SUMMARY
Nuclear transport of immune receptors, signal transducers, and transcription factors is an essential regulatory mechanism for immune activation. Whether and how this process is regulated at the level of the nuclear pore complex (NPC) remains unclear. Here we report that CPR5, which plays a key inhibitory role in effector-triggered immunity (ETI) and programmed cell death (PCD) in plants, is a novel transmembrane nucleoporin. CPR5 associates with anchors of the NPC selective barrier to constrain nuclear access of signaling cargos and sequesters Cyclin-dependent Kinase Inhibitors (CKIs) involved in ETI signal transduction. Upon activation by immunoreceptors, CPR5 undergoes an oligomer to monomer conformational switch, which coordinates CKI release for ETI signaling and reconfigures the selective barrier to allow significant influx of nuclear signaling cargos through the NPC. Consequently, these coordinated NPC actions result in simultaneous activation of diverse stress-related signaling pathways and constitute an essential regulatory mechanism specific for ETI/PCD induction.
Graphical Abstract
INTRODUCTION
Effector-triggered immunity (ETI) is a vital mechanism for host recognition of pathogen virulence effectors to trigger defense (Jones and Dangl, 2006; Stuart et al., 2013). In plants, ETI is activated by nucleotide-binding leucine-rich repeat (NB-LRR) receptors, which are divided into two major classes based on the presence of an N-terminal coiled-coil (CC) domain or a Toll-interlukin1 receptor (TIR) domain. NB-LRRs have been found to localize in various subcellular compartments, where they can detect activities of different pathogen effectors (Elmore et al., 2011). Although quantitative differences exist in outcomes of ETI mediated by different NB-LRRs, they all result in similar transcriptional reprogramming of the infected cells, which leads to restriction of pathogen growth and rapid programmed cell death (PCD). This suggests a common cellular regulatory mechanism connecting distinct NB-LRR activation events to a unified transcriptional response in the nucleus.
Genetic and molecular studies have shown that changes in the nucleocytoplasmic dynamics of NB-LRR receptor complexes, signal transducers and immune-related transcriptional regulators are crucial for defense gene expression and resistance during ETI (Garcia and Parker, 2009; Rivas, 2012). Screens for suppressors of an autoactivated TIR-NB-LRR protein mutant, snc1, led to the identification of mos (modifiers of snc1) mutants, including two (mos3 and mos7) in the nuclear pore complex (NPC) and one (mos6) in a nuclear transport receptor (NTR) (Cheng et al., 2009; Palma et al., 2005; Zhang and Li, 2005). These and other evidence suggested that the NPC- and NTR-directed nucleocytoplasmic transport are involved in subcellular defense coordination triggered by NB-LRRs (Wirthmueller et al., 2013). However, because these mos and other nucleoporin mutants are also compromised in resistance independent of NB-LRRs (Wiermer et al., 2012), whether the NPC plays a generic role in mediating transport of defense signals or a specific regulatory role for distinct immune mechanisms remains unclear.
In contrast to the mos mutants, which block immune responses, loss-of-function mutations in the putative nuclear envelope (NE) protein CPR5 (Constitutive Expresser of PR Genes 5) result in an ETI-like transcriptome and PCD (Wang et al., 2014). Consequently these mutants show resistance against multiple pathogens carrying effectors independent of cognate NB-LRR receptors (Boch, 1998; Bowling et al., 1997). These evidence suggest that CPR5 regulates an essential downstream inhibitory mechanism of ETI/PCD, possibly at the nucleocytoplasmic barrier.
We previously showed that two Cyclin-dependent Kinase Inhibitors (CKIs), SIM (SIAMESE) and SMR1 (SIAMESE-Related 1), are redundantly required for downstream ETI/PCD signaling in the cpr5 mutant. CPR5 sequesters CKIs in the NE and specifically releases them in response to NB-LRR activation to engage the Retinoblastoma (Rb) and the E2F–mediated cell cycle pathway to regulate defense gene expression and PCD (Wang et al., 2014). However, how are CKIs released remains unknown. Moreover, whether redirection of the cell cycle pathway is sufficient for cpr5-mediated ETI/PCD needs further investigation.
In this study, we report that CPR5 is a plant transmembrane nucleoporin that physically associates with the NPC core scaffold. CPR5 resides in the NPC as a homomeric complex, which is specifically disrupted in response to NB-LRR activation. This conformational change in the NPC plays a dual role during ETI/PCD activation: it enables dissociation of CKIs from the NPC to engage cell cycle regulators for defense gene expression and reconfigures the NPC selective barrier to allow massive nuclear influx of diverse stress-related signaling cargos. These CPR5-coordinated actions of the NPC are required for ETI/PCD induction and constitute a downstream regulatory mechanism specific for NB-LRR-mediated ETI/PCD.
RESULTS
CPR5 Is a Transmembrane Protein Enriched in the Nuclear Pore
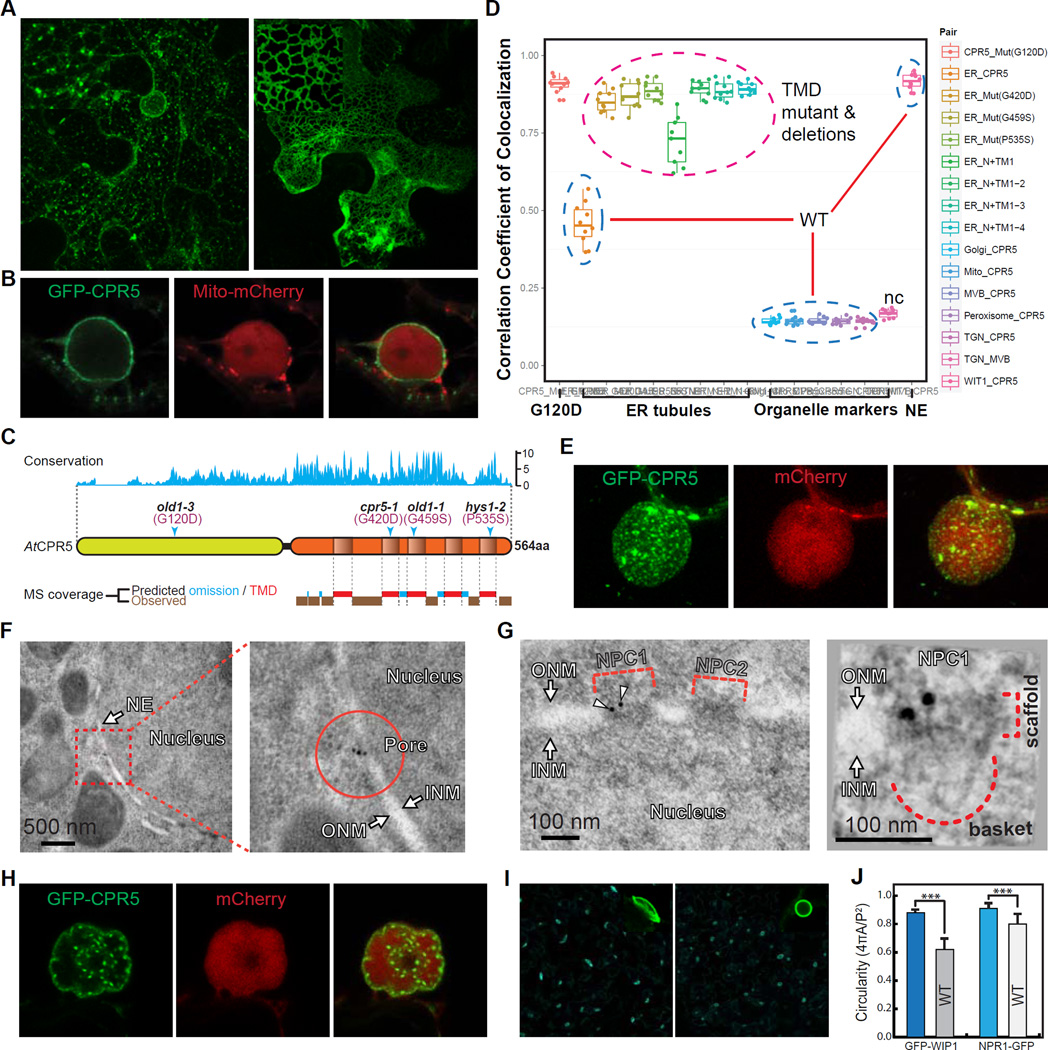
To define the molecular role of CPR5, we first determined its precise subcellular localization using a fusion to the green fluorescence protein (GFP-CPR5), which we have previously shown to be functional (Wang et al., 2014). Using both transient expression in Nicotiana benthamiana and stable expression in Arabidopsis, we found that CPR5 was exclusively associated with the endomembrane system, including the nuclear envelope (NE) and endoplasmic reticulum (ER)-associated large granules (Figures 1A, 1B, S1A and S1B). We next investigated its membrane targeting mechanism by first verifying its predicted transmembrane domains (TMDs). Using trypsin digestion followed by shotgun sequencing with mass spectrometry (MS), we found a highly biased peptide coverage pattern. In the conserved C-terminal region of AtCPR5, no peptide was detected within the predicted TMDs (Figures 1C and S1C), consistent with the MS profiling patterns found in other integral membrane proteins (Washburn et al., 2001). Null mutations within the TMDs (Bowling et al., 1997; Jing et al., 2007; Yoshida et al., 2002) (Figure 1C) and sequential deletions of TMDs from the C terminal end all caused the protein to be trapped in a tubular ER structure (Figures 1A, right panel, and 1D), highlighting the importance of these TMDs in functional targeting of the CPR5 protein.
Figure 1. CPR5 Is a Transmembrane Protein Enriched in the Nuclear Pore.
(A and B) Subcellular localization of GFP-tagged CPR5 when transiently expressed in N. benthamiana. Wild-type (WT) CPR5 (A, left), the G420D mutant (A, right), and WT CPR5 co-expressed with a mCherry-tagged marker labeling mitochondria and nucleoplasm (B) were shown. Images were obtained 24 hrs post Agrobacterium infiltration.
(C) CPR5 contains an evolutionarily conserved transmembrane (TM) region at the carboxyl terminus. Top, amino acid conservation map derived from multiple sequence alignments of CPR5 proteins from Micromonas pusilla, Chlorella variabilis, Physcomitrella patens, Sorghum bicolor, Selaginella moellendorffii, Vitis vinifera, Populus tricocarpa, Ricinus communis, Oryza sativa, Zea mays and Arabidopsis thaliana. Middle, schematic of AtCPR5 domain structure with transmembrane TM domains (TMDs) predicted by TMpred. Arrowheads indicate sites of loss-of-function missense mutations. Bottom, LC-MS/MS peptide coverage in the C-terminal half of AtCPR5 purified from transgenic Arabidopsis. Predicted omissions were calculated by PeptideMass for trypsin digestion (see Figure S1C).
(D) Pearson’s correlation coefficients of co-localization between CPR5 and endomembrane organelle markers. WT CPR5 (blue dashed circles) co-localized with both the nuclear envelope (NE) marker (WIT1) and ER-associated granules (see Figure S1B). CPR5 with missense mutations in the TMDs (Mut) and sequential truncations of individual TMDs (N+TM) all exclusively localized in tubular ER structures (magenta dashed circle). TGN (early endosome) and MVB (late endosome) markers were used as a negative control (nc).
(E) Three-dimensional image reconstruction of the nuclear surface in a GFP-CPR5 expressing cell. The nucleoplasm is labeled by free mCherry. Arrowheads indicate large ER-associated granules close to the nuclear surface.
(F and G) Immunoelectron microscopy and tomography analyses of GFP-CPR5 in root cells of transgenic Arabidopsis. Immunogold particles (arrowheads) labeled NPC1 but not NPC2 as antibodies detect only surface-exposed epitopes (G, left). The scaffold and nuclear basket of NPC1 were recognized together with two GFP-CPR5 specific immunogold particles in a projection of the tomographic volume (G, right). ONM/INM, outer/inner nuclear membrane.
(H) Hypolobulated NE and inner nuclear speckles resulted from prolonged overexpression of GFP-CPR5 (40 hrs after Agrobacterium infiltration). (I and J) Nuclear morphology in WT and cpr5 mutant plants. Epidermal cells of 5-day-old seedlings expressing the NE marker GFP-WIP1 were imaged (I). Quantification of the nuclear circularity was performed using GFP-WIP1 and NPR1-GFP as NE and nucleoplasm markers, respectively (J). Data are presented as mean ± SDM (n = 30 cells for each marker and genotype). Asterisks indicate significance (Student’s t-test, ***p-value < 0.001).
See also Figure S1.
