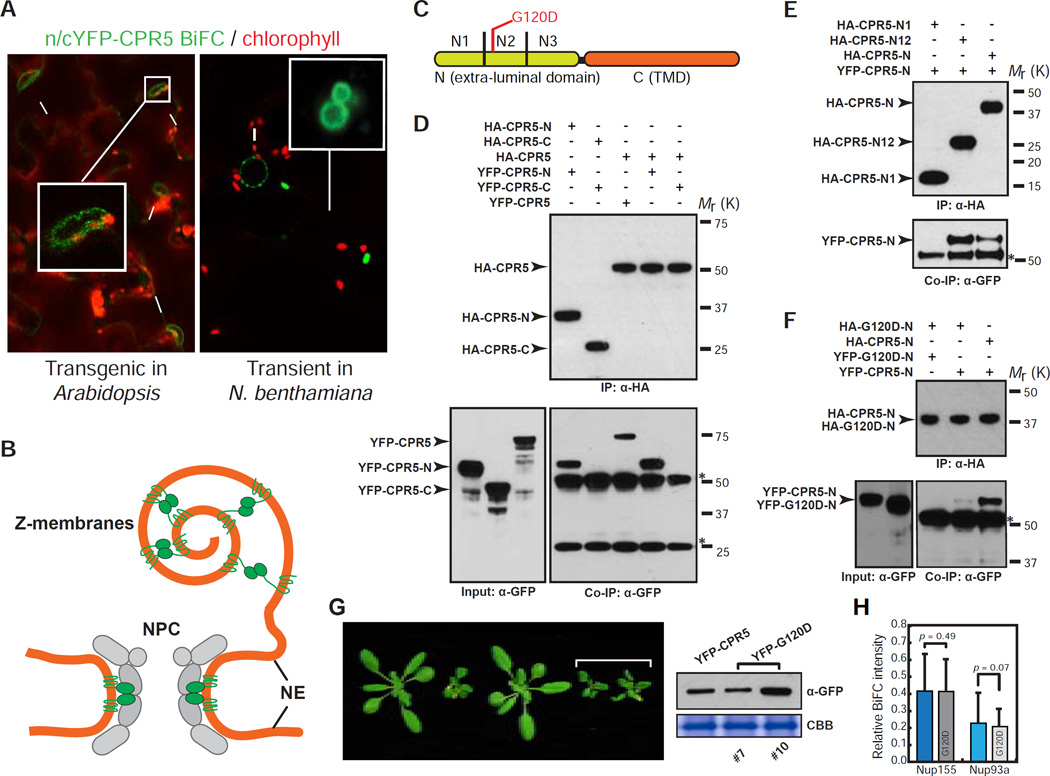
Figure 5. CPR5 Homomeric Interaction at the N-Terminal Extra-Luminal Domain Is Required for Suppressing ETI and PCD.
(A) CPR5 BiFC assay. nYFP-CPR5 and cYFP-CPR5 were constructed in two separate 35S promoter-driven expression cassettes within one vector (35S:n/c-YFP-CPR5). BiFC signals were observed in a stable transgenic Arabidopsis line (left) and transiently transformed N. benthamiana (right), respectively. Arrows and arrowheads indicate the nucleus and Z-membranes, respectively.
(B) A model proposed for CPR5 homomeric interaction in the NPC and Z-membranes.
(C) Schematic of CPR5 constructs used in in vitro pull-down assays. CPR5-N (1–274 aa) and CPR5-C (275–564 aa). CPR5-N was further divided into N1 (1–91 aa), N2 (92–182 aa) and N3 (183–274 aa).
(D–F) CPR5 homomeric interaction is mediated by its N-terminal extra-luminal domain. CPR5-N, rather than CPR5-C, mediated the homomeric interaction (D). The N1 and N2 (N12) domains of CPR5-C is sufficient and the N2 domain is necessary for the interaction (E). The G120D mutation compromised the homomeric interaction of CPR5-N (F). In vitro pull-down assays were performed using HA antibody-conjugated agarose beads. Stars indicate non-specific signals from immunoglobulins.
(G) Homomeric interaction is required for CPR5 function. 35S:YFP-CPR5 fully complemented the cpr5-1 mutant phenotype but 35S:YFP-G120D did not (left) when expressed at comparable levels (right). Two independent 35S:YFP-G120D transgenic lines (#7 and #10) are shown.
(H) The G120D mutation does not affect heteromeric interactions of CPR5 with other nucleoporins. BiFC was performed by transiently coexpressing nYFP-CPR5/G120D pairwise with Nup155/Nup93a–cYFP in N. benthamiana. The BiFC intensity was normalized using averaged expression levels of YFP-CPR5/G120D measured in separate experiments and plotted as relative values. Data are presented as mean ± SDM (n = 15 cells for each BiFC combination). p-values were calculated by Student’s t-test.
See also Figure S5.