Abstract
Pseudogulbenkiania is a relatively recently characterized genus within the order Neisseriales, class Betaproteobacteria. This genus contains several strains that are capable of anaerobic, nitrate-dependent Fe(II) oxidation (NDFO), a geochemically important reaction for nitrogen and iron cycles. In the present study, we examined denitrification functional gene diversities within this genus, and clarified whether other Pseudogulbenkiania sp. strains perform denitrification and NDFO. Seventy strains were analyzed, including two type strains, a well-characterized NDFO strain, and 67 denitrifying strains isolated from various rice paddy fields and rice-soybean rotation fields in Japan. We also attempted to identify the genes responsible for NDFO by mutagenesis. Our comprehensive analysis showed that all Pseudogulbenkiania strains tested performed denitrification and NDFO; however, we were unable to obtain NDFO-deficient denitrifying mutants in our mutagenesis experiment. This result suggests that Fe(II) oxidation in these strains is not enzymatic, but is caused by reactive N-species that are formed during nitrate reduction. Based on the results of the comparative genome analysis among Pseudogulbenkiania sp. strains, we identified low sequence similarity within the nos gene as well as different gene arrangements within the nos gene cluster, suggesting that nos genes were horizontally transferred. Since Pseudogulbenkiania sp. strains have been isolated from various locations around the world, their denitrification and NDFO abilities may contribute significantly to nitrogen and iron biogeochemical cycles.
Keywords: Pseudogulbenkiania, nitrate-dependent Fe(II) oxidation, denitrification, mutagenesis, comparative genomics
The genus Pseudogulbenkiania, which belongs to the family Chromobacteriaceae (1), order Neisseriales, class Betaproteobacteria, was first proposed by Lin et al. (27). Two species have since been reported: Pseudogulbenkiania subflava isolated from a cold spring in Taiwan (27) and Pseudogulbenkiania gefcensis isolated from soil in South Korea (26).
The genus Pseudogulbenkiania also contains the anaerobic, nitrate-dependent Fe(II)-oxidizing bacterium strain 2002 (also called “Pseudogulbenkiania ferrooxidans”), isolated from a freshwater lake in Illinois, USA (42, 44), and strain MAI-1, isolated from a freshwater lake in Indonesia (23). Anaerobic, nitrate-dependent Fe(II) oxidation (NDFO) is a process in which ferrous iron (Fe[II]) is oxidized to ferric iron (Fe[II]) coupled with the reduction of nitrate under anoxic, circumneutral conditions (5, 9, 38, 43, 45). This reaction is ecologically important (28, 29) and has great potential for biotechnological applications such as the bioremediation of toxic metals (11, 19, 24, 32, 33). However, Fe(II) oxidoreductase has not yet been identified among NDFO microbes (6), which limits our understanding of this geochemically important biological reaction. Since some Pseudogulbenkiania strains have the potential to be genetically engineered (23), their use is advantageous for the study of NDFO.
We previously isolated 67 Pseudogulbenkiania strains by using a functional single-cell isolation method (3) from rice paddy fields and rice-soybean rotation fields in Kumamoto, Niigata, and Yamagata in Japan (39). These strains showed strong denitrification and N2O reduction activities. The findings of a culture-independent RNA-based analysis also suggest that Pseudogulbenkiania spp. strongly contribute to denitrification and N2O reduction in rice paddy soils (46). Rice paddy fields are abundant in nitrate and Fe(II) (12, 16, 35) and NDFO activity has also been detected (12, 16, 35); therefore, Pseudogulbenkiania spp. may be involved in NDFO in the environment. However, the NDFO ability of Pseudogulbenkiania denitrifiers isolated from rice paddy soils has not yet been examined. Furthermore, relatedness among the Pseudogulbenkiania denitrifiers, NDFO strains 2002 and MAI-1, and other Pseudogulbenkiania species has not been analyzed to date. We targeted denitrification functional genes (nitrite reductase gene [nirS] and nitrous oxide reductase gene [nosZ]) in order to analyze diversity among strains.
The objectives of the present study are (i) to examine the NDFO abilities of Pseudogulbenkiania strains, (ii) to identify the genes responsible for NDFO, and (iii) to analyze denitrification functional gene diversities within the genus Pseudogulbenkiania.
Materials and Methods
Bacterial strains
Sixty-seven Pseudogulbenkiania strains were previously isolated from rice paddy fields and rice-soybean rotation fields in Kumamoto (29 strains), Niigata (33 strains), and Yamagata (5 strains) in Japan (39). Among these strains, Pseudogulbenkiania sp. strain NH8B was selected as a representative strain to sequence its whole genome (14). P. subflava strain BP-5T (= LMG 24211T), P. gefcensis strain yH16T (= JCM 17850T), and Pseudogulbenkiania sp. strain 2002 (= ATCC BAA-1479) were obtained from the Belgian Coordinated Collections of Microorganisms (BCCM), Japan Collection of Microorganisms (JCM), and American Type Culture Collection (ATCC), respectively. Bacterial cells were maintained in R2A medium (Wako Pure Chemical) at 30°C.
The plasmid vector pRL27, which contains a hyperactive Tn5 transposase gene (25), was used in transposon mutagenesis. Escherichia coli WM3064, a 2,6-diaminopimelic acid (DAP) auxotroph, was used as a donor organism for the pRL27 vector (20, 37). This strain was maintained in LB agar medium supplemented with DAP (300 μg mL−1) and kanamycin (100 μg mL−1).