Three-dimensional image reconstruction of the nuclear surface revealed that CPR5 was not distributed evenly in the NE, but enriched in punctate structures (Figures 1E and S1A, right panel). Distinct from the large mobile granules associated with the ER, these static puncta are smaller in size and densely distributed in the NE, resembling nuclear pores. Subsequent immunogold labeling of GFP-CPR5 followed by transmission electron microscopy (TEM) and tomography analyses confirmed that CPR5 is indeed associated with the NPC (Figures 1F and 1G). Structural integrity of the NPC is known to play a role in maintaining NE stability (Alber et al., 2007) and changes in levels of a number of NPC components (nucleoporins) cause NE membrane deformation (Jevtic et al., 2014). We found that prolonged overexpression of CPR5 could indeed elicit hypolobulated nuclei and inner nuclear speckles (Figure 1H), whereas loss of CPR5 resulted in abnormal spherical nuclei (Figures 1I and 1J), consistent with the NE morphology observed in multiple nucleoporin mutants (Parry, 2014; Parry et al., 2006; Tamura et al., 2010). In contrast to its NE localization, which clearly has a functional impact, CPR5-associated ER granules are distinct from any of the known membrane structures tested (Figures 1D and S1B). We hypothesize that this pool of CPR5 may represent a nonspecific membrane association due to overexpression.
CPR5 Is a Transmembrane Nucleoporin Associated with the NPC Core Scaffold
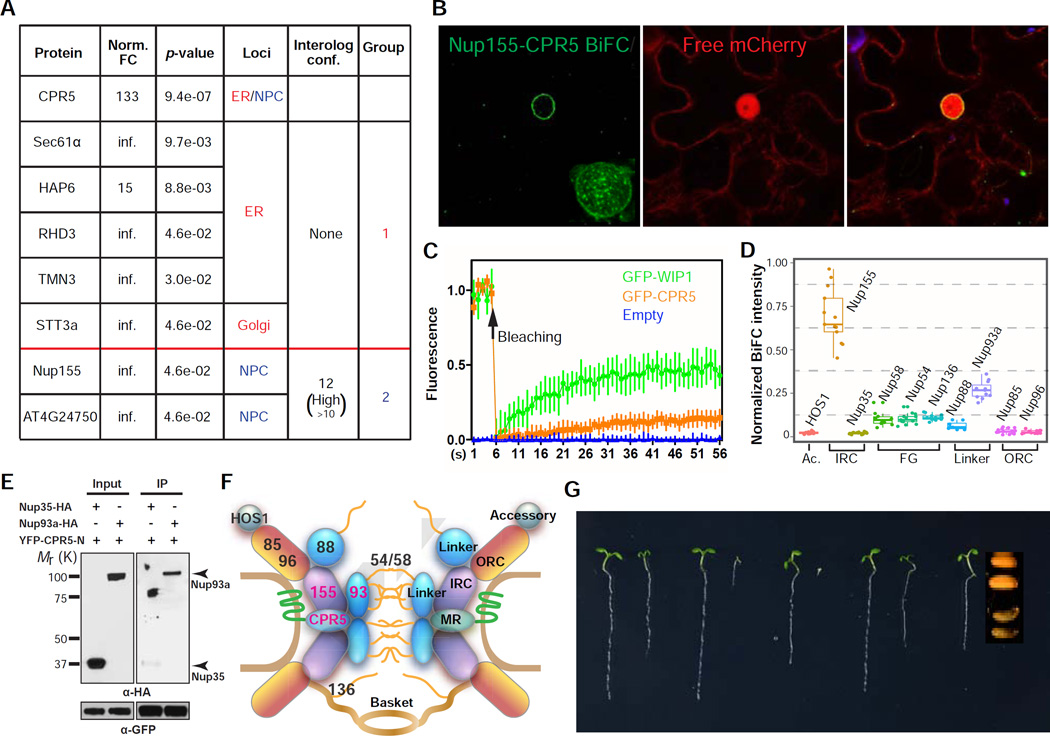
Proteomic analysis of the affinity-purified YFP-CPR5 protein complex identified a total of 28 potential binding partners of CPR5 (Figure S2A and S2B), which contain two functional groups that support our hypothesis of CPR5 being a membrane-bound nucleoporin (Figure 2A). The first group contains proteins which function in membrane protein synthesis and maturation. These interactors were likely captured by newly synthesized CPR5 protein in the ER on route to the NE. The second group consists two proteins, nucleoporin 155 (Nup155), a core scaffold component of NPC, and a putative cell cycle controlling phosphatase, both of which were previously identified as associated with the NPC in plants (Tamura et al., 2010). We subsequently verified that CPR5 and Nup155 interact specifically in the NPC using the Bimolecular Fluorescence Complementation (BiFC) assay (Figure 2B). In addition, a Fluorescence Recovery After Photobleaching (FRAP) assay indicated that CPR5 is anchored in the NE with low mobility, and thus likely forms a stable complex with Nup155 in the NPC (Figure 2C).
Figure 2. CPR5 Physically and Genetically Interacts with Nucleoporins as a Component of the NPC.
(A) CPR5 interactors identified by protein complex purification followed by LC-MS/MS. FC, fold change of spectrum counts in YFP-CPR5 vs GFP sample. Infinite (inf.) indicates that peptide was not detected in GFP samples. Interolog conf., confidence of predicted interaction between proteins.
(B) CPR5 and Nup155 interacts in the NE. Bimolecular fluorescence complementation (BiFC) assay was performed by transiently coexpressing nYFP-CPR5 and Nup155-cYFP in N. benthamiana. The interaction pattern on the nuclear surface was reconstructed by Z-stack images (inset).
(C) Fluorescence Recovery After Photobleaching (FRAP) analysis of GFP-CPR5 in transgenic Arabidopsis. A mobile NE protein GFP-WIP1 served as a control. Data are presented as mean ± SDM (n = 5 experimental replications).
(D) Interaction mapping of CPR5 with nucleoporins. BiFC was performed by transiently coexpressing nYFP-CPR5 with Nup-cYFP in N. benthamiana. The BiFC intensity was normalized using averaged expression levels of corresponding Nup-YFP measured in separate experiments. Ac, accessory nucleoporin; IRC, inner ring complex; FG, Phe-Gly repeat-containing nucleoporin; ORC, outer ring complex; Linker, linker nucleoporin.
(E) CPR5 interacts with the IRC-associated linker nucleoporin Nup93a. In vitro pulldown assay was performed using GFP-TrapA agarose beads. YFP-CPR5-N, YFP-tagged N-terminal half of CPR5.
(F) The structural modules of the nuclear pore complex (NPC) and the proposed position of CPR5 within the NPC.
(G) Genetic interaction between cpr5 and mutants of the ORC nucleoporins. 5-day-old seedlings were shown. Since the cpr5 nup160 double mutant did not germinate, the seed morphology of the homozygote was compared to that of a heterozygote.
See also Figure S2.
To gain further insight into the position of CPR5 within the NPC, we mapped the interactions of CPR5 with nucleoporins of different NPC subcomplexes. The core NPC contains eight copies of symmetric spokes, each consisting of interconnected subcomplexes: the outer ring complex (ORC) that coats the pore membrane, the inner ring complex (IRC) that forms the NPC core scaffold together with the ORC, the transmembrane ring (MR) that anchors the core scaffold to the pore membrane, and the linker nucleoporins that bridge the core scaffold and Phe-Gly (FG) proteins, which form the selective barrier for cargo transport (Alber et al., 2007). Using both BiFC in planta and an in vitro pull-down assay, we found that CPR5 interacts with not only Nup155 in the IRC but also the IRC-associated linker nucleoporin Nup93a through its N-terminus (Figures 2D and 2E). However, no robust interaction was detected with FG proteins, ORC components or other NPC accessory proteins tested (Figure 2D). Based on these observations, we propose that CPR5 is a transmembrane nucleoporin that is anchored at the equatorial plane of the NPC in the nuclear pore membrane by its C-terminal TMDs and physically interacts with the NPC core scaffold as well as an associated linker nucleoporin through its soluble N-terminus (Figure 2F).
CPR5 Is Involved for NPC Function
Consistent with the notion of CPR5 being a nucleoporin, CPR5 displays strong genetic interactions with the ORC nucleoporins Nup85, Nup96 and Nup160. Whereas the nup85, nup96 and nup160 single mutants did not exhibit obvious aberrations in early seedling development, double mutants with cpr5 all resulted in embryonic or seedling lethality (Figure 2G). This synergistic genetic relationship is likely due to a cooperative role between CPR5 and ORC components in maintaining the structural integrity of the NPC. We were unable to assess genetic interactions between CPR5 and IRC nucleoporins due to seedling or embryonic lethality of the single mutants (Parry, 2014).
Plants have sequence-homologs of almost all the vertebrate nucleoporins (Tamura et al., 2010). However, because transmembrane nucleoporins are not evolutionarily conserved (Mans et al., 2004), functional analogs of vertebrate transmembrane nucleoporins that interact with the IRC and anchor the NPC to the pore membrane have not been identified in plants. Our study suggests that CPR5 is a plant-specific transmembrane nucleoporin that physically associates with the IRC and may contribute to the stability of the NPC core scaffold even though it is not required for NPC anchoring (Figure S2C). Besides its potential structural role in the NPC, the cpr5 mutant phenotype suggests that this plant transmembrane nucleoporin may have evolved distinct functions, such as regulation of ETI/PCD.
CPR5 Modulates the Nucleocytoplasmic Transport Activity of the NPC
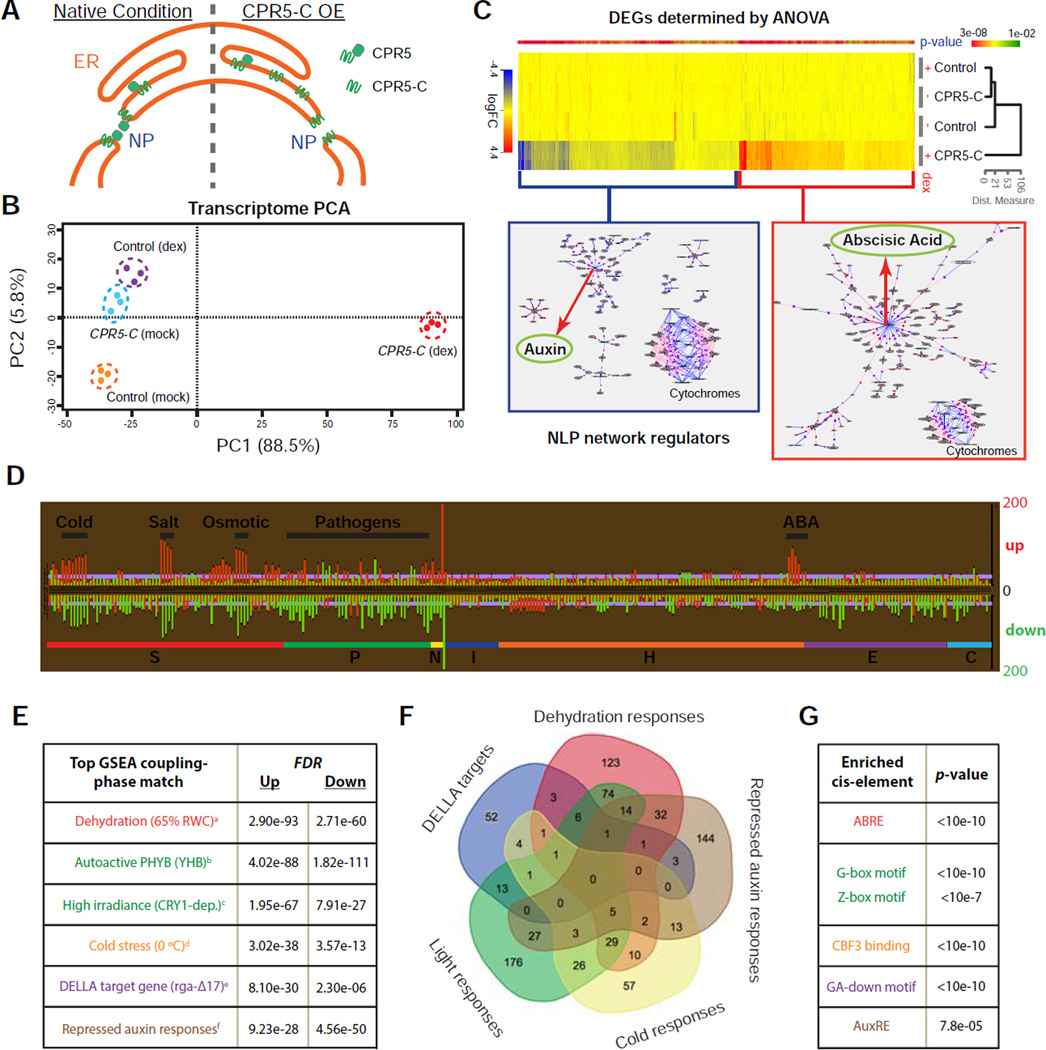
The NPC is a platform for multiple nuclear activities, including nucleocytoplasmic transport, genome maintenance, and regulation of gene expression (Strambio-De-Castillia et al., 2010). To understand how CPR5 regulates ETI in the NPC, we first investigated the cellular processes controlled by this nucleoporin using transcriptome profiling. To avoid the indirect effects of the stable cpr5 mutation, which is known to activate ETI as well as the downstream signaling pathways mediated by the immune signal salicylic acid (SA) (Bowling et al., 1997; Wang et al., 2014), a transient interference system was developed. Because of the low turnover rate of nucleoporins (Toyama et al., 2013), we designed a protein interference strategy that involved dexamethasone (dex)-inducible expression of the C-terminal half (275–564 aa) of CPR5 (CPR5-C). Although CPR5-C is not functional (Figure S3A), it is targeted to the NPC (Figure S3B) and therefore might compete with the wild type (WT) protein (Figure 3A). We first tested the system using a transient expression assay performed in N. benthamiana. We found that overexpression of CPR5-C led to tissue collapse similar to ETI-associated PCD induced by NB-LRR activation, whereas overexpression of the full-length CPR5 or CPR5-N had no such an effect (Figure S3C). This CPR5-C-induced PCD was likely due to interference with the function of endogenous CPR5 because it was suppressible by simultaneous overexpression of full-length CPR5, but not CPR5-N (Figure S3D). We subsequently validated the interference activity of YFP-CPR5-C in Arabidopsis, where constitutive or inducible expression of YFP-CPR5-C in the WT background resulted in the cpr5 mutant phenotypes, including growth arrest, PCD and increased expression of defense genes (Figure S3E–S3G).
