SCN neuronal networks undergo differential and redundant regulations of AVP and VIP during postnatal development in mice.
Keywords: AVP, VIP, ontogeny, neural network, suprachiasmatic nucleus
Abstract
The suprachiasmatic nucleus (SCN) is the site of the master circadian clock in mammals. The SCN neural network plays a critical role in expressing the tissue-level circadian rhythm. Previously, we demonstrated postnatal changes in the SCN network in mice, in which the clock gene products CRYPTOCHROMES (CRYs) are involved. Here, we show that vasoactive intestinal polypeptide (VIP) signaling is essential for the tissue-level circadian PER2::LUC rhythm in the neonatal SCN of CRY double-deficient mice (Cry1,2−/−). VIP and arginine vasopressin (AVP) signaling showed redundancy in expressing the tissue-level circadian rhythm in the SCN. AVP synthesis was significantly attenuated in the Cry1,2−/− SCN, which contributes to aperiodicity in the adult mice together with an attenuation of VIP signaling as a natural process of ontogeny. The SCN network consists of multiple clusters of cellular circadian rhythms that are differentially integrated by AVP and VIP signaling, depending on the postnatal period.
INTRODUCTION
The hypothalamic suprachiasmatic nucleus (SCN) plays an essential role in the expression of circadian rhythms and light entrainment in mammals (1, 2). The molecular mechanism of circadian oscillation is generally believed to consist of a negative feedback loop, in which clock genes Per(s), Cry(s), Clock, and Bmal1 and their protein products are involved (3). A unilateral SCN consists of approximately 10,000 neurons, and most of the cells show self-sustained circadian oscillations (4–6). The cellular circadian rhythms are integrated to express a coherent circadian rhythm through the neural network in the SCN (7–9). Perturbation of the SCN network changes the circadian rhythms in the SCN and behavior (10–13).
Cry1 and Cry2 double-deficient mice (Cry1,2−/− mice) fail to show circadian rhythms in behavior and clock gene expressions in the SCN under constant darkness (DD) (14, 15). Despite a lack of rhythmicity in the adult mice, the SCN of neonatal Cry1,2−/− mice was shown to exhibit robust circadian rhythms in culture (16, 17). The tissue-level circadian rhythms in the SCN disappeared by the time of weaning and were not detected in adulthood (17). It turned out that cellular circadian rhythms were still detectable in the adult Cry1,2−/− SCN, although they were sloppy and unstable. The disappearance of rhythmicity was due to the desynchronization of cellular circadian rhythms (17). The circadian rhythms in clock gene expression were rescued in the aperiodic adult Cry1,2−/− SCN by coculture with the neonatal SCN of the wild-type (WT) mice, suggesting the involvement of a diffusible molecule(s) secreted from the graft SCN for the rescue of circadian rhythm (16, 17). Because coculture with the adult WT SCN failed to express the tissue-level circadian rhythms in the Cry1,2−/− SCN, the responsible factor(s) was suggested to function in the neonatal WT SCN but not in the adult. These findings indicate that the postnatal changes in the neural network occur not only in the Cry1,2−/− but also in the WT SCN. In this respect, Cry1,2−/− mice would be a marvelous model to examine postnatal changes of the SCN neural network.
The SCN expresses a variety of neuropeptides that are topologically specific (18, 19). Among them, vasoactive intestinal polypeptide (VIP) expressed in the core region of the SCN and arginine vasopressin (AVP) in the shell region are the two major neuropeptides that have been suggested to mediate the networking in the SCN (13, 16, 20–22). AVP and VIP in the cultured SCN of neonatal rats show robust circadian rhythms that can be internally desynchronized under inhibition of glial cell overproliferation (23). The findings indicated that different mechanisms are involved in the clustering of the AVP- or VIP-containing cells and in integrating cell clusters. Importantly, VIP expression and release showed endogenous circadian oscillations in the neonatal SCN, but not in the adult SCN (24, 25), suggesting a postnatal change in the role of VIP signaling in the SCN network.
Abolishment of the circadian rhythms in VIP- or AVP-containing SCN cells alters the tissue-level circadian rhythms in the SCN and behavior (12, 13). Recently, competent AVP signaling was suggested to induce, but not maintain, the circadian rhythms rescued by genetic induction of CRYPTOCHROME 1 (CRY1) in the aperiodic Cry1,2-/- SCN (26). However, the mechanism of rhythm restoration by AVP signaling was not clarified. Here, we examined the roles of AVP and VIP signaling in the expression of the coherent circadian rhythm in the Cry1,2−/− SCN to elucidate the mechanism of postnatal changes in the SCN neural network.
RESULTS
A lack of VIP receptor (VPAC2) abolishes the tissue-level circadian PER2::LUC rhythm in the neonatal Cry1,2−/− SCN
To understand the role of VIP signaling in the expression of the tissue-level circadian rhythms in the SCN, we generated mice that lacked Cry1, Cry2, and Vipr2, a gene that encodes the VIP receptor VPAC2, by crossing Vipr2-null (Vipr2−/−) mice with Cry1,2−/− mice carrying a PER2::LUC bioluminescence reporter (Cry1,2−/−/Vipr2−/−). Locomotor activity in Cry1,2−/−/Vipr2−/− mice failed to show the circadian rhythm not only in DD but also in light-dark (LD) cycles (fig. S1), which is a marked contrast to the behavior of Cry1,2−/− and Vipr2−/− mice whose daily rhythms are mostly maintained in LD (14, 27).
The intensity of bioluminescence was continuously measured for more than 7 days in the cultured SCN slices from the WT, Vipr2−/−, Cry1,2−/−, and Cry1,2−/−/Vipr2−/− mice on the tissue level using a photomultiplier tube (PMT) (Fig. 1) or on the pixel level using a charge-coupled device (CCD) camera (Fig. 2). The tissue-level circadian rhythm was evaluated by a χ2 periodogram, and the pixel-level circadian rhythm was analyzed using a cosine curve fitting method (see Materials and Methods).
Fig. 1. Tissue-level circadian PER2::LUC rhythms in the cultured SCN slices of WT, Vipr2−/−, Cry1,2−/−, and Cry1,2−/−/Vipr2−/− mice.
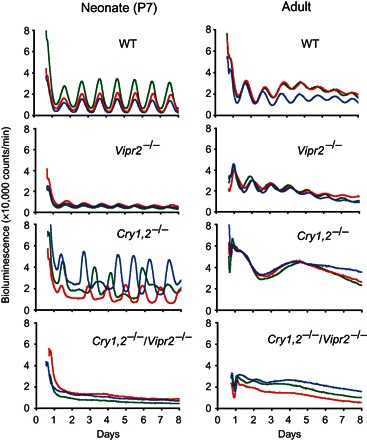
Representative tissue-level PER2::LUC bioluminescence in the neonatal [postnatal day 7 (P7); left] and adult (right) SCN slices of WT, Vipr2−/−, Cry1,2−/−, and Cry1,2−/−/Vipr2−/− mice measured by a PMT. Different colored lines in each panel indicate bioluminescence from the SCN of different mice. The number of days in the culture is indicated on the abscissa with day 0 as the day of harvest of the SCN. The ordinate indicates the intensity of bioluminescence in arbitrary units. The vertical divisions in the panels indicate a local time of 0000.
Fig. 2. Pixel-level circadian PER2::LUC rhythms in the cultured SCN slices of WT, Vipr2−/−, Cry1,2−/−, and Cry1,2−/−/Vipr2−/− mice.