Iron oxidation assays
Standard anaerobic culturing techniques with anoxic grove box (Coy laboratories) with an N2:CO2:H2 (80:10:10) atmosphere were used for this experiment. Cells grown on R2A agar were suspended in anoxic basal medium supplemented with 5 mM nitrate in test tubes with butyl rubber stoppers. Basal medium contained the following chemicals (L−1): 0.25 g of NH4Cl, 0.6 g of NaH2PO4, 0.1 g of KCl, 2.52 g of NaHCO3, and 10 mL each of the trace metal solution and vitamin solution (45). The cells were then incubated under anoxic conditions (N2:CO2:H2 = 80:10:10) at 30°C. This incubation was performed in order to deplete carbon in the cells. After a 3-d incubation, cells were pelleted and washed three times with anoxic PIPES (piperazine-N,N′-bis[2-ethanesulfonic acid]) buffer (10 mM, pH 7.0). Washed cells (final conc. 5 × 107 cells mL−1 as established by microscopy and OD600 measurements [15]) were inoculated into anoxic basal medium (20 mL) supplemented with 10 mM FeCl2 and 5 mM NaNO3 in test tubes with butyl rubber stoppers. After replacing the air phase of the test tubes with H2-free anoxic gas (N2:CO2 = 80:20), cells were incubated at 30°C. Samples (2 mL) were periodically collected using syringe needles. Aliquots (100 μL) of the samples were immediately mixed with 700 μL of 1 M HCl to stabilize Fe(II) and dissolve Fe(II) minerals. Twenty microliters was kept frozen at −20°C for a quantitative PCR (qPCR) analysis (see below). The remainders of the samples (ca. 1.9 mL) were filtered through 0.20-μm pore membrane filters and stored at −20°C until used for ion chromatography.
Fe(II) concentrations were measured spectrophotometrically using the ferrozine method as described by Hegler et al. (10). In order to measure total Fe concentrations (Fe[II] + Fe[III]), Fe(III) was reduced by 50% (w/v in 1M HCl) hydroxylamine hydrochloride prior to measurements by the ferrozine method. Nitrate and nitrite concentrations were measured using an ion chromatograph IC-2010 equipped with the TSKgel SuperIC-Anion HS column (Tosoh).
A high-throughput iron oxidation assay was performed using 96-well plates. In brief, cells grown on R2A agar were suspended in anoxic basal medium (150 μL) supplemented with 5 mM nitrate. After a 3-d anoxic incubation, cells (15 μL) were transferred to anoxic basal medium (135 μL) supplemented with 10 mM FeCl2 and 5 mM NaNO3, and incubated under anoxic conditions (N2:CO2:H2 = 80:10:10) at 30°C. Before and after a 1-week incubation, 25 μL of the cell culture was mixed with 175 μL of 1 M HCl solution in order to measure Fe(II) and total Fe concentrations, as described above. Pseudogulbenkiania sp. strain 2002 was used as a positive control.
Testing for autotrophy by NDFO
In an attempt to clarify whether cells have the ability to grow under NDFO conditions, we performed quantitative PCR by using strain-specific primers (IAC_23F and IAC_92R primers; Table S1) (15) and the KOD Sybr qPCR Mix (Toyobo). Culture medium (1 μL) in the NDFO assay was directly used as a template for qPCR. Standard DNA was prepared by diluting the linearized plasmid (15) with basal medium containing the same concentrations of potential PCR inhibitors (i.e., Fe2+) as the samples. The amplification efficiency (E) of the assay was 76% with a linear dynamic range from 20–2 × 106 copies μL−1.
We also performed stable isotope probing using 13C-labeled bicarbonate. Cells were prepared as described above, and incubated in anoxic basal medium (20 mL) supplemented with 10 mM FeCl2 and 5 mM NaNO3. All bicarbonate in this medium was labeled with 13C (Cambridge isotope laboratories). As a control, cells were incubated in medium with 12C bicarbonate. DNA was extracted after a 9-d incubation by using the PowerSoil DNA Isolation Kit (MoBio Laboratories). Cesium chloride density gradient ultracentrifugation and DNA collection were performed as previously described (13, 30). DNA concentrations in each of the density-resolved fractions were quantified using the Quant-iT PicoGreen ds DNA Assay Kit (ThermoFisher Scientific).
Transposon mutagenesis
Conjugation was performed as described by Kim et al. (20). In brief, E. coli pRL27 donor strain WM3064 and the recipient Pseudogulbenkiania sp. strain NH8B were mixed at a 1:4 ratio, and spotted on R2A agar supplemented with DAP (300 μg mL−1). After an overnight incubation, cells were suspended in R2A broth and spread onto R2A agar supplemented with kanamycin (100 μg mL−1). Colonies grown on this agar were selected and tested for their iron-oxidizing ability, as described above.
DNA extraction, PCR, and Sequencing
DNA was extracted by heating cells in 100 μL of 0.05 M NaOH at 95°C for 15 min (3). After centrifugation, the supernatant was diluted 10-fold and used for PCR as described below.
The 16S rRNA gene and nirS were amplified using m-27F and m-1492R primers (41) and m-cd3aF and m-R3cd primers (18), respectively. Regarding the amplification of nosZ, we slightly modified the sequences of the nosZ-F-1181 and nosZ-R-1880 primers (36). The nosZ-F-1181_NH8B and nosZ-R-1880_NH8B primers and nosZ-F-1181_2002 and nosZ-R-1880_2002 primers (Table S1) allow for the amplification of nosZ from strains NH8B and 2002, respectively. In addition, in order to amplify the region between nosZ (locus tag NH8B_3640) and the gene encoding the NosZ-like protein (locus tag NH8B_3641) of strain NH8B, PCR was performed using nosZ-R-1880_NH8B and IAC_92R (15) (Table S1).