Figure 3. Transient Interference with CPR5 Function Simultaneously Activates Diverse Nuclear Signaling Pathways.
(A) A proposed cellular mechanism for transient interference of CPR5 function by overexpression of CPR5-C. NP, nuclear pore.
(B) Principal component analysis (PCA) of transcriptome changes induced by transient expression of CPR5-C.
(C) Heat map of differentially expressed genes (DEGs) that depend on CPR5-C transgene induction (p-value < 0.01). Genes with fold change (dex vs mock) higher than 2 were subject to natural language processing (NLP)-based network regulator discovery analysis. Major network regulators are highlighted with red arrows.
(D) Comparative analysis of CPR5-C-mediated CPR5 interference data with 438 published Arabidopsis microarrays. Each row represents an analysis with a specific array dataset, and datasets are sub-categorized into sections according to treatments. Section code: S, stress; P, pathogen; N, nutrient; I, inhibitor; H, hormone; E, elicitor; C, chemical. The length of each bar represents the number of overlapping DEGs (up- or down-regulated) between the CPR5-C data and a specific treatment.
(E) Gene Set Enrichment Analysis (GSEA). CPR5-C-induced up- and down-regulated genes were used as separate inputs for GSEA and non-redundant top matches are listed. PubMed IDs for the listed datasets are: a-21050490, b-19529817, c-17478635, d-16214899, e-17933900, f-19392692.
(F) Venn diagram of the transcriptome signatures identified in (E).
(G) Cis-element enrichment analysis of CPR5-C-induced total DEGs. ABRE, ABA-responsive element; G/Z-box, light-responsive elements; CBF3, transcription factor for cold acclimation; GA-down, gibberellin down regulated d1 cluster; AuxRE, auxin response element.
The stable Dex:YFP-CPR5-C transgenic line was then used for transcriptome analysis. Using principal component analysis, we detected a major change in global gene expression 24 hours after YFP-CPR5-C was induced (Figure 3B), when SA-mediated response had yet to occur in this system (Figure S3H). About 1,800 differentially expressed genes (DEGs) were identified (Figure 3C and Table S1). Comparative analysis with 438 published Arabidopsis microarray datasets collected under the conditions of a board spectrum of chemical/stress/hormone treatments (Reina-Pinto et al., 2010) revealed that the CPR5-C-induced transcriptome has significant matches with a variety of stress responses, especially to cold, salt/osmotic stress, abscisic acid, and various pathogens (Figure 3D). We also performed Gene Set Enrichment Analysis (GSEA) using a even more comprehensive database (Yi et al., 2013), which revealed a transcriptome feature composed of distinct molecular signatures, including activation of cold/dehydration/ABA responses and PHYB/CRY1-dependent light responses, and repression of gibberellin and auxin responses (Figures 3E and 3F). The significance of these molecular signatures was further supported by the enrichment of cis-elements known for these responses in total DEGs (Figure 3G).
Such a composite transcriptome profile is most likely due to perturbation of a key cellular process shared by these corresponding pathways instead of crosstalk effects induced by a single signaling pathway. Nucleocytoplasmic protein transport is a critical rate-limiting step for hormones and light signal transduction. Nuclear translocation of photoreceptors PHYs and CRY1 is required for activation of light responses, whereas nuclear accumulation of DELLA and Aux/IAA proteins results in repression of gibberellin and auxin signaling, respectively (Lee et al., 2008). Notably, a number of mutants with altered responses to cold, drought/ABA and auxin have been genetically identified as components of the NPC and NTRs (Dong et al., 2006; Lee et al., 2001; Parry et al., 2006; Verslues et al., 2006), illustrating the sensitivity of these processes to the structural integrity and/or transport activity of the nuclear pore. Indeed, ABA and auxin were the most sensitive pathways detected by a natural language processing (NLP)-based network regulator discovery algorithm when applied to the DEGs caused by CPR5-C interference (Figure 3C). We hypothesize that with compromised CPR5 function, the NPC adopts a structure with significantly increased permeability and/or transport activity that allows deregulated nuclear influx of diverse signaling cargos, which normally undergo nuclear translocation only under stimulus-induced conditions.
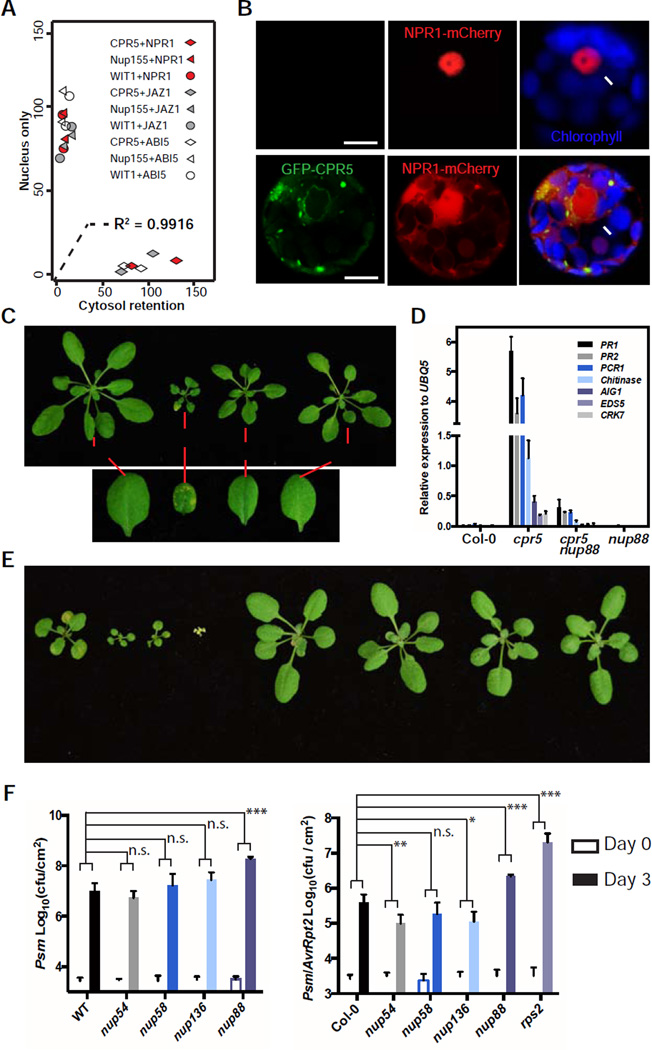
We next tested whether the WT CPR5 constrains the nuclear accumulation of signaling cargos. We found that overexpressing CPR5 indeed caused substantial cytoplasmic retention of stress- and hormone-related nuclear proteins NPR1, JAZ1 and ABI5 (Figures 4A, 4B, S4A and S4B). However, WIT1 (an NE protein), Nup155 or a mutant form of CPR5 (G420D) could not recapitulate this effect (Figures 4A, S4A and S4B), supporting a direct role for CPR5 in modulating nucleocytoplasmic transport activity of the NPC. Consistent with the cytoplasmic retention of these nuclear signaling molecules, overexpression CPR5 compromises ETI-associated PCD and resistance in Arabidopsis (Wang et al., 2014), suggesting that CPR5 is a direct rate-limiting regulator for ETI/PCD.
Figure 4. CPR5 Modulates Protein Nucleocytoplasmic Transport to Gate ETI.
(A) Overexpression of CPR5 resulted in cytosolic retention of nuclear proteins. NPR1, JAZ1 or ABI5 were co-expressed with NPC protein CPR5, Nup155 or WIT1 in Arabidopsis protoplasts. Each combination was repeated twice with around 100 transformed cells counted per repeat. Localization of nuclear proteins in each cell was recorded as binary data (‘nucleus only’ or ‘cytosol retention’). A logistic regression model using the NPC protein as the sole independent variable explains 99% of the data variance.
(B) Representative Arabidopsis protoplasts co-transformed with NPR1-mCherry and GFP-CPR5 constructs or the empty vector.
(C) Four-week-old WT, nup88 (also known as mos7-1), cpr5 and nup88 cpr5 plants are shown. The lower panel shows close-ups of leaves with or without spontaneous PCD.
(D) Expression levels of the most induced defense-related genes in the cpr5 mutant were measured using quantitative RT-PCR.
(E) Three-week-old WT, cpr5, cpr5 nup54, cpr5 nup58, cpr5 nup136, nup54, nup58 and nup136 plants.
(F) FG-Nup mutants nup54 and nup136 specifically enhanced ETI, but not basal immunity. Three-week-old plants were inoculated with bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) without (left) or with (right) the effector gene AvrRpt2. Data are presented as mean ± SDM (n = 6 biological replications for each genotype and treatment). Two-way ANOVA was used for statistical tests. p-values * < 0.05, ** < 0.01, *** < 0.001, n.s. not significant.
See also Figure S4.
CPR5 Inhibits Immune Signal Transport through the Selective Barrier of the NPC
To further evaluate the functional importance of CPR5-gated NPC cargo transport to ETI/PCD activation and to investigate the gating mechanism, we crossed cpr5 with stress-related karyopherin mutants as well as transport-related nucleoporin mutants (Figure S4C). We found that a partial loss-of-function mutant of Nup88, which associates with the NPC cytoplasmic filaments, could largely rescue the cpr5 phenotypes, including the stunted growth, spontaneous PCD and elevated defense gene expression (Figures 4C and 4D). This mutant has previously been found to block nuclear accumulation of multiple defense-related proteins (Cheng et al., 2009), and its suppressor activity on cpr5 suggests that NPC-mediated nuclear transport of immune cargos is required for CPR5-gated ETI/PCD activation. In contrast to nup88, crossing cpr5 with mutants of FG nucleoporin Nup54, Nup58 and Nup136 exacerbated the cpr5 phenotype (Figure 4E). More importantly, Nup54 and Nup136 display a specific inhibitory role in ETI, as their mutants were found to enhance ETI, but not basal immunity (Figure 4F). These evidence suggest that CPR5 may modulate the properties of the NPC selective barrier as a mechanism to regulate ETI. Indeed, it has been shown that loss of yeast IRC nucleoporins relaxes the NPC permeability barrier due to inappropriate anchoring of FG nucleoporins (Shulga et al., 2000). Given that CPR5 directly interacts with Nup93 (Figure 2D-2F), the molecular anchor of FG nucleoporins (Chug et al., 2015), we propose that loss-of-CPR5 activates ETI partly by perturbing the structural arrangement of FG meshwork to compromise its inhibitory role in immune signaling cargo transport.