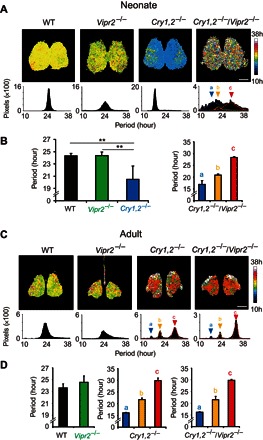
(A) Representative period maps of the circadian PER2::LUC rhythm on the pixel level in the neonatal SCN slices (top) and distributions of the pixel-level circadian periods (bottom). A color key to the right indicates units of the heat map from 10 hours (blue) to 38 hours (white). Scale bar, 200 μm. Superimposed curves in the period distribution of Cry1,2−/−/Vipr2−/− SCN indicate three Gaussian curves fitted to the data. Arrowheads a, b, and c indicate the peaks of a fitted Gaussian curve. (B) Mean circadian periods on the pixel level in the SCN of WT (n = 5), Vipr2−/− (n = 5), and Cry1,2−/− (n = 6) mice (left) and periods of three clusters in the Cry1,2−/−/Vipr2−/− mice (n = 3) (right). (C) Representative period maps of the circadian PER2::LUC rhythm on the pixel level in the adult SCN slices (top) and distributions of pixel-level circadian periods (bottom). See also the legend for (A). (D) Mean circadian periods on the pixel level at the cluster peak of the WT (n = 6) and Vipr2−/− (n = 4) SCN (left) and periods of the three clusters in the Cry1,2−/− (n = 4) (middle) and Cry1,2−/−/Vipr2−/− SCN (n = 4) (right). *P < 0.01, one-way ANOVA with post hoc Tukey-Kramer test.
In the neonatal SCN, significant circadian PER2::LUC rhythms were detected on the tissue level in Vipr2−/− mice (Fig. 1), but the amplitude of rhythmicity was significantly smaller than that of WT mice on the first day of culture [mean ± SD: WT, 0.75 ± 0.06 (n = 6); Vipr2−/−, 0.39 ± 0.06 (n = 8); P = 1.19 × 10−7]. Robust tissue-level circadian rhythms were also observed in the Cry1,2−/− SCN, as reported previously (17). The amplitude of rhythmicity in the Cry1,2−/− SCN was comparable to that in WT mice. On the other hand, PER2::LUC bioluminescence remained at the basal level in the Cry1,2−/−/Vipr2−/− SCN and failed to show robust circadian rhythms on the tissue level. These findings indicate a critical role for VIP signaling in the expression of the tissue-level circadian rhythm in the neonatal Cry1,2−/− SCN.
In the adult SCN, the tissue-level circadian rhythm was detected in Vipr2−/− mice, but the amplitude was significantly smaller than that in the WT mice on the first day of culture [WT, 0.58 ± 0.04 (n = 5); Vipr2−/−, 0.36 ± 0.06 (n = 5); P = 0.0001]. A robust circadian rhythm was detected neither in the Cry1,2−/− nor in the Cry1,2−/−/Vipr2−/− SCN. These results confirmed our previous finding that the neural network of the SCN changed in Cry1,2−/− mice during postnatal development (17).
The SCN neural network was substantially altered during postnatal development
The pixel-level analysis of bioluminescence images demonstrated different features of the neural network, in which approximately two pixels (37 μm2) represented a typical single SCN neuron (40 μm2) (19). Significant circadian PER2::LUC rhythms were detected in most of the pixels and in the cell-size regions of interest (ROIs) in the SCN of four genotypes (Fig. 2 and fig. S2, A to C). However, the distribution of circadian period on the pixel level (period map) was extremely different among the genotypes and developmental stages.
In the neonatal SCN, the period distribution of pixel-level circadian rhythms showed a single peak in the WT, Vipr2−/−, and Cry1,2−/− SCN, whereas it was multimodal, ranging from 10 to 38 hours, in the Cry1,2−/−/Vipr2−/− SCN (Fig. 2A and fig. S2D). In the Cry1,2−/− SCN, one of seven slices demonstrated three cell clusters with different circadian periods, despite the presence of the significant tissue-level circadian rhythm. This result was excluded from the calculation of the mean circadian period. Fitting of a triple Gaussian curve to the period distribution (see Materials and Methods) clarified three periodicities in Cry1,2−/−/Vipr2−/− mice at 16.9 ± 1.6 hours, 21.0 ± 0.4 hours, and 28.3 ± 0.4 hours (n = 3) (Fig. 2B, right, and fig. S2D). The circadian amplitude at the cluster peak was slightly but significantly different between the short and long clusters [P = 0.0341, one-way repeated-measures analysis of variance (ANOVA); P < 0.05, post hoc Tukey-Kramer test]. On the other hand, the circadian period of single cell cluster was 24.3 ± 0.4 hours (n = 5) in WT, 24.4 ± 0.6 hours (n = 5) in Vipr2−/−, and 20.5 ± 2.2 hours (n = 6) in Cry1,2−/− SCN, and it was significantly shorter in the Cry1,2−/− SCN than in the WT and Vipr2−/− mice. Regional specificity in the distribution of circadian period was evident neither in the Cry1,2−/− nor in the Cry1,2−/−/Vipr2−/− SCN.
In the adult mice, three cell clusters were detected in the distribution of circadian period in the Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN, whereas a single cell cluster was observed in the WT and Vipr2−/− SCN (Fig. 2, C and D). The circadian period of each cluster was 16.2 ± 0.1 hours, 21.8 ± 0.7 hours, and 29.9 ± 1.1 hours in the Cry1,2−/− SCN (n = 4) and 16.2 ± 0.2 hours, 21.6 ±1.4 hours, and 29.8 ± 0.5 hours in the Cry1,2−/−/Vipr2−/− SCN (n = 4). They were not significantly different when compared with respective short, middle, and long periods. On the other hand, the WT and Vipr2−/− SCN showed a single cluster at the circadian period of 23.9 ± 0.7 hours (n = 6) and 24.6 ± 1.1 hours (n = 4), which were not significantly different from each other and from those of the respective neonatal SCN.
Pharmacological evidence for the involvement of intracellular cAMP and Ca2+ in the expression of tissue-level circadian rhythms in the neonatal Cry1,2−/− SCN
Cyclic adenosine 3′,5′-monophosphate (cAMP) is a part of the intracellular VIP signaling cascade (28) and is suggested to have an important role in the synchronization of cellular circadian rhythms in the SCN (29). MDL-12330A, a specific inhibitor of adenylyl cyclase that converts adenosine 5′-triphosphate to cAMP, substantially attenuated and eventually abolished the tissue-level circadian PER2::LUC rhythm in the neonatal Cry1,2−/− SCN (Fig. 3A and fig. S3A). The circadian rhythm was restored by washing out the inhibitor. The attenuation was dose-dependent in terms of rhythm synchrony, as expressed by the level of Qp (fig. S3C). In the WT SCN, the amplitude of circadian rhythm was significantly reduced by 5 μM MDL-12330A (P = 2.7 × 10−7, one-way ANOVA; P < 0.01, post hoc Tukey-Kramer test), but not abolished. Qp at the peak period decreased in a dose-dependent manner (fig. S3C).
Fig. 3. Pharmacological manipulations of intracellular cAMP or Ca2+ abolish circadian PER2::LUC rhythms in the neonatal SCN of Cry1,2−/− mice.
