KCNE3 modulates the KCNQ1 K+ channel in epithelia by directly stabilizing the voltage-sensor S4 segment in its activated state.
Keywords: Structural biology, KCNQ1, Kv7.1, KCNE3, channel, cystic fibrosis, estrogen, potassium, NMR, integrative modeling
Abstract
The single-span membrane protein KCNE3 modulates a variety of voltage-gated ion channels in diverse biological contexts. In epithelial cells, KCNE3 regulates the function of the KCNQ1 potassium ion (K+) channel to enable K+ recycling coupled to transepithelial chloride ion (Cl−) secretion, a physiologically critical cellular transport process in various organs and whose malfunction causes diseases, such as cystic fibrosis (CF), cholera, and pulmonary edema. Structural, computational, biochemical, and electrophysiological studies lead to an atomically explicit integrative structural model of the KCNE3-KCNQ1 complex that explains how KCNE3 induces the constitutive activation of KCNQ1 channel activity, a crucial component in K+ recycling. Central to this mechanism are direct interactions of KCNE3 residues at both ends of its transmembrane domain with residues on the intra- and extracellular ends of the KCNQ1 voltage-sensing domain S4 helix. These interactions appear to stabilize the activated “up” state configuration of S4, a prerequisite for full opening of the KCNQ1 channel gate. In addition, the integrative structural model was used to guide electrophysiological studies that illuminate the molecular basis for how estrogen exacerbates CF lung disease in female patients, a phenomenon known as the “CF gender gap.”
INTRODUCTION
KCNE3 [MinK-related peptide 2 (MiRP2); Fig. 1] is a widely expressed (1–4) single-span integral membrane protein that modulates the function and trafficking of several voltage-gated potassium channels, including KCNQ1, KCNQ4, hERG, Kv2.1, Kv3.1, and Kv3.2 (2, 4–6). In intestinal and tracheal epithelia, KCNE3 complexes with KCNQ1 and plays a crucial role in potassium ion recycling, which is required for transepithelial chloride ion secretion (Fig. 1) (1, 6, 7). KCNE3 converts KCNQ1 into a voltage-independent and constitutively active “leak” channel with enhanced activity (2, 4, 8). KCNE3 has been implicated in disorders associated with salt and fluid homeostasis, such as cystic fibrosis (CF). Its proposed role as a potential modifier of CF is based on impaired transepithelial Cl− efflux observed in homozygous Kcne3 knockout mice (1). Epidemiological studies of CF have indicated that females generally suffer significantly higher mortality rates than do males and have shorter life spans (9, 10), and estrogen has been postulated to contribute to this gender gap in CF survival (11). Estrogen treatment leads to disruption of the KCNE3-KCNQ1 channel complex (12, 13) and reduced channel activity, with KCNE3 Ser82 being critically involved, possibly in a phosphorylation-dependent manner (1, 12, 14). Therefore, it is likely that the observed effect of estrogen on KCNQ1-KCNE3–dependent currents may also contribute to the CF gender gap. Here, we combined experimental structural biology and electrophysiology with computational modeling (fig. S1) to develop an atomically explicit integrative working structural model that suggests how KCNE3 modulates KCNQ1 function and provides an underlying mechanism for rapid estrogen-dependent reduction in KCNQ1-KCNE3 activity.
Fig. 1. The KCNE3 protein and its function.
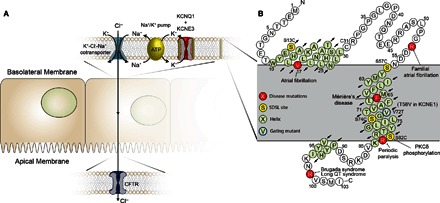
(A) Role of the KCNQ1-KCNE3 channel complex in chloride ion secretion. KCNE3 modulates the voltage-gated potassium channel KCNQ1, removing voltage-dependent gating, leading to a constitutively open leak channel. The KCNQ1-KCNE3 complex is expressed in basolateral epithelial membranes, where it plays a role in K+ recycling necessary for Cl− secretion across the apical membrane. Disruptions in transepithelial Cl− transport are involved in human pathologies, such as CF and cholera. (B) Sequence and membrane topology of KCNE3. The α-helical regions determined by NMR are highlighted in light green. Sites of disease-linked mutations are highlighted in red, whereas yellow sites were mutated to cysteine and spin-labeled to enable PRE NMR distance measurements. SDSL, site-directed spin labeling; PKCδ, protein kinase Cδ.
RESULTS
Structure and membrane topology of KCNE3
The structure of KCNE3 in isotropic dihexanoylphosphatidylcholine (DHPC)/dimyristoylphosphatidylglycerol (DMPG) bicelles was determined using solution nuclear magnetic resonance (NMR) spectroscopy (15), which was followed by molecular dynamics refinement in an explicit water-membrane medium using NMR-derived restraints (figs. S2 to S5 and table S1) (16–18). Structure determination was complemented by mapping the membrane topology of KCNE3 in bicelles by NMR paramagnetic relaxation enhancement (PRE) measurements using both water-soluble [Gd(III)-diethylenetriaminepentaacetic acid (Gd-DTPA)] and lipophilic [16-doxyl stearicacid (16-DSA)] paramagnetic probes (Fig. 2). NMR relaxation measurements that provided insight into the conformational dynamics of the protein were also conducted (fig. S6).
Fig. 2. Assessment of KCNE3 topology in bicelles from Gd3+ and 16-DSA paramagnet-induced NMR peak broadening.
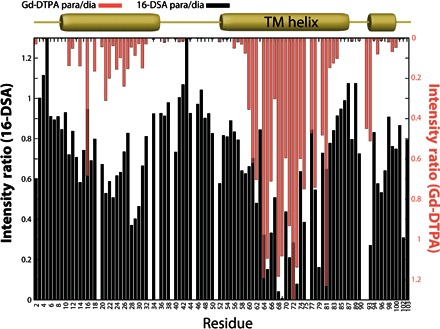
1H-15N TROSY NMR (800 MHz) peak intensity reductions induced by the lipophilic 16-DSA and hydrophilic Gd-DTPA paramagnetic probes relative to a matched paramagnet-free reference spectrum are shown as bar graphs in black and salmon, respectively. All three segments with helical secondary structure have some degree of protection from Gd-DTPA and access to 16-DSA. Measurements were carried out for KCNE3 solubilized in DHPC/DMPG bicelles at pH 6.5 and 40°C. TM, transmembrane.
The KCNE3 structure is composed of an extracellular N-terminal surface-associated amphipathic helix (residues 10 to 30) connected by a flexible loop to the α-helical transmembrane domain (TMD; 57 to 82), followed by a short juxtamembrane helix (90 to 95) and a disordered C terminus (96 to 103) (Figs. 1B and 3 and fig. S7). The α-helical TMD exhibits a moderate degree of curvature, with bending being most pronounced near the C-terminal end of the helical TMD. Residues Thr71, Ser74, and Gly78 line the concave face of the curved TMD (Fig. 3).
Fig. 3. Structure of KCNE3 in a bilayer membrane.
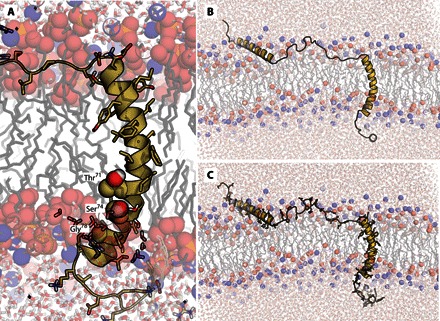
(A) Representative example from final KCNE3 structure ensemble output from experimentally restrained AMBER molecular dynamics calculations. Only the TMD is shown, as embedded in a dimyristoylphosphatidylcholine (DMPC) bilayer. Residues Thr71, Ser74, and Gly78 are depicted as spheres to highlight the concave face of KCNE3. (B) Same as (A) but with a different view, which includes the surface-associated helices flanking the TMD. (C) Same as (B) but with side chains also shown. The ensemble of 10 final models from the restrained dynamics simulation in a membrane environment is shown in fig. S7.
To confirm that the observed curvature of the KCNE3 TMD is an inherent property of the protein and not an artifact of the bicelle model membranes used to solubilize KCNE3 for NMR, we conducted pulsed double electron-electron resonance (DEER) (19–21) electron paramagnetic resonance (EPR) experiments on the protein in lipid bilayers. KCNE3 was nitroxide spin-labeled at both ends of the TMD and probed by DEER to compare the curvature-dependent probe-to-probe distances measured in micelles [lyso-myristoylphosphatidylcholine (LMPC)], bicelles (DMPG/DHPC), and 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG) lipid bilayers (vesicles; Fig. 4). The resulting distances for all three media are statistically identical, with the most populated end-to-end distances determined to be 35.8 ± 0.9 Å, 36.0 ± 1.0 Å, and 35.7 ± 1.1 Å, respectively. These results indicate that the curvature of the TMD is an intrinsic property of KCNE3 that is maintained in lipid bilayers, bicelles, and detergent micelles.
