Abstract
The syntrophic degradation of branched-chain fatty acids (BCFAs) such as 2-methylbutyrate and isobutyrate is an essential step in the production of methane from proteins/amino acids in anaerobic ecosystems. While a few syntrophic BCFA-degrading bacteria have been isolated, their metabolic pathways in BCFA and short-chain fatty acid (SCFA) degradation as well as energy conservation systems remain unclear. In an attempt to identify these pathways, we herein performed comparative genomics of three syntrophic bacteria: 2-methylbutyrate-degrading “Syntrophomonas wolfei subsp. methylbutyratica” strain JCM 14075T (=4J5T), isobutyrate-degrading Syntrophothermus lipocalidus strain TGB-C1T, and non-BCFA-metabolizing S. wolfei subsp. wolfei strain GöttingenT. We demonstrated that 4J5 and TGB-C1 both encode multiple genes/gene clusters involved in β-oxidation, as observed in the Göttingen genome, which has multiple copies of genes associated with butyrate degradation. The 4J5 genome possesses phylogenetically distinct β-oxidation genes, which may be involved in 2-methylbutyrate degradation. In addition, these Syntrophomonadaceae strains harbor various hydrogen/formate generation systems (i.e., electron-bifurcating hydrogenase, formate dehydrogenase, and membrane-bound hydrogenase) and energy-conserving electron transport systems, including electron transfer flavoprotein (ETF)-linked acyl-CoA dehydrogenase, ETF-linked iron-sulfur binding reductase, ETF dehydrogenase (FixABCX), and flavin oxidoreductase-heterodisulfide reductase (Flox-Hdr). Unexpectedly, the TGB-C1 genome encodes a nitrogenase complex, which may function as an alternative H2 generation mechanism. These results suggest that the BCFA-degrading syntrophic strains 4J5 and TGB-C1 possess specific β-oxidation-related enzymes for BCFA oxidation as well as appropriate energy conservation systems to perform thermodynamically unfavorable syntrophic metabolism.
Keywords: syntroph, branched-chain fatty acid, genomics, energy conservation
Under methanogenic conditions, the degradation of amino acids and proteinaceous materials inevitably generates fatty acids as byproducts (26). Fatty acid-oxidizing bacteria and methanogens are known to form syntrophic interactions in order to accomplish the endergonic oxidation of these fatty acids (9, 19, 26). Although the biochemical pathways and genes involved in the syntrophic degradation of short-chain fatty acids (SCFA; e.g., propionate and butyrate) have already been described, they have not yet been elucidated for branched-chain fatty acids (BCFAs; e.g., isobutyrate, isovalerate, and 2-methylbutyrate) derived from branched-chain amino acids (13, 26). Syntrophic BCFA degradation to acetate and propionate has been observed in isolates and mixed cultures (14, 26, 34, 35, 41). Only three strains of the family Syntrophomonadaceae are currently known to syntrophically degrade 2-methylbutyrate (“Syntrophomonas wolfei subsp. methylbutyratica” strain JCM 14075T (=4J5T) and S. bryantii strain CuCalT) and isobutyrate (Syntrophothermus lipocalidus strain TGB-C1T) (29, 33, 40). These Syntrophomonadaceae species are considered to be important for fatty acid degradation in anaerobic ecosystems, including the sludge digestion process (19), rice paddy fields (12), and the termite gut (42). Furthermore, an uncultivated Syntrophaceae member has been proposed to degrade BCFA syntrophically in a methanogenic bioreactor through metagenomic and metatranscriptomic approaches (21). However, the key catabolic enzymes and energy conservation systems necessary to drive thermodynamically unfavorable BCFA and SCFA degradation remain unclear.
In the present study, the genomes of strains 4J5 (20) and TGB-C1 (4) were investigated in order to identify the metabolic pathways for 2-methylbutyrate and isobutyrate catabolism and energy conservation systems for syntrophic metabolism. A comparative genomic analysis between BCFA-and non-BCFA-degrading syntrophs within the family Syntrophomonadaceae (i.e., S. wolfei subsp. wolfei strain GöttingenT [30]) provides genomic insights into the degradation of BCFA in methanogenic ecosystems.
