SUMMARY
Apicomplexan parasites are leading causes of human and livestock diseases—like malaria and toxoplasmosis—yet most of their genes remain uncharacterized. Here, we present the first genome-wide genetic screen of an apicomplexan. We adapted CRISPR/Cas9 to assess the contribution of each gene from the human parasite Toxoplasma gondii during infection of fibroblasts. Our analysis defines ~200 previously uncharacterized, fitness-conferring genes unique to the phylum, from which 16 were investigated, revealing essential functions during infection of human cells. Secondary screens identify as an invasion factor the claudin-like apicomplexan microneme protein (CLAMP), which resembles mammalian tight-junction proteins and localizes to secretory organelles, making it critical to the initiation of infection. CLAMP is present throughout sequenced apicomplexan genomes, and is essential during the asexual stages of the malaria parasite Plasmodium falciparum. These results provide broad-based functional information on T. gondii genes and will facilitate future approaches to expand the horizon of antiparasitic interventions.
Keywords: Apicomplexan parasites, host-pathogen interactions, genome-wide CRISPR screen, eukaryotic pathogen, toxoplasmosis, malaria, host-cell invasion
INTRODUCTION
Apicomplexans comprise a phylum of over 5,000 obligate parasites whose hosts span the animal kingdom (Levine, 1988). Several species are leading causes of infant mortality, such as Plasmodium and Cryptosporidium spp., which cause malaria and severe diarrhea, respectively (Checkley et al., 2015; World Health Organization, 2014). Toxoplasma gondii, predicted to establish life-long infections in a quarter of the world’s population, can cause life-threatening disease in immune-compromised individuals or when contracted congenitally (Pappas et al., 2009). Despite their importance to global health, apicomplexans remain enigmatic. Only a handful of species have been studied and fewer than half of their genes have been functionally annotated. The ease with which T. gondii can be cultured, along with the genetic tractability that comes with its balanced nucleotide composition and high transfection rates, present compelling arguments for using this parasite as a model apicomplexan. Scalable methods to assess gene function in T. gondii could therefore greatly extend our understanding of apicomplexan biology.
Genetic crosses have long been used to identify loci responsible for phenotypes ranging from drug resistance in Plasmodium falciparum (Wellems et al., 1991), to virulence in T. gondii (Saeij et al., 2006; Taylor et al., 2006). However, completing the sexual cycles of T. gondii or Plasmodium spp. in cats or mosquitoes is challenging, and the traits examined must vary within the species. Spontaneous mutations, or those induced chemically or by transposition, can sample a wider range of phenotypes (Crabb et al., 2011; Farrell et al., 2014; Flannery et al., 2013), but the population size required to achieve saturation is impractical and causal mutations are often difficult to identify.
Gene deletion collections, such as those available for fungi (Winzeler et al., 1999), can aid functional analysis of eukaryotic genomes. With this aim, large-scale efforts have generated collections of knockout vectors for P. falciparum (Maier et al., 2008) and Plasmodium berghei (Gomes et al., 2015), which have led to the functional annotation of dozens of genes in both species. However, similar approaches have not been adapted to T. gondii, despite the advantage of both high transfection rates and a continuous culture system. The recent adaptation of CRISPR/Cas9 has further enhanced the genetic tractability of T. gondii (Shen et al., 2014; Sidik et al., 2014). This technology has the advantage of being easily reprogrammable by changing the 20 bp of homology between the single guide RNA (sgRNA, or guide) and the genomic target (reviewed in; Sander and Joung, 2014). The endogenously high rates of non-homologous end-joining (NHEJ) in T. gondii make it well suited to CRISPR-mediated gene disruption by efficiently creating frame-shift mutations and insertions at the cleavage site (Sidik et al., 2014).
Several studies have developed CRISPR/Cas9-based genome-wide genetic screens for mammalian cells (Koike-Yusa et al., 2014; Shalem et al., 2014; Wang et al., 2014). These screens use lentiviral libraries of sgRNAs to generate pools of mutants that can be exposed to selective pressures. The integrated sgRNAs can be used as barcodes to measure the contribution of targeted genes to cell fitness. Despite the lack of viral transduction, we adapted CRISPR/Cas9 for pooled screening in T. gondii. We present the first genome-wide genetic screen performed in any apicomplexan. We demonstrate the power of this approach using both positive and negative selection strategies. This approach provides the first complete survey of contributions to parasite fitness, cataloguing the ~40% of genes needed during infection of human fibroblasts. Based on this analysis, we were able to pinpoint previously uncharacterized conserved apicomplexan proteins necessary for the T. gondii lytic cycle. We demonstrate that one of these proteins acts as an essential invasion factor, and is also required by the malaria parasite P. falciparum to complete its asexual replication cycle. This protein is conserved throughout the phylum, providing an important molecular link to the invasion process of distantly related apicomplexans. Our analysis demonstrates the potential of genetic screens in T. gondii to uncover conserved biological processes, and provides a transformative tool for parasitology.