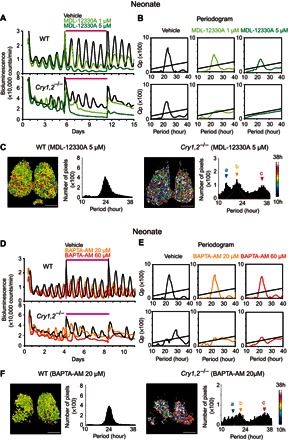
(A) Representative PER2::LUC bioluminescence in the neonatal SCN slices of the WT (top) and Cry1,2−/− (bottom) mice treated with a blocker of adenylyl cyclase (MDL-12330A) by exchanging with the culture medium containing each drug. Magenta lines indicate the duration of drug or vehicle treatment. Bioluminescence in the vehicle (black), MDL-12330A (1 μM; pale green), and MDL-12330A (5 μM; dark green) treatment. (B) χ2 periodogram during drug treatment. An oblique line in the periodogram indicates the significance level (P = 0.01). (C) Representative period maps on the pixel level and distributions of period in the WT (left) and Cry1,2−/− (right) SCN treated with 5 μM MDL-12330A. A color key of the heat map is indicated in the right margin from 10 hours (blue) to 38 hours (white). Scale bars, 200 μm. Superimposed curves in the period distribution of Cry1,2−/− indicate three Gaussian curves fitted to the data (far right). Arrowheads a, b, and c indicate the peaks of a fitted Gaussian curve. (D) Representative PER2::LUC bioluminescence in the neonatal SCN slices of the WT (top) and Cry1,2−/− (bottom) mice treated with Ca2+-chelating agent BAPTA-AM [20 μM (orange) or 60 μM (red)]. See also the legend for (A). (E) χ2 periodogram during drug treatment. See also the legend for (B). (F) Representative period maps on the pixel level and distributions of period in a WT (left) and Cry1,2−/− (right) SCN treated with 20 μM BAPTA-AM. See also the legend for (C).
The pixel-level analysis revealed that Cry1,2−/− SCN exhibited three cell clusters with different period by MDL-12330A treatment. The periods were similar to those of the respective clusters in the Cry1,2−/−/Vipr2−/− SCN (Fig. 3C and fig. S3D). On the other hand, 5 μM MDL-12330A did not disrupt the neural network of the WT SCN despite substantial damping of the tissue-level circadian rhythms (Fig. 3C).
VIP signaling in the SCN is also mediated by intracellular Ca2+, which cross-talks with cAMP (28, 30). BAPTA-AM, a common chelating agent for Ca2+, attenuated the amplitude and synchrony (Qp) of the circadian PER2::LUC rhythm in the neonatal Cry1,2−/− SCN in a dose-dependent manner (Fig. 3, D and E, and fig. S3, B and C). The circadian rhythm was restored by washing out the chelating agent. The pixel-level analysis revealed that 20 μM BAPTA-AM disrupted the integration of cellular circadian rhythms (Fig. 3F and fig. S3E) and again exhibited three cell clusters with different circadian periods, each similar to those observed by MDL-12330A treatment. On the other hand, 20 μM BAPTA-AM did not affect the neural network of the WT SCN (Fig. 3D).
These pharmacological findings indicate involvements of intracellular cAMP and Ca2+ in the clustering of cell groups in the Cry1,2−/− SCN and are consistent with the findings in the Cry1,2−/−/Vipr2−/− mice. This notion does not exclude an involvement of cAMP or Ca2+ in systems other than VIP signaling.
Coculture with the neonatal WT SCN rescued the tissue-level circadian PER2::LUC rhythms in the arrhythmic Cry1,2−/−/Vipr2−/− SCN
The neonatal or adult SCN slice of Cry1,2−/−/Vipr2−/− mice (recipient) was cocultured with the neonatal WT SCN slice (donor), which did not carry a bioluminescence reporter (Fig. 4A). The tissue-level circadian rhythm was rescued not only in the neonatal (Fig. 4B) but also in the adult Cry1,2−/−/Vipr2−/− SCN (Fig. 4C and fig. S4B). The tissue-level circadian rhythm was also rescued in the adult Cry1,2−/− SCN by coculture with neonatal WT SCN slice (Fig. 4C). Coculture of the neonatal cerebral cortex failed to restore circadian rhythms in the Cry1,2−/− SCN (fig. S4, C and D).
Fig. 4. Coculture with the neonatal WT SCN slice rescues the circadian PER2::LUC rhythm in the SCN of Cry1,2−/− and Cry1,2−/−/Vipr2−/− mice.
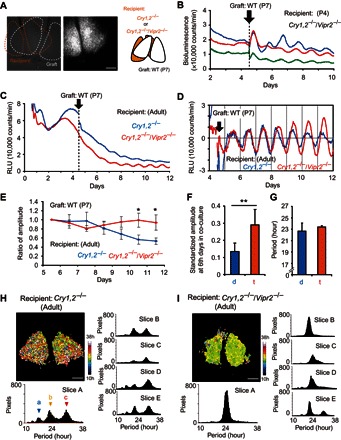
(A) Representative bright-field (left) and bioluminescent (middle) images of recipient and graft SCN slices in coculture and their schematic drawing (right). Scale bar, 200 μm. (B) Representative PER2::LUC rhythms in the neonate (P4) cocultured with a neonatal (P7) WT SCN slice are shown using three different colors. The abscissa indicates the days in culture. The day of coculture was indicated by an arrow. (C) Representative bioluminescence in the Cry1,2−/− (blue) and Cry1,2−/−/Vipr2−/− (red) adult SCN cocultured with a neonatal WT SCN slice. (D) Detrended PER2::LUC rhythms are the same as those indicated in (C). (E) Change in the amplitude of circadian PER2::LUC rhythm in the Cry1,2−/− (blue) and Cry1,2−/−/Vipr2−/− (red) adult SCN during the course of coculture with the neonatal WT SCN. The amplitudes are expressed as a ratio of the first amplitude after the coculture. Values are means and SD (n = 7 for both genotypes). *P < 0.05, two-way repeated-measures ANOVA with post hoc Student’s t test. (F) Standardized amplitude of rescued circadian PER2::LUC rhythm on the 12th culture day (6 days after coculture) of the Cry1,2−/− (d) (n = 7) and Cry1,2−/−/Vipr2−/− (t) (n = 7). **P < 0.01, Student’s t test. (G) Periods of rescued circadian PER2::LUC rhythm after coculture with the neonatal WT SCN, evaluated by a χ2 periodogram, showed no significant difference between Cry1,2−/− and Cry1,2−/−/Vipr2−/−. (H) Representative period map (slice A) and period distribution of circadian PER2::LUC rhythm in Cry1,2−/− SCN after coculture on the pixel level (slices A to E). Superimposed curves in the period distribution of slice A indicate three Gaussian curves fitted to the data. Arrowheads a, b, and c indicate the peaks of a Gaussian curve. (I) Representative period map and period distribution of circadian PER2::LUC rhythm in Cry1,2−/−/Vipr2−/− SCN after coculture on the pixel level (slices A to E).
The rescued circadian rhythm in the adult Cry1,2−/−/Vipr2−/− SCN persisted without obvious damping for up to 7 days after coculture (Fig. 4E), whereas the rescued circadian rhythm in the adult Cry1,2−/− SCN was gradually damped. The standardized amplitude on the sixth day in coculture and the ratio to that immediately after coculture were significantly higher in the Cry1,2−/−/Vipr2−/− SCN than in the Cry1,2−/− SCN (Fig. 4, E and F, and fig. S4, A and B). The circadian period of the rescued rhythm was not significantly different between the two genotypes (Fig. 4G).
The pixel-level analysis revealed a surprising difference in the neural network of rescued circadian rhythm between the Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN of adult mice. In most of the Cry1,2−/− SCN, three cell clusters were detected at the circadian period of 16.5 ± 0.2 hours, 21.8 ± 0.5 hours, and 30.0 ± 0.3 hours (n = 5), each of which was not significantly different from that of the respective cluster in the adult Cry1,2−/− SCN without coculture. By contrast, a single cell cluster was observed in the Cry1,2−/−/Vipr2−/− SCN at 22.6 ±1.1 hours (n = 5) except in one SCN (slice E) that showed three cell clusters (Fig. 4, H and I). These findings indicate that the neural network of the rescued tissue-level circadian rhythm differed between the Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN.
