Abstract
The mechanism that duplicates the nuclear genome during the trillions of cell divisions required to develop from zygote to adult is the same throughout the eukarya, but the mechanisms that determine where, when and how much nuclear genome duplication occur regulate development and differ among the eukarya. They allow organisms to change the rate of cell proliferation during development, to activate zygotic gene expression independently of DNA replication, and to restrict nuclear DNA replication to once per cell division. They allow specialized cells to exit their mitotic cell cycle and differentiate into polyploid cells, and in some cases, to amplify the number of copies of specific genes. It is genome duplication that drives evolution, by virtue of the errors that inevitably occur when the same process is repeated trillions of times. It is, unfortunately, the same errors that produce age-related genetic disorders such as cancer.
1. Introduction
The mechanism by which DNA is replicated is so highly conserved throughout the three domains of life—bacteria, archaea, and eukarya (DePamphilis & Bell, 2010)—that developmental biologists might think that genome duplication is an immutable mechanism that awakes at fertilization and then beats in an unchanging rhythm throughout development. But the real story is more complicated. All living organisms do encode their genomes in DNA, and they do replicate the nuclear/nucleoid portion of their DNA using the “replication fork” mechanism. One DNA strand serves as a template upon which a new complementary strand is synthesized. DNA unwinding is coupled to DNA synthesis that occurs continuously on one template and discontinuously on the other by the repeated synthesis and joining of short RNA-primed nascent DNA chains termed “Okazaki fragments.” To be sure, other strategies for genome duplication exist, but they are confined to bacteriophage, bacterial plasmids, animal and plant viruses, and mitochondria. The essential point is that during metazoan development, the mechanism for replicating nuclear DNA is the same from zygote to adult. In fact, the sequence of events at replication forks is the same in all living cells, and the proteins that execute these events in metazoa are orthologs or analogs of the proteins used in archaea and bacteria.
Now consider the problem. A single diploid human cell contains 2.09 m of DNA (6.16 billion base pairs, 6.3 pg) of nuclear DNA that encodes 99.9995% of the genome. The remaining 0.0005% is encoded in mitochondrial DNA, which is maternally inherited, replicates independently of nuclear DNA replication, and varies widely in copy number among cell types. This means that development of a fertilized human egg into an adult of 75 kg requires the production of 29 trillion cells or about 60 trillion meters of DNA. Do the math. That's enough DNA to stretch from the Earth to the Sun and back 200 ×! Include the approximately 1 trillion hematopoietic cells produced each day in adults, together with the turnover in epithelial cell populations, and the number of cell divisions in a human life span is truly astronomical!
Now consider the solution. Replication fork velocity did not increase to compensate for increasing genome size; in fact, replication forks are about 60 × slower in humans than in bacteria! Instead, the mechanisms that determine where and when DNA replication begins evolved amazing flexibility in order to accommodate the ever-increasing demands of large genomes encoding thousands of different genes that must be turned on and off at different times during cell proliferation and differentiation. In other words, the requirements for DNA replication evolved so that they did not compete with the requirements for DNA transcription. Replication of nuclear DNA became tightly restricted to one duplication for each cell division. This ensured that the trillions of cells in complex metazoa are genetically identical, but mechanisms also evolved to escape this restriction so that specialized cells could produce huge amounts of gene products. Unfortunately, this flexibility came at a price: DNA replication machines make frequent mistakes. The good news is that those mistakes ensure that evolution continues. The bad news is that genetic alterations lead to genomic instability, human disease, and aging.
2. The First Mitotic Cell Division is Universal
The mitotic cell cycle is a sequence of events in all eukarya that ensures that nuclear DNA replication (S phase) is always followed by mitosis (M phase) and then cytokinesis, and that the nuclear genome is duplicated once, but only once, during each cell division (Fig. 1). It applies to single-cell organisms as well as metazoa. It applies to zygotes, embryos, and adults. Progress through the mitotic cell cycle in mammals is regulated by at least 34 different genes (Ciemerych & Sicinski, 2005), but only eight protein kinases directly regulate DNA replication, of which only cyclin-dependent kinases Cdk1 and Cdk7, the Dbf4-dependent kinase Cdc7, and the DNA damage checkpoint kinase Chk1/Chek1 are essential for mammalian development (Depamphilis et al., 2012). The other four kinases have roles that can be fulfilled by other proteins. Finally, the mitotic cell cycle contains five major “checkpoints” where progress through the cycle is arrested if cells cannot grow in size, if DNA is damaged, or if mitosis is compromised.
Figure 1.
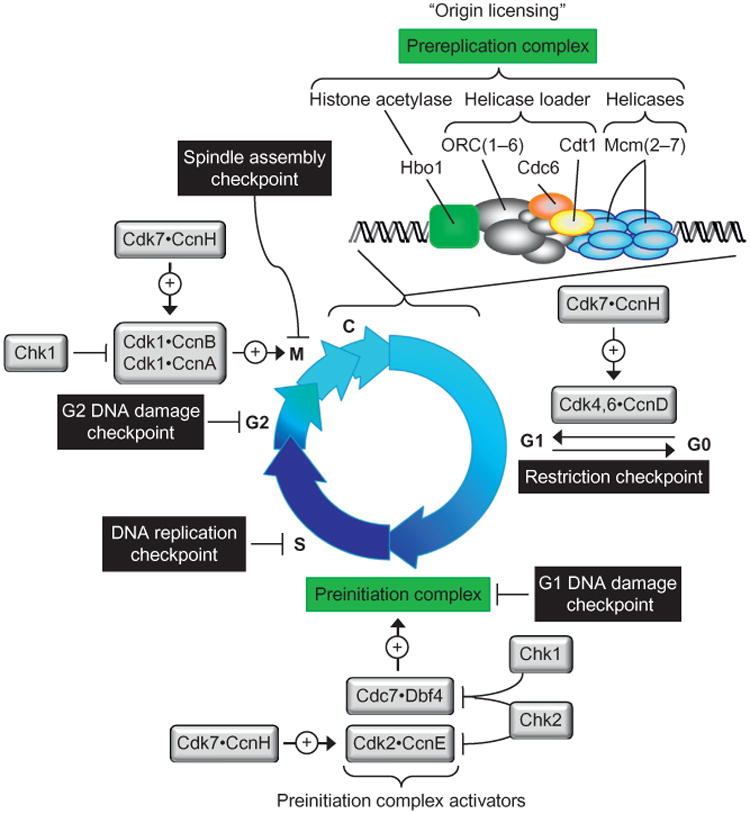
Regulation of DNA replication during mitotic cell cycles. Nuclear DNA replication in the metazoa begins during the anaphase to G1 phase transition of the mitotic cell cycle with the assembly of prereplication complexes onto the DNA in chromatin (Baldinger & Gossen, 2009; Noguchi, Vassilev, Ghosh, Yates, & DePamphilis, 2006). In mammals, this event, termed “origin licensing,” begins by assembly of the six subunit “origin recognition complex” [ORC(1–6)] through independent nuclear localization of Orc1, ORC(2–5), and Orc6 (Ghosh, Vassilev, Zhang, Zhao, & DePamphilis, 2011). The BAH domain in human Orc1 facilitates the ability of Orc1 to activate replication origins in vivo and triggers assembly of ORC(1–6) (Kara, Hossain, Prasanth, & Stillman, 2015; Noguchi et al., 2006). Mutations in the BAH domain of Orc1 link histone H4 methylation to origin licensing and to Meier–Gorlin syndrome, a genetic defect leading to primordial dwarfism (Kuo et al., 2012). Cdc6 stabilizes the binding of ORC(1–6) to DNA and allows recruitment of Cdt1 to form a “helicase loader.” The six subunit “minichromosome maintenance complex” (Mcm2–7) is the eukaryotic replicative DNA helicase. Each replication fork requires one MCM helicase; thus two or more are loaded at each origin of bidirectional replication. In vivo, the Hbo1/Kat7 histone acetylase activity is also essential origin licensing, because assembly of prereplication complexes is facilitated by histone acetylation (Miotto & Struhl, 2010; Unnikrishnan, Gafken, & Tsukiyama, 2010). However, prereplication complexes cannot unwind DNA; they must first be converted into preinitiation complexes by addition of 10 other protein (Bell & Kaguni, 2013; Tanaka & Araki, 2013). Only then can the MCM helicase unwind the double helix and allow DNA polymerases to synthesize the nascent DNA strand. DNA replication is regulated directly by eight protein kinases (Depamphilis, de Renty, Ullah, & Lee, 2012), five of which are cyclin-dependent kinases (Cdk) that require a cyclin (Ccn) partner. Cdk7CcnH is essential to activate Cdk1, Cdk2, Cdk4, and Cdk6. Cdk4CcnD and Cdk6CcnD drive cells from a quiescent state (G0) into a proliferative state. Cdk1 CcnB1 and Cdk1 CcnA2 initiate and maintain mitosis. Cdk2CcnE and the Dbf4-dependent kinase Cdc7 are the engines that drive the assembly and activation of preinitiation complexes. Cdk2CcnA2 prevents DNA rereplication during S phase. Cdk1CcnA2 prevents premature initiation during G2 phase. The remaining two protein kinases are components of the cell's DNA damage response. Five checkpoints ensure the accuracy of mitotic cell division (DePamphilis & Bell, 2010). In the absence of sufficient nutrients, growth factors or optimal temperature, cells in G1 phase enter a quiescent state (G0). This transition is regulated by the “restriction checkpoint,” which consists of two signal detection pathways (Foster, Yellen, Xu, & Saqcena, 2010). One senses growth factors, and the other senses nutrients. The G1 to G0 transition is regulated by action of Cdk4CcnD and Cdk6CcnD. When cells are arrested in G0, their DNA replication machines are dismantled, and they eventually enter a senescent state from which they cannot reenter the mitotic cell cycle (Campisi, 2013). Conversely, once a cell passes the restriction checkpoint, it is committed to division; it must divide, or terminally differentiate into a polyploid cell, or die. Nuclear DNA damage is assessed before beginning S phase by the G1 DNA damage checkpoint, during S phase by the DNA replication checkpoint and before beginning M phase by the G2 DNA damage checkpoint, but in reality there are only two DNA damage response systems that basically detect excess single-stranded DNA complexed with replication protein-A that results from different problems. DNA damage and stalled replication forks are detected by the ATR kinase which then activates the Chk1 kinase to phosphorylate and inhibit Cdc7 and Cdc25 (a phosphatase required for activation of Cdk1 and Cdk2). Double-stranded DNA breaks are detected by the ATM kinase, which then activates Cdk2 kinase to phosphorylate Cdc7, Cdc25, and tumor suppressor Tp53. Many other proteins are also targeted. Finally, the spindle assembly checkpoint prevents the onset of anaphase until the kinetochores are attached correctly to the mitotic spindle (Lara-Gonzalez, Westhorpe, & Taylor, 2012).
