Abstract
Pol γ, the only DNA repair and replication. During mtDNA base-excision repair, gaps are created after damaged base excision. Here we show that Pol γ efficiently gap-fills except when the gap is only a single nucleotide. Although wild-type Pol γ has very limited ability for strand displacement DNA synthesis, exo− (3′–5′ exonuclease-deficient) Pol γ has significantly high activity and rapidly unwinds downstream DNA, synthesizing DNA at a rate comparable to that of the wild-type enzyme on a primer-template. The catalytic subunit Pol γA alone, even when exo−, is unable to synthesize by strand displacement, making this the only known reaction of Pol γ holoenzyme that has an absolute requirement for the accessory subunit Pol γB.
Keywords: DNA repair and replication, strand displacement, molecular motor
1. INTRODUCTION
As a major location for metabolic redox reactions, mitochondria contain high concentrations of reactive oxygen species (ROS), a chief cause of mitochondrial DNA oxidative damage (Beckman and Ames, 1999; Driggers et al., 1997; Richter et al., 1988). Base-excision repair (BER) is a conserved DNA repair pathway for restoring oxidized or alkylated DNA. The basic steps of BER begin with damaged base excision by a glycosylase, AP-endonuclease cleavage to generate a 3′-OH, removal of the 5′-deoxyribose by lyase, followed by gap-filling synthesis by DNA polymerase (DNAP) and then nick sealing by DNA ligase (Hegde et al., 2008). The DNAP may be bi-functional and contain the 5′-dRP lyase activity. If only a single nucleotide is incorporated by the DNAP, the process is referred to as single-nucleotide BER (SN-BER). If the 5′-ribose is oxidized, often resulting in a deoxyribulonolactone, then the long-patch BER (LP-BER) pathway is activated, where downstream DNA containing the damaged residue is removed and is replaced by newly synthesized DNA.
DNA polymerases have an intrinsic motor activity that allows them to translocate along the template single-stranded DNA (Patel et al., 2012). The motor activity of some DNAPs, e.g., bacterial DNAP I is sufficiently strong that it can unwind downstream duplex DNA to provide a single-stranded template for primer extension in a process referred to as strand displacement synthesis. However, other enzymes require accessory proteins, such as a DNA helicase, to effect strand displacement synthesis.
In spite of an overall similarity, BER exhibits species-dependent characteristics. In mammalian cells, depending on the nature of the damage, different nuclear repair complexes contain distinct glycosylases, polymerases and ligases (reviewed in (Hegde et al., 2008)). Human nuclear and mitochondrial BER pathways also display distinct features. In the nucleus, a dedicated repair polymerase, Pol β, is exclusively involved in SN-BER, whereas LP-BER is performed by DNA polymerase δ and FEN1, which function together to perform stepwise strand displacement. Pol δ can displace 1–2 nt of downstream duplex after gap-filling to form a small flap that is subsequently removed by FEN1. These reactions are suggested to then cycle in order to remove additional 1–2 nt segments from the downstream DNA (Garg et al., 2004). A major difference between the two sub-pathways is that during LP-BER DNA synthesis extends beyond the lesion site, which requires removal of perfect duplex downstream DNA.
One notable difference between nuclear and mitochondrial BER is that the role of Pol β in SN-BER, and of Pol δ in LP-BER are both performed by Pol γ. LP-BER is required, for example when the deoxyribose is oxidized or the damage prevents formation of a 3′-OH primer and downstream 5′-P. However, even if this last requirement is met, the choice between SN-BER and LP-BER, and which sub-pathway is utilized, must be governed by the availability of specific repair proteins and by the relative activities of Pol γ on different templates. For example, the 5′-dRP lyase activity of Pol γ is weaker than Pol β (Longley et al., 1998a; Pinz and Bogenhagen, 2006), which may bias repair in mitochondria towards the LP-BER pathway.
Mitochondrial LP-BER is more complex than SN-BER. Several exo/endonucleases have been implicated in removing downstream DNA, but which specific enzyme is essential in coordinating with Pol γ and ligase III is still debated. Potential nucleases include DNA2, an ATP-dependent-helicase/nuclease; FEN1, a flap exo/endonuclease, and EXOG, an exo/endonuclease. By binding to a long single-stranded flap DNA, the helicase DNA2 could unwind the duplex downstream of the lesion site, providing a single-stranded template for Pol γ. In support of this idea, DNA2 and Pol γ have been shown to interact by co-immunoprecipitation (Zheng et al., 2008). However, DNA2 requires single-stranded DNA in order to initiate helicase activity (Balakrishnan et al., 2010; Stewart et al., 2009), but no single-stranded flap exists after damaged residue excision. Duplex DNA downstream of the gap therefore has to be unwound to allow DNA2 to bind. A likely candidate for this step would be DNA polymerase γ but in the absence of other replisomal components the enzyme lacks an intrinsic ability to synthesize significant lengths of DNA on a duplex template. FEN1, which has been found in mitochondria (Kalifa et al., 2009), could cooperate with Pol γ in an analogous manner to its activity with Pol δ to perform stepwise strand displacement synthesis in the nucleus (Garg et al., 2004). EXOG, which is exclusively located in mitochondria, is required for repair of single-stranded DNA breaks (Tann et al., 2011). EXOG can remove 2 nt from the 5′-end of a nick or gap in duplex DNA, thereby creating a gap extending beyond the lesion site for gap-filling synthesis by Pol γ. Although these nucleases are all capable of cooperating with Pol γ to perform stand displacement synthesis in vitro, which one is the primary nuclease during LP-BER remains unknown.
Pol γ is a holoenzyme that contains the catalytic subunit Pol γA and a dimeric accessory subunit Pol γB. Pol γA contains all the enzymatic activities of the holoenzyme, including polymerase, 3′–5′ exonuclease for proofreading, and a 5′-dRP lyase for DNA repair; Pol γB, although it displays no independent enzymatic activity, enhances all known functions of Pol γA, including BER (Johnson and Johnson, 2001; Longley and Mosbaugh, 1991; Longley et al., 1998a; Pinz and Bogenhagen, 2006; Prasad et al., 2009). Like most other replicases, Pol γA has low processivity, which is substantially increased after it associates with Pol γB to form holoenzyme (Lim et al., 1999). Dimeric Pol γB has a unique mode of processivity enhancement: one monomer accelerates the reaction rate while the second strengthens the DNA-binding affinity of the holoenzyme (Lee et al., 2010b). The crystal structure of Pol γ holoenzyme provided a framework for understanding how Pol γB alters both exonuclease (exo) and polymerase (pol) activities simultaneously: it biases the primer terminus towards the pol site, thereby reducing shuttling of the primer strand to the exo site during regular DNA synthesis (Lee et al., 2009). The structure of the Pol γ holoenzyme also reveals features suitable for a replicase, but provides little information of how it can also function as a repair enzyme.
