Abstract
Replicative DNA helicases expose the two strands of the double helix to the replication apparatus, but accessory helicases are often needed to help forks move past naturally occurring hard-to-replicate sites, such as tightly bound proteins, RNA/DNA hybrids, and DNA secondary structures. Although the Schizosaccharomyces pombe 5’-to-3’ DNA helicase Pfh1 is known to promote fork progression, its genomic targets, dynamics, and mechanisms of action are largely unknown. Here we address these questions by integrating genome-wide identification of Pfh1 binding sites, comprehensive analysis of the effects of Pfh1 depletion on replication and DNA damage, and proteomic analysis of Pfh1 interaction partners by immunoaffinity purification mass spectrometry. Of the 621 high confidence Pfh1-binding sites in wild type cells, about 40% were sites of fork slowing (as marked by high DNA polymerase occupancy) and/or DNA damage (as marked by high levels of phosphorylated H2A). The replication and integrity of tRNA and 5S rRNA genes, highly transcribed RNA polymerase II genes, and nucleosome depleted regions were particularly Pfh1-dependent. The association of Pfh1 with genomic integrity at highly transcribed genes was S phase dependent, and thus unlikely to be an artifact of high transcription rates. Although Pfh1 affected replication and suppressed DNA damage at discrete sites throughout the genome, Pfh1 and the replicative DNA polymerase bound to similar extents to both Pfh1-dependent and independent sites, suggesting that Pfh1 is proximal to the replication machinery during S phase. Consistent with this interpretation, Pfh1 co-purified with many key replisome components, including the hexameric MCM helicase, replicative DNA polymerases, RPA, and the processivity clamp PCNA in an S phase dependent manner. Thus, we conclude that Pfh1 is an accessory DNA helicase that interacts with the replisome and promotes replication and suppresses DNA damage at hard-to-replicate sites. These data provide insight into mechanisms by which this evolutionarily conserved helicase helps preserve genome integrity.
Author Summary
Progression of the DNA replication machinery is challenged in every S phase by active transcription, tightly bound protein complexes, and formation of stable DNA secondary structures. Using genome-wide analyses, we show that the evolutionarily conserved fission yeast Pfh1 DNA helicase promotes fork progression and suppresses DNA damage at natural sites of fork pausing, which occur at “hard-to-replicate” sites. Our data suggest that Pfh1 interacts with the replication apparatus. First, mass spectrometry revealed that Pfh1 interacts with many components of the replication machinery. Second, Pfh1 and the leading strand DNA polymerase occupy many common regions genome-wide, not only hard-to-replicate sites, but also sites whose replication is not Pfh1-dependent. The human genome encodes a Pfh1 homolog, hPIF1, and contains all of the same hard-to-replicate features that make fission yeast DNA replication dependent upon Pfh1. Thus, human cells likely also require replicative accessory DNA helicases to facilitate replication at hard-to-replicate sites, and hPIF1 is a good candidate for this role.
Introduction
Faithful and efficient replication of the genome is essential in every cell cycle, yet there are many naturally occurring obstacles that impede fork progression. These sites include stable protein complexes, DNA secondary structures, and ongoing transcription, each of which can challenge replication fork progression. Failure to circumvent these obstacles can cause DNA double strand breaks (DSBs) that impair genome integrity and increase the risk of cancer and other disorders that are associated with genome instability. As many of the proteins involved in DNA replication are highly conserved, model organisms such as S. pombe provide genetically tractable systems to identify and characterize genes with important roles in genome preservation whose human orthologs might have similar functions.
DNA replication is accomplished by the multi-subunit replisome, a complex that is assembled at and moves bi-directionally away from replication origins. Replicative helicases, such as the Escherichia coli DnaB and the eukaryotic hexameric MCM complex, are required to unwind the double helix to allow DNA polymerases access to the replication template. In addition, accessory helicases, such as the E. coli rep, dinG and UvrD proteins help the polymerase maneuver past protein complexes, RNA transcripts, and other naturally occurring impediments [1–5]. In Bacillus subtilis the essential accessory DNA helicase PcrA promotes fork movement through transcribed genes [6, 7]. E. coli rep physically interacts with the replicative DnaB helicase to bypass protein complexes on DNA [8].
The best-studied eukaryotic accessory DNA helicases are the two budding yeast enzymes, ScRrm3 and ScPif1, which are both members of the Pif1 family [9]. Although these two helicases have largely non-overlapping functions, they both promote progression past naturally occurring hard-to-replicate sites. ScRrm3 acts at over 1000 sites, including RNA polymerase III transcribed genes, the replication fork barrier (RFB) within ribosomal DNA (rDNA), inactive replication origins, silencers, telomeres, centromeres, and converged replication forks [10–14]. The diverse ScRrm3-sensitive sites have the common feature of being assembled into stable protein complexes. Removal of these proteins in trans or mutation of their binding sites in cis relieves the requirement for ScRrm3 at the affected site. In its absence, forks tend to stall and break at ScRrm3-sensitive sites. Rather than being recruited to its sites of action, ScRrm3 moves with the replisome through both ScRrm3-sensitive and insensitive sites [15] and interacts with leading strand DNA polymerase ε and PCNA [15, 16], suggesting that it is a replisome component.
ScPif1 also promotes fork progression, but so far this activity has been observed only at putative G-quadruplex (G4) structures [17–21]. G4 DNA is a stable, four-stranded secondary structure held together by non-canonical G-G base pairs [reviewed in 22]. In cells lacking ScPif1, DNA replication slows and DNA damage is detected at many G4 motifs. Current evidence suggests that ScPif1 is recruited to G4 motifs after their replication [18], and the abundance of ScPif1, unlike that of ScRrm3, is cell cycle regulated, peaking in late S phase [15, 23]. Thus, unlike ScRrm3, ScPif1 probably does not move with the leading strand DNA polymerase. ScRrm3 is a backup helicase for ScPif1 at G4 motifs [17]. As ScRrm3 and ScPif1 are both associated with stalled replication forks [24], ScPif1 might be a backup for ScRrm3 at some of its targets, such as RNA polymerase III transcribed genes. ScPif1 actions are not limited to its being an accessory DNA helicase, as it also inhibits telomerase by displacing it from DNA ends [25, 26], promotes formation of long flap Okazaki fragments [27, 28], and is needed for the stable maintenance of mitochondrial DNA [29] and break-induced replication [30–33].
Unlike budding yeast, most eukaryotes, including fission yeast and humans, encode a single Pif1 family helicase. While neither ScRrm3 nor ScPif1 is essential, the fission yeast Pfh1 DNA helicase is required for maintenance of both the mitochondrial and nuclear genomes [34]. Pfh1 also affects nuclear DNA repair: it localizes to DNA damage foci upon exogenous DNA damage, and its absence results in spontaneous DNA damage foci [34].
In earlier work, we and others used two-dimensional gels to show that Pfh1, like ScRrm3 promotes fork progression through specific stable protein complexes, including RNA polymerase II and III transcribed genes, silencers, converged replication forks [35, 36], and telomeres [37]. In addition, microscopic studies show that Pfh1 suppresses formation of ultrafine anaphase bridges that arise at incompletely replicated regions, such as Lac repressor bound LacO arrays [38], supporting the idea that it is needed to complete DNA replication. Like ScPif1, Pfh1 binds to and suppresses DNA damage at G4 motifs [39]. Also, ScPif1 and Pfh1 both unwind G4 structures in vitro [17, 40–43].
In this paper, we address two fundamental questions about Pfh1 function: where does Pfh1 act along the genome and how is it recruited to its sites of action? Earlier studies focused on Pfh1’s role in replication of a few examples of hard-to-replicate sites [35, 37, 39]. Here we used chromatin immunoprecipitation combined with genome-wide deep sequencing (ChIP-seq) to study the full range of Pfh1-sensitive sites. This analysis revealed hundreds of sites of Pfh1 binding where replication slows and DNA damage occurs, especially in the absence of Pfh1. These Pfh1-sensitive sites included all previously identified hard-to-replicate sites as well as novel sites, such as nucleosome depleted regions (NDR). Second, we assayed binding and fork progression to determine if Pfh1 is nearby the replisome during S phase or, if it is recruited to its sites of action. These analyses revealed that Pfh1 and the leading strand DNA polymerase bind both Pfh1-sensitive and Pfh1-insensitive sites to a similar extent. Likewise, mass spectrometry (MS) found that Pfh1 interacts with many key replisome components. Together these data argue that Pfh1 is not just recruited to its sites of action, but that it is in proximity to the replisome during DNA synthesis and functions as an accessory DNA helicase at all known classes of hard-to-replicate sites. These results inform our understanding of Pif1 helicases in higher eukaryotes, such as humans, which like S. pombe, encode only one Pif1 family helicase. Given that the helicase domains of Pfh1 and hPIF1 are related (36% sequence identity) [25], hPIF1 may have similar functions in preserving genome integrity.
Results
Many sites in the S. pombe genome display high Pfh1 occupancy
We analyzed 621 previously identified Pfh1 binding sites across the S. pombe genome from ChIP-seq on cycling WT cells (S1 Table) [39]. Given the low coverage of rDNA repeats and telomeres in the S. pombe annotated genome, the rest of this paper considers only non-telomeric, non-rDNA Pfh1 binding sites.
We determined if the peaks of Pfh1 binding correlated with any of sixteen annotated genomic features (Table 1; Methods). Because Pfh1 bound preferentially to GC-rich sites, we determined the significance of its association with features after accounting for GC content using random GC-matched controls (Methods). Pfh1 peaks were significantly associated with many known hard-to-replicate sites, such as tRNA genes, 5S ribosomal RNA (rRNA), and highly active RNA polymerase II transcribed genes (Table 1). For example, Pfh1 binding occurred ≤ 300 base pairs (bp), the shearing size of the ChIP DNA, from 80 out of 171 (47%) tRNA genes, 18 of 33 (55%) 5S rRNA genes, and 302 of the top 500 (60%) most highly transcribed RNA polymerase II genes (as defined in Rhind, Chen (44)). In addition, Pfh1 binding sites were significantly associated with meiotic double strand break (DSB) hotspots, nucleosome depleted regions (NDRs), 3’ untranslated regions (UTRs), and mating type loci (Table 1). In all following association analyses, associations with p-values less than the Bonferroni multiple testing adjusted threshold of 0.003 will be interpreted as significant.
Table 1. Genomic features associated with Pfh1 binding sites.
Association p-values were computed with an empirical permutation-based procedure that accounts for length and GC content (Methods). Associations significant after Bonferroni multiple testing correction (p < 0.003) are in bold. All p-values less than 10−50 are reported as ≈0.
| Genome Features | Number of Features | Fraction with Pfh1 | p-value |
|---|---|---|---|
| 500 Highest Expressed RNA polymerase II Genes | 500 | 0.60 (302) | ≈ 0 |
| 5S rRNA genes | 33 | 0.55 (18) | ≈ 0 |
| tRNA genes | 171 | 0.47 (80) | ≈ 0 |
| Meiotic Double Strand Break Hotspots | 288 | 0.37 (106) | ≈ 0 |
| 3’ UTRs | 5144 | 0.18 (939) | ≈ 0 |
| Nucleosome Depleted Regions | 2300 | 0.18 (406) | ≈ 0 |
| Mating Type Loci | 5 | 0.40 (2) | 0.002 |
| G4 motifs* | 446 | 0.20 (90) | 0.002 |
| RNA Pol II Transcribed Genes | 5144 | 0.23 (1188) | 0.005 |
| Origins of Replication | 741 | 0.23 (170) | 0.014 |
| Promoters | 3237 | 0.16 (504) | 0.042 |
| Protein Coding Sequence | 10244 | 0.14 (1459) | 0.093 |
| Dubious Genes | 71 | 0.21 (15) | 0.112 |
| 5’ UTRs | 5144 | 0.13 (693) | 0.139 |
| Long Terminal Repeat | 236 | 0.07 (16) | 0.982 |
| Centromeres | 3 | 0.33 (1) | 0.999 |
* from Sabouri, Capra [39].
