Abstract
Elevated fasting blood glucose (FBG) has been associated with increased risk for development of type 2 diabetes. Single nucleotide polymorphisms (SNPs) in G6PC2 are the most important common determinants of variations in FBG in humans. Studies using G6pc2 knockout mice suggest that G6pc2 regulates the glucose sensitivity of insulin secretion. G6PC2 and the related G6PC1 and G6PC3 genes encode glucose-6-phosphatase catalytic subunits. This study describes a functional analysis of 22 non-synonymous G6PC2 SNPs, that alter amino acids that are conserved in human G6PC1, mouse G6pc1 and mouse G6pc2, with the goal of identifying variants that potentially affect G6PC2 activity/expression. Published data suggest strong conservation of catalytically important amino acids between all four proteins and the related G6PC3 isoform. Because human G6PC2 has very low glucose-6-phosphatase activity we used an indirect approach, examining the effect of these SNPs on mouse G6pc1 activity. Using a novel in situ functional assay for glucose-6-phosphatase activity we demonstrate that the amino acid changes associated with the human G6PC2 rs144254880 (Arg79Gln), rs149663725 (Gly114Arg) and rs2232326 (Ser324Pro) SNPs reduce mouse G6pc1 enzyme activity without affecting protein expression. The Arg79Gln variant alters an amino acid mutation of which, in G6PC1, has previously been shown to cause glycogen storage disease type 1a. We also demonstrate that the rs368382511 (Gly8Glu), rs138726309 (His177Tyr), rs2232323 (Tyr207Ser) rs374055555 (Arg293Trp), rs2232326 (Ser324Pro), rs137857125 (Pro313Leu) and rs2232327 (Pro340Leu) SNPs confer decreased G6PC2 protein expression. In summary, these studies identify multiple G6PC2 variants that have the potential to be associated with altered FBG in humans.
Introduction
Elevated fasting blood glucose (FBG) has been associated with increased risk for the development of type 2 diabetes (T2D) and cardiovascular associated mortality (CAM) [1–3]. Previous studies have shown that an increase in FBG of 9–18 mg/dl is associated with a ~30% increased risk of CAM [2]. Conversely, a reduction in FBG of ~9 mg/dl is associated with a 25% reduction in risk of CAM [3]. Multiple groups have performed genome wide association studies (GWAS) in an effort to identify genes associated with variations in FBG. These studies have identified SNPs that effect glucose homeostasis in over 50 loci, many of which are also associated with T2D [4–7]. Notably, the rs560887 single nucleotide polymorphism (SNP) located in the third intron of the G6PC2 locus has been identified as the strongest common genetic determinant of FBG levels in terms of significance and effect size, accounting for ~1% of total variance in FBG [4,6–11]. Molecular studies have shown that this intronic SNP affects G6PC2 RNA splicing [12]. Further genetic and molecular analyses of this locus have shown that two common promoter SNPs, rs13431652 and rs2232316, in addition to the intronic rs560887 SNP, are potentially causative and may contribute to the association between G6PC2 and FBG [12,13].
G6PC2, formerly known as IGRP, encodes an islet-specific glucose-6-phosphatase catalytic subunit [14–16] that catalyzes the hydrolysis of glucose-6-phosphate (G6P) to glucose and inorganic phosphate (Pi) [11,17]. Studies on G6pc2 knockout (KO) mice show that G6pc2 is a negative regulator of glucose-stimulated insulin secretion (GSIS) [11,16]. G6pc2 acts by hydrolyzing G6P, thereby increasing glucose cycling and presumably decreasing glycolytic flux [16,18]. As such, G6pc2 opposes the action of the glucose sensor glucokinase [19,20]. Both male and female G6pc2 KO mice have significantly decreased FBG levels [15,16,21], consistent with GWAS data and molecular studies.
Common variants associated with variations in FBG were thought to account for a low percentage (~10%; Ref. [22]) of total heritable variation, leading to speculation that rare (minor allele frequency <5%), high impact variants undetected by GWAS might account for the remaining 90% of heritability [23]. However, more recent studies have suggested that the combined effects of multiple common variants have the potential to largely account for missing heritability [24–26]. Nevertheless, the identification of high impact rare variants has provided tremendous insight into beta cell biology [27,28]. For example, while common SNPs in the glucokinase (GCK) GCK locus have modest effects on FBG [8,9], rare inactivating variants in the GCK locus have been shown to cause neonatal diabetes mellitus or maturity-onset diabetes in youth while rare activating variants cause hyperinsulinemia [27,28]. Evolutionarily this is logical, as rare variants with significant detrimental effects on health would be selected against and therefore not be propagated in the human population. These data also highlight an important caveat in the interpretation of GWAS data: the effect size of common genetic variants does not necessarily reflect the importance of the gene in relation to the disease or phenotype being studied. As observed with GCK and G6PC2, despite the greater effect size of common G6PC2 variants on FBG, deletion of the Gck gene in mice is lethal [29] whereas deletion of G6pc2 results in a mild reduction in FBG [15,16].
Because the identification of high impact rare variants in GCK has provided tremendous insight into beta cell biology [27,28], we were interested in identifying variants in G6PC2 that affect G6PC2 protein expression. We were also interested in identifying variants that potentially have a significant effect on G6PC2 enzyme activity. However, because G6PC2 has much lower enzyme activity than G6PC1, we addressed this second question indirectly by analyzing the effect of 22 non-synonymous G6PC2 SNPs, that change amino acids that are conserved in human G6PC1, mouse G6pc1 and mouse G6pc2, on G6pc1 enzyme activity using a novel in situ functional assay for glucose-6-phosphatase activity.
Materials and Methods
Cell Culture
Rat islet-derived 832/13 cells and monkey kidney-derived COS 7 cells were passaged as sub-confluent cultures in RPMI medium supplemented with 10% (vol/vol) fetal bovine serum, 0.05 mM βmercaptoethanol, 100 U/ml penicillin and 100 μg/ml streptomycin.
SNP Databases
Human G6PC2 SNPs were identified using the UCSC Genome Browser (https://genome.ucsc.edu/), HumSAVR (http://omictools.com/humsavar-tool) or dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) databases.
G6PC Expression Vector Construction
The construction of plasmids encoding human G6PC2 (accession number NM_021176), mouse G6pc2 (accession number NM_021331), human G6PC1 (accession number NM_000151) and mouse G6pc1 (accession number NM_008061) in the pcDNA3.1D v5-His-TOPO vector with a C-terminal V5-His Tag has been previously described [30,31]. A common SNP, rs492594, switches a valine to a leucine at amino acid (AA) 219 in human G6PC2 [14]. The human G6PC2 sequence designated as wild type (WT) in this study contained a leucine at AA 219.
Human G6PC2:mouse G6pc2 and mouse G6pc2:human G6PC2 chimeras in the pcDNA3.1D v5-His-TOPO vector were constructed by ligating fragments of the respective open reading frames. This was achieved by sub-cloning using a combination of restriction enzyme sites in the pcDNA3.1D v5-His-TOPO vector and the internal Dra I, Stu I and BamH I restriction enzyme sites, which are common to both human G6PC2 and mouse G6pc2. These three sites truncate G6PC2/G6pc2 at AAs 72, 192 and 249, respectively, from the N terminus.
Site-directed mutagenesis was used to change specific codons in human G6PC2 and mouse G6pc1. This was achieved either by using the Quikchange II kit (Agilent Technologies, Santa Clara, CA) or a three-step PCR protocol [32]. DNA sequencing was used to verify all codon changes and the absence of secondary mutations. Two to three independent plasmid preparations were analyzed for each SNP variant described. Plasmids were purified by centrifugation through cesium chloride gradients [32].
G6pc1 and Pklr Fusion Gene Construction
A bacterial artificial chromosome (BAC) clone (CH230-220J5) containing the entire rat G6pc1 gene (Accession number AC123346) was purchased from BACPAC Resources, Children's Hospital Oakland Research Institute, Oakland, CA. This clone was digested with Kpn I to isolate a 7319 bp fragment, representing the rat G6pc1 promoter region between -7253 and +66 [33], that was then ligated into the Kpn I digested pGEM7 vector (Promega, Madison, WI). This fragment was then re-isolated, blunted ended using Klenow and ligated into the Xba I—Bgl II, digested and Klenow treated pCAT(An) vector, a gift from Dr. Howard Towle [34]. Fragments of the rat G6pc1 promoter, representing promoter sequence between -7248 and +62 and -1640 and +62, were then re-isolated from the pCAT(An) plasmid by digestion with Hind III and Xho I and ligated into the Hind III and Xho I digested pGL3 MOD luciferase vector [30].
Two fragments of the rat liver pyruvate kinase (Pklr) promoter representing sequences from -206 to +1 and -100 to +1 [35], were generated by PCR reaction using rat genomic DNA as the template in conjunction with the following primers:
(5'-CCCAAGCT(-206)TCTGCAGACAGGCCAAAGGGGATCC-3'),
(5'-CCCAAGCT(-100)TGCTAGCTGGTTATACTTTAAC-3'), and
(5'-CCGCTCGAGA(+1)CCTGCTGTGTCTGTGGGTCTGCT-3'); Hind III and Xho I cloning sites underlined. The PCR fragments generated were digested with Hind III and Xho I, ligated into the Hind III and Xho I digested pGEM7 vector and then sequenced to ensure the absence of polymerase errors. The fragments were then re-isolated from the pGEM7 plasmid and ligated into the Hind III and Xho I digested pGL3 MOD luciferase vector [30].
RNA Isolation and Quantification
To compare wild type human G6PC2 and mouse G6pc2 RNA expression, plasmids encoding human G6PC2 and mouse G6pc2 (3 μg) were transiently transfected in semi-confluent COS 7 cells in 10 cm diameter dishes using the lipofectamine reagent (InVitrogen, Waltham, MA) as previously described [36]. Following transfection, cells were incubated for 18–20 hours in serum-containing medium. RNA was then harvested and purified using the RNAqueous® kit (Ambion, Carlsbad, CA). Gene expression was then quantitated by using the Turbo DNA-free DNAse Treatment Kit (Ambion, Carlsbad, CA) to remove trace genomic DNA followed by cDNA generation using the iScript DNA Synthesis Kit (Bio-Rad, Hercules, CA) and then PCR using the dUTP-containing FastStart SYBR Green Master Mix in conjunction with Uracil-Glycosylase (Roche, Nutley, NJ). PCR products were analyzed by electrophoresis on 1% agarose gels.
