Abstract
The nitrogen fixation (nif) genes of nodule-forming Bradyrhizobium strains are generally located on symbiosis islands or symbiosis plasmids, suggesting that these genes have been transferred laterally. The nif genes of rhizobial and non-rhizobial Bradyrhizobium strains were compared in order to infer the evolutionary histories of nif genes. Based on all codon positions, the phylogenetic tree of concatenated nifD and nifK sequences showed that nifDK on symbiosis islands formed a different clade from nifDK on non-symbiotic loci (located outside of symbiosis islands and plasmids) with elongated branches; however, these genes were located in close proximity, when only the 1st and 2nd codon positions were analyzed. The guanine (G) and cytosine (C) content of the 3rd codon position of nifDK on symbiosis islands was lower than that on non-symbiotic loci. These results suggest that nif genes on symbiosis islands were derived from the non-symbiotic loci of Bradyrhizobium or closely related strains and have evolved toward a lower GC content with a higher substitution rate than the ancestral state. Meanwhile, nifDK on symbiosis plasmids clustered with nifDK on non-symbiotic loci in the tree representing all codon positions, and the GC content of symbiotic and non-symbiotic loci were similar. These results suggest that nif genes on symbiosis plasmids were derived from the non-symbiotic loci of Bradyrhizobium and have evolved with a similar evolutionary pattern and rate as the ancestral state.
Keywords: nitrogen fixation, Bradyrhizobium, nodule symbiosis, evolution, guanine and cytosine content
Rhizobial bacteria establish nodule symbiosis with legumes. Inside nodules, rhizobia fix atmospheric dinitrogen and provide fixed nitrogen to the plant. It is generally hypothesized that rhizobial genes related to nodulation (nod) and symbiotic nitrogen fixation (nif) were acquired by lateral gene transfer in Bradyrhizobium because these genes are clustered in a large genomic island known as a symbiosis island, which is characterized by a lower GC content than the rest of the genome (10, 11).
To date, the complete genome sequences of four nod-lacking Bradyrhizobium strains have been elucidated: Bradyrhizobium spp. S23321 (19), S58 (20), ORS278, and BTAi1 (5). These strains do not possess symbiosis islands. However, nif gene clusters (nif–clusters) are present in their chromosomes, which were likely to have been vertically transferred from their ancestors (19). The organization of nif–clusters is very similar among these strains (17, 19, 20), but differ from those on symbiosis islands of other strains such as B. diazoefficiens USDA110 (19). Hereafter, we designate the location of genes outside of the symbiotic region (symbiosis island or symbiosis plasmid) as “non-symbiotic loci.” A phylogenetic analysis of nifH (encoding nitrogenase iron protein) showed that nifH on non-symbiotic loci formed a different clade from those on symbiosis islands (19). These results suggest that the evolutionary history of nif genes is different between those found on non-symbiotic loci and on symbiosis islands.
A previous study showed that symbiotic genes (e.g., nod and nif) in Bradyrhizobium sp. DOA9 were located on a large-sized plasmid (approximately 736 kb), known as the symbiosis plasmid, providing the first evidence for symbiotic genes being located on a plasmid in Bradyrhizobium strains (17, 23). Strain DOA9 possesses two nif–clusters in its genome. One is located on the symbiosis plasmid (symbiotic loci) and the other is located on the chromosome (non-symbiotic loci) (17). The nucleotide sequences of nifH on the plasmid and chromosome share 92% sequence identity (17). A phylogenetic analysis revealed that nifH in the plasmid and chromosome of strain DOA9 clustered together with nifH genes located in the non-symbiotic loci of other Bradyrhizobium strains: S23321, S58, ORS278, and BTAi1 (17). These results indicate that the evolutionary histories of nif genes on symbiosis islands and plasmids are distinct.
In the present study, the nif genes of rhizobial and non-rhizobial Bradyrhizobium strains were compared to infer the evolutionary histories of nif genes using various Bradyrhizobium strains including newly genome-sequenced strains: Bradyrhizobium sp. DOA1 (a rhizobial strain possessing divergent nodulation genes [15]) and Bradyrhizobium sp. MAFF107635 (a synonym of AT1; a non-rhizobial strain isolated from the storage roots of sweet potato [24]).
Materials and Methods
Genome sequencing and analyses of strains DOA1 and MAFF107635
Strains DOA1 and MAFF107635 were cultured to the stationary phase at 30°C in HM salt medium (3) containing 0.1% arabinose and 0.025% yeast extract. The cells were harvested by centrifugation, and total DNA was prepared as previously described (14). The 8-kb paired-end 454 libraries of strains DOA1 and MAFF107635 were then constructed from total DNA by Takara Bio (Shiga, Japan). Genome sequencing was performed using a 454 GS FLX+ sequencer (Roche Diagnostics; Tokyo, Japan). The generated sequences were assembled using Roche GS De Novo Assembler version 2.5p1 (Roche Diagnostics). Two potentially contaminated regions from primer or adaptor sequences used in 454 sequencing were detected in the strain DOA1 genome at positions 1625647–1625666 and 6325190–6325209, both of which were located at the edge of contigs. The nucleotides of these regions were changed into Ns. Predictions of gene regions and annotations were performed using NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAP) (1). A circular genome map showing the GC skew and GC content was created using arcWithColor version 1.3.8 (http://www.ige.tohoku.ac.jp/joho/gmProject/gmdownload.html). GC content and skew were calculated with sliding windows along the genome. The window size was set to 20,000 bases and the step size to 500 bases. The chromosome sequences of strains DOA1 and MAFF107635 containing runs of Ns that represent gaps were used as inputs. All predicted gap lengths were less than 5 kb. Organizations of nif–clusters were compared using GenomeMatcher version 1.861 (16).