Materials and Methods
Genome sequencing and annotation
This study analyzed the “S. wolfei subsp. methylbutyratica” strain JCM 14075T (=4J5T) draft genome (DDBJ/GenBank/EMBL accession: BBQT01000001–BBQT01000092) (20), S. lipocalidus strain TGB-C1T complete genome (CP002048) (4), and S. wolfei strain Göttingen complete genome (CP000448) (30). As reported previously (20), the genomic DNA of strain 4J5 was provided by the RIKEN BRC through the National Bio-Resource Project of MEXT, Japan, and sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA) at FASMAC (Atsugi, Japan). Briefly, we constructed and sequenced a 300-bp paired-end library totaling ca. 2.2 Gb of MiSeq data. Assemblies were performed using SPAdes version 3.1.1 (2). The strain 4J5 draft genome comprises 89 scaffolds and has an estimated genome size of 3.2 Mbp with an average G+C content of 45.55%. The quality of the genome sequence was evaluated using the Check M version 1.0.5 program with a marker gene set of the class Clostridia (23). A total of 2,964 protein coding genes were annotated with Prokka version 1.11 (see Supplemental Information) (28). Basic local alignment search tool (BLAST) (ver. 2.2.30) with a non-redundant protein sequence database (nr) and the protein sequence database of Göttingen and TGB-C1 (11) and BLASTKoala of Kyoto Encyclopedia of Genes and Genomes (KEGG) (8) were used to search for functional domains and characterize potential protein functions. Proteins associated with energy conservation systems were identified by criteria based on the genomic and physiological information of previously reported energy conservation pathways (21). Transport systems were identified using TransportDB (25).
Results and Discussion
BCFA degradation
The Syntrophomonadaceae strains 4J5 (40), TGB-C1 (29), and Göttingen (1, 15, 16) have very similar genetic compositions (see Supplemental information, Table S1, S2, and S3), reflecting previously observed physiological similarities, such as central metabolic pathways (e.g., glycolysis and the TCA cycle) and pilus and flagellum formation. Syntrophic BCFA degradation is considered to proceed through β-oxidation (13, 21), as observed for butyrate degradation by the strain Göttingen (16, 27, 30, 38). These Syntrophomonadaceae genomes all encode multiple genes for β-oxidation (Fig. 1, Table S4), potentially with varying specificities to particular fatty acid structures (i.e., alkyl branching, chain length, and saturation), affinities to specific concentration ranges, or adaptation to other environmental conditions (30). Strain 4J5 possesses several β-oxidation genes with high similarities (>90%) to those in Göttingen, presumably involved in butyrate metabolism (Table S4, Fig. S1) (27, 32). However, we also identified several strain 4J5 β-oxidation-related genes with relatively low similarities to those of Göttingen (42.3%–62.6% amino acid identity), which may specifically be involved in 2-methylbutyrate degradation—acyl-CoA dehydrogenase (Swmb_1942), enoyl-CoA hydratases (Swmb_01703 and Swmb_03023), 3-hydroxybutyryl-CoA dehydrogenase (Swmb_01947), and acetyl-CoA C-acetyltransferase (Swmb_02509). The TGB-C1 genome encodes several β-oxidation genes not only with relatively high similarities (30–85%), but also with no significant similarity (<30%) to those in the mesophilic strains Göttingen and 4J5 (Table S5), implying that TGB-C1 drives the β-oxidation pathway by using thermostable/thermophilic enzymes. Further biochemical, transcriptomic, and/or proteomic analyses are needed in order to clarify the activities and specificities of the β-oxidation-related enzymes of these strains.
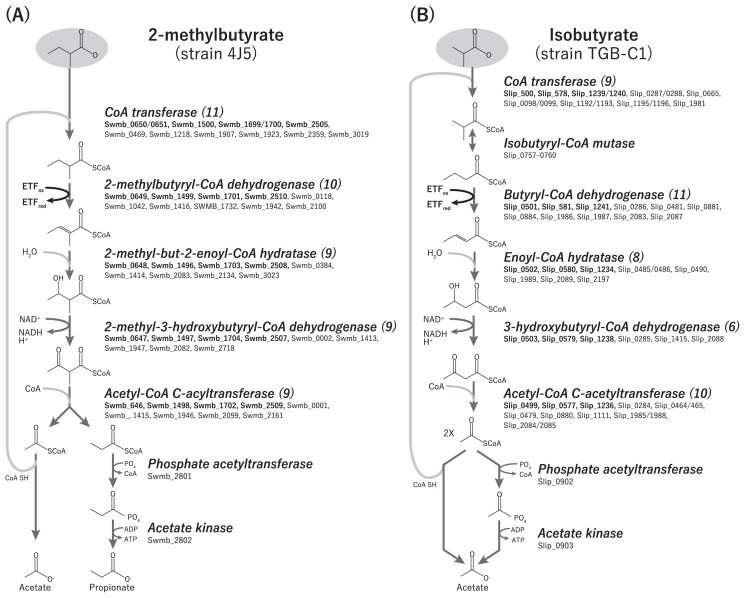
Fig. 1.
