Abstract
Collective cell migration has emerged in the recent decade as an important phenomenon in cell and developmental biology and can be defined as the coordinated and cooperative movement of groups of cells. Most studies concentrate on tightly connected epithelial tissues, even though collective migration does not require a constant physical contact. Movement of mesenchymal cells is more independent, making their emergent collective behaviour less intuitive and therefore lending importance to computational modelling. Here we focus on such modelling efforts that aim to understand the collective migration of neural crest cells, a mesenchymal embryonic population that migrates large distances as a group during early vertebrate development. By comparing different models of neural crest migration, we emphasize the similarity and complementary nature of these approaches and suggest a future direction for the field. The principles derived from neural crest modelling could aid understanding the collective migration of other mesenchymal cell types.
Keywords: chemotaxis, contact inhibition of locomotion, co-attraction, in silico modelling, self-propelled particle, alignment
Introduction
Research in the last decade has implicated collective cell migration as one of the important contributors to fundamental processes such as morphogenesis, organ formation, wound healing, and cancer metastasis [1–11]. Collective migration is not limited to cells; it is a general phenomenon observed in, for example, bacterial and fish colonies, amoeba, humans, and even in non-living systems such as shaken metallic rods [5, 12–15]. The common feature of these systems is that the movement of individuals within the collective depends on cooperation with the others (Figure 1a, blue arrows). This cooperation distinguishes collective migration from simply coordinated movements where movement is directed entirely by factors external to the collective such as long-distance chemotaxis of cells. Consequently, behaviour of cells during collective motion is markedly different from the behaviour of isolated cells lacking cell-cell interactions, while during externally coordinated motion individual and group cell behaviours are similar (Figure 1b). Therefore, in order to understand how collective movement is achieved, it is important to study the structure of the collective and the interactions therein.
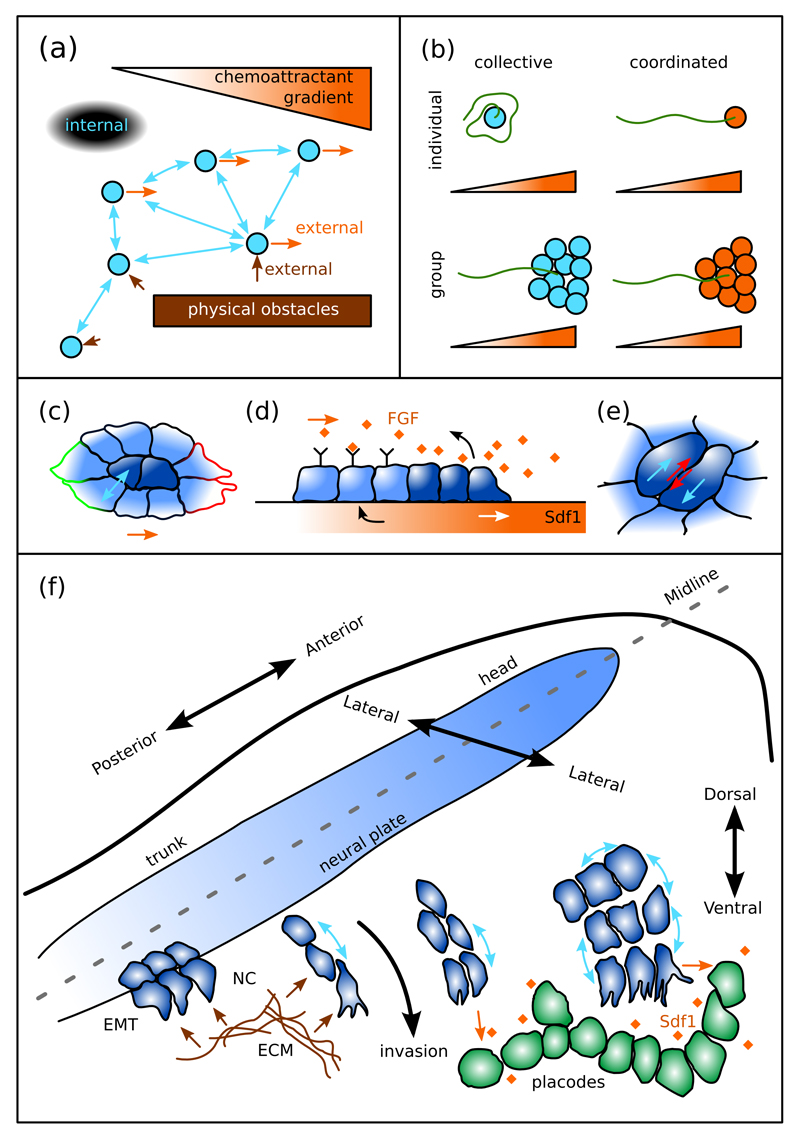
Figure 1. Collective migration depends on internal and external factors.