To further characterize how human Pol γ functions in mitochondrial BER, here we investigate its properties in gap-filling and in strand displacement using templates with defined gap-sizes. We found that Pol γ can efficiently fill large gaps; however, unlike the dedicated repair polymerase Pol β, Pol γ fills a single nucleotide gap, a major BER intermediate, slowly and at a relatively low efficiency. Using small angle X-ray scattering and dynamic light scattering, we show that the structural basis for the inefficient repair is a reduced affinity of Pol γ for duplex DNA containing a single nucleotide gap. We also show that when the proofreading exonuclease activity is abrogated, although there is little or no change in synthetic activity for Pol γA alone, holoenzyme gains the ability for robust strand displacement synthesis.
2. MATERIALS AND METHODS
2.1 Preparations of Pol γ subunits and their variants
Pol γA wild type and its exonuclease-deficient variant, both containing a C-terminal Histag, were expressed in Sf9 cells. The exonuclease-deficiency is due to substitution of catalytic residues D198 and E200 with alanine (Lee et al., 2009). Cells infected with a Pol γA-baculovirus construct were harvested 72 hours post-infection. Pol γB wild type and the mutant ΔI4-D129K were expressed at 25 °C in Rosetta (DE3), and the mutants RK and RKK in BL21 Tuner™ (DE3). Pol γB protein expression was induced at A600=0.6 with 0.4 mM isopropyl 1-thio-β-D-galactopyranoside and incubation continued for 6 hr at 25 °C before harvesting. All proteins were purified following a modified procedure of Yakubovskaya et al. (Yakubovskaya et al., 2006). Briefly, the proteins were purified on a Ni-NTA column, followed by Source S and gel filtration chromatography. Proteins were stored at −20°C in 30% glycerol. Protein concentrations were determined by their respective molar extinction coefficient (243,790 L·mol−1·cm−1 for Pol γA and 71,940 L·mol−1·cm−1 for monomeric Pol γB). Clones of the wild-type, and the Pol γB RK and RKK mutants were kind gifts from Dr. Dan Bogenhagen.
DNA substrates
Gapped DNA templates were constructed by mixing primer (P) and blocker (B) strands to the 3′ and 5′ ends, respectively, of a template (T) strand at a molar ratio of 1:1.2:1.2 (P:B:T), and annealing by heating at 95°C for 5 min followed by slow cooling to 20°C. Constructs used in this study are listed in Table 1. Except where noted, primer P was labeled with [γ-32P]ATP using T4 polynucleotide kinase, and blocker B contained a 5′-OH.
Table 1.
The structure and sequences of oligonucleotide complexes
| Substrate name | Construct sequence |
|---|---|
| T45/P25-B19 (1nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG-5′ 5′-TCCTCGCAGCCGTCCAACCAACTCA CCTCGATCCAATGCCGTCC-3′ |
| T45/P25-B19-RD (1nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG-5′ |
| T45/P25-B18 (2nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG-5′ 5′-TCCTCGCAGCCGTCCAACCAACTCA CTCGATCCAATGCCGTCC-3′ |
| T45/P22-B18 (5nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG-5′ 5′-TCCTCGCAGCCGTCCAACCAAC CTCGATCCAATGCCGTCC-3′ |
| T45/P45 (duplex) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG-5′ 5′-TCCTCGCAGCCGTCCAACCAACTCAACCTCGATCCAATGCCGTCC-3′ |
| T55/P25-B29 (1nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGGTTACGGCAGG-5′ 5′-TCCTCGCAGCCGTCCAACCAACTCA CCTCGATCCAATGCCGTCCAATGCCGTCC-3′ |
| T′55/P25-B′29 (1nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGATACGGGTTACGGCAGGTTACGGCAGG-5′ 5′-TCCTCGCAGCCGTCCAACCAACTCA CCTATGCCCAATGCCGTCCAATGCCGTCC-3′ |
| T55/P25-B29-RD (1nt-gap) |
3′-AGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGGTTACGGCAGG-5′ |
2.2 Gap-filling and strand displacement assay
Pol γA exhibits maximal activity in the absence of monovalent ions, but the Pol γA: γB2 holoenzyme has a broad salt optimum (Graves et al., 1998; Johnson et al., 2000; Kaguni, 2004; Lee et al., 2010a; Lee et al., 2010b; Lim et al., 1999; Longley et al., 1998b). In order to allow a straightforward comparison of the contribution of the Pol γB accessory subunit, we assayed both Pol γA and holoenzyme under conditions that approximate the natural physiological state in mitochondria. Unless otherwise specified, reactions contained 50 nM Pol γ, 250 nM Pol γB (wild type or mutant) and 500 nM DNA substrate in 60 μl of 20 mM HEPES, pH 7.5, 5% glycerol, 100 mM KCl, 100 μg/ml bovine serum albumin (BSA), and 1 mM β-mercaptoethanol (BME). After pre-incubation at 25°C for 15 min, reactions were initiated by addition of MgCl2 (10 mM) and dNTPs (1.0 mM each), and incubated at 37°C. Aliquots (6 μl) were taken at various times and the reaction was stopped by the addition of an equal volume of buffer containing 5% SDS and 250 mM EDTA. Samples were heat-denatured at 95°C for 15 min in gel-loading buffer (80% formamide, 10 mM EDTA, 0.025% xylene cyanol, 0.025% bromophenol blue), and analyzed on a 25% polyacrylamide-7 M urea gel. Reaction products were visualized by autoradiography, and were quantified using Quantity One (Bio-Rad) and Image J (NIH).
2.3 Electrophoresis mobility shift assay (EMSA)
The affinity of Pol γ holoenzyme containing either wild-type or mutant ΔI4-D129K, RK or RKK Pol γB for gapped DNA was determined by EMSA. Reactions contained 50 nM gapped 32P-labeled DNA, and varying molar ratios of Pol γ in 20 mM HEPES, pH 7.5, 5% glycerol, 100 mM KCl, 10 mM MgCl2, 100 μg/ml BSA, and 1 mM BME. Reactions were incubated at 37 °C for 10 min, and products resolved on a native 6% polyacrylamide gel. Reaction products were visualized by autoradiography and were quantified.