The following primer pairs, that recognize non-coding sequences in the pcDNA3.1D v5-His-TOPO vector, were used for the analysis of both human G6PC2 and mouse G6pc2 RNA expression:
pcDNA Forward 5’ CCCAAGCTTGGTACCGAGCTCGGATCCAGT
pcDNA Reverse 3’ CCCGTTTAAACTCAATGGTGATGGTGATGATGACCGGTA
The pcDNA forward primer recognizes 5’ untranslated leader sequence in the mRNA. This sequence represents part of the polylinker 3’ of the CMV transcription start site. The pcDNA reverse primer recognizes 3’ untranslated sequence in the mRNA. This sequence represents the region immediately 3’ of the V5 His tag.
Monkey cyclophilin A (PPIA) expression was quantitated as an internal control using the following primers:
Monkey PPIA Forward 5’-AATGGCACTGGTGGCAAGTC -3’
Monkey PPIA Reverse 5’- GCTCCATGGCCTCCACAATA -3’
Protein Expression, Western blotting and Luciferase Assays
To determine whether wild type and variant proteins were expressed at similar levels, plasmids encoding human G6PC2 and mouse G6pc1 variants (2 μg) were transiently transfected in semi-confluent 832/13 or COS 7 cells in 3.5 cm diameter dishes using the lipofectamine reagent (InVitrogen, Waltham, MA) as previously described [36]. Following transfection, cells were incubated for 18–20 hours in serum-containing medium. Cells were then harvested using 50 mM Tris, pH 8.0, 150 mM NaCl, 5.8 mM PMSF, and 1% NP-40. Protein samples were quantified using the Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA). 20 μg of cell extract was electrophoresed on 10% SDS-polyacrylamide gels and the proteins transferred to PVDF membrane (Perkin Elmer, Waltham, MA). Protein expression was determined by immunoblotting with a conjugated mouse monoclonal Anti-V5-horseradish peroxidase (HRP) antibody (1:100–1:5000, InVitrogen, Waltham, MA). A primary anti-beta actin monoclonal antibody (1:10,000, Sigma, St. Louis, MO) with an Anti-Mouse HRP secondary antibody (1:10,000, Promega, Madison, WI) was used to determine beta actin expression as a loading control. HRP activity was assayed using the Pierce™ ECL reagent (Thermo Fisher Scientific, Waltham, MA). Protein expression data were normalized by scanning both V5 and actin signals on Western blots. The ratio of V5 to actin expression obtained with the variants shown was expressed as a percentage relative to the ratio obtained with WT human G6PC2 or mouse G6pc1.
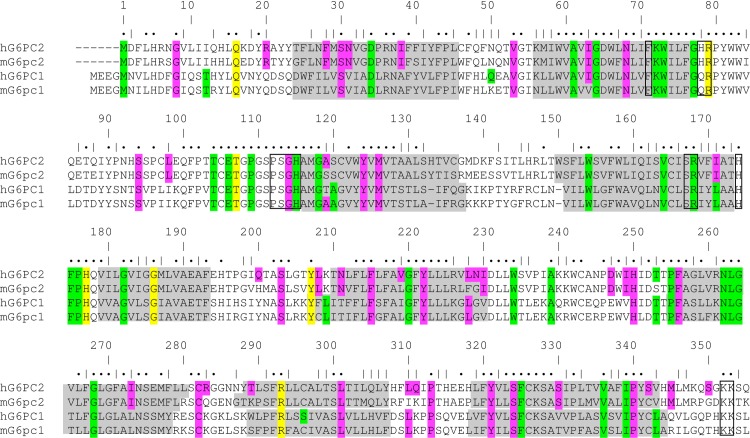
The expected sizes of human G6PC2, mouse G6pc2, human G6PC1 and mouse G6pc1 with V5 His tags are 45.60, 45.71, 45.54 and 45.51 kDa. As previously observed, both the human G6PC1 [30] and mouse G6pc1 [31] expression plasmids generate doublets, possibly though the use of alternate methionine start codons (Fig 1).
Fig 1. Conservation of Amino Acids Between Human 12, Mouse G6pc2, Human G6PC1 and Mouse G6pc1.
Sequence alignment showing the conservation of AAs between human (h) G6PC2, mouse (m) G6pc2, human G6PC1 and mouse G6pc1. Residues highlighted in green represent AAs mutation of which in G6PC1 causes GSD type 1a [40]. Residues highlighted in pink represent AAs that are changed by human G6PC2 SNPs that were identified using the UCSC Genome Browser (https://genome.ucsc.edu/) and HumSAVR (http://omictools.com/humsavar-tool) databases. Residues highlighted in yellow represent conserved AAs in human G6PC2, mouse G6pc2, human G6PC1 and mouse G6pc1 that are changed by a human G6PC2 SNP and where mutation in G6PC1 can cause GSD type 1a. Identities are indicated by filled circles and similarities by vertical bars.
For fusion gene analyses, semi-confluent 832/13 cells in 3.5 cm diameter dishes were co-transfected with 2 μg of a G6pc1- or Pklr firefly luciferase fusion gene construct, 0.5 μg of SV40-Renilla luciferase (Promega, Madison, WI) and the indicated amount of a wild type or variant G6pc1 expression vector, using the lipofectamine reagent (InVitrogen, Waltham, MA) as previously described [36]. Following transfection, cells were incubated for 18–20 hours in serum-free medium supplemented with 2 or 30 mM glucose. Cells were then harvested using passive lysis buffer (Promega, Madison, WI). Firefly and Renilla luciferase activity were assayed using the Dual Luciferase Assay kit (Promega, Madison, WI). To correct for variations in transfection efficiency, the results were calculated as a ratio of firefly to Renilla luciferase activity. As indicated in the Table and Figure Legends, results were presented either as this ratio or relative to the ratio obtained with 30 mM glucose or relative to the ratio obtained at 30 mM glucose in the presence of either catalytically dead G6pc1 or WT G6pc1. Fusion gene expression was assessed in multiple transfections using at least two independent preparations of each plasmid, as indicated in the Table and Figure Legends.
Statistical Analysis
All data were analyzed using the Student’s t-test: two sample assuming equal variance. The level of significance was as indicated (two-sided test).
Results
Analysis of Glucose-6-Phosphatase Activity
We have previously shown that glucose-6-phosphate activity is abolished in G6pc2 KO mouse islets strongly suggesting that G6pc2 has phosphohydrolase activity [16]. However, while several groups have attempted to detect G6P hydrolysis following overexpression of human G6PC2 or mouse G6pc2 [14,37,38], only one group has been successful [39]. Petrolonis et al. demonstrated that the rate of G6P hydrolysis by G6PC2 overexpressed in COS7 cells was 20–40 fold lower than that of G6PC1 [39]. This suggests that there are inherent technical difficulties in demonstrating G6P hydrolysis following overexpression of G6PC2.
Because of the low enzyme activity of G6PC2 in this study we decided to focus on non-synonymous human G6PC2 SNPs that alter AAs that are conserved in the highly related [14] and much more enzymatically active human G6PC1 and mouse G6pc1 isoforms of the glucose-6-phosphatase catalytic subunit (Fig 1; Table 1). Specifically, we decided to analyze the effect of these G6PC2 SNPs indirectly by examining their effect on mouse G6pc1 enzyme activity. We hypothesize that G6PC2 SNPs that affect the function of mouse G6pc2 are highly likely to affect the function of human G6PC2 because of the strong conservation of catalytically important amino acids between these proteins [14]. Supporting this hypothesis is the observation that of the 56 AAs in human G6PC1 mutation of which gives rise to glycogen storage disease (GSD) type 1a [40], 51 are conserved or represent conserved changes in human G6PC2 (S1 Table). Based on this logic, we searched available databases for non-synonymous human G6PC2 SNPs that alter AAs that are conserved in mouse G6pc2 and the highly related and much more enzymatically active human G6PC1 and mouse G6pc1 isoforms of the glucose-6-phosphatase catalytic subunit. We identified 22 such non-synonymous human G6PC2 SNPs (Fig 1; Table 1). These SNPs change AAs in a number of different regions of G6PC2 (Table 1), based on the predicted membrane topology of human G6PC1 [41]. We analyzed the effect of these G6PC2 SNPs indirectly by examining their effect on mouse G6pc1 enzyme activity using a novel in situ assay.
Table 1. Analysis of the Effect of Amino Acids Changed by Human G6PC2 SNPs on Human G6PC2 and Mouse G6pc1 Protein Expression and Activity.
Amino acids (AAs) changed by human G6PC2 SNPs that are conserved in human G6PC2, mouse G6pc2, human G6PC1 and mouse G6pc1 were identified using the UCSC Genome Browser (https://genome.ucsc.edu/) and HumSAVR (http://omictools.com/humsavar-tool) databases. The G6PC2 domain affected by each AA change was predicted by comparison with the proposed structure of G6PC1 [41]. The Table shows the effect of these SNPs on G6pc1 enzyme activity based on comparison with wild type (WT) G6pc1 as assessed using a novel in situ enzyme assay (Figs 2 & 3). This assay measures the ability of G6pc1 to suppress glucose-stimulated fusion gene expression (Figs 2 & 3). Results for each variant represent the mean ± S.E.M. of 3 experiments using two independent preparations of each expression vector construct in which each experimental condition was assayed in triplicate. While all of these G6PC2 SNPs change AAs that are conserved in G6PC1, mutation of some of these AAs in G6PC1 causes GSD type 1a [40]. In each case the AA associated with GSD type 1a is shown in parentheses. In each case the G6PC2 SNP changes the AA to one distinct from that associated with GSD type 1a. For simplicity and comparisons between human G6PC2 and mouse G6pc1 the AAs in mouse G6pc1 are numbered based on the position of the equivalent conserved AA in human G6PC2 (Fig 1). N.D., not determined; N.C., no change. **, these residues have been associated with variations in FBG in healthy individuals who do not have diabetes [50,52,53].