(a) Collective migration depends on interactions within the migrating collective (blue arrows) although external factors may also influence the movement, such as physical obstacles (brown arrows) or external gradients (orange arrows). (b) Collective versus coordinated migration: coordinated movement is simply the sum of the parts while collective movement depends on interactions within the group. (c-e) Examples of epithelial collective migration: (c) Border cells (light blue) during collective migration in the Drosophila ovary acquire outwards polarity due to interactions with “polar cells” (dark blue) within the cluster, while the whole cluster polarizes in the direction of an external chemoattractant gradient (indicated by red/green outlines). (d) Internal structure and interactions of the posterior lateral line primordium during collective migration in zebrafish development (lateral view of the cluster). The trailing population (light blue) sequesters the underlying chemoattractant (black arrow) generating a gradient that stimulates forward chemotaxis of the leader population (dark blue). In return the leaders secrete FGF that attracts the trailing cells. (e) Cell-cell interactions within epithelial sheets: during plithotaxis, cells move (blue arrows) within an epithelial sheet to minimize intercellular shear (red arrows). (f) Schematic representation of neural crest (NC) migration within the embryo. The NC differentiates and undergoes EMT at the borders of the neural plate and then invades the surrounding tissues, including placodes. During its migration the NC interacts with the ECM and external chemoattractants and maintains interactions within the migrating cluster. Migration occurs in the head first, where streams are wider and larger than at more posterior locations.
Studies of collective cell migration have mainly focused on epithelial tissues, including the in vivo migration of border cells (Figure 1c), the posterior lateral line primordium (Figure 1d), and in vitro epithelia (Figure 1e), where adhesions play a major role in organizing the collective [2, 3, 8, 16, 17]. In contrast, collectively migrating mesenchymal cells move more independently and rely more on other modes of cell interactions, similar to collectively migrating animals. How these interactions give rise to collective movement is less intuitive, making computational modelling an indispensable tool for understanding such behaviours.
Here we focus on one such mesenchymal collective migration system, the neural crest (NC), which has been addressed by various in silico studies [18–24]. In all vertebrates, development of most organs depends on the efficient migration of these loosely connected cells that invade the developing embryo to reach their target regions, not unlike metastatic cancer cells invade the adult organisms. Below we provide an overview of the most important features of NC migration and review recent in silico studies aiming at understanding the internal structure and interactions leading to the collective migration of the NC.
The migrating neural crest
During vertebrate development the NC forms at the lateral edges of the neural plate (Figure 1f). Soon after differentiation, NC cells delaminate and undergo epithelial-to-mesenchymal transition (EMT) in an anterior to posterior order along the midline. Cells invade the neighbouring tissues, including placodes, in distinct streams stereotypic within species. Width and size of the streams decrease from the head to the trunk where cells migrate in single cell wide chains. The NC also colonize the gut [25–28], however we will only focus on the head and trunk NC for the purpose of this review. The microenvironment has been shown to present molecular cues restricting migration, such as ephrins, semaphorins, proteoglycans, Slit/Robo [29–33] or promoting migration, such as VEGF and Sdf1 [34]. Indeed, it is now well established that chemotaxis is vital for NC migration [35], although it is unlikely that it would simply provide a guiding gradient for the streams along their long and complex paths.
Leaders and followers
A series of high throughput studies has revealed heterogeneity of gene expression profiles within the NC streams of the chick embryo [18–20]. Genes preferentially expressed at the leading edge of the NC cluster (“trailblazer” cells) include metalloproteinases (MMP2, ADAM33), integrins (ITGB5), and guidance-related genes (FGFR2, EPHB3). Expression of some “trailblazer” genes can be triggered by addition of VEGF in vitro within minutes of application [20]. Likewise, “trailblazer” genes are expressed in the trailing cells following the “trailblazers” at the back of the stream in vivo when they are exposed to exogenous VEGF [20].