Branched-chain fatty acid metabolism in (A) “Syntrophomonas wolfei subsp. methylbutyratica” strain 4J5 and (B) Syntrophothermus lipocalidus strain TGB-C1. Each reaction is labeled with the protein name (italicized) of the gene encoding the function and locus tag(s). Figures in brackets indicate the number of multiple gene copies. Four (Swmb_00646–00651, Swmb1699–1704, Swmb_1496–1500, and Swmb_2505–02510) and three (Slip_0499–0503, Slip_0577–0581, and Slip_1236–1241) β-oxidation gene clusters were identified in strains 4J5 and TGB-C1, respectively (shown in bold face).
In syntrophic isobutyrate degradation, strain TGB-C1 has been reported to perform alkyl isomerization in order to form butyrate for subsequent β-oxidation (29). Strain TGB-C1 encodes an isobutyryl-CoA mutase gene cluster including the C- and N-terminal domain proteins, cob(I)alamin adenosyltransferase and MeaB-like protein (Slip_0757–0760) to facilitate the rearrangement step (Fig. 1B, Table S5). Homologous gene clusters have been found in “Ca. Caldisyntrophus multiacidovorans”, a syntrophic BCFA degrader in a lab-scale anaerobic bioreactor (21) and other prokaryotes (Fig. S2). The isobutyryl-CoA mutase genes of TGB-C1 and “Ca. Caldisyntrophus” are phylogenetically related to a group associated with sulfate-reducing bacteria (Fig. S2), suggesting potential evolutionary relationships between isobutyrate-isomerizing syntrophs and sulfate reducers.
As final products of the predicted β-oxidation pathways, the syntrophic degradation of isobutyrate generates two acetyl-CoA and 2-methylbutyrate produces acetyl-CoA and propionyl-CoA (Fig. 1A). Acetyl-CoA yields ATP through dethiolation to acetate by phosphate acetyltransferase (Swmb_02801 and Slip_0902) and acetate kinase (Swmb_02802 and Slip_0903). Regarding strain 4J5, these enzymes may perform the dethiolation of 2-methylbutyrate-derived propionyl-CoA because the active site structure of previously known propionate kinase resembles those of acetate kinase and butyrate kinase (7). AMP-dependent acyl-CoA synthetases found in the 4J5 and TGB-C1 genomes (Swmb_02363 and Swmb_02710; Slip_0475, Slip_0583, and Slip_1686) potentially serve as an alternative acyl-CoA degradation pathway, as suggested by McInerney et al. (17). However, Swmb_02710 and Slip_1686 have high identities (>64% by amino acid sequences) to that of strain Göttingen (Swol_1180), which has been predicted to function in biosynthesis (30). The other homolog found in 4J5 (Swmb_02363) may be involved in poly-β-hydroxybutyrate metabolism due to an association with the poly-β-hydroxybutyrate polymerase gene, as observed in strain Göttingen (Swol_1144). The remaining TGB-C1 homologs (Slip_0475 and Slip_0583) have low amino acid sequence identities (<32%) with the biosynthesis-associated acyl-CoA synthetase (Table S4), implying that these acyl-CoA synthetase genes are responsible for the production of acetate from acetyl-CoA through the degradation of isobutyrate.