Pfh1 binding to highly transcribed genes is not an artifact of their high transcription
A recent study reported that highly transcribed S. cerevisiae genes are over-represented in ChIP experiments carried out with diverse nuclear proteins, suggesting that their presence might be a technical artifact caused by their high transcription rate [45]. The specific cause of the “hyper-ChIPability” of these regions has not been resolved. It has been proposed that, because highly transcribed genes are more accessible during the pull-down, they may be more likely to interact with beads or antibodies during the IP, and therefore be subject to nonspecific precipitation by the antibody.
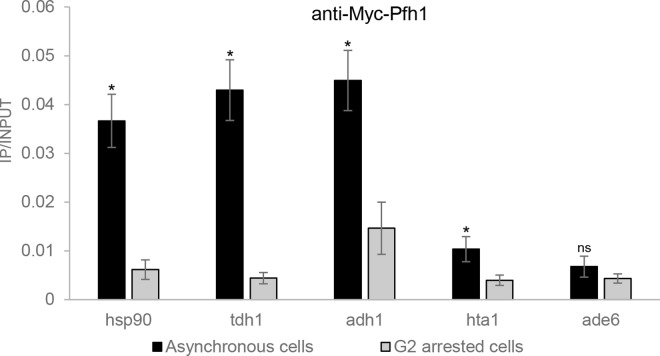
To ensure that the association with highly transcribed RNA polymerase II genes was not due to this artifact, we used ChIP combined with quantitative PCR (qPCR) to examine Pfh1 association to four highly transcribed genes, hsp90+, tdh1+, adh1+, and hta1+, which are among the top 500 most highly transcribed genes and were all Pfh1-associated sites in the genome-wide analysis. Transcription of hsp90+, tdh1+, adh1+ occurs throughout the cell cycle, while hta1+ transcription peaks in S phase [46]. In fission yeast, the G2 phase comprises about 75% of the cell cycle, so the majority of cells in an asynchronous culture are in G2 phase, and most genes are transcribed in this interval [47]. Thus, if the association of Pfh1 with highly transcribed genes was non-specific, it should occur to a similar extent in asynchronous and G2-arrested cells for hsp90+, tdh1+, adh1+, but not hta1+.
We performed ChIP-qPCR experiments in both asynchronous and G2 arrested cells in a temperature sensitive cdc25-22 strain that expressed Pfh1-13Myc. Pfh1 was significantly associated to all four genes in asynchronous cells grown at 25°C, compared to an untagged control strain (S1 Fig), which validated the peaks found in the ChIP-seq data. To arrest the cells in G2 phase, cells growing logarithmically at 25°C were shifted to 37°C for 4h.
Consistent with the expectation in the absence of bias, high Pfh1 binding to all four of the highly transcribed genes varied across the cell cycle; it was approximately four times higher in asynchronous compared to G2 arrested cells in the ChIP-qPCR assay (p ≤ 0.016; Fig 1). In contrast, Pfh1 binding to the much less frequently transcribed ade6+ gene was not significantly different in asynchronous versus G2 arrested cells (p = 0.14, Fig 1). Thus, the ChIP-qPCR experiment confirmed the high Pfh1 binding to these highly transcribed genes seen by ChIP-seq and established that this binding is not an artifact of the ChIP.
Fig 1. Pfh1 is enriched at highly transcribed genes in asynchronous cells compared to G2 arrested cells.
Samples from asynchronous or G2 arrested cells expressing Pfh1-13Myc were chromatin immunoprecipitated using an anti-Myc antibody. The immunoprecipitated DNA was analyzed using quantitative PCR with primers specific for four highly transcribed genes, hsp90+, tdh1+, adh1+, hta1+, and a low/medium transcribed control gene, ade6+. Pfh1 association is shown as immunoprecipitated DNA divided by input DNA. Data are means of three independent replicates. Error bars are the standard deviation. The p-value was determined by two-tailed Student’s t-test. An “*” indicates significant (p < 0.05), and “ns” indicates a non-significant difference between asynchronous cells compared to G2 arrested cells.
Replisome pausing is increased genome-wide at hard-to-replicate sites in Pfh1-depleted cells
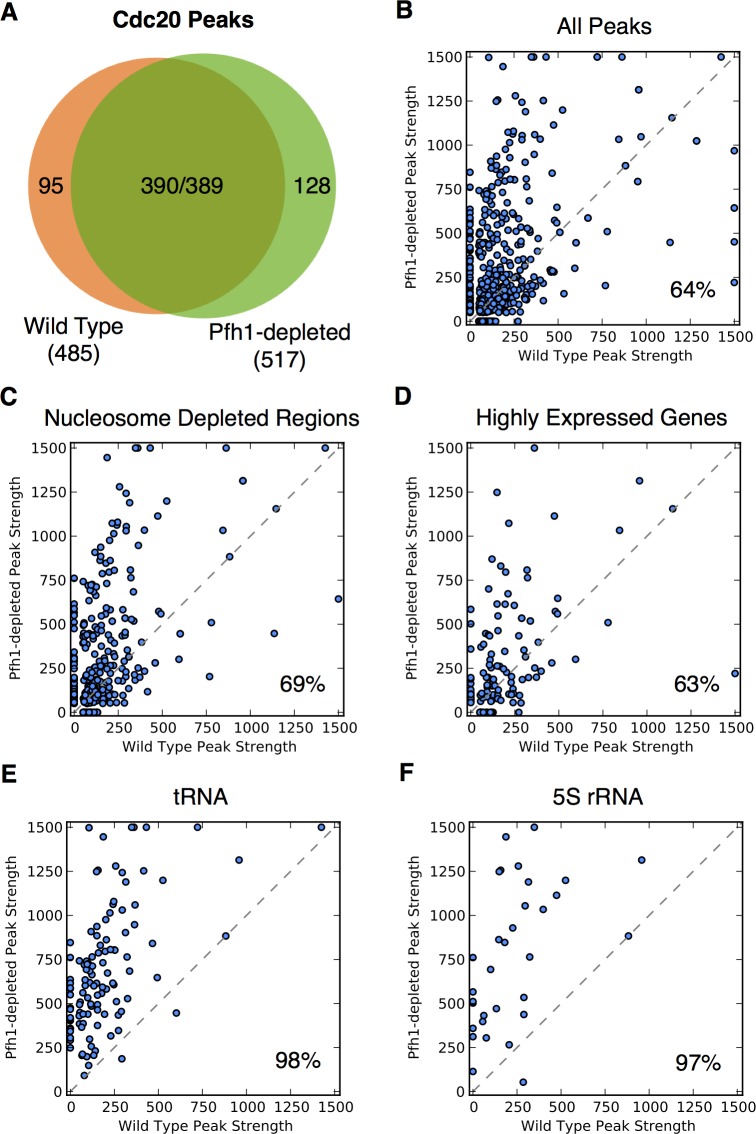
To identify genomic sites that require Pfh1 for their timely replication, we analyzed genome-wide occupancy of Cdc20, the catalytic subunit of the leading strand DNA polymerase ε [48] in WT and Pfh1-depleted cells. As previously reported [39], there are 485 peaks of high Cdc20 occupancy in WT cells and 517 in Pfh1-depleted cells (S1 Table). Although all genomic sites are Cdc20-associated when they are being replicated, high DNA polymerase occupancy correlates with replication fork slowing in both S. pombe and S. cerevisiae [39, 49]. Most of the high Cdc20 occupancy sites in WT cells were also found in Pfh1-depleted cells (390/485, 80%) and vice versa (389/517, 75%) (Fig 2A).
Fig 2. Fork pausing as marked by Cdc20 occupancy is increased in the absence of Pfh1.
(A) Venn diagram showing the overlap of genome-wide Cdc20 peaks in the presence and absence of Pfh1. The two numbers in the intersection are the number of WT peaks that overlap a Pfh1-depleted peak and vice versa. (B) Scatter plot comparing Cdc20 peak strength (-10*log10(p-value)) in WT and Pfh1-depleted cells. Each point represents a genomic region with a Cdc20 occupancy peak in at least one context. If a peak was not present in a context, it is plotted at 0 on the corresponding axis. The number in the bottom right of each plot gives the percentage of peaks stronger in Pfh1-depleted cells. When all peaks were considered, 64% of peaks were significantly stronger in Pfh1-depleted cells (p ≈ 0, Wilcoxon signed-rank test). Likewise, peaks at most tested genomic features were significantly stronger in Pfh1-depleted cells (S3 Table), as shown for (C) NDRs (69%, p ≈ 0) and (D) highly expressed genes (63%, p = 2.4x10-5). The difference was particularly large for (E) tRNA (98%, p ≈ 0) and (F) 5S rRNA genes (97%, p = 1.4x10-6).
In an earlier study, we used these data to demonstrate that many G4 motifs bind Pfh1 and that fork slowing and DNA breakage is more frequent at G4 motifs than at other G-rich regions in Pfh1-depleted cells (S2 Fig) [39]. Here we extend this analysis from G4 motifs to the rest of the genome. This analysis showed that in addition to G4 motifs, tRNA genes, 5S rRNA genes, NDRs, replication origins, RNA polymerase II promoters, RNA polymerase II transcribed genes, and meiotic DSB hotspots were significantly enriched among high Cdc20 occupancy sites in both WT and Pfh1-depleted cells (Table 2 and S2 Table; p < 0.001). The sets of high Cdc20 occupancy were identical in the two conditions except that dubious genes were enriched in Pfh1-depleted but not in WT cells. Despite this similarity, the evidence for elevated Cdc20 occupancy was significantly greater in Pfh1-depleted cells (p ≈ 0, Wilcoxon signed-rank test, p-values < 10−50 are reported as ≈0; Fig 2B). This pattern held for all genomic features tested (Fig 2C–2F; S3 Table). For example, 69% of Cdc20 peaks near NDRs (p = 2.4x10-5; Fig 2C) and 62% of peaks near highly transcribed RNA polymerase II genes (p ≈ 0; Fig 2D) were significantly stronger in Pfh1-depleted cells. This effect was particularly striking at tRNA (p ≈ 0; Fig 2E) and 5S rRNA (p = 1.4x10-6; Fig 2F) genes, where over 97% of the peaks showed evidence of significantly elevated Cdc20 occupancy when cells were Pfh1-depleted compared to WT cells. These findings argue that these genomic features, all of which had significant Pfh1 occupancy in WT cells (Table 1), were particularly dependent on Pfh1 for timely replication.
Table 2. Association of genomic features with Cdc20 and γ-H2A in WT and Pfh1-depleted cells.
Only features significantly associated with Pfh1 binding after correction for GC content and multiple testing (Bonferroni, p < 0.003) are shown. Each row gives the fraction of each feature associated with Cdc20 or γ-H2A; p-values for the association are in parentheses. Associations significant after Bonferroni multiple testing correction are bold. See S3 Table and S6 Table for full association results.
| WT | Pfh1-depleted | |||
|---|---|---|---|---|
| Genome Features | Cdc20 | γ-H2A | Cdc20 | γ-H2A |
| 5S rRNA genes | 0.73 (≈ 0) | 0.24 (0.08) | 0.97 (≈ 0) | 0.85 (≈ 0) |
| tRNA genes | 0.68 (≈ 0) | 0.28 (0.08) | 0.80 (≈ 0) | 0.75 (≈ 0) |
| Meiotic Double Strand Break Hotspots | 0.34 (≈ 0) | 0.22 (0.11) | 0.41 (≈ 0) | 0.47 (≈ 0) |
| 500 Highest Expressed RNA Polymerase II Genes | 0.22 (≈ 0) | 0.14 (0.83) | 0.22 (≈ 0) | 0.36 (0.85) |
| Nucleosome Depleted Regions | 0.16 (≈ 0) | 0.15 (0.17) | 0.19 (≈ 0) | 0.38 (≈ 0) |
| G4 motifs* | 0.11 (0.03) | 0.17 (0.01) | 0.17 (≈ 0) | 0.40 (≈ 0) |
| 3’ UTRs | 0.08 (0.05) | 0.13 (0.53) | 0.09 (0.21) | 0.33 (0.004) |
*from Sabouri, Capra [39]
To compare the Pfh1-dependent effects in more detail, we analyzed the genomic features associated with the 95 peaks of high Cdc20 binding unique to WT cells versus those associated with the 128 peaks of high Cdc20 binding that were found only in Pfh1-depleted cells. No features were enriched among the unique WT peaks. In sharp contrast, 5S rRNA and tRNA genes, meiotic DSB hotspots, NDRs, and dubious genes were all significantly enriched amongst the Cdc20 peaks unique to Pfh1-depleted cells (S2 Table).