Tested strains for phylogenetic and molecular evolutionary analyses
The tested strains for phylogenetic and molecular evolutionary analyses are listed in Table 1. In addition to the newly genome-sequenced strains MAFF107635 and DOA1, we used Bradyrhizobium strains whose complete or high-quality draft (less than 10 scaffolds) genome sequences have been deposited in the NCBI genome database (last accessed April 2015). Since complete or high-quality draft genomes were limited to a narrow phylogenetic range of Bradyrhizobium species (Table 1), B. yuanmingense CCBAU 35157 and B. liaoningense CCBAU83689 were added to the tested strains even though their genomes are comprised of more than 10 scaffolds. The genomes of other Bradyrhizobium species were unavailable in the NCBI genome database.
Table 1.
Genomic locations of nitrogen fixation gene clusters in strains tested in the present study.
| Strain | Type of symbiosis region | Location of the nif–cluster | nod gene | Accession number |
|---|---|---|---|---|
| Bradyrhizobium sp. DOA1 | symbiosis island | symbiosis island | + | JXJM01000000 |
| Bradyrhizobium diazoefficiens USDA110 | symbiosis island | symbiosis island | + | BA000040 |
| Bradyrhizobium japonicum USDA6 | symbiosis island | symbiosis island | + | AP012206 |
| Bradyrhizobium japonicum E109 | symbiosis island | symbiosis island | + | CP010313 |
| Bradyrhizobium japonicum SEMIA 5079 | symbiosis island | symbiosis island | + | CP007569 |
| Bradyrhizobium elkanii USDA76 | symbiosis island | symbiosis island | + | KB900701, KB900702 |
| Bradyrhizobium sp. WSM471 | symbiosis island | symbiosis island | + | CM001442 |
| Bradyrhizobium sp. WSM1253 | symbiosis island | symbiosis island | + | JH600072, JH600073 |
| Bradyrhizobium genosp. SA-4 str. CB756 | symbiosis island | symbiosis island | + | AXBC00000000 |
| Bradyrhizobium sp. WSM1417 | symbiosis island | symbiosis island | + | AZXU00000000 |
| Bradyrhizobium sp. SUTN9-2 | symbiosis island | symbiosis island | + | LAXE00000000 |
| Bradyrhizobium liaoningense CCBAU 83689 | unknowna | unknowna | + | AJQD00000000 |
| Bradyrhizobium yuanmingense CCBAU 35157 | unknowna | unknowna | + | AJQL00000000 |
| Bradyrhizobium sp. DOA9 | symbiosis plasmid | non-symbiotic loci symbiosis plasmid | + | DF820425, DF820426 |
| Bradyrhizobium sp. MAFF107635 | — | non-symbiotic loci | − | JXDL01000000 |
| Bradyrhizobium sp. S23321 | — | non-symbiotic loci | − | AP012279 |
| Bradyrhizobium japonicum 22 | — | non-symbiotic loci | − | AXVG00000000 |
| Bradyrhizobium oligotrophicum S58 | — | non-symbiotic loci | − | AP012603 |
| Bradyrhizobium sp. BTAi1 | — | non-symbiotic loci | − | CP000494, CP000495 |
| Bradyrhizobium sp. ORS278 | — | non-symbiotic loci | − | CU234118 |
Information was not obtained because the nif–cluster was located on a small contig.
A symbiosis island is defined as a DNA segment containing a cluster of symbiotic genes (nif and/or nod) with a lower GC content than the entire genome (Table 1) (10). The genomic locations of nif genes are elucidated based on these criteria.
Phylogenetic analysis of the 16S rRNA gene and concatenated housekeeping genes
16S rRNA gene sequences were aligned using the CLUSTAL W program in MEGA 6.06 (22). The phylogenetic tree was constructed using nucleotide sequences via the maximum likelihood method with 1,000 bootstrap replicates in MEGA 6.06. The model with the lowest Bayesian Information Criterion (BIC) score by the “Find Best DNA/Protein Models” procedure of MEGA 6.06 was applied. The same analyses were conducted using the concatenated nucleotide sequences of four housekeeping genes (rpoB, recA, gyrB, and atpD).
Phylogenic and molecular evolutionary analysis of concatenated nifDK sequences
The concatenated sequences of nifD (encoding the alpha subunit of the nitrogenase molybdenum-iron protein) and nifK (encoding the beta subunit of the nitrogenase molybdenum-iron protein) were analyzed in order to examine the phylogenetic relationships of nif genes in the tested strains. nifH was not used in the phylogenetic analysis because the tested strains, S23321, S58, ORS278, and BTAi1, possess two copies of nifH in the nif–cluster, which may affect the substitution rates of nifH sequences and lead to the incorrect estimation of phylogenetic relationships. The phylogenetic tree was constructed using nucleotide sequences by the maximum likelihood method as described above. The same analyses were performed using the 1st + 2nd codon positions. Rhodopseudomonas palustris CGA009 was used as an outgroup because it is phylogenetically close to Bradyrhizobium and possesses a nif-cluster, which may be descended from a common ancestor of the nif-cluster loci of Bradyrhizobium strains on non-symbiotic loci. The neighbor-joining method was also applied to confirm the tree topology. Evolutionary distances were computed using the Tamura 3-parameter method. Differences in the composition bias among sequences were considered in evolutionary comparisons.