Fig. 4. DEER EPR–based distance measurements for KCNE3 in different model membrane conditions confirm the persistently curved nature of its TMD.
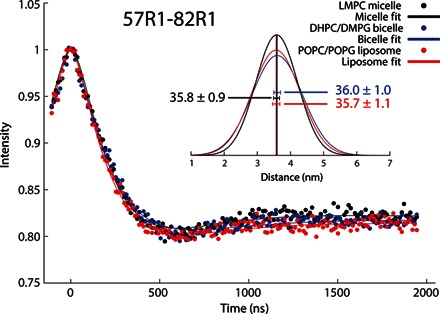
X-band DEER spectroscopy time evolutions are shown for double spin-labeled KCNE3 at the TMD ends (residues Ser57Cys and Ser82Cys) in different model membranes. (Inset) Distance distributions between the spin labels calculated from the DEER data. The average distances in the different membrane environments are shown with the given uncertainties reflecting the SEs from fitting of the data with a 95% confidence interval. DEER EPR data show similar TMD distance measurements taken in LMPC micelles, DHPC/DMPG bicelles, or POPC/POPG bilayers.
Additional insight into KCNE3-membrane interactions was gleaned from an NMR data–restrained molecular dynamics (rMD) simulation of the protein in an explicit bilayer. Analysis of the time-evolved rMD trajectory suggests that the curvature of the KCNE3 TMD helix is stabilized by transient contact with water (Fig. 3), a feature supported by experimental NMR-based backbone hydrogen-deuterium exchange measurements (fig. S8). The backbone amide nitrogen for residues Thr71, Ser74, and Leu75 all experience elevated water access compared to neighboring residues, as can be seen from the backbone hydrogen-deuterium exchange measurements and frequency of water contact throughout the rMD trajectory (fig. S8). The presence of water likely stabilizes the observed curvature by reducing the energetic penalty of breaking backbone hydrogen bonds (22).
The rMD trajectory also offers insight into both the N-terminal and C-terminal helix interactions with the lipid bilayer (Fig. 3). Both helices have an amphipathic sequence that does not deeply embed into the membrane, allowing for a dynamic interaction with the membrane surface. Experimentally, this dynamic interaction with the membrane is suggested by modest protection from the soluble probe Gd-DTPA for each of these helices (Fig. 2) and by NMR 15N relaxation profiles for each of these segments (fig. S6). This is in contrast to the flexible linking segments connecting the N- and C-terminal amphipathic helices to the TMD, which are largely unstructured, flexible, and water-accessible (Fig. 2 and figs. S6 and S8).
KCNQ1 structural elements required for KCNE3 modulation
We next exploited differences in the KCNQ1 and KCNQ4 channels using mutagenesis followed by electrophysiology to provide insight into how KCNE3 interacts with KCNQ1. The KCNQ4 potassium channel is homologous with KCNQ1 (fig. S9) but is inhibited by KCNE3 rather than being constitutively activated as for KCNQ1 (Fig. 5, A and B) (2, 4, 8). We expanded upon an earlier study of the interaction of KCNE3 with KCNQ1/KCNQ4 chimeras (23) by swapping each individual transmembrane helix of KCNQ4 into KCNQ1 to identify which segments of KCNQ1 are required for the constitutive activation by KCNE3. Each KCNQ1/4 chimera was studied using whole-cell patch-clamp recording in the absence and presence of KCNE3 (Fig. 5, C to H). The data identify the S4 helix as the critical segment that determines the KCNE3-dependent modulatory differences between KCNQ1 and KCNQ4. Transplanting the S4 segment from KCNQ4 into KCNQ1 yields a channel, KCNQ1[Q4S4], that is KCNQ4-like in the absence of KCNE3 (Fig. 5F, middle panel). Moreover, the channel function of KCNQ1[Q4S4] is inhibited by KCNE3, thereby functionally resembling wild-type (WT) KCNQ4 (Fig. 5F, right panel). We further refined the functional determinants of KCNE3 modulation to two pairs of KCNQ1-to-KCNQ4 mutations, His240Arg/Gln244Arg and Val241Met/Gln244Arg, either pair of which was sufficient to convert KCNQ1 into a KCNQ4-like channel that is inhibited by KCNE3 (Fig. 6). These results strongly suggest that KCNE3 is proximal to KCNQ1 residues His240, Val241, and Gln244 in the channel complex.
Fig. 5. The S4 domain of KCNQ4 causes KCNQ1 to be inhibited by KCNE3.
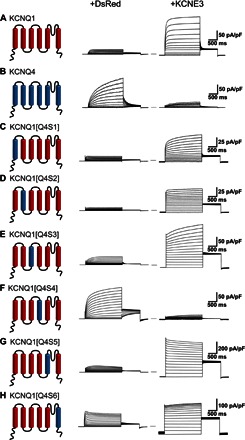
Average whole-cell currents recorded from cells transiently expressing KCNQ1/KCNQ4 chimeras alone (+DsRed) or with KCNE3: (A) KCNQ1 + KCNE3, (B) KCNQ4 + KCNE3, (C) KCNQ1 with the S1 domain of KCNQ4, (D) KCNQ1 with the S2 domain of KCNQ4, (E) KCNQ1 with the S3 domain of KCNQ4, (F) KCNQ1 with the S4 domain of KCNQ4, (G) KCNQ1 with the S5 domain of KCNQ4, and (H) KCNQ1 with the S6 domain of KCNQ4.
Fig. 6. Pairwise replacement of sites 240 + 244 or 241 + 244 in the S4 domain of KCNQ1 with the corresponding residues of KCNQ4 causes KCNQ1 to be inhibited by KCNE3.
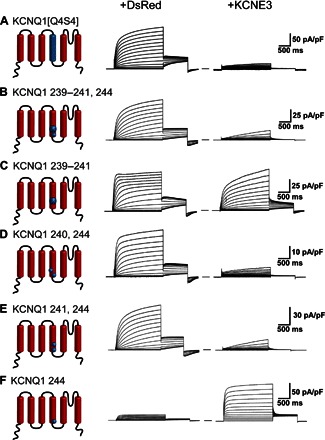
Average whole-cell currents recorded from cells transiently expressing KCNQ1 mutants or KCNQ4 chimeras, both alone (+DsRed) and with KCNE3: (A) KCNQ1 with the S4 domain of KCNQ4 (KCNQ1[Q4S4]); (B) Leu239Val,His240Arg,Val241Met,Gln244Arg KCNQ1; (C) Leu239Val,His240Arg,Val241Met KCNQ1; (D) His240Arg,Gln244Arg KCNQ1; (E) Val241Met,Gln244Arg KCNQ1; and (F) Gln244Arg KCNQ1. We note that the His240Arg, Gln244Arg KCNQ1, and Gln244Arg KCNQ1 mutants have been previously examined (36), with somewhat different results being obtained. However, our studies were conducted using CHO cells, which are bereft of endogenous expression of KCNE subunits, whereas the previous studies were conducted using KCNQ1 expressed in oocytes, in which the expression of either endogenous KCNE subunits or endogenous K+ channels can confound studies of potassium channels (75–77).
With the above results as restraints, preliminary models for the KCNE3-KCNQ1 complex led to two experimentally testable predictions: (i) The extracellular N-terminal end of the KCNE3 TMD interacts with the extracellular end of the KCNQ1 S1 transmembrane segment, and (ii) the C-terminal cytosolic end of the KCNE3 TMD interacts with the intracellular C terminus of the S4 segment in the voltage-sensing domain (VSD) located at the membrane/cytosol interface. We tested the model-predicted proximity between the KCNQ1 and KCNE3 sites by introducing a cysteine into each protein and then testing for disulfide bond formation, a positive result for which would be taken as evidence for interprotein site-site proximity and experimental validation of the preliminary KCNE3-KCNQ1 model complex. Double mutations of (i) KCNQ1 Leu142Cys (near the extracellular end of S1) plus KCNE3 Met59Cys (near the N-terminal end of the TMD) and (ii) KCNQ1 Gln244Cys (at the cytosolic end of S4) plus KCNE3 Ser82Cys (near the C-terminal end of the TMD) in both cases led to highly conductive channels under reducing conditions but displayed lower current amplitude under oxidizing conditions (Fig. 7). The most direct explanation of these results is that disulfide bond formation between either pair of residues traps the KCNQ1-KCNE3 channel in a conformation that resembles the fully activated channel state but that is slightly perturbed by disulfide bond formation and therefore has lower conductance. This is consistent with the notion that the WT side chains for these pairs of sites are proximal to each other but have suboptimal geometry for disulfide bond formation.