| hG6PC2 SNP | Base # | AA# | GSD Type1a Mutation | Domain Location | hG6PC2 Expression | mG6pc1 Expression | % WT mG6pc1 Activity | P Value |
|---|---|---|---|---|---|---|---|---|
| rs368382511 | GGA23GAA | Gly8Glu | No | N terminus | Decreased | Decreased | 90.13 ± 2.97 | 0.03 |
| rs372008743 | CAG48CAT | Gln16His | Yes (Arg) | N terminus | N.D. | N.C. | 110.04 ± 1.71 | 0.001 |
| rs142189264 | TCC89TTC | Ser30Phe | No | In membrane 1 | N.D. | N.C. | 67.22 ± 1.39 | 0.00002 |
| rs375874967 | GTT157ATT | Val53Ile | No | In loop | N.D. | N.C. | 109.98 ± 2.07 | 0.01 |
| rs199682245 | AAT203ATT | Asn68Ile | No | In membrane 2 | N.D. | N.C. | 79.45 ± 4.46 | 0.01 |
| rs144254880 | CGA236CAA | Arg79Gln | Yes (His or Cys) | In membrane 2 | N.D. | N.C. | 52 ± 1.07 | 0.000001 |
| rs371234742 | ACA320AGA | Thr107Arg | Yes (Ile) | In loop | N.D. | N.C. | 84.25 ± 4.99 | 0.03 |
| rs149663725 | GGC340CGC | Gly114Arg | No | In loop | N.D. | N.C. | 53.32 ± 0.88 | 0.000001 |
| rs187707963 | TAT371TGT | Tyr124Cys | No | In membrane 3 | N.D. | N.C. | 104 ± 0.92 | 0.01 |
| rs367930047 | ATG376GTG | Met126Val | No | In membrane 3 | N.D. | N.C. | 103.96 ± 5.75 | 0.53 |
| rs138726309 | CAT529TAT | His177Tyr** | Yes (Pro) | In membrane 5 | Decreased | N.C. | 92 ± 2.69 | 0.03 |
| rs201094274 | AGT609AGA | Ser203Arg | No | In loop | N.D. | N.C. | 115.33 ± 3.85 | 0.003 |
| rs2232323 | TAC620TCC | Tyr207Ser** | Yes (Cys) | In loop | Decreased | N.C. | 88.19 ± 6.53 | 0.02 |
| rs139587795 | TTC644TCC | Phe215Ser | No | In membrane 6 | N.D. | N.C. | 105 ± 5.09 | 0.42 |
| rs375527806 | TAC664CAC | Tyr222His | No | In membrane 6 | N.D. | N.C. | 94.59 ± 2.37 | 0.08 |
| rs147360987 | CAC748TAC | His250Tyr | No | In loop | N.D. | N.C. | 94.04 ± 5.08 | 0.31 |
| rs150538801 | TTT766CTT | Phe256Leu | No | In loop | N.D. | N.C. | 78.96 ± 1.95 | 0.0004 |
| rs374055555 | CGG877TGG | Arg293Trp | Yes (Cys) | In membrane 8 | Decreased | Decreased | 83.55 ± 2.56 | 0.003 |
| rs141041285 | TTG902TCG | Leu301Ser | No | In membrane 8 | N.D. | N.C. | 86.94 ± 5.85 | 0.02 |
| rs137857125 | CCG938CTG | Pro313Leu | No | In loop | Decreased | N.C. | 83 ± 5.48 | 0.04 |
| rs2232326 | TCT970CCT | Ser324Pro** | No | In membrane 9 | Decreased | N.C. | 58.72 ± 6.27 | 0.03 |
| rs2232327 | CCC1019CTC | Pro340Leu | No | In membrane 9 | Decreased | Decreased | 55.31 ± 4.93 | 0.001 |
**, indicates SNPs that have been association with variations in fasting plasma glucose in non-diabetic individuals.
Characterization of a Novel Assay for the Measurement of Glucose-6-Phosphatase Activity In Situ
Because the activity of G6pc1 appears to be regulated by unknown factors [42], it is unclear whether glucose-6-phosphatase activity assayed in vitro truly reflects activity in intact cells. Therefore, before beginning the functional analysis of non-synonymous human G6PC2 SNPs on mouse G6pc1 activity, we first developed a novel assay for the measurement of glucose-6-phosphatase activity in situ.
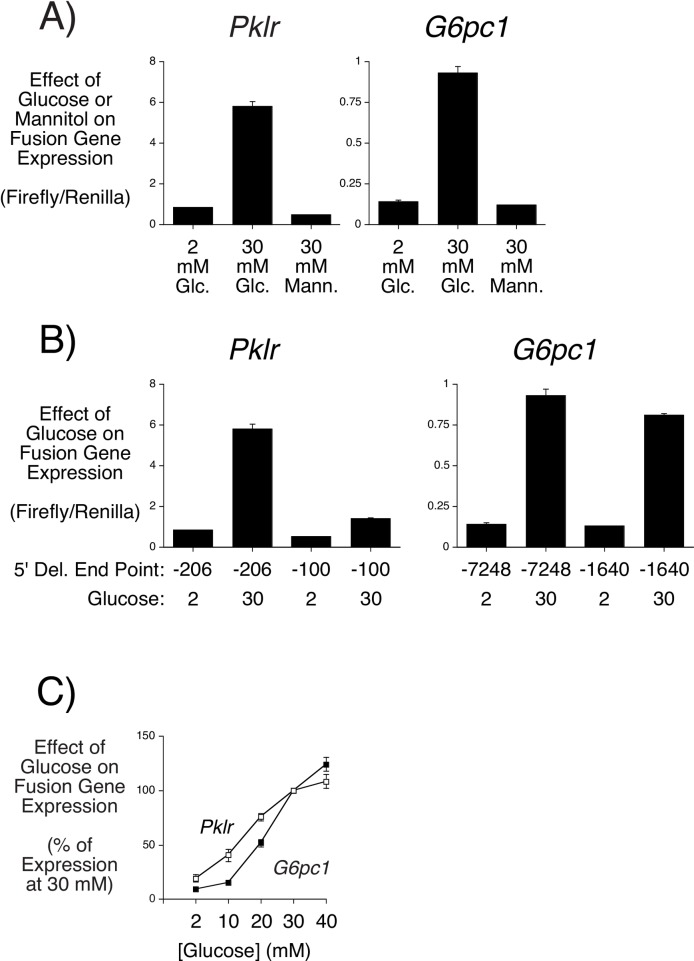
Newgard and colleagues [43,44] have previously described a highly glucose responsive INS-1 cell line variant, designated 832/13. Rat G6pc1 [45] and liver pyruvate kinase (Pklr) [46] fusion gene expression are robustly induced by glucose in 832/13 cells. Fig 2A shows that, following transient transfection of 832/13 cells with luciferase fusion genes containing Pklr promoter sequence between -206 and +1 or G6pc1 promoter sequence between -7248 and +62, glucose markedly stimulated reporter gene expression, confirming published reports [45,46]. Mannitol, a control for the osmotic effect of glucose, had no effect (Fig 2A). Deletion of the Pklr promoter region between -206 and -101 markedly reduced the effect of glucose on Pklr-luciferase fusion gene expression whereas deletion of the G6pc1 promoter region between -7248 and -1641 had little effect on glucose-stimulated G6pc1-luciferase fusion gene expression (Fig 2B). A comparison of the EC50 for glucose-stimulated Pklr-luciferase and G6pc1-luciferase expression showed that Pklr-luciferase fusion gene expression was more sensitive to glucose (Fig 2C).
Fig 2. Glucose-Regulated Fusion Gene Expression in 832/13 Cells.
832/13 cells were transiently co-transfected, as described in Materials and Methods, with an expression vector encoding Renilla luciferase (0.5 μg) and Pklr-luciferase or G6pc1-luciferase fusion genes (2 μg) containing the promoter regions from -206 to +1 and -7253 to +66, respectively, (Panels A and C) or the indicated promoter regions (Panel B). Following transfection, cells were incubated for 18–20 hr in serum-free medium in the presence of the indicated concentrations of glucose (Glc) or mannitol (Mann). Cells were then harvested and luciferase activity assayed as described in Materials and Methods. Results are presented as the ratio of firefly:Renilla luciferase activity (Panels A and B) or a percentage of the induction achieved with 30 mM glucose (Panel C). Results represent the mean ± S.E.M. of 3 experiments using independent preparations of all fusion gene constructs in which each experimental condition was assayed in triplicate.
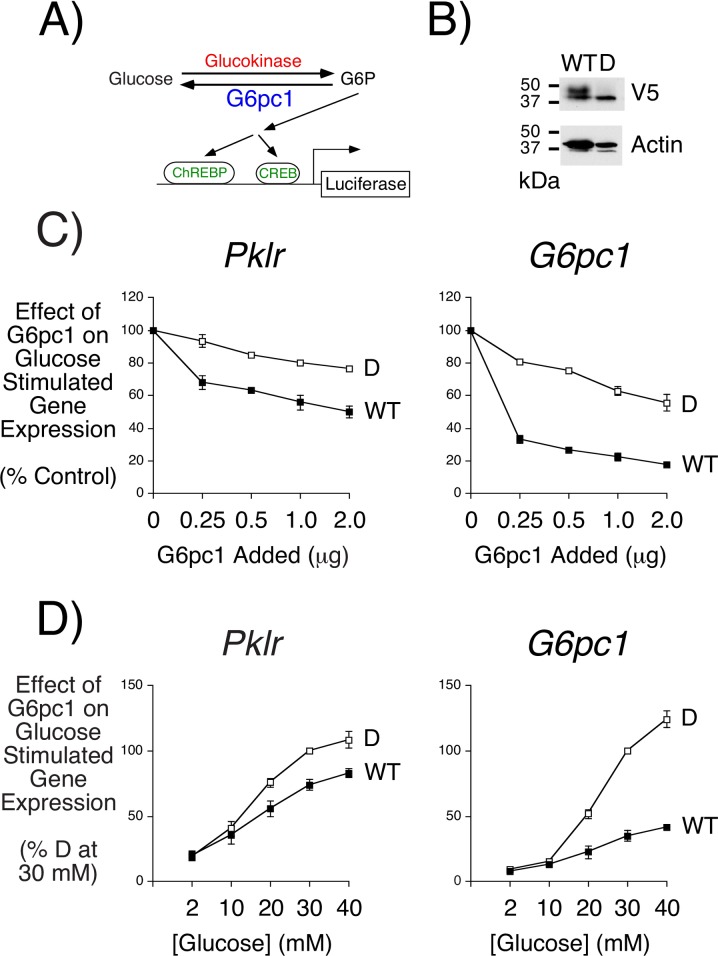
Since the 832/13 cell line is derived from rat islets [43,44] these cells do not express endogenous G6pc2 because, in contrast to all other species examined to date, G6pc2 is a pseudogene in rats [14]. It therefore occurred to us that these cells could be used to assay glucose-6-phosphatase enzyme activity in situ by measuring the ability of G6pc1 expression to blunt glucose-stimulated Pklr-luciferase or G6pc1-luciferase fusion gene expression (Fig 3A). We hypothesized that G6pc1 would repress glucose-stimulated fusion gene expression by stimulating G6P hydrolysis [47], therefore opposing the action of the endogenous glucokinase in these cells [48] and thereby reducing glycolytic flux. To test this hypothesis plasmids encoding wild type (WT) G6pc1 or a catalytically dead (D) variant were co-transfected with the Pklr-luciferase and G6pc1-luciferase fusion genes. WT and catalytically dead G6pc1 were expressed at similar levels (Fig 3B). In the catalytically dead variant AA 83 was changed from arginine to alanine, which abolishes G6P hydrolysis [49]. Fig 3C and 3D show that WT G6pc1 repressed glucose-stimulated Pklr-luciferase and G6pc1-luciferase fusion gene expression relative to the expression obtained in the presence of catalytically dead G6pc1.