AVP signaling is critical for the restoration of circadian PER2::LUC rhythm in the Cry1,2−/−/Vipr2−/− SCN by coculture: Pharmacological evidence
The role of AVP from the graft SCN in the rhythm restoration was examined for the neonatal (n = 4) and adult Cry1,2−/−/Vipr2−/− SCN (n = 4), in addition to the adult Cry1,2−/− SCN (n = 6). A cocktail of SR49059 (AVP receptor V1a antagonist) and SSR149415 (AVP receptor V1a and V1b antagonists) was applied to the cultured SCN slices of Cry1,2−/− and Cry1,2−/−/Vipr2−/− mice, in which the tissue-level circadian rhythm had been rescued by coculturing with neonatal WT SCN.
The cocktail of AVP receptor antagonists significantly reduced the amplitude of the rescued circadian PER2::LUC rhythm in the adult (P = 0.006, Student’s t test) and neonatal (P = 0.017, Student’s t test) Cry1,2−/−/Vipr2−/− SCN (Fig. 5B and fig. S5, B and E). Unexpectedly, the cocktail significantly enhanced the amplitude of rescued circadian rhythm in the adult Cry1,2−/− SCN (P = 0.0001, Student’s t test) (Fig. 5A and fig. S5A). However, the circadian rhythm of the donor (WT neonatal) SCN was not affected by the AVP receptor antagonists (fig. S5, C and D).
Fig. 5. AVP in the graft SCN is responsible for the rescue of circadian PER2::LUC rhythms in the adult Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN.
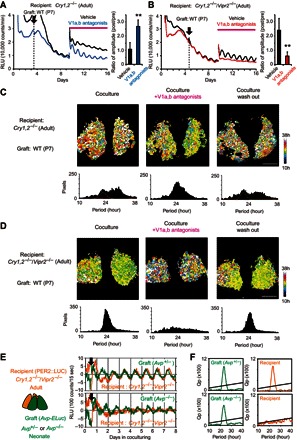
(A and B) Representative circadian PER2::LUC rhythms rescued in the arrhythmic adult SCN of Cry1,2−/− (A) and Cry1,2−/−/Vipr2−/− (B) mice by coculture of a neonatal WT SCN before and after treatment of AVP receptor (V1a and V1b) antagonists (blue and red) or vehicle (black). An arrow indicates the day of coculture, and a horizontal magenta line indicates the days under treatment. Coculture was done without exchanging culture medium, whereas the drug treatment was done by exchanging into a new medium containing drug or vehicle. The histogram shown on the right side demonstrates the mean ratios of circadian amplitude on the third day after drug treatment to that immediately before the treatment (Cry1,2−/−, n = 6; Cry1,2−/−/Vipr2−/−, n = 4; each for vehicle and antagonists). **P < 0.01, Student’s t test, significant difference between antagonists and vehicle treatments. (C and D) Representative period maps and period distributions of the rescued circadian PER2::LUC rhythm (left), after AVP antagonist treatment (middle) and washout of the antagonists (right) in the adult SCN of Cry1,2−/− (C) and Cry1,2−/−/Vipr2−/− (D) mice by coculture with a neonatal WT SCN on the pixel level. Superimposed curves in the period distribution indicate three Gaussian curves fitted to the data. Scale bars, 200 μm. (E) Schematic drawing of the coculture of the SCN slices and representative circadian PER2::LUC rhythm of the adult SCN slice of Cry1,2−/−/Vipr2−/− mice (orange) cocultured with the SCN from the heterozygote (top) and homozygote (bottom) of Avp knockout mice (green). Recipient adult SCN exhibits PER2::LUC bioluminescence (orange) and graft neonatal (P4 + 3 days culture) SCN exhibits Avp-ELuc bioluminescence (green), which were calculated after alternatively monitoring total and filtered bioluminescence in the same cocultured SCN slices. The abscissa indicates days in coculture. (F) χ2 periodograms for the circadian Avp-ELuc rhythms of graft SCN (left) and PER2::LUC rhythms of recipient SCN (right) in culture. An oblique line in the periodogram indicates the level of significance (P < 0.01).
The pixel-level analysis revealed different features of the cell clusters in the Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN of adult mice (Fig. 5, C and D, and fig. S5, F and G). In the Cry1,2−/− SCN, multiple cell clusters detected by coculture disappeared, and a single cell cluster appeared at 23.6 ± 0.1 hours (n = 3) by AVP receptor antagonists (Fig. 5C, middle, and fig. S5F, middle). Washout of the antagonists returned the network profile to the previous bimodality (Fig. 5C, right, and fig. S5F, right). By contrast, in the Cry1,2−/−/Vipr2−/− SCN, a single cell cluster detected by coculturing at 22.7 ± 0.2 hours (n = 3) (Fig. 5D, left, and fig. S5G, left) was completely abolished by AVP receptor antagonists. Washout of the antagonists returned the profile to the previous single cell cluster peaking at 23.2 ± 0.9 hours (n = 3) (Fig. 5D, right, and fig. S5G, right). These findings indicated that AVP from the graft SCN rescued the tissue-level circadian rhythm in the arrhythmic adult Cry1,2−/−/Vipr2−/− SCN.
The AVP-null SCN graft fails to rescue the tissue-level circadian PER2::LUC rhythm in the adult Cry1,2−/−/Vipr2−/− SCN
To confirm the role of AVP in the rescue of tissue-level circadian rhythms in the adult Cry1,2−/−/Vipr2−/− SCN, we examined the effects of the AVP-null SCN graft (Fig. 5, E and F). The AVP-null SCN was obtained from the newborn of Avp knockout mice carrying a bioluminescence reporter for the expression of the AVP gene (Avp-ELuc) (31). In these mutant mice, the AVP coding region was blocked by knocking-in the ELuc complementary DNA, which was expressed by activation of the AVP promoter. The homozygote mice are indistinguishable from the heterozygote or WT mice at birth, but they lose body weight thereafter and do not survive beyond postnatal day 7. As for the control, the SCN from the heterozygote mice was used. The heterozygote mice are indistinguishable from the WT mice with respect to body weight and circadian rhythms (31).
The AVP-null SCN was obtained on postnatal days 3 and 4 and was precultured for 3 to 4 days before coculture. The AVP-null SCN failed to rescue the tissue-level circadian rhythm in the adult Cry1,2−/−/Vipr2−/− SCN slice, whereas the heterozygote SCN succeeded in rescuing the rhythmicity (Fig. 5, E and F). The graft SCN of both the homozygote and heterozygote mice showed significant circadian Avp-ELuc rhythms (Fig. 5, E and F).
Avp expression in the neonatal and adult SCN is attenuated in Cry1,2−/− mice
Because AVP from the graft SCN is important for the rescue of the tissue-level circadian rhythm in the Cry1,2−/−/Vipr2−/− SCN, dysfunction of AVP signaling is possibly involved in an aperiodicity of the adult Cry1,2−/− SCN. We tested this hypothesis by measuring the circadian rhythm in the AVP gene (Avp) expression in the SCN of Cry1,2−/− mice. Cry1,2−/− mice were cross-mated with Avp-ELuc mice (31). As expected, the circadian Avp-ELuc expression rhythm was significantly attenuated in both the neonatal and adult Cry1,2−/− SCN (Fig. 6A). The 24-hour gene expression on the second day of culture was significantly smaller in the Cry1,2−/− SCN than in the WT, regardless of the age (Fig. 6B). When comparing between the two ages, the gene expression in the WT SCN significantly increased in adults than in neonates (P < 0.05, n = 3), but this age-related increase was not detected in the Cry1,2−/− SCN.
Fig. 6. Cry1,2−/− mice show attenuated circadian rhythms in Avp expression in the cultured SCN.