2.4 Exonuclease assays
Reactions were carried out at 37°C for 10 min using 200 nM gapped DNA, 100 nM Pol γ holoenzyme, 300 μM each dATP, dGTP and dTTP, 30 μM dCTP plus 1.5 μM [α-32P]dCTP (3000Ci/mol) in 20 mM HEPES, pH 7.5, 5% glycerol, 100 mM KCl, 10 mM MgCl2, 100 μg/ml BSA, and 1 mM BME. The [α-32P]dCTP and 32P-dCMP in the reaction mixture were resolved on PEI TLC plates; and the DNA products from the same reaction were resolved by electrophoresis through a 20% polyacrylamide gel. TLC plates and gels were exposed to phosphoimager screens and quantified.
2.5 Small angle scattering (SAXS) analysis
All experiments were performed on a BioSAXS-1000 (Rigaku). To determine the optimal concentration for scattering measurements, Pol γ was serially diluted in GF buffer (20mM HEPES pH 7.5, 140 mM KCl, 1mM EDTA, 5% glycerol, 5mM BME, 1mM PMSF), with and without 10 mM MgCl2, to 10.0, 5.0, 2.5, 1.25 and 0.625 mg/mL. 1.25 mg/mL is the minimal concentration that gave an adequate Radius of gyration measurement and was used for further analysis. Pol γ and the buffer alone were each analyzed twenty 30-min scans per analytical scan, with no indication of radiation-induced aggregation. Complexes of Pol γ and gapped DNA were formed by mixing enzyme at 1.25 mg/ml (5.2 μM) with T45/P25-B19 or T45/P25-B18 in a 1:1 molar ratio. Six 30-min scans were obtained, respectively, for T45/P25-B19- and T45/P25-B18-polymerase complexes. Six 30-min scans of apo Pol γ were obtained for structural comparisons. Prior to each measurement, Six 30 min-scans of GF buffer were collected for buffer subtraction. Scattering data were processed, with images converted to averaged and buffer-subtracted 1-D profiles, using the program SAXSLab (Rigaku). Guinier plots and pair density distribution functions P(r) were calculated using program ATSAS (Konarev, 2006). Ab initio Dammin & Dammif ‘consensus’ bead models (Svergun, 2001) were generated from 10 models each.
2.6 Dynamic light scattering
All experiments were performed on a Malvern Zetasizer μV Dynamic Light Scattering instrument. Samples of Pol γ holoenzyme, Pol γ complexed with 1-nt gap (T45/P25-B19), 2-nt gap (T45/P25-B18), primer/template (T45/P25) and the 45 bp duplex DNA (Table 1) were prepared at 2.5 μM in GF buffer; each sample was measured three times at 830 nm at 10°C in a 2-μL cuvette. The measurements were averaged and molecular dimensions and molecular weights were then calculated.
3. RESULTS
3.1 The efficiency of gap-filling by Pol γ is affected by gap size
To evaluate the efficiency of Pol γ in BER, we tested for gap-filling synthesis activity. A gapped template DNA was constructed by annealing three oligos to form a duplex with a central gap. Experimental data and the resulting conclusions were the same whether or not the blocker DNA contained a 5′-P, but for clarity only one set of reactions are shown. For convenience, the constructs are denoted Tx*/Py-Bz, where Tx is the template and Py and Bz are primer and blocker oligos complementary to the 3′ and 5′ end of the template, respectively. Subscripts denote oligo length, with * indicating the 5′-32P used to monitor primer extension. Thus the template T45/P25*-B19 denotes a single nucleotide gap duplex comprised of a 45-nt template annealed to a labeled 25-nt primer and a 19-nt blocker.
Pol γA alone is able to fill the single nucleotide gap (Fig. 1A); adding Pol γB increases the efficiency and rate of the gap-filling reaction but it does not significantly alter the distribution of product sizes. Pol γB thus does not confer a new property to the catalytic subunit. In both cases, the filled-in duplex (26-nt) is accompanied by limited displacement of the blocker DNA to form slightly longer products, an observation consistent with the known role of Pol γ in LP-BER (Zheng et al., 2008). Significantly, with respect to data we show below, wild-type Pol γ is incapable of displacing the entire blocker oligonucleotide to make a perfect 45 bp duplex. Note that these reactions are slow and inefficient, relative to those using a simple primer-template (Fig. 1C). There are several possible explanations: Pol γ may have a lower affinity for a single nucleotide gap in duplex DNA, a reduced rate of incorporation in filling that size gap, or a slow dissociation from the gap-filled product. As a first step in attempting to understand the basis for the inefficient reaction, which is central to BER, we enlarged the gap size to 2-nt and 5-nt with the constructs T45/P25*-B18 and T45/P22*-B18, respectively.
Fig 1.

Pol γ (exo+) gap-filling activity. Assays were performed with 45 nucleotide duplex templates containing a 1-nt (A), 2-nt, and 5-nt gap (B). C. Rate of formation of the gap-filled product by Pol γ holenzyme; P25/T45: primer-template (gel data not shown).
On larger gapped DNAs, both Pol γA (not shown) and holoenzyme synthesize much more efficiently (Fig. 1B): the rate of synthesis by holoenzyme on both 2-nt and 5-nt gapped templates is comparable to that on a simple primer-template (Fig. 1C). Significantly, there is no accumulation of a P25*+1 product on the 2-nt gapped template or a P22*+4-nt product on the 5-nt gapped template (Fig. 1B), directly showing that the rate of incorporation of the last nucleotide in the gap-filling process is no slower than in previous steps. Furthermore, the more efficient utilization of the 2- and 5-nt gapped templates suggest that neither the reaction rate of the last nucleotide insertion nor polymerase dissociation is the cause of the inefficient 1-nt gap filling. The low efficiency is therefore likely due to a low affinity of the enzyme for the template.
We examined the binding of Pol γ to various gapped constructs by EMSA. To be consistent with the activity assays, EMSA was performed at comparable concentrations of enzyme and DNA, except that their ratio was reversed in order to maintain a dynamic range of analysis. We observed different DNA binding patterns between Pol γ binding to 1-nt and 2-nt gapped templates. At a 3:1 ratio, 150 nM Pol γ and 50 nM 2-nt gapped DNA, neither Pol γA nor Pol γB show detectable binding to the 1-nt gapped DNA (data not shown), but using holoenzyme a single homogenous shifted band, containing 31% of the DNA present, was observed (Fig. 2A). Because Pol γ efficiently utilizes a 2-nt gapped DNA template for synthesis, this band probably represents the complex that is competent to synthesize DNA. In addition to this band, at a 9:1 enzyme:DNA ratio, a super-shifted band was visible. This band presumably contains more than one enzyme molecule bound to DNA. In contrast, using the 1-nt gapped DNA with a 3-fold molar excess of enzyme, three shifted species, together totaling 26% of the DNA, are visible. The fastest of these, which has the same mobility as found with the 2-nt gap DNA and by analogy may contain enzyme bound correctly to the 1-nt gap, contains ~11% of the DNA, about 3-fold less than found using the 2-nt gapped DNA. EMSA therefore supports the idea that Pol γ cannot efficiently recognize a 1-nt gap DNA. We show below that two molecules of Pol γ can bind to the 1-nt gapped DNA oligonucleotide, and the middle shifted band, which is the majority species in the EMSA assay at high enzyme:DNA ratios, likely contains two copies of Pol γ.