RESULTS
Constitutive Cas9 Expression Maximizes Gene Disruption in T. gondii
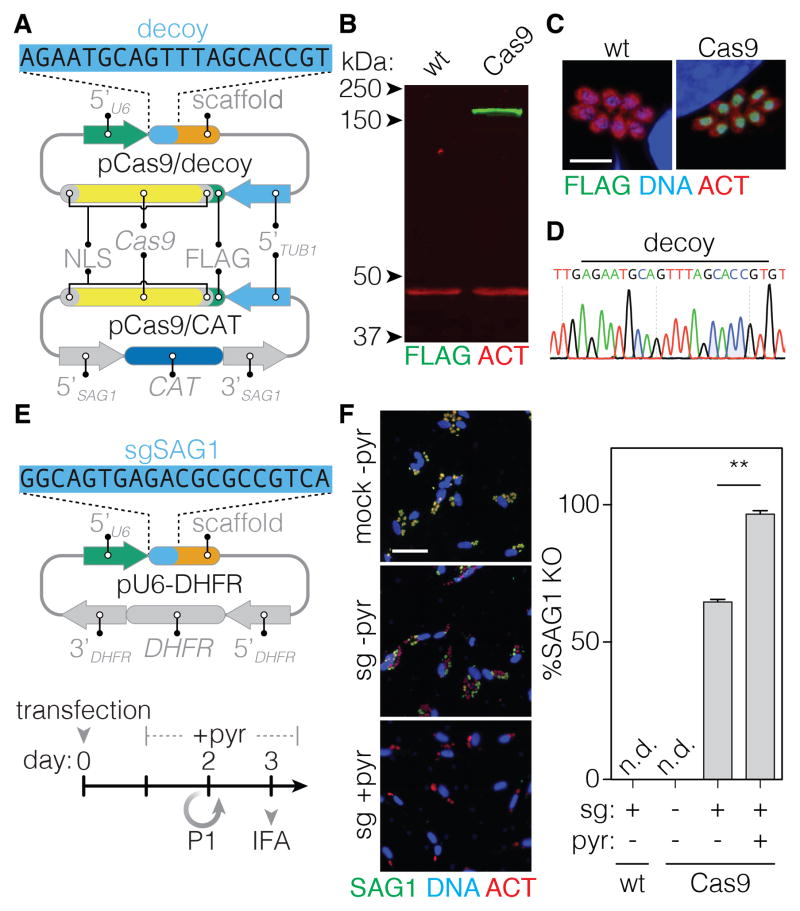
Highly efficient gene disruption and stable integration of the sgRNA are necessary to develop large-scale CRISPR screens. Transient expression of SpCas9 and an sgRNA in T. gondii can disrupt a targeted gene in ~20% of parasites (Sidik et al., 2014). We reasoned that constitutive Cas9 expression, prior to introducing the sgRNA, might increase the likelihood of gene disruption. We transfected parasites with a Cas9-expression plasmid carrying a chloramphenicol acetyltransferase (CAT) selectable marker (pCas9/CAT; Figure 1A). However, repeated attempts failed to isolate Cas9-expressing parasites, suggesting that Cas9 expression is detrimental to T. gondii, as has been suggested for other microorganisms (Jiang et al., 2014; Peng et al., 2015). We hypothesized that expression of a “decoy” sgRNA (pCas9/decoy; Figure 1A) could prevent toxicity that might arise from unintended Cas9 activity directed by endogenous RNAs. For this purpose, we used an sgRNA that appeared non-functional against the 3′ untranslated region of NHE1. Co-transfection of pCas9/CAT and pCas9/decoy readily yielded Cas9-expressing parasites, confirmed by immunoblotting (Figure 1B) and immunofluorescence (Figure 1C). As predicted, the Cas9-expressing strain retained the decoy locus (Figure 1D), reinforcing its requirement for constitutive Cas9 expression.
Figure 1. Expression of Cas9 Maximizes Gene-Disruption in T. gondii.