Fig. 7. Oxidation state–dependent electrophysiology measurements confirm that KCNQ1 Leu142 and KCNE3 Met59 residues are in close contact, as are KCNE3 Ser82 and KCNQ1 Gln244.
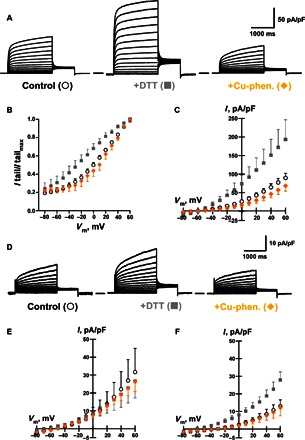
(A) Average whole-cell currents recorded from cells transiently expressing KCNQ1 Leu142Cys plus KCNE3 Met59Cys exposed to control bath solution, +DTT, or Cu-phenanthroline (Cu-phen.). The solid line between the two traces indicates the zero current. (B) Voltage dependence of activation for currents recorded from cells expressing KCNQ1 Leu142Cys plus KCNE3 Met59Cys exposed to control bath solution (○), +DTT (▪), or Cu-phenanthroline (♦). (C) Average current-voltage (I-V) relationships normalized by membrane capacitance measured from cells expressing KCNQ1 Leu142Cys plus KCNE3 Met59Cys exposed to control bath solution (○), +DTT (▪), or Cu-phenanthroline (♦). (D) The KCNQ1 Cys122Ser/Gln244Cys + KCNE3 Ser82Cys complex is sensitive to DTT, suggesting that KCNQ1 Gln244Cys and KCNE3 Ser82Cys are in close enough contact with each other to spontaneously form a disulfide bond. (E) Average I-V normalized by membrane capacitance measured from cells expressing WT KCNQ1 plus KCNE3 Ser82Cys exposed to control bath solution (○), +DTT (▪), or Cu-phenanthroline (♦). (F) Average I-V normalized by membrane capacitance measured from cells expressing KCNQ1 Cys122Ser/Gln244Cys plus KCNE3 Ser82Cys exposed to control bath solution (○), +DTT (▪), or Cu-phenanthroline (♦). I tailmax, tail current maximum.
Development of a data-validated integrative model of the KCNE3-KCNQ1 complex
The above results suggest close proximity of sites in the KCNE3 TMD with His240, Val241, and Gln244 of KCNQ1. They also indicate close proximity between the side chains of KCNQ1 Leu142 and KCNE3 Met59 as well as between the side chains of KCNQ1 Gln244 and KCNE3 Ser82. These results, in conjunction with additional structural data available from the literature (23–28), were used as restraints in developing experimentally supported structural models for the KCNE3-KCNQ1 complex.
We first developed a new ensemble of homology models for the KCNQ1 open state using Rosetta Membrane and Rosetta Symmetry potentials (29, 30) and the open-state Kv1.2/2.1 chimera structure (31) as a template [figs. S10 and S11 and supplemental KCNQ1 Protein Data Bank (PDB)–format atomic coordinate file]. The ensemble of the 10 lowest-energy KCNE3 TMD (residues 56 to 84) NMR structures determined in this work was then docked onto the open-state channel ensemble using Rosetta (32). We focused on the isolated KCNE3 TMD for the rigid-body docking because previous structure-function studies had demonstrated that this domain alone is sufficient to induce the expected constitutive activation of KCNQ1 (33). Model complexes were generated by iterative docking followed by flexible loop reconstruction using Rosetta (figs. S12 to S15; see Methods and the Supplementary Materials for details). The generated structures (fig. S16) satisfied most of the experimental restraints (table S2). The most favorably scoring KCNE3-KCNQ1 structure complex exhibited an overall MolProbity score of 1.5 (95th percentile) and is illustrated in Fig. 8, with its atomic coordinate file included with the Supplementary Materials for this paper. This model was used in the subsequent studies of this work, as described below.
Fig. 8. Activated-state KCNE3-KCNQ1 channel complex derived from experimentally restrained docking.
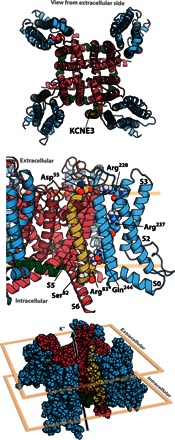
The lowest-scoring complex is displayed with KCNQ1 colored blue at residues 120 to 244 (voltage-sensor domain), green at residues 245 to 259 (S4-S5 linker), and red at residues 260 to 370 (pore); KCNE3 is colored gold. The top image represents the view looking from the extracellular space toward the membrane plane. The middle image is the view from within the membrane plane and is zoomed in on the site of KCNE3-KCNQ1 interaction. The bottom image is viewed from slightly above the membrane plane, with the extracellular environment above and the intracellular environment below.
Within the integrative model of the complex (Fig. 8), it is observed that the structure of the channel, even in the vicinity of KCNE3, remains largely similar to the Kv1.2/2.1 template. On the other hand, whereas the KCNE3 TMD retains helicity and transmembrane orientation, the curvature, as observed for the free KCNE3 structure in bicelles, is slightly altered when in complex with KCNQ1, where the curvature at the middle of the TMD is more tempered (fig. S17). This change indicates that upon binding to KCNQ1, KCNE3 adapts to the contours of the KCNQ1 crevice in which it sits. Additional description of the complex is provided in Discussion.
Among the experimental restraints used (table S2), there were three that were not satisfied by the integrative KCNE3-KCNQ1 model shown in Fig. 8. One of these was a minor violation (1 Å) of a tight (5 Å) restraint and was therefore deemed insignificant. The other two involve pairs of residues originally believed to be in direct contact based on double-mutant cycle experiments. One pair is KCNE3 Val72 and KCNQ1 Ser338 (26), and the other is either KCNE3 Asp54 or Asp55 to KCNQ1 Arg237 (27). Asp54 and Asp55 are located just outside of the membrane, N-terminal to the start of the KCNE3 TMD helix. Unlike Arg228 on S4, which is proximal to Asp55 in the KCNE3-KCNQ1 model (Fig. 8), Arg237 is located much further down the S4 segment where it is buried deeply within the four-helix bundle of the VSD in the activated-state KCNQ1 channel model—and is therefore inaccessible to direct contact with KCNE3 Asp54 or Asp55. Similarly, KCNQ1 residue 338 in the activated-state KCNQ1 model is deeply buried and would not be accessible to KCNE3. Satisfying either of the above restraints would require a marked global conformational rearrangement in the structure of KCNQ1 upon formation of a complex with KCNE3. This seems unlikely, suggesting that the energy coupling between the KCNQ1 and KCNE3 residue pairs documented in the double-mutant cycle studies (26, 27) involves allosteric networks rather than direct contact. The allosteric coupling of distal residues is a well-established phenomenon in protein science (34).
Structural basis for dissociation of the KCNQ1-KCNE3 channel complex caused by estrogen
It has previously been shown that estrogen stimulates PKCδ-derived inhibition of KCNQ1 function by two mechanisms: a rapid decrease in current caused by disassociation of KCNQ1 and KCNE3 and a sustained reduction in current density due to internalization of KCNQ1 and KCNE3 (13, 14). The decrease in current due to disassociation of KCNE3 from KCNQ1 is dependent on KCNE3 Ser82, a residue phosphorylated by PKC (12, 35).
The integrative KCNE3-KCNQ1 model developed in this work (Fig. 8) offers insight for how estrogen-stimulated phosphorylation of KCNE3 Ser82 results in the dissociation of KCNE3 from the channel (14). In this model, KCNE3 Ser82 is proximal to KCNQ1 Gln244 at the cytosolic end of S4 (Fig. 8), a key residue in the activation of KCNQ1 by KCNE3 (Fig. 6). In light of the observed sensitivity of KCNE3-KCNQ1 channel function to the charge on the cytosolic end of the S4 helix [Fig. 6 and the study of Panaghie and Abbott (36)], we tested whether mutating Gln244 to arginine (the corresponding residue in KCNQ4) would reverse the inhibitory effect of estrogen on the channel complex. As shown in Fig. 9A, application of estrogen (4-min exposure) induces a significant decrease in KCNQ1-KCNE3 currents. Figure 9B shows that the KCNQ1 Gln244Arg mutation negates the inhibitory effect of estrogen on KCNQ1-KCNE3 currents. These results suggest that estrogen-stimulated phosphorylation of KCNE3 Ser82 destabilizes the KCNE3-KCNQ1 complex, an effect that can be reversed by the Gln244Arg mutation, possibly due to favorable electrostatic attraction between the phosphoryl-serine moiety and the introduced Arg244 side chain (Fig. 9C).