Based on the observed heterogeneity, a line of computational models emerged that aim to explain NC migration through the interaction between follower and leader cells (Figure 2a) [18–20]. The key difference between leaders and followers in the model is assumed to be the ability of leaders to move up VEGF gradients [36] (Table 1). Followers, on the other hand, move randomly until they contact a leader cell, or another follower in a chain of followers connected to a leader, after which they move in the direction of their contact cell (Figure 2a). The model assumes a homogeneous VEGF concentration throughout the stream at the onset of migration based on in vivo observations [36] and a significant internalization of VEGF by the NC. Finally, the models incorporate the expansion of the domain, which both conveys cells and dilutes VEGF concentrations. To mimic repulsive stream borders, VEGF concentrations are forced to be zero at these locations resulting in chemorepulsion.
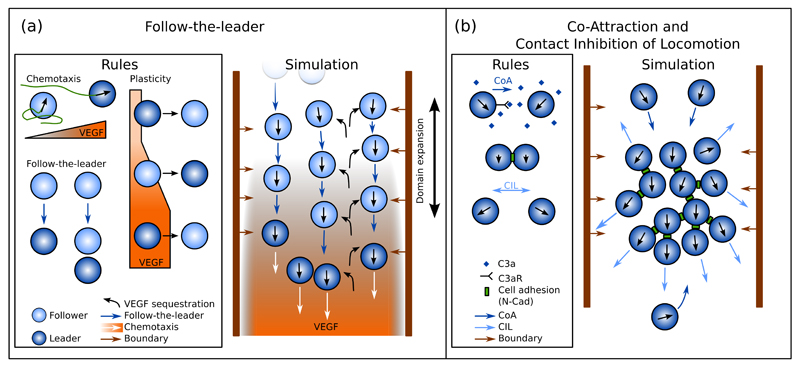
Figure 2. Main models of cranial neural crest migration.
(a) Follow-the-leader model of NC migration. Leader cells (dark blue) chemotax towards VEGF, unlike followers (light blue) that are moving towards the closest leader cell or a chain of followers led by a leader cell (Follow-the-leader, dark blue arrows). If a follower is not contacted by any other cell, it moves randomly until it contacts a leader of a chain. Sufficient exposure to VEGF gradient triggers follower-to-leader phenotype switch, while lack of a VEGF gradient leads to leader-to-follower switch. All cells sequester VEGF (black curved arrows), leading to a self-generated gradient from the initial uniformly high concentrations. (b) NC migration model based on co-attraction (CoA, dark blue arrows) and contact inhibition of locomotion (CIL, light blue arrows). The persistently moving cells secrete the chemoattractant C3a (dark blue rhomboids) leading to CoA. After contact the cells form adhesions via N-Cadherin (green boxes), align their movement, and subsequently repolarise and move away from one another (CIL). These interactions lead to a coherently migrating NC cluster.
Table 1. Main assumptions in the follow-the-leader (FtL) model and CIL-CoA model, their experimental basis, and their consequences.