Fig 3. Overexpression of G6pc1 Suppresses Glucose-Stimulated Fusion Gene Expression in 832/13 Cells.
Panel A: Schematic illustrating how the extent of glucose cycling catalyzed by glucokinase and G6pc1 determines intracellular G6P levels and hence activation of fusion gene expression through ChREBP and CREB. Panel B: Western blot showing that wild type (WT) and catalytically dead (D) G6pc1 are expressed at similar levels following expression in 832/13 cells. A representative blot is shown. Panel C: 832/13 cells were transiently co-transfected, as described in Materials and Methods, with the -206/+1 Pklr-luciferase or -7248/+62 G6pc1-luciferase fusion genes (2 μg), an expression vectors encoding Renilla luciferase (0.5 μg) and the indicated amounts of expression vectors encoding either wild type (WT) or catalytically dead (D) G6pc1. The total DNA added was kept constant using the empty pcDNA3 vector. Following transfection, cells were incubated for 18–20 hr in serum-free medium in the presence of 30 mM glucose. Cells were then harvested and luciferase activity assayed as described in Materials and Methods. Results were calculated as the ratio of firefly:Renilla luciferase activity and are presented as a percentage relative to that in 30 mM glucose-treated cells transfected with the empty pcDNA3 vector (2 μg). Results represent the mean ± S.E.M. of 3 experiments using independent preparations of both fusion gene constructs in which each experimental condition was assayed in triplicate. Panel D: 832/13 cells were transiently co-transfected, as described in Materials and Methods, with the -206/+1 Pklr-luciferase or -7248/+62 G6pc1-luciferase fusion genes (2 μg), an expression vectors encoding Renilla luciferase (0.5 μg) and 2 μg of expression vectors encoding either wild type (WT) or catalytically dead (D) G6pc1. Following transfection, cells were incubated for 18–20 hr in serum-free medium in the presence of the indicated glucose concentrations. Cells were then harvested and luciferase activity assayed as described in Materials and Methods. Results were calculated as the ratio of firefly:Renilla luciferase activity and are presented as a percentage relative to that in 30 mM glucose-treated cells in the presence of catalytically dead G6pc1. Results represent the mean ± S.E.M. of 3 experiments using independent preparations of both fusion gene constructs in which each experimental condition was assayed in triplicate.
Most importantly, Fig 3C and 3D demonstrate that the effect of WT G6pc1 on glucose-stimulated Pklr-luciferase and G6pc1-luciferase fusion gene expression was not equivalent with G6pc1 mediating a greater repression of the latter. For the purpose of studying the impact of SNPs on glucose-6-phosphatase enzyme activity, subsequent experiments therefore examined the repression of glucose-stimulated G6pc1-luciferase fusion gene expression by glucose-6-phosphatase.
Analysis of the Effect of Human G6PC2 SNPs on the Glucose-6-Phosphatase Activity of Mouse G6pc1
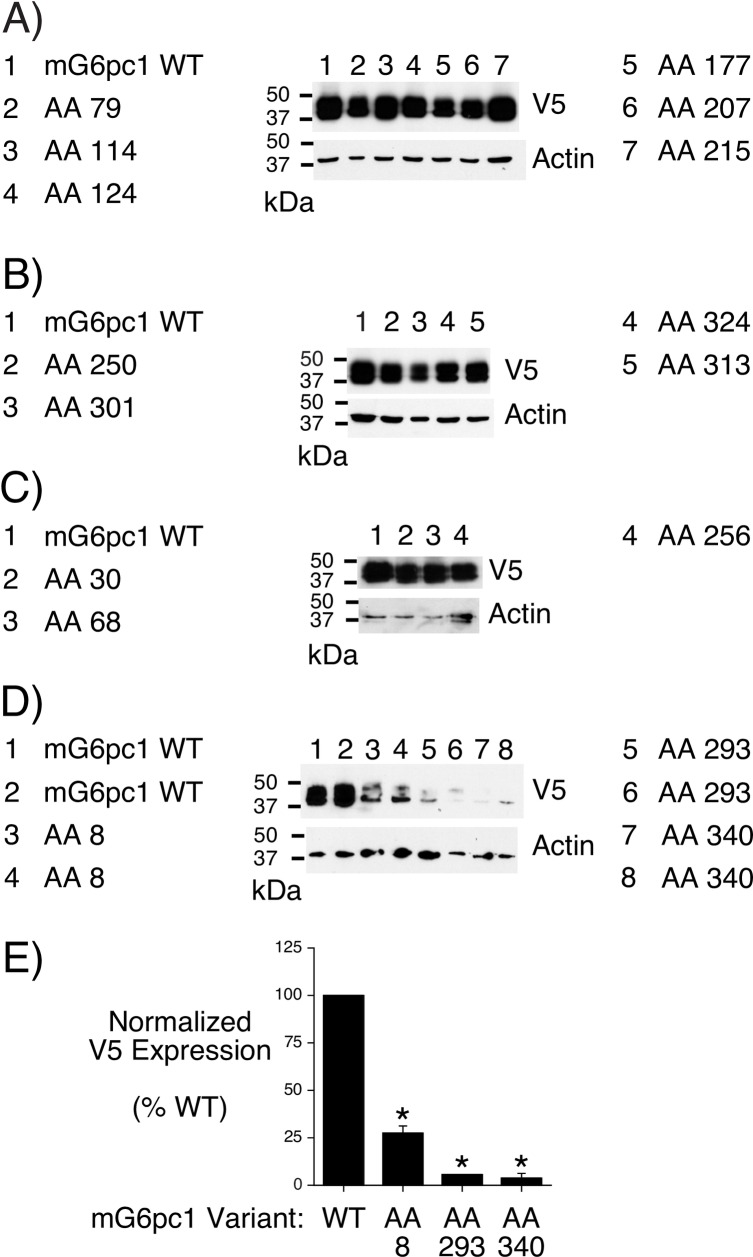
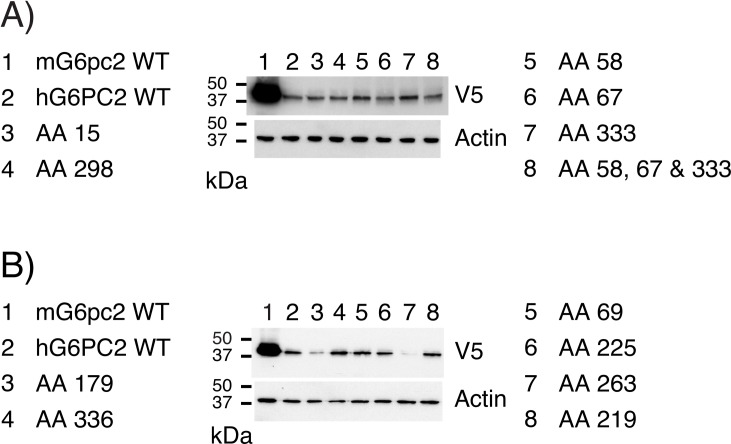
For these experiments we transfected 0.05 μg of plasmids encoding various G6pc1 variants, which confers a sub-maximal repression of glucose-stimulated G6pc1-luciferase fusion gene expression (Fig 3C). This approach allowed for the identification of both inhibitory and activating variants. Using this assay we determined that the AA changes associated with the rs144254880 (Arg79Gln), rs149663725 (Gly114Arg) and rs2232326 (Ser324Pro) SNPs markedly reduce G6pc1 enzyme activity (Table 1) without affecting protein expression (Fig 4A and 4B). For simplicity and comparison with the effect of these variants on human G6PC2 protein expression, these conserved AAs are numbered based on the position of the AA in human G6PC2 rather than their actual location in mouse G6pc1 (Fig 1). The AA changes associated with the rs142189264 (Ser30Phe), rs199682245 (Asn68Ile) and rs150538801 (Phe256Leu) SNPs also reduced G6pc1 enzyme activity (Table 1), though to a lesser degree, but again without affecting protein expression (Fig 4C). The AA changes associated with several other SNPs had statistically significant though minor effects on G6pc1 enzyme activity (Table 1), without affecting protein expression (S1 Fig). Interestingly, the AA changes associated with the rs368382511 (Gly8Glu), rs374055555 (Arg293Trp) and rs2232327 (Pro340Leu) SNPs markedly reduced G6pc1 protein expression (Fig 4D and 4E) but only the latter had a marked effect on enzyme activity (Table 1). This result suggests that these AA changes may actually be increasing the specific activity of G6pc1.
Fig 4. Analysis of the Effect of Amino Acid Changes on Mouse G6pc1 Protein Expression.
832/13 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type (WT) mouse (m) G6pc1 or G6pc1 variants in which the indicated amino acid (AA) had been changed as shown in Table 1. Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and protein expression assayed as described in Materials and Methods. In some instances these AA changes did not affect G6pc1 protein expression (Panels A-C). In other cases they resulted in reduced expression (Panel D); these data were quantitated by scanning with the results in Panel E showing the mean ± S.E.M. of 4 experiments. For simplicity and comparison with Fig 8, these AAs are numbered based on the position of the equivalent conserved AA in human G6PC2 (Fig 1).
Table 1 and Fig 1 show that there are six SNPs in G6PC2 that alter AAs that are conserved in G6PC1 and where mutation of these AAs in G6PC1 causes GSD type 1a [40]. However, these SNPs change the residue associated with GSD type 1a to an AA distinct from that that causes GSD type 1a. For example, rs372008743 changes a glutamine at residue 16 to a histidine whereas the mutation associated with GSD type 1a involves a change from a glutamine at residue 16 to an arginine (Table 1; Ref. [40]). For four of these 6 SNPs the AA change associated with the G6PC2 SNP had little effect on G6pc1 enzyme activity or protein expression (Table 1) suggesting that the change is silent. However, for two of these 6 SNPs the AA change associated with the G6PC2 SNP markedly affected G6pc1 enzyme activity (rs144254880; Arg79Gln) (Table 1) or expression (rs374055555; Arg293Trp) (Fig 4D and 4E).