Fig. 9. Reversal of estrogen-induced inhibition of KCNQ1-KCNE3 association by Q244R KCNQ1 mutation and structural model to explain this effect.
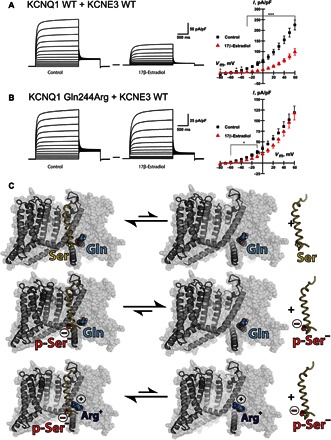
(A) (Left) Average whole-cell currents recorded from cells transiently expressing KCNQ1 WT plus KCNE3 exposed to control solution (ethanol) or estrogen (17β-estradiol, 100 μM). The solid line between the two traces is at zero current. (Right) Average I-V normalized by membrane capacitance measured from cells coexpressing KCNQ1 WT plus KCNE3 WT in the absence (▪, n = 16) or presence of estrogen (▴, n = 14). ***P ≤ 0.001, *P ≤ 0.05. (B) (Left) Average whole-cell currents recorded from cells transiently coexpressing KCNQ1 Gln244Arg and KCNE3 WT exposed to control solution or to estrogen (right). (Right) Average I-V normalized by membrane capacitance measured from cells coexpressing KCNQ1 Gln244Arg with KCNE3 WT in the absence (▪, n = 13) or presence of estrogen (▴, n = 13). Note that Fig. 6F provides key control data showing that the Q244R mutation in KCNQ1 induces a marked loss of current when KCNE3 is not present. (C) Cartoon representation of the channel, less one voltage sensor, set on the background of the complete channel shown in translucent spheres. KCNE3 WT, shown as a yellow ribbon, forms an energetically favorable complex with KCNQ1 WT (top). Phosphorylated KCNE3 Ser82 reduces the affinity to the WT channel (middle). When the KCNQ1 Gln244Arg mutation is introduced, the binding affinity to phosphorylated KCNE3 Ser82 is restored (bottom).
DISCUSSION
Structure of KCNE3: Comparison with KCNE1 and KCNE2 and functional implications
The structure determined in this work of KCNE3 in bicelle model membranes can be compared to previous structures of KCNE1 and KCNE2 (19, 37, 38). Both proteins exhibit a modest degree of amino acid sequence identity with each other and with KCNE3 (figs. S17 and S18) but modulate KCNQ1 channel function very differently. Although KCNE2 also constitutively activates KCNQ1, it reduces current levels to approximately 50% of KCNQ1 alone (39, 40). In contrast, KCNE3 markedly increases the KCNQ1-dependent current density by ~10-fold (Fig. 5A). On the other hand, KCNE1 shares with KCNE3 the property of increasing current density but differs in that KCNE1 delays channel opening in response to membrane depolarization (6, 41, 42).
The KCNE3 structure shares several important structural features with the previously determined structures of KCNE1 and KCNE2 (fig. S17) (19, 37, 38). First, all three proteins have an N-terminal amphipathic helix that is flexibly linked to the TMD. In the absence of a channel partner, this helix sits on the membrane surface and is only loosely tethered to the TMD of each KCNE family member [this work and previous studies (38, 43)]. When complexed with KCNQ1 or another Kv channel, the N-terminal amphipathic helix present in these three KCNE family members almost certainly binds to the extracellular face of the channel, involving a structural rearrangement that is enabled by the length and flexibility of the tether between this N-terminal helix and the start of the TMD. The N-terminal domains of KCNE1, KCNE2, and KCNE3 are glycosylated (44) and likely contribute to the assembly and stabilities of their KCNQ1 complexes and their drug sensitivities as well as proper trafficking (45). However, this domain does not seem to be critical for modulating channel gating.
The TMDs of the KCNE1, KCNE2, and KCNE3 proteins are composed of a curved/kinked helix, although the curvature of KCNE2 is reduced compared to KCNE1 and KCNE3 (fig. S17). For both KCNE3 (this work) and KCNE1 (19), it has been confirmed that this curvature is an intrinsic conformational property that is present even in lipid bilayer membranes. The fact that these three proteins bind to the open channel state through their TMDs, which are critical to their functions, indicates that this curvature is likely an evolutionarily conserved trait and important for binding to the activated-state channel. In this context, the observation that the curvature measured for free KCNE3 differs from the one calculated for KCNE3 as part of the KCNE3-KCNQ1 channel complex model (Fig. 8 and fig. S17) suggests that the TMD curvature of free KCNE3 may be important for the kinetics of initial binding of KCNE3 to KCNQ1, not for the stability of the complex, once formed. This idea agrees with a previous study, which suggests that the energetic cost associated with shifting hydrogen bonds in flexible transmembrane helices is surprisingly low (22).
The highest degree of sequence identity between these three KCNE proteins occurs at the beginning and ends of their TMDs (fig. S18), suggesting that these segments play a critical role in anchoring these subunits to the KCNQ1 channel. The fact that these same residues are conserved in KCNE4, which inhibits channel function and likely binds preferentially to the closed channel state, suggests that these residues may be critical for KCNE subunit association to both active and resting KCNQ1 channel states. The significantly different patterns seen for the locations of the helices in the C-terminal cytosolic domains of these three KCNEs likely reflect variations in the roles played by this domain in the varying function of each KCNE family member (46–49).
Working model for the KCNE3-KCNQ1 complex and implications for how KCNE3 modulates KCNQ1 function
A series of elegant studies of KCNE1 and KCNE3 have converged on an appreciation that KCNE3 acts by stabilizing the voltage-sensor domain of KCNQ1, especially the S4 helix, in its fully activated configuration [see previous studies (27, 28, 36, 50, 51)]. At physiologically relevant potentials, association of KCNE3 with the channel results in constitutive activation. However, if the transmembrane potential is shifted to supra-hyperpolarizing potentials, the channel closes (28). Therefore, KCNE3 stabilizes the activated state of the voltage sensor but does not lock it in this state at all potentials. It is now thought that a key interaction within the fully activated configuration of the KCNQ1 voltage sensor is ion pairing between Glu160 on the S2 segment and Arg237 of the S4 segment (52–54). This interaction is seen in the activated-state KCNQ1 model developed as part of this work (atomic coordinate file is included with the Supplementary Materials).
The integrative model for the KCNQ1-KCNE3 complex developed in this work (Fig. 8) offers a structurally explicit model for how KCNE3 stabilizes the activated state of the KCNQ1 channel. The extracellular end of the KCNE3 TMD interacts with the cleft between the extracellular end of the S1 domain helix in one subunit and the “turret” (S5/P-loop/helix) of the channel domain of an adjacent subunit (52). Several constitutive gain-of-function mutations that result in atrial fibrillation have previously been localized to this cleft, where it has been proposed that they promote the activated state by stabilizing direct interactions between S1 and the turret (52). Placement of KCNE3 in this cleft suggests that it may resemble such mutations by also stabilizing S1-turret interactions. There is precedence for this type of modulation in other channels by peptide toxins that link separate subunits in the TRPV1 (55) and ASIC1a channels (56). Possibly even more important, this configuration locates an anionic KCNE3 residue, Asp55, in a position where it can favorably interact with the positively charged side chain of Arg228 located at the extracellular end of S4 in the fully activated VSD state (Fig. 8). This interaction has previously been identified on the basis of structure-function electrophysiological data (28) and was invoked as a restraint in the algorithms used to generate this integrative model. The role of this interaction in stabilizing the activated state is very reasonable because the Arg228 side chain will be buried from KCNE3 interaction in both the intermediate-activated and resting (“down”) state VSD conformations.
Within the integrative model, the central segment of the KCNE3 TMD interacts with sites in S1 and S5 from different KCNQ1 monomers, whereas the cytosolic end sits in a cleft that allows it to make contacts with S6 in one subunit, the end of the S4-S5 linker of a second subunit, and with the beginning of the S4-S5 linker, as well as with S1 and S4 helices from a third subunit (Figs. 8 and 10). The intimate interaction of the C-terminal end of KCNE3 with several elements of the KCNQ1 channel that must undergo large amplitude movement for the channel to return to the resting state—the S6 gate helix, S4-S5 linkers, and S4—seems well suited to help stabilize the fully activated channel state.
Fig. 10. Location of key KCNQ1 functional determinants of KCNE3 modulation.