| Model | Assumption | Experimental basis | Consequence |
|---|---|---|---|
| FtL | Heterogeneous NC population | Gene expression profiling | Population of two cell types with two independent phenotypic behaviours |
| FtL | Leader NC cells chemotax towards VEGF | NC move towards ectopic VEGF sources | Directional cluster movement |
| FtL | Follower NC cells do not chemotax towards VEGF | None | |
| FtL | Follower NC cells move towards the nearest leader or towards collectives headed by a leader (similar to CoA) | None | Alignment and cohesion of the cluster |
| FtL | VEGF is homogeneously distributed along the NC path before migration | Immunohistochemistry on sections from avian embryos | VEGF gradient made possible through sequestration |
| FtL | VEGF is internalized by the NC cells | VEGF internalized in in vitro wounding assay of endothelial cells [37] | VEGF gradient generated by sequestration |
| FtL | Leader phenotype is triggered by exposure to VEGF gradient | Leader expression profile observed in trailer cells when exposed to VEGF | Adaptive cluster behaviour able to respond to ectopic VEGF |
| CIL-CoA | Co-Attraction: NC cells secrete a chemoattractant | Complement component C3 is expressed by NC cells in Xenopus laevis, and cells chemotax towards C3a. | Cluster cohesion |
| CIL-CoA | Homogeneous NC population | Similar response to signals from leader and followers (potential indication of plasticity) | Simplifying assumption |
| CIL-CoA | CIL | NC cells exhibit CIL in vivo and in vitro across species | Polarized cell protrusions driving cells away from the cluster |
| CIL-CoA | Cell velocities are aligned after contact | Phenomenological observation based on trajectory analysis of NC cells | Synchronization of cell movements in the cluster |
NC clusters in this model are directed by the VEGF gradient generated by the group itself (Figure 2a), similar to what has been proposed for the posterior lateral line primordium in zebrafish. This is supported by simulations of graft experiments: leaders introduced to the trailing edge (together with their VEGF rich environment) stall due to the backward VEGF gradient; followers transplanted in front of the leaders in silico (together with their VEGF-depleted environment) block the migration of the cluster due to the disrupted VEGF gradient [18]. Since VEGF is depleted inside the NC cluster, a homogeneous population of chemotaxing cells proves to be insufficient to give rise to cohesive cluster migration in this model [18]. Furthermore, the leader population has to be restricted to the leading edge of the cluster as suggested by gene expression profiling; otherwise invasion efficiency drops due to loss of directional cues within the cluster [19].
Plasticity has been incorporated in the model to explore switching of leader and follower fates in experiments altering VEGF levels [20]. Switching is implemented as an integrate-and-switch mechanism: a follower is assumed to turn into a leader if it is exposed to a detectable VEGF gradient for a sufficiently long time; conversely, a leader is turned into a follower if it fails to sense a gradient for a given time even if VEGF levels remain high but without a gradient (Figure 2a). For efficient migration, the time required for follower-to-leader and leader-to-follower switches are required to be similar, and based on the in vitro observations of the study, are selected to be on the order of minutes. With this addition, the model successfully approximates the movement of NC in the presence of an ectopic VEGF source near the trailing edge of the cluster. Switching in this model is required to explain how trailing cells are able to break the cohesion of the group and respond to VEGF [20].
Although heterogeneity within the streams has been established experimentally, some aspects of this model remain to be explored. The main assumption of the model that leaders and not followers respond to VEGF lacks experimental evidence. Furthermore, cluster cohesion in the model is achieved by an ad-hoc mechanism, whereby followers distinguish and move towards connections that are part of a collective headed by a leader. How follower cells are able to make this distinction, and how movement is mediated, remains unresolved by these studies, but alternative investigations detailed below may provide the answer.
Cohesion and emergent leaders via co-attraction and contact inhibition of locomotion
Another line of research focusing on the behaviour of individual NC cells revealed two cellular interactions that together provide cohesion and an emergent group polarity for the NC clusters [21, 22, 38–41]. Cohesion is provided by co-attraction (CoA), whereby NC cells secrete the complement factor C3a, which acts as a NC chemoattractant [21] (Figure 2b). To date, CoA has been assumed to act in all cells of the NC cluster, which would lead to a collapsing cluster with no outward protrusive activity. However, NC cells have also been shown to undergo contact inhibition of locomotion (CIL) [38, 39, 42, 43], whereby contacting cells collapse their protrusions at the region of cell contact, repolarise away from one another and eventually separate [44]. The CIL mechanism provides outward polarity for cells in the cluster (Figure 2b). Explorations of the molecular mechanisms of CIL in this system have implicated PCP signalling [38] and N-Cadherin-mediated cell-cell contacts [41]. These components required for CIL have also been shown to be necessary for the efficient collective chemotaxis of NC clusters towards Sdf1 sources and induce a more polarised cell motion within the cluster [41]. Although these mechanisms are expected to be at work in all cells within the migrating NC, interaction within the cluster and with the microenvironment are proposed to give rise to a distinctive segregation of roles among the collective.