Analysis of the Effect of Human G6PC2 Codon Variation on G6PC2 Protein Expression
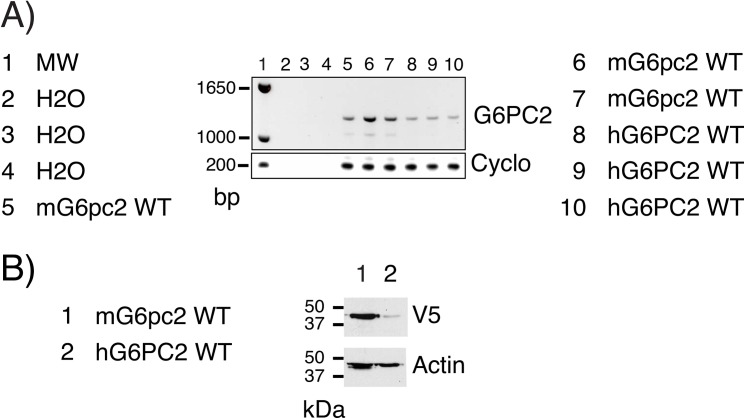
Before beginning the analysis of non-synonymous human G6PC2 SNPs on G6PC2 protein expression we sought to maximize human G6PC2 protein expression in transient transfection assays. Plasmids encoding V5 His-tagged variants of human G6PC2 and mouse G6pc2 [14,30,31] were transiently transfected into COS cells. Fig 5A shows that human G6PC2 and mouse G6pc2 RNA were expressed at similar levels but mouse G6pc2 protein expression was much higher than human G6PC2 (Fig 5B).
Fig 5. Analysis of Human G6PC2 and Mouse G6pc2 mRNA and Protein Expression.
COS 7 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type (WT) mouse (m) G6pc2, human (h) G6PC2 or the empty pcDNA3 vector. Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and either RNA (Panel A) or protein (Panel B) expression were assayed as described in Materials and Methods. A representative agarose gel (Panel A) or Western blot (Panel B) are shown. The faint band of the same size in the empty vector transfected cells represents background plasmid contamination of our PCR reagents/tubes.
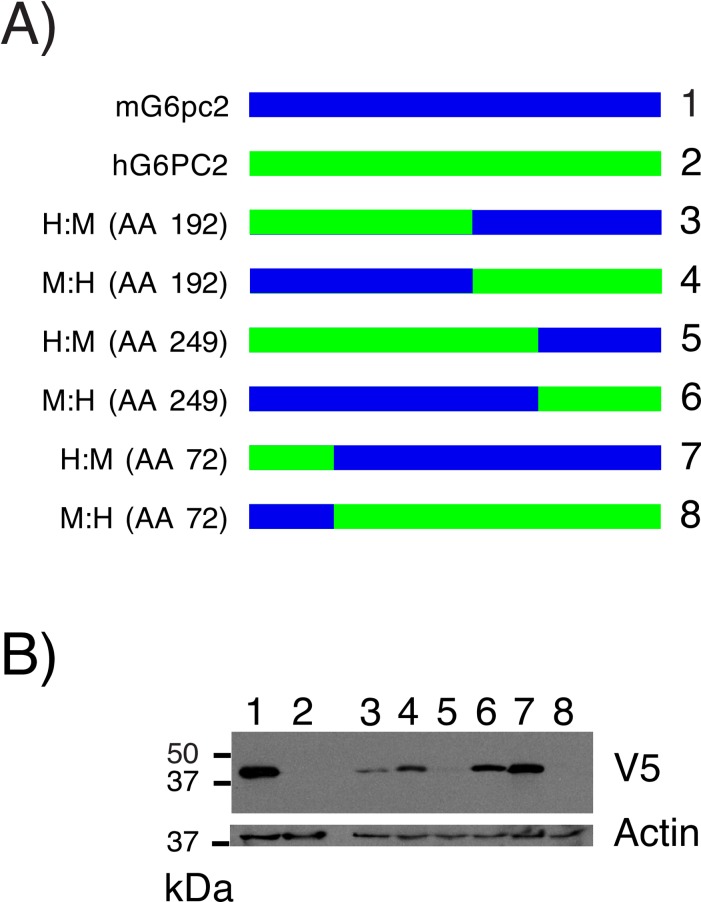
Chimeras of human G6PC2 and mouse G6pc2 were generated to investigate whether this difference in protein expression was associated with a particular region of the human G6PC2 or mouse G6pc2 proteins (Fig 6A). The results show that chimeric protein expression decreased as the proportion of human G6PC2 coding sequence increased (Fig 6B). The region of G6PC2 between AAs 72 and 192 appeared to have the greatest impact on the difference in expression between mouse G6pc2 and human G6PC2 (Fig 6B). In this region 110/121 AAs are conserved between mouse G6pc2 and human G6PC2. This suggested that multiple differences between human and mouse codons potentially explained the difference in protein expression. We therefore next investigated the effect of switching individual human G6PC2 codons that are rarely present in human mRNAs, with codons that code for the same amino acid (AA) but are more commonly found in human mRNAs. In some instances this resulted in a switch to the same codon used to encode the equivalent AA in mouse G6pc2 (Fig 7A; S2 Table). In other cases this resulted in a switch to a codon that was distinct from the codon used to encode the equivalent AA in mouse G6pc2 (Fig 7B; S3 Table). Switching the codons encoding three AAs, 58, 67 and 333, resulted in a slight improvement in human G6PC2 expression but the effect of combining these codon changes was not additive (Fig 7A). Changing two other codons, encoding AAs 179 and 263, further reduced human G6PC2 expression (Fig 7B). These data suggest that the molecular basis for the increased expression of mouse G6pc2 versus human G6PC2 is complex and involves differences in translation efficiency and/or stability that are conferred by multiple codons and/or AAs, respectively.
Fig 6. Analysis of Human G6PC2:Mouse G6pc2 Chimeric Protein Expression.
832/13 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type mouse (m) G6pc2, human (h) G6PC2 or the indicated chimeric proteins (Panel A). Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and protein expression assayed as described in Materials and Methods (Panel B). A representative blot is shown.
Fig 7. Analysis of the Effect of Human G6PC2 Codon Variation on Protein Expression.
832/13 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type (WT) mouse (m) G6pc2, human (h) G6PC2 or G6PC2 variants in which the codon used to encode the indicated AAs had been optimized as shown in S2 Table and S3 Table. Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and protein expression assayed as described in Materials and Methods. In some instances codon optimization resulted in a switch to the same codon used to encode the equivalent AA in mouse G6pc2 (Panel A; S2 Table). In other cases this resulted in a switch to a codon that was distinct from the codon used to encode the equivalent AA in mouse G6pc2 (Panel B; S3 Table). Representative blots are shown.
Analysis of the Effect of Human G6PC2 SNPs on G6PC2 Protein Expression
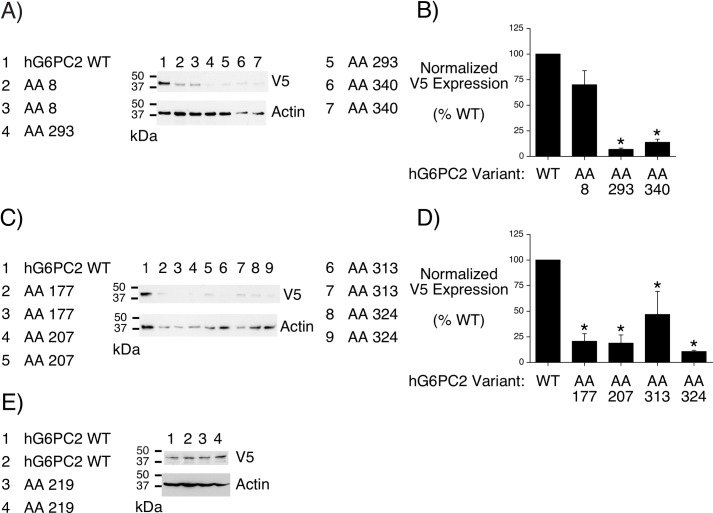
We next analyzed the effect of several human non-synonymous G6PC2 SNPs on human G6PC2 protein expression (Table 1). We began by analyzing the 3 SNPs that were associated with reduced mouse G6pc1 protein expression (Fig 4D and 4E), namely rs368382511 (Gly8Glu), rs374055555 (Arg293Trp) and rs2232327 (Pro340Leu). Fig 8A and 8B show that rs374055555 (Arg293Trp) and rs2232327 (Pro340Leu) also confer reduced expression of human G6PC2 in COS cells, with a trend towards reduced expression observed with rs368382511 (Gly8Glu). We next analyzed three SNPs, namely rs138726309 (His177Tyr), rs2232323 (Tyr207Ser) and rs492594 (Val219Leu) that Mahajan et al. [50] recently showed reduced human G6PC2 protein expression in 832/13 cells. Fig 8C and 8D show that rs138726309 (His177Tyr) and rs2232323 (Tyr207Ser) also confer reduced expression of human G6PC2 in COS cells. In contrast, the rs492594 (Val219Leu) variant had little effect on human G6PC2 expression in COS cells (Fig 8E). The AA changes associated with rs138726309 (His177Tyr) and rs2232323 (Tyr207Ser) did not affect mouse G6pc1 protein expression (Fig 4A). The rs492594 (Val219Leu) variant alters an AA that is not conserved in mouse G6pc2, human G6PC1 or mouse G6pc1 (S4 Table).
Fig 8. Analysis of the Effect of Human G6PC2 SNPs on Human G6PC2 Protein Expression.
COS 7 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type (WT) human (h) G6PC2 or G6PC2 variants in which the indicated amino acid (AA) had been changed as shown in Table 1. Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and protein expression assayed as described in Materials and Methods. The variants shown either reduced (Panels A-D) or had little effect (Panel E) on human G6PC2 protein expression. Data were quantitated by scanning with the results in Panels B and D showing the mean ± S.E.M. of 4 experiments. Representative blots are shown.
Finally, we analyzed two additional SNPs at the C terminus of human G6PC2, namely rs137857125 (Pro313Leu) and rs2232326 (Ser324Pro). Fig 8C and 8D show that these SNPs also confer reduced expression of human G6PC2 in COS cells. In contrast, the AA changes associated with these SNPs did not affect mouse G6pc1 protein expression (Fig 4B). Strikingly, these results suggest that despite the conservation of key AAs involved in enzyme activity between mouse G6pc1, mouse G6pc2, human G6PC1 and human G6PC2 [40,49,51] the mutation of conserved AAs has variable effects on human G6PC2 and mouse G6pc1 protein expression.
Discussion
This study focused on 22 non-synonymous SNPs in human G6PC2 that change AAs that are conserved between human G6PC2, mouse G6pc2, human G6PC1 and mouse G6pc1 (Table 1) (Fig 1), though database analyses identified multiple additional non-synonymous G6PC2 SNPs that affect AAs in G6PC2 that are not conserved across all four isoforms (S4 Table). We show that the AA changes associated with the rs144254880 (Arg79Gln), rs149663725 (Gly114Arg), rs2232326 (Ser324Pro), rs142189264 (Ser30Phe), rs199682245 (Asn68Ile) and rs150538801 (Phe256Leu) SNPs reduced G6pc1 enzyme activity in situ (Table 1) without affecting protein expression (Fig 4A–4C). We also show that the AA changes associated with the rs368382511 (Gly8Glu), rs374055555 (Arg293Trp) and rs2232327 (Pro340Leu) SNPs markedly reduced G6pc1 protein expression (Fig 4D and 4E). Finally, we show that the rs368382511 (Gly8Glu), rs374055555 (Arg293Trp), rs2232327 (Pro340Leu), rs138726309 (His177Tyr), rs2232323 (Tyr207Ser), rs137857125 (Pro313Leu) and rs2232326 (Ser324Pro) SNPs confer reduced expression of human G6PC2 (Fig 8A–8D).