The PCR reaction mixture (50 μL) contained 1 × Ex Taq buffer (Takara Bio), 0.2 μM of each primer, 0.2 mM of each dNTP, 1 U of Ex Taq DNA polymerase (Takara Bio), and 1 μL of a DNA template. PCR was performed using a Veriti 96-well thermal cycler (Applied Biosystems) with the following conditions: initial annealing at 96°C for 5 min, followed by 35 cycles at 95°C for 30 s, the annealing temperature shown in Table S1 for 30 s, and 72°C for the extension time shown in Table S1. After the final extension at 72°C for 7 min, the PCR mixtures were stored at 4°C. The sizes of the PCR products were verified by agarose gel electrophoresis.
PCR products were purified using the FastGene Gel/PCR Purification Kit (Nippon Genetics) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and ABI 3730xl capillary sequencer (Applied Biosystems) with the primers shown in Table S1 and Fig. S1.
Transcription analysis
Pseudogulbenkiania sp. strain 2002 and strain NH8B were grown in basal medium (5 mL) supplemented with 10 mM acetate under oxic or anoxic conditions. When grown under anoxic conditions, 5 mM nitrate was added to this medium as an electron acceptor. Total RNA was extracted using the RiboPure Bacteria kit (Ambion). After the DNase treatment, complementary DNA (cDNA) was synthesized using random hexamers and a PrimeScript RT reagent kit (Perfect Real Time) (Takara Bio). The resulting cDNA was used to detect gene transcription by PCR (Table S2). While genomic DNA was used as a positive control, RNA samples were used as negative controls in order to verify the absence of contamination by genomic DNA. Experiments were performed in triplicate (i.e., three test tubes per condition for each strain).
Genomic and phylogenetic analyses
Multiple reads obtained by sequencing reactions were assembled using the Phred-Phrap program (7). The nucleotide sequences of multiple clones were aligned using ClustalW, and used to generate phylogenetic trees. Phylogenetic trees were constructed based on the maximum likelihood method by using MEGA ver. 6 (40).
The genome of Pseudogulbenkiania sp. strain NH8B was previously sequenced (14). The genome sequences of Pseudogulbenkiania sp. strain 2002 (4) and Chromobacterium violaceum ATCC 12472 were retrieved from the GenBank database. These genomes were compared using GenomeMatcher (31).
Nucleotide sequence accession numbers
The nucleotide sequences of the 16S rRNA gene, nirS, and nosZ were deposited in the DDBJ/EMBL/GenBank databases under the accession numbers KU175358–KU175423, KU175424–KU175493, and KU175494–KU175563, respectively (Table S3).
Results and Discussion
Anaerobic nitrate-dependent Fe(II) oxidation
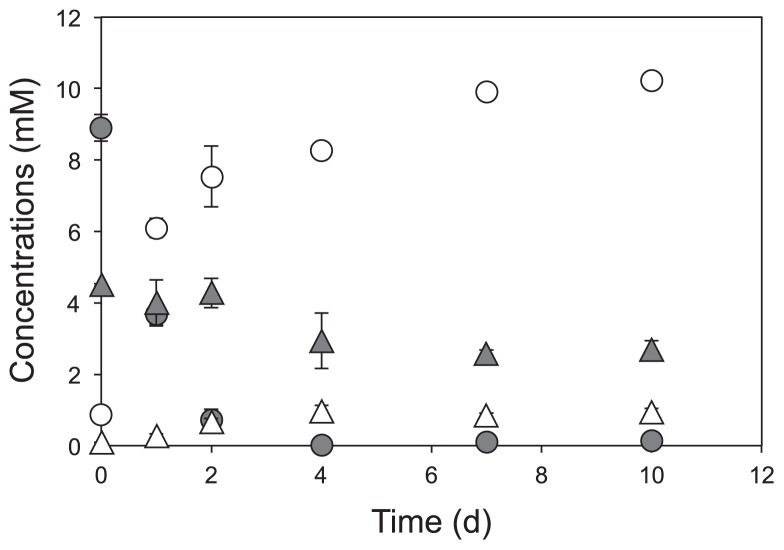
Pseudogulbenkiania sp. strain NH8B exhibited NDFO activity: Fe(II) was rapidly oxidized to Fe(III) along with the reduction of nitrate to nitrite (Fig. 1), similar to previous findings obtained with Pseudogulbenkiania sp. strain 2002 (42, 44). In contrast, the oxidation of Fe(II) was not observed when no cells or heat-killed cells were added to the test tubes (data not shown). These results indicate that Fe(II) was oxidized as a result of biological activity, either by a direct or indirect reaction.
Fig. 1.
Occurrence of anaerobic, nitrate-dependent Fe(II) oxidation by Pseudogulbenkiania sp. strain NH8B. Washed cells (5 × 107 cells) were inoculated into anoxic basal medium supplemented with 10 mM FeCl2 and 5 mM NaNO3, and incubated under anoxic conditions at 30°C for 10 d. Legends: Concentrations of Fe(II) (
 ), Fe(III) (○), NO3
− (
), Fe(III) (○), NO3
− (
 ), and NO2
− (△). The mean ± SD is shown.