(A) Constructs used to constitutively express Cas9 in T. gondii. The sequence of the decoy sgRNA is highlighted (blue), followed by the Cas9-binding scaffold (orange).
(B) Immunoblot showing expression of FLAG-tagged Cas9 (green) in the strain constitutively expressing the transgene. ACT1 serves as a loading control (red).
(C) Cas9 localizes to the parasite nucleus. ACT1 provides a counterstain and DAPI stains for host-cell and parasite nuclei. Scale bar = 10 μm.
(D) Chromatogram showing the presence of the decoy in the Cas9-expressing strain.
(E) The sgRNA expression construct with the pyrimethamine-resistance selectable marker (DHFR). The targeting sequence of the SAG1 sgRNA is highlighted. The timeline indicates the period of pyrimethamine (pyr) selection (if applied), passaging to new host cells (P1), and the immunofluorescence assay (IFA).
(F) Representative micrographs showing intracellular parasites 3 days post transfection. Parasites were stained for SAG1 (green), and ACT1 (red). Host-cell and parasite nuclei were stained with DAPI (blue). Scale bar = 60 μm. The efficiency of SAG1 disruption in WT and Cas9-expressing parasites was measured following different treatments. Mean ± SD for n = 2 independent experiments, ** p < 0.005.
We assessed the efficiency of gene disruption in the Cas9-expressing strain by expressing an sgRNA against the surface antigen SAG1. Pyrimethamine treatment of the population selected for stable integration of the sgRNA expression vector (pU6-DHFR), which carries the resistant allele of dihydrofolate reductase (DHFR; Figure 1E). SAG1 provides a reliable measure of gene disruption because it is dispensable, yet stably maintained in cultured parasites (Kim and Boothroyd, 1995). Three days after transfection with the sgRNA construct, 70% of Cas9-expressing parasites had lost SAG1 expression. Pyrimethamine selection further improved SAG1 disruption to 97% over the same time period (Figure 1F). The high efficiency of CRISPR-mediated gene disruption in Cas9-expressing parasites provided the platform for large-scale genetic screens in T. gondii.
A Genome-Scale Genetic Screen Identifies Genes Involved in Drug Sensitivity
We designed a library of sgRNAs containing ten guides against each of the 8,158 predicted T. gondii protein-coding genes using previously described criteria (Wang et al., 2014). The library was cloned into the sgRNA expression vector (Figure 1E). 40% of the parasites that survived transfection integrated the vector into their genomes (data not shown). We could therefore measure the relative abundance of each integrated sgRNA by next-generation sequencing. Since the frequency of a given sgRNA corresponds to the relative abundance of parasites carrying the targeted disruption, the change in relative abundance from the composition of the plasmid library before transfection indicates the enrichment or depletion of a given mutant. We defined the average log2 fold-change in abundance for sgRNAs targeting a given gene as the “phenotype” score for that gene (Figure 2A). To determine whether we could maintain diversity over time, we transfected the library into both wild-type and Cas9-expressing parasites and sampled the populations after each of three lytic cycles (Figure 2B, left). The representation of guides against all genes remained stable over the course of the experiment in the absence of Cas9. In contrast, sgRNAs against specific genes were lost from the Cas9-expressing population (Figure 2C), indicating that a diverse set of mutants had been generated.
Figure 2. Using Pooled Screens to Identify genes responsible for drug sensitivity.
(A) Schematic depiction of the pooled CRISPR screen. Cas9-expressing parasites are transfected with the sgRNA library and grown in human fibroblasts (HFFs). At various time-points, sgRNAs are amplified and enumerated by sequencing to determine relative abundance and phenotype scores for individual genes.
(B) Time-line for the generation of mutant populations and subsequent selection in the presence or absence of FUDR. Times at which parasites were passaged (P) are indicated.
(C) Heat-map showing the phenotype score of genes at different time-points following transfection of the library into wild-type (wt) or Cas9-expressing parasites.
(D) Relative abundance of sgRNAs following growth of the population in the presence or absence of FUDR. Mean log2(normalized abundance) for each sgRNA in three independent experiments; sgRNAs against UPRT (blue).
(E) Phenotype score calculated for each gene comparing growth ± FUDR. Mean ± SEM for n = 3 independent experiments; UPRT (blue).
(F) Comparison of phenotypic scores in untreated samples after three (P3) or six (P6) passages. Pearson’s correlation coefficient (r) is shown.