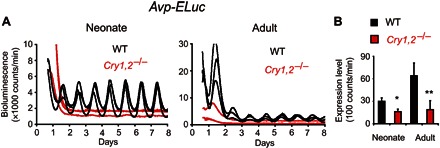
(A) Representative Avp-ELuc rhythms in the neonatal and adult SCN slices of WT (black) and Cry1,2−/− (red) mice. (B) Avp expression levels in the SCN of WT (black) and Cry1,2−/− (red) mice. Mean intensity of bioluminescence between 24 and 48 hours after culture was calculated as the Avp expression level in each SCN. Values are means and SD (n = 3 to 5). *P < 0.05; **P < 0.01, Student’s t test, significant difference between the WT and Cry1,2−/− mice.
DISCUSSION
Here, we found that a loss of VIP signaling abolished the tissue-level circadian PER2::LUC rhythm in the neonatal Cry1,2−/− SCN, which otherwise showed a robust circadian rhythm. The finding suggests that aperiodicity of the adult Cry1,2−/− SCN is due to the dysfunction of VIP signaling. However, genetic deletion of VIP signaling itself did not affect the tissue-level circadian rhythm in the SCN, indicating an involvement of another factor(s) in the aperiodicity of adult Cry1,2−/− SCN. Coculture with a neonatal WT SCN rescued the tissue-level circadian rhythm in the arrhythmic neonatal and adult Cry1,2−/−/Vipr2−/− SCN. Pharmacological experiments revealed that AVP from the graft SCN was primarily responsible for the rescue of tissue-level circadian rhythm in Cry1,2−/−/Vipr2−/− SCN.
Redundant but differential roles of VIP and AVP signaling in the integration of SCN neural network were revealed by pixel-level analyses of PER2::LUC bioluminescence. Multiple clusters of cellular circadian rhythms with different periodicities were identified in the arrhythmic neonatal Cry1,2−/−/Vipr2−/− SCN as well as in the adult Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN. These cell clusters appeared to be the fundamental units of the SCN neural network, because they frequently appear when the tissue-level circadian rhythm is disrupted.
Neural network for the expression of tissue-level circadian PER2::LUC rhythm in the SCN
CRY1 and CRY2 are necessary for the expression of the tissue-level circadian rhythm in the adult SCN but not in the neonatal SCN (Fig. 1) (17). We hypothesized that the difference was due to the postnatal change in the SCN neural network for coherent circadian rhythm (17). The present study shows that VIP signaling is critical in expressing the tissue-level circadian rhythm in the neonatal Cry1,2−/− SCN (Fig. 2).
A lack of VPAC2 in the neonatal Cry1,2−/− mice markedly changed the neural network from single to three cell clusters of different periods (Fig. 2). Each of the cell clusters was located at approximately 16, 22, and 30 hours. This finding was confirmed in the neonatal Cry1,2−/− SCN by pharmacological depletion of cAMP and Ca2+, major intracellular signal transmitters of the VIP signaling (Fig. 3, C and F). Three cell clusters were also detected in the adult Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN (Fig. 2C). Here, we assume that these three cell clusters are the fundamental units of SCN neural network. They may be integrated to a single cell cluster to express the tissue-level circadian rhythm. Three cell clusters have been observed in the SCN of WT mice under a photoperiodic long day (32).
In the neonatal Cry1,2−/− SCN, VIP signaling may integrate these cell clusters through oscillatory couplings, because the integrated single period was not a simple mathematical mean of the circadian periods. In the oscillatory coupling, rhythmic impacts of VIP are required for the constituent cellular oscillations to mutually synchronize (33). The requirement is fulfilled for the neonatal SCN but not for the adult SCN (23, 24). However, VIP signaling is not the only mechanism to integrate the cell clusters, because the tissue-level circadian rhythm was detected even without VPAC2 (Fig. 1). The circadian parameters, such as period of PER2::LUC rhythm in the Vipr2−/− mice, were not different from those in the WT mice except for reduced amplitude, and the tissue-level circadian rhythms were detected in the SCN regardless of whether the behavior rhythm was disrupted or not (34). Multiple cell clusters in the Vipr2−/− SCN are possibly integrated by other mechanisms, such as AVP signaling.
The circadian period was reported to differ in some subregions of the SCN (35, 36). In this respect, it is interesting to note that regional specificity was not clear in the period distribution of the three clusters (Fig. 2, A and C). Thus, clustering of cellular circadian rhythms and integrating the cell clusters to the tissue-level circadian rhythm represent different dynamics of the neural network.
A single cell cluster is associated with the appearance of the tissue-level circadian rhythm in the SCN (Fig. 2), but this relationship is not always the case, as demonstrated by coculture experiments in the adult Cry1,2−/− SCN (Fig. 4H). The number of cell clusters may reflect the robustness of the tissue-level circadian rhythm and, in turn, the strength of integration.
AVP and VIP signaling show redundancy in the expression of the tissue-level circadian rhythm in the SCN
The tissue-level circadian rhythm is not expressed in the neonatal Cry1,2−/− SCN without VIP signaling (Figs. 1 and 2). However, coculture with the neonatal WT SCN rescues the tissue-level circadian rhythm in the aperiodic neonatal and adult Cry1,2−/−/Vipr2−/− SCN (Fig. 4 and figs. S4 and S5). AVP signaling is most likely the mechanism of this restoration, because AVP receptor antagonists attenuate or abolish the restoration in the Cry1,2−/−/Vipr2−/− SCN (Fig. 5). Thus, there is redundancy in the function of the neural network to express the tissue-level circadian rhythm in the Cry1,2−/− SCN, in which signaling of both AVP and VIP is involved. AVP receptor antagonists failed to influence the tissue-level circadian rhythm in the neonatal WT SCN (fig. S5, C and D), which could be explained by the redundancy.
Curiously, the AVP receptor antagonists showed an opposite effect on the rescued circadian rhythm: suppression in the Cry1,2−/−/Vipr2−/− SCN and enhancement in the Cry1,2−/−SCN (Fig. 5, A and B). This paradoxical effect of the AVP receptor antagonists is explained by a difference in the neural network between the adult Cry1,2−/− and Cry1,2−/−/Vipr2−/− SCN (Fig. 5, C and D). After coculture, the Cry1,2−/− SCN showed essentially three cell clusters that were integrated into one cluster by the AVP receptor antagonists, whereas the Cry1,2−/−/Vipr2−/− SCN demonstrated only one cell cluster that was completely abolished by AVP receptor antagonists. The three cell clusters in the Cry1,2−/− SCN could have been integrated differentially by AVP and VIP signaling. The integration due to AVP signaling was abolished by AVP receptor antagonists, resulting in the enhancement of the tissue-level circadian rhythm (Fig. 5A). The competitive interactions between AVP and VIP signaling may diminish the robustness of the tissue-level circadian rhythm (Fig. 4, D to F).
Recently, Edwards et al. (26) reported that AVP receptor antagonists blocked the tissue-level circadian rhythms rescued by genetic induction of CRY1 in the aperiodic Cry1,2−/− SCN. The antagonist was effective when treated at the time of CRY1 induction, but failed to attenuate the rescued circadian rhythms, which rather enhanced the circadian amplitude, when treated several days later. They interpreted the results as that AVP signaling was required for the induction, but not for the maintenance, of CRY-dependent circadian rhythms. The dual effects of AVP receptor antagonists in their study could be alternatively explained by assuming delayed activation of VIP signaling by CRY1 induction [Fig. 4F from the study of Edward et al. (26)]. Attenuation of circadian rhythm by AVP receptor antagonists could be due to disintegration of oscillating cell clusters similar to Cry1,2−/−/Vipr2−/− SCN in the present study (Fig. 5, B and D), and persistence or enhancement of rhythmicity could be due to the integration of cell clusters by VIP signaling similar to Cry1,2−/− SCN (Fig. 5, A and C).