), and NO2
− (△). The mean ± SD is shown.
The amount of nitrate consumed (ΔNO3 − = −1.89 ± 0.08 mM; Table S4) in the 7-d incubation was larger than the amount of nitrite produced (ΔNO2 − = 0.76 ± 0.08 mM), indicating that nitrite was further reduced to gaseous nitrogen oxides and dinitrogen. Based on the electron balance of the NDFO reaction (eq.1 and 2), two and five moles of Fe(II) are required to reduce one mole of nitrate to nitrite and to N2 gas, respectively.
| (eq. 1) |
| (eq. 2) |
If 0.76 mM and 1.13 mM (= 1.89 – 0.76 [mM]) of nitrate were reduced to nitrite and N2 gas, respectively, 1.52 mM and 5.65 mM of Fe2+ were oxidized according to eq. 1 and 2, respectively. The amount of Fe(II) oxidized in this study (ΔFe2+ = −8.78 ± 0.37 mM) was larger than the theoretical amount of Fe(II) required to reduce nitrate to nitrite and N2 gas (7.17 mM) as calculated above. This result suggested the occurrence of the abiotic oxidation of Fe(II) by reactive nitrogen species (e.g., nitrous acid [HNO2], nitrogen dioxide [NO2], and nitric oxide [NO]) formed during the acidic Fe extraction step (eq. 3–7) (21).
| (eq. 3) |
| (eq. 4) |
| (eq. 5) |
| (eq. 6) |
| (eq. 7) |
| (eq. 8) |
The reactions shown in eq. 3–7 proceed abiotically, particularly under acidic conditions (e.g., during the HCl extraction of Fe minerals) (21). Furthermore, NO may be oxidized back to NO2 under oxic conditions (eq. 8). Although our HCl extraction was performed in the anoxic grove box, the ferrozine assay was conducted under a 20% O2 atmosphere. Thus, some Fe(II) may have been abiotically oxidized during sample processing. This may partly explain the discrepancy between the rates of Fe(II) oxidation and nitrate reduction: i.e., most Fe(II) was oxidized within the first 2 d, while nitrate was reduced for a longer time period (ca. 4–7 d) in Fig. 1. The accumulation of nitrite may be due to the depletion of electron donors (e.g., Fe[II] and organic substrates stored in cells).
Cell growth was not observed during the incubation period (Fig. S2). In addition, 13C-bicarbonate was not incorporated into nucleic acids based on the SIP analysis (Fig. S3), suggesting that NDFO does not support the autotrophic growth of strain NH8B. Thus, NDFO-dependent growth may differ according to the strains (33, 38, 44) or test conditions.
The oxidation of Fe(II) was also observed in all other Pseudogulbenkiania sp. strains based on our high-throughput iron oxidation assay (Table S3), suggesting that NDFO is a common characteristic among Pseudogulbenkiania sp. strains.
In order to identify the genes associated with Fe(II) oxidoreductase, we performed transposon mutagenesis using strain NH8B. A total of 1,440 mutants were obtained and screened based on NDFO activity. As expected, mutants unable to grow under anoxic conditions on R2A agar supplemented with nitrate did not exhibit NDFO activity. We were unable to obtain mutants that did not oxidize iron, but had the ability to grow by anaerobic nitrate reduction, indicating that mutants disrupted with Fe(II) oxidoreductase-associated genes were not obtained in this study. In addition, the genome of strain NH8B did not contain homologs to mtrAB and pioAB, which are involved in respiratory iron reduction and phototrophic iron oxidation, respectively (17, 34). The homologs to mtrAB and pioAB are candidates for a potential Fe(II) oxidoreductase gene.
Similar to the present study, a previous attempt to identify specific Fe(II) oxidoreductase in the NDFO strain, Acidovorax ebreus strain TPSY, using a proteomic approach was not successful (6). These authors showed that respiratory complexes such as nitrate reductase (Nar) directly mediated NDFO, indicating that a specific Fe(II) oxidoreductase is not necessarily required for NDFO (6, 24). In addition, biotically or abiotically formed NO2 or NO may also react with Fe(II) to form Fe(III) oxides (6, 21, 22). These results suggest that NDFO exhibits an intrinsic ability in all nitrate-reducing bacteria, and may explain the failure to obtain the mutants of interest (i.e., NDFO-incapable, nitrate-reducing mutants). Strong NDFO abilities among Pseudogulbenkiania spp. may depend on their toxicity tolerance of Fe(II) and nitrogen oxides (e.g., NO2 − and NO), the extent of the accumulation of nitrogen oxides, and/or the nature of their respiratory proteins (6).
Genome comparison
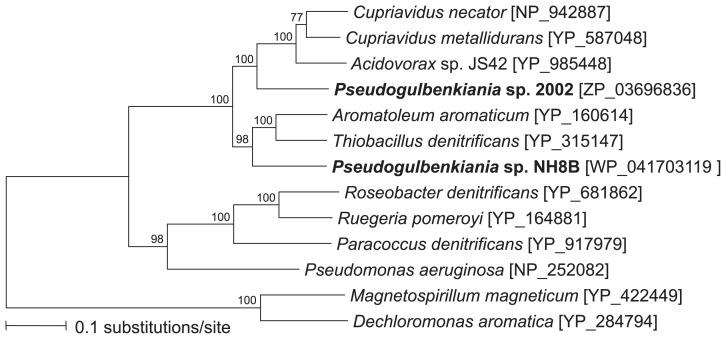
Genome structures and gene contents were similar between Pseudogulbenkiania sp. strains NH8B and 2002 (Fig. S4A), although there were several inverted genome fragments. In contrast, genome structures differed between Pseudogulbenkiania sp. strains NH8B and C. violaceum ATCC 12472 (Fig. S4B), a strain phylogenetically close to Pseudogulbenkiania sp. strains (Fig. S5).