Energy conservation and electron flow
A syntrophic substrate metabolizer uses energy conservation systems, such as reverse election transfer and electron bifurcation, to conduct thermodynamically unfavorable proton (H+) reduction to hydrogen (H2) (26, 31). Since the 4J5, Göttingen, and TGB-C1 genomes lack the Rhodobacter nitrogen fixation (Rnf) complex, these Syntrophomonadaceae syntrophs may not perform reverse electron transport from NADH to oxidized ferredoxin (Fdox) through the Rnf found in several syntrophs (e.g., Syntrophus and Syntrophobacter) (17, 39). Instead, we identified other types of reverse electron transport systems employing electron transfer flavoprotein (ETF) dehydrogenases described in the Göttingen genome: ETF-linked iron-sulfur binding (Fe-S) reductase and ETF-oxidizing hydrogenase complex (FixABCX) systems (6, 10, 22, 30) (Fig. 2A, Table S6). These results support a previous finding on the expression of Fe-S reductase with strain Göttingen during syntrophic butyrate degradation (18, 27). Although these Syntrophomonadaceae genomes also encode an electron-bifurcating ETF-associated butyryl-CoA dehydrogenase (Bcd/EtfAB), this system may support the capacities of these organisms to perform crotonate reduction (10, 18). In addition, the Syntrophomonadaceae syntrophs encode a syntroph-associated flavin oxidoreductase-heterodisulfide reductase (Flox-Hdr) system (22, 24) expressed by the BCFA-degrading syntroph “Ca. Caldisyntrophus multiacidovorans” and aromatic compound-degrading syntrophs (Pelotomaculum spp. and Syntrophorhabdus spp.) along with FixABCX (21), suggesting the involvement of these energy conservation systems in the syntrophic degradation of various organic compounds including BCFAs.
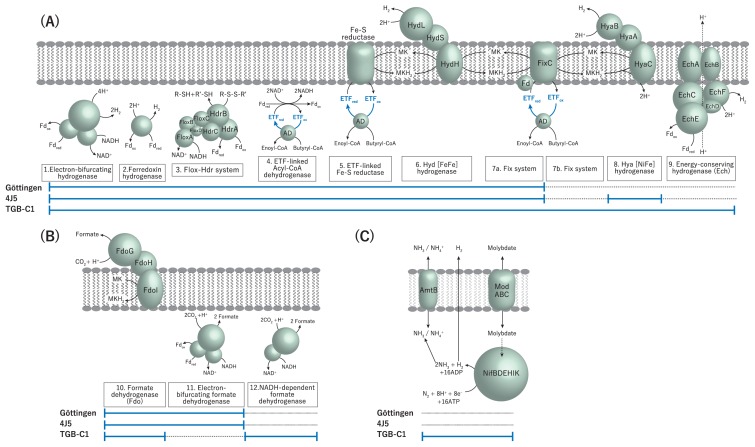
Fig. 2.
Hydrogenases, formate dehydrogenases, and energy conservation systems of Syntrophomonas wolfei subsp. wolfei strain Göttingen, “Syntrophomonas wolfei subsp. methylbutyratica” strain 4J5, and Syntrophothermus lipocalidus strain TGB-C1. (A) Hydrogenases and reverse electron transport systems including (1) electron-bifurcating hydrogenase, (2) ferredoxin hydrogenase, (3) flavin oxidoreductase-heterodisulfide reductase (flox-Hdr), (4) electron-transfer-flavoprotein (ETF)-linked acyl-CoA dehydrogenase, (5) ETF-linked iron-sulfur binding (Fe-S) reductase, (6) [FeFe] hydrogenase (Hyd), (7) ETF-oxidizing hydrogenase complex (FixABCX), (8) [NiFe] hydrogenase (Hya), and (9) energy-conserving hydrogenase (Ech). (B) Formate dehydrogenases including (10) formate dehydrogenase (Fdo), (11) electron-bifurcating formate dehydrogenase, and (12) NADH-dependent formate dehydrogenase. (C) A possible nitrogenase-mediated hydrogen generation system. The solid line indicates the existence of corresponding genes in the strains, while the dotted line indicates the absence of these genes.