Computational models exploring the CIL and CoA mechanisms during NC migration assume a homogeneous cell population confined into a migratory stream by reflective or repulsive lateral boundary conditions similar to the self-generated gradient models [21, 22]. Cells are modelled as self-propelled particles that periodically change their migration direction (tumble), and are attracted towards each other through CoA. CoA is modelled by either assisting the cells to move towards the centre of mass of nearby cells [21], or as a force proportional to the gradient of a diffusing and decaying chemoattractant secreted by all cells [22]. The attractant field is assumed to have reached a quasi-steady state due to the low molecular weight of C3a, and therefore is approximated as the sum of exponentials [22]. Upon contact, cells align their velocities either as a result of turning towards the local average of velocities [21] or due to a soft volume exclusion force based on contact mechanics [22]. During contact, the cells do not tumble and after a given time they repolarise: they either take on a new, random direction [21], or experience a force pushing them in a random but biased direction away from the contact [22].
These two cellular interactions, together with the boundary constraints, are sufficient to generate directionally and collectively migrating NC clusters [21, 22]. After an initial lag, a common direction emerges within the group with leader cells at the front keeping movement direction fairly constant. Trailing cells occasionally separate from the cluster but are attracted back via CoA (Figure 2b). Simulations reproduce the phenotypes observed in vivo and in vitro: lack of CoA leads to dispersion of NC clusters [21, 22], while lack of CIL leads to disrupted cluster migration [22].
In these models, cluster polarity emerges as a result of alignment and the interactions with the bounding environment. The molecular basis for this alignment is not yet clear, however, experimental observations of NC cell collisions and trajectories support this notion [41].
Conclusions and future directions
In summary, here we reviewed two main models of collective NC cell migration. The first model explains directional migration of the NC by a self-generated gradient of VEGF created as a result of heterogenetic composition of the population. These studies demonstrate a remarkable plasticity in the NC population by rapidly changing gene expression profiles. Understanding how this plasticity is achieved could provide invaluable insight for understanding cancer recurrence where plasticity is thought to play an important role [45, 46]. Although an attractive option, the main assumption that only leader cells react to VEGF gradients remains to be demonstrated experimentally. Moreover, cluster cohesion in the model is based on a model assumption lacking experimental basis. This assumption may be explained by the second model where the CoA process could represent the follow-the-leader activity of the first model. In other words, the follower cells move towards the leaders because the leaders secrete a chemoattractant that could be the molecule C3a. Importantly, order in this model emerges as a consequence of movement alignment during CIL. Alignment plays an important role in emergent collective migration [47, 48] and has been suggested to result from cell-cell collisions with or without repolarization at high cell densities [4, 49, 50, 51, 52, 53]. While alignment during CIL of NC cells is observed experimentally, its molecular basis is still under investigation.
Combining the two approaches could lead to a deeper understanding of collective chemotaxis [54]. A current modelling study shows that an external chemoattractant may induce collective NC chemotaxis by enhancing the effect of CIL-induced polarity in a CoA-CIL type model [55].
Another promising integration of the chemotaxis-driven follow-the-leader model and the self-organizing CIL-CoA model is provided by a recent discovery of a novel “chase-and-run” interaction between the NC and placodal cells in the NC microenvironment [56, 57]. Placodes are ectodermal structures fated to become cranial nerves and sensory organs, and they secrete Sdf1 that attract the NC [41, 58] (Figure 1f). Upon contact, the NC cell and the placode cell undergo CIL by which both cells retract from the contact, followed by the repeated attraction of the NC cell. The displacement of the placode leads to a unidirectional migration of the NC-placode collective, a phenomenon observed both in vivo and in vitro [56]. Exploring whether placode-derived Sdf1 polarises the NC cluster in a similar way as VEGF could unify the two approaches.
Highlights.
Collective cell migration emerges from factors both internal and external to the collective
Stable cell adhesion is not essential for collective cell migration
A self-generated gradient drives NC migration in a follow-the-leader type model
Cohesion and polarisation result from co-attraction and contact inhibition of locomotion
Integration of complementing models could explain NC-placode interactions
Acknowledgements
We thank Adam Shellard and András Czirók for comments on the manuscript. Work in R. Mayor’s laboratory is supported by grants from the Medical Research Council (M010465 and J000655), Biotechnology and Biological Sciences Research Council (M008517), the Wellcome Trust, and Marie Curie Fellowship (329968) to A Szabó.