Fig 2.

Pol γ interaction with gapped DNAs. A) EMSA analyses. Templates containing a 1-nt gap (T45/P25-B19) and a 2-nt gap (T45/P25-B18) were mixed with Pol γ holoenzyme at 1:1, 1:3 and 1:9 ratios, and resolved on a 10% native acrylamide gel. B) SAXS structures. The molecular envelope of Pol γ complex containing a 2-nt gap DNA is similar to the apo enzyme, whereas that containing a 1-nt gapped DNA is doubled, indicating that it contains two copies of Pol γ.
3.2 Solution structures of Pol γ-gapped DNA complexes
To obtain structural information for the gap-size dependency, we determined solution structures of Pol γ complexes with gapped DNAs using small angle X-ray scattering (SAXS). The DNA constructs used have identical sequences except that the gap sizes are 1-nt and 2-nt, T45/P25-B19 and T45/P25-B18. The concentration of Pol γ required to achieve an optimal signal in SAXS was found to be 2.5 μM, 10–100 fold higher than we used in the EMSA, and thus data from the two techniques cannot be directly compared. The radius of gyration (Rg) for Pol γ apo-protein was determined by Guinier approximation to be 48.2 Å (± 1.1), in good agreement with its crystal structure (pdb: 3KLM) with a CRYSOL (Svengun, 1995) χ2 value of 2.2. When Pol γ complexes with the 2-nt gapped DNA T45/P25-B18 at a 1:1 ratio, the enzyme undergoes a significant conformational change. The complex becomes elongated with Rg increasing to 52.3 (± 1.3) Å but ab initio molecular envelope calculations indicate that the complex contains a single copy of holoenzyme (Fig. 2B). As this complex is competent for DNA synthesis, Pol γ must be directly recognizing the 3′-OH in the 2-nt gap. The Rg value of Pol γ-primer/template (T45/P25) complex was determined to be 49.8 (±2.3) Å, a value close to that of the 2-nt gap DNA complex, suggesting therefore that the two complexes have a very similar structure.
In contrast, when Pol γ binds to the 1-nt gapped template T45/P25-P19 at the same 1:1 molar ratio, the Rg value was increased to 65 (±2.3) Å and the complex appears to contain two holoenzyme molecules. Comparison of the I0 values also indicate that the larger Rg of the 1-nt gapped DNA complex corresponds to an approximate doubling of the molecular weight. Since SAXS directly interrogates the thermodynamic ensemble of conformations, the structures clearly illustrate that at equimolar concentrations, two molecules of Pol γ combine with one 1-nt gap DNA to form the majority species in the complex. Pol γ holenzyme has a mass of ~250 kDa and is estimated to bind to ~25 bp primer-template DNA (Lee et al., 2009). The length of the oligos used, 44 bp + a 1-nt gap, can therefore maximally accommodate two non-specifically bound holenzyme molecules. This indicates that the Pol γ is less discriminating in binding to 1-nt gap versus 2-nt gap DNAs. Complexes formed between Pol γA perfect 45 bp duplex were highly dependent on concentration when analyzed by SAXS and estimated values of both Rg and I0 were indicative of extensive heterogeneity. Importantly, however, because Pol γ binds differently to equivalent DNAs that differ only in the size of the gap (1-nt vs. 2-nt), these SAXS data provide direct evidence that the inefficient synthesis reaction of Pol γ on a 1-nt gapped template (Fig. 1) is due to the failure of the enzyme to efficiently make a productive complex at a single nucleotide gap in duplex DNA.
We also determined the molecular sizes of Pol γ-DNA complexes using dynamic light scattering. Both apo Pol γ and its complex with primer-template DNA (T45/P25) are similarly sized at 6.8 r.nm, whereas the 2nt gap complex is 7.4 r.nm (Table 2). However, complexes of Pol γ with the 1nt-gap DNA and the 45 bp duplex are considerably larger: 7.8 and 8.3, respectively, values that correspond to twice the molecular weight of Pol γ complexed to a 2-nt gapped DNA. In addition, apo enzyme and its complex with primer-template or 2-nt gapped DNA are all monodisperse, whereas complexes with 1-nt gapped and duplex DNA are polydisperse, suggesting that the former complexes are more structurally homogeneous. In summary, our EMSA, SAXS and DLS data strongly indicate that Pol γ is unable to effectively recognize DNA containing a single nucleotide gap, and that the enzyme requires a gap of at least two nucleotides in order to catalyze efficient DNA repair.
Table 2.
Dynamic light scattering data
| Enz | DNA | % int | Mean (r.nm) | std | MW(kDa) | dispersity |
|---|---|---|---|---|---|---|
| Pol γ | – | 100 | 6.8 | 304 | Mono | |
| 1nt-gap | 100 | 8.3 | 2.3 | 480 | poly | |
| 2nt-gap | 99 | 7.4 | 0.9 | 365 | mono | |
| p/t | 100 | 6.7 | 1 | 291 | mono | |
| 45 bp | 100 | 8.7 | 2.1 | 507 | poly |
3.3 Exonuclease-deficient Pol γ efficiently performs strand displacement synthesis
Examination of gap-filling products reveals that Pol γ synthesizes DNA that is 1–2 nt longer than the length of the gap (Fig 1), suggesting that Pol γ can perform limited strand displacement synthesis; the enzyme thus possesses an intrinsic motor function that can separate the strands of a duplex DNA using energy obtained during DNA synthesis. This raises the question why Pol γ only displaces one or two nucleotides and then stops. The persistence of the gap-filled product, even after long reaction times, indicates that the rate of strand displacement is slower than that of gap filling. When a proof-reading DNAP stalls, the primer is expected to be shuttled to the exo site, and we therefore examined the role of the 3′–5′ exonuclease on strand displacement synthesis. On a normal primer-template, exo− Pol γ synthesizes DNA at the same rate as its exo+ parent (Lee et al., 2010a).