Once the challenge of achieving high human G6PC2 expression is overcome (Figs 5–7), future studies will aim to determine whether the SNPs that affect mouse G6pc1 enzyme activity also affect human G6PC2 enzyme activity. This seems highly likely since site directed mutagenesis studies [49,51] and the analysis of mutations causing GSD type 1a [40] have shown AAs that are essential for high G6PC1 enzyme activity are conserved between mouse G6pc1, mouse G6pc2, human G6PC1 and human G6PC2. Indeed, of the 56 AAs in human G6PC1 mutation of which gives rise to GSD type 1a [40], 51 are conserved or represent conserved changes in human G6PC2 (S1 Table). It is striking that the G6PC2 SNPs rs138726309 (His177Tyr) and rs2232323 (Tyr207Ser), that have been linked to variations in FBG [50,52,53], affect AAs mutation of which in G6PC1 cause GSD type 1a (Table 1) [40]. Similarly, site directed mutagenesis studies [54] have shown a conservation of catalytically important AAs between G6PC1 and the G6PC3 isoform of glucose-6-phosphatase, initially referred to as UGRP, even though they share only a 36% overall AA conservation [30]. In humans mutations in G6PC3 cause Dursun syndrome [55,56]. Several mutations of residues that are identical between G6PC1 and G6PC3 have been associated with GSD type 1a and Dursun syndrome, respectively [55,56], supporting the notion that SNPs that alter conserved residues in all three G6PC1 isoforms will likely have similar effects on enzyme activity because catalytically important residues are conserved in all three isoforms.
Future studies will also use Vanderbilt University’s BioVU biobank to examine whether SNPs that affect human G6PC2 protein expression or activity are associated with altered phenotypic characteristics in humans, such as FBG and T2D risk. BioVU is a DNA biobank linked to a de-identified version of the Vanderbilt electronic health records, called the Synthetic Derivative (SD) [57,58]. The SD can be screened, using a procedure referred to as a PheWAS, to identify associations between specific SNPs and human diseases as well as associations with altered plasma hormone/metabolite levels [59–63]. Of particular interest will be the medical records of individuals with G6PC2 SNPs that result in frameshift mutations or premature termination (S5 Table).
Mahajan et al. [50] recently showed that the rs138726309 (His177Tyr) and rs2232323 (Tyr207Ser) SNPs result in reduced human G6PC2 protein expression in HEK293 and INS-1E cells, which we confirmed in COS cells (Fig 8C and 8D). They also showed that another SNP rs492594 (Val219Leu), that changes an AA that is not conserved in mouse G6pc2, human G6PC1 or mouse G6pc1 (Fig 1) (S1 Table), also results in reduced human G6PC2 protein expression in HEK293 cells though not in INS-1E cells [50]. We observed that this SNP also does not appear to affect human G6PC2 protein expression in COS cells (Fig 8E). This suggests that for this particular SNP, unknown cell line-dependent factors influence its action on G6PC2 expression. As with the initially described GWAS SNP, rs560887 [8,9], Mahajan et al. [50] showed that all three of these SNPs are associated with variations in fasting plasma glucose (FPG). Horikoshi et al. [53] have also shown that the rs138726309 (His177Tyr) is associated with variations in FPG. In addition, Wessel et al. [52] have shown that rs138726309 (His177Tyr) and rs2232323 (Tyr207Ser), as well as two additional non-synonymous SNPs, rs2232326 (Ser324Pro) and rs146779637 (Arg283STOP) are associated with variations in FPG. We showed that the rs2232326 (Ser324Pro) SNP results in altered G6PC2 protein expression in COS cells (Fig 8C & 8D). Interestingly, for reasons that are unclear, Mahajan et al. [50], in contrast to Wessel et al. [52], did not observe an association between rs146779637 (Arg283STOP) and FPG. Mahajan et al. [50] speculated that the lack of association with FPG was because this variant might retain activity despite the removal of the terminal 72 AAs of G6PC2. This variant clearly merits further study, especially since the data suggest a potential difference with human G6PC1, whose activity is susceptible to C terminal truncation [49].
Previous studies have suggested a complex relationship between G6PC2 and T2D risk with apparently conflicting results in different populations [4,7,64,65]. Interestingly, Mahajan et al. [50] showed that the rs492594 (Val219Leu) SNP is associated with altered risk for T2D. In contrast, in their study of non-synonymous G6PC2 SNPs, Wessel et al. [52] reported no association between G6PC2 and T2D risk. The rs492594 (Val219Leu) G6PC2 variant was not included in the studies of Wessel et al. [52] so potential reasons for this apparent discrepancy between G6PC2 variation and T2D risk remain unclear. These results may indicate that the rs492594 (Val219Leu) G6PC2 variant has a unique effect on beta cell function, unrelated to the control of glycolytic flux, especially since our results (Fig 8) and the results of Mahajan et al. [50] suggest that the rs492594 (Val219Leu) variant reduces G6PC2 protein expression less than the rs138726309 (His177Tyr) and rs2232323 (Tyr207Ser) variants that were included in the studies of Wessel et al. [52]. Indeed, we have previously speculated that G6PC2 may affect beta cell endoplasmic reticulum calcium retention, in addition to its action on glycolytic flux [11]. Indirect support for such a function for G6PC2 was recently suggested by the observation that deletion of the sorcin gene, which regulates endoplasmic reticulum calcium retention, resulted in elevated G6pc2 expression [66].
Our study also describes a novel assay for the measurement of glucose-6-phosphatase activity in situ (Figs 2 and 3). G6pc1 is unstable [67] and much remains unknown about the factors regulating G6pc1 activity [42] so this assay has the advantage that G6pc1 activity can be studied in an endogenous environment rather than in isolated and/or permeabilized microsomes. However, there are several caveats associated with this assay. Firstly, even though the amount of G6pc1 expressed was sufficient to achieve a sub-maximal repression of glucose-stimulated G6pc1-luciferase gene expression (Fig 3), this assay will not have the same linearity relative to an in vitro assay given the influence of other intracellular factors on G6pc1 activity. Secondly, in this assay apparent changes in G6pc1 activity could arise indirectly due to a change in sub-cellular distribution. Finally, because G6pc1 is located in the endoplasmic reticulum with its active site directed towards the lumen, the glucose-6-phosphatase activity of G6pc1 in situ is dependent on transport of its substrate G6P into the lumen by a G6P/Pi transporter, encoded by the SLC37A4 gene [11,17]. Pan et al. [68] have shown that G6PC1 and SLC37A4 are functionally coupled. Therefore, AA changes that affect this coupling will also appear to affect the inherent glucose-6-phosphatase activity of G6pc1 in the in situ assay. Future studies comparing the activity of specific G6pc1 variants in this in situ assay and the standard in vitro assay may lead to the identification of variants that affect aspects of G6pc1 function other than G6P hydrolysis.
In the course of developing our novel assay we made several interesting observations about glucose-regulated gene expression. Collier et al. [46] demonstrated that the effect of glucose on rat Pklr gene expression in 832/13 cells is mediated by the carbohydrate response element binding protein (ChREBP), which binds a carbohydrate response element (ChoRE) located between -188 and -172 in the rat Pklr promoter [69]. Consistent with this observation, deletion of the promoter region between -206 and -101 markedly reduced the effect of glucose on Pklr-luciferase fusion gene expression (Fig 2B). In contrast, Pederson et al. [45] demonstrated that the effect of glucose on rat G6pc1 expression in 832/13 cells is mediated by two promoter elements, a ChoRE located between -3616 and -3600 that binds ChREBP [45], and a cAMP response element (CRE) located between -163 and -156 that binds CRE binding protein (CREB) [70]. Pederson et al. [45] found that deletion of the G6PC1 ChoRE reduced the glucose response by ~80%. However, Fig 2B shows that deletion of the promoter region between -7248 and -1641 had little effect on glucose-stimulated G6pc1-luciferase fusion gene expression, suggesting that differences possibly related to cell passage number or growth conditions have altered the relative importance of ChREBP and CREB in glucose signaling to the G6pc1 promoter in our 832/13 cells. Interestingly, Pklr-luciferase fusion gene expression was more sensitive to glucose suggesting that the signaling pathways used by glucose to regulate ChREBP and CREB are distinct (Fig 2C). Consistent with this idea, the effect of WT G6pc1 on glucose-stimulated Pklr-luciferase and G6pc1-luciferase fusion gene expression was not equivalent with G6pc1 mediating a greater repression of the latter (Fig 3C and 3D). This result not only suggests that the glucose-signaling pathways to ChREBP and CREB are distinct but that G6pc1 preferentially influences the latter. Finally, we also observed that catalytically dead G6pc1 partially represses glucose-stimulated fusion gene expression (Fig 3C and 3D). This may reflect activation of the endoplasmic reticulum stress response [71] and could also explain why catalytically dead G6pc1 has a greater effect on G6pc1 versus Pklr fusion gene expression since the former promoter contains a stress response element [72]. Interestingly, over expression of G6PC2 in mice causes diabetes due to activation of the ER stress response [73].
Supporting Information
832/13 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type (WT) mouse (m) G6pc1 or G6pc1 variants in which the indicated amino acid (AA) had been changed as shown in Table 1. Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and protein expression assayed as described in Materials and Methods. With the exception of AA 8, these AA changes did not markedly affect G6pc1 protein expression. Representative blots are shown. For simplicity and comparison with Fig 8, these AAs are numbered based on the position of the equivalent AA in human G6PC2 (Fig 1).
(PDF)
The Table shows AAs in human G6PC1 whose mutation causes glycogen storage disease (GSD) type 1a [40] and whether these AAs are conserved or similar in mouse G6pc1, mouse G6pc2 and human G6PC2.
(PDF)
The Table shows that the codons used to encode the indicated AAs in human G6PC2 are not the most commonly used codons to encode these AAs in human proteins. The Table also shows that the codons that are commonly used to encode these AAs in human proteins are the same as the codons used to encode these AAs in mouse G6pc2. The effect on human G6PC2 protein expression of changing these codons to the most frequently used codon was assessed as described in Fig 7A. Codon usage in human mRNAs is described at the following website: http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=9606&aa=1&style=N
(PDF)
The Table shows that the codons used to encode the indicated AAs in human G6PC2 are not the most commonly used codons to encode these AAs in human proteins. The Table also shows that the codons that are commonly used to encode these AAs in human proteins are also different to the codons used to encode these AAs in mouse G6pc2. The effect on human G6PC2 protein expression of changing these codons to the most frequently used codon was assessed as described in Fig 7B. In this analysis we just focused on codons for AAs that are conserved between mouse G6pc2 and human G6PC2. In other words, we did not optimize codons that encode AAs that are unique to human G6PC2.