There may be multiple hydrogenase and formate dehydrogenase genes in the 4J5 and TGB-C1 genomes (Fig. 2, Table S6). Both strains harbor systems for hydrogen generation through electron-bifurcating Fe-only hydrogenase (NADH/Fd-oxidizing), Hyd hydrogenase (quinol), and cytoplasmic hydrogenase (Fd) with relatively high similarities (>68%) to those of strain Göttingen (30). In addition, the genomes of strains 4J5 and TGB-C1 encode the Hya hydrogenase gene cluster (Swmb_01823–01933 and Slip_0383–0392), which are lacking in the Göttingen genome. Strain TGB-C1 also has an energy-conserving hydrogenase (Ech; Fd-oxidizing) gene cluster (Slip_0655–0660). Regarding formate generation, we identified putative electron-bifurcating formate dehydrogenases (37) in strain 4J5, cytoplasmic formate dehydrogenase (NADH-oxidizing) in strain TGB-C1, and Fdo formate dehydrogenase (quinol-oxidizing) in both. These results suggest that BCFA-degrading syntrophs flexibly employ multiple energy conservation systems to maintain a syntrophic lifestyle in thermodynamically limited environments.
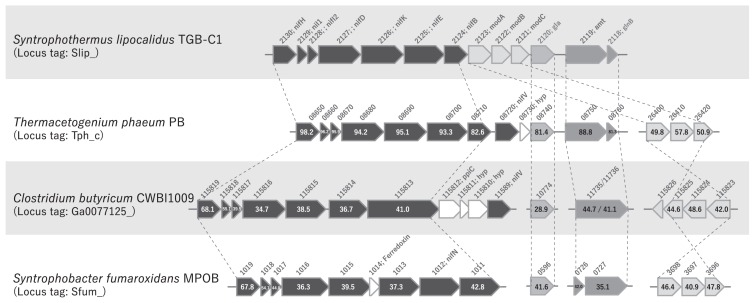
A nitrogenase gene cluster (NifBDEHIK) was detected in the genome of strain TGB-C1 (Slip_2124–2130) along with a molybdate transporter (ModABC, Slip_2121–2123) and ammonia transporter (AmtB, Slip_2119) (Fig. 2C, Table S7). A previous microbial genome survey revealed that nitrogen fixation-related proteins (Nif) are distributed in phylogenetically diverse microbes including some fermentative bacteria and syntrophic substrate metabolizers (5). The nitrogenase activity of the fermentative H2-producing organism Clostridium butyricum strain CWBI1009 has been proposed to generate H2 and enhance tolerance to acidification through the consumption of protons in the reaction, which produces ammonia as a base (3). We observed high amino acid sequence similarities (up to 98%) between the nitrogenase, molybdate transporter, and ammonia transporter genes of TGB-C1 and CWBI1009, and also moderate similarities (up to 68%) with two other Nif-encoding syntrophs (Thermacetogenium phaeum strain PB and Syntrophobacter fumaroxidans strain MPOB) (Fig. 3, Table S7). Among syntrophs, nitrogen fixation may serve as a mechanism to tolerate acidification and provide hydrogen and ammonia for partner hydrogenotrophic methanogens to survive under hydrogen/ammonia-limited conditions.
Fig. 3.
Comparison of nitrogenase-like gene cassettes found in Syntrophothermus lipocalidus strain TGB-C1, Thermacetogenium phaeum strain PB, Clostridium butyricum strain CWBI1009, and Syntrophobacter fumaroxidans strain MPOB. Strain TGB-C1 nitrogenase-associated cassette encodes nitrogen fixation proteins (NifBDEHIK), glutamine amidotransferase class-I (gla), a molybdate transporter (ModABC), and ammonia transporter (amt) with a nitrogen regulatory protein P-II family (glnB). Other strains harbor NifV (strains PB and CWBI1009), peptidyl-prolyl cis-trans isomerase C (ppiC) (strain PB), ferredoxin (strain MPOB), and hypothetical protein (hyp) (strains PB and CWBI1009) within the cassette. Abbreviated locus tags are shown (e.g., ‘Slip_2130’ as ‘2130’ in the row of strain TGB-C1). Figures in gene boxes indicate amino acid sequence identity to the corresponding gene of strain TGB-C1.
In summary, a comparative genomic analysis of the Syntrophomonadaceae strains revealed multiple genes for the β-oxidation of BCFA/SCFA and energy conservation systems employing various types of electron carriers for electron disposal. In theory, the syntrophic degradation of 2-methylbutyrate and isobutyrate have Gibbs free energy values identical to butyrate under standard conditions (calculations based on Thauer et al. [36]):
Thus, while Syntrophomonadaceae strains degrading the above fatty acids have unique substrate-specific genes (e.g., β-oxidation), they share similar energy conservation systems reflecting the thermodynamic similarity shown above. TGB-C1 has several distinct energy conservation pathways from Göttingen and 4J5 potentially due to differences in their temperature preferences. These genome-based results further provide insights into how syntrophic BCFA-degraders interact with H2/formate-utilizing methanogen in an anaerobic ecosystem.