In the absence of Pol γB, exo− Pol γA fills a 1-nt gap with a similar low efficiency as the wild-type catalytic subunit, including the ability to strand displace a couple of nucleotides (Fig. 3A). Remarkably, however, exo− holoenzyme exhibits strong strand displacement synthesis ability, including on the 1-nt gap template (Fig. 3B–D). These data suggest that the exo− mutations, although they do not affect DNA synthesis on a standard primer-template (Lee et al., 2010b), considerably stimulate the intrinsic motor function of the holoenzyme. In contrast to the displacement of only 2–3 nt by wild-type, exo-holoenzyme fully extends the primer to the end of the template, displacing the entire blocker DNA. Furthermore, few intermediate products (27–44 nts) are observed. However, a small amount of gap-filled product is always visible, indicating that the rate of strand displacement synthesis is slightly slower than gap-filling.
Fig 3.

Strand displacement by exonuclease-deficient Pol γ. A. exo− Pol γA. B–D. exo− Pol γ holenzyme with 1-nt (B), 2-nt (C) and 5-nt (D) gapped templates. E). Rates of strand displacement product formation. Primer-template gel data are not shown.
The intermediate products that are less than full-length do not represent a complete ladder. Suspecting that specific sequences, particularly GC base-pairs, may cause pauses during strand displacement synthesis, we altered a tetranucleotide sequence on the template and blocker DNAs to alter the pattern of dCMP/dGMP incorporation in the 30–33 nt size range. Strand displacement synthesis by exonuclease-deficient Pol γ on the gapped templates, T55/P25*-B29 and T′55/P25*-B′29 are similar, except that the former yields a prominent 29-mer and the latter a 32-mer (Fig. S1). These data are consistent with GC base-pairs being harder for exo− Pol γ to unwind during strand displacement synthesis.
A unique feature of the repair polymerase Pol β is that it binds directly to the 5′-phosphate of the duplex DNA downstream of a gapped (Prasad et al., 1994). This enables the polymerase to gain processivity when synthesizing DNA in a small gap, in contrast to distributive synthesis on larger gaps or on singly primed templates. Binding is due to the 8 kb domain of Pol β, but as the structure of Pol γ bound to DNA is not yet known we examined the effect of a downstream 5′-phosphate on Pol γ gap-filling synthesis. Exonuclease-deficient Pol γ acts indistinguishably when the downstream DNA contains a 5′-P or 5′-OH (Fig. S2), suggesting that Pol γ strand displacement synthesis is performed differently than Pol β, i.e., not by direct association with the 5′-P of the blocker DNA.
Strand displacement activity may also be important during lagging strand replication, where RNA primers need to be removed. To determine whether Pol γ is able to displace downstream RNA, we substituted the 5′-end of the blocker DNA with nine ribonucleotides (T55/P25*-B29-RD, Table 1). Exonuclease-deficient Pol γ displaces the modified blocker as effectively as DNA (Fig. S3), indicating that neither the geometry of the blocker duplex (A- or B-form) nor the nature of the sugar affects Pol γ strand displacement activity.
3.4 Exonuclease activity stalls the polymerase at the nick
Exonuclease-deficient Pol γ continues to synthesize DNA by strand displacement after reaching a blocker DNA. There is, however, a slight reduction in synthesis rate after the gap is completely filled that may cause the 3′-end of the DNA to switch to the exo site of the wild-type enzyme – the same mechanism used to proofread erroneously incorporated mismatched nucleotides. We hypothesized that the negative impact of the exo+ domain on the motor function of Pol γ when catalyzing strand displacement is due to elevated nucleolytic degradation at the 3′-end of the primer. We therefore examined exonucleolytic activity at a nick, using an unlabeled 5-nt gapped DNA: T45/P22-B18, as a template to ensure efficient binding by Pol γ. The second and last (fifth) nucleotides to be incorporated in the gap-filling reaction are dCMP, and we used [α-32P]dCTP with other dNTPs unlabeled. If Pol γ fills the gap completely, the initially unlabeled primer will become a labeled 27-nt. If the 27-nt is hydrolyzed to 26-nt by the 3′–5′ exonuclease activity of Pol γ, the 32P-dCMP formed is directly correlated to exonuclease activity. Because most proofreading DNAPs exhibit nucleolytic activity at a blunt end (also known as an exchange reaction), and the last two nucleotides at the 3′-end of the blocker DNA are ‘C’s, dCMP is also expected from the exchange reaction. A control reaction of a singly primed template, T45/P22, was used to measure the exchange reaction on the perfect 45 bp duplex product that will also occur on the blocker DNA in T45/P22-B18. Note that the [α-32P]dCTP in these reactions is only 30 μM whereas other dNTPs are present at 300 μM and thus the exchange reaction may involve more than the terminal dCMP.
Wild-type Pol γ indeed catalyzes the exchange reaction after extension of the singly primed T45/P22 template, as evidenced by the appearance of 32P-dCMP (Fig. 4A). However, on the T45/P22-B18 gapped DNA, the wild-type enzyme produces 40% more dCMP, directly showing exonucleolytic activity above that generated by the exchange reaction at the duplex terminus.
Fig 4.

Exonucleolytic activity of wild-type Pol γ on the T45/P22-B18 gapped DNA template. A) 32P-dCMP formation using T45/P22 and T45/P22-B18 templates. B) Products synthesized by wild-type (exo+) and exo− Pol γ.
Examination of the reaction products from the gapped DNA confirms that nucleolytic (exchange) reactions occur when using wild-type Pol γ on both the extended primer and the blocker DNA: both the labeled 27-nt gap-filled product and its 26-nt nucleolytic product are present (Fig.4B). The 18-nt blocker oligonucleotide and its 17-nt nucleolytic product also become radioactively labeled as a result of exchange reactions. The radioactivity of the 27-nt primer extension product is comparable to that of the 17-nt blocker DNA, but as the 27-nt extension product contains two 32P-dCMP residues and the 17-nt blocker DNA contains only one, the efficiency of the exonucleolytic reaction by the stalled Pol γ is ~50% of the exchange reaction. This value is in good agreement with the 40% additional turnover estimated from the production of 32P-dCMP that was measured by TLC. No exchange reaction products are seen from the blocker DNA in the reaction with exo-Pol γ (Fig.4B), confirming that the 3′–5′ nuclease activity of wild-type Pol γ is responsible for the conversion of dCTP into dCMP.
In an attempt to quantify whether exonucleolytic activity is sufficient to prevent wild-type Pol γ from strand displacement synthesis, we estimated net dCMP production at the nick. The amount of dCMP released in the T45/P22 singly primed reaction (the exchange reaction at the duplex termini) was subtracted from the dCMP released using T45/P22-B18 (Fig. 4A), and the net difference is converted into molar units. The quantity of the gap-filled 27-nt product by wild-type Pol γ was then calculated (Fig. 4B). The amount of dCMP generated at the gap-junction is ~15 fold more than the primer extension 27-nt product, i.e., for each nucleotide successfully added to complete gap-filling, ~15 rounds of synthesis-exonucleolytic digestion had taken place. Stated in a different way, only 6% of the incorporated dCTP is used in productive primer extension, the remaining 94% is excised by the exonuclease. The fact that dCMP continues to be produced over long reaction times suggests that the 3′-terminus of the primer continuously shuttles between the exo and pol sites despite incorporation of the correct nucleotide. Repetitive hydrolysis and re-incorporation then occurs, achieving little in terms of productive synthesis.