Differential integration of the SCN cell clusters by AVP and VIP signaling
The cell cluster in the neonatal Vipr2−/− and WT SCN showed the circadian period slightly but significantly longer than that in the neonatal Cry1,2−/− SCN (Fig. 2B), suggesting differential integration between the Cry1,2−/− and Vipr2−/− SCN. AVP and VIP signaling may integrate the cell clusters of these mutant mice in a different manner. This notion is based on three observations. First, three cell clusters detected in the neonatal Cry1,2−/−/Vipr2−/− SCN (Fig. 2) are largely due to AVP signaling (Fig. 5D). Second, a single cell cluster detected in the Vipr2−/− SCN (Fig. 2) is possibly due to AVP signaling and to other transmitters, such as gastrin-releasing peptide (16), and its circadian period is significantly different from that in the neonatal Cry1,2−/− SCN in which VIP signaling is involved. Third, the number of cell clusters after coculture and responses to the AVP receptor antagonists are different in the adult Cry1,2−/− SCN and Cry1,2−/−/Vipr2−/− SCN, in which VIP signaling and AVP signaling are differently involved (Fig. 5). The differential integration could be due to different distributions of VIP and AVP receptors in the SCN (22, 37).
VIP signaling is not involved in the buildup of each cell cluster, because three cell clusters are still evident in the neonatal Cry1,2−/−/Vipr2−/− SCN (Fig. 2A). On the other hand, VIP signaling is involved in the integration of cell clusters to express the tissue-level circadian rhythm (Figs. 2 and 3). By contrast, AVP signaling is involved in both the buildup of each cell cluster and the integration of cell clusters (Fig. 5, C and D). Figure S6 shows the proposed conceptual model for differential but redundant roles of AVP and VIP signaling in the SCN network for coherent circadian rhythms.
Postnatal changes of the neural network in the Cry1,2−/− SCN
Aperiodicity of the adult Cry1,2−/− SCN is explained by postnatal changes of the neural network in which attenuation of both VIP and AVP signaling is involved. VIP signaling in the adult Cry1,2−/− SCN is attenuated and loses the capability of integrating the SCN cell clusters. The notion is based on the finding that the neonatal Cry1,2−/− SCN showed three cell clusters similar to those in the adult SCN when Vipr2 was knocked out or VIP signaling was pharmacologically blocked (Figs. 2 and 3). The attenuation of VIP signaling is related with a loss of circadian rhythm, which is the natural course of development (24). The postnatal attenuation of VIP signaling may be affected by environmental lights, because continuous exposure of lights to neonatal Cry1,2−/− mice antagonized the age-related disruption of tissue-level circadian rhythm in the SCN and behavior (38).
AVP signaling is also involved in the integration of cell clusters, and attenuated AVP signaling may contribute to aperiodicity in the adult Cry1,2−/− SCN. Avp expression in the SCN was significantly reduced in the Cry1,2−/− mice, and the reduction was much larger in adults than in neonates (Fig. 6B). The age-related attenuation of AVP expression is specific to the Cry1,2−/− SCN.
Together, loss of the tissue-level circadian rhythm in the adult Cry1,2−/− SCN is attributable to two factors: attenuated circadian rhythms of VIP release as a natural course of development and attenuated AVP expression specific to the Cry1,2−/− SCN. In the neonatal Cry1,2−/− SCN, VIP signaling integrates the fundamental units of neural network—three cell clusters—that are built-up by AVP signaling, and possibly by others. On the other hand, in the adult Cry1,2−/− SCN, VIP signaling loses its potency to integrate the cell clusters, probably because of attenuated circadian nature. Tissue-level circadian rhythms are rescued in the Cry1,2−/− SCN by coculturing the WT SCN, probably through rhythmic releases of VIP and AVP from the graft. AVP signaling is a redundant mechanism for the integration of cell clusters, especially in the adult SCN. The integration by AVP signaling may target different cell populations, resulting in the tissue-level circadian rhythm of different periods. Thus, the cell network in the SCN undergoes ontogenic changes for coherent expression of circadian rhythm.
MATERIALS AND METHODS
Animals
Cry1,2−/− and Vipr2−/− mice of C57BL/6J background were acquired from Tohoku University (27) and Medical Research Council UK (20), respectively. The bioluminescence reporter system was introduced to each of the knockout mice by breeding with PER2::LUC homozygote mice carrying a PER2 fusion luciferase reporter (39). The mice were back-crossed with C57BL/6J mice for more than 13 generations. Cry1, Cry2, and Vipr2 triple-knockout mice (Cry1,2−/−/Vipr2−/−) were generated by crossing Cry1,2−/− mice carrying a PER2::LUC bioluminescence reporter with Vipr2−/− mice. To monitor Avp expression in Cry1,2−/− mice, we crossed them to Avp-ELuc heterozygote mice of C57BL/6J background (31).
Mice were bred and reared in our animal quarters, where environmental conditions were controlled (lights on, 0600 to 1800; light intensity, approximately 100 lux at the bottom of cage; humidity, 60 ± 10%). Both male and female mice were used for the experiments. The day of birth was defined as postnatal day 0. Experiments were conducted in compliance with the rules and regulations established by the Animal Care and Use Committee of Hokkaido University under the ethical permission of the Animal Research Committee of Hokkaido University (approval no. 08-0279).
Measurement of behavioral activity
Circadian rhythms in locomotor activity were continuously monitored in mice under LD for at least 1 week and under DD for at least 2 weeks. Mice were individually housed in a polycarbonate cage (182 × 260 × 128 mm) placed in a light-tight and air-conditioned box (40 × 50 × 50 cm). Spontaneous movements were measured by a passive infrared sensor, which detects a change in thermal radiation from an animal due to movements (40). The amount of movement was recorded every 1 min by a computer software program (The Chronobiology Kit, Stanford Software Systems). Circadian rhythms were evaluated by a χ2 periodogram with a significance level of P = 0.01.
SCN slice preparation for culture
For the measurement of PER2::LUC bioluminescence, animals were euthanized to harvest SCNs between 0800 and 1600. For the tissue level measurement, 300-μm-thick coronal SCN slices were made with a tissue chopper (McIlwain) from neonatal mice (postnatal day 7) and with a Microslicer DTK-1000 (Dosaka EM) from adult mice (2 to 6 months old). For the pixel-level measurement, 150- or 200-μm-thick coronal SCN slices were made with the tissue chopper or the Microslicer. The SCN tissue was dissected at the mid-rostrocaudal region, and a paired SCN was cultured on a Millicell-CM culture insert (Millipore Corporation). The culture conditions were the same as those described previously (17). Briefly, the slice was cultured in air at 36.5°C with 1.2 ml of Dulbecco’s modified Eagle’s medium (Invitrogen) with 0.1 mM d-luciferin K and 5% supplement solution.
In the coculture experiment, the 150-μm-thick SCN slices were obtained from adult mice carrying the PER2::LUC reporter (recipient). The slice was precultured for 3 or 4 days and then cocultured with an SCN slice from mice without the reporter system (donor). The 200-μm-thick donor SCN slice was obtained from 7-day-old WT mice and precultured for 1 day before coculture. When cocultured, the graft SCN slice was placed inside out on the surface of the recipient SCN slice. We measured bioluminescence from the beginning of culturing of the recipient SCN until at least 5 days after coculture.
An inhibitor of adenylyl cyclase MDL-12330A (Sigma), Ca2+-chelating agent BAPTA-AM (Sigma), and AVP receptor antagonists (V1a receptor antagonist: SR49059, Tocris; V1b receptor antagonist: SSR149415, Axon Medchem) were dissolved in water (SR49059 and SSR149415; final concentration, 2.5 μM) or dimethyl sulfoxide (DMSO) (MDL-12330A and BAPTA-AM). The chemicals were either directly added to the culture medium (bath application) or dissolved in the culture medium to exchange with the whole medium in culture.