To investigate the compatibility of our screen with positive-selection strategies, we treated pools of mutants with 5-fluorodeoxyuridine (FUDR), which is toxic to parasites through its incorporation into pyrimidine pools. Three lytic cycles after transfection with the library, we split the Cas9-expressing parasites into cultures with or without FUDR (Figure 2B, right). As expected, FUDR-treated cultures recovered slowly, and untreated cultures were passaged 2–3 times over the same period. Measuring the sgRNAs in the two populations revealed that FUDR strongly selected against uracil phosphoribosyltransferase (UPRT) activity, observed as an increased abundance of sgRNAs against UPRT and the highly reproducible phenotype score for the gene (Figure 2D & E). Since loss of UPRT—a component of the pyrimidine salvage pathway—is known to confer FUDR resistance (Donald and Roos, 1995), this experiment demonstrates the power of this approach to rapidly and efficiently identify positively selected mutants from a T. gondii population.
A Genome-Scale Genetic Screen Identifies Fitness-Conferring Genes in T. gondii
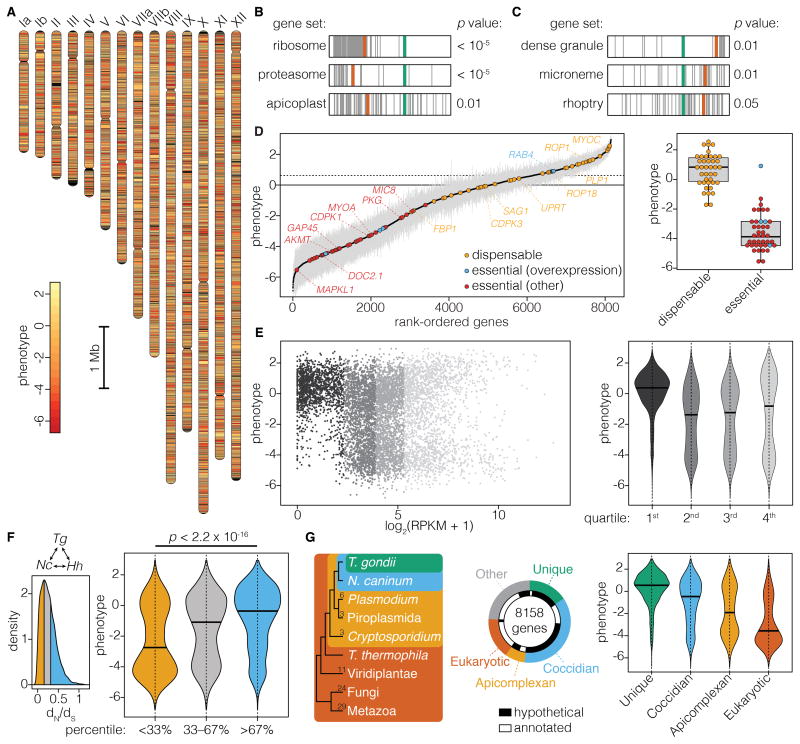
Loss of sgRNAs from a population of mutants can serve to identify genes that contribute to cellular fitness (Wang et al., 2015). In the context of our T. gondii screen, the changes in sgRNA representation observed after the third lytic cycle provided a convenient measure of a gene’s contribution to fitness. This time point resembled the gene rankings of later cycles (Figure 2F) while minimizing the chance of stochastic guide loss. We calculated the mean phenotype score for each parasite gene from four biological replicates of the screen. Genes that contribute to parasite fitness, represented by negative scores, were distributed throughout the genome and did not segregate by gene length or position on the chromosome (Figure 3A). Gene set enrichment analysis (GSEA) (Croken et al., 2014; Subramanian et al., 2005) showed that genes predicted to be essential, like those encoding ribosomal and proteasomal constituents, were enriched in low phenotype scores (Figure 3B). Genes that encode components of the apicoplast—a plastid common to most apicomplexans—showed a similar enrichment, in accordance with the essential metabolic functions performed by this organelle (Seeber and Soldati-Favre, 2010). In contrast, specialized secretory organelles like the micronemes, dense granules, and rhoptries had fewer genes with low phenotype scores, possibly reflecting functional redundancy or dispensability in cell culture (Figure 3C).
Figure 3. A Genome-Scale Screen Measures the Contribution of Each Parasite Gene to Fitness in Human Fibroblasts.
(A) Diagram of T. gondii chromosomes with genes colored according to phenotype.
(B–C) Significantly enriched (B) or depleted (C) gene sets identified by GSEA. Genes belonging to each category (gray) are plotted according to their rank in the screen, relative to the maximum enrichment score (red) and zero phenotype (green). Phenotype scores for a given set were compared to the entire set by a Komolgorov-Smirnov test (FDR corrected) to calculate the p values.