Disparity index tests of substitution pattern homogeneity were performed for each pair of concatenated nucleotide sequences of nifDK using MEGA 6.06 (22). A Monte Carlo test (1,000 replicates) was used to estimate P-values. P-values smaller than 0.01 were considered significant. The nucleotide compositions of each of the concatenated nifDK sequences were computed using MEGA 6.06. The same analyses were performed for the 1st + 2nd codon position. The molecular clock test was performed for tree topologies representing all and the 1st + 2nd codon positions by comparing the maximum likelihood value with and without the molecular clock constraints using MEGA 6.06. Codon-based tests of positive selection were performed using MEGA 6.06 in order to compare the number of synonymous and non-synonymous nucleotide substitutions.
Accession numbers
The BioProject for genome sequencing has been deposited at DDBJ/EMBL/GenBank under the accession numbers PRJNA271717 for strain DOA1 and PRJNA271718 for strain MAFF107635. The BioSample has been deposited at DDBJ/EMBL/GenBank under the accession number SAMN03276511 for strain DOA1 and SAMN03276517 for strain MAFF107635. The genome sequences have been deposited at DDBJ/EMBL/GenBank under the accession number JXJM01000000 for strain DOA1 and JXDL01000000 for strain MAFF107635.
Results and discussion
Phylogenetic diversity of tested strains
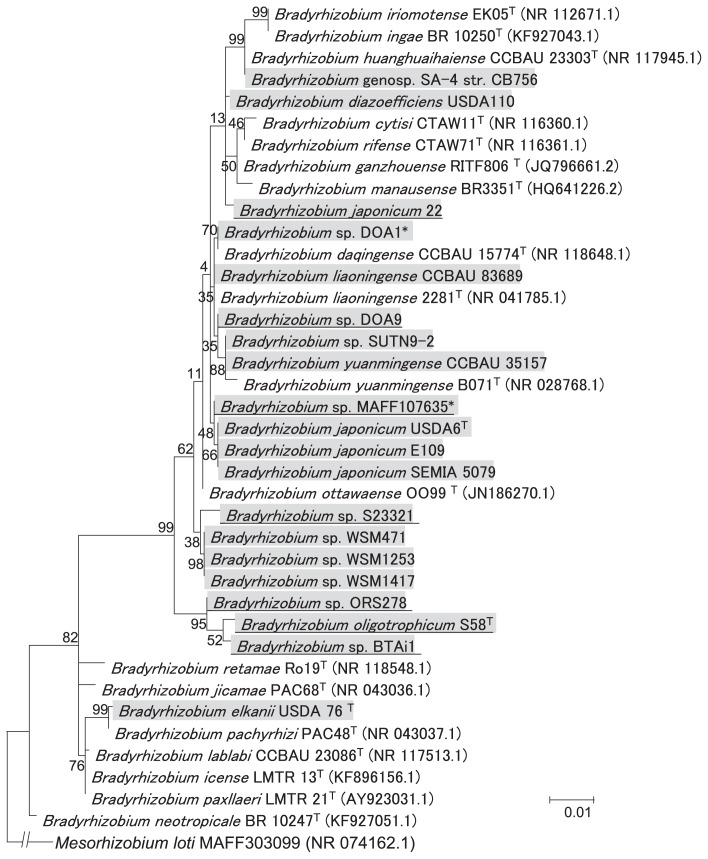
A phylogenetic tree of the 16S rRNA gene sequences was constructed using the tested strains and type strains of Bradyrhizobium species (Fig. 1). The tested strains (grey shadow in Fig. 1) appear to cover a clade including B. diazoefficiens, B. japonicum, B. yuanmingense, and B. oligotrophicum (Fig. 1). Thus, our data set provides some insights into the evolution of nif genes in Bradyrhizobium, as described below.
Fig. 1.
Phylogenetic tree by the maximum likelihood method for the 16S rRNA gene of tested and type strains in Bradyrhizobium species. Numbers at the nodes are the percentages of 1,000 bootstrap replications supporting the partition. The tested strains are shaded in gray. The tested strains possessing nifDK on non-symbiotic loci are underlined. The numbers in parentheses show sequence ID in public databases. Mesorhizobium loti MAFF303099 (NR_074162.1) was used as an outgroup. The newly genome-sequenced strains DOA1 and MAFF107635 are marked with asterisks.
Genome sequencing of strains DOA1 and MAFF107635
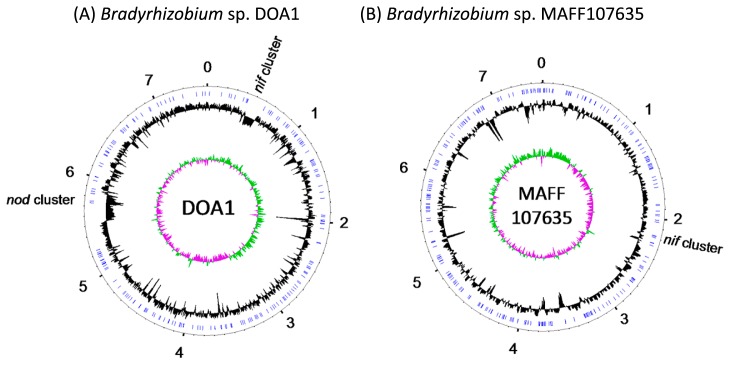
The draft genome sequences of strains DOA1 and MAFF107635 were determined using 8-kb paired-end 454 pyrosequences. The read assemblies of strain DOA1 produced one circular scaffold consisting of 201 contigs. Two large low-GC regions were found in the 450–550-kb and 5,600–5,800-kb regions and contained the nif–cluster and nod genes, respectively (Fig. 2A). These characteristics strongly suggest that these regions are symbiosis islands.