Supplementary Information
Acknowledgements
This research was supported by JSPS KAKENHI (Grant Number 26630250, 26710012, 26106004, and 15H05331). We are grateful to Aya Akiba (AIST) for her technical assistance. MKN was supported by JSPS Fellowship Programs for Overseas Researchers.
References
- 1.Amos D.A., McInerney M.J. Poly-β-hydroxyalkanoate in Syntrophomonas wolfei. Arch Microbiol. 1989;152:172–177. [Google Scholar]
- 2.Bankevich A., Nurk S., Antipov D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calusinska M., Hamilton C., Monsieurs P., et al. Genome-wide transcriptional analysis suggests hydrogenase- and nitrogenase-mediated hydrogen production in Clostridium butyricum CWBI 1009. Biotechnol Biofuels. 2015;8:27. doi: 10.1186/s13068-015-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djao O.D.N., Zhang X.J., Lucas S., et al. Complete genome sequence of Syntrophothermus lipocalidus type strain (TGB-C1(T)) Stand Genomic Sci. 2010;3:267–275. doi: 10.4056/sigs.1233249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dos Santos P.C., Fang Z., Mason S.W., Setubal J.C., Dixon R. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics. 2012;13:162. doi: 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann G., Jayamani E., Mai G., Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingram-Smith C., Gorrell A., Lawrence S.H., Iyer P., Smith K., Ferry J.G. Characterization of the acetate binding pocket in the Methanosarcina thermophila acetate kinase. J Bacteriol. 2005;187:2386–2394. doi: 10.1128/JB.187.7.2386-2394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato S., Watanabe K. Ecological and evolutionary interactions in syntrophic methanogenic consortia. Microbes Environ. 2010;25:145–151. doi: 10.1264/jsme2.me10122. [DOI] [PubMed] [Google Scholar]
- 10.Li F., Hinderberger J., Seedorf H., Zhang J., Buckel W., Thauer R.K. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P.F., Qiu Q.F., Lu Y.H. Syntrophomonadaceae- affiliated species as active butyrate-utilizing syntrophs in paddy field soil. Appl Environ Microbiol. 2011;77:3884–3887. doi: 10.1128/AEM.00190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massey L.K., Sokatch J.R., Conrad R.S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976;40:42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthies C., Schink B. Reciprocal isomerization of butyrate and isobutyrate by the strictly anaerobic bacterium strain WoG13 and methanogenic isobutyrate degradation by a defined triculture. Appl Environ Microbiol. 1992;58:1435–1439. doi: 10.1128/aem.58.5.1435-1439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McInerney M.J., Bryant M.P., Hespell R.B., Costerton J.W. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty-acid oxidizing bacterium. Appl Environ Microbiol. 1981;41:1029–1039. doi: 10.1128/aem.41.4.1029-1039.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McInerney M.J., Wofford N.Q. Enzymes involved in crotonate metabolism in Syntrophomonas wolfei. Arch Microbiol. 1992;158:344–349. [Google Scholar]
- 17.McInerney M.J., Rohlin L., Mouttaki H., et al. The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller N., Schleheck D., Schink B. Involvement of NADH:acceptor oxidoreductase and butyryl coenzyme A dehydrogenase in reversed electron transport during syntrophic butyrate oxidation by Syntrophomonas wolfei. J Bacteriol. 2009;191:6167–6177. doi: 10.1128/JB.01605-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narihiro T., Nobu M.K., Kim N.K., Kamagata Y., Liu W.T. The nexus of syntrophy-associated microbiota in anaerobic digestion revealed by long-term enrichment and community survey. Environ Microbiol. 2015;17:1707–1720. doi: 10.1111/1462-2920.12616. [DOI] [PubMed] [Google Scholar]