The first mitotic cell division is universal. Proliferating cells prepare for genome duplication by assembling prereplication complexes on chromatin as the cell exits mitosis (Fig. 1). The same event occurs in zygotes (Fig. 2). Oocytes are terminally differentiated cells that are arrested after completing the first stage of meiosis. They support RNA and protein synthesis but not DNA replication. When oocytes complete meiosis, the resulting eggs are not only competent for fertilization; they are competent for DNA replication. Eggs are transcriptionally silent, but translationally active. Cdc6 and Orc6 are the only DNA replication protein missing in the oocytes of flies, frogs, and mice whose translation during meiotic maturation is necessary and sufficient to confer DNA replication competence before fertilization (Lemaitre, Bocquet, & Mechali, 2002; Lemaitre et al., 2004; Murai, Stein, Buffone, Yamashita, & Schultz, 2010; Whitmire, Khan, & Coue, 2002). In addition to their role in origin licensing, Cdc6 also behaves as a Cdk1 inhibitor that regulates entry into and progression through mitosis by limiting the level of Cdk1 activity (El Dika et al., 2014). Fertilization triggers formation of two pronuclei, followed by nuclear DNA replication, and then mitosis, all of which happens in the absence of DNA transcription. Here, the first cell division cycles diverge. Whereas the zygotes of frogs and mice undergo cytokinesis to form a two-cell embryo, the zygotes of flies simply continue nuclear DNA replication and mitosis within a syncytium.
Figure 2.
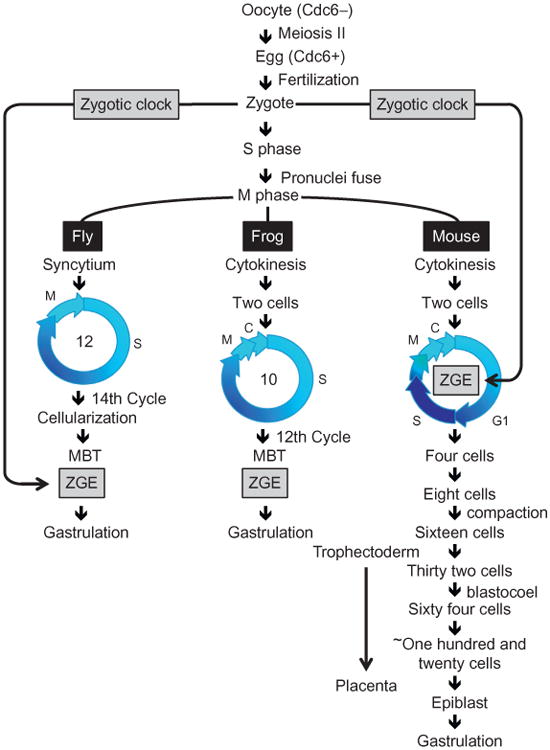
Developmentally programmed changes in the mitotic cell cycle. The first mitotic cell division is universal, as exemplified in Drosophila melanogaster (fly), Xenopus laevis (frog), and Mus musculus (mouse). Oocyte maturation into eggs includes expression of the Cdc6 protein that is essential for initiation of nuclear DNA replication. The second mitotic cell division distinguishes flies from frogs and mammals from non-mammals. Flies and other insects continue to rapidly undergo cycles of DNA replication (S phase) followed by mitosis (M phase) without undergoing cell cleavage. Frogs, fish, and echinoderms continue to rapidly undergo cycles of DNA replication followed by mitosis followed by cell cleavage (cell division in the absence of cell growth). Mice and other mammals continue to undergo slow cycles of DNA replication followed by mitosis followed by cell cleavage. However, mammalian cell cleavage appears to include G phases. The most significant change is that zygotic gene expression (ZGE), the onset of transcription and the degradation of maternally inherited RNA, occurs in two-cell embryos of mice and the 4- to 16-cell embryos of rabbits, cows, pigs, and humans. Nevertheless, activation of the bulk of ZGE in both flies and mice is determined by the time elapsed after fertilization (a “zygotic clock”), rather than by DNA replication or mitosis. The differences in cell division cycles between mammals and nonmammals might well be related to the fact that mammals need to produce both a placenta and an embryo concurrently (O'Farrell, Stumpff, & Su, 2004).
3. The Second Mitotic Cell Division Distinguishes Mammals from Nonmammals
The embryos of flies and frogs, like representatives of all the major phyla that deposit their eggs externally, undergo a series of rapid cleavage cell cycles in the absence of cell growth, so that cells become progressively smaller (Lee, Bonneau, & Giraldez, 2014; O'Farrell et al., 2004). Rapid cell cleavages lack G1 and G2 phases and DNA damage checkpoints. When DNA replication in either Xenopus or Drosophila embryos is blocked, cells progress to mitosis with catastrophic consequences (Glover et al., 1989; Newport & Dasso, 1989). Mammalian zygotes also undergo a series of cleavage cell cycles, but the first cell cleavage event in mouse zygotes is about 123 × longer than in the zygotes of flies and 37 × longer than those of frogs (Table 1 and Fig. 2). Similarly, fly genomes (represented by Drosophila melanogaster) can be replicated in as few as 3.4 min, frog genomes (represented by Xenopus laevis) in 20 min, and rodent genomes in 120 min, but the fastest replication rates in flies and frogs occur prior to gastrulation whereas in mammals they occur during gastrulation. As a rule, external fertilization leads to rapid cell cleavage cycles, whereas internal fertilization leads to slow cell cleavage cycles.
Table 1. Changes in the Time Required for Cell Division During Animal Development.
| Organism | Time | Velocity | ||
|---|---|---|---|---|
| Cell Cycle | S Phase | Replication (Mb/min) | Fork (Kb/min) | |
| Flies (180 Mbp DNA) | ||||
| Drosophila syncytial embryo | 8.3→23 m | 3.4 m | 53 | 2.6 |
| Kc cells | 8h | 6h | 0.8 | |
| Frogs (3100 Mbp DNA) | ||||
| Xenopus cleavage embryos | 30 m | 20 m | 155 | 0.3→ 1.1 |
| Rodents (2600 Mbp DNA) | ||||
| Mouse preimplantation embryo | ||||
| Cycles 1 and 2 | 17→20 h | 6h | 7.2 | |
| Cycles 3 and 4 | 10→14 h | 6h | ||
| Mouse gastrula | ||||
| Proliferative zone | 2.2 h | ∼1.9 h | ∼23 | |
| Rat gastrula | ||||
| Anterior primitive streak | 3→3.5 h | 2→2.75 h | 16→22 | |
| Mouse fibroblasts | 1.8 | |||
| Human fibroblasts | 18→25 h | 6→8 h | 6.4→ 8.6 | 0.6→3.6 |
Abbreviations: Mega (106) bases (Mb), mega base pairs. Kb=“kilo (103) bases.” Embryo data are from Blumenthal et al. (1974), Bolton et al. (1984), Howlett (1986), Howlett and Bolton (1985), Mac Auley, Werb and Mirkes (1993), O'Farrell et al. (2004), Snow (1977), and references therein. Replication fork data are from Cayrou et al. (2011), Conti, Sacca, et al. (2007), and Conti, Seiler and Pommier (2007). The 3.4 min S phase in flies is for the most rapid early cell cycles. Duration of metaphase/anaphase in mice is ∼20 min. Note that DNA replication rates depend strongly on temperature in both animals and cell cultures. Cultured Xenopus cells appear similar to cultured mammalian cells, although specific data were not available.