Abbreviations:
- NC
neural crest
- FtL
follow-the-leader
- CoA
co-attraction
- CIL
contact inhibition of locomotion
- EMT
epithelial-to-mesenchymal transition
References
- 1.Méhes E, Vicsek T. Collective motion of cells: from experiments to models. Integr Biol. 2014;6:831–854. doi: 10.1039/c4ib00115j. [* A systematic and wide overview of studies in collective cell migration.] [DOI] [PubMed] [Google Scholar]
- 2.Scarpa E, Mayor R. Collective cell migration in development. J Cell Biol. 2016;212:143–55. doi: 10.1083/jcb.201508047. [* An up-to-date review on collective cell migration phenomena during development.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nat Rev Mol Cell Biol. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- 4.Coburn L, Cerone L, Torney C, Couzin ID, Neufeld Z. Tactile interactions lead to coherent motion and enhanced chemotaxis of migrating cells. Phys Biol. 2013;10:046002. doi: 10.1088/1478-3975/10/4/046002. [DOI] [PubMed] [Google Scholar]
- 5.Chang WK, Carmona-Fontaine C, Xavier JB. Tumour-stromal interactions generate emergent persistence in collective cancer cell migration. Interface Focus. 2013;3:20130017–20130017. doi: 10.1098/rsfs.2013.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaritsky A, Kaplan D, Hecht I, Natan S, Wolf L, Gov NS, Ben-Jacob E, Tsarfaty I. Propagating Waves of Directionality and Coordination Orchestrate Collective Cell Migration. PLoS Comput Biol. 2014;10:e1003747. doi: 10.1371/journal.pcbi.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haeger A, Wolf K, Zegers MM, Friedl P. Collective cell migration: guidance principles and hierarchies. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Vedula SRK, Ravasio A, Lim CT, Ladoux B. Collective Cell Migration: A Mechanistic Perspective. Physiology. 2013;28:370, 379. doi: 10.1152/physiol.00033.2013. [DOI] [PubMed] [Google Scholar]
- 9.Ladoux B, Mège R-M, Trepat X. Front–Rear Polarization by Mechanical Cues: From Single Cells to Tissues. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.02.002. pii: S0962-8924(16)00014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pocha SM, Montell DJ. Cellular and Molecular Mechanisms of Single and Collective Cell Migrations in Drosophila : Themes and Variations. Annu Rev Genet. 2014;48:295–318. doi: 10.1146/annurev-genet-120213-092218. [DOI] [PubMed] [Google Scholar]
- 11.Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol. 2012;13:631–45. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicsek T, Zafeiris A. Collective motion. Phys Rep. 2012;517:71–140. [Google Scholar]
- 13.Rosenthal SB, Twomey CR, Hartnett AT, Wu HS, Couzin ID. Revealing the hidden networks of interaction in mobile animal groups allows prediction of complex behavioral contagion. Proc Natl Acad Sci. 2015 doi: 10.1073/pnas.1420068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chraibi M, Ezaki T, Tordeux A, Nishinari K, Schadschneider A, Seyfried A. Jamming transitions in force-based models for pedestrian dynamics. Phys Rev E. 2015;92:042809. doi: 10.1103/PhysRevE.92.042809. [DOI] [PubMed] [Google Scholar]
- 15.Attanasi A, Cavagna A, Del Castello L, Giardina I, Melillo S, Parisi L, Pohl O, Rossaro B, Shen E, Silvestri E, et al. Collective behaviour without collective order in wild swarms of midges. PLoS Comput Biol. 2014;10:e1003697. doi: 10.1371/journal.pcbi.1003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theveneau E, Mayor R. Can mesenchymal cells undergo collective cell migration? The case of the neural crest. Cell Adh Migr. 2011;5:490–8. doi: 10.4161/cam.5.6.18623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theveneau E, Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cell Mol Life Sci. 2013;70:3481–3492. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLennan R, Dyson L, Prather KW, Morrison JA, Baker RE, Maini PK, Kulesa PM. Multiscale mechanisms of cell migration during development: theory and experiment. Development. 2012;139:2935–44. doi: 10.1242/dev.081471. [* A study combining computational and experimental approaches to introduce the follow-the-leader model described in the present review.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLennan R, Schumacher LJ, Morrison JA, Teddy JM, Ridenour DA, Box AC, Semerad CL, Li H, McDowell W, Kay D, et al. Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development. 2015 doi: 10.1242/dev.117507. [** A recent in study combining computational and experimental approaches to show the heterogeneity of the migrating NC and emphasize the need for the leader population to be confined to the very leading edge of the cluster.] [DOI] [PubMed] [Google Scholar]