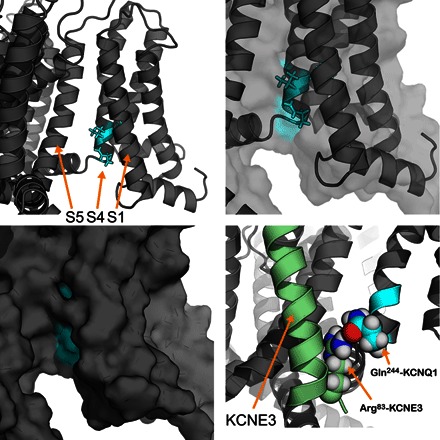
The integrative models of the KCNE3-KCNQ1 complex are useful to visualize the S4 functional determinants of KCNE3 modulation in KCNQ1. The upper left panel highlights His240, Val241, and Gln244 in cyan that are key to KCNE3 modulation. The upper right panel shows the same residues, zoomed in, with both a cartoon and transparent surface representation of the same region of KCNQ1. A surface representation (lower left) of these KCNQ1 residues (cyan) shows that these residues are surface-accessible in the KCNE3 binding cleft. The lower right panel shows the predicted hydrogen bond between KCNE3 Arg83 and KCNQ1 Gln244, which suggest a potential mechanism for the distinct KCNE3 modulation of KCNQ1 and KCNQ4.
Our electrophysiology experiments identified three mutations—His240Arg, Val241Met, and Gln244Arg—in the KCNQ1 S4 helix that converted the KCNE3-modulated KCNQ1 channel to a KCNQ4-like phenotype (Fig. 6). The KCNE3-KCNQ1 model shows that these S4 residues are surface-accessible and in the KCNE3 binding cleft (Fig. 10). Previous studies have shown that KCNE3 stabilizes the KCNQ1 S4 helix in an active (“up”) state, leading to constitutive activation at all physiological potentials (28, 50).
In validating the model, we showed that KCNE3 Ser82 is close in space to KCNQ1 Gln244 (Fig. 7). The integrative model predicts a hydrogen bond between KCNE3 Arg83 and KCNQ1 Gln244 as indicated in the bottom right panel of Fig. 10. This feature and alignments of the KCNQ1 and KCNQ4 S4 helices suggest a possible mechanism for the differential modulation by KCNE3 of KCNQ1 and KCNQ4, resulting in either channel activation or inhibition, respectively. Specifically, the residue corresponding to Gln244 in KCNQ4 is Arg220. Residue Arg83 of KCNE3 cannot form a hydrogen bond to this KCNQ4-equivalent site in the Gln244Arg mutant form of KCNQ1. We suggest that the KCNE3 Arg83/KCNQ1 Gln244 hydrogen bond is likely to be replaced by ion pairing of KCNE3 Arg83 with KCNQ4 Asp218 in the KCNE3-KCNQ4 complex, which would shift the KCNQ4 S4 helix down toward the cytosol by two residues relative to its position in the KCNE3-KCNQ1 complex. Given the features of α helices, a two-residue shift in an S4 helix would result in a backbone translocation of ~3.0 Å. Coupling this shift with the relatively long length of arginine side chains (~6 to 7 Å), this shift would be close in magnitude to the structural biology–determined movements of the S4 helix between an active (up) state and a resting (down) state of the voltage sensor of a voltage-regulated phosphatase (57). In that work, it was shown that in the VSD resting (down) state, the S4 helix shifts intracellularly (down) by about 5 Å. Therefore, the integrative models of the KCNQ1-KCNE3 complex also suggest a structural mechanism whereby KCNE3 may suppress KCNQ4 conductance by stabilizing the S4 helix in a resting/closed state.
Structural basis for modulation of KCNE3-KCNQ1 interactions by estrogen
The results of this work support a model in which estrogen-stimulated phosphorylation of KCNE3 Ser82 results in disruption of its complex with KCNQ1 and reduced channel activity. The negatively charged phosphorylated KCNE3 Ser82 has a reduced affinity to the pocket-containing KCNQ1 Gln244, which causes dissociation of KCNE3 from KCNQ1 (Fig. 9C). This is structurally reasonable because Ser82 occupies a central location on the cytosolic C-terminal end of the KCNE3 TMD, where it interacts with functionally significant segments located on multiple subunits of KCNQ1. We hypothesize that estrogen-induced phosphorylation of KCNE3 resulting in disruption of the KCNE3-KCNQ1 complex is a factor contributing to the gender difference in CF patients, in which the average age of death is significantly lower for females than for males (9–11). A central etiological component of CF is a reduction in normal chloride ion secretion in airway epithelial cells, the normal functioning of which depends on KCNE3-KCNQ1 channel function in potassium ion recycling for transepithelial chloride ion secretion (Fig. 1) (1, 6, 7). When coupled with a deleterious cystic fibrosis transmembrane conductance regulator mutation, an estrogen-stimulated reduction in potassium efflux would lead to an exacerbated susceptibility to lethal infection in the affected epithelial tissues.
CONCLUSIONS
The channel complex model we present in this work was developed by integrating experimental data, molecular dynamics, homology modeling, and state-of-the-art protein structure predictive modeling. This atomically explicit integrative model provides a structural framework for explaining how KCNE3 exerts its distinctive modulation of KCNQ1 channel function and illuminates how estrogen causes the dissociation of the KCNE3-KCNQ1 channel complex. This modeling approach was validated by the ability to predict interactions between KCNE3 and KCNQ1 at the juxtamembrane regions for both the intracellular and extracellular sides of the membrane. The KCNE3-KCNQ1 model can be used to explain previous experimental and physiological observations at atomic-level detail. In this context, the channel complex model successfully predicted the proximity between KCNE3 Ser82 and KCNQ1 Gln244, identifying the region of KCNQ1 that plays a role in estrogen modulation (that is, subunit disassociation) of the channel complex. These two results point to a major strength of this type of integrative approach, which is the ability to inspire testable hypotheses while refining and updating the models as additional structural and functional data emerge. This work also highlights the utility of hybrid functional-structural approaches to address vexing structural biological problems. These approaches are likely to be of increasing importance as personal exome/genome sequencing unveils millions of new protein-impacting gene variants of unknown significance, many of which will beg structural and functional illumination for the purposes of precision medical care (58).
METHODS
Fully detailed materials and methods are presented in the Supplementary Materials. Summaries are presented as follows.
Recombinant expression and purification of the KCNE3 protein
Human KCNE3 was expressed in Escherichia coli in various isotopically labeled forms and purified as described previously (59) into 2% bicelles (w/v total DHPC + DMPG) containing a lipid-to-detergent molar ratio (q) of 0.33. Samples were concentrated 10 times for NMR. NMR assignments for KCNE3 in DHPC/DMPG bicelles were reported previously (59) and are available in the Biological Magnetic Resonance Data Bank (BMRB Entry 16621).
Determination of the KCNE1 structure in bicelles using NMR spectroscopy
Restraints for use in backbone structure determination were collected for KCNE3 in bicelle model membranes under previously described sample conditions, which were used for all of the NMR experiments of this work (37) [0.8 mM, 250 mM imidazole (pH 6.5), 2 mM dithiothreitol (DTT) (not present in PRE samples, see below), 0.5 mM EDTA, 22% (w/v) DHPC/DMPC/DMPG bicelles with 3:1 DHPC/DMPG, and q = 0.33 (three DMPC + DMPG lipid molecules per DHPC detergent molecule), 40°C]. The high bicelle-to-protein concentration ratio was designed to avoid oligomerization or aggregation of the natively monomeric KCNE3. The following classes of NMR restraints were collected: backbone 13C and 15N chemical shifts [used to derive torsion angle restraints (60, 61)], backbone 1H-15N residual dipolar couplings (RDCs; orientational restraints), nuclear Overhauser effect (NOE) measurements (source of backbone amide proton-proton distances), and nitroxide free radical/backbone amide proton PREs [source of long-range distances (62, 63)]. RDCs were measured for samples in which the KCNE3-bicelle complexes were aligned using strained polyacrylamide gels. PREs were measured using samples prepared by spin-labeling single-cysteine mutant forms of KCNE3 [Ser13Cys, Ser57Cys, Ser74Cys, or Ser82Cys, all prepared using a cysteine-less (Cys31Ala) parent mutant form of KCNE3] with 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl-methanethiosulfonate (MTSL; Toronto Research Chemicals). For PRE measurements, diamagnetic reference spectra were acquired using the same sample as for paramagnetic conditions but following quenching of the paramagnet with 20 mM ascorbic acid. All NMR data were collected at 600 to 900 MHz using Bruker spectrometers. NMR spectra were processed using TopSpin and NMRPipe (64) and analyzed using NMRview (65) and SPARKY (66).