Rapid cell cleavage cycles occur in the absence of DNA transcription. Therefore, DNA replication and mitosis depend entirely on maternally inherited proteins and translation of maternally inherited mRNA. Rapid cleavage cycles terminate with the “midblastula transition” (MBT), which is recognized by the onset of transcription and translation of zygotic genes [zygotic gene expression (ZGE)], degradation of maternally inherited RNAs, and an extended cell division cycle (Fig. 1). Frog embryos undergo 12 cleavage events before the MBT. Fly embryos cycle through a series of 14 synchronous S and M phases in the absence of cytokinesis before cellularization occurs, followed quickly by the MBT.
Slow cell cleavages occur in mammals. Fertilization of mouse eggs triggers pronuclei formation, followed by DNA replication, mitosis, and cytokinesis. These events require 17–20 h with subsequent cleavage cell cycles occurring every 10–14 h. These slow cleavage cell cycles exhibit G1, S, G2, and M phases as well as DNA damage checkpoints. When DNA replication is blocked in mouse preimplantation embryos, cells do not undergo mitosis, cytokinesis, or apoptosis (Bolton, Oades, & Johnson, 1984). ZGE occurs in the two-cell embryos of mice and in the 4- to 16-cell embryos of other mammals. Despite this rapid transition from maternal to zygotic control, cell cleavage continues after activation of ZGE to produce smaller cells with an increased nuclear to cytoplasmic ratio until implantation (Aiken, Swoboda, Skepper, & Johnson, 2004). This change in strategy allows mammals to begin cell differentiation prior to implantation, which insures that placenta and fetus develop concurrently.
4. Activation of ZGE does not Depend on DNA Replication
A small group of zygotic genes are expressed in the embryos of flies and frogs when the nuclear to cytoplasmic ratio reaches a critical stage, presumably because the exponential increase in DNA content during cell cleavages results in the titration and inactivation of a hypothetical cytoplasmic factor that inhibits the MBT (Lee et al., 2014). However, most zygotic transcription, as well as the accompanying maternal RNA degradation, does not rely on the nuclear to cytoplasmic ratio in fly embryos, but instead occurs at a strict time interval following fertilization, indicating the presence of a timing mechanism (Lu, Li, Elemento, Tavazoie, & Wieschaus, 2009). The presence of this “zygotic clock” was demonstrated earlier in mice (Aoki, Worrad, & Schultz, 1997; Bolton et al., 1984; Wiekowski, Miranda, & DePamphilis, 1991). Activation of ZGE occurs in mouse embryos at the same time postfertilization (the time required to form a two-cell embryo) regardless of whether or not DNA replication is inhibited. Zygotes remained morphologically as one-cell embryos, but they activate ZGE as if they had reached the two-cell stage. This clock applies to genes on plasmid expression vectors as well as endogenous genes, to promoters driven by any one of the three RNA polymerases, and to both transcription and translation (Nothias, Miranda, & DePamphilis, 1996).
The zygotic clock delays transcription until both paternal and maternal genomes are replicated and remodeled from a postmeiotic state to one in which transcription is repressed by chromatin structure in a manner that can be relieved by enhancers at appropriate times during development (Nothias, Majumder, Kaneko, & DePamphilis, 1995). Injection of DNA into the nuclei of mouse oocytes and embryos revealed that transcription promoters and origins of DNA replication are repressed in the maternal pronucleus of oocytes and zygotes, and in the nuclei of two-cell embryos, regardless of their parental origin or ploidy. This repression is linked to changes in chromatin, and the ability to relieve this repression with embryo-responsive transcription enhancers requires a unique coactivator that first appears with formation of a two-cell embryo (Majumder, Miranda, & DePamphilis, 1993; Majumder, Zhao, Kaneko, & DePamphilis, 1997). A TATA box was required for enhancer stimulation of promoter activity only after cells began to differentiate (Majumder & DePamphilis, 1994). These enhancers respond to TEAD transcription factors present in preimplantatconventions, genes have toion embryos and embryonic stem cells (Melin, Miranda, Montreau, DePamphilis, & Blangy, 1993). Tead2 is expressed at the beginning of preimplantation development (Kaneko, Cullinan, Latham, & DePamphilis, 1997), and Tead4 is expressed at the eight-cell stage where it is essential for trophectoderm development (Kaneko & DePamphilis, 2013; Yagi et al., 2007). All four TEAD transcription factors bind the same DNA sequence, utilize the Yap1/Yap65 transcriptional coactivator to activate transcription, and are regulated through cytoplasmic localization of Yap1 by 14-3-3 protein (Vassilev, Kaneko, Shu, Zhao, & DePamphilis, 2001). In this manner, Yap1 and its cousin Taz regulate TEAD-dependent transcription in response to mitogenic signals via the HIPPO signal transduction pathway, a regulator of tissue regeneration and stem cell biology (Hansen, Moroishi, & Guan, 2015).
5. Replication Origins are Developmental Regulated
In 1974, David Hogness and Harold Callan discovered that the density of replication origins throughout the chromosomes of flies, newts, and frogs decreased significantly as rapidly cleaving nuclei in the fertilized eggs of these organisms developed into adults. At the beginning, replication bubbles were closely spaced, but as cells differentiated, the number of active replication origins in the genome decreased dramatically. With the development of origin mapping techniques, it eventually became clear that selection of initiation sites for DNA replication can change during metazoan development from virtually random initiation throughout the nuclear DNA to site-specific replication origins that can be activated either in clusters or individually. Replication bubbles in fly embryos or frog eggs or egg extracts are distributed throughout the DNA with periodic spacing of 5–15 kbp that depend on the amount of initiator protein [ORC(1–6)] available (Blow, Gillespie, Francis, & Jackson, 2001; Blumenthal, Kriegstein, & Hogness, 1974; Hyrien & Mechali, 1993). This allows the frog genome to be duplicated at a rate of ∼155 Mbp/min (Table 1). As development proceeds, however, some initiation sites disappear while others are enhanced, resulting in a clear pattern of preferred initiation sites at specific genomic loci (Hyrien, Maric, & Mechali, 1995; Sasaki, Sawado, Yamaguchi, & Shinomiya, 1999). Developmental acquisition of origin specification also occurs during mammalian development, although the changes are less dramatic (Borowiec & Schildkraut, 2011).
6. Many Replication Origins Exist, but Few are Activated
In 1990, two independent efforts to locate specific DNA loci that acted as replication origins in hamster chromosomes resulted in paradoxical conclusions: one study concluded that initiation events were essentially “randomly” distributed throughout the 55 kbp intergenic region adjacent to the DHFR gene(Vaughn, Dijkwel, & Hamlin, 1990), whereas the other study concluded that most initiation events began at a specific “origin of bidirectional replication” located within this intergenic region (Burhans, Vassilev, Caddle, Heintz, & DePamphilis, 1990). How might such disparate results be reconciled? The simplest explanation was that replication complexes assembled at many sites along the chromosomes, but that only a subset of these sites were activated at the onset of S phase (DePamphilis, 1993a, 1993b). This hypothesis was termed the “Jesuit Model,” because it is the Jesuits who remind us that “many are called, but few are chosen” (Matthew 22:14). Now, 25 years later, the “Jesuit Model” provides an accurate description of how metazoan cells regulate when and where to initiate DNA replication throughout animal development. Cells derived from human and mouse tissues contain 30,000–50,000 fully active replication origins, but deep sequencing of short nascent DNA strands can detect 10 × as many replication sites with a mean interorigin distance of 11 kbp (Besnard et al., 2012; Leonard & Mechali, 2013). Similarly, cells derived from flies contain 62 initiation sites with a mean size of 32 kbp distributed along 22 Mbp of chromatin (MacAlpine, Rodriguez, & Bell, 2004), but the median distance between ORC-binding sites is also 11 kbp (MacAlpine, Gordan, Powell, Hartemink, & MacAlpine, 2010). The spacing of replication origins in cells from mammals and insects is comparable to the spacing during rapid cleavage cycles in frogs. Thus, the bulk of DNA replication is driven by a small subset (∼10%) of the total number of available replication origins. Moreover, most of the fully active origins exist in several different cell lines, and more than 80% of the fully active origins used in one mitotic cell cycle are reused in the following cell cycle (Li, Chen, Solessio, & Gilbert, 2003).
The existence of a plethora of minor replication origins provides cells with the ability to respond to genotoxic stress. Replication forks frequently stall, for example, when encountering tightly bound protein–DNA complexes, transcription machinery, repetitive sequences, or DNA lesions. Even changes in deoxyribonucleotide pools can change the frequency of initiation at minor replication origins (Anglana, Apiou, Bensimon, & Debatisse, 2003). Minor replication origins remain dormant until activated in response to replication stress (Ge et al., 2015; McIntosh & Blow, 2012). Dormant replication origins are licensed origins that become fully active only under conditions that inhibit ongoing DNA replication forks. Thus, dormant origins ensure genome integrity by replicating regions between stalled replication forks. Dormant origins confer two important advantages to the development of complex organisms. First, they allow flexibility in determining where DNA replication begins, thereby avoiding conflicts between the need to replicate the entire genome and the need to express specific genes, at specific sites, at a specific time. Second, they allow flexibility in determining how many replication origins are necessary to complete the job.