Measurement of bioluminescence
Bioluminescence on the SCN tissue level was measured using a PMT (LumiCycle, Actimetrics; Kronos; ATTO) at 10-min intervals with an exposure time of 1 min. Bioluminescence on the pixel level in the cultured SCN slice was measured using an electron-multiplying CCD (ImagEM, Hamamatsu Photonics; iXon, Andor) or CCD (ORCA-II, Hamamatsu Photonics) camera. Bioluminescence was measured every 60 min with an exposure time of 59 min. The intensity of bioluminescence was expressed in relative light units (RLU; counts/hour). The measurement was continued typically for 3 to 10 days as previously described (17).
Bioluminescence of Avp-ELuc and PER2::LUC from the same SCN was monitored alternatively by a dish-type luminometer (Kronos, ATTO). Bioluminescence emitted from firefly luciferase (F-luc) and enhanced green-emitting luciferase (E-luc) was separated with a 600-nm long-pass filter (R60 filter, Hoya). We measured the transmittance ratio of bioluminescence from each luciferase and calculated the contributions of different luciferases to the total emission using the following equation (41, 42)
where E and F are the light intensity values for E-luc and F-luc, respectively; F0 and F1 are the total RLU and the RLU passed through the filter; and KE and KF are the optical filter’s transmission coefficients for E-luc and F-luc, respectively. Both transmission coefficients were very stable under culture conditions throughout the day and for, as far as we examined, up to 17 days. Bioluminescence was measured for 15 s in the presence of the long-pass filter and for the following 15 s in the absence of the filter. The measurement was repeated at 10-min intervals. The intensities of Avp-ELuc and PER2::LUC bioluminescence were calculated as previously described by Nakajima et al. (41) and Noguchi et al. (42).
Data analysis
The property of circadian rhythm in bioluminescence signals on the tissue level was analyzed as previously described (17). Bioluminescence records of the first 12 hours were not used for rhythm analyses because of an initial high level of bioluminescence. The raw bioluminescence data were smoothed using a five-point moving average method and then detrended using a 24-hour running average subtraction method (17). The significance of a circadian rhythm and the rhythm parameters were evaluated by two methods, depending on the stability of recording. We used a χ2 periodogram (ClockLab) for the tissue-level bioluminescent rhythm. The periodogram analysis was applied for a record of seven consecutive days in a period range of 10.0 to 38.0 hours with a significance level of P = 0.01. When more than one significant peak was detected in the periodogram, a cosine curve fitting method was performed to exclude the harmonics. The significance of the curve fitting was evaluated using the percent rhythm analysis (P < 0.05). The circadian period was calculated by a χ2 periodogram. The circadian amplitude was defined as the difference between the peak and trough in a circadian cycle and standardized by the peak level as previously described (17).
The significance and property of circadian rhythm in bioluminescence signals on the pixel level were analyzed using a cosine curve fitting method as previously described by Mieda et al. (13) and Yoshikawa et al. (31). Because an SCN slice is slowly deformed during culturing, the position of a pixel is affected in the long run, and the spatial accuracy of circadian rhythms is reduced. By contrast, bioluminescence from the whole SCN was not significantly affected by this deformation of the SCN structure. On this reason, the pixel-level circadian rhythms are not analyzed by χ2 periodogram, which requires several cycles of circadian rhythms. The 48- or 72-hour raw data were applied to the curve fitting after subtracting the background level of bioluminescence. The background level was calculated using ROI outside the SCN for each slice and was defined as mean + 5× SD. The significance of the curve fitting was evaluated using the percent rhythm analysis (P < 0.01) (43). The parameters (period, amplitude, and phase) of circadian rhythm were estimated from the best fitted cosine curve and expressed as heat maps. The distribution of period lengths was analyzed by fitting a Gaussian equation to a period histogram using the least squares method. The equation contains three Gaussian formulas with respective baseline levels (triple Gaussian), which was based on the assumption that the SCN network contains three fundamental cell clusters (see “Neural network for the expression of tissue-level circadian PER2::LUC rhythm in the SCN” in Discussion). Each Gaussian formula contains a different median (μ) (μ1 < μ2 < μ3). Variance (σ) was assumed to be less than 10 hours for each formula, and the baseline was larger than zero. The range of period analyzed was set from 10 to 38 hours.
The peak of a fitted Gaussian curve was regarded as the period of a cluster peak. Bioluminescence signals on the cell-size ROI level were also analyzed for the property of circadian rhythm using Aquacosmos software (Hamamatsu Photonics). The significance of circadian rhythm was examined by χ2 periodogram using a record of five consecutive days.
Statistics
Student’s t test was used when two independent group means were compared, and Welch’s t test was used when the variances of two group means were different. A paired t test was used when dependent group means were compared. A one-way ANOVA with a post hoc Tukey-Kramer test was applied to data of a single time series, and Kruskal-Wallis test was used when the variances of group means were different. A two-way ANOVA was adapted when two independent time series data were compared (StatView or Statcel 3).
Supplementary Material
Acknowledgments
We thank G. T. van der Horst and A. Yasui for supplying the Cry1,2−/− mice, J. S. Takahashi for supplying the PER2::LUC mice, M. H. Hastings for giving the Vipr2−/− mice, and Y. Nakajima for supplying the Avp-ELuc mice. We also thank S. Kuroda and T. Ueda for the analysis program and M. P. Butter for the intensive discussion. Funding: This work was supported in part by the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program, Ministry of Education, Culture, Sports, Science and Technology, Japan and the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) (nos. 24390055, 26860156, and 15H04679). Author contributions: D.O., S.H., and K.-i.H. designed the study. D.O. performed the experiments and analyzed data. D.O., S.H., and K.-i.H. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/9/e1600960/DC1
fig. S1. Circadian rhythms in locomotor activity in the WT, Vipr2−/−, Cry1,2−/−, and Cry1,2−/−/Vipr2−/− mice.
fig. S2. Circadian rhythms in PER2::LUC in the cultured SCN of Cry1,2−/−/Vipr2−/− mice.
fig. S3. Circadian PER2::LUC rhythms in the cultured SCN slice of Cry1,2−/− mice applied with MDL-12330A or BAPTA-AM of different doses.
fig. S4. Rescue of the circadian PER2::LUC rhythm in the arrhythmic adult SCN slice of Cry1,2−/− or Cry1,2−/−/Vipr2−/− mice by coculture with a neonatal WT SCN.
fig. S5. AVP receptor antagonists modify rescued circadian PER2::LUC rhythms in the arrhythmic adult Cry1,2−/− or Cry1,2−/−/Vipr2−/− SCN.
fig. S6. A differential cell cluster model for the mouse SCN neural network.