Homomeric Interaction of CPR5 in the NPC Is Required to Inhibit ETI/PCD
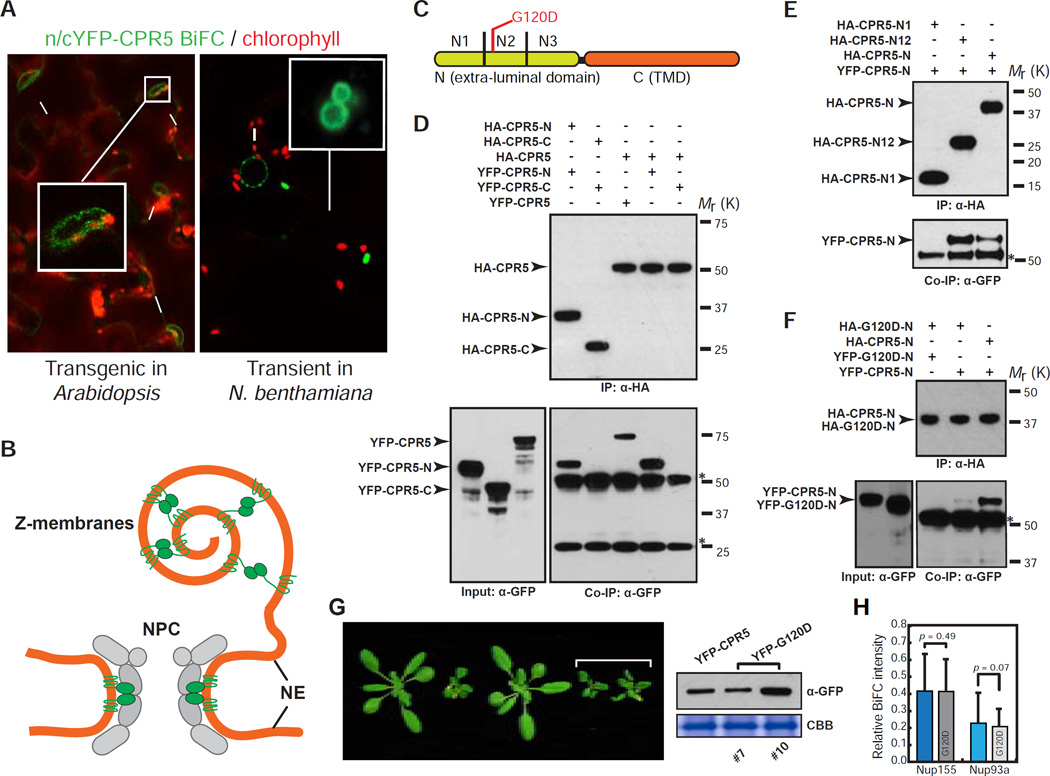
Since previous genetic data clearly showed that CPR5 is a negative regulator of ETI/PCD (Boch, 1998; Wang et al., 2014), we hypothesize that its repression must be alleviated upon ETI activation. To test this, we first examined, but ruled out a significant reduction in CPR5 transcription or translation during ETI triggered by the bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) carrying the effector AvrRpt2 (Figure S5A–S5C). Based on the knowledge that homo- and hetero-oligomerization of nucleoporins is crucial for their functions, we then tested whether CPR5 activity is regulated through homo-oligomerization. We performed BiFC assay in 35S:n/c-YFP-CPR5 transgenic plants coexpressing n-YFP-CPR5 and c-YFP-CPR5 and found that CPR5 indeed formed a homomeric complex in the NE (Figure 5A, left). In a transient assay, the BiFC signal was also observed in the so-called Z-membranes (Figure 5A, right), which are artificial organelles formed when integral membrane proteins oligomerize through their extra-luminal domains (Gong et al., 1996). Since Z-membranes were only observed when n/cYFP were fused to the N-terminus of CPR5, we hypothesized that the CPR5 N-terminal domain is extra-luminal and mediates the homomeric interaction (Figure 5B). Indeed, in vitro pull-down assays mapped the homomeric interaction domain to the first two thirds of the CPR5-N (1–274 aa), with the middle region (N2) being essential (Figures 5C-5E and S5D). Notably, a known loss-of-function mutation (G120D, old1–3) resides in the CPR5 N2 region (Jing et al., 2007). We found that the G120D mutation significantly compromised CPR5 homomeric interaction (Figures 5F and S5E). Furthermore, at similar protein levels, the monomeric G120D allele could not complement the cpr5-1 phenotypes as the WT CPR5 (Figure 5G). Since the G120D mutation affected neither CPR5 localization (Figure 1D) nor CPR5 heteromeric interactions with Nup155 and Nup93a (Figure 5H), it appears to specifically affect CPR5 homomeric interaction required for its function.
Figure 5. CPR5 Homomeric Interaction at the N-Terminal Extra-Luminal Domain Is Required for Suppressing ETI and PCD.
(A) CPR5 BiFC assay. nYFP-CPR5 and cYFP-CPR5 were constructed in two separate 35S promoter-driven expression cassettes within one vector (35S:n/c-YFP-CPR5). BiFC signals were observed in a stable transgenic Arabidopsis line (left) and transiently transformed N. benthamiana (right), respectively. Arrows and arrowheads indicate the nucleus and Z-membranes, respectively.
(B) A model proposed for CPR5 homomeric interaction in the NPC and Z-membranes.
(C) Schematic of CPR5 constructs used in in vitro pull-down assays. CPR5-N (1–274 aa) and CPR5-C (275–564 aa). CPR5-N was further divided into N1 (1–91 aa), N2 (92–182 aa) and N3 (183–274 aa).
(D–F) CPR5 homomeric interaction is mediated by its N-terminal extra-luminal domain. CPR5-N, rather than CPR5-C, mediated the homomeric interaction (D). The N1 and N2 (N12) domains of CPR5-C is sufficient and the N2 domain is necessary for the interaction (E). The G120D mutation compromised the homomeric interaction of CPR5-N (F). In vitro pull-down assays were performed using HA antibody-conjugated agarose beads. Stars indicate non-specific signals from immunoglobulins.
(G) Homomeric interaction is required for CPR5 function. 35S:YFP-CPR5 fully complemented the cpr5-1 mutant phenotype but 35S:YFP-G120D did not (left) when expressed at comparable levels (right). Two independent 35S:YFP-G120D transgenic lines (#7 and #10) are shown.
(H) The G120D mutation does not affect heteromeric interactions of CPR5 with other nucleoporins. BiFC was performed by transiently coexpressing nYFP-CPR5/G120D pairwise with Nup155/Nup93a–cYFP in N. benthamiana. The BiFC intensity was normalized using averaged expression levels of YFP-CPR5/G120D measured in separate experiments and plotted as relative values. Data are presented as mean ± SDM (n = 15 cells for each BiFC combination). p-values were calculated by Student’s t-test.
See also Figure S5.
CPR5 Homomeric Interaction Is Specifically Disrupted upon NB-LRR Immune Receptor Activation
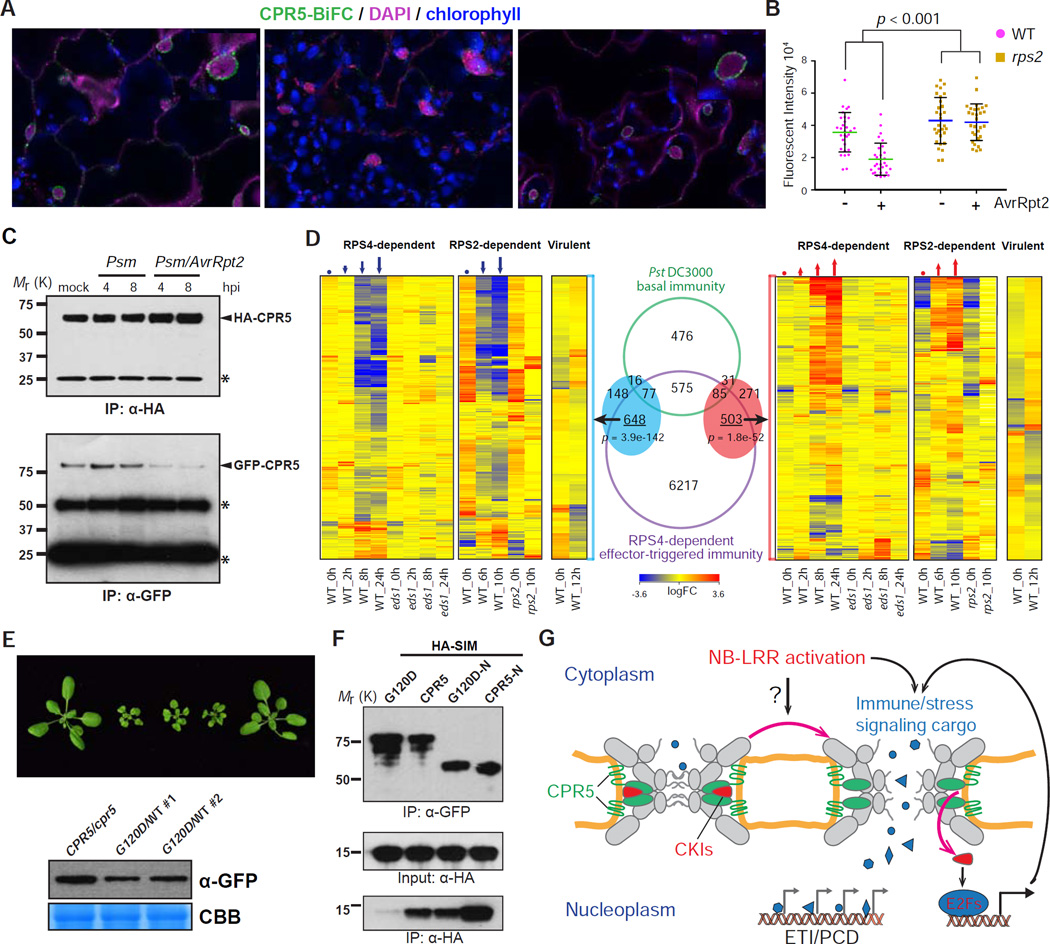
We next investigated how CPR5 homomeric interaction might be regulated upon NB-LRR activation. We introduced the 35S:n/c-YFP-CPR5 into Arabidopsis lines carrying a dex-inducible AvrRpt2 construct in both WT and the cognate immune receptor mutant rps2 by genetic crosses. We found a significant reduction in the BiFC signal as early as 6 hours after dex provision when the morphology of nucleus (stained by DAPI) was still intact (Figure 6A). This reduction in CPR5 homomeric interaction was dependent on both AvrRpt2 and RPS2 (Figure 6B), but independent of CPR5 protein levels (Figure S5C). Similar results were also obtained in the coimmunoprecipitation experiment using pathogen-challenged Arabidopsis plants carrying the 35S:GFP-CPR5 and 35S:HA-CPR5 double transgenes (Figure 6C). A significant reduction in CPR5 homomeric interaction was observed in response to Psm carrying AvrRpt2, but not Psm without the effector. These results suggest that in response to effector-triggered NB-LRR activation, the CPR5 homomeric complex in the NPC is disrupted to release its inhibition on ETI activation. This hypothesis was supported by comparing the transient CPR5-interference transcriptome described in Figure 3 with time course ETI transcriptional responses mediated by RPS4 (a TIR-NB-LRR) and RPS2 (a CC-NB-LRR). We found that the majority of the genes differentially expressed upon functional interference of CPR5 (Figure 6D, red and blue ovals) overlapped with the transcriptome changes mediated by RPS4 and RPS2 (p-value < 1e-50 in both cases), and displayed concordant expression patterns with the ETI response in WT, but not in ETI-deficient eds1 and rps2 mutants (Figure 6D). In contrast, those genes had limited overlap with the host transcriptome changes induced by a virulent strain that activates the basal immunity and displayed a random distribution. Lastly, we showed that overexpression of the monomeric mutant form of CPR5 (G120D) resulted in a cpr5-like phenotype in WT plants (Figure 6E), suggesting that disruption of CPR5 homomeric interaction is sufficient to activate ETI/PCD downstream of NB-LRR activation.
Figure 6. Disruption of CPR5 Homomeric Interaction Is Triggered by NB-LRR Activation and Coordinates CKI-Mediated ETI Signal Transduction and NPC Permeabilization.
(A–C) ETI-triggered disruption of CPR5 homomeric interaction in the NPC. Leaves of 35S:n/c-YFP-CPR5IDex:AvrRpt2 double transgenic line were incubated with mock or 50 uM dex for 6–8 hrs before imaging (A). DAPI stained nuclei (arrowheads). BiFC intensities in the NE were measured (B). Data are presented as mean ± SDM (n = 30 cells for each genotype and treatment). Statistical test was performed using 2-way ANOVA. Total protein was extract from 35S:GFP-CPR5/HA-CPR5 double transgenic plants treated with Psm at indicated time points. Immunoprecipitation was performed using anti-HA agarose beads in the presence of high concentration of detergents (C). hpi, hours post inoculation.