7. Both Genetic and Epigenetic Parameters Define Replication Origins
DNA replication origins in bacteria, archaea, plasmids, bacteriophage, and viruses are specific sequences essential for initiation of DNA replication, because they contain binding sites for either their cognate helicase loader or cognate DNA helicase (DePamphilis & Bell, 2010; Leonard & Mechali, 2013). In contrast, DNA replication origins in the metazoa are not essential for initiation of replication. Artificial assembly of a transcription complex or binding of an ORC or Cdc6 protein to DNA is sufficient to create a site-specific replication origin in frog eggs and mammalian cells (Crevel & Cotterill, 2012; Danis et al., 2004; Takeda, Shibata, Parvin, & Dutta, 2005). Otherwise, DNA replication in either rapid cleavage embryos or egg extract from either frogs or flies begins at virtually any sequence, foreign or domestic, introduced into these cells (Hyrien & Mechali, 1993; Shinomiya & Ina, 1991). In contrast, DNA injected into the nucleus of mouse zygotes does not replicate unless it contains a known replicator (poly-omavirus), and the cognate origin recognition protein (polyomavirus T-antigen) is provided (Martinez-Salas, Cupo, & DePamphilis, 1988). The simplest explanation is that the ratio of replication proteins to DNA in rapid cleavage zygotes is much greater than in mouse zygotes, thereby negating any advantage of specific DNA sequences. The volume of a frog egg (1.2 mm diameter), for example, is about 2800 × greater than a mouse egg (0.085 mm diameter). As cell cleavage reduces the ratio of maternally inherited replication proteins to DNA, some sites will be more effective as replication origins than others.
Metazoan replication origins are simply chromosomal loci that permit assembly of prereplication complexes that can later be converted into preinitiation complexes and activated by Cdk2-CcnE and Cdc7-Dbf4 (Fig. 1). Such sites are determined by both genetic and epigenetic parameters (Aladjem, Falaschi, & Kowalski, 2006; DePamphilis, 1996; Gilbert, 2001; Leonard & Mechali, 2013). Genetic parameters include easily unwound DNA sequences (Aladjem et al., 2006) such as asymmetric A:T-rich sequences (Stanojcic, Lemaitre, Brodolin, Danis, & Mechali, 2008) and consensus G-quadruplex-forming motifs (Besnard et al., 2012; Cayrou et al., 2011; Foulk, Urban, Casella, & Gerbi, 2015; Picard et al., 2014). Epigenetic parameters include nuclear structure (Gilbert, Miyazawa, & DePamphilis, 1995), histone modifications (Dorn & Cook, 2011), chromosome structure, and transcription.
The effect of epigenetic parameters on regulatory DNA sequences begins in mammals during the onset of ZGE when changes in chromatin structure result in the need for enhancers to activate DNA replication replicators as well as transcription promoters. The polyomavirus replicator is active in the paternal pronucleus of a mouse zygote as long as the polyomavirus origin recognition protein (the T-antigen DNA helicase) is provided (Martinez-Salas, Linney, Hassell, & DePamphilis, 1989). However, beginning with the two-cell embryo, the polyomavirus replicator is inactive unless linked to a cell-specific enhancer and provided with T-antigen. Enhancers are required to relieve chromatin-mediated repression of both promoters and replicators (Majumder & DePamphilis, 1995; Majumder et al., 1997; Wiekowski, Miranda, Nothias, & DePamphilis, 1997). Thus, as changes in chromatin structure prevent access to some DNA sites but not to others, the ability to license and then activate replication origins becomes more restricted, as postulated by the “Jesuit Model.”
8. Restricting Genome Duplication to Once Per Cell Division
Proliferating cells must produce one, but only one, copy of their nuclear DNA prior to cell division. Therefore, once S phase begins, origin licensing is blocked until mitosis is completed. If DNA is rereplicated prior to mitosis, the result is DNA damage, particularly in the form of double-stranded breaks that arise from stalled replication forks. If the DNA damage response cannot correct the problem, then apoptosis ensues. To prevent such a catastrophe, at least seven pathways exist that can inactivate the helicase loader during S phase, thereby preventing both the reloading of MCM helicases at activated replication origins, and the licensing of new replication origins. These pathways, which can be grouped into two biological cycles, exist in flies, frogs, nematodes, and mammals (Blow & Dutta, 2005; DePamphilis et al., 2006; Siddiqui, On, & Diffley, 2013; Sonneville, Querenet, Craig, Gartner, & Blow, 2012).
Helicase loaders consist of ORC(1–6), Cdc6, and Cdt1 (Fig. 1). The “ORC Cycle” refers to the inactivation of Orc1 and Cdc6 during S phase by posttranslational CDK-dependent phosphorylation and ubiquitin-dependent degradation, followed by origin licensing during the anaphase to G1 phase transition (Fig. 3). The loss of Orc1 destabilizes the remaining ORC subunits. The “Cdt1 Cycle” refers to the inactivation of Cdt1 by CDK-dependent phosphorylation, ubiquitination of Cdt1-P as well as PCNA– DNA-dependent ubiquitination of Cdt1, and inactivation of Cdt1 by binding to geminin protein. Helicase loader proteins become available again during the anaphase to G1 phase transition followed by origin licensing.
Figure 3.
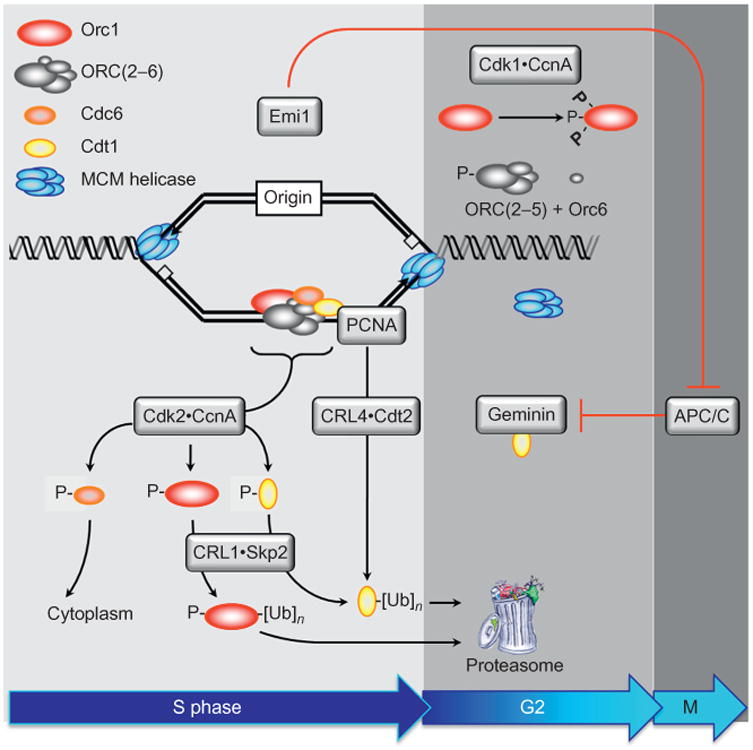
Multiple concerted pathways prevent DNA rereplication. Once metazoan cells have entered S phase, seven concerted pathways can inactivate the helicase loader, thereby preventing rereplication of DNA prior to mitosis. Upon S phase entry, Cdk2CcnA phosphorylates Orc1, Cdc6, and Cdt1, thereby inhibiting their ability to form helicase loaders. In addition, Cdc6-P is localized to the cytoplasm where it cannot facilitate origin licensing. Orc1-P is ubiquitinated and targeted for destruction, with subsequent dissociation of ORC(2–5) and Orc6from chromosomes. Cdt1-P is ubiquitinated by the CRL1 Skp2 ubiquitin ligase. Cdt1 that is not phosphorylated is ubiquitinated by the CRL4Cdt2 ubiquitin ligase bound to PCNA and DNA. PCNA is the eukaryotic sliding clamp protein that facilitates DNA synthesis by DNA polymerases δ and ε. The ubiquitinated proteins are then degraded by the 26S proteasome. Geminin is a protein that binds specifically to Cdt1 and inhibits its activity. These activities are available from S through early M phase. As cells exit mitosis, geminin and cyclin A are ubiquitinated by the anaphase promoting complex [originally termed cyclosome (APC/C)], an activity that is inhibited during S to early M phase by the APC/C-specific inhibitor Emi1. Reassembly of the helicase loader occurs by reassembly of Orc1, ORC(2-5), Orc6, and Cdc6 onto chromatin (Ghosh et al., 2011; Kara et al., 2015; Sonneville et al., 2012), an event that appears to be triggered by the Orc1 subunit (Noguchi et al., 2006). Geminin binding to Cdt1 and CDK-dependent phosphorylation of Cdc6 prevents targeting of Cdt1 and Cdc6 for degradation by the APC/C, thereby allowing them to participate in origin licensing (Ballabeni et al., 2004; Mailand & Diffley, 2005).
The importance of any one pathway in a particular cell type depends on the relative levels of their respective components. Thus, during early development of frogs and mammals only geminin and the CRL4 ubiquitin ligase are essential to prevent DNA rereplication. Cdt1 is a stable abundant protein in Xenopus egg extracts. It is not degraded substantially until cycle 10 (Kisielewska & Blow, 2012). Geminin is only fully degraded by the APC/ C after the MBT (cell cycle 12) (Hodgson, Li, Tada, & Blow, 2002; Li & Blow, 2005). Therefore, two concerted pathways prevent DNA rereplication in cleavage stage embryos by regulating both Cdt1 activity and Cdt1 proteolysis. In the beginning, when the nuclear to cytoplasmic ratio is low, Cdt1 activity is blocked by geminin and regulation of geminin activity appears to involve both the APC/C and nuclear import of geminin. As the ratio of nuclei to cytoplasm increases, Cdt1 binds to CRL4-PCNADNA complexes that assemble at newly minted replication forks and at sites of DNA repair where Cdt1 is then targeted for ubiquitin-dependent degradation (Arias & Walter, 2005, 2007). Thus, suppressing geminin RNA levels in Xenopus egg extracts or two-cell embryos cannot immediately deplete the supply of geminin protein. Consequently, DNA rereplication and apoptosis are not manifested until the MBT and beyond (Kerns, Schultz, Barry, Thorne, & McGarry, 2012; Kerns, Torke, Benjamin, & McGarry, 2007).