Fig. 2.
Circular view of whole-genome sequences of Bradyrhizobium sp. DOA1 (A) and MAFF107635 (B) chromosomes. The outermost circle represents genome positions in megabases. The second-outermost circle illustrates the positions of gap regions (blue). The innermost and second-innermost circles illustrate the GC (guanine-cytosine) skew (green and purple) and GC content (black), respectively.
The read assemblies of strain MAFF107635 produced one circular scaffold consisting of 260 contigs. The nif genes were located in the 2,193–2,226-kb region (Fig. 2B). The GC content of this region was similar to that of the whole genome. No symbiosis island or plasmid was found in the genome of strain MAFF107635 (Fig. 2B).
Phylogenic analysis of 16S rRNA and housekeeping gene sequences
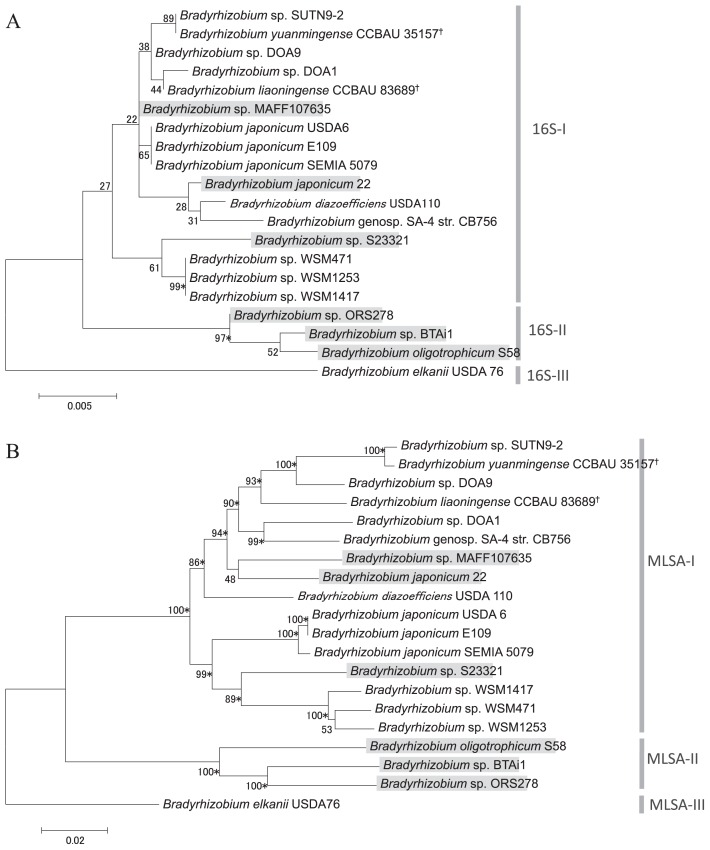
The phylogenetic tree of 16S rRNA gene sequences separated the tested Bradyrhizobium strains into three groups (Fig. 3A). Group 16S-I was comprised of three non-rhizobial strains including the newly sequenced strain MAFF107635 and 13 rhizobial strains including the newly sequenced strain DOA1. Group 16S-II was comprised of three strains, all of which have the ability to establish nodule symbiosis on some species of Aeschynomene plants (e.g., Aeschynomene indica and A. evenia) without nodulation factor signals (5, 18, 20); none of the strains in this group possessed symbiosis islands or plasmids. The rhizobial strain USDA76 formed a separate group from the other strains. The tree of 16S rRNA genes suggested that nod gene-lacking and-possessing strains were not clearly separated because the nod gene-lacking strains MAFF107635, S23321, and 22 were sporadically distributed in group 16S-I along with nod gene-possessing strains. However, the phylogenetic tree of 16S rRNA genes showed less than 70% bootstrap support for all branches in group 16S-I. In order to achieve better phylogenetic resolution, we applied a multilocus sequence analysis (MLSA) approach (4). The tree of the concatenated sequences of four housekeeping genes (rpoB, recA, gyrB, and atpD) showed 70% bootstrap support for most branches (Fig. 3B). The MLSA tree separated the tested strains into three groups consisting of the same members as the 16S rRNA gene tree. The non-rhizobial strains MAFF107635, S23321, and 22 were clustered together with nod gene-possessing strains in the group MLSA-I. These results show that nod gene-lacking and-possessing strains are closely related based on the sequences of the marker genes used for the phylogenetic analysis.
Fig. 3.
Phylogenetic trees by the maximum likelihood method for the 16S rRNA gene (A) and for concatenated sequences of four housekeeping genes (rpoB, recA, gyrB, and atpD) (B). Numbers at the nodes are the percentages of 1,000 bootstrap replications supporting the partition. Strains possessing nifDK on non-symbiotic loci are shaded in gray. Asterisks following the bootstrap values show that the branches are supported by the neighbor-joining method with ≥70% bootstrap values. Strains whose genomic locations of nifDK are unknown are marked with †.