3.5 Accessory protein Pol γB is essential for Pol γA motor function
Neither wild-type holoenzyme nor exo− mutant Pol γA is capable of strand displacement synthesis. Strand displacement activity is only observed when exo− Pol γA associates with Pol γB to form a mutant holoenzyme. We attempted to delineate which subunit or which subdomain of Pol γB is important. The monomeric Pol γB (ΔI4-D129K) enhances DNA-binding of holoenzyme normally but cannot accelerate the synthesis rate on a primer-template (Lee et al., 2010b). Pol γB binds to dsDNA directly; mutant holoenzymes lacking this property (RK and RKK) display normal DNA synthesis activity on a primer-template but are defective in replisomecatalyzed synthesis on duplex DNA (Carrodeguas et al., 2002).
Using exo− Pol γA, the three mutants were tested on both 1-nt and 5-nt gapped templates. All mutant holoenzymes display lower strand displacement activities than the wild-type enzyme on the 1-nt gapped template; Pol γB RKK is the most defective with the rate of synthesis of the 45-nt strand displacement product being 2-fold lower (Fig. 5A). On the 5-nt gapped template, holenzyme containing Pol γB RK is as active as wild-type, and although all Pol γB variants increased reaction rates of the holoenzymes, relative to the 1-nt gapped template, ΔI4-D129K and RKK mutants still exhibit a defect (Fig. 5A). We then employed EMSA to ask whether a particular mutant Pol γB could be correlated with a reduced affinity for gapped DNA. However, on a given DNA, the affinities of the mutant holoenzymes are not significantly different from wild-type, retaining the same preference as wild-type for the larger gap (Fig. 5B). We conclude that the proximal Pol γB subunit, which enhances the affinity of holoenzyme for DNA (Lee et al., 2010b) (Fig. 6A), is necessary to promote strand displacement synthesis by exo− Pol γ, and that contacts between the RKK region of Pol γB and the thumb domain of Pol γA have an important role in this process.
Fig 5.
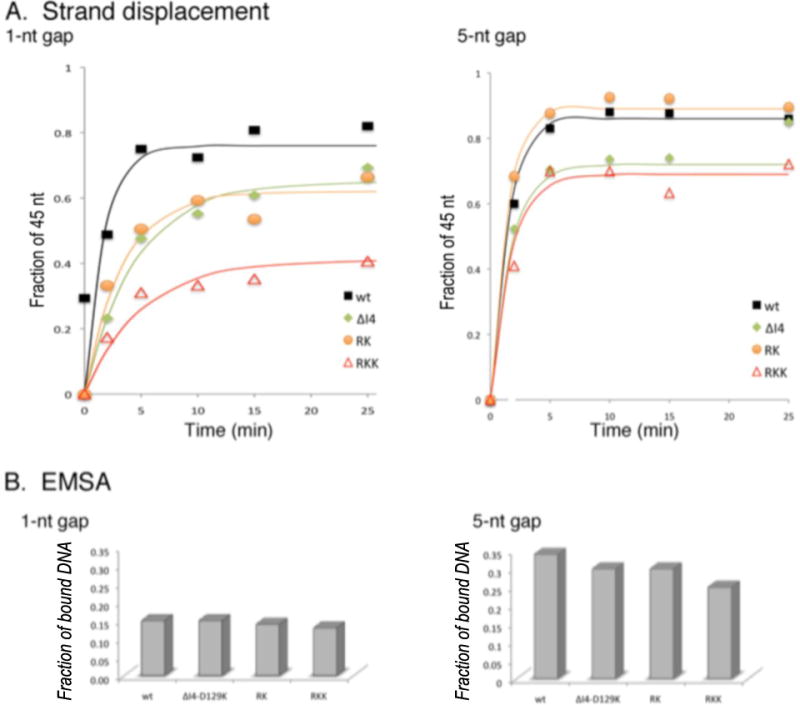
Effects of Pol γB variants on exo− holoenzyme activities. A. Strand displacement syntheses on the T45/P25*-B19 (1-nt gap) and T45/P20*-B18 (5-nt gap) templates by Pol γ containing wt or a variant Pol γB. B. EMSA using Pol γ holoenzyme containing a Pol γB variant in the presence of gapped DNAs. Experiments were performed under identical conditions to those in Fig. 2 using a Pol γ:DNA molar ratio of 3:1.
Fig 6.

Modeled Pol γ in strand displacement synthesis. A. Pol γ apo enzyme crystal structure docked with a duplex DNA with a primer. Constructed using the T7 RNAP elongation complex after superimposing the active sites of the enzymes. Strand displacement requires two elements: the O1 helix and a ssDNA-binding site in the exo domain. The displaced blocker strand could bind either to Pol γB RKK region or to the exo domain of Pol γA, near the exo− mutations. B. A proposed mechanism for how exonuclease activity affects Pol γ motor function. The rate of hydrolysis in exo is comparable to the rate of synthesis in pol, and the primer strand idles between the two sites. For exo− Pol γ, the frayed primer 3′-OH is isoenergetic with the displaced blocker. In the presence of dNTP, the equilibrium shifts to the primer binding in the pol site.
4. DISCUSSION
Extensive mitochondrial DNA (mtDNA) oxidative damage results from the high content of endogenous reactive oxygen species and the tendency of mitochondria to accumulate toxic xenobiotics (Richter et al., 1988). The situation is aggravated because radical scavengers, such as histones, that protect the chromosome are absent in mitochondria, which further increases the vulnerability of mtDNA to oxidative damage. BER is the predominant mtDNA repair mechanism, and both SN-BER and LP-BER have been detected in mitochondria (Liu et al., 2008; Szczesny et al., 2008).
We have shown here that Pol γ is inefficient in gap-filling during SN-BER; its activity is lowest on a 1-nt gap. In contrast, the nuclear repair polymerase Pol β is especially suited for SN-BER as it has a higher affinity for a small (<5 nt) gapped DNA than even a simple primer-template; Pol β most efficiently fills a 1-nt gap (Brown et al., 2010; Chagovetz et al., 1997), due to direct contacts of an 8 kDa domain with the 5′-P of downstream DNA (Prasad et al., 1994). SAXS and DLS analyses show that Pol γ cannot effectively distinguish a single nucleotide gap from a perfect duplex: the predominant complex on a 1-nt gapped duplex contains two Pol γ molecules, where at least one molecule has to bind nonspecifically, i.e., other than to the 3′-OH at the gap. In contrast, the complex with a 2-nt gapped DNA contains only a single Pol γ. This complex is competent for DNA synthesis, and thus Pol γ must recognize the 3′-OH directly.