Once the developmental program activates ZGE, totipotent and pluripotent cells depend primarily, if not exclusively, on Geminin to prevent DNA rereplication. Geminin is essential for gastrulation in frogs (Kerns et al., 2012), and probably in flies as well (Takada, Kwak, Koppetsch, & Theurkauf, 2007). Most notably, geminin is essential to prevent DNA rereplication-dependent apoptosis in mouse preimplantation embryos (Hara, Nakayama, & Nakayama, 2006; Huang, Kaneko, Pan, & DePamphilis, 2015), in the epiblast (Huang et al., 2015; Patterson, Waller, & Kroll, 2014), and in embryonic stem cells (Huang et al., 2015), which are derived from the epiblast. However, as pluripotent cells differentiate, geminin is no longer essential for viability, although it continues to contribute to preventing DNA rereplication (Dorn, Chastain, Hall, & Cook, 2009; Huang et al., 2015). In effect, other pathways, such as Cdk2•CcnA (Fig. 3), contribute significantly (Zhu & Depamphilis, 2009). Thus, whether or not a particular cell, such as a cancer cell, requires geminin to prevent DNA rereplication depends on the relative levels of relevant gene products. For example, suppression of CRL4 in HeLa cells induces DNA rereplication (Jin, Arias, Chen, Harper, & Walter, 2006), whereas suppression of geminin does not, unless Cdt1 is overexpressed (Nishitani et al., 2006). Thus, geminin is essential for viability when the cellular levels of Cdt1 are high, as in totipotent blastomeres, pluripotent stem cells, some cancer cells, and perhaps some rapidly proliferating cells in adult animals, but the role of geminin in most cells of the adult organism facilitates proliferation without being essential for viability. Similarly, the primary role of CRL4 in mammals and presumably other animals is to facilitate DNA repair by inhibiting origin licensing (Havens & Walter, 2011).
9. Developmentally Programmed Polyploidy
Given the extraordinary problem of preventing excess genome duplication during metazoan development, it is remarkable that some cells are developmentally programmed to exit their mitotic cell cycle in response to environmental signals, injury or stress, and differentiate into nonproliferating, viable, polyploid cells. In fact, developmentally programmed polyploidy is a normal part of animal and plant development that occurs frequently in ferns, flowering plants, mollusks, arthropods, amphibians, and fish, but infrequently in mammals (Lacroix & Maddox, 2012; Ullah, Lee, & Depamphilis, 2009). Some cells become multinucleated by not undergoing cytokinesis, or by fusing together during G1 phase (Gentric & Desdouets, 2014). In contrast, other cells are programmed to undergo multiple S phases in the absence of an intervening mitosis and cytokinesis. These cells become giant cells with one enlarged nucleus containing multiples of 4C DNA, in some cases reaching hundreds of copies (Orr-Weaver, 2015). DNA replication-dependent polyploidy occurs via endoreplication and endomitosis.
In Drosophila, the Notch signal transduction induces endoreplication by inhibiting Cdk1, an activity essential for initiating and maintaining mitosis (Fig. 1). Notch induces expression of Cdh1/Fzr, an activator of the APC/C that triggers ubiquitin-mediated destruction of mitotic cyclins, and Notch suppresses Cdc25/String, a phosphatase that activates the Cdk1•Cyclin complexes essential for mitosis (Lee, Davidson, & Duronio, 2009). Similarly, endoreplication occurs in the trophectoderm of mouse peri-implantation blastocysts in response to the absence of fibroblast growth factor-4 (FGF4) that is produced by the inner cell mass (Roberts & Fisher, 2011). This phenomenon is recapitulated by trophoblast stem cells (TSCs). TSCs undergo self-renewal when cultured in the presence of FGF4, and differentiate into giant cells when cultured in the absence of FGF4 (Fig. 4). Selective inhibition of Cdk1 by RO3306 triggers endoreplication in TSCs undergoing self-renewal. FGF4-deprivation induces expression of the CDK-specific inhibitor p57, which inhibits Cdk1. Since p57 also inhibits Cdk2, p57 levels in giant cells must oscillate during multiple rounds of endoreplication; high during G phase but low during S phase. That permits Cdk2-CcnE to initiate S phase (Fig. 1) and Cdk2•CcnA to prevent DNA rereplication (Fig. 3). FGF4 regulates expression of p57 in TSCs via control of Chk1 levels. Chk1 phosphorylates p57, thereby targeting it for ubiquitin-dependent degradation. FGF4 deprivation in TSCs results in downregulation of geminin, which results in the loss of Chk1 protein with concomitant upregulation of p57 (Fig. 5). Thus, TSCs will also differentiate into giant cells as a result of DNA damage activating Chk1, the DNA damage response kinase that inhibits Cdc25 (required for Cdk1 activity), thereby inhibiting Cdk1 activity (Fig. 4).
Figure 4.
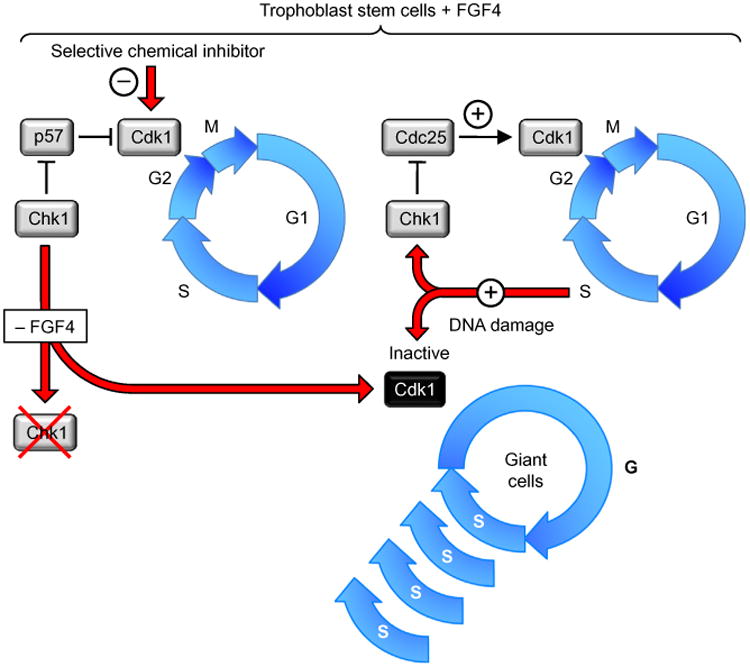
Endoreplication is triggered in trophoblast cells by selective inhibition of Cdk1 activity. Developmentally programmed endoreplication in the metazoa occurs when Cdk1 activity is suppressed so that cells cannot enter mitosis (Edgar, Zielke, & Gutierrez, 2014; Fox & Duronio, 2013; Zielke, Edgar, & DePamphilis, 2013). This results in multiple S phases without an intervening mitosis or cytokinesis to produce giant cells with a single enlarged nucleus containing as many as several hundred copies of each chromosome. In the absence of FGF4, trophoblast cells in mouse blastocysts are triggered to differentiate into giant cells. In the presence of FGF4, the same result can be elicited either by selective inhibition of Cdk1 with RO3306, or by suppressing or inactivating geminin (de Renty, Kaneko, & DePamphilis, 2014; Ullah, Kohn, Yagi, Vassilev, & DePamphilis, 2008). The effect of FGF4 deprivation is to reduce the level of geminin, which results in the loss of checkpoint kinase-1 (Chek1/Chk1). Since Chk1 phosphorylation of the CDK-specific inhibitor Cdkn1c/p57 targets it for ubiquitin-dependent degradation, loss of Chk1 results in p57 inhibition of Cdk1 activity, which leads to endoreplication (Ullah, de Renty, & Depamphilis, 2011). Ironically, DNA damage in trophoblast cells also results in endoreplication, because DNA damage activates Chk1, which then phos-phorylates and inactivates Cdc25, a phosphatase that is essential to maintain Cdk1 in its active form.
Figure 5.