Phylogenetic analyses of concatenated nifDK sequences
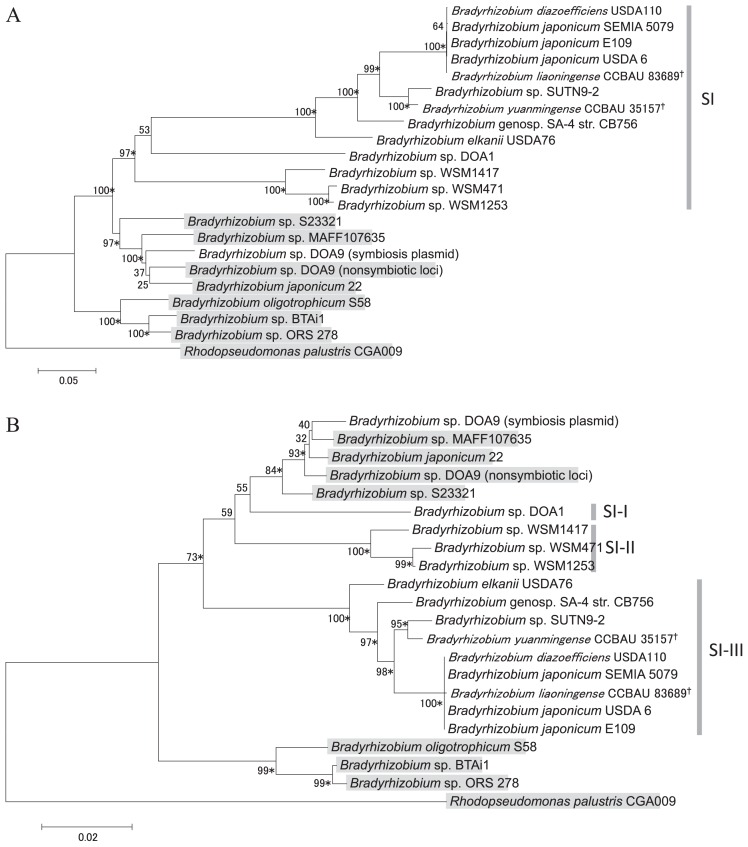
In order to infer the phylogenetic relationships of nif genes in the tested strains, we analyzed the concatenated sequences of nifD and nifK. nifDK on the symbiosis plasmid of strain DOA9 was clustered with those on the non-symbiotic loci of strain DOA9 and other strains (Fig. 4A), suggesting that the nif genes found on the symbiosis plasmid of strain DOA9 were derived from non-symbiotic loci of Bradyrhizobium.
Fig. 4.
Phylogenetic trees by the maximum likelihood method for concatenated nifDK sequences representing all codon positions (A) and representing only the 1st and 2nd codon positions (B). Numbers at the nodes are the percentages of 1,000 bootstrap replications supporting the partition. nifDK on non-symbiotic loci are shaded in gray. Asterisks following the bootstrap values show that the branches are supported by the neighbor-joining method with ≥70% bootstrap values. Strains whose genomic locations of nifDK are unknown are marked with †.
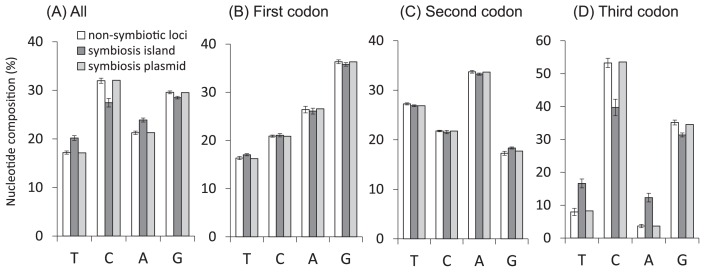
nifDK genes on symbiosis islands formed a monophyletic group with 97% bootstrap support (Fig. 4A), whereas the same strains were more distinctly distributed according to 16S rRNA (Fig. 3A) and concatenated housekeeping gene (Fig. 3B) trees. The branches of nifDK on symbiosis islands were elongated (Fig. 4A). These results indicate the different evolutionary histories of nifDK on symbiosis islands and non-symbiotic loci. The nucleotide composition analysis of nifDK showed that nifDK on symbiosis islands had a lower GC content than on non-symbiotic loci (Fig. 5A). In addition, the molecular clock test rejected the equal evolutionary rate model throughout the tree (P<0.001). These results suggest two possible scenarios for nifDK on symbiosis islands. One scenario is that nifDK on symbiosis islands was inherited from a different ancestor from non-symbiotic loci with a low GC content. The other scenario is that nifDK on symbiosis islands was inherited from the same ancestor with non-symbiotic loci; however, it has since evolved toward a lower GC content with a higher substitution rate than nifDK on non-symbiotic loci.
Fig. 5.
The average nucleotide composition for concatenated nifDK sequences located on non-symbiotic loci, symbiosis islands, and symbiosis plasmids. The average base compositions of all positions (A), the first codon position (B), second codon position (C), and third codon position (D) are shown in each panel. The error bars represent standard deviations. n=7 for non-symbiotic loci, n=11 for symbiosis islands, n=1 for symbiosis plasmids.