The structure of duplex DNA containing a 1-nt gap displays a 12–17° anisotropic bend from the canonical B-form but is not otherwise significantly distorted (Guo and Tullius, 2003; Lin et al., 2009), whereas 2-nt or larger gaps have much greater flexibility (Roll et al., 1998), In principle, a polymerase can employ at least two methods to locate a gap in duplex DNA: it can either recognize the gap structure directly and bind to the 3′-OH of the primer, or it may bind non-specifically to duplex DNA and then slide until it encounters the gap. DNAPs that bend dsDNA readily expose the 3′-OH in a gap. Indeed, a crystal structure of Pol β clearly demonstrates that it bends DNA by ~90° at a gap (Batra et al., 2008; Sawaya et al., 1997), providing structural support to Pol β’s high efficiency in filling 1-nt gaps. The relatively low activity of Pol γ at a 1-nt gap could simply result from either a lower mobility in sliding along duplex DNA or a reduced ability to bend DNA., Pol γ may therefore require the greater flexibility of a ≥2-nt gap DNA in order to bind efficiently to a primer 3′-OH. However, there is experimental evidence supporting the presence of SN-BER (Bogenhagen et al., 2001; Hansen et al., 2006; Liu and Demple, 2010; Pinz and Bogenhagen, 1998; Richter et al., 1988), suggesting that other mitochondrial BER proteins may actively recruit Pol γ to the damage site.
LP-BER requires Pol γ to synthesize DNA beyond the damage site. This can be achieved either by degradation of downstream DNA or directly by strand displacement DNA synthesis, a process that requires unwinding the downstream duplex. Several exonucleases, including DNA2, FEN1 and EXOG have all been implicated in LP-BER, and depending on their immediate availability different pathways may be activated. After gap-filling, Pol γ does displace 1–2nt of downstream DNA, which could be removed by FEN1 or EXOG, both of which show the highest activity on ssDNA (Mimitou and Symington, 2011; Sykora et al., 2011; Tomlinson et al., 2010). However, our data showing that exo− Pol γ can perform robust strand displacement synthesis reveals another potential pathway, one that provides a long single-strand flap. The exonuclease activity of Pol γ may be modulated by interaction with another BER protein, including the aforementioned nucleases, and the enzymes may coordinate their activities. For example, strand displacement synthesis by Pol γ would provide a single-strand flap that could be efficiently degraded by FEN1 or EXOG or that would allow DNA2 to load.
The two seemingly unrelated activities of Pol γ – exonuclease and strand displacement motor function - are mutually exclusive; only one can be manifest at any one time. The correlation between the lack of exonuclease and strand displacement is surprising, as the exo mutations are 30–40 Å away from the strand displacement synthesis pol site in DNAPs (Fig 6A). The structural explanation for a polymerase motor function in strand displacement was first revealed using another member of the Pol A family: T7 RNA polymerase. The T7 RNAP elongation complex structure shows that strand separation is mediated by a helix (O1) situated in the major grove of the downstream duplex DNA. A bulky residue (F644) of the helix stacks against the base-pair to be unwound and facilitates strand separation (Yin and Steitz, 2002); consequently, a F644 mutant enzyme is defective in transcription (Huang et al., 2000). The functions of the equivalent helix and bulky residue were also tested using E. coli DNA Pol I: mutating the corresponding F771 residue in the O1 helix abrogates strand displacement activity (Singh et al., 2007). However, although possessing similar structural features to E. coli DNA Pol I, other members of the Pol A DNAP family are not able to strand displace and thus do not appear to maintain the motor function of the E. coli enzyme. T7 DNAP and Pol γ are two examples: both contain the O1 helix that is poised to unwind downstream template duplex but neither wild-type enzyme is able to synthesize DNA by strand displacement. However, when their intrinsic 3′–5′exonuclease activity is abrogated, strand displacement motor activity is revealed ((Lechner et al., 1983) and this study.
The critical modulation of the two activities is the distribution of the primer terminus and the relative reaction rates of excision in the exo site and synthesis in the pol site. On ssDNA templates, Pol γ exhibits a forward synthesis rate in pol greater than the excision rate in exo. Only when misincorporation slows down the synthesis reaction does excision in exo usually occur. However, the rate of exo− Pol γ repair synthesis on duplex DNA, where strand displacement is necessary, appears to be slightly slower than on a simple primer-template, so that the 3′-OH terminus of a wild-type enzyme can switch to the exo site more readily. When the enzyme is exo+, the 3′-nucleotide would be excised, resulting in idling of wild-type Pol γ at a nick. We have shown here that only 6% of the nucleotides incorporated escape immediate excision. However, the 3′-OH terminus remains intact after traveling to the exo site in exo- Pol γ, no idling occurs and a rapid switch of the terminus back to the pol site allows continued primer extension. A lack of excision activity in Pol γ creates two isoenergetic flapped DNA complexes, the flap being either the 3′ end of the primer or the 5′-end of the downstream DNA (Fig. 6B). In the presence of incoming dNTP, the affinity of pol for the primer 3′-end is enhanced, allowing primer extension by the polymerase via strand displacement.
Almost all replicases possess a proof-reading 3′-exonuclease domain or associated subunit. Replicases are also unable to catalyze extensive strand displacement synthesis by themselves, for the lack of strand displacement motor activity. However, the results of this study as well as those on T7 DNAP suggest that replicating polymerases may contain intrinsic strand displacement ability, but one that is hindered by their exo activity. Interestingly, the 3′–5′exonuclease activity of E. coli DNA Pol I, relative to its synthetic activity, is much lower than for either T7 DNAP or Pol γ. Perhaps the evolutionary trade-off of becoming a high-fidelity DNA replicase by acquiring a proof-reading exonuclease function is to suppress the ability to synthesize by strand displacement, and thus to become dependent on a separate DNA helicase to unwind downstream DNA. However, both Pol γ and T7 DNAP are not only replicases, they also function in repair processes that may require strand displacement activities; these enzymes may therefore have retained a process by which their exonuclease activity can be temporarily disabled.