Initial structure calculations were conducted within XPLOR-NIH v2.24 (15) using rMD with simulated annealing. This led to an ensemble of KCNE3 structures consistent with the NMR data. Ten of the top-scoring 1% XPLOR-NIH–generated KCNE3 structures were selected for further structure refinement in an explicit DMPC membrane bilayer (generated via the DMPC bilayer with the CHARMM-GUI server) using NMR data rMD based on the LIPID14 force field and AMBER14. This ultimately led to 10 production trajectories. A representative structure for each of the 10 production trajectories was chosen by determining which frame contains the KCNE3 structure that is closest to the mean for that trajectory. The resulting final 10 representative KCNE3 structures have been deposited to the PDB under accession code 2M9Z.
Mapping the dynamics and membrane topology of KCNE3 using NMR methods
To probe KCNE3 backbone dynamics in bicellar model membranes, we used sensitivity- and gradient-enhanced NMR pulse sequences to measure heteronuclear single-quantum coherence (HSQC)–based T1, T2, and 15N-(1H)-NOE parameters on a 15N-labeled KCNE3 sample at 600 MHz (67, 68). KCNE3 backbone amide site accessibility to lipophilic and aqueous probes was determined in a site-directed manner for KCNE3 in bicelles at 800 MHz by comparing parameter-matched pairs of two-dimensional 1H-15N TROSY-HSQC diamagnetic/paramagnetic samples, in which the broadening of amide peaks due to protein site access to either water-soluble (Gd-DTPA) or lipophilic (16-DSA) probes was measured. The access of backbone amide sites of KCNE3 in bicelles to water was probed in a site-specific manner by using 1H-15N NMR to quantitate amide hydrogen-deuterium exchange, which was probed by examining the time-dependent disappearance of TROSY-HSQC peaks following dilution of a fully protonated KCNE3 sample in D2O.
Four-pulse DEER assays to determine the distances between spin labels on opposite ends of the KCNE3 TMD in various model membranes
Double spin-labeled KCNE3 was prepared by chemically conjugating MTSL to a double-cysteine mutant form of KCNE3 (Ser57Cys/Ser82Cys) via disulfide bonds using the same spin-labeling procedure as described above for PRE experiments. The protein was then reconstituted into three sets of conditions: bicelles (same as NMR conditions), 5% LMPG micelles, and POPC/POPG vesicles (3:1 POPC/POPG). Four-pulse DEER experiments were performed using an X-Band (9.5 GHz) Bruker spectrometer (ELEXSYS E580) equipped with a Bruker split ring resonator (ER 4118X-MD5) at 80 K using an Oxford CF935 cryostat coupled with an Oxford ITC4 temperature controller. A standard four-pulse sequence was used with a 32-ns π pulse and a 16-ns π/2 pulse (69). All DEER data were analyzed to determine interprobe distances using GLADD (70).
Expression of KCNE3 and KCNQ channels in mammalian cells
The complementary DNAs for KCNQ1-KCNQ4 TMD chimeras were generated by megaprimer polymerase chain reaction of whole plasmids (MEGAWHOP). Chinese hamster ovary (CHO-K1) cells were transiently transfected with plasmids encoding WT or mutant human KCNE3 (pIRES2-DsRed-MST) and either full-length human KCNQ1 (WT or mutant), KCNQ4, or a KCNQ1-KCNQ4 chimera (pIRES2-EGFP) (37, 71, 72). After transfection, cells were incubated for 48 hours before use in electrophysiological or biochemical experiments.
Analysis of KCNQ1 protein in CHO-K1 cells
Biochemical experiments to analyze KCNQ1 protein at the plasma membrane, including the preparation of cellular lysates, immunoprecipitation, and Western blotting, were completed as previously described (71, 73). Cell surface membrane proteins were biotinylated and then solubilized with detergent. Biotinylated proteins were then pulled down with streptavidin and probed using an anti-KCNQ1 antibody (compare with fig. S19).
Electrophysiological analysis of KCNQ channel function
Whole-cell currents were recorded at room temperature using Axopatch 200B amplifiers (Molecular Devices Corp.) in the whole-cell configuration of the patch-clamp technique (74). Pulse generation was conducted with ClampEx 8.0 (Molecular Devices Corp.), and currents were filtered at 1 kHz and acquired at 5 kHz. Whole-cell currents were not leak-subtracted. Whole-cell currents were measured from −80 to +60 mV (in 10-mV increments) 1.99 s after the start of the voltage pulse from a holding potential of −80 mV.
Data were collected for each experimental condition from at least three transient transfections and analyzed and plotted using a combination of Clampfit (Molecular Devices) and SigmaPlot 2000 (Systat Software). Statistical analyses were carried out using SigmaStat 2.03. Whole-cell currents were normalized for membrane capacitance, and results are expressed as means ± SEM.
Homology/Rosetta modeling of KCNQ1
A model for the open-state structure of the human KCNQ1 channel TMD (S0 to S6, residues 100 to 370) was developed using a combination of homology modeling based on the template provided by the Kv1.2/2.1 chimera crystal structure [PDB 2R9R (66)] combined with Rosetta modeling to define parts of the protein that homology modeling does not directly illuminate. An ensemble of structures deemed to be energetically reasonable based on the Rosetta scoring function was the output. The lowest-energy structure from the final ensemble of structures was chosen as the “representative structure” used in this study. The overall MolProbity score is 1.18 (upper 99th percentile).
Docking of KCNE3 to KCNQ1
The transmembrane spanning segment of the 10 lowest-energy NMR-derived KCNE3 structures was placed near the transmembrane region of the 23 representative homology models of KCNQ1. Each of the 230 starting complexes was docked 100 times. The resulting models were filtered on the basis of a sum of experimental distance-restraint violations squared. The 1000 best-scoring structures were then used to reseed a subsequent iteration of docking to generate additional 25,000 model complexes. The docking-reseeding steps were iterated until the change in interface energy from iteration to iteration converged to less than 5% (fig. S13). The Rosetta scores of the interface between KCNQ1 and KCNE3 were used to evaluate model complexes. Once the convergence criterion was met, flexible extramembrane regions of KCNE3 and KCNQ1 were rebuilt, refined, and energy-minimized, with restraints applied, within the LoopRebuild module of Rosetta. Representative structures were chosen by clustering the 1000 best-scoring, loop-rebuilt/relaxed structures, followed by selection of the lowest-energy member from each family cluster with more than 20 members.
Supplementary Material
Acknowledgments
We thank A. Hadziselimovic for help with molecular biology and E. Hustedt for his assistance with EPR. Funding: This work was supported by the U.S. NIH (grants R01 DC007416 and R01 HL122010), the American Heart Association Greater Southeast Affiliate Postdoctoral Fellowship (grant 10POST4210008 to W.D.V.H.), and an early career Bisgrove fellowship from the Science Foundation Arizona (grant 12081999 to W.D.V.H.). B.M.K. was supported by the NIH (grant F32 GM113355-01). Author contributions: B.M.K., W.D.V.H., J.S., C.K., R.C.W., K.C.T., N.J.S., Y.S., and D.P.N. conducted the experiments and calculations of this work. All authors participated in data analysis. B.M.K., W.D.V.H., C.G.V., and C.R.S. wrote the paper with input from all authors. W.D.V.H., A.L.G., J.M., C.G.V., and C.R.S. conceived this work and directed the approaches used. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The atomic coordinates of the KCNE3 backbone structure have been deposited in the PDB under accession code 2M9Z. The coordinates of the open-state KCNQ1 structural model and the KCNE3-KCNQ1 integrative model are included in the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/9/e1501228/DC1
Supplementary Materials and Methods
fig. S1. Integrative structural biology to generate experimentally restrained structural models of the KCNE3-KCNQ1 channel complex.
fig. S2. Chemical shift index analysis for KCNE3 in bicelles.
fig. S3. RDC NMR data for KCNE3 in bicelles.
fig. S4. Dipolar wave analysis of bicellar KCNE3 1H-15N RDCs.
fig. S5. Examples of PRE NMR data for KCNE3 in bicelles.
fig. S6. 15N NMR relaxation measurements for KCNE3 in bicelles.
fig. S7. Representative structures of KCNE3 amphipathic and transmembrane helices from AMBER restrained molecular dynamics (rMD) simulations.
fig. S8. Water access to the TMD of KCNE3.
fig. S9. Sequence conservation between KCNQ1 and KCNQ4.
fig. S10. Homology modeling of the open state of KCNQ1.
fig. S11. Homology/Rosetta modeling of the KCNQ1 channel open state.
fig. S12. Flowchart displaying the process used to dock KCNE3 to open-state KCNQ1.
fig. S13. Calculated binding energies (ddG) versus interface root mean squared deviation (Irms) of interface α-carbon positions compared to the lowest-scoring KCNE3-KCNQ1 (open) complex.
fig. S14. Same plot as fig. S13 with color displaying the total distance restraint violations.
fig. S15. Rebuilding flexible regions within the KCNQ1-KCNE3 complex.
fig. S16. Representative 23 KCNE3-KCNQ1 models based on satisfaction of experimental restraints and Rosetta scoring function.
fig. S17. Comparison between structurally characterized KCNE family members.
fig. S18. Sequence conservation within the entire KCNE family and within KCNE3 from different organisms.
fig. S19. KCNE3 reduces the KCNQ1[Q4S4] current amplitude without reducing channel protein levels at the membrane.
table S1. Statistics for restraints, structural calculations, and structural quality for the 10 lowest-energy structures of 9764 calculated using XPLOR and further refined in AMBER.
table S2. KCNQ1-KCNE3 residue pairs predicted to be proximal based on experimental work.