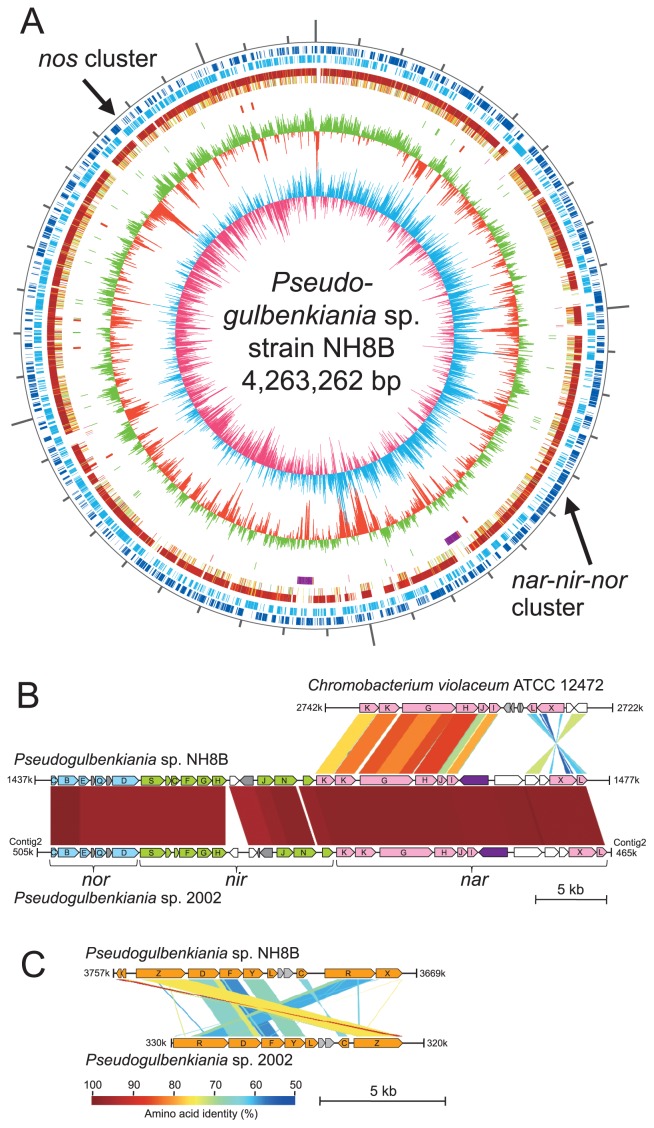
Complete sets of denitrification functional genes were identified on the genomes of the strains NH8B and 2002. Strains NH8B and 2002 both possessed gene clusters for membrane-bound Nar, cytochrome cd1-containing nitrite reductase (Nir), nitric oxide reductase (Nor), and nitrous oxide reductase (Nos). While nar-nir-nor operons were located next to each other, the nos operon was distantly located on the genome in strains NH8B and 2002 (Fig. 2A).
Fig. 2.
Comparison of genomes of Pseudogulbenkiania sp. strain NH8B, strain 2002, and Chromobacterium violaceum ATCC 12472. (A) Circular representation of the Pseudogulbenkiania sp. NH8B genome. From the outside in: circles 1 and 2 of the chromosome show the positions of protein-coding sequences on the positive and negative strands, respectively. Circles 3 and 4 show the positions of protein-coding sequences that have orthologs in Pseudogulbenkiania sp. 2002 and Chromobacterium violaceum ATCC 12472, respectively. Circle 5 shows the positions of the prophages (purple) and integrative elements (pink). Circle 6 shows the positions of tRNA genes (green) and rRNA genes (red). Circle 7 shows a plot of the G + C content (higher values outward). Circle 8 shows a plot of the GC skew ([G − C]/[G + C]; light blue indicates values > 0; red indicates values < 0). (B) Schematic diagram of nitrate reductase (Nar), nitrite reductase (Nir), and nitric oxide reductase (Nor) gene clusters from strain NH8B, strain 2002, and C. violaceum ATCC 12472. (C) Schematic diagram of the nitrous oxide reductase (Nos) gene cluster from strain NH8B and strain 2002.
The gene arrangement within the nar-nir-nor cluster was similar between strains NH8B and 2002 (Fig. 2B). In addition, gene sequences were highly conserved between the two strains. For example, the nirS sequence was 98.6% similar between strain NH8B and strain 2002. In contrast, the gene arrangement within the nos cluster was different between the two strains, and nos gene sequence similarities were relatively low (Fig. 2C). These results suggest that one of the two nos clusters was horizontally acquired. Further studies (e.g., comparative genome analysis with many other strains) may be necessary in order to obtain concrete evidence of the horizontal gene transfer event.