Taken together, our results show that several classes of genomic features, especially RNA polymerase III transcribed genes, depend on Pfh1 for normal fork progression. In cases where sites were hard to replicate even in WT cells, fork pausing at these sites was significantly more pronounced in Pfh1-depleted cells.
Pfh1 protects the genome from DNA damage
In virtually all eukaryotes, including S. pombe, phosphorylation of H2A (γ-H2A) marks sites of DNA damage, typically DSBs [50]. To determine if the site-specific increases in replication pausing detected in Pfh1-depleted cells were associated with DNA damage, we analyzed peaks from previous ChIP-seq experiments using anti-γ-H2A antibodies in WT or Pfh1-depleted cells (S1 Table) [39]. We quantified the genomic distribution and Pfh1-dependence of the γ-H2A peaks with the same methods used for Cdc20.
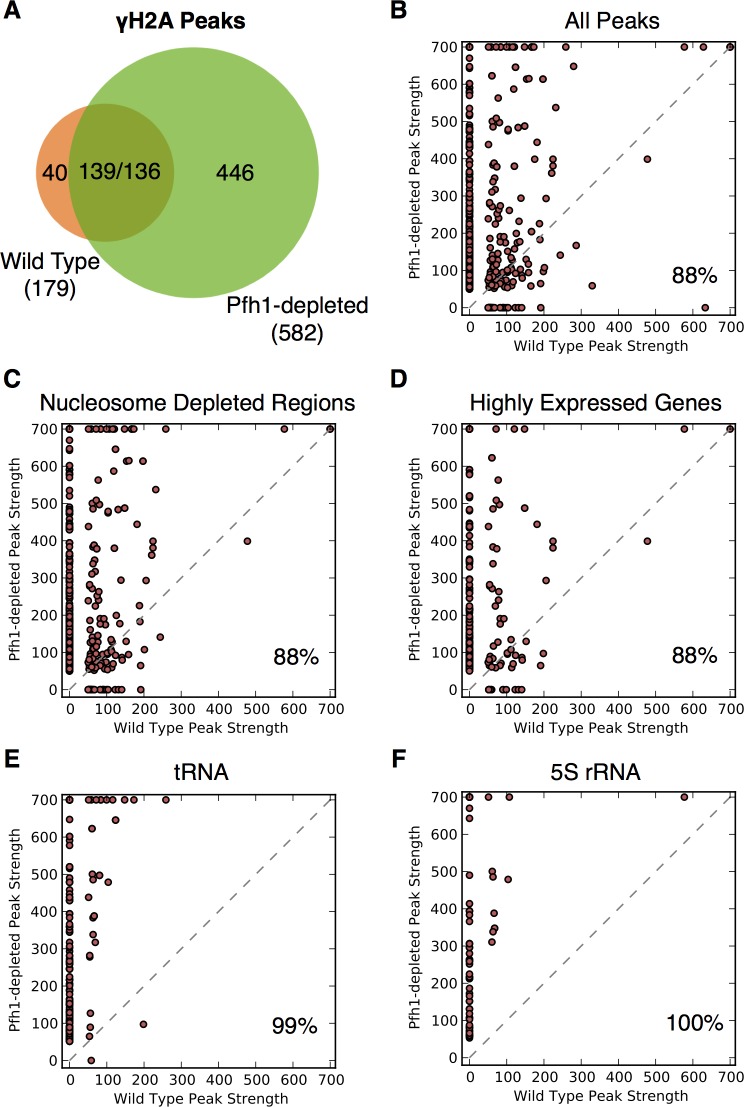
As demonstrated in previous work, WT cells had 179 γ-H2A peaks and Pfh1-depleted cells had 582 peaks (Fig 3A) [39]. Only two genomic features, the mating type loci and origins of replication, were significantly enriched near high occupancy γ-H2A sites in WT cells (p < 0.001 for both; S5 Table). However, in Pfh1-depleted cells, 5S rRNA and tRNA genes, meiotic DSB hotspots, NDRs, the mating type loci, and origins of replication all overlapped significantly with γ-H2A peaks (p < 0.001; Table 2 and S5 Table).
Fig 3. DNA damage as marked by phosphorylated histone H2A (γ-H2A) levels is increased in the absence of Pfh1.
(A) Venn diagram showing the overlap of genome-wide γ-H2A occupancy peaks in the presence and absence of Pfh1. The two numbers in the intersection are the number of WT peaks that overlap a Pfh1-depleted peak and vice versa. (B) Scatter plot comparing γ-H2A peak strength in WT and Pfh1-depleted cells. The layout is the same as in Fig 2B. Overall, γ-H2A peaks were significantly stronger in Pfh1-depleted cells (88%, p ≈ 0, Wilcoxon signed-rank test). Peaks near most tested genomic features were significantly stronger in Pfh1-depleted cells (S3 Table), e.g., (C) NDRs (88%, p ≈ 0) and (D) highly expressed genes (88%, p ≈ 0). γ-H2A peaks associated with (E) tRNA (99%, p ≈ 0) and (F) 5S rRNA genes (100%, p ≈ 0) were almost universally stronger in Pfh1-depleted cells.
We also determined the degree of overlap between Cdc20 and γ-H2A peaks in Pfh1-depleted cells. Of the 582 γ-H2A peaks in Pfh1-depleted cells, 71% (411) also had high Cdc20 occupancy; this number is significantly more than expected by chance (p < 0.001). γ-H2A sites that were high occupancy for both γ-H2A and Cdc20 were enriched for multiple genomic features including 5S rRNA and tRNA genes, origins of replication, meiotic DSB hotspots, 3’ and 5’ UTRs, and promoters (p < 0.001 for all; S6 Table). These features include most of those with significant Pfh1 association in WT cells. In contrast, γ-H2A peaks without corresponding Cdc20 peaks were enriched only in 3’ and 5’ UTRs and promoters (p < 0.001 for both). The significant overlap between peaks of Cdc20 and γ-H2A binding supports the connection between Pfh1-dependent fork slowing (as marked by Cdc20 peaks) and DNA damage (as marked by nearby γ-H2A) at several classes of hard-to-replicate sites.
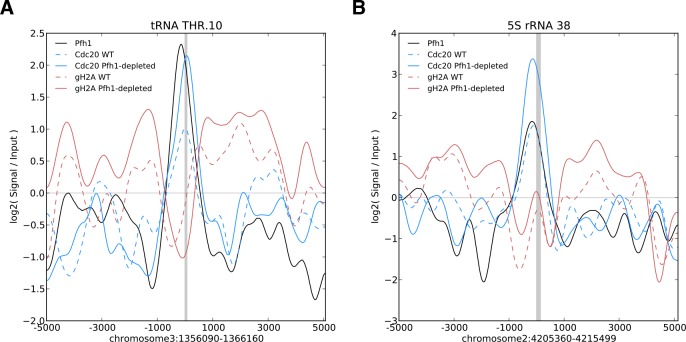
These patterns are illustrated for two specific sites, a tRNA gene (Fig 4A) and a 5S rRNA gene (Fig 4B). The strengths of the Pfh1, Cdc20, and γ-H2A binding are shown relative to input for the different strains in a 10 kilobase (kb) window around each gene. At both sites, a Pfh1 peak overlapped the gene (grey vertical lines mark centers of genes). A Cdc20 peak showed a similar overlap with the gene in both WT (dashed blue line) and Pfh1-depleted cells (solid blue line), but the peak was stronger in Pfh1-depleted cells. Broad peaks of γ-H2A flanked both genes in Pfh1-depleted cells, consistent with the 40 kb domains of γ-H2A that flank DSB sites [50]. Plots for all tRNA and 5S rRNA genes are shown in S3 and S4 Figs.
Fig 4.
ChIP-seq signal surrounding two representative RNA-PolIII transcribed genes: a threonine tRNA gene (A) and a 5S rRNA gene (B). Each plot displays the smoothed base 2 logarithm of the ChIP-seq reads mapping to each position in the experimental context over the corresponding input only read count. The raw signal was smoothed by convolution with a 1 kb Hanning window. The coordinates on the x-axis give the genomic location of the gene (gray box). These examples highlight a common pattern for tRNA and 5S rRNA genes: a peak of Pfh1 occupancy centered on the gene (black), with overlapping Cdc20 binding that is increased in the absence of Pfh1 (blue), and elevated γ-H2A in the regions flanking the genes in Pfh1-depleted cells (red). Plots for all tRNA and 5S rRNA genes are given in S3 and S4 Figs.
When the strengths of all the γ-H2A peaks between WT and Pfh1-depleted cells were compared, 88% of all peaks were higher in Pfh1-depleted cells (p ≈ 0, Wilcoxon signed-rank test; Fig 3B). This pattern held for the subsets of γ-H2A peaks associated with nearly all classes of genomic features (Fig 3C–3F; S3 Table). However, as seen for Cdc20, the magnitude of the difference was strongest for the RNA polymerase III transcribed genes: 190 of 192 γ-H2A peaks near tRNA genes (p ≈ 0, Fig 3E) and all 67 γ-H2A 5S rRNA peaks (p ≈ 0, Fig 3F) were greater in Pfh1-depleted cells.
During S phase, Pfh1 associates with Pfh1-sensitive and Pfh1-insensitive sites throughout the genome
Our data indicate that Pfh1 promotes replication and suppresses DNA damage at many discrete sites in the genome. We considered two models to explain this pattern of Pfh1 action. First, Pfh1 could act by binding nearby the replisome and mitigating replication obstacles as the replisome moves past Pfh1-sensitive sites. Alternatively, Pfh1 could be recruited only to sites that are hard to replicate or to stalled replication forks. In that case, Pfh1 would have low or no binding to sites that are Pfh1-insensitive. To distinguish between the two possibilities, we used ChIP-qPCR to monitor association of Pfh1 and Cdc20 in synchronized cells. For these experiments, we used a cdc25-22 strain that expressed Pfh1-13Myc inserted at the leu1+ locus under the control of the pfh1+ promoter (the endogenous pfh1+ locus was unaltered). This strain also expressed Cdc20-3HA from its endogenous location.
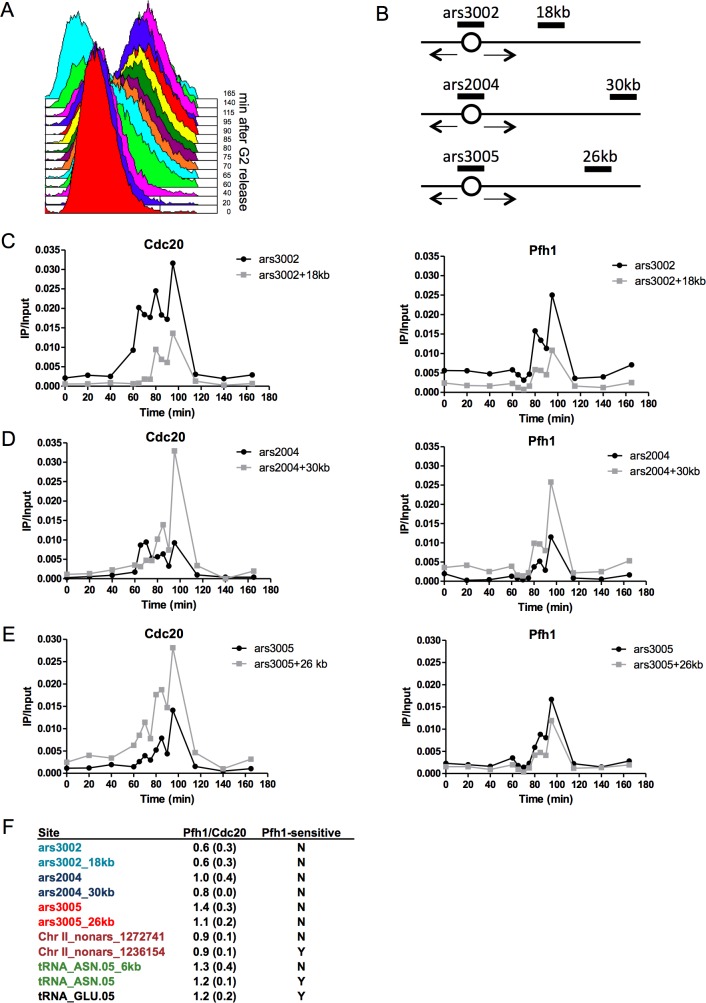
Cells were arrested in G2 phase by incubation at the non-permissive temperature (37°C) and then released at permissive temperature (25°C). The position in the cell cycle and the quality of the synchrony were determined by FACS analysis (Fig 5A). To determine association and movement of the replication fork, samples were taken throughout one synchronous cell cycle. At each time point, we examined association of Pfh1-13Myc and Cdc20-3HA to three origins of replication and their adjacent regions. We examined binding to the efficient ars3002 and a region18 kb away from ars3002 (ars3002_18kb), ars2004 and a region 30 kb from ars2004 (ars2004_30kb), and ars3005 and a region 26 kb from ars3005 (ars3005_26kb) (Fig 5B).