(D) T. gondii genes rank-ordered based on their phenotype. Genes previously reported are highlighted, indicating whether they are dispensable (yellow), or indispensable as inferred from overexpression (blue) or another method (red). Dotted line represents the median phenotype score for the dispensable genes. Mean ± SEM for n = 4 independent experiments.
(E) Correlation of phenotype scores to gene expression based on maximum RPKM values. The distribution of phenotypes in each expression quartile are plotted in the violin graph. Bars indicate the group median.
(F) Analysis of selective pressure for 5,897 syntenic genes found in all three coccidian genomes (Tg, T. gondii; Nc, N. caninum; Hh, H. hammondi). Histogram shows the distribution of dN/dS values, highlighting the top and bottom third. Genes binned according to dN/dS show higher phenotype scores for genes under purifying selection (orange). Bars indicate the group median. The distributions were compared using a Kolmogorov-Smirnov test.
(G) Correlation between phenotype scores and depth of conservation. The phylogenetic relationship between T. gondii and the other genomes used in the analysis is illustrated by the dendrogram, with number of genomes in each taxon indicated. The proportion of T. gondii genes and, within this, the proportion that are functionally annotated, are shown for each category. The distribution of phenotype scores for each category is plotted. Bars indicate the group median.
See also Figures S1 and S2, and Tables S1, S2 and S3.
We analyzed the screen results for 81 genes previously reported to be either dispensable or essential for T. gondii growth in human fibroblasts (Table S1). The two groups of genes were clearly segregated on the basis of their phenotype scores (p = 6.7 × 10−16), with lower scores for the essential genes (Figure 3D). The most prominent outlier was RAB4, which appeared to be essential based on overexpression of a dominant-negative allele (Kremer et al., 2013). However, we readily obtained RAB4 knockouts that grew normally (Figure S1), demonstrating its dispensability in cell culture. We therefore excluded RAB4 and, for consistency, other genes classified by overexpression experiments from subsequent analyses. To predict which genes might contribute to parasite fitness, we compared the phenotype score and distribution of sgRNAs for each gene to the values of 40 known dispensable genes. Using ten-fold cross validation on the set of control genes, we estimate this method can classify genes with >95% accuracy. Based on these results, we expect approximately 40% of T. gondii genes significantly contribute to parasite fitness under the conditions tested.
To further classify the fitness-conferring genes, we compared our predictions to other measures of gene function. Genes that are not expressed during the examined developmental stage are more likely to appear dispensable. Accordingly, only 6.9% of the fitness-conferring genes were found in the lowest quartile of expression, in contrast to 38.6% of genes predicted to be dispensable (Figure 3E). Genes under purifying selection are also more likely to be essential (Jordan et al., 2002). Low ratios of non-synonymous to synonymous mutation rates (dN/dS) are consistent with purifying selection. Comparison of syntenic genes between related species or other T. gondii strains revealed the expected enrichment for low phenotype scores among genes with low dN/dS values (Figure 3F and Figure S2). As an extension of the same principle, genes found in a greater proportion of eukaryotic genomes are more likely to be essential. To test this prediction, we assessed the depth of conservation of T. gondii genes using ortholog groupings of 79 eukaryotic genomes available through OrthoMCL DB (Chen et al., 2006). The distribution of phenotype scores within each category followed the predicted trend, which correlated depth of conservation with contribution to fitness and functional annotation (Figure 3G). The strong agreement of our results with published observations and expected trends allows us to confidently predict which genes will contribute to parasite fitness.