Interestingly, strain NH8B possessed 183-bp-long nosZlike pseudogenes (locus tag NH8B_3641 and NH8B_3642) downstream of the full-length nosZ (locus tag NH8B_RS17660), which were 84% similar to the nosZ of strain 2002.
nirS and nosZ diversity
In order to further investigate nirS and nosZ sequence diversities within the genus Pseudogulbenkiania, we sequenced nirS and nosZ from 70 Pseudogulbenkiania sp. strains (i.e., 67 Pseudogulbenkiania sp. strains isolated in our previous study, Pseudogulbenkiania sp. strain 2002, P. subflava strain BP-5T, and P. gefcensis strain yH16T). Since commonly used PCR primers for nosZ (nosZ-F-1181 and nosZ-R-1880) did not amplify nosZ from strains NH8B and 2002, we designed new primers.
The nirS sequences from all Pseudogulbenkiania sp. strains were relatively highly conserved among the 70 strains, with >90.4% similarity (367 bp). Pseudogulbenkiania sp. strains isolated from different locations were clustered together based on their nirS sequences (Fig. S6).
In contrast, we observed greater dissimilarity among nosZ from Pseudogulbenkiania sp. strains (> 59.4% sequence similarities among 633-bp fragments). Based on nosZ sequence similarities, the 70 Pseudogulbenkiania sp. strains were divided into two groups: NH8B-type (15 strains) and 2002-type (55 strains) groups (Fig. 3).
Fig. 3.
Phylogenetic relationships of deduced NosZ amino acid sequences between Pseudogulbenkiania sp. strain NH8B and Pseudogulbenkiania sp. strain 2002. A phylogenetic tree was constructed using the maximum likelihood method. Bootstrap values (>70%) from 1,000 replicates are indicated next to the branches. The accession numbers of the reference strains in the DDBJ/EMBL/GenBank databases are indicated in the brackets.
All 15 NH8B-type strains had a nosZ-like pseudogene similar to strain NH8B, based on NH8B_3641-specific qPCR (15) and an analysis of the sequences downstream of nosZ. The intergenic region between nosZ and the nosZ-like pseudogene was relatively well conserved among the 15 strains (Fig. S7). The intergenic region is known to be generally more variable than protein-coding sequences (8). The highly conserved intergenic regions among the 15 strains indicate that these strains may have diversified relatively recently from a common ancestor, and were then distributed to different locations.
In this region, we detected a FNR-binding motif (FNR box; TTGAT----ATCAA), which is recognized by FNR-like transcription regulators and reported to be essential for the transcription of nosZ in Pseudomonas aeruginosa (2). We detected the transcription of nosZ and nosZ-like pseudogenes in the cells of Pseudogulbenkiania sp. strain NH8B under oxic and anoxic conditions (Fig. S8A and S8B). In contrast, the transcription of nosZ was only detected in the cells of Pseudogulbenkiania sp. strain 2002 (Fig. S8C) grown under anoxic conditions, suggesting that transcription regulation may differ between strains NH8B and 2002. The role of FNR-like transcription regulators in the transcription of nosZ and related proteins is currently unknown. Since Pseudogulbenkiania sp. strain NH8B reduces exogenous N2O (14), a deeper understanding of the transcription mechanism of nosZ will be useful for mitigating N2O emissions.
Conclusions
Our comprehensive analysis showed that diverse strains within the genus Pseudogulbenkiania perform denitrification and NDFO. The failure to detect NDFO-deficient denitrifying mutants in our mutagenesis experiment suggests that NDFO is linked to nitrate-reducing activity (i.e., abiotic Fe[II] oxidation by reactive nitrogen species formed during nitrate reduction) without the involvement of specific Fe(II) oxidoreductase. The NDFO abilities of Pseudogulbenkiania spp. may depend on their toxicity tolerance of Fe(II) and nitrogen oxides (e.g., NO2 − and NO), the extent of the accumulation of nitrogen oxides, and/or the nature of their respiratory proteins (6). Different gene arrangements within the nos cluster and low sequence similarity within nos gene sequences among Pseudogulbenkiania sp. strains indicate the occurrence of horizontal gene transfer. Since Pseudogulbenkiania sp. strains have been isolated from various locations (e.g., agricultural soils and freshwater lakes) around the world, their denitrification and NDFO abilities may contribute significantly to nitrogen and iron biogeochemical cycles.
Supplementary Information
Acknowledgements
We thank Reiko Hirano (Hokkaido University), Daichi Fujii, and Naofumi Somekawa (University of Tokyo) for their technical assistance, and Kanako Tago (National Institute for Agro-Environmental Sciences) and Yoshitomo Kikuchi (National Institute of Advanced Industrial Science and Technology) for providing Pseudogulbenkiania sp. strains and E. coli WM3064, respectively. We also thank Daisuke Sano (Hokkaido University) and Tomoyasu Nishizawa (Ibaraki University) for their valuable comments, and Nora Powers for scientific editing.
This work was supported, in part, by the Steel Foundation for Environmental Protection Technology and by Minnesota’s Discovery, Research and InnoVation Economy (MnDRIVE) initiative of the University of Minnesota.