Fig 5. Pfh1 and DNA Pol ε both bind Pfh1-sensitive and insensitive sites.
(A) FACS analysis of cell synchrony. (B) Schematic of the regions examined by qPCR. Open circles represent origins of replication. Boxes mark the position of primer-pairs for qPCR that detect ars3002, ars3002_18kb, ars2004, ars2004_30kb, ars3005, and ars3005_26kb for ChIP-qPCR experiments. (C-E) Samples from each time point of the synchronized Cdc20-3HA Pfh1-13Myc cdc25-22 strain were immunoprecipitated with either anti-HA (left) or anti-Myc (right) antibody. The DNA was analyzed with qPCR using primer pairs for (C) ars3002 and ars3002_18kb, (D) ars2004 and ars2004_30kb, and (E) ars3005 and ars3005_26kb. Although cells appeared synchronous by FACS analysis, and Pfh1 and Cdc20 showed similar temporal patterns with peak binding at 95 min, we did not detect progression of the replisome from the ars2004 and ars3005 origins to their adjacent regions (ars2004_30kb and ars3005_26kb). This is likely due to the documented heterogeneity of origin usage in different cells in the same population [66] and to inefficient origins. The experiments were performed at least two times and the graphs show one representative biological replicate. (F) The ratio of Pfh1 IP/input to Cdc20 IP/input at peak binding after G2 release for eleven regions. Samples from Cdc20-3HA Pfh1-13Myc cdc25-22 synchronized cells were immunoprecipitated with either anti-HA or anti-Myc antibody and analyzed by qPCR. The experiments were repeated two times and the reported ratio is the average of two biological replicates; the standard deviation is given in parentheses. The names of adjacent regions are in the same color. Y: yes, N: no.
If Pfh1 were nearby or associated with the replisome, Pfh1 and Cdc20 would have similar temporal patterns of binding to the three origins and their adjacent sites. If Pfh1 were recruited only to hard-to-replicate sites, then Cdc20 and Pfh1 would have different binding patterns. In fact, under the second model, Pfh1 should not bind to any of these sites, as none of them were Pfh1-dependent in the whole genome analysis. Consistent with the first model, Cdc20 and Pfh1 displayed similar association patterns at all three origins and their adjacent regions (Fig 5C–5E). We first examined the binding to ars3002 and its adjacent region ars3002_18kb (Fig 5C). Although Cdc20 bound in early S phase to ars3002, earlier than Pfh1, the peak binding for Cdc20 and Pfh1 was reached at 95 min (Fig 5C). Both proteins had their start of binding to ars3002_18kb at 80 min after release from G2 phase and their peak binding at 95 min (Fig 5C). Thus, Pfh1 and Cdc20 bound to the Pfh1-insensitive site located 18 kb downstream of ars3002 similarly. However, while clear movement of Cdc20 was detected in this region, movement from this origin to the downstream regions was not visible for Pfh1.
Next, we examined the binding of Pfh1 and Cdc20 to the four other Pfh1-insensitive sites ars2004, ars2004_30kb, ars3005, and ars3005_26kb. Pfh1 bound to all these four regions, similarly to Cdc20 (Fig 5D and 5E). However, movement of neither Cdc20 nor Pfh1 was detected at any of these origins to their adjacent sites.
Because the dynamics of Pfh1 and the replisome were not clear from the above experiments, we further investigated the binding pattern of Pfh1 and Cdc20 at five other regions, including both Pfh1-sensitive and Pfh1-insensitve sites (Fig 5F). We tested two regions that were not origins of replication on chromosome II: one Pfh1-sensitive (Chr II_nonars_1236154), and a Pfh1-insensitive site 36 kb away (Chr II_nonars_1272741). Finally, we examined two Pfh1-sensitive tRNAs (tRNAGLU.05 and tRNAASN.05) and one Pfh1-insensitive site 6 kb away from tRNAASN.05 (tRNAASN.05_6kb).
At all five regions, both Pfh1 and Cdc20 had peak binding at 95 min after release from the G2 arrest, which by FACS analysis is mid-S phase (Fig 5). To determine if Pfh1 was recruited only to Pfh1-sensitive sites, we calculated the ratio of Pfh1 to Cdc20 binding at Pfh1-sensitive and -insensitive sites (IP/input of Pfh1 divided by IP/input for Cdc20) at the peak of replication for all sites (Fig 5F). If Pfh1 were recruited solely to Pfh1-sensitive sites, the ratio of Pfh1/Cdc20 would be higher at Pfh1-sensitive sites compared to Pfh1-insensitive sites. If Pfh1 were in proximity to the replisome, the ratios should be similar at the two classes of sites. Indeed, the Pfh1/Cdc20 ratio was on average 1.1 and 0.9 for Pfh1-sensitive and Pfh1-insensitive sites, respectively (Fig 5F). Thus, Pfh1 binds similarly to both Pfh1-sensitive and -insensitive sites. These data suggest that Pfh1 is near the replisome during S phase, rather than being recruited to its sites of action at Pfh1-sensitive sites. However, we cannot rule out the possibility that additional Pfh1 molecules are recruited to some or even all Pfh1-sensitive sites upon replisome pausing.
Mass spectrometry identifies Pfh1 interacting proteins
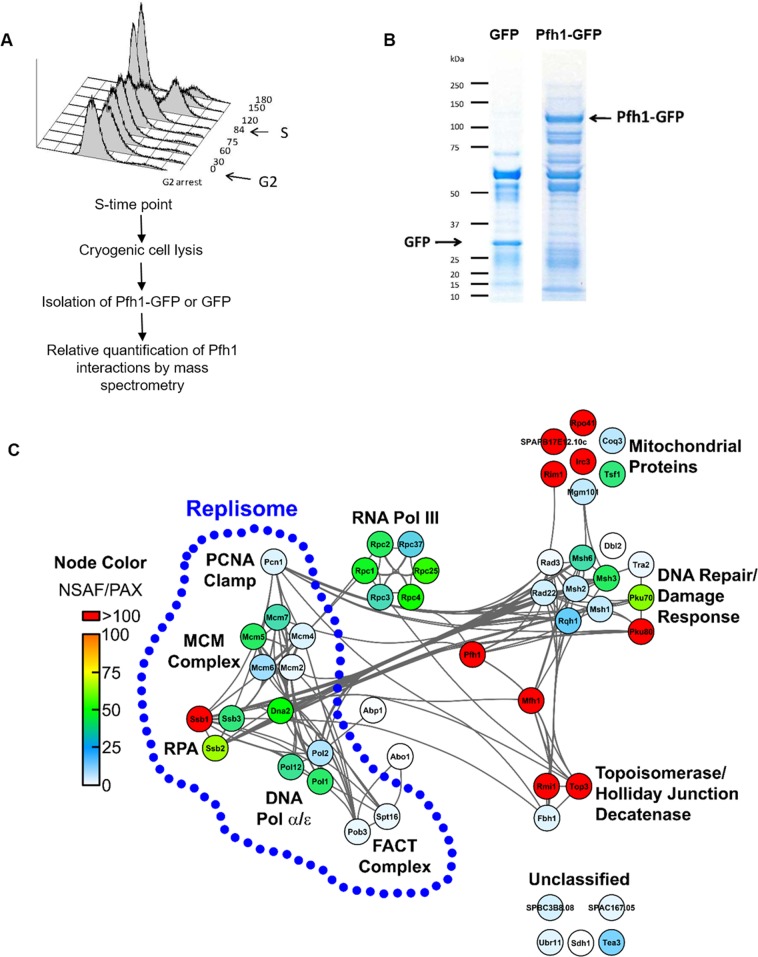
If Pfh1 maintains proximity to the replisome, as suggested by its pattern of binding to chromosomal DNA (Fig 5), then Pfh1 should be associated in vivo with known replisome subunits. To address this possibility, we used immunoaffinity purification mass spectrometry (IP-MS) to identify the Pfh1 protein interactions in S phase cells (Fig 6A). In these experiments, Pfh1 was expressed under its endogenous promoter as a GFP fusion (Pfh1-GFP), allowing immunoaffinity isolation of Pfh1 and its associated proteins through the GFP tag [51]. Cells expressing a SV40 nuclear localization signal-GFP fusion (NLS-GFP) were used as a negative control for non-specific association of proteins to GFP. Two biological replicates of both Pfh1-GFP and NLS-GFP were isolated in parallel from S phase cells (Fig 6A) using anti-GFP antibodies (Fig 6B).
Fig 6. Identification of Pfh1 protein interactions during S phase.
(A) Experimental design for parallel immunoaffinity purifications of Pfh1-GFP and NLS-GFP from cells harvested at S phase of the cell cycle and relative quantification of Pfh1 specific interactions by MS. As determined by FACS analysis, cdc25-22 cells were collected at the start of S phase, 84 minutes after G2 phase arrest. (B) Immunoaffinity purifications of GFP or Pfh1-GFP from S phase resolved by SDS-PAGE gel with the target and associated proteins visualized by Coomassie Blue staining. (C) Interaction network of high confidence Pfh1 protein associations during S phase, as identified by MS and assessed for specificity of binding by SAINT (p ≥ 0.80, n = 2). Nodes represent individual proteins interacting with Pfh1; node color represents relative enrichment compared to their abundance in the proteome (NSAF/PAX); and edges represent known interactions curated by the STRING database. Replisome components are highlighted by the blue dotted line.
Following mass spectrometry analysis of Pfh1-GFP and NLS-GFP immunoisolates, the interaction specificity of individual co-isolating proteins was assessed using the SAINT (significance analysis of interactome) algorithm [52]. SAINT determines confidence scores (ranging from 0 to 1) for protein-protein interactions based on the spectrum count distributions obtained from bait (Pfh1-GFP) isolations relative to the negative control (NLS-GFP). High confidence Pfh1 interactions were defined as proteins having a SAINT score ≥ 0.80, a threshold used previously to identify functional protein interactions [53, 54]. By this metric, there were 50 high confidence Pfh1 protein associations that comprise the Pfh1 S phase interactome (Table 3 and S7 Table). Although five of the Pfh1 interacting proteins are uncharacterized, there is functional data for 45 of the 50 proteins. Table 3 lists these proteins.
Table 3. Pfh1-GFP interacting proteins.