Molecular evolutionary analyses of concatenated nifDK sequences
In order to examine whether the pattern of nucleotide substitution is constant in nifDK on symbiosis islands and non-symbiotic loci, the disparity index tests of substitution pattern homogeneity (12) were performed for each pair of nifDK sequences (Table 2). In the disparity index tests using all (1st+2nd+3rd) codon positions, the null hypothesis that sequences have evolved with the same pattern of substitutions was rejected for all pairs of nifDK sequences in the comparison of symbiosis islands and non-symbiotic loci, suggesting that nifDK on symbiosis islands have evolved via markedly different evolutionary processes from those on non-symbiotic loci. In contrast, the acceptance rate of the null hypothesis was 100% in the comparison of symbiosis plasmids and non-symbiotic loci, suggesting that nifDK on non-symbiotic loci and symbiosis plasmids have evolved via the same or similar evolutionary processes. The acceptance rate of the null hypothesis was 87% among symbiosis islands, suggesting that the substitution pattern may not be constant among nifDK on symbiosis islands. The acceptance rate of the null hypothesis for all pairs of tested strains was 44%, which increased to 99% when only the 1st or 2nd codon position was analyzed, suggesting that nifDK on symbiosis islands and non-symbiotic loci have evolved with the same or similar pattern of substitutions in the 1st and 2nd codon positions only. A nucleotide composition analysis of nifDK showed that nifDK on symbiosis islands had a lower GC content than those in non-symbiotic loci (Fig. 5A). This difference was particularly notable at the 3rd codon position (Fig. 5B, C, and D). The results of disparity index tests and nucleotide composition analyses suggested that nifDK in symbiosis islands and non-symbiotic loci have evolved via similar evolutionary processes at the 1st and 2nd codon positions.
Table 2.
Number of concatenated nifDK sequence pairs for which the null hypothesis was accepted or rejected that sequences have evolved with the same pattern of substitution, as judged from a disparity index test
| Comparison pair | All codon positions (1st+2nd+3rd) | 1st and 2nd codon position | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Number of pairs | Acceptance rate (%) | Number of pairs | Acceptance rate (%) | |||
|
|
|
|||||
| Accept | Reject | Accept | Reject | |||
| Symbiosis island vs. Non-symbiotic loci | 0 | 77 | 0 | 77 | 0 | 100 |
| Symbiosis plasmid vs. Non-symbiotic loci | 7 | 0 | 100 | 7 | 0 | 100 |
| Symbiosis island vs. Symbiosis island | 48 | 7 | 87 | 55 | 0 | 100 |
| Non-symbiotic loci vs. Non-symbiotic loci | 20 | 1 | 95 | 20 | 1 | 95 |
| Symbiosis plasmid vs. Symbiosis island | 0 | 11 | 0 | 11 | 0 | 100 |
|
| ||||||
| Total | 75 | 96 | 44 | 170 | 1 | 99 |
P-values less than 0.01 were considered significant.
We next analyzed the concatenated sequences of nifDK using only the 1st and 2nd codon positions to alleviate the effects of heterogeneous sequence evolution among the tested strains. The tree representing the 1st and 2nd codon positions (Fig. 4B) showed a different topology from the tree representing all positions (Fig. 4A). In the 1st and 2nd codon positions, nifDK on symbiosis islands were clustered into three groups and formed a polyphyly (Fig. 4B). The branch length elongation of nifDK on symbiosis islands was alleviated (Fig. 4A and B), suggesting that the long branches of nifDK on symbiosis islands in the tree representing all codon positions mainly resulted from heterogeneous sequence evolution in the 3rd codon position. nifDK on symbiosis islands was located close to nifDK on non-symbiotic loci, suggesting that nifDK on symbiosis islands was derived from the non-symbiotic loci of Bradyrhizobium or closely related strains. It is also noteworthy that the branches of nifDK on symbiosis islands were still slightly elongated, suggesting that heterogeneous sequence evolution may occur not only in the 3rd codon position, but also in the 1st and 2nd codon positions, which is supported by the molecular clock test showing that the evolutionary rate is not equal throughout the tree in the 1st and 2nd codon positions (P<0.001). Codon-based tests of positive selection showed that the number of synonymous nucleotide substitutions exceeds the number of non-synonymous nucleotide substitutions in all sequence pairs (P>0.05), which will lead to a higher substitution rate in the 3rd codon position containing a large number of synonymous sites. Although nifDK on symbiosis islands formed a polyphyly in the phylogenetic tree representing the 1st and 2nd codon positions (Fig. 4B), this does not necessary indicate a polyphyletic origin of nif–clusters on symbiosis islands because we cannot exclude the possibility that heterogeneous evolution occurred among nifDK genes located on symbiosis islands, which may also lead to a polyphyletic relationship. The hypothesis of the monophyletic origin of nif–clusters in symbiosis islands is supported by previous studies showing highly congruent trees for nifH and nod genes (nodYK, nodA, and nodZ) (13) and high linkage disequilibrium values for nifD, nifH, and nodC in symbiosis islands (21).
The disparity index (Table 2) and nucleotide composition (Fig. 5) analyses indicated that nifDK on symbiosis plasmids are more closely related to nifDK on non-symbiotic loci than to those on symbiosis islands, suggesting different evolutionary histories for nifDK on symbiosis islands and plasmids. However, this hypothesis needs to be carefully evaluated using more strains because only one of the strains tested herein possessed a symbiosis plasmid (Table 1).
