287(39):33001–13. 10.1074/jbc.M112.383430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gu DM, Lu PH, Zhang K, et al. : EGFR mediates astragaloside IV-induced Nrf2 activation to protect cortical neurons against in vitro ischemia/reperfusion damages. Biochem Biophys Res Commun. 2015;457(3):391–7. 10.1016/j.bbrc.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 65. Dahlhoff M, Emrich D, Wolf E, et al. : Increased activation of the epidermal growth factor receptor in transgenic mice overexpressing epigen causes peripheral neuropathy. Biochim Biophys Acta. 2013;1832(12):2068–76. 10.1016/j.bbadis.2013.07.011 [DOI] [PubMed] [Google Scholar]
- 66. Knudsen SL, Mac AS, Henriksen L, et al. : EGFR signaling patterns are regulated by its different ligands. Growth Factors. 2014;32(5):155–63. 10.3109/08977194.2014.952410 [DOI] [PubMed] [Google Scholar]
- 67. Ronan T, Macdonald-Obermann JL, Huelsmann L, et al. : Different Epidermal Growth Factor Receptor (EGFR) Agonists Produce Unique Signatures for the Recruitment of Downstream Signaling Proteins. J Biol Chem. 2016;291(11):5528–40. 10.1074/jbc.M115.710087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilson KJ, Gilmore JL, Foley J, et al. : Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol Ther. 2009;122(1):1–8. 10.1016/j.pharmthera.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Leahy DJ: Structure and function of the epidermal growth factor (EGF/ErbB) family of receptors. Adv Protein Chem. 2004;68:1–27. 10.1016/S0065-3233(04)68001-6 [DOI] [PubMed] [Google Scholar]
- 70. Macdonald-Obermann JL, Pike LJ: Different epidermal growth factor (EGF) receptor ligands show distinct kinetics and biased or partial agonism for homodimer and heterodimer formation. J Biol Chem. 2014;289(38):26178–88. 10.1074/jbc.M114.586826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doğaner BA, Yan LK, Youk H: Autocrine Signaling and Quorum Sensing: Extreme Ends of a Common Spectrum. Trends Cell Biol. 2016;26(4):262–71. 10.1016/j.tcb.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 72. Miller MA, Meyer AS, Beste MT, et al. : ADAM-10 and -17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc Natl Acad Sci U S A. 2013;110(22):E2074–83. 10.1073/pnas.1222387110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Freeman M: The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol. 2014;30:235–54. 10.1146/annurev-cellbio-100913-012944 [DOI] [PubMed] [Google Scholar]
- 74. Zettl M, Adrain C, Strisovsky K, et al. : Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145(1):79–91. 10.1016/j.cell.2011.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Christova Y, Adrain C, Bambrough P, et al. : Mammalian iRhoms have distinct physiological functions including an essential role in TACE regulation. EMBO Rep. 2013;14(10):884–90. 10.1038/embor.2013.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dang M, Armbruster N, Miller MA, et al. : Regulated ADAM17-dependent EGF family ligand release by substrate-selecting signaling pathways. Proc Natl Acad Sci U S A. 2013;110(24):9776–81. 10.1073/pnas.1307478110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li X, Maretzky T, Weskamp G, et al. : iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. Proc Natl Acad Sci U S A. 2015;112(19):6080–5. 10.1073/pnas.1505649112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maretzky T, McIlwain DR, Issuree PD, et al. : iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. Proc Natl Acad Sci U S A. 2013;110(28):11433–8. 10.1073/pnas.1302553110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brooke MA, Etheridge SL, Kaplan N, et al. : iRHOM2-dependent regulation of ADAM17 in cutaneous disease and epidermal barrier function. Hum Mol Genet. 2014;23(15):4064–76. 10.1093/hmg/ddu120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Siggs OM, Grieve A, Xu H, et al. : Genetic interaction implicates iRhom2 in the regulation of EGF receptor signalling in mice. Biol Open. 2014;3(12):1151–7. 10.1242/bio.201410116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hosur V, Johnson KR, Burzenski LM, et al. : Rhbdf2 mutations increase its protein stability and drive EGFR hyperactivation through enhanced secretion of amphiregulin. Proc Natl Acad Sci U S A. 2014;111(21):E2200–9. 10.1073/pnas.1323908111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Johnson KR, Lane PW, Cook SA, et al. : Curly bare ( cub), a new mouse mutation on chromosome 11 causing skin and hair abnormalities, and a modifier gene ( mcub) on chromosome 5. Genomics. 2003;81(1):6–14. 10.1016/S0888-7543(02)00013-7 [DOI] [PubMed] [Google Scholar]
- 83. Zheng Y, Li X, Qian X, et al. : Secreted and O-GlcNAcylated MIF binds to the human EGF receptor and inhibits its activation. Nat Cell Biol. 2015;17(10):1348–55. 10.1038/ncb3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Klein DE, Nappi VM, Reeves GT, et al. : Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430(7003):1040–4. 10.1038/nature02840 [DOI] [PubMed] [Google Scholar]
- 85. Schweitzer R, Howes R, Smith R, et al. : Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature. 1995;376(6542):699–702. 10.1038/376699a0 [DOI] [PubMed] [Google Scholar]
- 86. Liao HW, Hsu JM, Xia W, et al. : PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125(12):4529–43. 10.1172/JCI82826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pike LJ: Negative co-operativity in the EGF receptor. Biochem Soc Trans. 2012;40(1):15–9. 10.1042/BST20110610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Baeyens L, Lemper M, Leuckx G, et al. : Transient cytokine treatment induces acinar cell reprogramming and regenerates functional beta cell mass in diabetic mice. Nat Biotechnol. 2014;32(1):76–83. 10.1038/nbt.2747 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89. Scafidi J, Hammond TR, Scafidi S, et al. : Intranasal epidermal growth factor treatment rescues neonatal brain injury. Nature. 2014;506(7487):230–4. 10.1038/nature12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Johnson NR, Wang Y: Coacervate delivery of HB-EGF accelerates healing of type 2 diabetic wounds. Wound Repair Regen. 2015;23(4):591–600. 10.1111/wrr.12319 [DOI] [PMC free article] [PubMed] [Google Scholar]