Pfh1-associated proteins during S phase (SAINT score ≥ 0.80) presented in alphabetical order. Names in bold are replisome components or replication related; italics indicate repair/recombination proteins; underlining indicates mitochondrial proteins. Functional annotations are from PomBase [68].
| Protein | Accession number | Function | SAINT score |
|---|---|---|---|
| Abp1 | P49777 | ARS-binding protein, CENP-B homologue, less abundant in G2 | 0.83 |
| Coq3 | O74421 | Mitochondrial hexaprenyldihydroxybenzoate methyltransferase | 0.86 |
| Dna2 | Q9URU2 | ATP-dependent helicase-nuclease, processes Okazaki fragments | 0.95 |
| Fbh1 | Q9USU3 | F-box DNA helicase, modulates homologous recombination | 0.97 |
| Irc3 | Q1MTR1 | Putative mitochondrial ATP-dependent helicase | 1.00 |
| Mcm2 | P40377 | Subunit of MCM replicative DNA helicase | 0.89 |
| Mcm4 | P29458 | Subunit of MCM replicative DNA helicase | 0.92 |
| Mcm5 | P41389 | Subunit of MCM replicative DNA helicase | 0.90 |
| Mcm6 | P49731 | Subunit of MCM replicative DNA helicase | 0.83 |
| Mcm7 | O75001 | Subunit of MCM replicative DNA helicase | 0.94 |
| Mfh1 | Q9UT23 | FANCM-like DNA helicase, repairs inter-strand crosslinks, promotes meiotic recombination | 0.99 |
| Mgm101 | O14354 | Mitochondrial genome maintenance protein | 0.87 |
| Msh1 | O13921 | DNA mismatch repair protein | 0.98 |
| Msh2 | O74773 | DNA mismatch repair protein | 0.99 |
| Msh3 | P26359 | DNA mismatch repair protein | 0.99 |
| Msh6 | O74502 | DNA mismatch repair protein | 0.99 |
| Pcn1 | Q03392 | Subunit of homotrimeric PCNA (Proliferating cell nuclear antigen) | 0.90 |
| Pku70 | O94395 | Subunit of Ku heterodimer | 1.00 |
| Pku80 | Q9HGM8 | Subunit of Ku heterodimer | 1.00 |
| Pob3 | O94529 | FACT complex subunit, remodels histones | 0.86 |
| Pol1 | P28040 | Catalytic subunit of DNA polymerase alpha | 0.82 |
| Pol12 | O74946 | Subunit B of DNA polymerase alpha | 0.84 |
| Pol2 | P87154 | Catalytic subunit of DNA polymerase epsilon; also called Cdc20 | 0.99 |
| Rad22 | P36592 | DNA repair and recombination protein, homologue budding yeast Rad52 | 0.83 |
| Rad3 | Q02099 | ATR-like checkpoint protein kinase | 0.98 |
| Rim1 | O14087 | Mitochondrial single-stranded DNA-binding protein | 0.99 |
| Rmi1 | Q10160 | Subunit of Rqh1/Top3 complex, suppresses DNA damage | 0.89 |
| Rpc1 | O94666 | Subunit of RNA polymerase III | 0.98 |
| Rpc2 | Q10233 | Subunit of RNA polymerase III | 0.83 |
| Rpc25 | O94285 | Subunit of RNA polymerase III | 0.82 |
| Rpc3 | Q9C106 | Subunit of RNA polymerase III | 0.86 |
| Rpc37 | O74883 | Subunit of RNA polymerase III | 0.83 |
| Rpc4 | O74857 | Subunit of RNA polymerase III | 0.89 |
| Rpo41 | O13993 | Mitochondrial RNA polymerase, priming mtDNA replication | 0.89 |
| Rqh1 | Q09811 | RecQ family DNA helicase subunit of Rqh1/Top3 complex, suppresses DNA damage | 1.00 |
| Sdh1 | Q9UTJ7 | Mitochondrial protein, probable succinate dehydrogenase flavoprotein subunit | 0.83 |
| SPAC167.05 | P87132 | Largely uncharacterized but implicated in meiotic chromosome segregation | 0.84 |
| SPAC1F5.11c | Q8TFH4 | Tra2 subunit of NuA4 complex phosphatidylinositol pseudokinase | 0.99 |
| SPAC31G5.19 | O14114 | Uncharacterized AAA domain-containing protein | 1.00 |
| SPAPB17E12.10c | Q8TFH4 | Mitochondrial protein, affects RNA processing | 0.99 |
| SPBC3B8.08 | O59716 | Uncharacterized protein | 0.90 |
| SPCC553.01c | O74939 | Meiotic chromosome segregation protein Dbl2 | 0.99 |
| Spt16 | O94267 | FACT complex subunit, remodels histones | 0.86 |
| Ssb1 | Q92372 | Replication factor A protein 1, RPA subunit | 1.00 |
| Ssb2 | Q92373 | Replication factor A protein 2, RPA subunit | 0.99 |
| Ssb3 | Q92374 | Replication factor A protein 3, RPA subunit | 0.87 |
| Tea3 | O14248 | Tip elongation aberrant protein 3, cell polarity | 0.94 |
| Top3 | O60126 | DNA topoisomerase 3, interacts with Rqh1 helicase, suppresses DNA damage | 0.98 |
| Tsf1 | Q9HGL5 | Mitochondrial translation elongation factor | 0.85 |
| Ubr11 | O13731 | E3 ubiquitin-protein ligase ubr11, affects chromosome stability, perhaps affects kinetochore | 0.97 |
We also assessed the relative abundance of individual Pfh1 interacting proteins within the interaction network by calculating the normalized spectrum abundance factor (NSAF) for each protein relative to its proteome abundance value (PAX) [55]. NSAF values provide a measure of protein abundance by accounting for factors such as protein length and sample complexity that can influence the number of spectra acquired for a given protein within a sample. Normalizing NSAF values to PAX values, as described in [56], provides insight into proteins and functional protein classes that are enriched in the Pfh1 isolation relative to their abundances in the cellular proteome. These data are presented in Fig 6C, which also categorizes interacting proteins by function.
Pfh1 interacts with multiple replisome components
The replisome is the multi-protein complex that is present at the replication fork as it moves through the chromosome. Multiple replisome components interacted with Pfh1 with high specificity (SAINT score ≥ 0.80; Fig 6C and Table 3). These proteins were: (1) five of the six subunits of the replicative helicase, the MCM complex (Mcm2 and Mcm4-7); (2) catalytic subunits of two of the three replicative polymerases (DNA Pol1 from DNA polymerase α; DNA Pol2/Cdc20, from DNA polymerase ε); (3) Pol12, the β subunit of DNA Pol1; (4) proliferating cell nuclear antigen (PCNA, Pcn1), a processivity factor that encircles and slides along the DNA; (5) the three subunits of the single-strand binding replication factor A (RPA, Ssb1, 2 and 3); (6) the Dna2 helicase-nuclease that is required for Okazaki fragment maturation; and (7) the two subunits of the FACT complex (facilitates chromatin transcription), Pob3 and Spt1, which facilitates nucleosome remodeling during both transcription and DNA replication. The association of FACT subunits with Pfh1 suggests that FACT and Pfh1 might act synergistically to promote replication through hard-to-replicate sites. Four mismatch repair (MMR) proteins, Msh1, 2, 3 and 6, were also Pfh1-associated. The Msh2/6 and Msh2/3 heterodimers interact directly with DNA for the recognition of base pair mismatches. Because MMR and DNA replication are strongly coupled in budding yeast, MMR proteins are proposed to track with the replisome and hence can also be considered replisome components [57, 58].
Additional replisome components were present in the Pfh1-GFP isolations but did not meet our SAINT score criterion. These proteins were: (1) the sixth Mcm subunit (Mcm3; SAINT score, 0.68); (2) the catalytic subunit of the lagging strand DNA polymerase; Pol3 (SAINT score, 0.15); (3) Dpb2, the second largest subunit of Pol ε (SAINT score 0.31); (4) Pri1 and Pri2, the primase subunits that function together with DNA polymerase α to synthesize the primers on the leading and lagging strand (SAINT scores of 0.65 and 0.33, respectively); and (5) Mcl1, the S. cerevisiae Ctf4 homologue that interacts with DNA polymerase α (SAINT score 0.70). While SAINT scores point to high confidence interactions, being based on detected protein spectral counts, they are influenced by sample complexity and the dynamic range of the co-isolated proteins, and thereby weighted towards large and abundant proteins, and stable interactions. Pfh1-associated replisome components with lower SAINT scores may be smaller proteins, have lower cellular abundances, and/or form transient interactions [59].
We performed two additional experiments to confirm the association of Pfh1 with the replisome. First, we isolated Pfh1 from asynchronous cells both in the presence and absence of DNase (S7 Table and S5 and S6 Figs). Because this experiment was performed with an asynchronous population, only a subset of the proteins that interacted with Pfh1 in S phase was detected (S6 Fig), even without DNase treatment. Of the 19 replisome/replication proteins that passed our stringent SAINT score threshold, ten were found in the DNase untreated sample, and nine of these retained their Pfh1 association after DNase treatment: Ssb1 and 2, Msh2, Mcm4, 5, and 6, Cdc20 (Pol2), Pol12, and Spt16. Because these interactions were not DNA-dependent, they are likely due to protein-protein interactions. Second, we isolated Pfh1 and its associated proteins from G2 arrested cells. We detected eight Pfh1-associated replication/replisome proteins in G2, and all eight were detected with fewer spectral counts in G2 extracts than in S phase extracts. The remaining eleven were not detected at all as Pfh1-interacting proteins in G2 phase (Table 3; S7 Table). Thus, as expected for a replisome component, Pfh1 association with known replisome subunits was either lost or diminished in G2 phase.
Together, these results show that Pfh1 associated in vivo with numerous replisome proteins, and that replisome and replication-related proteins represent a substantial subset of specific Pfh1 interactions (19 out of 50 proteins (38%) with SAINT score of ≥0.8) (Table 3; in bold; Fig 6C). Almost all of these associations were S phase-limited or S phase-enriched as well as DNA independent.
Pfh1 interacts with multiple mitochondrial and repair proteins
Pfh1 is a multi-functional protein: in addition to its role in nuclear DNA replication, it promotes DNA repair and is essential for maintenance of mitochondrial (mt) DNA [34]. Consistent with Pfh1 having mt function, 8 of the 50 high confidence Pfh1 interaction proteins have mt annotations (Fig 6C; mt proteins are underlined in Table 3). This subset includes several proteins implicated in mtDNA replication, such as (1) Rim1, the mt single-strand DNA binding protein (MS analyses reveal that ScPif1 is also ScRim1-associated; [60]), (2) Rpo1, the mtRNA polymerase that is thought to prime mtDNA replication, and (3) Mgm101, which is required for maintenance of mtDNA by an unknown mechanism.
Consistent with the reported DNA repair functions of Pfh1 [34], we observed multiple repair proteins among the high confidence Pfh1 interactions (Table 3, italics), including Rad22, the S. pombe homolog of budding yeast Rad52, which is required for homologous recombination [61], Rad3, the ATR-like checkpoint kinase [62], and both subunits of the non-homologous end joining Ku complex, pKu70 and 80. In addition, Rqh1 (homolog of human BLM) DNA helicase and its two interacting partners, the topoisomerase Top3 and Rmi1, were Pfh1-associated. This highly conserved heterotrimeric complex has multiple functions, but is best known for suppressing DNA damage at hard-to-replicate sites, such as converged forks [63] and/or collapsed replication forks—functions relevant to those of Pfh1.
Finally, six subunits of the 26 subunit RNA polymerase III complex were Pfh1-associated with high confidence (Rpc1, 2, 3, 4, 25 and 37), while four others were Pfh1-associated but had SAINT scores <0.8 (Rpc6, 0.71; Rpc9 and 10, 0.30; Rpc19, 0.76) [64]. This finding is probably related to RNA polymerase III transcribed genes being among the most potent replication impediments in Pfh1-depleted cells (Figs 2–4; see Discussion).