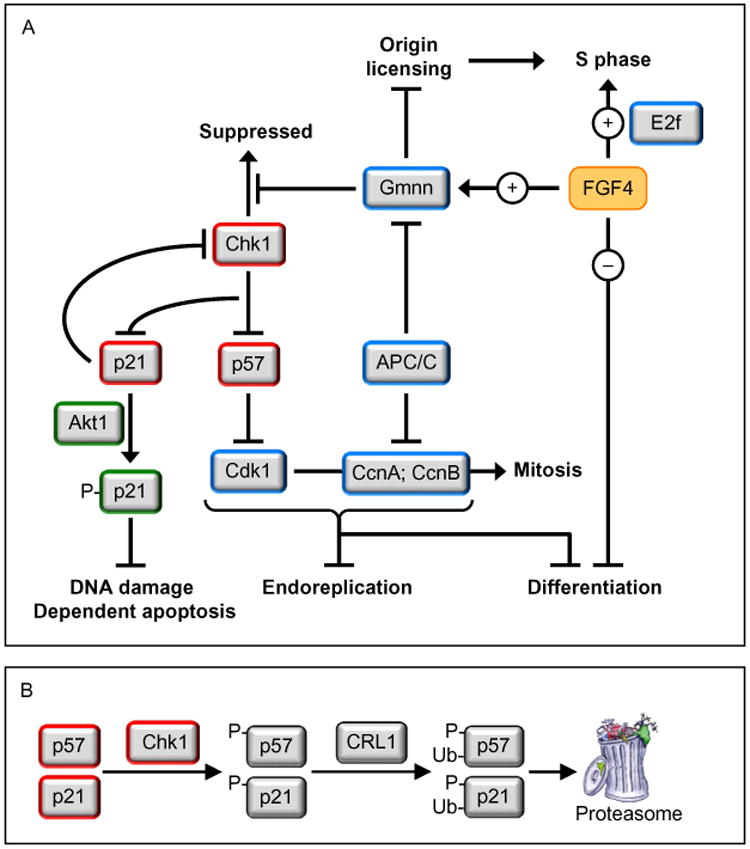
The FGF4 signal transduction pathway governs trophoblast proliferation and differentiation. FGF4 (and probably other mitogenic proteins as well) is essential for trophoblast proliferation. This mitogenic activity is likely mediated by E2F-dependent gene expression (Chen et al., 2010), and possibly directed at regulating the activity of the anaphase-promoting complex (APC) (Yang et al., 2012). FGF4 deprivation results in down-regulation of geminin activity to a level that maintains endocycles (de Renty, Kaneko, & DePamphilis, 2014), but that does not prevent downregulation of Chk1 protein. The loss of Chk1 kinase activity results in expression of two CDK-specific inhibitors, p57 and p21 (Ullah et al., 2011). The p57 protein prevents the onset of mitosis by selectively inhibiting Cdk1 activity, thereby triggering the first round ofendoreplication (Hattori, Davies, Anson-Cartwright, & Cross, 2000; Ullah et al., 2008). This event activates the G1-phase APCCdh1 ubiquitin ligase, which targets geminin, cyclin B, and cyclin A for degradation, thereby allowing licensing of replication origins and the onset of S-phase without passing through mitosis (Ullah, Lee, Lilly, & DePamphilis, 2009). Inhibition of Cdk1 triggers both endoreplication and TSC differentiation. In the absence of p57, FGF4 deprivation produces multinucleated TGCs, revealing the existence of alternative mechanisms to trigger TSC differentiation (Ullah et al., 2008). Endocycles also require p57, which is expressed during G-phase and then suppressed during S-phase to allow sequential assembly and activation of pre-replication complexes (Ullah et al., 2008). Geminin maintains endocycles by preventing DNA rereplication. The p21/Cdkn1a protein localizes to the cytoplasm in TGCs where it prevents DNA damage induced apoptosis (de Renty, DePamphilis, & Ullah, 2014). It might also maintain suppression of Chk1 by reducing Chk1 RNA levels (Gottifredi, Karni-Schmidt, Shieh, & Prives, 2001), as observed during FGF4 deprivation (Fig. 3).
Terminally differentiated cells must remain viable, but since they cannot proliferate, they cannot pass on genetic mutations, so repairing DNA damage is not a concern. Induction of DNA rereplication induced DNA damage in Drosophila does not induce apoptosis in follicle cells undergoing endoreplication, whereas it does in cells undergoing proliferation (Mehrotra, Maqbool, Kolpakas, Murnen, & Calvi, 2008). Loss of Chk1 protein in TSCs during FGF4-deprivation induces both p57 and p21 expression by the same mechanism (Fig. 5), but p57 localizes to the nucleus where it inhibits Cdk1, and p21 localizes to the cytoplasm where it suppresses DNA damage-dependent apoptosis (de Renty, DePamphilis, et al., 2014).
Endomitosis is similar to endoreplication, except that the nuclear envelope breaks down and parts of anaphase occur without nuclear division, thereby producing mononucleated cells with separated chromatids. The mechanism responsible remains speculative, but selective inactivation of Cdk1 readily converts endomitosis into endoreplication (Trakala et al., 2015). Selective inactivation of Cdk1 also induces endoreplication in mouse embryonic fibroblasts and chicken DT40 cells (Diril et al., 2012; Hochegger et al., 2007). Suppression of Cdk1 activity induces endoreplication.
10. Developmentally Programmed Gene Amplification
Genes encoding products needed in high amounts at specific stages during development are sometimes amplified by selectively replicating those genes multiple times. Developmentally programmed gene amplification is an example of site-specific initiation of DNA replication in terminally differentiated cells (Calvi, 2006; Tower, 2004). Specialized cis-acting sequences (amplification origins) allow specific genes to be selectively rereplicated multiple times. In response to ecdysone, the master regulator of insect development, the larval salivary glands of Sciara coprophila produce a parallel array of hundreds of chromatids termed “polytene chromosomes.” Gene amplification then produces DNA puffs in these chromosomes (not to be confused with RNA puffs, which arise by multiple rounds of transcription). The genes encoding the proteins required for pupation are then amplified many fold. A larval-specific ecdysone receptor binds to a specific DNA sequence adjacent to the origin recognition complex-binding site within a site-specific origin of bidirectional replication (Foulk et al., 2013). Similarly, the follicle cells that envelop each oocyte in Drosophila amplify the four genes that produce the proteins for chorion (eggshell) formation. Amplification depends on the interaction of prereplication complexes assembled at two specific DNA sites, ACE3 and Ori-β, within the chorion gene cluster. It appears that amplification origins in flies/gnats use the same proteins required to initiate DNA replication during normal cell proliferation, but in contrast to replication origins activated during S phase of mitotic cell division cycles, these specialized “amplification origins” continue to initiate DNA replication in the absence of mitosis or cytokinesis.
Xenopus oocytes also amplify their rRNA genes, apparently by excising one or more copies from their chromosomes and then replicating them several thousand folds via a “rolling circle” mechanism (Hourcade, Dressler, & Wolfson, 1973). Although the details of Xenopus rRNA amplification remain speculative, the mechanism for rRNA gene amplification in the ciliate protozoan Tetrahymena thermophila has been well characterized. This single-cell organism has two nuclei: a transcriptionally silent, diploid, germline micronucleus and a transcriptionally active, polyploid, somatic macronucleus. The chromosomes in the macronucleus are first endoreplicated about 45-fold, and then genome-wide replication ceases, and a 21 kbp linear minichromosome encoding the three rRNAs is generated by excision and rearrangement of the micronuclear rRNA gene locus. The minichromosome is amplified up to 10,000 copies in a single S phase during development, but it is replicated, on average, only once per cell cycle during vegetative growth. DNA replication begins at a specific site within the rRNA gene locus of Tetrahymena, as it does within the rRNA gene loci in yeast and mammalian cells (Donti, Datta, Sandoval, & Kapler, 2009).
11. Evolution and Human Disease
The vast majority of mutations are caused by misincorporation of the wrong nucleotide when DNA polymerases replicate DNA and when damaged DNA is repaired. The two human DNA polymerases (δ and ε) responsible for the bulk of nuclear DNA replication have error rates ∼1 to 2 per 100,000 nucleotides synthesized, and eight human DNA repair polymerases have errorrates of 23–72,000 (DePamphilis & Bell, 2010). Thus, the very act of duplicating the genome creates the basis for genomic instability, and the simple fact that natural selection has never completely suppressed the accumulation of errors during genome duplication attests to the theory that biological evolution relies upon these errors. If evolution by natural selection produced error free DNA polymerases, then the evolution of species would cease. It has not, DNA replication continues to drive evolution.
The spontaneous mutation rate in human cells is one mutation in every 10 billion base pairs that are replicated during human development. The reason the rate is not higher is because nature added a “proofreading” mechanism to polymerases δ and ε in the form of an exonuclease that degrades one strand of duplex DNA beginning at its 3′-OH terminus, thereby allowing the DNA polymerase to try again. This reduces the error rate for these enzymes by 4- to 24-fold. In addition, nature developed “mismatch repair” systems to correct single mistakes in base pairing during DNA synthesis. Nevertheless, at an error rate of 1/109 bp, every zygote will receive about 138 new mutations. There are about 400 cell divisions between zygote and spermatocytes and about 30 cell divisions between zygote and oocytes (Crow, 2000).
Unfortunately, the price for continued evolution is disease. Genetic mutations just in human DNA damage response genes, restriction checkpoint genes, DNA helicases, DNA replication proteins, and DNA repair genes are associated with at least 60 different cancers and disease syndromes (DePamphilis, 2006; DePamphilis & Bell, 2010). Mutations are also produced by aberrant events at DNA replication forks that result in synthesis of multiple repeats of the same nucleotide sequence within a gene. This produces a mutated protein containing a series of additional amino acids. At least 19 “expanded repeat diseases” have been identified, including several neurodegenerative disorders.