5. Conclusions
Pol γ can fill gaps at high efficiency except those containing only a single nucleotide. Although wild-type Pol γ holoenzyme has very limited strand displacement activity, a 3′–5′ exonuclease deficient (exo−) enzyme replicates through duplex DNA at a rate comparable to that on a simple primer-template. Strand displacement activity is completely dependent on the presence of the accessory subunit Pol γB. Pol γ is thus intrinsically capable of strand displacement DNA synthesis, but the activity is normally suppressed by its exonuclease, suggesting that nucleolytic activity modulates both DNA synthesis fidelity and the motor function of human Pol γ. Our data suggest the hypothesis that another BER protein must assist Pol γ in SN-BER by increasing its affinity of the enzyme for a 1-nt gap, or by modulating its exonuclease activity to enable the repair complex to catalyze strand displacement synthesis.
Highlights.
The paper characterizes functions of mitochondrial DNA polymerase (Pol γ) in base excision repair (BER).
The 3′ exonuclease activity regulates Pol γ strand displacement DNA synthesis ability.
The findings provide an alternative pathway for long-patch BER.
Acknowledgments
This work was supported by grants from the Welch Foundation (H-1592) and the National Institutes of Health Grant (GM083703) to YWY, and by an endowment from the Sealy and Smith Foundation to Sealy Center for Structural Biology at University of Texas Medical Branch.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balakrishnan L, Polaczek P, Pokharel S, Campbell JL, Bambara RA. Dna2 exhibits a unique strand end-dependent helicase function. J Biol Chem. 2010;285:38861–38868. doi: 10.1074/jbc.M110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. Endogenous oxidative damage of mtDNA. Mutat Res. 1999;424:51–58. doi: 10.1016/s0027-5107(99)00007-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Pinz KG, Perez-Jannotti RM. Enzymology of mitochondrial base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:257–271. doi: 10.1016/s0079-6603(01)68105-4. [DOI] [PubMed] [Google Scholar]
- Brown JA, Pack LR, Sanman LE, Suo Z. Efficiency and fidelity of human DNA polymerases lambda and beta during gap-filling DNA synthesis. DNA Repair (Amst) 2010;10:24–33. doi: 10.1016/j.dnarep.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrodeguas JA, Pinz KG, Bogenhagen DF. DNA binding properties of human pol gammaB. J Biol Chem. 2002;277:50008–50014. doi: 10.1074/jbc.M207030200. [DOI] [PubMed] [Google Scholar]
- Chagovetz AM, Sweasy JB, Preston BD. Increased activity and fidelity of DNA polymerase beta on single-nucleotide gapped DNA. J Biol Chem. 1997;272:27501–27504. doi: 10.1074/jbc.272.44.27501. [DOI] [PubMed] [Google Scholar]
- Driggers WJ, Holmquist GP, LeDoux SP, Wilson GL. Mapping frequencies of endogenous oxidative damage and the kinetic response to oxidative stress in a region of rat mtDNA. Nucleic Acids Res. 1997;25:4362–4369. doi: 10.1093/nar/25.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves SW, Johnson AA, Johnson KA. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry. 1998;37:6050–6058. doi: 10.1021/bi972685u. [DOI] [PubMed] [Google Scholar]
- Guo H, Tullius TD. Gapped DNA is anisotropically bent. Proc Natl Acad Sci U S A. 2003;100:3743–3747. doi: 10.1073/pnas.0737062100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AB, Griner NB, Anderson JP, Kujoth GC, Prolla TA, Loeb LA, Glick E. Mitochondrial DNA integrity is not dependent on DNA polymerase-beta activity. DNA Repair (Amst) 2006;5:71–79. doi: 10.1016/j.dnarep.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brieba LG, Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J Biol Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- Johnson AA, Tsai Y, Graves SW, Johnson KA. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry. 2000;39:1702–1708. doi: 10.1021/bi992104w. [DOI] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun D. ATSAS 2.1, a program package for small-angle scattering data analysis. Journal of Applied Crystallography. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner RL, Engler MJ, Richardson CC. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J Biol Chem. 1983;258:11174–11184. [PubMed] [Google Scholar]
- Lee YS, Johnson KA, Molineux IJ, Yin YW. A single mutation in human mitochondrial DNA polymerase Pol gammaA affects both polymerization and proofreading activities of only the holoenzyme. J Biol Chem. 2010a;285:28105–28116. doi: 10.1074/jbc.M110.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Kennedy WD, Yin YW. Structural insights into human mitochondrial DNA replication and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Lee S, Demeler B, Molineux IJ, Johnson KA, Yin YW. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J Biol Chem. 2010b;285:1490–1499. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- Lin S, Horning DP, Szostak JW, Chaput JC. Conformational analysis of DNA repair intermediates by time-resolved fluorescence spectroscopy. J Phys Chem A. 2009;113:9585–9587. doi: 10.1021/jp906746w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of Oxidative DNA Damage via FEN1-Dependent Long-Patch Base Excision Repair in Human Cell Mitochondria. Mol Cell Biol. 2008 doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Mosbaugh DW. Properties of the 3′ to 5′ exonuclease associated with porcine liver DNA polymerase gamma. Substrate specificity, product analysis, inhibition, and kinetics of terminal excision. J Biol Chem. 1991;266:24702–24711. [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A. 1998a;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Ropp PA, Lim SE, Copeland WC. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998b;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection–unraveling the tail. DNA Repair (Amst) 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Pandey M, Nandakumar D. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr Opin Chem Biol. 2012;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinz KG, Bogenhagen DF. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol Cell Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinz KG, Bogenhagen DF. The influence of the DNA polymerase gamma accessory subunit on base excision repair by the catalytic subunit. DNA Repair (Amst) 2006;5:121–128. doi: 10.1016/j.dnarep.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase beta. Dependence on 5′-phosphate group. J Biol Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll C, Ketterle C, Faibis V, Fazakerley GV, Boulard Y. Conformations of nicked and gapped DNA structures by NMR and molecular dynamic simulations in water. Biochemistry. 1998;37:4059–4070. doi: 10.1021/bi972377w. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- Singh K, Srivastava A, Patel SS, Modak MJ. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J Biol Chem. 2007;282:10594–10604. doi: 10.1074/jbc.M611242200. [DOI] [PubMed] [Google Scholar]
- Stewart JA, Campbell JL, Bambara RA. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 2009;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svengun D, Barberato C, Koch MHJ. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. J Biophys. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Wilson DM, 3rd, Bohr VA. Repair of persistent strand breaks in the mitochondrial genome. Mech Ageing Dev. 2011;133:169–175. doi: 10.1016/j.mad.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J Biol Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson CG, Atack JM, Chapados B, Tainer JA, Grasby JA. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem Soc Trans. 2010;38:433–437. doi: 10.1042/BST0380433. [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