Discussion
Pfh1 acts at diverse sites genome-wide
We used genome-wide assays to determine sites where replication and genome integrity are Pfh1-dependent. The most striking aspect of these data is the strong dependence of RNA polymerase III transcribed genes on Pfh1. 2D gel analyses showed previously that replication of five of five tested tRNA genes is Pfh1-dependent, and this dependence is seen regardless of whether replication is co-directional or opposite to the direction of transcription through the gene [35]. Here we show that close to 50% (80/171) of the tRNA genes bound Pfh1 (Table 1). Moreover, nearly all of the tRNA genes that bound Pfh1 were sites of fork pausing and DNA damage in both WT and Pfh1-depleted cells (Table 2; high Cdc20 binding: WT: 74/80, Pfh1-depleted: 80/80: high γ-H2A: WT: 25/80, Pfh1-depleted: 69/80). However, both Cdc20 (Fig 2E) and γ-H2A binding (Fig 3E) were significantly higher at virtually all (99%) tRNA genes in Pfh1-depleted compared to WT cells. Likewise, genome-wide analyses revealed that 55% (18/32) of 5S rRNA genes bound Pfh1 (Table 1), and again the majority of these genes were sites of fork pausing (Table 2; WT: 16/18, Pfh1-depleted: 18/18) and/or DNA damage (WT: 5/18, Pfh1-depleted: 14/18), and both features were significantly higher in Pfh1-depleted compared to WT cells (Figs 2F and 3F). The independent MS analysis also supports the importance of Pfh1 at RNA polymerase III transcribed genes, as Pfh1 interacted with multiple subunits of RNA polymerase III (Fig 6C; Table 3). The association of these subunits with Pfh1 is consistent with a model where Pfh1 promotes replication past genes by displacing these proteins from DNA.
In addition to RNA polymerase III genes, 60% of the 500 most highly expressed protein-coding genes over a range of growth conditions were bound by Pfh1 (Table 1), and 48% were sites of fork slowing and/or DNA damage in Pfh1-depeleted cells (Figs 2D and 3D; S2 and S3 Tables). The likelihood of Pfh1 association was significantly greater for highly expressed genes than expected from other RNA polymerase II expressed genes (p≈0; hypergeometric test) as only 23% of all genes were Pfh1 associated.
A recent paper suggested that association of proteins with highly expressed genes in S. cerevisiae might be an artifact of the ChIP assay [45]. This interpretation is unlikely for our S. pombe analyses as a non-ChIP method, 2D gel analysis of replication intermediates, also showed that replication of three of three tested highly expressed RNA polymerase II transcribed genes is Pfh1-dependent [35]. Also, the peak strength at the highly transcribed genes was elevated in Pfh1-depleted cells compared to WT cells, suggesting that the association is not a ChIP artifact. Moreover, the genome-wide approach in this paper showed that high association of Pfh1 to highly transcribed genes was S phase specific (Fig 1), even though transcription of the genes is not S phase-limited. In addition, Pfh1, Cdc20, and γ-H2A all associate with ribosomal DNA (rDNA), probably the most highly transcribed region in all organisms, but high binding of all three does not occur over the 18 or 28S coding regions [39]. Also, the patterns of γ-H2A occupancy—low at the gene site and highest in flanking regions—are inconsistent with transcription artifacts (Fig 4), because the artifactual enrichment was observed to be high across the gene body. Thus, high binding of anti-Myc (used for Pfh1-13Myc), anti-HA (used for Cdc20-3HA), and anti-γ-H2A to highly transcribed genes is unlikely an artifact of their high transcription.
Fork pausing at highly transcribed genes as marked by high Pol2 occupancy is also demonstrated in budding yeast [49]. In contrast to S. pombe, where Pfh1 depletion increased fork pausing, pausing at these sites is not affected in the absence of ScRrm3 [49] or ScPif1 [18]. However, cells with a double deletion have not been tested, so it is possible that ScRrm3 and ScPif1 have overlapping functions in promoting replication past these genes.
Replication of multiple classes of genomic features is Pfh1-dependent. In addition to highly transcribed RNA polymerase II and III genes, this study identified several novel sites of Pfh1 association, such as NDRs, 3’ UTRs, and preferred meiotic DSB sites. In previous work, we showed that Pfh1 promotes fork movement past G4 motifs [39]. We analyzed whether the novel Pfh1 associations could be explained by overlap with G4 motifs or other Pfh1-associated features (S7 Fig). For example, NDRs often overlapped other significantly Pfh1-associated features, such as highly transcribed genes (32%), but none were sufficient to explain the association completely. Given their open state, it is likely that nucleosome-free regions are enriched for tightly bound proteins, other interactions, or formation of stable secondary structures that pause replication. The association with meiotic DSB hotspots in mitotically growing cells is largely driven by overlap with other elements that challenge replication; 92% (265 of 289 DSB) overlap another hard-to-replicate site identified in this study, most notably highly expressed genes and 3’ UTRs.
As noted above, several lines of evidence point to RNA polymerase III genes as the most Pfh1-dependent set of sites. Significant, but relatively smaller, fractions of the other Pfh1-associated features ultimately produce fork stalling and DNA damage in the absence of Pfh1 (Table 2). However, given the greater length of RNA polymerase II transcribed genes, if some cause fork stalling throughout their entire length, they may have a greater impact on fork progression genome-wide than the shorter tRNA genes.
The comprehensive analysis of sites of Pfh1 activity provided here demonstrates that, in addition to G4 motifs [39], there are many classes of hard-to-replicate sites that depend on Pfh1 for their proper replication. Many of these other sites, in particular RNA polymerase III transcribed genes, exhibit even stronger Pfh1-dependent effects than G4 motifs (Tables 1 and 2 and S3 Table). Altogether, Pfh1 supports DNA replication at thousands of diverse sites across the genome.
Pfh1 is an accessory helicase that maintains proximity to the replisome
Although accessory helicases are well studied in bacteria, it is not clear for any of the bacterial enzymes if they are recruited to their sites of action or if they move with the replisome through the genome. Here, we present several lines of evidence that support the association of Pfh1 with the replisome in S. pombe. First, using a strain in which Pfh1 and Cdc20 were both epitope tagged, Pfh1 had strong binding to three different origins during S phase (Fig 5), even though these origins were not a peak of Pfh1 binding in asynchronous cells. Pfh1 was also bound to adjacent regions of the origins, although neither of these sites were Pfh1-dependent sites. The temporal patterns of binding to the sites adjacent to the origins were similar to those of Cdc20 in the same cells; however, we could not detect a movement of Pfh1 from an origin to its adjacent region, as we detected for Cdc20 at ars3002. This may be due to the role of DNA polymerase ε in replication initiation [65], technical challenges, and/or stochastic origin usage in different cells in the synchronized population [66]. If Pfh1 were recruited only to Pfh1-dependent sites, we would have expected to see low binding to these Pfh1-independent sites compared to Cdc20. Second, the levels of Pfh1 and Cdc20 binding to five other sites were similar, regardless of whether the site was Pfh1-dependent or independent. This again argues against a model in which Pfh1 is only recruited and associated with sites that require it for their normal replication. Third, genome wide analyses show that many sites of high Cdc20 binding overlap with Pfh1 peaks (230 of the 485 high Cdc20 binding sites are also sites of high Pfh1 binding). These data are most consistent with Pfh1 being in close proximity to the replisome throughout the genome.
Our MS analyses also provide independent evidence in support of the hypothesis that Pfh1 maintains proximity to the replisome and is likely a replisome component. During S phase, Pfh1 was associated with many replisomal proteins, including the MCM replicative helicase, subunits of the replicative polymerases, the processivity clamp PCNA, RPA, and four MMR proteins (Fig 6C). The association of Pfh1 with replisome proteins was either S phase limited or S phase enhanced and almost all of these associations were DNase-resistant. While it is possible that Pfh1 would have interactions with replisome subunits if it were recruited after replisome pausing, in the context of the evidence presented above, we conclude that Pfh1’s strong associations with replication proteins are likely due to protein-protein interactions, as expected if it is a replisome subunit.
Pfh1 interacts with the FACT complex
Pfh1 also strongly associated with Spt16 and Pob3 (homologs of human SPT16 and SSRP1), two subunits of the heterodimeric evolutionarily conserved chromatin remodeling FACT complex (Table 3). In budding yeast, FACT promotes replication at replication conflicts past transcribed regions, and when FACT is depleted, ScRrm3 occupancy increases at highly transcribed RNA polymerase II and III genes [67]. Also, R-loop formation is elevated in FACT depleted cells, suggesting that the FACT complex resolves R-loop-mediated transcription-replication conflicts. The S phase association of FACT with Pfh1 suggests that the two cooperate to promote replication through these genes in S. pombe as they do in budding yeast. Pfh1 may facilitate replication at these genes by removing R-loops, as does budding yeast ScPif1 [27, 41]. In combination with its biochemical activities, the enrichment of Pfh1 at 3’ UTRs may also reflect a role in resolving R-loop-mediated transcription-replication conflicts.
Summary
The genomes of all organisms are littered with hard-to-replicate sites. Accessory helicases promote the movement of the replisome past these natural impediments. Although multiple bacterial accessory helicases have been characterized, much less is known about accessory helicases in eukaryotes. Our genomic and proteomic analyses, in combination with previous work, show that Pfh1 promotes replication and suppresses DNA damage at hundreds of diverse, naturally occurring hard-to-replicate sites. The pattern of binding of Pfh1 through the genome combined with its association with many replisome components argues that it is in close proximity with the replisome to help it maneuver past these obstacles. For many of the Pfh1-sensitive sites (e.g., tRNA and other highly transcribed genes), replication slowed at these sites even in WT cells, but usually did not result in significant DNA damage. However, when Pfh1 was depleted, fork slowing intensified and DNA damage increased dramatically at hard-to-replicate sites. In budding yeast, the two Pif1 family helicases, ScPif1 and ScRrm3, collaborate to promote fork progression past replication impediments. Our results establish the importance of this helicase family in S. pombe, a eukaryotic organism that is deeply diverged from S. cerevisiae and shares many genomic characteristics in common with higher eukaryotes. As the replication-impeding obstacles found in budding and fission yeasts are ubiquitous across genomes of other organisms, accessory helicases are likely to be required in all organisms, even though helicases that act at most of these sites have not been identified in higher eukaryotes. Thus, we propose that Pif1 family helicases present in multicellular eukaryotes also act as accessory helicases to promote fork progression and preserve genome stability.
Materials and Methods
ChIP analysis
ChIP-qPCR were performed in cdc25-22 leu1-32::PJK148-Pfh1-13MYC-kanmx6 cdc20::cdc20-3HA-kanmx6 cells (S8 Table). Asynchronous samples were grown at 25°C. The G2 phase cells were arrested at 37°C and collected after 4h arrest. To perform ChIP-qPCR in synchronized cells, cells were arrested at 37°C for 4h and released at permissive temperature at 25°C. Samples from the cell synchrony were collected for FACS analysis and ChIP-qPCR at 0, 20, 40, 60, 65, 70, 75, 80, 85, 90, 95, 115, 140, and 165 min. The ChIP experiments were performed as described previously [35]. Briefly, cells were cross-linked in 1% formaldehyde at 25°C for 5 min. The chromatin was sheared to an average of ~300 bp with a Covaris E220 system. G2 phase and asynchronous cells were immunoprecipitated with anti-Myc antibody (Clontech Cat. nr 631206). The synchronous cells were divided (3/4 of sample for Pfh1-Myc and 1/4 of sample for Cdc20-HA) and immunoprecipitated with either anti-Myc antibody or anti-HA antibody (Santa Cruz Biotechnology Cat. nr Sc7392x). Both immunoprecipitated DNA and the corresponding input amount for each sample were purified and quantified by real-time PCR, using the primer pairs described in S9 Table. At least two biological replicates were performed for each synchronous ChIP analysis. To calculate the average ratio of the peak binding time in Fig 5F, the IP/Input of two biological replicates were used. The time for peak binding (highest binding) in replicate 1 was 95 min and 70 min for replicate 2. The difference between the highest peak binding for the two synchronies was due to different start times of S phase after G2 release, which was detected by FACS analysis.
Flow cytometric analysis (FACS)
S. pombe cells were collected in 165 mM EDTA, 0.1% sodium azide 70% EtOH. Cells were pelleted, washed in 100% EtOH, and stored at 4°C. In preparation for FACS analysis, approximately 2 x 106 cells were washed in 3 ml of 50 mM Na citrate, pH 7.2, and incubated overnight at 37°C in 0.5 ml 50 mM Na citrate plus 0.1 mg/ml RNaseA. Following sonication, cells were incubated in 1 μM Sytox Green (Molecular Probes) at room temperature for 30 minutes. Cells were analyzed using a FACScan single laser fixed-alignment benchtop analyzer.