12. EPILOG
Genome duplication, mitosis, and cytokinesis are all essential for cell proliferation. Changing patterns of gene expression are essential for metazoan development. However, metazoan development could not have evolved without genome duplication becoming flexible enough to accommodate the ever-changing demands of gene expression during cell differentiation and reliable enough to sustain the trillions of cell divisions required from zygote to adult. Although much remains to be discovered, one question has far reaching implications: Are some genes essential for genome duplication in some cell types, but not in others, and if so, can those genes provide therapeutic targets in the treatment of human disease?
References
- Aiken CE, Swoboda PP, Skepper JN, Johnson MH. The direct measurement of embryogenic volume and nucleo-cytoplasmic ratio during mouse preimplantation development. Reproduction. 2004;128:527–535. doi: 10.1530/rep.1.00281. [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Falaschi A, Kowalski D. Eukaryotic DNA replication origins. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 31–62. [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental Biology. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes and Development. 2005;19:114–126. doi: 10.1101/gad.1255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes and Development. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Baldinger T, Gossen M. Binding of Drosophila ORC proteins to anaphase chromosomes requires cessation of mitotic cyclin-dependent kinase activity. Molecular and Cellular Biology. 2009;29:140–149. doi: 10.1128/MCB.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabeni A, Melixetian M, Zamponi R, Masiero L, Marinoni F, Helin K. Human geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO Journal. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Kaguni JM. Helicase loading at chromosomal origins of replication. Cold Spring Harbor Perspectives in Biology. 2013;5:61–80. doi: 10.1101/cshperspect.a010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nature Structural and Molecular Biology. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nature Reviews Molecular Cell Biology. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract are 5-15 kilobases apart and are activated in clusters that fire at different times. Journal of Cell Biology. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symposia on Quantitative Biology. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Bolton VN, Oades PJ, Johnson MH. The relationship between cleavage, DNA replication, and gene expression in the mouse 2-cell embryo. Journal of Embryology and Experimental Morphology. 1984;79:139–163. [PubMed] [Google Scholar]
- Borowiec JA, Schildkraut CL. Open sesame: Activating dormant replication origins in the mouse immunoglobulin heavy chain (Igh) locus. Current Opinion in Cell Biology. 2011;23:284–292. doi: 10.1016/j.ceb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans WC, Vassilev LT, Caddle MS, Heintz NH, DePamphilis ML. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Calvi BR. Developmental gene amplification. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 233–255. [Google Scholar]
- Campisi J. Aging, cellular senescence, and cancer. Annual Review of Physiology. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Research. 2011;21:1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes and Development. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemerych MA, Sicinski P. Cell cycle in mouse development. Oncogene. 2005;24:2877–2898. doi: 10.1038/sj.onc.1208608. [DOI] [PubMed] [Google Scholar]
- Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Molecular Biology of the Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: Implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6:2760–2767. doi: 10.4161/cc.6.22.4932. [DOI] [PubMed] [Google Scholar]
- Crevel G, Cotterill S. Forced binding of the origin of replication complex to chromosomal sites in Drosophila S2 cells creates an origin of replication. Journal of Cell Science. 2012;125:965–972. doi: 10.1242/jcs.094409. [DOI] [PubMed] [Google Scholar]
- Crow JF. The origins, patterns and implications of human spontaneous mutation. Nature Reviews Genetics. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nature Cell Biology. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Eukaryotic DNA replication: Anatomy of an origin. Annual Review of Biochemistry. 1993a;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Origins of DNA replication in metazoan chromosomes. Journal of Biological Chemistry. 1993b;268:1–4. [PubMed] [Google Scholar]
- DePamphilis ML. Origins of DNA replication. In: DePamphilis ML, editor. DNA replication in eukaryotic cells. New York: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- DePamphilis ML. DNA replication and human disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. p. 814. [Google Scholar]
- DePamphilis ML, Bell SD. Genome duplication (concepts, mechanisms, evolution and disease) London and New York: Garland Science; 2010. [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Current Opinion in Cell Biology. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Depamphilis ML, de Renty CM, Ullah Z, Lee CY. “The octet”: Eight protein kinases that control mammalian DNA replication. Frontiers in Physiology. 2012;3:368. doi: 10.3389/fphys.2012.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renty C, DePamphilis ML, Ullah Z. Cytoplasmic localization of p21 protects trophoblast giant cells from DNA damage induced apoptosis. PLoS ONE. 2014;9:e97434. doi: 10.1371/journal.pone.0097434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renty C, Kaneko KJ, DePamphilis ML. The dual roles of geminin during trophoblast proliferation and differentiation. Developmental Biology. 2014;387:49–63. doi: 10.1016/j.ydbio.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diril MK, Ratnacaram CK, Padmakumar VC, Du T, Wasser M, Coppola V, et al. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3826–3831. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donti TR, Datta S, Sandoval PY, Kapler GM. Differential targeting of Tetrahymena ORC to ribosomal DNA and non-rDNA replication origins. EMBO Journal. 2009;28:223–233. doi: 10.1038/emboj.2008.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn ES, Chastain PD, 2nd, Hall JR, Cook JG. Analysis of re-replication from deregulated origin licensing by DNA fiber spreading. Nucleic Acids Research. 2009;37:60–69. doi: 10.1093/nar/gkn912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn ES, Cook JG. Nucleosomes in the neighborhood: New roles for chro-matin modifications in replication origin control. Epigenetics. 2011;6:552–559. doi: 10.4161/epi.6.5.15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nature Reviews Molecular Cell Biology. 2014;15:197–210. doi: 10.1038/nrm3756. [DOI] [PubMed] [Google Scholar]
- El Dika M, Laskowska-Kaszub K, Koryto M, Dudka D, Prigent C, Tassan JP, et al. CDC6 controls dynamics of the first embryonic M-phase entry and progression via CDK1 inhibition. Developmental Biology. 2014;396:67–80. doi: 10.1016/j.ydbio.2014.09.023. [DOI] [PubMed] [Google Scholar]
- Foster DA, Yellen P, Xu L, Saqcena M. Regulation of G1 cell cycle progression: Distinguishing the restriction point from a nutrient-sensing cell growth check-point(s) Genes & Cancer. 2010;1:1124–1131. doi: 10.1177/1947601910392989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulk MS, Urban JM, Casella C, Gerbi SA. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Research. 2015;25:725–735. doi: 10.1101/gr.183848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulk MS, Waggener JM, Johnson JM, Yamamoto Y, Liew GM, Urnov FD, et al. Isolation and characterization of the ecdysone receptor and its heterodimeric partner ultraspiracle through developmentinSciara coprophila. Chromosoma. 2013;122:103–119. doi: 10.1007/s00412-012-0395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: Insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Han J, Cheng EC, Yamaguchi S, Shima N, Thomas JL, et al. Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Reports. 2015;5(2):185–194. doi: 10.1016/j.stemcr.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric G, Desdouets C. Polyploidization in liver tissue. American Journal of Pathology. 2014;184:322–331. doi: 10.1016/j.ajpath.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Vassilev AP, Zhang J, Zhao Y, DePamphilis ML. Assembly of the human origin recognition complex occurs through independent nuclear localization of its components. Journal of Biological Chemistry. 2011;286:23831–23841. doi: 10.1074/jbc.M110.215988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Miyazawa H, DePamphilis ML. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Molecular and Cellular Biology. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover DM, Alphey L, Axton JM, Cheshire A, Dalby B, Freeman M, et al. Mitosis in Drosophila development. Journal of Cell Science. 1989;(12):277–291. doi: 10.1242/jcs.1989.supplement_12.22. [DOI] [PubMed] [Google Scholar]
- Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Molecular and Cellular Biology. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Moroishi T, Guan KL. YAP and TAZ: A nexus for Hippo signaling and beyond. Trends in Cell Biology. 2015;25(9):499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Nakayama KI, Nakayama K. Geminin is essential for the development of preimplantation mouse embryos. Genes to Cells. 2006;11:1281–1293. doi: 10.1111/j.1365-2443.2006.01019.x. [DOI] [PubMed] [Google Scholar]
- Hattori N, Davies TC, Anson-Cartwright L, Cross JC. Periodic expression of the cyclin-dependent kinase inhibitor p57(Kip2) in trophoblast giant cells defines a G2-like gap phase of the endocycle. Molecular Biology of the Cell. 2000;11:1037–1045. doi: 10.1091/mbc.11.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes and Development. 2011;25:1568–1582. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Dejsuphong D, Sonoda E, Saberi A, Rajendra E, Kirk J, et al. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. Journal of Cell Biology. 2007;178:257–268. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Li A, Tada S, Blow JJ. Geminin becomes activated as an inhibitor of Cdt1/RLF-B following nuclear import. Current Biology. 2002;12:678–683. doi: 10.1016/s0960-9822(02)00778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourcade D, Dressler D, Wolfson J. The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:2926–2930. doi: 10.1073/pnas.70.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett SK. A set of proteins showing cell cycle dependent modification in the early mouse embryo. Cell. 1986;45:387–396. doi: 10.1016/0092-8674(86)90324-7. [DOI] [PubMed] [Google Scholar]
- Howlett SK, Bolton VN. Sequence and regulation of morphological and molecular events during the first cell cycle of mouse embryogenesis. Journal of Embryology and Experimental Morphology. 1985;87:175–206. [PubMed] [Google Scholar]