References
- 1.Adeolu M., Gupta R. Phylogenomics and molecular signatures for the order neisseriales: proposal for division of the order neisseriales into the emended family Neisseriaceae and Chromobacteriaceae fam. Nov. Antonie van Leeuwenhoek. 2013;104:1–24. doi: 10.1007/s10482-013-9920-6. [DOI] [PubMed] [Google Scholar]
- 2.Arai H., Mizutani M., Igarashi Y. Transcriptional regulation of the nos genes for nitrous oxide reductase in pseudomonas aeruginosa. Microbiology. 2003;149:29–36. doi: 10.1099/mic.0.25936-0. [DOI] [PubMed] [Google Scholar]
- 3.Ashida N., Ishii S., Hayano S., Tago K., Tsuji T., Yoshimura Y., Otsuka S., Senoo K. Isolation of functional single cells from environments using a micromanipulator: application to study denitrifying bacteria. Appl Microbiol Biotechnol. 2010;85:1211–1217. doi: 10.1007/s00253-009-2330-z. [DOI] [PubMed] [Google Scholar]
- 4.Byrne-Bailey K.G., Weber K.A., Coates J.D. Draft genome sequence of the anaerobic, nitrate-dependent, Fe(II)-oxidizing bacterium Pseudogulbenkiania ferrooxidans strain 2002. J Bacteriol. 2012;194:2400–2401. doi: 10.1128/JB.00214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson H.K., Clark I.C., Melnyk R.A., Coates J.D. Towards a mechanistic understanding of anaerobic nitrate dependent iron oxidation: balancing electron uptake and detoxification. Front Microbiol. 2012;3:57. doi: 10.3389/fmicb.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson H.K., Clark I.C., Blazewicz S.J., Iavarone A.T., Coates J.D. Fe(II) oxidation is an innate capability of nitrate-reducing bacteria that involves abiotic and biotic reactions. J Bacteriol. 2013;195:3260–3268. doi: 10.1128/JB.00058-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces usingphred. I. accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 8.García-Martínez J., Acinas S.G., Antón A.I., Rodríguez- Valera F. Use of the 16S–23S ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods. 1999;36:55–64. doi: 10.1016/s0167-7012(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 9.Hafenbradl D., Keller M., Dirmeier R., Rachel R., Roßnagel P., Burggraf S., Huber H., Stetter K.O. Ferroglobus placidus gen. nov., sp. nov., a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch Microbiology. 1996;166:308–314. doi: 10.1007/s002030050388. [DOI] [PubMed] [Google Scholar]
- 10.Hegler F., Posth N.R., Jiang J., Kappler A. Physiology of phototrophic iron(II)-oxidizing bacteria: implications for modern and ancient environments. FEMS Microbiol Ecol. 2008;66:250–260. doi: 10.1111/j.1574-6941.2008.00592.x. [DOI] [PubMed] [Google Scholar]
- 11.Hohmann C., Winkler E., Morin G., Kappler A. Anaerobic Fe(II)-oxidizing bacteria show as resistance and immobilize as during Fe(III) mineral precipitation. Environ Sci Technol. 2010;44:94–101. doi: 10.1021/es900708s. [DOI] [PubMed] [Google Scholar]
- 12.Ishii S., Ikeda S., Minamisawa K., Senoo K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ. 2011;26:282–292. doi: 10.1264/jsme2.me11293. [DOI] [PubMed] [Google Scholar]
- 13.Ishii S., Ohno H., Tsuboi M., Otsuka S., Senoo K. Identification and isolation of active N2O reducers in rice paddy soil. ISME J. 2011;5:1936–1945. doi: 10.1038/ismej.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii S., Tago K., Nishizawa T., Oshima K., Hattori M., Senoo K. Complete genome sequence of the denitrifying and N2O reducing bacterium Pseudogulbenkiania sp. strain NH8B. J Bacteriol. 2011;193:6395–6396. doi: 10.1128/JB.06127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii S., Segawa T., Okabe S. Simultaneous quantification of multiple food- and waterborne pathogens by use of microfluidic quantitative pcr. Appl Environ Microbiol. 2013;79:2891–2898. doi: 10.1128/AEM.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh H., Ishii S., Shiratori Y., Oshima K., Otsuka S., Hattori M., Senoo K. Seasonal transition of active bacterial and archaeal communities in relation to water management in paddy soils. Microbes Environ. 2013;28:370–380. doi: 10.1264/jsme2.ME13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao Y., Newman D.K. The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J Bacteriol. 2007;189:1765–1773. doi: 10.1128/JB.00776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandeler E., Deiglmayr K., Tscherko D., Bru D., Philippot L. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microbiol. 2006;72:5957–5962. doi: 10.1128/AEM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato S. Biotechnological aspects of microbial extracellular electron transfer. Microbes Environ. 2015;30:133–139. doi: 10.1264/jsme2.ME15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J.K., Jang H.A., Won Y.J., Kikuchi Y., Heum Han S., Kim C.-H., Nikoh N., Fukatsu T., Lee B.L. Purine biosynthesisdeficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J. 2014;8:552–563. doi: 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klueglein N., Kappler A. Abiotic oxidation of Fe(II) by reactive nitrogen species in cultures of the nitrate-reducing Fe(II) oxidizer Acidovorax sp. BoFeN1—questioning the existence of enzymatic Fe(II) oxidation. Geobiology. 2013;11:180–190. doi: 10.1111/gbi.12019. [DOI] [PubMed] [Google Scholar]