(PDF)
Human G6PC2 SNPs that change AAs that are not uniformly conserved in human G6PC2, mouse G6pc2, human G6PC1 and mouse G6pc1 were identified using the UCSC Genome Browser (https://genome.ucsc.edu/) and HumSAVR (http://omictools.com/humsavar-tool) databases. The G6PC2 domain affected by each AA change was predicted by comparison with the proposed structure of G6PC1 [41]. **, this residue has been associated with variations in FBG in healthy individuals who do not have diabetes [50].
(PDF)
Human G6PC2 SNPs that result in frameshift mutations or premature termination were identified using the UCSC Genome Browser (https://genome.ucsc.edu/) and HumSAVR (http://omictools.com/humsavar-tool) databases. The rs35259259 (Gly186Ala) SNP represents a deletion of a single base pair (G) at the G6PC2 exon five 5’ splice junction causing a shift in the open reading frame. The G6PC2 domain affected by each AA change was predicted by comparison with the proposed structure of G6PC1 [41].
(PDF)
Acknowledgments
We are indebted to John Hutton (1948–2012) for the numerous insights he contributed to this project. The following graduate students contributed to these studies: Courtney Copeland, Peter Kropp, Bethany Carboneau, Elijah Trefts and Brandon Bout. We thank Dr. Chris Newgard for providing the 832/13 cell line. Research in the laboratory of R.O’B. was supported by NIH grant DK092589 to R.O’B. and by NIH grant P60 DK20593, which supports the Vanderbilt Diabetes Research Training Center. K. E. S. and L. D. P. were supported by the Vanderbilt Molecular Endocrinology Training Program grant 5T32 DK07563. R.O’B. is the guarantor of this work, had full access to all the data, and takes full responsibility for the integrity of data and the accuracy of data analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Research in the laboratory of RO’B was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant DK092589 to RO’B and by NIH grant P60 DK20593, which supports the Vanderbilt Diabetes Research Training Center. KES and LDP were supported by the Vanderbilt Molecular Endocrinology Training Program grant 5T32 DK07563. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Abdul-Ghani MA, DeFronzo RA (2009) Plasma glucose concentration and prediction of future risk of type 2 diabetes. Diabetes Care 32 Suppl 2: S194–198. 10.2337/dc09-S309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coutinho M, Gerstein HC, Wang Y, Yusuf S (1999) The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes Care 22: 233–240. [DOI] [PubMed] [Google Scholar]
- 3.Lawes CM, Parag V, Bennett DA, Suh I, Lam TH, Whitlock G, et al. (2004) Blood glucose and risk of cardiovascular disease in the Asia Pacific region. Diabetes Care 27: 2836–2842. [DOI] [PubMed] [Google Scholar]
- 4.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116. 10.1038/ng.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. (2012) Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 44: 991–1005. 10.1038/ng.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. (2012) A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44: 659–669. 10.1038/ng.2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Liu L, Zhao J, Cui G, Chen C, Ding H, et al. (2013) Large Scale Meta-Analyses of Fasting Plasma Glucose Raising Variants in GCK, GCKR, MTNR1B and G6PC2 and Their Impacts on Type 2 Diabetes Mellitus Risk. PLoS One 8: e67665 10.1371/journal.pone.0067665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proenca C, et al. (2008) A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 320: 1085–1088. 10.1126/science.1156849 [DOI] [PubMed] [Google Scholar]
- 9.Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, Silver KD, et al. (2008) Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest 118: 2620–2628. 10.1172/JCI34566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiling E, van 't Riet E, Groenewoud MJ, Welschen LM, van Hove EC, Nijpels G, et al. (2009) Combined effects of single-nucleotide polymorphisms in GCK, GCKR, G6PC2 and MTNR1B on fasting plasma glucose and type 2 diabetes risk. Diabetologia 52: 1866–1870. 10.1007/s00125-009-1413-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien RM (2013) Moving on from GWAS: functional studies on the G6PC2 gene implicated in the regulation of fasting blood glucose. Curr Diab Rep 13: 768–777. 10.1007/s11892-013-0422-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baerenwald DA, Bonnefond A, Bouatia-Naji N, Flemming BP, Umunakwe OC, Oeser JK, et al. (2013) Multiple functional polymorphisms in the G6PC2 gene contribute to the association with higher fasting plasma glucose levels. Diabetologia 56: 1306–1316. 10.1007/s00125-013-2875-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouatia-Naji N, Bonnefond A, Baerenwald DA, Marchand M, Bugliani M, Marchetti P, et al. (2010) Genetic and Functional Assessment of the Role of the rs13431652-A and rs573225-A Alleles in the G6PC2 Promoter that Strongly Associate With Elevated Fasting Glucose Levels. Diabetes 59: 2662–2671. 10.2337/db10-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin CC, Bischof LJ, Bergman B, Hornbuckle LA, Hilliker C, Frigeri C, et al. (2001) Cloning and Characterization of the Human and Rat Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein (IGRP) Genes. J Biol Chem 276: 25197–25207. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Martin CC, Oeser JK, Sarkar S, McGuinness OP, Hutton JC, et al. (2007) Deletion of the Gene Encoding the Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein Autoantigen Results in a Mild Metabolic Phenotype. Diabetologia 50: 774–778. [DOI] [PubMed] [Google Scholar]
- 16.Pound LD, Oeser JK, O'Brien TP, Wang Y, Faulman CJ, Dadi PK, et al. (2013) G6PC2: A Negative Regulator of Basal Glucose-Stimulated Insulin Secretion. Diabetes 62: 1547–1556. 10.2337/db12-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutton JC, O'Brien RM (2009) The glucose-6-phosphatase catalytic subunit gene family. J Biol Chem 284: 29241–29245. 10.1074/jbc.R109.025544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wall ML, Pound LD, Trenary I, O'Brien RM, Young JD (2015) Novel Stable Isotope Analyses Demonstrate Significant Rates of Glucose Cycling In Mouse Pancreatic Islets. Diabetes 64 2129–2137. 10.2337/db14-0745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matschinsky FM (2005) Glucokinase, glucose homeostasis, and diabetes mellitus. Curr Diab Rep 5: 171–176. [DOI] [PubMed] [Google Scholar]
- 20.Iynedjian PB (2009) Molecular physiology of mammalian glucokinase. Cell Mol Life Sci 66: 27–42. 10.1007/s00018-008-8322-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boortz KA, Syring KE, Dai C, Pound LD, Oeser JK, Jacobson DA, et al. (2016) G6PC2 Modulates Fasting Blood Glucose In Male Mice in Response to Stress. Endocrinology 157: 3002–8. 10.1210/en.2016-1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visscher PM, Brown MA, McCarthy MI, Yang J (2012) Five years of GWAS discovery. Am J Hum Genet 90: 7–24. 10.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. (2009) Finding the missing heritability of complex diseases. Nature 461: 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llewellyn CH, Trzaskowski M, Plomin R, Wardle J (2013) Finding the missing heritability in pediatric obesity: the contribution of genome-wide complex trait analysis. Int J Obes (Lond) 37: 1506–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46: 1173–1186. 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, et al. (2015) Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47: 1114–1120. 10.1038/ng.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell GI, Pilkis SJ, Weber IT, Polonsky KS (1996) Glucokinase mutations, insulin secretion, and diabetes mellitus. Annu Rev Physiol 58: 171–186. [DOI] [PubMed] [Google Scholar]
- 28.Osbak KK, Colclough K, Saint-Martin C, Beer NL, Bellanne-Chantelot C, Ellard S, et al. (2009) Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat 30: 1512–1526. 10.1002/humu.21110 [DOI] [PubMed] [Google Scholar]
- 29.Grupe A, Hultgren B, Ryan A, Ma YH, Bauer M, Stewart TA (1995) Transgenic knockouts reveal a critical requirement for pancreatic beta cell glucokinase in maintaining glucose homeostasis. Cell 83: 69–78. [DOI] [PubMed] [Google Scholar]
- 30.Martin CC, Oeser JK, Svitek CA, Hunter SI, Hutton JC, O'Brien RM (2002) Identification and Characterization of a Human cDNA and Gene Encoding a Ubiquitously Expressed Glucose-6-Phosphatase Catalytic Subunit-Related Protein. J Mol Endocrinol 29: 205–222. [DOI] [PubMed] [Google Scholar]
- 31.Boustead JN, Martin CC, Oeser JK, Svitek CA, Hunter SI, Hutton JC, et al. (2004) Identification and characterization of a cDNA and the gene encoding the mouse ubiquitously expressed glucose-6-phosphatase catalytic subunit-related protein. J Mol Endocrinol 32: 33–53. [DOI] [PubMed] [Google Scholar]
- 32.Streeper RS, Eaton EM, Ebert DH, Chapman SC, Svitek CA, O'Brien RM (1998) Hepatocyte nuclear factor-1 acts as an accessory factor to enhance the inhibitory action of insulin on mouse glucose-6-phosphatase gene transcription. Proc Natl Acad Sci U S A 95: 9208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornbuckle LA, Edgerton DS, Ayala JE, Svitek CA, Oeser JK, Neal DW, et al. (2001) Selective, Tonic Inhibition of G6Pase Catalytic Subunit but not G6P Transporter Gene Expression by Insulin in Vivo. Am J Physiol 281: E713–725. [DOI] [PubMed] [Google Scholar]
- 34.Jacoby DB, Zilz ND, Towle HC (1989) Sequences within the 5'-flanking region of the S14 gene confer responsiveness to glucose in primary hepatocytes. J Biol Chem 264: 17623–17626. [PubMed] [Google Scholar]
- 35.Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T (1987) The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem 262: 14366–14371. [PubMed] [Google Scholar]
- 36.Bischof LJ, Martin CC, Svitek CA, Stadelmaier BT, Hornbuckle LA, Goldman JK, et al. (2001) Characterization of the Mouse Islet-Specific Glucose-6-Phosphatase Catalytic Subunit-Related Protein Gene Promoter by In Situ Footprinting. Correlation with Fusion Gene Expression in the Islet Derived bTC-3 and Hamster Insulinoma Tumor Cell Lines. Diabetes 50: 502–514. [DOI] [PubMed] [Google Scholar]
- 37.Lei KJ, Chen H, Pan CJ, Ward JM, Mosinger B Jr., Lee EJ, et al. (1996) Glucose-6-phosphatase dependent substrate transport in the glycogen storage disease type-1a mouse. Nat Genet 13: 203–209. [DOI] [PubMed] [Google Scholar]
- 38.Arden SD, Zahn T, Steegers S, Webb S, Bergman B, O'Brien RM, et al. (1999) Molecular cloning of a pancreatic islet-specific glucose-6-phosphatase catalytic subunit-related protein. Diabetes 48: 531–542. [DOI] [PubMed] [Google Scholar]