PDB coordinates
REFERENCES AND NOTES
- 1.Preston P., Wartosch L., Günzel D., Fromm M., Kongsuphol P., Ousingsawat J., Kunzelmann K., Barhanin J., Warth R., Jentsch T. J., Disruption of the K+ channel β-subunit KCNE3 reveals an important role in intestinal and tracheal Cl– transport. J. Biol. Chem. 285, 7165–7175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroeder B. C., Waldegger S., Fehr S., Bleich M., Warth R., Greger R., Jentsch T. J., A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature 403, 196–199 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Abbott G. W., Butler M. H., Bendahhou S., Dalakas M. C., Ptacek L. J., Goldstein S. A. N., MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell 104, 217–231 (2001). [DOI] [PubMed] [Google Scholar]
- 4.McCrossan Z. A., Lewis A., Panaghie G., Jordan P. N., Christini D. J., Lerner D. J., Abbott G. W., MinK-related peptide 2 modulates Kv2.1 and Kv3.1 potassium channels in mammalian brain. J. Neurosci. 23, 8077–8091 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis A., McCrossan Z. A., Abbott G. W., MinK, MiRP1, and MiRP2 diversify Kv3.1 and Kv3.2 potassium channel gating. J. Biol. Chem. 279, 7884–7892 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Abbott G. W., KCNE1 and KCNE3: The yin and yang of voltage-gated K+ channel regulation. Gene 576, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hazza A., Linley J., Aziz Q., Hunter M., Sandle G., Upregulation of basolateral small conductance potassium channels (KCNQ1/KCNE3) in ulcerative colitis. Biochem. Biophys. Res. Commun. 470, 473–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strutz-Seebohm N., Seebohm G., Fedorenko O., Baltaev R., Knirsch E. J., Functional coassembly of KCNQ4 with KCNE-β- subunits in Xenopus oocytes. Cell. Physiol. Biochem. 18, 57–66 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Harness-Brumley C. L., Elliott A. C., Rosenbluth D. B., Raghavan D., Jain R., Gender differences in outcomes of patients with cystic fibrosis. J. Women’s Health 23, 1012–1020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweezey N. B., Ratjen F., The cystic fibrosis gender gap: Potential roles of estrogen. Pediatr. Pulmonol. 49, 309–317 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Chotirmall S. H., Smith S. G., Gunaratnam C., Cosgrove S., Dimitrov B. D., O’Neill S. J., Harvey B. J., Greene C. M., McElvaney N. G., Effect of estrogen on pseudomonas mucoidy and exacerbations in cystic fibrosis. N. Engl. J. Med. 366, 1978–1986 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Alzamora R., O’Mahony F., Bustos V., Rapetti-Mauss R., Urbach V., Pablo Cid L., Sepúlveda F. V., Harvey B. J., Sexual dimorphism and oestrogen regulation of KCNE3 expression modulates the functional properties of KCNQ1 K+ channels. J. Geophys. Res. 589, 5091–5107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rapetti-Mauss R., O’Mahony F., Sepulveda F. V., Urbach V., Harvey B. J., Oestrogen promotes KCNQ1 potassium channel endocytosis and postendocytic trafficking in colonic epithelium. J. Physiol. 591, 2813–2831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Mahony F., Alzamora R., Betts V., LaPaix F., Carter D., Irnaten M., Harvey B. J., Female gender-specific inhibition of KCNQ1 channels and chloride secretion by 17β-estradiol in rat distal colonic crypts. J. Biol. Chem. 282, 24563–24573 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Case D. A., Cheatham T. E. III, Darden T., Gohlke H., Luo R., Merz K. M. Jr, Onufriev A., Simmerling C., Wang B., Woods R. J., The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickson C. J., Madej B. D., Skjevik Å. A., Betz R. M., Teigen K., Gould I. R., Walker R. C., Lipid14: The Amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salomon-Ferrer R., Case D. A., Walker R. C., An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 3, 198–210 (2013). [Google Scholar]
- 19.Sahu I. D., Kroncke B. M., Zhang R., Dunagan M. M., Smith H. J., Craig A., McCarrick R. M., Sanders C. R., Lorigan G. A., Structural investigation of the transmembrane domain of KCNE1 in proteoliposomes. Biochemistry 53, 6392–6401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam S. M., Roux B., Simulating the distance distribution between spin-labels attached to proteins. J. Phys. Chem. B 119, 3901–3911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeschke G., Polyhach Y., Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 9, 1895–1910 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Cao Z., Bowie J. U., Shifting hydrogen bonds may produce flexible transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 109, 8121–8126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melman Y. F., Um S. Y., Krumerman A., Kagan A., McDonald T. V., KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron 42, 927–937 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Nakajo K., Nishino A., Okamura Y., Kubo Y., KCNQ1 subdomains involved in KCNE modulation revealed by an invertebrate KCNQ1 orthologue. J. Gen. Physiol. 138, 521–535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seebohm G., Sanguinetti M. C., Pusch M., Tight coupling of rubidium conductance and inactivation in human KCNQ1 potassium channels. J. Physiol. 552, 369–378 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panaghie G., Tai K.-K., Abbott G. W., Interaction of KCNE subunits with the KCNQ1 K+ channel pore. J. Physiol. 570, 455–467 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi E., Abbott G. W., A shared mechanism for lipid- and β-subunit-coordinated stabilization of the activated K+ channel voltage sensor. FASEB J. 24, 1518–1524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barro-Soria R., Perez M. E., Larsson H. P., KCNE3 acts by promoting voltage sensor activation in KCNQ1. Proc. Natl. Acad. Sci. U.S.A. 112, E7286–E7292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yarov-Yarovoy V., Schonbrun J., Baker D., Multipass membrane protein structure prediction using Rosetta. Proteins 62, 1010–1025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiMaio F., Leaver-Fay A., Bradley P., Baker D., André I., Modeling symmetric macromolecular structures in Rosetta3. PLOS One 6, e20450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long S. B., Tao X., Campbell E. B., MacKinnon R., Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann K. W., Lemmon G. H., DeLuca S. L., Sheehan J. H., Meiler J., Practically useful: What the Rosetta protein modeling suite can do for you. Biochemistry 49, 2987–2998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melman Y. F., Domènech A., de la Luna S., McDonald T. V., Structural determinants of KvLQT1 control by the KCNE family of proteins. J. Biol. Chem. 276, 6439–6444 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Dokholyan N. V., Controlling allosteric networks in proteins. Chem. Rev. 116, 6463–6487 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Abbott G. W., Butler M. H., Goldstein S. A. N., Phosphorylation and protonation of neighboring MiRP2 sites: Function and pathophysiology of MiRP2-Kv3.4 potassium channels in periodic paralysis. FASEB J. 20, 293–301 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Panaghie G., Abbott G. W., The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J. Gen. Physiol. 129, 121–133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang C., Tian C., Sönnichsen F. D., Smith J. A., Meiler J., George A. L. Jr, Vanoye C. G., Kim H. J., Sanders C. R., Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry 47, 7999–8006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P., Liu H., Lai C., Sun P., Zeng W., Wu F., Zhang L., Wang S., Tian C., Ding J., Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2. Sci. Rep. 4, 4973 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCrossan Z. A., Abbott G. W., The MinK-related peptides. Neuropharmacology 47, 787–821 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Tinel N., Diochot S., Borsotto M., Lazdunski M., Barhanin J., KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO J. 19, 6326–6330 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G., KVLQT1 and lsK (minK) proteins associate to form the IKS cardiac potassium current. Nature 384, 78–80 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Sanguinetti M. C., Curran M. E., Zou A., Shen J., Specter P. S., Atkinson D. L., Keating M. T., Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKS potassium channel. Nature 384, 80–83 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Sahu I. D., Craig A. F., Dunagan M. M., Troxel K. R., Zhang R., Meiberg A. G., Harmon C. N., McCarrick R. M., Kroncke B. M., Sanders C. R., Lorigan G. A., Probing structural dynamics and topology of the KCNE1 membrane protein in lipid bilayers via site-directed spin labeling and electron paramagnetic resonance spectroscopy. Biochemistry 54, 6402–6412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malaby H. L. H., Kobertz W. R., Molecular determinants of co- and post-translational N-glycosylation of type I transmembrane peptides. Biochem. J. 453, 427–434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girmatsion Z., Biliczki P., Takac I., Schwerthelm C., Hohnloser S. H., Ehrlich J. R., N-terminal arginines modulate plasma-membrane localization of Kv7.1/KCNE1 channel complexes. PLOS One 6, e26967 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J., Zheng R., Melman Y. F., McDonald T. V., Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PLOS One 4, e5143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lvov A., Gage S. D., Berrios V. M., Kobertz W. R., Identification of a protein-protein interaction between KCNE1 and the activation gate machinery of KCNQ1. J. Gen. Physiol. 