- 20.McLennan R, Schumacher LJ, Morrison JA, Teddy JM, Ridenour DA, Box AC, Semerad CL, Li H, McDowell W, Kay D, et al. VEGF signals induce trailblazer cell identity that drives neural crest migration. Dev Biol. 2015;407:12–25. doi: 10.1016/j.ydbio.2015.08.011. [** A combined computational and experimental study showing phenotypic switching between followers and leaders.] [DOI] [PubMed] [Google Scholar]
- 21.Carmona-Fontaine C, Theveneau E, Tzekou A, Tada M, Woods ML, Page KM, Parsons M, Lambris JD, Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell. 2011;21:1026–37. doi: 10.1016/j.devcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woods ML, Carmona-Fontaine C, Barnes CP, Couzin ID, Mayor R, Page KM. Directional Collective Cell Migration Emerges as a Property of Cell Interactions. PLoS One. 2014;9:e104969. doi: 10.1371/journal.pone.0104969. [** Mostly computational study of the CIL-CoA model based on the discreet element method.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wynn ML, Rupp P, Trainor PA, Schnell S, Kulesa PM. Follow-the-leader cell migration requires biased cell-cell contact and local microenvironmental signals. Phys Biol. 2013;10:035003. doi: 10.1088/1478-3975/10/3/035003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wynn ML, Kulesa PM, Schnell S. Computational modelling of cell chain migration reveals mechanisms that sustain follow-the-leader behaviour. J R Soc Interface. 2012;9:1576–1588. doi: 10.1098/rsif.2011.0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newgreen DF, Dufour S, Howard MJ, Landman KA. Simple rules for a “simple” nervous system? Molecular and biomathematical approaches to enteric nervous system formation and malformation. Dev Biol. 2013;382:305–319. doi: 10.1016/j.ydbio.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheeseman BL, Zhang D, Binder BJ, Newgreen DF, Landman KA. Cell lineage tracing in the developing enteric nervous system: superstars revealed by experiment and simulation. J R Soc Interface. 2014;11:20130815–20130815. doi: 10.1098/rsif.2013.0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young HM, Bergner AJ, Simpson MJ, McKeown SJ, Hao MM, Anderson CR, Enomoto H. Colonizing while migrating: how do individual enteric neural crest cells behave? BMC Biol. 2014;12:23. doi: 10.1186/1741-7007-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D, Ighaniyan S, Stathopoulos L, Rollo B, Landman K, Hutson J, Newgreen DF. The neural crest: A versatile organ system. Birth Defects Res C Embryo Today. 2014 doi: 10.1002/bdrc.21081. [DOI] [PubMed] [Google Scholar]
- 29.Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7:561–570. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- 30.Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr Biol. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- 31.Gammill LS. Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- 32.De Bellard ME, Rao Y, Bronner-Fraser M. Dual function of Slit2 in repulsion and enhanced migration of trunk, but not vagal, neural crest cells. J Cell Biol. 2003;162:269–279. doi: 10.1083/jcb.200301041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerr RS, Newgreen DF. Isolation and characterization of chondroitin sulfate proteoglycans from embryonic quail that influence neural crest cell behavior. Dev Biol. 1997;192:108–24. doi: 10.1006/dbio.1997.8731. [DOI] [PubMed] [Google Scholar]
- 34.Theveneau E, Mayor R. Neural crest migration: interplay between chemorepellents, chemoattractants, contact inhibition, epithelial-mesenchymal transition, and collective cell migration. Wiley Interdiscip Rev Dev Biol. 2012;1:435–45. doi: 10.1002/wdev.28. [DOI] [PubMed] [Google Scholar]
- 35.Shellard A, Mayor R. Chemotaxis during neural crest migration. Semin Cell Dev Biol. 2016 doi: 10.1016/j.semcdb.2016.01.031. [DOI] [PubMed] [Google Scholar]