(D) CPR5 interference transcriptome shows a significant overlap and concordant expression pattern with those of ETI. Overlaps between CPR5-C-induced (red oval) and repressed (blue oval) genes with .KRS^-dependent ETI genes and basal immunity genes are shown in Venn diagram. Hypergeometric tests were used to calculated-values.
(E) 35S:YFP-G120D caused dominant negative phenotype in WT plants. Two independent transgenic lines (#1 and #2) and their expression levels are shown.
(F) CPR5 homomeric interaction is required for interaction with SEVI. CPR5/G120D was tagged with YFP and SEVI was tagged with HA. In vitro pull-down assay was performed using GFP-TrapA beads.
(G) Permeabilization of the NPC is a specific induction mechanism of ETI and PCD. Upon NB-LRR activation, an unknown intracellular signal is generated and transduced to the NPC to promote the disruption of CPR5 oligomer. This NPC change coordinates CKIs release for ETI signaling and reconfigures the selective barrier to allow significant influx of nuclear signaling cargos through the NPC. These combined effects result in simultaneous activation of diverse stress-related nuclear signaling pathways that contribute to ETI/PCD.
See also Figure S6.
Disruption of CPR5 Homomeric Complex Coordinates ETI Signaling by Cell Cycle Regulators
Our previous study showed that physical association of CPR5 with the CKI, SIM (SIAMESE), is diminished upon Psm/AvrRpt2 challenge, resulting in activation of a non-canonical pathway involving cell-cycle regulators that contributes to ETI and PCD (Wang et al., 2014). To test whether the interaction between CPR5 and CKIs is regulated by the homomeric interaction of CPR5, we performed an in vitro pull-down assay to examine the affinity of SIM to WT CPR5 and the monomeric G120D mutant. We found that the G120D mutation diminished the protein’s affinity to SIM (Figures 6F and S6A). This result suggests that the homomeric CPR5-N is necessary for association with SIM, and disruption of this interaction results in the release of SIM, allowing it to engage in downstream ETI signaling. Although CKIs are required for ETI signaling, overexpressing SIM is not sufficient for activating ETI/PCD unless the CPR5 function is compromised at the same time (Figure S6B), consistent with our hypothesis that elimination of CPR5’s inhibitory activity in nuclear transport is also necessary for ETI/PCD activation.
Nucleoporins have been reported to regulate cell cycle progression through their effect on expression levels of cell cycle regulators in mammals (Chakraborty et al., 2008). The direct sequestration of the cell cycle regulators in the NPC by a transmembrane nucleoporin is a surprising finding, whose significance in basic plant cell biology is currently not known. However, this association allows the NPC to play a dual role in response to ETI induction in redirecting certain cell cycle regulators for defense gene expression and permeabilizing the NPC transport activity for simultaneous activation of diverse stress-related nuclear signaling, both of which are necessary for the activation of ETI/PCD in plants (Figure 6G).
DISCUSSION
NPC-dependent nuclear transport of immune receptors, signal transducers, and transcription factors represents a prevalent regulatory mechanism for immune activation in both plants and animals (Deslandes and Rivas, 2011; Garcia and Parker, 2009; Gilmore, 2006; Rivas, 2012). To promote nuclear translocation, protein cargos usually undergo changes in affinity to nuclear transport receptors or piggybacks on molecules with mechanisms for nuclear translocation (Wirthmueller et al., 2013). However, whether the NPC itself has a direct role in immune signaling or undergoes any specific structural change to support these immune-activated transport events was previously unknown. Our study revealed that rather than a passive conduit, the NPC is both a signaling platform and a dynamic regulator of nucleocytoplasmic cargo transport. This dual function of the NPC is directed by CPR5 and plays a necessary and sufficient role for ETI/PCD induction.
CPR5 Is a Transmembrane Nucleoporin that Inhibits ETI through Modulating Selective Transport of the NPC
Multiple cpr5 loss-of-function mutants have been reported to confer spontaneous cell death and NB-LRR-independent resistance to pathogens carrying virulence effectors (Boch, 1998; Bowling et al., 1997), yet a direct inhibitory role for CPR5 in ETI has only recently been demonstrated with overexpression of the wild-type CPR5 showing inhibition on effector-triggered PCD and resistance (Wang et al., 2014). Here we demonstrate that CPR5 is a bona fide nucleoporin based on its NPC localization as well as physical and genetic interaction with the NPC core scaffold (Figures 1 and 2).
Cellular, genetic and genomic analyses showed that CPR5 inhibits ETI/PCD by constraining nuclear accumulation of a diverse array of signaling cargos (Figures 3 and 4). The enhanced defense observed in cpr5 is the opposite of the compromised basal and ETI resistance reported for the NPC scaffold nucleoporin mutants, which are defective in bulk mRNA export (Du et al., 2016; Wiermer et al., 2012; Zhang and Li, 2005). CPR5 appears to regulate nuclear transport of signaling cargos by affecting the selective barrier composed of FG proteins, because it is physically connected with the FG protein anchor Nup93 (Figure 2D-2F) and mutants of three individual FG proteins Nup54, Nup58 and Nup136 each exacerbated the cpr5 phenotype (Figure 4E). Importantly, mutating the FG proteins Nup54 and Nup136 alone led to enhancement of ETI, but not basal resistance (Figure 4F), suggesting that these FG proteins specifically inhibit ETI.
Disruption of CPR5 Homomeric Interaction Is an Induction Mechanism of ETI
CPR5 normally inhibits ETI at the NPC as a homomeric complex formed via its N-terminal extra-luminal domain (Figure 5). This inhibition is specifically “turned off” when CPR5 homomers are disrupted upon activation of an NB-LRR receptor RPS2 (Figures 6A-6C). Therefore, this conformational change of CPR5 in the NPC is a signaling event downstream of NB-LRRs. Moreover, this event appears to be sufficient for ETI/PCD activation as overexpression of either a N-terminus-truncated CPR5 or the G120D mutant compromised in oligomer formation resulted in the cpr5 phenotype in the WT background (Figure S3 and Figure 6E).
Disrupting the CPR5 oligomer significantly reduces its affinity to CKIs (Figure 6F), which provides a mechanistic explanation for our previous observation that CKIs are released from CPR5 for ETI induction (Wang et al., 2014). However, ectopic expression of CKIs alone is not sufficient for mounting ETI/PCD (Figure S6B), consistent with our genetic data indicating that NPC-gated cargo transport is also necessary. Therefore, disruption of CPR5 oligomerization coordinates the two NPC actions that are collaboratively required for ETI/PCD induction.
CPR5-Mediated CKI Release and NPC Permeabilization Is a Convergent Induction Mechanism by both CC-NB-LRR and TIR-NB-LRR
We propose that during ETI, in addition to activation of distinct receptors and their signaling complexes, a common signal is generated and transduced to the NPC, leading to a disruption of CPR5 oligomer. The resulting activation of the noncanonical CKI-Rb-E2F signaling module together with the influx of nuclear signaling cargos through the NPC leads to transcriptomic changes bearing signatures of diverse hormone and stress responses (Figure 3 and 4), which overlap significantly with those induced by both a CC-NB-LRR (RPS2) and a TIR-NB-LRR (RPS4) (Figure 6D). These CPR5-regulated and NPC-gated cellular responses were barely observed in basal immunity (Figure 6D), suggesting that ETI is more than a quantitative ramping-up of basal immunity even though the two defense mechanisms have some overlap in signaling networks (Tao et al., 2003; Tsuda et al., 2009). Our study suggests that the NPC contributes a specific regulatory mechanism for simultaneous activation of diverse stress responses, which ultimately lead to the extreme outcome of ETI, i.e., PCD.
It is tempting to speculate that the NB-LRR-triggered NPC alterations in plants bear certain similarity to cellular changes associated with two distinct types of PCD in animals, apoptosis and pyroptosis. Activation of both types of PCD involves specialized mechanisms for membrane permeabilization to release pro-death factors. Activation of apoptosis requires formation of permeability transition pore (PTP) on mitochondrial membranes in order to release cytochrome c and other proteins for caspase-9 activation (Tait and Green, 2010), whereas activation of pyroptosis requires cleaved Gasdermin D to form pores on the plasma membrane to promote cell lysis and release of IL-1β (Ding et al., 2016). Our study showed that during ETI induction, the selective barrier of the NPC becomes more permeable through the conformational change of CPR5 to simultaneously activate diverse nuclear signaling events required for the activation of ETI and the execution of PCD in plants.
METHODS AND RESOURCES
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GFP (JL-8, for western blot) | Clontech | Cat. #632381; RRID:AB_2313808 |
| Rabbit polyclonal anti-GFP (for immuno-EM) | Santa Cruz | Cat. #sc-8334; RRID:AB_641123 |
| Mouse monoclonal anti-HA (16B12) | Biolegend | Cat. #901502; RRID:AB_2565007 |
| Alpaca anti-GFP coupled to agarose beads | Chromotek | Cat. #GFP-Trap®_A |
| Pierce® Anti-HA Agarose | Thermofisher | Cat. #26181; RRID:AB_2537081 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dexamethasone | Sigma-Aldrich | Cat. #D1756–25MG; CAS: 50-02-2 |
| DAPI | Sigma-Aldrich | Cat. #D9542-5MG; CAS: 28718-90-3 |
| LR Clonase® II Plus enzyme | Thermofisher | Cat. #12538120 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat. #P9599-5ML; EC: 200-664-3 |
| TRIzol Reagent | Thermofisher | Cat. #15596026 |
| Critical Commercial Assays | ||
| QuikChange II site-directed mutagenesis kit | Agilent | Cat. #200524 |
| Wheat germ in vitro translation system | BioSieg | N/A |
| SuperScript® III First-Strand Synthesis System | Thermofisher | Cat. #18080050 |
| FastStart Universal SYBR Green Master Kit | Roche | Cat. # 04913850001 |
| Deposited Data | ||
| Microarray raw and analyzed data | This paper | GEO: GSE72742, GSE72743 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Arabidopsis: 35S::GFP-CPR5 | Wang et al., 2014 | N/A |
| Arabidopsis: 35S::YFP-CPR5/G120D | This paper | N/A |
| Arabidopsis: 35S/Dex::YFP-CPR5-C | This paper | N/A |
| Arabidopsis: 35S::(nYFP-CPR5+cYFP-CPR5) | This paper | N/A |
| Arabidopsis: 35S::(GFP-CPR5+3HA-CPR5) | This paper | N/A |
| Arabidopsis: GFP-WIP1 | The Meier Laboratory | N/A |
| Arabidopsis: Nup155-YFP | This paper | N/A |
| Arabidopsis: MOS7-GFP | Cheng et al., 2009 | N/A |
| nup88 (mos7-1), nup96 (mos3-1), importin-α3 (mos6-2) | The Li Laboratory, Cheng et al., 2009; Zhang and Li, 2005; Palma et al., 2005 |
N/A |
| All other nucleoporin and transport receptor mutants Used in this study, see Table S2 |
Arabidopsis Biological Resource Center |
see Table S2 for Salk line ID |
| Arabidopsis: 35S::YFP-SIM | This paper | N/A |
| Arabidopsis: Dex::AvrRpt2 | McNellis et al., 1998 | N/A |
| Recombinant DNA | ||
| pMDC43-GFP-CPR5 | Wang et al., 2014 | N/A |
| pEG100-YFP-CPR5 (WT and mutant constructs) | This paper | N/A |
| pEG100-n/cYFP-CPR5 | This paper | N/A |
| pEG100-Nups-YFP/cYFP | This paper | N/A |
| pBAV154/pEG100-YFP-CPR5-C | This paper | N/A |
| pEG100-NPR1/JAZ1/ABI5-mCherry | This paper | N/A |
| pEG100-mCherry-WIT1 | This paper | N/A |
| Sequence-Based Reagents | ||
| Primers used in this study, see Table S3 | This paper | N/A |
| Software and Algorithms | ||
| Mascot (v2.5.0) | Matrix Science | N/A |
| Scaffold (v4.4.1.1) | Proteome Software, Inc. |
N/A |
| R (v3.0.1) | N/A | N/A |
| GeneSpring (v13.0) | Agilent | N/A |
| MASTA | Reina-Pinto et al., 2010 | N/A |
| PlantGSEA | Yi et al., 2013 | http://structuralbiology.cau.edu.cn/PlantGSEA/ |
| Athena | O’Connor et al., 2005 | http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl |
| Other | ||
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| Mouse monoclonal anti-GFP (JL-8, for western blot) | Clontech | Cat. #632381; RRID:AB_2313808 |
| Rabbit polyclonal anti-GFP (for immuno-EM) | Santa Cruz | Cat. #sc-8334; RRID:AB_641123 |
| Mouse monoclonal anti-HA (16B12) | Biolegend | Cat. #901502; RRID:AB_2565007 |
| Alpaca anti-GFP coupled to agarose beads | Chromotek | Cat. #GFP-Trap®_A |
| Pierce® Anti-HA Agarose | Thermofisher | Cat. #26181; RRID:AB_2537081 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dexamethasone | Sigma-Aldrich | Cat. #D1756-25MG; CAS: 50-02-2 |
| DAPI | Sigma-Aldrich | Cat. #D9542-5MG; CAS: 28718-90-3 |
| LR Clonase® II Plus enzyme | Thermofisher | Cat. #12538120 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat. #P9599-5ML; EC: 200-664-3 |
| TRIzol Reagent | Thermofisher | Cat. #15596026 |
| Critical Commercial Assays | ||
| QuikChange II site-directed mutagenesis kit | Agilent | Cat. #200524 |
| Wheat germ in vitro translation system | BioSieg | N/A |
| SuperScript® III First-Strand Synthesis System | Thermofisher | Cat. #18080050 |
| FastStart Universal SYBR Green Master Kit | Roche | Cat. # 04913850001 |
| Deposited Data | ||
| Microarray raw and analyzed data | This paper | GEO: GSE72742, GSE72743 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Arabidopsis: 35S::GFP-CPR5 | Wang et al., 2014 | N/A |
| Arabidopsis: 35S::YFP-CPR5/G120D | This paper | N/A |
| Arabidopsis: 35S/Dex::YFP-CPR5-C | This paper | N/A |
| Arabidopsis: 35S::(nYFP-CPR5+cYFP-CPR5) | This paper | N/A |
| Arabidopsis: 35S::(GFP-CPR5+3HA-CPR5) | This paper | N/A |
| Arabidopsis: GFP-WIP1 | The Meier Laboratory | N/A |
| Arabidopsis: Nup155-YFP | This paper | N/A |
| Arabidopsis: MOS7-GFP | Cheng et al., 2009 | N/A |
| nup88 (mos7-1), nup96 (mos3-1), importin-α3 (mos6-2) | The Li Laboratory, Cheng et al., 2009; Palma et al., 2005; Zhang and Li, 2005 |
N/A |
| All other nucleoporin and transport receptor mutants Used in this study, see Table S2 |
Arabidopsis Biological Resource Center |
see Table S2 for Salk line ID |
| Arabidopsis: 35S::YFP-SIM | This paper | N/A |
| Arabidopsis: Dex::AvrRpt2 | McNellis et al., 1998 | N/A |
| Recombinant DNA | ||
| pMDC43-GFP-CPR5 | Wang et al., 2014 | N/A |
| pEG100-YFP-CPR5 (WT and mutant constructs) | This paper | N/A |
| pEG100-n/cYFP-CPR5 | This paper | N/A |
| pEG100-Nups-YFP/cYFP | This paper | N/A |
| pBAV154/pEG100-YFP-CPR5-C | This paper | N/A |
| pEG100-NPR1/JAZ1/ABI5-mCherry | This paper | N/A |
| pEG100-mCherry-WIT1 | This paper | N/A |
| Sequence-Based Reagents | ||
| Primers used in this study, see Table S3 | This paper | N/A |
| Software and Algorithms | ||
| Mascot (v2.5.0) | Matrix Science | N/A |
| Scaffold (v4.4.1.1) | Proteome Software, Inc. |
N/A |
| R (v3.0.1) | N/A | N/A |
| GeneSpring (v13.0) | Agilent | N/A |
| MASTA | Reina-Pinto et al., 2010 | N/A |
| PlantGSEA | Yi et al., 2013 | http://structuralbiology.cau.edu.cn/PlantGSEA/ |
| Athena | O’Connor et al., 2005 | http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl |
| Other |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Xinnian Dong (xdong@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis
All Arabidopsis plants used in this study were in the Col-0 background. Wild type (WT), mutant, and transgenic Arabidopsis seeds were stratified at 4°C for two days and plants were grown under a 12-hrs light and 12-hrs dark cycle at 22°C. GFP-WIP1, NPR1-GFP, Nup155-YFP and MOS7-GFP were introduced into the cpr5 mutant background through genetic crosses. Nucleoporin and transport receptor mutants used in this study were Salk T-DNA insertion lines obtained from Arabidopsis Biological Resource Center (see Table S2 for Salk line information) or mos mutants (Cheng et al., 2009; Palma et al., 2005; Zhang and Li, 2005) provided by Dr. Xin Li’s laboratory (see Table S2). cpr5 nup double mutants were obtained through genetic crosses. 35S:YFP-CPR5-C and Dex:YFP-CPR5-C were transformed into WT background for functional interference of CPR5 and subsequently crossed to the cpr5 mutant as controls to test the specificity of the interference effect. To obtain 35S:GFP-CPR5/HA-CPR5 double transgenic line, a previously reported 35S:GFP-CPR5 line (Wang et al., 2014) was transformed with 35S:HA-CPR5. T3 progeny homozygous for both transgenes were used for experiments. Isogenic 35S:n/cYFP-CPR5 line and Dex:AvrRpt2 line (McNellis et al., 1998) were generated in WT and rps2 backgrounds, respectively. The 35S:n/cYFP-CPR5/Dex:AvrRpt2 double transgenic lines were obtained by genetic crosses.
METHOD DETAILS
Plasmid Construction
All point mutations of CPR5 were generated using the QuikChange II site-directed mutagenesis kit (Agilent). The full-length cDNA of CPR5, WIT1, SIM and all CPR5 mutant constructs including the single-site mutations and truncations were cloned into pBSDONR p4r–p2, a multisite gateway donor vector for N-terminal tagging (Gu and Innes, 2011). The full-length cDNA of Nup136, Nup93a, Nup35, NPR1, JAZ1, ABI5 and the genomic DNA fragments (from start codon to stop codon) of Nup155, Nup96, Nup88, Nup85, Nup62, Nup58, Nup54, HOS1 were cloned into pBSDONR p1-p4, a multisite gateway donor vector for C-terminal tagging. Those clones were then paired with fluorescent tags, n/c-YFP (for BiFC) or 3xHA tag cloned in pBSDONR p1-p4 or pBSDONR p4r-p2, to generate fusion constructs in pEG100 or pBAV154 destination vector by LR reaction (LR clonase II plus, Thermofisher) for constitutive or dexamethasone inducible expression, respectively. To generate 35S:n/cYFP-CPR5 construct used in transgenic line, nYFP-CPR5 was first cloned in pGWB414 by LR reaction and a fragment including the promoter, the fusion construct and the terminator was amplified, cut by AseI and ligated into pEG100, which already contains an independent expression cassette for the cYFP-CPR5 fusion. All constructs were verified by sequencing before use.
Transient Expression Assays
Agrobacterium-mediated transient expression in N. benthamiana and transformation of Arabidopsis protoplasts were performed as described (Gu and Innes, 2011).
Confocal Laser Scanning Microscopy
Intracellular fluorescence was observed using a Leica SP8 upright confocal microscope with high sensitivity hybrid detectors. Intracellular membrane organelle markers used were described previously (Brkljacic et al., 2009; Gu and Innes, 2011, 2012; Nelson et al., 2007). Three-dimensional image reconstruction and co-localization statistics were carried out using IMARIS 8.0 (Bitplane). FRAP experiment was performed using Leica SP8 FRAP Wizard. Fluorescence intensity was normalized to the average expression level defined by random sampling before photobleaching. Recovery curves were plotted with 50 frames (1 frame/sec) recorded after photobleaching.
Immunoelectron Microscopy and Electron Tomography
Root tip samples were dissected from Arabidopsis seedlings expressing GFP-CPR5 and cryofixed by an HPM100 (Leica Microsystems). The frozen specimens were freeze-substituted at −80°C and embedded in HM20 resin (Electron Microscopy Sciences) at −45°C. After polymerization at −45°C, the root tip samples were sectioned and immunolabeled with a GFP antibody (Santa Cruz) as described previously (Kang, 2010).
Co-Immunoprecipitation
All tagged proteins for in vitro pull-down assays were synthesized using a wheat germ-based transcription/translation system (BioSieg). Synthesized proteins were mixed and incubated with GFP-TrapA (Chromo Tek) or Pierce anti-HA agarose beads (Thermofisher) overnight at 4°C in the pull-down buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, plant protease inhibitor cocktail (Sigma) and 40 µM MG115). Following immunoprecipitation (IP), beads were precipitated and washed five times with the pull-down buffer before eluted with the SDS sample buffer. For in vivo co-immunoprecipitation, leaf tissues of four-week-old transgenic plants were collected. Total protein was extracted using IP buffer with high concentration of detergents to completely solubilize membrane protein (50 mM Tris, pH7.5, 150 mM NaCl, 0.5% Triton X-100, 0.5% Nonidet P-40, 0.25% Na-deoxycholate, plant protease inhibitor cocktail, and 40 mM MG115) followed by immunoprecipitation with GFP-TrapA beads overnight at 4°C. After IP, beads were washed five times with the IP buffer. Samples were boiled with loading buffer for 10 min before separated by SDS-PAGE.
LC-MS/MS and Data Analysis
A total of 5 g leaf tissues from four-week-old transgenic plants were collected. Total protein was extracted with IP buffer (50 mM Tris, pH7.5, 150 mM NaCl, 0.5% Triton X-100, 0.5% Nonidet P-40, plant protease inhibitor cocktail, and 40 mM MG115) and immunoprecipitated with GFP-TrapA beads overnight at 4°C. After IP, samples were washed five times with the IP buffer and three times with 50 mM NH4HCO3 before on-bead trypsin digestion. Following immunoprecipitation, on-bead trypsin digestion, peptide lyophilization and LC-MS/MS were performed by the Duke Proteomics Core Facility. MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.5.0) and searched with a fragment ion mass tolerance of 0.020 Da and a parent ion tolerance of 5.0 PPM. Scaffold (version 4.4.1.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide/protein identifications were accepted if they could be established at greater than 95% probability by the Peptide/Protein Prophet algorithm, which yielded a false-discovery rate of 0.1% and 0.5% on the peptide and protein match level, respectively. Data from 35S:GFP samples ran in parallel were used as controls for a statistical model-based selection of CPR5-specific interactors. A total of 41 candidates were identified and this list was shortened to 28 by further excluding proteins that are abundant in chloroplasts and mitochondria. Predicted interactions between CPR5 interactors were based on an interolog method (Geisler-Lee et al., 2007).
Quantitative PCR
Arabidopsis RNA was extracted using TRIzol Reagent (Thermofisher), and cDNA was synthesized using the SuperScript III cDNA Synthesis (Thermofisher). Quantitative PCR (qPCR) was performed using FastStart Universal SYBR Green Master Kit (Roche) in Mastercycler ep realplex (Eppendorf).
Microarray Procedure and Data Analysis
For RPS2-dependent ETI response, four-week-old WT and rps2 mutant plants were inoculated with PsmlAvrRpt2. At 0, 6 and 10 hrs post inoculation, leaf tissues were collected for RNA preparation and microarray. The resulting data set was deposited to Gene Expression Omnibus (GEO: GSE72742). For transient interference of the CPR5 function, four-week-old WT and T3 homozygous Dex:YFP-CPR5-C transgenic plants were sprayed with water or 50 µM dexamethasone (Sigma). After 24 hrs, leaf tissues were collected for RNA preparation and microarray. The resulting data set was deposited to GEO: GSE72743. RNA quality control, cDNA synthesis, aRNA purification and fragmentation, hybridization, washing and scanning of Arabidopsis ATH1 Genome Arrays (Affymetrix) chips were performed by the Duke Microarray Facility. For CPR5-C interference microarray, differentially expressed genes were further filtered by subtracting genes whose expression are affected by the empty vector control upon dexamethasone induction (GSE8741). The resulting gene list was provided in Table S1. The RPS4-dependent ETI genes, RPS2-dependent ETI genes and basal immunity genes were determined by microarray analysis of dataset GSE50019, GSE73742 and GSE17464, respectively. Statistical analysis and natural language processing-based network regulator discovery were performed using GeneSpring 13.0 (Agilent, 2014) and R 3.0.1 (2013). Networks were established based on published transcriptional regulatory relationships between genes and physical interactions between proteins. Comparative microarray analysis and GSEA were performed using MASTA (Reina-Pinto et al., 2010) and PlantGSEA (Yi et al., 2013), respectively. Cis-element enrichment was analyzed using total differentially expressed genes with Athena (O’Connor et al., 2005).