- Huang YY, Kaneko KJ, Pan H, DePamphilis ML. Geminin is essential to prevent DNA re-replication-dependent apoptosis in pluripotent cells, but not in differentiated cells. Stem Cells. 2015;33(11):3239–3253. doi: 10.1002/stem.2092. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M. Transition in specification of embryonic meta-zoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Mechali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO Journal. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Molecular Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, Cullinan EB, Latham KE, DePamphilis ML. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development. 1997;124:1963–1973. doi: 10.1242/dev.124.10.1963. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ, DePamphilis ML. TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development. 2013;140:3680–3690. doi: 10.1242/dev.093799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara N, Hossain M, Prasanth SG, Stillman B. Orc1 binding to mitotic chromosomes precedes spatial patterning during G1 phase and assembly of the origin recognition complex in human cells. Journal of Biological Chemistry. 2015;290:12355–12369. doi: 10.1074/jbc.M114.625012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Schultz KM, Barry KA, Thorne TM, McGarry TJ. Geminin is required for zygotic gene expression at the Xenopus mid-blastula transition. PLoS ONE. 2012;7:e38009. doi: 10.1371/journal.pone.0038009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Torke SJ, Benjamin JM, McGarry TJ. Geminin prevents rereplication during xenopus development. Journal of Biological Chemistry. 2007;282:5514–5521. doi: 10.1074/jbc.M609289200. [DOI] [PubMed] [Google Scholar]
- Kisielewska J, Blow JJ. Dynamic interactions of high Cdt1 and geminin levels regulate S phase in early Xenopus embryos. Development. 2012;139:63–74. doi: 10.1242/dev.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Maddox AS. Cytokinesis, ploidy and aneuploidy. Journal of Pathology. 2012;226:338–351. doi: 10.1002/path.3013. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Current Biology. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annual Review of Cell and Developmental Biology. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. Endoreplication: Polyploidy with purpose. Genes and Development. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre JM, Bocquet S, Mechali M. Competence to replicate in the unfertilized egg is conferred by Cdc6 during meiotic maturation. Nature. 2002;419:718–722. doi: 10.1038/nature01046. [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Bocquet S, Terret ME, Namdar M, Ait-Ahmed O, Kearsey S, et al. The regulation of competence to replicate in meiosis by Cdc6 is conserved during evolution. Molecular Reproduction and Development. 2004;69:94–100. doi: 10.1002/mrd.20153. [DOI] [PubMed] [Google Scholar]
- Leonard AC, Mechali M. DNA replication origins. Cold Spring Harbor Perspectives in Biology. 2013;5:a010116. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Blow JJ. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO Journal. 2005;24:395–404. doi: 10.1038/sj.emboj.7600520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen J, Solessio E, Gilbert DM. Spatial distribution and specification of mammalian replication origins during G1 phase. Journal of Cell Biology. 2003;161:257–266. doi: 10.1083/jcb.200211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136:2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Research. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes and Development. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Auley A, Werb Z, Mirkes PE. Characterization of the unusually rapid cell cycles during rat gastrulation. Development. 1993;117:873–883. doi: 10.1242/dev.117.3.873. [DOI] [PubMed] [Google Scholar]
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML. TATA-dependent enhancer stimulation of promoter activity in mice is developmentally acquired. Molecular and Cellular Biology. 1994;14:4258–4268. doi: 10.1128/mcb.14.6.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. Bioessays. 1995;17:879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO Journal. 1993;12:1131–1140. doi: 10.1002/j.1460-2075.1993.tb05754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Zhao Z, Kaneko K, DePamphilis ML. Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO Journal. 1997;16:1721–1731. doi: 10.1093/emboj/16.7.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Cupo DY, DePamphilis ML. The need for enhancers is acquired upon formation of a diploid nucleus during early mouse development. Genes and Development. 1988;2:1115–1126. doi: 10.1101/gad.2.9.1115. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, Linney E, Hassell J, DePamphilis ML. The need for enhancers in gene expression first appears during mouse development with formation of the zygotic nucleus. Genes and Development. 1989;3:1493–1506. doi: 10.1101/gad.3.10.1493. [DOI] [PubMed] [Google Scholar]
- McIntosh D, Blow JJ. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harbor Perspectives in Biology. 2012;10(4):33–42. doi: 10.1101/cshperspect.a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes and Development. 2008;22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin F, Miranda M, Montreau N, DePamphilis ML, Blangy D. Transcription enhancer factor-1 (TEF-1) DNA binding sites can specifically enhance gene expression at the beginning of mouse development. EMBO Journal. 1993;12:4657–4666. doi: 10.1002/j.1460-2075.1993.tb06154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Molecular Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai S, Stein P, Buffone MG, Yamashita S, Schultz RM. Recruitment of Orc6l, a dormant maternal mRNA in mouse oocytes, is essential for DNA replication in 1-cell embryos. Developmental Biology. 2010;341:205–212. doi: 10.1016/j.ydbio.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Dasso M. On the coupling between DNA replication and mitosis. Journal of Cell Science. 1989;(12):149–160. doi: 10.1242/jcs.1989.supplement_12.13. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteol-ysis. EMBO Journal. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO Journal. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development. Journal of Biological Chemistry. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. EMBO Journal. 1996;15:5715–5725. [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH, Stumpff J, Su TT. Embryonic cleavage cycles: How is a mouse like a fly? Current Biology. 2004;14:R35–R45. doi: 10.1016/j.cub.2003.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL. When bigger is better: The role of polyploidy in organogenesis. Trends in Genetics. 2015;31:307–315. doi: 10.1016/j.tig.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson ES, Waller LE, Kroll KL. Geminin loss causes neural tube defects through disrupted progenitor specification and neuronal differentiation. Developmental Biology. 2014;393(1):44–56. doi: 10.1016/j.ydbio.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Cadoret JC, Audit B, Arneodo A, Alberti A, Battail C, et al. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genetics. 2014;10:e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Fisher SJ. Trophoblast stem cells. Biology of Reproduction. 2011;84:412–421. doi: 10.1095/biolreprod.110.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Molecular and Cellular Biology. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya T, Ina S. Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Research. 1991;19:3935–3941. doi: 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harbor Perspectives in Biology. 2013;5(9):361–380. doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow MHL. Gastrulation in the mouse: Growth and regionalization of the epiblast. Journal of Embryology and Experimental Morphology. 1977;42:293–303. [PubMed] [Google Scholar]
- Sonneville R, Querenet M, Craig A, Gartner A, Blow JJ. The dynamics of replication licensing in live Caenorhabditis elegans embryos. Journal of Cell Biology. 2012;196:233–246. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojcic S, Lemaitre JM, Brodolin K, Danis E, Mechali M. In Xenopus egg extracts, DNA replication initiates preferentially at or near asymmetric AT sequences. Molecular and Cellular Biology. 2008;28:5265–5274. doi: 10.1128/MCB.00181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Kwak S, Koppetsch BS, Theurkauf WE. grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development. 2007;134:1737–1744. doi: 10.1242/dev.02831. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes and Development. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harbor Perspectives in Biology. 2013;5:a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J. Developmental gene amplification and origin regulation. Annual Review of Genetics. 2004;38:273–304. doi: 10.1146/annurev.genet.37.110801.143851. [DOI] [PubMed] [Google Scholar]
- Trakala M, Rodriguez-Acebes S, Maroto M, Symonds CE, Santamaria D, Ortega S, et al. Functional reprogramming of polyploidization in megakaryocytes. Developmental Cell. 2015;32:155–167. doi: 10.1016/j.devcel.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Ullah Z, de Renty C, Depamphilis ML. Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Molecular and Cellular Biology. 2011;31:4129–4143. doi: 10.1128/MCB.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes and Development. 2008;22:3024–3036. doi: 10.1101/gad.1718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Lee CY, Depamphilis ML. Cip/Kip cyclin-dependent protein kinase inhibitors and the road to polyploidy. Cell Div. 2009a;4:10. doi: 10.1186/1747-1028-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Lee CY, Lilly MA, DePamphilis ML. Developmentally programmed endoreduplication in animals. Cell Cycle. 2009b;8:1501–1509. doi: 10.4161/cc.8.10.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nature Structural and Molecular Biology. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes and Development. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JP, Dijkwel PA, Hamlin JL. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990;61:1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- Whitmire E, Khan B, Coue M. Cdc6 synthesis regulates replication competence in Xenopus oocytes. Nature. 2002;419:722–725. doi: 10.1038/nature01032. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: Effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Developmental Biology. 1991;147:403–414. doi: 10.1016/0012-1606(91)90298-h. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, Nothias JY, DePamphilis ML. Changes in his-tone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. Journal of Cell Science. 1997;110(Pt. 10):1147–1158. doi: 10.1242/jcs.110.10.1147. [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yang VS, Carter SA, Ng Y, Hyland SJ, Tachibana-Konwalski K, Fisher RA, et al. Distinct activities of the anaphase-promoting complex/cyclosome (APC/C) in mouse embryonic cells. Cell Cycle. 2012;11:846–855. doi: 10.4161/cc.11.5.19251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Depamphilis ML. Selective killing of cancer cells by suppression of geminin activity. Cancer Research. 2009;69:4870–4877. doi: 10.1158/0008-5472.CAN-08-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N, Edgar BA, DePamphilis ML. Endoreplication. Cold Spring Harbor Perspectives in Biology. 2013;5:a012948. doi: 10.1101/cshperspect.a012948. [DOI] [PMC free article] [PubMed] [Google Scholar]