- 36.McLennan R, Teddy JM, Kasemeier-Kulesa JC, Romine MH, Kulesa PM. Vascular endothelial growth factor (VEGF) regulates cranial neural crest migration in vivo. Dev Biol. 2010;339:114–125. doi: 10.1016/j.ydbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Keller G. VEGF nuclear accumulation correlates with phenotypical changes in endothelial cells. J Cell Sci. 2000;113(Pt 9):1525–34. doi: 10.1242/jcs.113.9.1525. [DOI] [PubMed] [Google Scholar]
- 38.Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn Ga, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–61. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roycroft A, Mayor R. Forcing contact inhibition of locomotion. Trends Cell Biol. 2015 doi: 10.1016/j.tcb.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scarpa E, Szabó A, Bibonne A, Theveneau E, Parsons M, Mayor R. Cadherin Switch during EMT in Neural Crest Cells Leads to Contact Inhibition of Locomotion via Repolarization of Forces. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teddy JM, Kulesa PM. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development. 2004;131:6141–6151. doi: 10.1242/dev.01534. [DOI] [PubMed] [Google Scholar]
- 43.Blasky AJ, Pan L, Moens CB, Appel B. Pard3 regulates contact between neural crest cells and the timing of Schwann cell differentiation but is not essential for neural crest migration or myelination. Dev Dyn. 2014;243:1511–1523. doi: 10.1002/dvdy.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roycroft A, Mayor R. Molecular basis of contact inhibition of locomotion. Cell Mol Life Sci. 2015;25:373–375. doi: 10.1007/s00018-015-2090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pisco AO, Huang S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: “What does not kill me strengthens me”. Br J Cancer. 2015;112:1725–1732. doi: 10.1038/bjc.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doherty M, Smigiel J, Junk D, Jackson M. Cancer Stem Cell Plasticity Drives Therapeutic Resistance. Cancers (Basel) 2016;8:8. doi: 10.3390/cancers8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicsek T, Czirók A, Ben-Jacob E, Cohen I, Shochet O. Novel Type of Phase Transition in a System of Self-Driven Particles. Phys Rev Lett. 1995;75:1226–1229. doi: 10.1103/PhysRevLett.75.1226. [DOI] [PubMed] [Google Scholar]
- 48.Grégoire G, Chaté H, Tu Y. Moving and staying together without a leader. Phys D Nonlinear Phenom. 2003;181:157–170. [Google Scholar]
- 49.Szabó B, Szöllösi GJ, Gönci B, Jurányi Z, Selmeczi D, Vicsek T. Phase transition in the collective migration of tissue cells: experiment and model. Phys Rev E. 2006;74:061908. doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
- 50.Szabó A, Ünnep R, Méhes E, Twal WO, Argraves WS, Cao Y, Czirók A. Collective cell motion in endothelial monolayers. Phys Biol. 2010;7:046007. doi: 10.1088/1478-3975/7/4/046007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabla AJ. Collective cell migration: leadership, invasion and segregation. J R Soc Interface. 2012;9:3268–78. doi: 10.1098/rsif.2012.0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Löber J, Ziebert F, Aranson IS. Collisions of deformable cells lead to collective migration. Sci Rep. 2015;5:9172. doi: 10.1038/srep09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vedel S, Tay SS, Johnston DM, Bruus H, Quake SR. Migration of cells in a social context. Proc Natl Acad Sci U S A. 2012;110:129–34. doi: 10.1073/pnas.1204291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rappel W-J. Cell–cell communication during collective migration. Proc Natl Acad Sci. 2016;113:1471–1473. doi: 10.1073/pnas.1524893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Camley BA, Zimmermann J, Levine H, Rappel W. Emergent collective chemotaxis without single-cell gradient sensing. arXiv Prepr. 2015 doi: 10.1103/PhysRevLett.116.098101. arXiv ID: 1506.06698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–72. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szabó A, Mayor R. Cell traction in collective cell migration and morphogenesis: The chase and run mechanism. Cell Adh Migr. 2015 doi: 10.1080/19336918.2015.1019997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasemeier-Kulesa JC, McLennan R, Romine MH, Kulesa PM, Lefcort F. CXCR4 Controls Ventral Migration of Sympathetic Precursor Cells. J Neurosci. 2010;30:13078–13088. doi: 10.1523/JNEUROSCI.0892-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]