Genomic annotation enrichment analysis
We tested for enrichment between the genomic locations of the ChIP-seq peaks with sixteen sets of genomic annotations (Table 1). The ChIP-seq data for Pfh1, Cdc20, and γ-H2A are available in GEO data set GSE59178 [39]. All peaks analyzed are available in S1 Table. We took the location of genes, coding sequences, essential genes, dubious genes, 3’ and 5’ UTRs, promoters, tRNAs, centromeres, 5S rRNAs, long terminal repeats, and the mating type loci from PomBase [68]. We also considered the locations of meiotic DSB hotspots [69], NDRs [70] using podbat [71], origins of replication [72], the 500 most highly expressed genes from expression data collected across several growth conditions [44], and G4 motifs [39]. For Pfh1 and Cdc20 sites, regions within 300 bp were considered associated, and for γ-H2A peaks, we considered a window of 5 kb, since DNA damage results in elevated phosphorylation in a window of roughly this size around the break [50]. Overlaps between sets of genomic locations were calculated using BEDTools [73].
To determine if the observed association between two sets of genomic features, such as Pfh1 binding peaks and tRNA genes, was significantly greater than expected, we followed our previously described procedure for generating GC-content-aware empirical p-values [18, 39, 74]. In brief, we compared the observed number of overlaps to the number obtained when scrambling the peak locations 1,000 times in a manner that preserved the length, chromosome, and GC content of the regions. The number of randomized sets of peaks that obtain as many or more overlaps with the annotation (e.g., tRNA) as the actual peaks is the empirical p-value. If no random sets meet the level of overlap with the actual peaks, then the p-value is reported as < 0.001. We accounted for the testing of each set of ChIP-seq peaks with multiple genomic features using the Bonferroni correction.
Immunoaffinity purification (IP)
Strains expressing Pfh1-GFP were previously described (S8 Table) [34]. Briefly, the pJK148-integrating vector was used to express Pfh1-GFP from the leu1 locus using the endogenous Pfh1 promoter. The control IP strain expressed GFP-NLS from the leu1 locus under the control of the P3nmt promoter. The GFP-NLS construct was generated in pJK148 using the plasmid pFA6a-kanMX6-P3nmt1-GFP tagging construct as a PCR template with the addition of two SV40 nuclear localization signals introduced by PCR primers.
For cell synchronization, cdc25-22 strains were grown to early mid-log (0.5 x 107 cells/ml) at the permissive temperature of 25°C. The cells were collected by filtration and shifted to 37°C for G2 arrest. After 4 hours of incubation at 37°C, the media was quickly cooled (2 minutes by swirling in an ice bath) to 25°C for synchronized growth. Cells harvested at the G2 time point were collected at the end of the 4 hour 37°C incubation. Cells harvested at the S phase time point were collected at 84 minutes, corresponding to the start of replication. Cell cycle progression and the timing of DNA replication were confirmed by FACS analysis. Strains expressing either Pfh1-GFP (yKM333) or GFP alone (yKM346) from the S. pombe leu1-32 locus in a strain background containing the cdc25-22 mutant were confirmed to progress normally through the cell cycle by FACS analysis.
Two liters of S. pombe cells were either synchronized or grown to mid-log and harvested by centrifugation at 4°C for 10 minutes at 4,000 rpm (2,704 x g). Cell pellets were resuspended in freezing buffer (20 mM Na-HEPES, 1.2% polyvinylpyrrolidone (W/V), pH 7.4 containing a protease inhibitor cocktail (v/v 1/100) (Sigma) and frozen as cell droplets in liquid nitrogen [75]. Frozen cell droplets were cryogenically ground using a Retsch MM301 Mixer Mill (20 cycles x 2.5 min at 30 Hz) (Retsch, Newtown, PA) to achieve greater than 85% cell lysis, as assessed using light microscopy. Approximately 12 grams of ground, frozen cells were resuspended in lysis buffer (100mM Hepes KOH, pH 7.9, 300mM potassium acetate, 10mM magnesium acetate, 10% glycerol, 0.1% NP-40, 2mM EDTA, 2mM B-glycerophosphate, 50mM NaF, 10mM NaVO4, 1mM DTT, protease inhibitor cocktail (Roche) in a ratio of 5ml of lysis buffer per 1 gram of cells. Cells were gradually added to the lysis buffer with continuous mixing to avoid cell clumps. Lysis buffer conditions of varying salt concentrations (50–900 mM potassium acetate) were optimized for efficiency of Pfh1-GFP purification. Cell lysate was homogenized using a PT 10–35 Poyltron (Kinematica) for 3 sets of 10 seconds (30 seconds total) with 1 minute on ice in between each set. Insoluble material from the cell lysate was removed by centrifugation at 8000 rpm (9265 x g) for 10 minutes at 4°C. For immunoaffinity purification of Pfh1-GFP, the cell lysate supernatant was incubated for 30 minutes at 4°C with approximately 20 mg of M-270 epoxy magnetic beads (Invitrogen Dynal) conjugated with 50 μg of in-house developed rabbit polyclonal anti-GFP [51]. Following incubation, the beads were collected and washed six times with 1ml of lysis buffer. Proteins were eluted from the beads by incubation with 40 μl of 1x LDS sample buffer (Life Technologies) by shaking for 10 minutes at room temperature, followed by 10 minutes at 70°C. Eluted proteins were alkylated with 50 mM chloroacetamide for 30 minutes at room temperature in the dark.
To determine if DNA mediates the interactions of Pfh1-GFP, chromosomal DNA of the cell lysate was incubated with an excess of DNaseI (640 U/g of cell or ~70 ug/ml, 30 min. at 4°C) during the IP experiment immediately before the addition of conjugated beads (S6 Fig). DNaseI digestion was assessed by precipitation of DNA from the cell lysate before and after DNaseI treatment and was visualized on an agarose gel by ethidium bromide staining (S6B Fig). Low molecular weight DNA was observed in samples of cell lysate taken before and after DNaseI treatment (S6B Fig, lanes 1 and 2), suggesting that chromosomal DNA was degraded during earlier steps of the IP experiment before the addition of DNaseI. Enzymatic activity of DNaseI was not affected in the cell lysate, as demonstrated by the digestion of plasmid DNA (pDNA) that was added to a sample of the cell lysate prior to DNaseI digestion (S6B Fig, lanes 3 and 4).
Mass spectrometry analysis
Following immunoaffinity purifications (n = 2) of Pfh1-GFP or NLS-GFP from S phase cells, the isolated protein complexes were separated by gel electrophoresis. Samples were digested in-gel with trypsin and peptides were extracted from gel pieces using 0.5% formic acid. Peptides were concentrated by vacuum centrifugation to approximately 12 μl. 4uL of the sample was injected for nanoscale liquid chromatography tandem mass spectrometry (nLC-MS/MS) analysis on a Dionex Ultimate 3000 RSLC directly coupled to an LTQ-Orbitrap Velos with (ETD (ThermoFisher Scientific) instrument. Data were automatically acquired with MS2 fragmentation of the top 20 most intense precursor ions by collision-induced dissociation (CID). Parameters for data processing were also followed as described previously [56]. Briefly, raw files containing MS2 data were extracted by Proteome Discoverer (version 1.3; Thermo Scientific) and uploaded to SEQUEST (version 1.20) and searched against a compiled database of the yeast protein sequences from S. cervisiae and S. pombe. Post-search validation of the SEQUEST data was conducted by an X! Tandem algorithm in Scaffold (version Scaffold_3_00_04; Proteome Software) using the following filter selections to reduce peptide and protein global false discovery rate to < 1%: 99% protein confidence, 95% peptide confidence, and a minimum of two unique peptides per protein.
Assessing specificity of interactions
Protein interactions were accessed for specificity and enrichment (Pfh1-GFP vs. NLS-GFP control) using the SAINT (significance analysis of interactome) algorithm [52]. A SAINT confidence score cutoff of 0.80 was selected to retain high confidence Pfh1 interactions.
Building protein interaction networks
The protein interaction partners of Pfh1 were placed in the context of known protein interaction data from STRING (v9.1, medium confidence level, text mining = OFF) [76] and visualized using Cytoscape [77]. Within Cytoscape, nodes represent proteins that interact with Pfh1, and edges represent previously reported interactions among proteins in the network. To determine enrichment of protein interactions within the network relative to the background proteome, NSAF (normalized spectrum abundance factor) values were calculated to take into account protein length and the total number of spectra present in each individual IP experiment. NSAF values were normalized to proteome abundance (PAX) values for S phase S. pombe (pax-db.org), and the NSAF/PAX ratio was mapped onto each node (node color).
Supporting Information
Samples from asynchronous cells either expressing Pfh1-13Myc or an untagged control were chromatin immunoprecipitated using an anti-Myc antibody. Data for Pfh1-13Myc were also shown in Fig 1. The immunoprecipitated DNA was quantified using quantitative PCR with primers specific for hsp90+, tdh1+, adh1+, hta1+, and ade6+. Data are shown as immunoprecipitated DNA divided by input DNA. Data represent the mean of three independent replicates and error bars are standard deviation. The p-value was determined by two-tailed Student’s t-test and “*” indicates p < 0.05.
(PDF)
(A) Scatter plot comparing Cdc20 peak strength (-10*log10(p-value)) at G4 motifs in WT and Pfh1-depleted cells. Each point represents a genomic region with a Cdc20 occupancy peak in at least one context. If a peak was not present in a context, it is plotted at 0 on the corresponding axis. The number in the bottom right of each plot gives the percentage of peaks stronger in Pfh1-depleted cells. (B) Scatter plot comparing γ-H2A peak strength at G4 motifs in WT and Pfh1-depleted cells. The layout is the same as in part (A).
(PDF)
Immunoaffinity-purification of Pfh1-GFP from asynchronous cells. Proteins were resolved by SDS-PAGE and visualized by Coomassie stain. Peptides of identified proteins were confirmed by MS with nanoLC LTQ Orbitrap CID analyses. Pfh1 interaction was observed with proteins involved in DNA replication and repair such as the replicative DNA polymerase Pol2, members of the replicative Mcm helicase complex, and the Ku70/Ku80 heterodimer required for DNA repair and telomere maintenance in eukaryotic cells.
(PDF)
(A) Proteins were resolved by SDS-PAGE and visualized by Coomassie stain. (B) Ethidium bromide stained agarose gel of precipitated DNA from an aliquot of the cell lysate before and after DNAseI treatment (lanes 1 and 2) and with the addition of plasmid DNA (lanes 3 and 4) as a control for DNase I activity in the experimental lysis buffer.
(PDF)
(A) Fraction of Pfh1-bound features of a given type that overlap Pfh1-bound instances of each other tested feature type. (B) Fraction of Pfh1-sensitive (in terms of Cdc20) features of a given type that overlap Pfh1-bound instances of each other tested feature type. In each panel, the fraction is given in terms of the y-axis feature. For example, the 0.09 in the second entry in the first row of (A), indicates that 9% of the Pfh1-bound 3’ UTRs overlap a Pfh1-bound 5’ UTR.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We thank C. Redon for the kind gift of the γ-H2A antibody and K. Labib for his input on the MS data.
Data Availability
Sequencing data are available at GEO, data set GSE59178.