- 39.Petrolonis AJ, Yang Q, Tummino PJ, Fish SM, Prack AE, Jain S, et al. (2004) Enzymatic characterization of the pancreatic islet-specific glucose-6-phosphatase-related protein (IGRP). J Biol Chem 279: 13976–13983. [DOI] [PubMed] [Google Scholar]
- 40.Chou JY, Mansfield BC (2008) Mutations in the glucose-6-phosphatase-alpha (G6PC) gene that cause type Ia glycogen storage disease. Hum Mutat 29: 921–930. 10.1002/humu.20772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan CJ, Lei KJ, Annabi B, Hemrika W, Chou JY (1998) Transmembrane topology of glucose-6-phosphatase. J Biol Chem 273: 6144–6148. [DOI] [PubMed] [Google Scholar]
- 42.Mithieux G (1997) New knowledge regarding glucose-6 phosphatase gene and protein and their roles in the regulation of glucose metabolism. Eur J Endocrinol 136: 137–145. [DOI] [PubMed] [Google Scholar]
- 43.Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB (2000) Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49: 424–430. [DOI] [PubMed] [Google Scholar]
- 44.Newgard CB, Clark S, BeltrandelRio H, Hohmeier HE, Quaade C, Normington K (1997) Engineered cell lines for insulin replacement in diabetes: current status and future prospects. Diabetologia 40 Suppl 2: S42–47. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen KB, Zhang P, Doumen C, Charbonnet M, Lu D, Newgard CB, et al. (2007) The promoter for the gene encoding the catalytic subunit of rat glucose-6-phosphatase contains two distinct glucose-responsive regions. Am J Physiol Endocrinol Metab 292: E788–801. [DOI] [PubMed] [Google Scholar]
- 46.Collier JJ, Zhang P, Pedersen KB, Burke SJ, Haycock JW, Scott DK (2007) c-Myc and ChREBP regulate glucose-mediated expression of the L-type pyruvate kinase gene in INS-1-derived 832/13 cells. Am J Physiol Endocrinol Metab 293: E48–56. [DOI] [PubMed] [Google Scholar]
- 47.Trinh K, Minassian C, Lange AJ, O'Doherty RM, Newgard CB (1997) Adenovirus-mediated expression of the catalytic subunit of glucose-6- phosphatase in INS-1 cells. Effects on glucose cycling, glucose usage, and insulin secretion. J Biol Chem 272: 24837–24842. [DOI] [PubMed] [Google Scholar]
- 48.Bain JR, Schisler JC, Takeuchi K, Newgard CB, Becker TC (2004) An adenovirus vector for efficient RNA interference-mediated suppression of target genes in insulinoma cells and pancreatic islets of langerhans. Diabetes 53: 2190–2194. [DOI] [PubMed] [Google Scholar]
- 49.Lei KJ, Pan CJ, Liu JL, Shelly LL, Chou JY (1995) Structure-function analysis of human glucose-6-phosphatase, the enzyme deficient in glycogen storage disease type 1a. J Biol Chem 270: 11882–11886. [DOI] [PubMed] [Google Scholar]
- 50.Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, Highland HM, et al. (2015) Identification and Functional Characterization of G6PC2 Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the G6PC2-ABCB11 Locus. PLoS Genet 11: e1004876 10.1371/journal.pgen.1004876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh A, Shieh JJ, Pan CJ, Sun MS, Chou JY (2002) The catalytic center of glucose-6-phosphatase. HIS176 is the nucleophile forming the phosphohistidine-enzyme intermediate during catalysis. J Biol Chem 277: 32837–32842. [DOI] [PubMed] [Google Scholar]
- 52.Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, et al. (2015) Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Commun 6: 5897 10.1038/ncomms6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horikoshi M, Mgi R, van de Bunt M, Surakka I, Sarin AP, Mahajan A, et al. (2015) Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation. PLoS Genet 11: e1005230 10.1371/journal.pgen.1005230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh A, Shieh JJ, Pan CJ, Chou JY (2004) Histidine 167 is the phosphate acceptor in glucose-6-phosphatase-beta forming a phosphohistidine enzyme intermediate during catalysis. J Biol Chem 279: 12479–12483. [DOI] [PubMed] [Google Scholar]
- 55.Banka S, Newman WG (2013) A clinical and molecular review of ubiquitous glucose-6-phosphatase deficiency caused by G6PC3 mutations. Orphanet J Rare Dis 8: 84 10.1186/1750-1172-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin SR, Pan CJ, Mansfield BC, Chou JY (2015) Functional analysis of mutations in a severe congenital neutropenia syndrome caused by glucose-6-phosphatase-beta deficiency. Mol Genet Metab 114: 41–45. 10.1016/j.ymgme.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR (2010) Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci 3: 42–48. 10.1111/j.1752-8062.2010.00175.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, et al. (2008) Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 84: 362–369. 10.1038/clpt.2008.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. (2010) PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26: 1205–1210. 10.1093/bioinformatics/btq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. (2013) Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 31: 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. (2014) A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet 133: 95–109. 10.1007/s00439-013-1355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, et al. (2013) Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 127: 1377–1385. 10.1161/CIRCULATIONAHA.112.000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, et al. (2011) Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 89: 529–542. 10.1016/j.ajhg.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu C, Zhang R, Wang C, Ma X, Wang C, Fang Q, et al. (2009) A genetic variant of G6PC2 is associated with type 2 diabetes and fasting plasma glucose level in the Chinese population. Diabetologia 52: 451–456. 10.1007/s00125-008-1241-3 [DOI] [PubMed] [Google Scholar]
- 65.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44: 981–990. 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marmugi A, Parnis J, Chen X, Carmichael L, Hardy J, Mannan N, et al. (2016) Sorcin Links Pancreatic beta-Cell Lipotoxicity to ER Ca2+ Stores. Diabetes 65: 1009–1021. 10.2337/db15-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Speth M, Schulze HU (1986) Is thermostability of glucose-6-phosphatase indeed dependent on a stabilizing protein? FEBS Lett 202: 32–36. [DOI] [PubMed] [Google Scholar]
- 68.Pan CJ, Chen SY, Jun HS, Lin SR, Mansfield BC, Chou JY (2011) SLC37A1 and SLC37A2 are phosphate-linked, glucose-6-phosphate antiporters. PLoS One 6: e23157 10.1371/journal.pone.0023157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Towle HC (2005) Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab 16: 489–494. [DOI] [PubMed] [Google Scholar]
- 70.Hornbuckle LA, Everett CA, Martin CC, Gustavson SS, Svitek CA, Oeser JK, et al. (2004) Selective stimulation of G-6-Pase catalytic subunit but not G-6-P transporter gene expression by glucagon in vivo and cAMP in situ. Am J Physiol Endocrinol Metab 286: E795–808. [DOI] [PubMed] [Google Scholar]
- 71.Marciniak SJ, Ron D (2006) Endoplasmic reticulum stress signaling in disease. Physiol Rev 86: 1133–1149. [DOI] [PubMed] [Google Scholar]
- 72.Wang D, Wei Y, Schmoll D, Maclean KN, Pagliassotti MJ (2006) Endoplasmic reticulum stress increases glucose-6-phosphatase and glucose cycling in liver cells. Endocrinology 147: 350–358. [DOI] [PubMed] [Google Scholar]
- 73.Shameli A, Yamanouchi J, Thiessen S, Santamaria P (2007) Endoplasmic reticulum stress caused by overexpression of islet-specific glucose-6-phosphatase catalytic subunit-related protein in pancreatic Beta-cells. Rev Diabet Stud 4: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
832/13 cells were transiently transfected, as described in Materials and Methods, with expression vectors encoding either wild type (WT) mouse (m) G6pc1 or G6pc1 variants in which the indicated amino acid (AA) had been changed as shown in Table 1. Following transfection, cells were incubated for 18–20 hr in serum-containing medium. Cells were then harvested and protein expression assayed as described in Materials and Methods. With the exception of AA 8, these AA changes did not markedly affect G6pc1 protein expression. Representative blots are shown. For simplicity and comparison with Fig 8, these AAs are numbered based on the position of the equivalent AA in human G6PC2 (Fig 1).
(PDF)
The Table shows AAs in human G6PC1 whose mutation causes glycogen storage disease (GSD) type 1a [40] and whether these AAs are conserved or similar in mouse G6pc1, mouse G6pc2 and human G6PC2.
(PDF)
The Table shows that the codons used to encode the indicated AAs in human G6PC2 are not the most commonly used codons to encode these AAs in human proteins. The Table also shows that the codons that are commonly used to encode these AAs in human proteins are the same as the codons used to encode these AAs in mouse G6pc2. The effect on human G6PC2 protein expression of changing these codons to the most frequently used codon was assessed as described in Fig 7A. Codon usage in human mRNAs is described at the following website: http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=9606&aa=1&style=N
(PDF)
The Table shows that the codons used to encode the indicated AAs in human G6PC2 are not the most commonly used codons to encode these AAs in human proteins. The Table also shows that the codons that are commonly used to encode these AAs in human proteins are also different to the codons used to encode these AAs in mouse G6pc2. The effect on human G6PC2 protein expression of changing these codons to the most frequently used codon was assessed as described in Fig 7B. In this analysis we just focused on codons for AAs that are conserved between mouse G6pc2 and human G6PC2. In other words, we did not optimize codons that encode AAs that are unique to human G6PC2.
(PDF)
Human G6PC2 SNPs that change AAs that are not uniformly conserved in human G6PC2, mouse G6pc2, human G6PC1 and mouse G6pc1 were identified using the UCSC Genome Browser (https://genome.ucsc.edu/) and HumSAVR (http://omictools.com/humsavar-tool) databases. The G6PC2 domain affected by each AA change was predicted by comparison with the proposed structure of G6PC1 [41]. **, this residue has been associated with variations in FBG in healthy individuals who do not have diabetes [50].
(PDF)
Human G6PC2 SNPs that result in frameshift mutations or premature termination were identified using the UCSC Genome Browser (https://genome.ucsc.edu/) and HumSAVR (http://omictools.com/humsavar-tool) databases. The rs35259259 (Gly186Ala) SNP represents a deletion of a single base pair (G) at the G6PC2 exon five 5’ splice junction causing a shift in the open reading frame. The G6PC2 domain affected by each AA change was predicted by comparison with the proposed structure of G6PC1 [41].
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.