Bacterial Growth Assay
Infection of Arabidopsis plants with Psm ES4326 (with or without AvrRpt2) was performed as described previously (Wang et al., 2014). Bacterial suspension of OD600nm = 0.001 was infiltrated into 2 leaves per plant and 12 plants per genotype. Each experimental replication contained four leaf discs from two plants. Bacterial growth was quantified at 0 and 3 days post infiltration.
Primers
All primers used in this study were listed in Table S3.
QUANTIFICATION AND STATISTICAL ANALYSIS
The nuclear circularity index is defined as 4πA/P2, where A and P are the cross-sectional area and perimeter of the nucleus, respectively. A and P were measured for each nucleus using Fiji. Bacterial growth was reported as the number of colony forming units (cfu), which was subject to log transformation. For fluorescence quantification using Fiji, the fluorescence intensity was calculated using Integrated Density – (Area of selected cell × Mean fluorescence of background). Nuclear circularity, log(cfu) and fluorescence intensity data were assumed to follow normal distributions and were subjected to two-tailed Student’s t-test or ANOVA, where appropriate. Statistical tests were performed in GraphPad Prism 6. Statistical parameters including the exact value of n, the definition of center, dispersion and precision measures (mean ± SDM) and statistical significance can be found in the Figure Legends. In Figures, asterisks denote statistical significance test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001) as compared to untreated controls, unless otherwise specified by lines connecting the compared pieces of data. For LC-MS/MS analysis, exclusive spectrum count data were assumed to follow a Gamma-Poisson distribution. After normalized by size factors, negative binomial regression models were built with the normalized data using DESeq2 package in R, which provided the cutoff to select for CPR5-specific interactors (p-value < 0.05, fold change > 2.5, CPR5 vs GFP). For microarray analysis, array data were summarized using the Robust Multiarray Average method using GeneSpring and the normal distribution of the expression data were verified using EMA package in R. CPR5-C-induced differentially expressed genes were selected using 2-way ANOVA model (p-value < 0.01, fold change > 2, dex vs water); the RPS4-dependent ETI genes were determined by 2-way ANOVA (p-value < 0.01, fold change > 2) using the signaling mutant eds1 as a control (GSE50019); the RPS2-dependent ETI genes induced by Psm/AvrRpt2 was determined by 2-way ANOVA (p-value < 0.01, fold change > 2) using the immune receptor mutant rps2 as a control (GSE72742); the basal immunity induced by Pst DC3000 was determined by Moderated t-test (p-value < 0.05 and fold change > 2, GSE17464).
DATA AND SOFTWARE AVAILABILITY
Data Resources
Raw data files for the microarray analysis have been deposited in the NCBI Gene Expression Omnibus under accession number GSE72742 and GSE72743.
Supplementary Material
(A) Leaf epidermal cells of two-week-old 35S:GFP-CPR5/cpr5-1 transgenic Arabidopsis seedlings were imaged using confocal laser scanning microscopy. Three-dimensional image of the nuclear surface was reconstructed from z-stack images using Imaris (right). Arrowheads indicate the nuclear envelope (NE) or the nucleus (N).
(B) Co-localization of GFP-CPR5 with endomembrane markers. GFP-CPR5 was transiently co-expressed with mCherry-tagged endomembrane markers in N. benthamiana. The nuclear envelope (NE) marker is mCherry-WIT1. TGN/EE, trans-Golgi network/early endosome. MVB/LE, multi-vesicular body/late endosome.
(C) Peptide coverage of the carboxyl terminal half of the AtCPR5 protein. GFP-CPR5 was affinity purified from transgenic plants and subject to LC-MS/MS. The predicted transmembrane domains in CPR5 are labeled in red. Predicted omission regions by trypsin digestion are labeled in cyan. Individual peptides detected by LC-MS/MS are labeled in brown and listed below the reference sequence.
(A) CPR5 protein complex purification. Total protein extracts from four-week-old 35S:YFP-CPR5 and 35S:GFP transgenic plants were subject to immunoprecipitation using GFP-TrapA agarose beads. Before LC-MS/MS, sample aliquots were loaded onto SDS-PAGE and stained with Coomassie Blue, silver nitrate or blotted with the GFP antibody.
(B) The complete list of CPR5-specific interactors identified by protein complex purification. Subcellular localization of the proteins were color coded and determined by published fluorescent imaging studies or by transient expression of YFP-tagged protein in this study. Proteins with or without putative transmembrane domains are labeled with black and cyan in text, respectively. Interactions between CPR5 interactors (attached circles) were predicted by the interolog method (Geisler-lee et al., 2007), indicating identification of putative protein complexes.
(C) Localization of Nup155 and Nup88 in WT and the cpr5 mutant backgrounds. Isogenic 35S:Nup155-YFP and 35S:Nup88-GFP lines were generated in WT and cpr5-1 backgrounds. Root cells of five-day-old seedlings were imaged.
(A) CPR5-C did not complement the cpr5 mutant phenotypes. Two independent transgenic lines are shown. WT, the cpr5 mutant and the complemented line expressing the full-length CPR5 are shown as control. Expression levels of transgenes are shown in the right panel.
(B) CPR5-C co-localizes with CPR5. YFP-CPR5-C and mCherry-CPR5 were transiently co-expressed in N. benthamiana and epidermal cells were imaged 24 hours after Agrobacterium infiltration. The arrowhead indicates the nucleus (N).
(C and D) Transient expression of HA-CPR5-C but not HA-CPR5-N or HA-CPR5 induced PCD in N. benthamiana (C). PCD induced by transient expression of YFP-CPR5-C could be suppressed by coexpression of YFP-CPR5 but not YFP-CPR5-N (D, left) at comparable protein levels (D, right). All constructs are driven by the 35S promoter. Representative leaves were photographed at 36 hours after Agrobacterium infiltration.
(E) Segregating T2 progeny of 35S:YFP-CPR5-C transgenic Arabidopsis lines in WT background. The presence of the transgene was determined by PCR. Two independent lines (#3 and #4) are shown and cpr5-1 mutant served as a control.
(F) Quantitative RT-PCR analysis of SA-dependent defense gene expression in 35S:YFP-CPR5-C transgenic lines. The transcript levels of the endogenous CPR5 were not affected in the 35S:YFP-CPR5-C lines (right). Data are presented as mean ± SDM (n = 3 biological replicates).
(G) Inducible expression of CPR5-C is innocuous in the cpr5 mutant. Isogenic Dex:YFP-CPR5-C lines in WT and the cpr5 mutant background were imaged 5 days after dexamethasone treatment (upper panel). Relative expression levels of most induced defense genes in the cpr5 mutant were measured 3 days after dexamethasone treatment using quantitative RT-PCR (lower panel). Data are presented as mean ± SDM (n = 2 biological replicates).
(H) Time course quantitative RT-PCR analysis of SA-responsive genes in Dex:YFP-CPR5-C transgenic plants (WT background) after dexamethasone application. Relative expression levels of SA responsive (PR1) and transport (EDS5) genes were measured. A transgenic line with the empty vector and the cpr5 mutant served as controls. Data are presented as mean ± SDM (n = 3 technical replicates). Similar results were obtained in two independent experiments.
(A) Transient coexpression of mCherry-tagged NPR1/JAZ1/ABI5 with YFP-tagged WT (left panel) or G420D mutant (right panel) CPR5 in N. benthamiana.
(B) Quantification of results from experiments performed in panel A and B. Localization of nuclear proteins in each cell was recorded as binomial data and categorized as ‘nucleus only’ or with significant ‘cytosol retention’. A logistic regression model using CPR5/G420D as the sole independent variable can explain 95% of the data variance, illustrating the predominant effect of the functional CPR5 on protein nuclear transport.
(C) Phenotypes observed when cpr5 was crossed with stress-related karyopherin and transport-related nucleoporin mutants.
(A) Quantitative RT-PCR determined relative expression levels of CPR5 during ETI triggered by Psm/AvrRpt2 at OD600nm = 0.1. Samples were collected until 8 hours post inoculation (hpi) before ETI-induced PCD occurred. Data are presented as mean ± SDM (n = 3 biological replicates). Similar results were obtained in two independent experiments.
(B) GFP-CPR5 protein levels in 35S:GFP-CPR5 transgenic plants during ETI triggered by Psm/AvrRpt2 at OD600nm = 0.1 as measured by western blot using the GFP antibody.
(C) Fluorescence intensity measurement of GFP-CPR5 in the NE in 35S:GFP-CPR5 transgenic plants during ETI triggered by Psm/AvrRpt2 at OD600nm = 0.1. Data are presented as mean ± SDM (n = 15 cells for each time point and treatment).
(D) CPR5 homomeric interaction is mediated by its N-terminal domain. CPR5-N, rather than CPR5-C, mediated the homomeric interaction. In vitro pull-down assays were performed using wheat germ-synthesized recombinant proteins. Immunoprecipitation was performed using HA antibody-conjugated agarose beads. Immunoblotting was performed using GFP and HA antibodies. Stars indicate non-specific signals from immunoglobulins.
(E) The G120D mutation compromises heteromeric interactions of CPR5. BiFC was performed by transiently coexpressing nYFP-CPR5/cYFP-CPR5 or nYFP-G120D/cYFP-G120D in N. benthamiana. The BiFC intensity in the NE was normalized using averaged expression levels of YFP-CPR5/G120D measured in separate experiments and plotted as relative values (upper panel). Data are presented as mean ± SDM (n = 15 cells for each BiFC combination). The number of whorl structures in each cell when WT CPR5 or G120D mutant was transiently expressed in N. benthamiana (lower panel). Data are presented as mean ± SDM (n = 15 cells for each construct). Student’s t-test and non-parametric test were used, **** p-value < 0.0001.
(A) The G120D mutation compromises CPR5 interaction with SIM. BiFC was performed by transiently coexpressing SIM-nYFP and cYFP-CPR5/G120D in N. benthamiana. The BiFC intensity was normalized using averaged expression levels of YFP-CPR5/G120D measured in separate experiments. Data were plotted as relative values to the average BiFC intensity of CPR5-SIM and are presented as mean ± SDM (n = 15 cells for each BiFC combination).
(B) Three-week-old WT, cpr5, sim smr1 and cpr5 sim smr1 plants together with isogenic lines constitutively expressing YFP-SIM in sim smr1, and cpr5 sim smr1 backgrounds (upper panel). Overexpression of SIM in the sim smr1 background caused serrated leaf morphology without PCD, but in the cpr5 sim smr1 background led to the cpr5-like dwarfism and PCD (lower panel). Nuclear YFP-SIM is pseudo-colored in green and chlorophyll autofluorescence is in red. Arrowheads indicate enlarged nuclei as a result of SIM overactivation (lower panel).
Table S1. CPR5-C-Induced Differentially Expressed Genes, Related to Figure 3
Table S2. Nucleoporin and Transport Receptor Mutants Used in This Study, Related to Methods and Resources
Table S3. Primers Used in This Study, Related to Methods and Resources
In Brief.
Rather than being a passive conduit, the nuclear pore complex in response to immune signals undergoes a conformational switch to reconfigure the selective barrier and promote stress responses.
Highlights.
CPR5 is a component of the nuclear pore complex (NPC)
CPR5 regulates nuclear transport through the selective barrier of the NPC
CPR5 homomer is disrupted upon induction of effector-triggered immunity (ETI).
Conformational change in CPR5 leads to CKI release and NPC permeabilization for ETI.
Acknowledgments
The authors thank Dr. Roger Innes for providing membrane organelle markers and Dr. Iris Meier for sharing the 35S:GFP-WIP1 and 35S:RanGAP1-GFP transgenic lines. We also thank Drs. Iris Meier, Paul Zwack and John Withers for critical reading of the manuscript. This work was supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through grant GBMF3032) and by grants from the National Institutes of Health (NIH) (R01-GM069594) to X.D., the National Natural Science Foundation of China (Grant No. 31571254) to S.W., and Rural Development Administration, Republic of Korea (Project No. 10953092015) and Research Grants Council of Hong Kong (AoE/M-05/12, C4011-14R) to B.K..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes six figures and two tables and can be found with this article online at XXX.
AUTHOR CONTRIBUTIONS
Y.G., S.W., and X.D. designed the research. Y.G. performed most of the experiments. S.G.Z contributed dataset GSE72742. Z.L. and B.K. performed the immunoelectron microscopy. S.W. helped with the protein complex purification. Y.G. and X.D. wrote the paper with comments from all authors.