Funding Statement
Work in the Zakian lab was supported by NIH grant GM26938 (to VAZ), The Wenner-Gren Foundations (to NS), Swedish Society for Medical Research (to NS), and the New Jersey Commission on Cancer Research (to KRM). Work in the Sabouri lab was supported by the Swedish Research Council, The Wenner-Gren Foundations, and The Kempe Foundations (JCK-1325). Work in the Cristea lab was supported by NIH grants DA026192 and GM114141. AJG was supported by an NSF graduate research fellowship. JAC was supported by institutional funds from Vanderbilt University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Molecular cell. 2009;36(4):654–66. 10.1016/j.molcel.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. The EMBO journal. 2010;29(1):145–57. 10.1038/emboj.2009.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson J, Gupta MK, Rudolph CJ, Bell H, Lloyd RG, McGlynn P. Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication. Nucleic acids research. 2011;39(3):949–57. 10.1093/nar/gkq889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidnenko V, Lestini R, Michel B. The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Molecular microbiology. 2006;62(2):382–96. 10.1111/j.1365-2958.2006.05382.x . [DOI] [PubMed] [Google Scholar]
- 5.Gupta MK, Guy CP, Yeeles JT, Atkinson J, Bell H, Lloyd RG, et al. Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(18):7252–7. 10.1073/pnas.1303890110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merrikh CN, Brewer BJ, Merrikh H. The B. subtilis Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units. PLoS genetics. 2015;11(6):e1005289 10.1371/journal.pgen.1005289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagerburg MV, Schauer GD, Thickman KR, Bianco PR, Khan SA, Leuba SH, et al. PcrA-mediated disruption of RecA nucleoprotein filaments—essential role of the ATPase activity of RecA. Nucleic acids research. 2012;40(17):8416–24. 10.1093/nar/gks641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson J, Gupta MK, McGlynn P. Interaction of Rep and DnaB on DNA. Nucleic acids research. 2011;39(4):1351–9. 10.1093/nar/gkq975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA repair. 2010;9(3):237–49. 10.1016/j.dnarep.2010.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Molecular cell. 2010;39(4):595–605. 10.1016/j.molcel.2010.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Molecular cell. 2003;12(6):1525–36. . [DOI] [PubMed] [Google Scholar]
- 12.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5' to 3' DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes & development. 2002;16(11):1383–96. 10.1101/gad.982902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100(4):479–89. . [DOI] [PubMed] [Google Scholar]
- 14.Torres JZ, Bessler JB, Zakian VA. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes & development. 2004;18(5):498–503. 10.1101/gad.1154704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes & development. 2006;20(22):3104–16. 10.1101/gad.1478906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt KH, Derry KL, Kolodner RD. Saccharomyces cerevisiae RRM3, a 5' to 3' DNA helicase, physically interacts with proliferating cell nuclear antigen. The Journal of biological chemistry. 2002;277(47):45331–7. 10.1074/jbc.M207263200 . [DOI] [PubMed] [Google Scholar]
- 17.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–62. 10.1038/nature12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145(5):678–91. 10.1016/j.cell.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS genetics. 2009;5(5):e1000475 10.1371/journal.pgen.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piazza A, Serero A, Boule JB, Legoix-Ne P, Lopes J, Nicolas A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS genetics. 2012;8(11):e1003033 10.1371/journal.pgen.1003033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, et al. G-quadruplex-induced instability during leading-strand replication. The EMBO journal. 2011;30(19):4033–46. 10.1038/emboj.2011.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nature reviews Genetics. 2012;13(11):770–80. 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vega LR, Phillips JA, Thornton BR, Benanti JA, Onigbanjo MT, Toczyski DP, et al. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS genetics. 2007;3(6):e105 10.1371/journal.pgen.0030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossi SE, Ajazi A, Carotenuto W, Foiani M, Giannattasio M. Rad53-Mediated Regulation of Rrm3 and Pif1 DNA Helicases Contributes to Prevention of Aberrant Fork Transitions under Replication Stress. Cell reports. 2015;13(1):80–92. 10.1016/j.celrep.2015.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289(5480):771–4. . [DOI] [PubMed] [Google Scholar]
- 26.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438(7064):57–61. 10.1038/nature04091 . [DOI] [PubMed] [Google Scholar]
- 27.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic acids research. 2007;35(17):5809–18. 10.1093/nar/gkm613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. The Journal of biological chemistry. 2009;284(37):25170–80. 10.1074/jbc.M109.023325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1983;80(17):5345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502(7471):389–92. 10.1038/nature12584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502(7471):393–6. 10.1038/nature12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stafa A, Donnianni RA, Timashev LA, Lam AF, Symington LS. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics. 2014;196(4):1017–28. 10.1534/genetics.114.162297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasianovich Y, Harrington LA, Makovets S. Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening. PLoS genetics. 2014;10(10):e1004679 10.1371/journal.pgen.1004679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Molecular and cellular biology. 2008;28(21):6594–608. 10.1128/MCB.00191-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes & development. 2012;26(6):581–93. 10.1101/gad.184697.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes & development. 2012;26(6):594–602. 10.1101/gad.184663.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald KR, Sabouri N, Webb CJ, Zakian VA. The Pif1 family helicase Pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening. DNA repair. 2014. 10.1016/j.dnarep.2014.09.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sofueva S, Osman F, Lorenz A, Steinacher R, Castagnetti S, Ledesma J, et al. Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic acids research. 2011;39(15):6568–84. 10.1093/nar/gkr340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabouri N, Capra JA, Zakian VA. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC biology. 2014;12(1):101 10.1186/s12915-014-0101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallgren M, Mohammad JB, Yan KP, Pourbozorgi-Langroudi P, Ebrahimi M, Sabouri N. G-rich telomeric and ribosomal DNA sequences from the fission yeast genome form stable G-quadruplex DNA structures in vitro and are unwound by the Pfh1 DNA helicase. Nucleic acids research. 2016. 10.1093/nar/gkw349 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014;3:e02190 10.7554/eLife.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrd AK, Raney KD. A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase. The Journal of biological chemistry. 2015;290(10):6482–94. 10.1074/jbc.M114.630749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendoza O, Gueddouda NM, Boule JB, Bourdoncle A, Mergny JL. A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic acids research. 2015;43(11):e71 10.1093/nar/gkv193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332(6032):930–6. 10.1126/science.1203357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(46):18602–7. 10.1073/pnas.1316064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos A, Wernersson R, Jensen LJ. Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucleic acids research. 2015;43(Database issue):D1140–4. 10.1093/nar/gku1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman CS, Wood V, Fantes PA. An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System. Genetics. 2015;201(2):403–23. 10.1534/genetics.115.181503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS genetics. 2011;7(12):e1002407 10.1371/journal.pgen.1002407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Molecular cell. 2009;34(6):722–34. 10.1016/j.molcel.2009.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rozenzhak S, Mejia-Ramirez E, Williams JS, Schaffer L, Hammond JA, Head SR, et al. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS genetics. 2010;6(7):e1001032 10.1371/journal.pgen.1001032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Molecular & cellular proteomics: MCP. 2005;4(12):1933–41. 10.1074/mcp.M500227-MCP200 . [DOI] [PubMed] [Google Scholar]
- 52.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8(1):70–3. Epub 2010/12/07. 10.1038/nmeth.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, et al. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Molecular systems biology. 2015;11(1):787 10.15252/msb.20145808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin KW, McDonald KR, Guise AJ, Chan A, Cristea IM, Zakian VA. Proteomics of yeast telomerase identified Cdc48-Npl4-Ufd1 and Ufd4 as regulators of Est1 and telomere length. Nature communications. 2015;6:8290 10.1038/ncomms9290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. Journal of proteome research. 2006;5(9):2339–47. 10.1021/pr060161n . [DOI] [PubMed] [Google Scholar]
- 56.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Molecular & cellular proteomics: MCP. 2012;11(5):60–76. 10.1074/mcp.A111.015156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334(6063):1713–6. 10.1126/science.1210770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haye JE, Gammie AE. The Eukaryotic Mismatch Recognition Complexes Track with the Replisome during DNA Synthesis. PLoS genetics. 2015;11(12):e1005719 10.1371/journal.pgen.1005719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, et al. The functional interactome landscape of the human histone deacetylase family. Molecular systems biology. 2013;9:672 10.1038/msb.2013.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramanagoudr-Bhojappa R, Blair LP, Tackett AJ, Raney KD. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic acids research. 2013;41(2):1029–46. 10.1093/nar/gks1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ostermann K, Lorentz A, Schmidt H. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic acids research. 1993;21(25):5940–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, et al. The Schizosaccharomyces pombe rad3 checkpoint gene. The EMBO journal. 1996;15(23):6641–51. [PMC free article] [PubMed] [Google Scholar]
- 63.Hickson ID, Mankouri HW. Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell cycle. 2011;10(18):3078–85. . [DOI] [PubMed] [Google Scholar]
- 64.Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic acids research. 2001;29(13):2675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D'Urso G, Nurse P. Schizosaccharomyces pombe cdc20+ encodes DNA polymerase epsilon and is required for chromosomal replication but not for the S phase checkpoint. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(23):12491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Molecular biology of the cell. 2006;17(1):308–16. 10.1091/mbc.E05-07-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes & development. 2014;28(7):735–48. 10.1101/gad.234070.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic acids research. 2012;40(Database issue):D695–9. 10.1093/nar/gkr853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fowler KR, Gutierrez-Velasco S, Martin-Castellanos C, Smith GR. Protein determinants of meiotic DNA break hot spots. Molecular cell. 2013;49(5):983–96. 10.1016/j.molcel.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nature structural & molecular biology. 2010;17(2):251–7. 10.1038/nsmb.1741 . [DOI] [PubMed] [Google Scholar]
- 71.Sadeghi L, Bonilla C, Stralfors A, Ekwall K, Svensson JP. Podbat: a novel genomic tool reveals Swr1-independent H2A.Z incorporation at gene coding sequences through epigenetic meta-analysis. PLoS computational biology. 2011;7(8):e1002163 10.1371/journal.pcbi.1002163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siow CC, Nieduszynska SR, Muller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic acids research. 2012;40(Database issue):D682–6. 10.1093/nar/gkr1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS computational biology. 2010;6(7):e1000861 10.1371/journal.pcbi.1000861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cristea IM, Chait BT. Affinity purification of protein complexes. Cold Spring Harb Protoc. 2011;2011:pdb. prot5611. [DOI] [PMC free article] [PubMed]
- 76.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41(Database issue):D808–15. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. Epub 2010/12/15. 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples from asynchronous cells either expressing Pfh1-13Myc or an untagged control were chromatin immunoprecipitated using an anti-Myc antibody. Data for Pfh1-13Myc were also shown in Fig 1. The immunoprecipitated DNA was quantified using quantitative PCR with primers specific for hsp90+, tdh1+, adh1+, hta1+, and ade6+. Data are shown as immunoprecipitated DNA divided by input DNA. Data represent the mean of three independent replicates and error bars are standard deviation. The p-value was determined by two-tailed Student’s t-test and “*” indicates p < 0.05.
(PDF)
(A) Scatter plot comparing Cdc20 peak strength (-10*log10(p-value)) at G4 motifs in WT and Pfh1-depleted cells. Each point represents a genomic region with a Cdc20 occupancy peak in at least one context. If a peak was not present in a context, it is plotted at 0 on the corresponding axis. The number in the bottom right of each plot gives the percentage of peaks stronger in Pfh1-depleted cells. (B) Scatter plot comparing γ-H2A peak strength at G4 motifs in WT and Pfh1-depleted cells. The layout is the same as in part (A).
(PDF)
Immunoaffinity-purification of Pfh1-GFP from asynchronous cells. Proteins were resolved by SDS-PAGE and visualized by Coomassie stain. Peptides of identified proteins were confirmed by MS with nanoLC LTQ Orbitrap CID analyses. Pfh1 interaction was observed with proteins involved in DNA replication and repair such as the replicative DNA polymerase Pol2, members of the replicative Mcm helicase complex, and the Ku70/Ku80 heterodimer required for DNA repair and telomere maintenance in eukaryotic cells.
(PDF)
(A) Proteins were resolved by SDS-PAGE and visualized by Coomassie stain. (B) Ethidium bromide stained agarose gel of precipitated DNA from an aliquot of the cell lysate before and after DNAseI treatment (lanes 1 and 2) and with the addition of plasmid DNA (lanes 3 and 4) as a control for DNase I activity in the experimental lysis buffer.
(PDF)
(A) Fraction of Pfh1-bound features of a given type that overlap Pfh1-bound instances of each other tested feature type. (B) Fraction of Pfh1-sensitive (in terms of Cdc20) features of a given type that overlap Pfh1-bound instances of each other tested feature type. In each panel, the fraction is given in terms of the y-axis feature. For example, the 0.09 in the second entry in the first row of (A), indicates that 9% of the Pfh1-bound 3’ UTRs overlap a Pfh1-bound 5’ UTR.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
Sequencing data are available at GEO, data set GSE59178.