Origin of nif genes on the symbiosis island
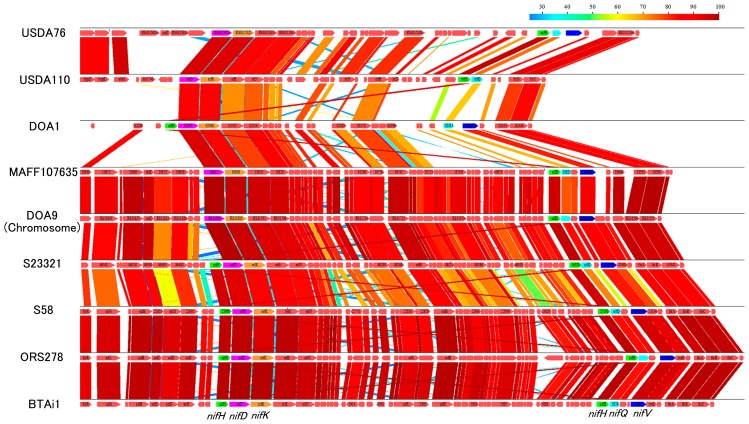
In order to test the plausibility that the origin of the nif–cluster in symbiosis islands is the non-symbiotic loci of Bradyrhizobium, we compared the nif–cluster of strains DOA1, USDA110, and USDA76 in the symbiosis island with those in the non-symbiotic loci of the other Bradyrhizobium strains MAFF107635, DOA9, S23321, S58, ORS278, and BTAi1. The organization of nif–clusters in symbiosis islands was very similar to those on non-symbiotic loci in other strains (Fig. 6), which supports the hypothesis of an origin from the non-symbiotic loci of Bradyrhizobium. In strain DOA1, nifH was located in an operon with nifDK, whereas in strains MAFF107635 and DOA9, nifH was located in an operon with nifQ (Fig. 6). In strains S23321, S58, ORS278, and BTAi1, nifH was located in operons with nifDK and nifQ (Fig. 6). These results suggest that the nif–cluster in the symbiosis island of strain DOA1 was laterally transferred from a strain possessing only the nifHDK operon. However, none of the tested strains only possess the nifHDK operon in non-symbiotic loci. These results suggest two possibilities: a rearrangement of gene organization occurred in the nif–cluster of strain DOA1 or we were unable to identify the strains possessing only the nifHDK operon because of the limited number of strains tested. Strains USDA110 and USDA76 only possess the nifHQ operon (Fig. 6). This feature was observed in the nif–clusters of strains MAFF107635 and DOA9 on non-symbiotic loci. The nif–cluster of strains DOA1 and USDA76 possesses nifV, which encodes homocitrate synthase (Fig. 6). Homocitrate is a component of the iron-molybdenum cofactor of nitrogenase (9). Most rhizobial strains do not possess nifV, which is complemented by homocitrate provided from the host plant during nodule symbiosis (7). The presence of nifV in strains DOA1 and USDA76 partially supports the hypothesis that the nif–cluster on symbiosis islands was derived from the non-symbiotic loci of Bradyrhizobium or closely related strains. However, we cannot absolutely rule out the possibility that nif genes on symbiosis islands and plasmids serve as the donors of nif genes on non-symbiotic loci. This possibility needs to be carefully examined by studying more Bradyrhizobium strains that possess nif genes at non-symbiotic loci.
Fig. 6.
Blastp comparisons of nif–clusters. The color scale indicates the percent similarity. The arrows show each gene. nifH genes are shown in green, nifD genes are shown in magenta, nifK genes are shown in orange, nifQ genes are shown in cyan, nifV genes are shown in blue.
Conclusion
The findings of previous studies suggest that nif genes in symbiosis islands were acquired by lateral gene transfer because these genes are located in a symbiosis island, which is characterized by a lower GC content than the rest of the genome (10, 11). In the present study, we inferred the origin and evolutionary process of nif genes in Bradyrhizobium strains. Our analyses indicate that nif genes on symbiosis islands and plasmids were derived from the non-symbiotic loci of Bradyrhizobium or closely related strains. Following gene transfer, nif genes on symbiosis islands evolved faster than those in the ancestral state toward a lower GC content; however, we cannot absolutely rule out the possibility that nif genes on symbiosis islands were transferred from bacteria with a low genomic GC content, closely related to Bradyrhizobium. In contrast, no apparent substitution bias was observed in the nif genes on symbiosis plasmids. Previous studies (2, 6, 8) analyzed nif genes across diverse taxonomic groups of α- and β-proteobacteria, and reported that rhizobial and non-rhizobial nitrogen-fixing strains were clustered in the same clade in phylogenetic trees. However, the nif genes of nodulation bacteria are closely related to those of non-rhizobial strains of the same or taxonomically-related groups. These results suggest that the translocation or lateral transmission of nif genes from non-symbiotic loci to symbiosis islands or plasmids occurred not only in Bradyrhizobium, but also in other rhizobial groups. Due to the limited number of complete or high-quality draft Bradyrhizobium genomes, our data set does not cover all Bradyrhizobium species. Further studies using diverse strains and genes will contribute to developing a greater understanding of the origin and evolution of legume–rhizobia symbiosis.