135, 607–618 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gage S. D., Kobertz W. R., KCNE3 truncation mutants reveal a bipartite modulation of KCNQ1 K+ channels. J. Gen. Physiol. 124, 759–771 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocheleau J. M., Gage S. D., Kobertz W. R., Secondary structure of a KCNE cytoplasmic domain. J. Gen. Physiol. 128, 721–729 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakajo K., Kubo Y., KCNE1 and KCNE3 stabilize and/or slow voltage sensing S4 segment of KCNQ1 channel. J. Gen. Physiol. 130, 269–281 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocheleau J. M., Kobertz W. R., KCNE peptides differently affect voltage sensor equilibrium and equilibration rates in KCNQ1 K+ channels. J. Gen. Physiol. 131, 59–68 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith J. A., Vanoye C. G., George A. L. Jr, Meiler J., Sanders C. R., Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry 46, 14141–14152 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu D., Delaloye K., Zaydman M. A., Nekouzadeh A., Rudy Y., Cui J., State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 135, 595–606 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Restier L., Cheng L., Sanguinetti M. C., Mechanisms by which atrial fibrillation-associated mutations in the S1 domain of KCNQ1 slow deactivation of IKs channels. J. Physiol. 586, 4179–4191 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao E., Liao M., Cheng Y., Julius D., TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baconguis I., Gouaux E., Structural plasticity and dynamic selectivity of acid-sensing ion channel—Spider toxin complexes. Nature 489, 400–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q., Wanderling S., Paduch M., Medovoy D., Singharoy A., McGreevy R., Villalba-Galea C. A., Hulse R. E., Roux B., Schulten K., Kossiakoff A., Perozo E., Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 21, 244–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kroncke B. M., Vanoye C. G., Meiler J., George A. L. Jr, Sanders C. R., Personalized biochemistry and biophysics. Biochemistry 54, 2551–2559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang C., Vanoye C. G., Welch R. C., Van Horn W. D., Sanders C. R., Functional delivery of a membrane protein into oocyte membranes using bicelles. Biochemistry 49, 653–655 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wishart D. S., Sykes B. D., The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Battiste J. L., Wagner G., Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry 39, 5355–5365 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Solomon I., Bloembergen N., Nuclear magnetic interactions in the HF molecule. J. Chem. Phys. 25, 261–266 (1956). [Google Scholar]
- 64.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Johnson B. A., Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004). [DOI] [PubMed] [Google Scholar]
- 66.T. D. Goddard, D. G. Kneller, Sparky 3 (University of California, San Francisco, 2008). [Google Scholar]
- 67.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E., Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 (1994). [DOI] [PubMed] [Google Scholar]
- 68.Kay L. E., Keifer P., Saarinen T., Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 (1992). [Google Scholar]
- 69.Jeschke G., Distance measurements in the nanometer range by pulse EPR. Chemphyschem 3, 927–932 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Brandon S., Beth A. H., Hustedt E. J., The global analysis of DEER data. J. Magn. Reson. 218, 93–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vanoye C. G., Welch R. C., Tian C., Sanders C. R., George A. L. Jr, KCNQ1/KCNE1 assembly, co-translation not required. Channels 4, 108–114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lundquist A. L., Manderfield L. J., Vanoye Carlos G., Rogers Christopher S., Donahue Brian S., Chang Paul A., Drinkwater Davis C., Murray Katherine T., George Alfred L. Jr, Expression of multiple KCNE genes in human heart may enable variable modulation of IKs. J. Mol. Cell. Cardiol. 38, 277–287 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Manderfield L. J., George A. L. Jr, KCNE4 can co-associate with the IKs (KCNQ1–KCNE1) channel complex. FEBS J. 275, 1336–1349 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J., Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Archiv. 391, 85–100 (1981). [DOI] [PubMed] [Google Scholar]
- 75.Anantharam A., Lewis A., Panaghie G., Gordon E., McCrossan Z. A., Lerner D. J., Abbott G. W., RNA interference reveals that endogenous Xenopus MinK-related peptides govern mammalian K+ channel function in oocyte expression studies. J. Biol. Chem. 278, 11739–11745 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Gordon E., Roepke T. K., Abbott G. W., Endogenous KCNE subunits govern Kv2.1 K+ channel activation kinetics in Xenopus oocyte studies. Biophys. J. 90, 1223–1231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Terhag J., Cavara N. A., Hollmann M., Cave Canalem: How endogenous ion channels may interfere with heterologous expression in Xenopus oocytes. Methods 51, 66–74 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Mesleh M. F., Veglia G., DeSilva T. M., Marassi F. M., Opella S. J., Dipolar waves as NMR maps of protein structure. J. Am. Chem. Soc. 124, 4206–4207 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weigelt J., Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 120, 10778–10779 (1998). [Google Scholar]
- 80.Van Horn W. D., Kim J., Ellis C. D., Hadziselimovic A., Sulistijo E. S., Karra M. D., Tian C., Sönnichsen F. D., Sanders C. R., Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science 324, 1726–1729 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G., Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Viklund H., Elofsson A., OCTOPUS: Improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24, 1662–1668 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Van Horn W. D., Vanoye C. G., Sanders C. R., Working model for the structural basis for KCNE1 modulation of the KCNQ1 potassium channel. Curr. Opin. Struct. Biol. 21, 283–291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/9/e1501228/DC1
Supplementary Materials and Methods
fig. S1. Integrative structural biology to generate experimentally restrained structural models of the KCNE3-KCNQ1 channel complex.
fig. S2. Chemical shift index analysis for KCNE3 in bicelles.
fig. S3. RDC NMR data for KCNE3 in bicelles.
fig. S4. Dipolar wave analysis of bicellar KCNE3 1H-15N RDCs.
fig. S5. Examples of PRE NMR data for KCNE3 in bicelles.
fig. S6. 15N NMR relaxation measurements for KCNE3 in bicelles.
fig. S7. Representative structures of KCNE3 amphipathic and transmembrane helices from AMBER restrained molecular dynamics (rMD) simulations.
fig. S8. Water access to the TMD of KCNE3.
fig. S9. Sequence conservation between KCNQ1 and KCNQ4.
fig. S10. Homology modeling of the open state of KCNQ1.
fig. S11. Homology/Rosetta modeling of the KCNQ1 channel open state.
fig. S12. Flowchart displaying the process used to dock KCNE3 to open-state KCNQ1.
fig. S13. Calculated binding energies (ddG) versus interface root mean squared deviation (Irms) of interface α-carbon positions compared to the lowest-scoring KCNE3-KCNQ1 (open) complex.
fig. S14. Same plot as fig. S13 with color displaying the total distance restraint violations.
fig. S15. Rebuilding flexible regions within the KCNQ1-KCNE3 complex.
fig. S16. Representative 23 KCNE3-KCNQ1 models based on satisfaction of experimental restraints and Rosetta scoring function.
fig. S17. Comparison between structurally characterized KCNE family members.
fig. S18. Sequence conservation within the entire KCNE family and within KCNE3 from different organisms.
fig. S19. KCNE3 reduces the KCNQ1[Q4S4] current amplitude without reducing channel protein levels at the membrane.
table S1. Statistics for restraints, structural calculations, and structural quality for the 10 lowest-energy structures of 9764 calculated using XPLOR and further refined in AMBER.
table S2. KCNQ1-KCNE3 residue pairs predicted to be proximal based on experimental work.
PDB coordinates