- 20.Narihiro T., Nobu M.K., Tamaki H., Kamagata Y., Liu W.T. Draft genome sequence of Syntrophomonas wolfei subsp. methylbutyratica strain 4J5 (JCM 14075), a mesophilic butyrate and 2-methybutyrate degrading syntroph. Genome Announc. 2016;4:e00047-16. doi: 10.1128/genomeA.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nobu M.K., Narihiro T., Rinke C., Kamagata Y., Tringe S.G., Woyke T., Liu W.T. Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J. 2015;9:1710–1722. doi: 10.1038/ismej.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobu M.K., Narihiro T., Tamaki H., et al. The genome of Syntrophorhabdus aromaticivorans strain UI provides new insights for syntrophic aromatic compound metabolism and electron flow. Environ Microbiol. 2015;17:4861–4872. doi: 10.1111/1462-2920.12444. [DOI] [PubMed] [Google Scholar]
- 23.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira I.A., Ramos A.R., Grein F., Marques M.C., da Silva S.M., Venceslau S.S. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol. 2011;2:69. doi: 10.3389/fmicb.2011.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren Q., Chen K., Paulsen I.T. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schink B., Stams A.J.M. Syntrophism among prokaryotes. In: Rosenberg E., DeLong E., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes. 4th ed. Springer-Verlag; Berlin Heidelberg: 2013. pp. 471–493. [Google Scholar]
- 27.Schmidt A., Muller N., Schink B., Schleheck D. A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PLoS One. 2013;8:e56905. doi: 10.1371/journal.pone.0056905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 29.Sekiguchi Y., Kamagata Y., Nakamura K., Ohashi A., Harada H. Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int J Syst Evol Microbiol. 2000;50:771–779. doi: 10.1099/00207713-50-2-771. [DOI] [PubMed] [Google Scholar]
- 30.Sieber J.R., Sims D.R., Han C., et al. The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production. Environ Microbiol. 2010;12:2289–2301. doi: 10.1111/j.1462-2920.2010.02237.x. [DOI] [PubMed] [Google Scholar]
- 31.Sieber J.R., McInerney M.J., Gunsalus R.P. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 32.Sieber J.R., Crable B.R., Sheik C.S., Hurst G.B., Rohlin L., Gunsalus R.P., McInerney M.J. Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei. Front Microbiol. 2015;6:115. doi: 10.3389/fmicb.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stieb M., Schink B. Anaerobic oxidation of fatty-acids by Clostridium bryantii sp. nov., a sporeforming, obligately syntrophic bacterium. Arch Microbiol. 1985;140:387–390. [Google Scholar]
- 34.Stieb M., Schink B. Anaerobic degradation of isovalerate by a defined methanogenic coculture. Arch Microbiol. 1986;144:291–295. [Google Scholar]
- 35.Stieb M., Schink B. Anaerobic degradation of isobutyrate by methanogenic enrichment cultures and by a Desulfococcus multivorans strain. Arch Microbiol. 1989;151:126–132. [Google Scholar]
- 36.Thauer R.K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Huang H., Kahnt J., Thauer R.K. Clostridium acidurici electron-bifurcating formate dehydrogenase. Appl Environ Microbiol. 2013;79:6176–6179. doi: 10.1128/AEM.02015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wofford N.Q., Beaty P.S., McInerney M.J. Preparation of cell-free extracts and the enzymes involved in fatty acid metabolism in Syntrophomonas wolfei. J Bacteriol. 1986;167:179–185. doi: 10.1128/jb.167.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worm P., Stams A.J., Cheng X., Plugge C.M. Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiology. 2011;157:280–289. doi: 10.1099/mic.0.043927-0. [DOI] [PubMed] [Google Scholar]
- 40.Wu C., Dong X., Liu X. Syntrophomonas wolfei subsp. methylbutyratica subsp. nov., and assignment of Syntrophomonas wolfei subsp. saponavida to Syntrophomonas saponavida sp. nov. comb. nov. Syst Appl Microbiol. 2007;30:376–380. doi: 10.1016/j.syapm.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Wu W.M., Jain M.K., Zeikus J.G. Anaerobic degradation of normal- and branched-chain Fatty acids with four or more carbons to methane by a syntrophic methanogenic triculture. Appl Environ Microbiol. 1994;60:2220–2226. doi: 10.1128/aem.60.7.2220-2226.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H., Bodington D., Zhang C., Miyanaga K., Tanji Y., Hongoh Y., Xing X.H. Comprehensive phylogenetic diversity of [FeFe]-hydrogenase genes in termite gut microbiota. Microbes Environ. 2013;28:491–494. doi: 10.1264/jsme2.ME13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.