REFERENCES AND NOTES
- 1.Moore R. Y., Eichler V. B., Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42, 201–206 (1972). [DOI] [PubMed] [Google Scholar]
- 2.Stephan F. K., Zucker I., Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U.S.A. 69, 1583–1586 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reppert S. M., Weaver D. R., Coordination of circadian timing in mammals. Nature 418, 935–941 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Welsh D. K., Logothetis D. E., Meister M., Reppert S. M., Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Honma S., Nakamura W., Shirakawa T., Honma K.-i., Diversity in the circadian periods of single neurons of the rat suprachiasmatic nucleus depends on nuclear structure and intrinsic period. Neurosci. Lett. 358, 173–176 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Herzog E. D., Aton S. J., Numano R., Sakaki Y., Tei H., Temporal precision in the mammalian circadian system: A reliable clock from less reliable neurons. J. Biol. Rhythms 19, 35–46 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi S., Isejima H., Matsuo T., Okura R., Yagita K., Kobayashi M., Okamura H., Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Maywood E. S., Reddy A. B., Wong G. K. Y., O’Neill J. S., O’Brien J. A., McMahon D. G., Harmar A. J., Okamura H., Hastings M. H., Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Liu A. C., Welsh D. K., Ko C. H., Tran H. G., Zhang E. E., Priest A. A., Buhr E. D., Singer O., Meeker K., Verma I. M., Doyle F. J. III, Takahashi J. S., Kay S. A., Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwell C. S., Michel S., Itri J., Rodriguez W., Tam J., Lelievre V., Hu Z., Liu X., Waschek J. A., Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285, R939–R949 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Hughes A. T., Fahey B., Cutler D. J., Coogan A. N., Piggins H. D., Aberrant gating of photic input to the suprachiasmatic circadian pacemaker of mice lacking the VPAC2 receptor. J. Neurosci. 24, 3522–3526 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee I. T., Chang A. S., Manandhar M., Shan Y., Fan J., Izumo M., Ikeda Y., Motoike T., Dixon S., Seinfeld J. E., Takahashi J. S., Yanagisawa M., Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85, 1086–1102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mieda M., Ono D., Hasegawa E., Okamoto H., Honma K.-i., Honma S., Sakurai T., Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85, 1103–1116 (2015). [DOI] [PubMed] [Google Scholar]
- 14.van der Horst G. T. J., Muijtjens M., Kobayashi K., Takano R., Kanno S.-i., Takao M., de Wit J., Verkerk A., Eker A. P. M., van Leenen D., Buijs R., Bootsma D., Hoeijmakers J. H. J., Yasui A., Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Okamura H., Miyake S., Sumi Y., Yamaguchi S., Yasui A., Muijtjens M., Hoeijmakers J. H. J., van der Horst G. T. J., Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286, 2531–2534 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Maywood E. S., Chesham J. E., O’Brien J. A., Hastings M. H., A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. U.S.A. 108, 14306–14311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono D., Honma S., Honma K.-i., Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat. Commun. 4, 1666 (2013). [DOI] [PubMed] [Google Scholar]
- 18.van den Pol A. N., Tsujimoto K. L., Neurotransmitters of the hypothalamic suprachiasmatic nucleus: Immunocytochemical analysis of 25 neuronal antigens. Neuroscience 15, 1049–1086 (1985). [DOI] [PubMed] [Google Scholar]
- 19.Abrahamson E. E., Moore R. Y., Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Harmar A. J., Marston H. M., Shen S., Spratt C., West K. M., Sheward W. J., Morrison C. F., Dorin J. R., Piggins H. D., Reubi J.-C., Kelly J. S., Maywood E. S., Hastings M. H., The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Aton S. J., Colwell C. S., Harmar A. J., Waschek J., Herzog E. D., Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 8, 476–483 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi Y., Suzuki T., Mizoro Y., Kori H., Okada K., Chen Y., Fustin J.-M., Yamazaki F., Mizuguchi N., Zhang J., Dong X., Tsujimoto G., Okuno Y., Doi M., Okamura H., Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342, 85–90 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Shinohara K., Honma S., Katsuno Y., Abe H., Honma K., Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc. Natl. Acad. Sci. U.S.A. 92, 7396–7400 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ban Y., Shigeyoshi Y., Okamura H., Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J. Neurosci. 17, 3920–3931 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinohara K., Funabashi T., Kimura F., Temporal profiles of vasoactive intestinal polypeptide precursor mRNA and its receptor mRNA in the rat suprachiasmatic nucleus. Mol. Brain Res. 63, 262–267 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Edwards M. D., Brancaccio M., Chesham J. E., Maywood E. S., Hastings M. H., Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 2732–2737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honma S., Yasuda T., Yasui A., van der Horst G. T. J., Honma K.-i., Circadian behavioral rhythms in Cry1/Cry2 double-deficient mice induced by methamphetamine. J. Biol. Rhythms 23, 91–94 (2008). [DOI] [PubMed] [Google Scholar]
- 28.An S., Irwin R. P., Allen C. N., Tsai C., Herzog E. D., Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J. Neurophysiol. 105, 2289–2296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H., cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–953 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen H. S., Hannibal J., Fahrenkrug J., Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur. J. Neurosci. 15, 570–574 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Yoshikawa T., Nakajima Y., Yamada Y., Enoki R., Watanabe K., Yamazaki M., Sakimura K., Honma S., Honma K.-i., Spatiotemporal profiles of arginine vasopressin transcription in cultured suprachiasmatic nucleus. Eur. J. Neurosci. 42, 2678–2689 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Inagaki N., Honma S., Ono D., Tanahashi Y., Honma K.-i., Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc. Natl. Acad. Sci. U.S.A. 104, 7664–7669 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ananthasubramaniam B., Herzog E. D., Herzel H., Timing of neuropeptide coupling determines synchrony and entrainment in the mammalian circadian clock. PLOS Comput. Biol. 10, e1003565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes A. T. L., Guilding C., Piggins H. D., Neuropeptide signaling differentially affects phase maintenance and rhythm generation in SCN and extra-SCN circadian oscillators. PLOS One 6, e18926 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noguchi T., Watanabe K., Ogura A., Yamaoka S., The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur. J. Neurosci. 20, 3199–3202 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Koinuma S., Asakawa T., Nagano M., Furukawa K., Sujino M., Masumoto K.-H., Nakajima Y., Hashimoto S., Yagita K., Shigeyoshi Y., Regional circadian period difference in the suprachiasmatic nucleus of the mammalian circadian center. Eur. J. Neurosci. 38, 2832–2841 (2013). [DOI] [PubMed] [Google Scholar]
- 37.An S., Tsai C., Ronecker J., Bayly A., Herzog E. D., Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J. Comp. Neurol. 520, 2730–2741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono D., Honma S., Honma K.-i., Postnatal constant light compensates Cryptochrome1 and 2 double deficiency for disruption of circadian behavioral rhythms in mice under constant dark. PLOS One 8, e80615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo S.-H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H.-K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S., PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe H., Honma S., Ohtsu H., Honma K.-i., Circadian rhythms in behavior and clock gene expressions in the brain of mice lacking histidine decarboxylase. Mol. Brain Res. 124, 178–187 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Nakajima Y., Ikeda M., Kimura T., Honma S., Ohmiya Y., Honma K.-i., Bidirectional role of orphan nuclear receptor RORα in clock gene transcriptions demonstrated by a novel reporter assay system. FEBS Lett. 565, 122–126 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Noguchi T., Ikeda M., Ohmiya Y., Nakajima Y., Simultaneous monitoring of independent gene expression patterns in two types of cocultured fibroblasts with different color-emitting luciferases. BMC Biotechnol. 8, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson W., Tong Y. L., Lee J. K., Halberg F., Methods for cosinor-rhythmometry. Chronobiologia 6, 305–323 (1979). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/9/e1600960/DC1
fig. S1. Circadian rhythms in locomotor activity in the WT, Vipr2−/−, Cry1,2−/−, and Cry1,2−/−/Vipr2−/− mice.
fig. S2. Circadian rhythms in PER2::LUC in the cultured SCN of Cry1,2−/−/Vipr2−/− mice.
fig. S3. Circadian PER2::LUC rhythms in the cultured SCN slice of Cry1,2−/− mice applied with MDL-12330A or BAPTA-AM of different doses.
fig. S4. Rescue of the circadian PER2::LUC rhythm in the arrhythmic adult SCN slice of Cry1,2−/− or Cry1,2−/−/Vipr2−/− mice by coculture with a neonatal WT SCN.
fig. S5. AVP receptor antagonists modify rescued circadian PER2::LUC rhythms in the arrhythmic adult Cry1,2−/− or Cry1,2−/−/Vipr2−/− SCN.
fig. S6. A differential cell cluster model for the mouse SCN neural network.