Acknowledgements
We thank Dr. Junko Terakado-Tonooka for her constructive comments and the GenBank submission staff performing the automatic annotation of DOA1 and MAFF107635 genomes by PGAP. This work was supported by JSPS KAKENHI Grant Number 269473 to T. Okubo and (A) 23248052 and 26252065 to K. Minamisawa, and by Suranaree University of Technology (SUT) and the Office of the Higher Education Commission under NRU project of Thailand. T. Okubo is a Research Fellow of the Japan Society for the Promotion of Science.
Abbreviations
- GC
guanine and cytosine
- nif genes
nitrogen fixation gene
- nod genes
nodulation genes
- MLSA
multilocus sequence analysis
References
- 1.Angiuoli S.V., Gussman A., Klimke W., et al. Toward an online repository of Standard Operating Procedures (SOPs) for (meta) genomic annotation. Omics. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki S., Ito M., Iwasaki W. From β- to α-proteobacteria: the origin and evolution of rhizobial nodulation genes nodIJ. Mol Biol Evol. 2013;30:2494–2508. doi: 10.1093/molbev/mst153. [DOI] [PubMed] [Google Scholar]
- 3.Cole M.A., Elkan G.H. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother. 1973;4:248–253. doi: 10.1128/aac.4.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delamuta J.R., Ribeiro R.A., Menna P., Bangel E.V., Hungria M. Multilocus sequence analysis (MLSA) of Bradyrhizobium strains: revealing high diversity of tropical diazotrophic symbiotic bacteria. Braz J Microbiol. 2012;43:698–710. doi: 10.1590/S1517-83822012000200035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraud E., Moulin L., Vallenet D.V., et al. Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 6.Gyaneshwar P., Hirsch A.M., Moulin L., et al. Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol Plant Microbe Interact. 2011;24:1276–1288. doi: 10.1094/MPMI-06-11-0172. [DOI] [PubMed] [Google Scholar]
- 7.Hakoyama T., Niimi K., Watanabe H., et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature. 2009;462:514–517. doi: 10.1038/nature08594. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann L.S., Barnum S.R. Inferring the evolutionary history of Mo-dependent nitrogen fixation from phylogenetic studies of nifK and nifDK. J Mol Evol. 2010;71:70–85. doi: 10.1007/s00239-010-9365-8. [DOI] [PubMed] [Google Scholar]
- 9.Hoover T.R., Imperial J., Ludden P.W., Shah V.K. Homocitrate is a component of the iron-molybdenum cofactor of nitrogenase. Biochemistry. 1989;28:2768–2771. doi: 10.1021/bi00433a004. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko T., Nakamura Y., Sato S., et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko T., Maita H., Hirakawa H., Uchiike N., Minamisawa K., Watanabe A., Sato S. Complete genome sequence of the soybean symbiont Bradyrhizobium japonicum strain USDA6T. Genes. 2011;2:763–787. doi: 10.3390/genes2040763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S., Gadagkar S.R. Disparity index: a simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics. 2001;158:1321–1327. doi: 10.1093/genetics/158.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menna P., Hungria M. Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int J Syst Evol Microbiol. 2011;61:3052–3067. doi: 10.1099/ijs.0.028803-0. [DOI] [PubMed] [Google Scholar]
- 14.Minamisawa K. Division of rhizobitoxine-producing and hydrogen-uptake positive strains of Bradyrhizobium japonicum by nifDKE sequence divergence. Plant Cell Physiol. 1990;31:81–89. [Google Scholar]
- 15.Noisangiam R., Teamtisong K., Tittabutr P., Boonkerd N., Toshiki U., Minamisawa K., Teaumroong N. Genetic diversity, symbiotic evolution, and proposed infection process of Bradyrhizobium strains isolated from root nodules of Aeschynomene americana L. in Thailand. Appl Environ Microbiol. 2012;78:6236–6250. doi: 10.1128/AEM.00897-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okazaki S., Noisangiam R., Okubo T., et al. Genome analysis of a novel Bradyrhizobium sp. DOA9 carrying a symbiotic plasmid. PLoS One. 2015;10:e0117392. doi: 10.1371/journal.pone.0117392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okubo T., Fukushima S., Minamisawa K. Evolution of Bradyrhizobium-Aeschynomene mutualism: living testimony of the ancient world or highly evolved state? Plant Cell Physiol. 2012;53:2000–2007. doi: 10.1093/pcp/pcs150. [DOI] [PubMed] [Google Scholar]
- 19.Okubo T., Tsukui T., Maita H., et al. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ. 2012;27:306–315. doi: 10.1264/jsme2.ME11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okubo T., Fukushima S., Itakura M., et al. Genome analysis suggests that the soil oligotrophic bacterium Agromonas oligotrophica (Bradyrhizobium oligotrophicum) is a nitrogen-fixing symbiont of Aeschynomene indica. Appl Environ Microbiol. 2013;79:2542–2551. doi: 10.1128/AEM.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker M.A. Legumes select symbiosis island sequence variants in Bradyrhizobium. Mol Ecol. 2012;21:1769–1778. doi: 10.1111/j.1365-294X.2012.05497.x. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teamtisong K., Songwattana P., Noisangiam R. Divergent nod-containing Bradyrhizobium sp. DOA9 with a megaplasmid and its host range. Microbes Environ. 2014;29:4. doi: 10.1264/jsme2.ME14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terakado-Tonooka J., Fujihara S., Ohwaki Y. Possible contribution of Bradyrhizobium on nitrogen fixation in sweet potatoes. Plant Soil. 2013;367:639–650. [Google Scholar]