REFERENCES
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Boch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN. Analysis of Resistance Gene-Mediated Defense Responses in Arabidopsis thaliana Plants Carrying a Mutation in CPR5. Mol. Plant Microbe Interact. 1998;11:1196–1206. [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic J, Zhao Q, Meier I. WPP-domain proteins mimic the activity of the HSC70-1 chaperone in preventing mistargeting of RanGAP1-anchoring protein WIT1. Plant Physiol. 2009;151:142–154. doi: 10.1104/pp.109.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Wang Y, Wei JH, van Deursen J, Yu H, Malureanu L, Dasso M, Forbes DJ, Levy DE, Seemann J, Fontoura BM. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev. Cell. 2008;15:657–667. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, Wirthmueller L, Despres C, Parker JE, Zhang Y, Li X. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D. Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science. 2015 doi: 10.1126/science.aac7420. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Rivas S. The plant cell nucleus: a true arena for the fight between plants and pathogens. Plant Signal. Behav. 2011;6:42–48. doi: 10.4161/psb.6.1.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH, Zhu JK. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell Biol. 2006;26:9533–9543. doi: 10.1128/MCB.01063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Gao Y, Zhan Y, Zhang S, Wu Y, Xiao Y, Zou B, He K, Gou X, Li G, Lin H, Li J. Nucleocytoplasmic trafficking is essential for BAK1- and BKK1-mediated cell-death control. Plant J. 2016;85:520–531. doi: 10.1111/tpj.13125. [DOI] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJ, Coaker G. Plant NB-LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol. 2011;14:365–371. doi: 10.1016/j.pbi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AV, Parker JE. Heaven’s Gate: nuclear accessibility and activities of plant immune regulators. Trends Plant Sci. 2009;14:479–487. doi: 10.1016/j.tplants.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145:317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- Gong FC, Giddings TH, Meehl JB, Staehelin LA, Galbraith DW. Z-membranes: artificial organelles for overexpressing recombinant integral membrane proteins. Proc. Natl. Acad. Sci. U S A. 1996;93:2219–2223. doi: 10.1073/pnas.93.5.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Innes RW. The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 2011;155:1827–1838. doi: 10.1104/pp.110.171785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Innes RW. The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell. 2012;24:4717–4730. doi: 10.1105/tpc.112.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P, Edens LJ, Vukovic LD, Levy DL. Sizing and shaping the nucleus: mechanisms and significance. Curr. Opin. Cell Biol. 2014;28:16–27. doi: 10.1016/j.ceb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing HC, Anderson L, Sturre MJ, Hille J, Dijkwel PP. Arabidopsis CPR5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J. Exp. Bot. 2007;58:3885–3894. doi: 10.1093/jxb/erm237. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kang BH. Electron microscopy and high-pressure freezing of Arabidopsis. Methods Cell Biol. 2010;96:259–283. doi: 10.1016/S0091-679X(10)96012-3. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo--cytoplasmic partitioning. Genes. Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Lee HS, Lee JS, Kim SK, Kim SH. Hormone- and light-regulated nucleocytoplasmic transport in plants: current status. J. Exp. Bot. 2008;59:3229–3245. doi: 10.1093/jxb/ern200. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- McNellis TW, Mudgett MB, Li K, Aoyama T, Horvath D, Chua NH, Staskawicz BJ. Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 1998;14:247–257. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Parry G. Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J. Exp. Bot. 2014;65:6057–6067. doi: 10.1093/jxb/eru346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18:1590–1603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Pinto JJ, Voisin D, Teodor R, Yephremov A. Probing differentially expressed genes against a microarray database for in silico suppressor/enhancer and inhibitor/activator screens. Plant J. 2010;61:166–175. doi: 10.1111/j.1365-313X.2009.04043.x. [DOI] [PubMed] [Google Scholar]
- Rivas S. Nuclear dynamics during plant innate immunity. Plant Physiol. 2012;158:87–94. doi: 10.1104/pp.111.186163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 2000;149:1027–1038. doi: 10.1083/jcb.149.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat. Rev. Immunol. 2013;13:199–206. doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, Hetzer MW., 3rd Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Guo Y, Dong CH, Ma W, Zhu JK. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant J. 2006;47:776–787. doi: 10.1111/j.1365-313X.2006.02833.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X. A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe. 2014;16:787–794. doi: 10.1016/j.chom.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wiermer M, Cheng YT, Imkampe J, Li M, Wang D, Lipka V, Li X. Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 2012;70:796–808. doi: 10.1111/j.1365-313X.2012.04928.x. [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Roth C, Banfield MJ, Wiermer M. Hop-on hop-off: importin-alpha-guided tours to the nucleus in innate immune signaling. Front. Plant Sci. 2013;4:149. doi: 10.3389/fpls.2013.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Du Z, Su Z. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41:W98–W103. doi: 10.1093/nar/gkt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 2002;29:427–437. doi: 10.1046/j.0960-7412.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Leaf epidermal cells of two-week-old 35S:GFP-CPR5/cpr5-1 transgenic Arabidopsis seedlings were imaged using confocal laser scanning microscopy. Three-dimensional image of the nuclear surface was reconstructed from z-stack images using Imaris (right). Arrowheads indicate the nuclear envelope (NE) or the nucleus (N).
(B) Co-localization of GFP-CPR5 with endomembrane markers. GFP-CPR5 was transiently co-expressed with mCherry-tagged endomembrane markers in N. benthamiana. The nuclear envelope (NE) marker is mCherry-WIT1. TGN/EE, trans-Golgi network/early endosome. MVB/LE, multi-vesicular body/late endosome.
(C) Peptide coverage of the carboxyl terminal half of the AtCPR5 protein. GFP-CPR5 was affinity purified from transgenic plants and subject to LC-MS/MS. The predicted transmembrane domains in CPR5 are labeled in red. Predicted omission regions by trypsin digestion are labeled in cyan. Individual peptides detected by LC-MS/MS are labeled in brown and listed below the reference sequence.
(A) CPR5 protein complex purification. Total protein extracts from four-week-old 35S:YFP-CPR5 and 35S:GFP transgenic plants were subject to immunoprecipitation using GFP-TrapA agarose beads. Before LC-MS/MS, sample aliquots were loaded onto SDS-PAGE and stained with Coomassie Blue, silver nitrate or blotted with the GFP antibody.
(B) The complete list of CPR5-specific interactors identified by protein complex purification. Subcellular localization of the proteins were color coded and determined by published fluorescent imaging studies or by transient expression of YFP-tagged protein in this study. Proteins with or without putative transmembrane domains are labeled with black and cyan in text, respectively. Interactions between CPR5 interactors (attached circles) were predicted by the interolog method (Geisler-lee et al., 2007), indicating identification of putative protein complexes.
(C) Localization of Nup155 and Nup88 in WT and the cpr5 mutant backgrounds. Isogenic 35S:Nup155-YFP and 35S:Nup88-GFP lines were generated in WT and cpr5-1 backgrounds. Root cells of five-day-old seedlings were imaged.
(A) CPR5-C did not complement the cpr5 mutant phenotypes. Two independent transgenic lines are shown. WT, the cpr5 mutant and the complemented line expressing the full-length CPR5 are shown as control. Expression levels of transgenes are shown in the right panel.
(B) CPR5-C co-localizes with CPR5. YFP-CPR5-C and mCherry-CPR5 were transiently co-expressed in N. benthamiana and epidermal cells were imaged 24 hours after Agrobacterium infiltration. The arrowhead indicates the nucleus (N).
(C and D) Transient expression of HA-CPR5-C but not HA-CPR5-N or HA-CPR5 induced PCD in N. benthamiana (C). PCD induced by transient expression of YFP-CPR5-C could be suppressed by coexpression of YFP-CPR5 but not YFP-CPR5-N (D, left) at comparable protein levels (D, right). All constructs are driven by the 35S promoter. Representative leaves were photographed at 36 hours after Agrobacterium infiltration.
(E) Segregating T2 progeny of 35S:YFP-CPR5-C transgenic Arabidopsis lines in WT background. The presence of the transgene was determined by PCR. Two independent lines (#3 and #4) are shown and cpr5-1 mutant served as a control.
(F) Quantitative RT-PCR analysis of SA-dependent defense gene expression in 35S:YFP-CPR5-C transgenic lines. The transcript levels of the endogenous CPR5 were not affected in the 35S:YFP-CPR5-C lines (right). Data are presented as mean ± SDM (n = 3 biological replicates).
(G) Inducible expression of CPR5-C is innocuous in the cpr5 mutant. Isogenic Dex:YFP-CPR5-C lines in WT and the cpr5 mutant background were imaged 5 days after dexamethasone treatment (upper panel). Relative expression levels of most induced defense genes in the cpr5 mutant were measured 3 days after dexamethasone treatment using quantitative RT-PCR (lower panel). Data are presented as mean ± SDM (n = 2 biological replicates).
(H) Time course quantitative RT-PCR analysis of SA-responsive genes in Dex:YFP-CPR5-C transgenic plants (WT background) after dexamethasone application. Relative expression levels of SA responsive (PR1) and transport (EDS5) genes were measured. A transgenic line with the empty vector and the cpr5 mutant served as controls. Data are presented as mean ± SDM (n = 3 technical replicates). Similar results were obtained in two independent experiments.
(A) Transient coexpression of mCherry-tagged NPR1/JAZ1/ABI5 with YFP-tagged WT (left panel) or G420D mutant (right panel) CPR5 in N. benthamiana.
(B) Quantification of results from experiments performed in panel A and B. Localization of nuclear proteins in each cell was recorded as binomial data and categorized as ‘nucleus only’ or with significant ‘cytosol retention’. A logistic regression model using CPR5/G420D as the sole independent variable can explain 95% of the data variance, illustrating the predominant effect of the functional CPR5 on protein nuclear transport.
(C) Phenotypes observed when cpr5 was crossed with stress-related karyopherin and transport-related nucleoporin mutants.
(A) Quantitative RT-PCR determined relative expression levels of CPR5 during ETI triggered by Psm/AvrRpt2 at OD600nm = 0.1. Samples were collected until 8 hours post inoculation (hpi) before ETI-induced PCD occurred. Data are presented as mean ± SDM (n = 3 biological replicates). Similar results were obtained in two independent experiments.
(B) GFP-CPR5 protein levels in 35S:GFP-CPR5 transgenic plants during ETI triggered by Psm/AvrRpt2 at OD600nm = 0.1 as measured by western blot using the GFP antibody.
(C) Fluorescence intensity measurement of GFP-CPR5 in the NE in 35S:GFP-CPR5 transgenic plants during ETI triggered by Psm/AvrRpt2 at OD600nm = 0.1. Data are presented as mean ± SDM (n = 15 cells for each time point and treatment).
(D) CPR5 homomeric interaction is mediated by its N-terminal domain. CPR5-N, rather than CPR5-C, mediated the homomeric interaction. In vitro pull-down assays were performed using wheat germ-synthesized recombinant proteins. Immunoprecipitation was performed using HA antibody-conjugated agarose beads. Immunoblotting was performed using GFP and HA antibodies. Stars indicate non-specific signals from immunoglobulins.
(E) The G120D mutation compromises heteromeric interactions of CPR5. BiFC was performed by transiently coexpressing nYFP-CPR5/cYFP-CPR5 or nYFP-G120D/cYFP-G120D in N. benthamiana. The BiFC intensity in the NE was normalized using averaged expression levels of YFP-CPR5/G120D measured in separate experiments and plotted as relative values (upper panel). Data are presented as mean ± SDM (n = 15 cells for each BiFC combination). The number of whorl structures in each cell when WT CPR5 or G120D mutant was transiently expressed in N. benthamiana (lower panel). Data are presented as mean ± SDM (n = 15 cells for each construct). Student’s t-test and non-parametric test were used, **** p-value < 0.0001.
(A) The G120D mutation compromises CPR5 interaction with SIM. BiFC was performed by transiently coexpressing SIM-nYFP and cYFP-CPR5/G120D in N. benthamiana. The BiFC intensity was normalized using averaged expression levels of YFP-CPR5/G120D measured in separate experiments. Data were plotted as relative values to the average BiFC intensity of CPR5-SIM and are presented as mean ± SDM (n = 15 cells for each BiFC combination).
(B) Three-week-old WT, cpr5, sim smr1 and cpr5 sim smr1 plants together with isogenic lines constitutively expressing YFP-SIM in sim smr1, and cpr5 sim smr1 backgrounds (upper panel). Overexpression of SIM in the sim smr1 background caused serrated leaf morphology without PCD, but in the cpr5 sim smr1 background led to the cpr5-like dwarfism and PCD (lower panel). Nuclear YFP-SIM is pseudo-colored in green and chlorophyll autofluorescence is in red. Arrowheads indicate enlarged nuclei as a result of SIM overactivation (lower panel).
Table S1. CPR5-C-Induced Differentially Expressed Genes, Related to Figure 3
Table S2. Nucleoporin and Transport Receptor Mutants Used in This Study, Related to Methods and Resources
Table S3. Primers Used in This Study, Related to Methods and Resources