- 22.Klueglein N., Zeitvogel F., Stierhof Y.-D., Floetenmeyer M., Konhauser K.O., Kappler A., Obst M. Potential role of nitrite for abiotic Fe(II) oxidation and cell encrustation during nitrate reduction by denitrifying bacteria. Appl Environ Microbiol. 2014;80:1051–1061. doi: 10.1128/AEM.03277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopf S.H., Henny C., Newman D.K. Ligand-enhanced abiotic iron oxidation and the effects of chemical versus biological iron cycling in anoxic environments. Environ Sci Technol. 2013;47:2602–2611. doi: 10.1021/es3049459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lack J.G., Chaudhuri S.K., Kelly S.D., Kemner K.M., O’Connor S.M., Coates J.D. Immobilization of radionuclides and heavy metals through anaerobic bio-oxidation of Fe(II) Appl Environ Microbiol. 2002;68:2704–2710. doi: 10.1128/AEM.68.6.2704-2710.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen R., Wilson M., Guss A., Metcalf W. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee D.-G., Im D.-M., Kang H., Yun P., Park S.-K., Hyun S.-S., Hwang D.-Y. Pseudogulbenkiania gefcensis sp. nov., isolated from soil. Intl J Syst Evol Microbiol. 2013;63:187–191. doi: 10.1099/ijs.0.040873-0. [DOI] [PubMed] [Google Scholar]
- 27.Lin M.-C., Chou J.-H., Arun A.B., Young C.-C., Chen W.-M. Pseudogulbenkiania subflava gen. nov., sp. nov., isolated from a cold spring. Intl J Syst Evol Microbiol. 2008;58:2384–2388. doi: 10.1099/ijs.0.65755-0. [DOI] [PubMed] [Google Scholar]
- 28.Mattes A., Gould D., Taupp M., Glasauer S. A novel autotrophic bacterium isolated from an engineered wetland system links nitrate-coupled iron oxidation to the removal of As, Zn and S. Water Air Soil Pollut. 2013;224:1–15. [Google Scholar]
- 29.Melton E.D., Stief P., Behrens S., Kappler A., Schmidt C. High spatial resolution of distribution and interconnections between Fe- and N-redox processes in profundal lake sediments. Environ Microbiol. 2014;16:3287–3303. doi: 10.1111/1462-2920.12566. [DOI] [PubMed] [Google Scholar]
- 30.Neufeld J.D., Vohra J., Dumont M.G., Lueders T., Manefield M., Friedrich M.W., Murrell J.C. DNA stable-isotope probing. Nat Protoc. 2007;2:860–866. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- 31.Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omoregie E.O., Couture R.-M., Van Cappellen P., Corkhill C.L., Charnock J.M., Polya D.A., Vaughan D., Vanbroekhoven K., Lloyd J.R. Arsenic bioremediation by biogenic iron oxides and sulfides. Appl Environ Microbiol. 2013;79:4325–4335. doi: 10.1128/AEM.00683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshiki M., Ishii S., Yoshida K., Fujii N., Ishiguro M., Satoh H., Okabe S. Nitrate-dependent ferrous iron oxidation by anaerobic ammonium oxidation (anammox) bacteria. Appl Environ Microbiol. 2013;79:4087–4093. doi: 10.1128/AEM.00743-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitts K.E., Dobbin P.S., Reyes-Ramirez F., Thomson A.J., Richardson D.J., Seward H.E. Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome mtra: expression in Escherichia coli confers the ability to reduce soluble Fe(III) chelates. J Biol Chem. 2003;278:27758–27765. doi: 10.1074/jbc.M302582200. [DOI] [PubMed] [Google Scholar]
- 35.Ratering S., Schnell S. Nitrate-dependent iron(II) oxidation in paddy soil. Environ Microbiol. 2001;3:100–109. doi: 10.1046/j.1462-2920.2001.00163.x. [DOI] [PubMed] [Google Scholar]
- 36.Rich J.J., Heichen R.S., Bottomley P.J., Cromack K., Myrold D.D. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl Environ Microbiol. 2003;69:5974–5982. doi: 10.1128/AEM.69.10.5974-5982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saltikov C.W., Newman D.K. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straub K.L., Benz M., Schink B., Widdel F. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl Environ Microbiol. 1996;62:1458–1460. doi: 10.1128/aem.62.4.1458-1460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tago K., Ishii S., Nishizawa T., Otsuka S., Senoo K. Phylogenetic and functional diversity of denitrifying bacteria isolated from various rice paddy and rice-soybean rotation fields. Microbes Environ. 2011;26:30–35. doi: 10.1264/jsme2.me10167. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.Tyson G.W., Chapman J., Hugenholtz P., et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 42.Weber K., Hedrick D., Peacock A., Thrash J., White D., Achenbach L., Coates J. Physiological and taxonomic description of the novel autotrophic, metal oxidizing bacterium, Pseudogulbenkiania sp. strain 2002. Appl Microbiol Biotechnol. 2009;83:555–565. doi: 10.1007/s00253-009-1934-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber K.A., Achenbach L.A., Coates J.D. Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol. 2006;4:752–764. doi: 10.1038/nrmicro1490. [DOI] [PubMed] [Google Scholar]
- 44.Weber K.A., Pollock J., Cole K.A., O’Connor S.M., Achenbach L.A., Coates J.D. Anaerobic nitrate-dependent iron(II) bio-oxidation by a novel lithoautotrophic betaproteobacterium, strain 2002. Appl Environ Microbiol. 2006;72:686–694. doi: 10.1128/AEM.72.1.686-694.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber K.A., Coates J.D. Microbially mediated anaerobic iron(II) oxidation at circumneutral pH. In: Hurst C.J., Crawford R.L., Garland J.L., Lipson D.A., editors. Manual of Environmental Microbiology. ASM Press; Washington, DC: 2007. pp. 1147–1154. [Google Scholar]
- 46.Yoshida M., Ishii S., Fujii D., Otsuka S., Senoo K. Identification of active denitrifiers in rice paddy soil by DNA- and rna-based analyses. Microbes Environ. 2012;27:456–461. doi: 10.1264/jsme2.ME12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.