Functional Characterization of Fitness-Conferring Genes Conserved in Apicomplexans
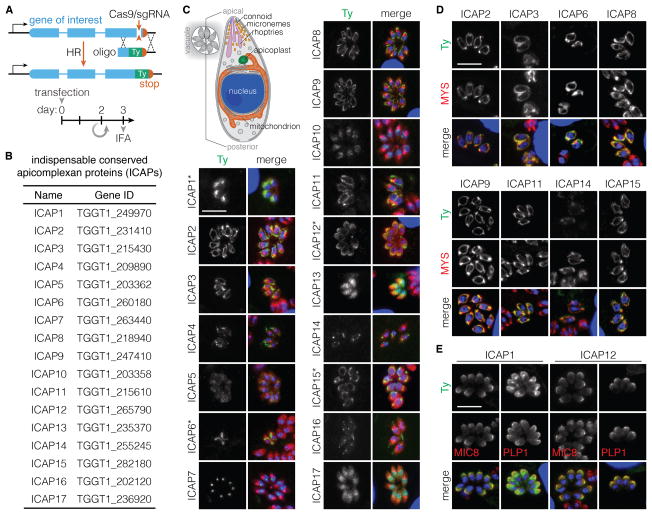
We focused our efforts on the ~200 fitness-conferring genes that lacked functional annotation and were only present in apicomplexans, which we called indispensable conserved apicomplexan proteins (ICAPs). We examined the subcellular localization of 28 ICAPs using CRISPR-mediated endogenous tagging to introduce a C-terminal Ty epitope into each targeted gene (Figure 4A). Three days post transfection, ~60% of the populations displayed Ty expression in 5–20% of parasite vacuoles (Figure 4B–C). Several proteins displayed characteristic structures including secretory vesicles like the micronemes (ICAPs 1 & 12), organelles like the mitochondrion (ICAPs 2, 3, 6, 8, 9, 11, 14 & 15), and compartments like the nucleolus (ICAP7) and conoid (ICAP16) (Figure 4C). Overlap with a known marker of the mitochondrion (Macrae et al., 2012) confirmed the localization of the putative mitochondrial ICAPS (Figure 4D). Both micronemal proteins co-localized with the microneme markers MIC8 (Kessler et al., 2008) and PLP1 (Kafsack et al., 2009) (Figure 4E). ICAP1 was shown to be essential for regulated exocytosis during the preparation of this work (Bullen et al., 2016), confirming our predictions regarding its importance and localization.
Figure 4. Subcellular Localization of Indispensable Conserved Apicomplexan Proteins.
(A) CRISPR was used to introduce a C-terminal Ty tag into the endogenous locus of individual genes through homologous recombination (HR) following the indicated timeline.
(B) List of successfully tagged indispensable conserved apicomplexan proteins (ICAPs), numbered according to their phenotype scores from lowest (ICAP1) to highest (ICAP17).
(C) Three days after transfection, intracellular parasites were fixed and stained for Ty (green), ACT1 (red), and DAPI (blue). * denotes fixation in methanol instead of formaldehyde. Scale bar = 10 μm. The diagram illustrates the relative position of various organelles within the parasite.
(D–E) Colocalization ICAPs (green) with a mitochondrial marker (MYS; red) (D), or micronemal proteins (MIC8 or PLP1; red) (E). Nuclei stained with DAPI (blue) are shown in the merged image. Scale bar = 10 μm.
See also Table S2.
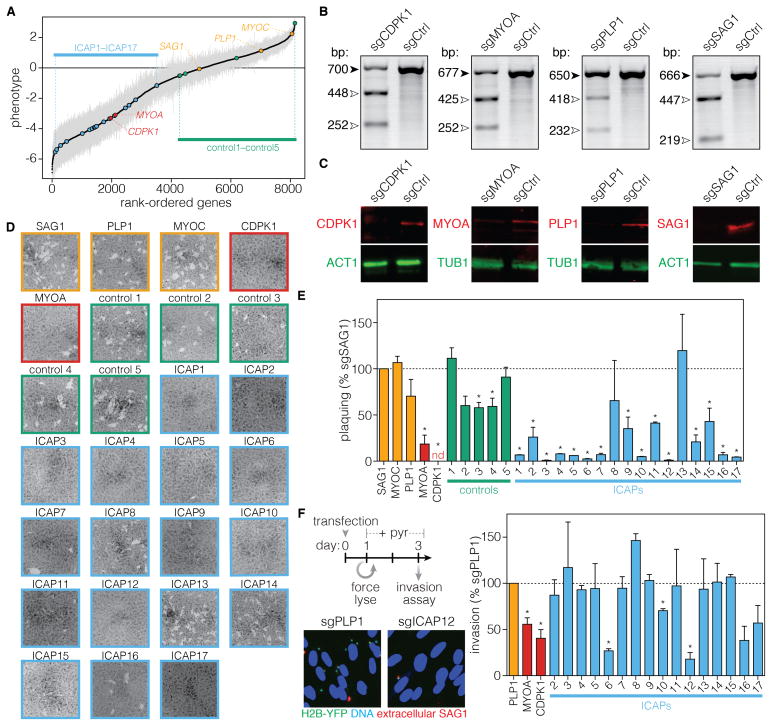
To assist the functional characterization of ICAPs, we engineered parasites that stably expressed both Cas9 and a nuclear YFP marker (H2B-YFP) (Hu et al., 2004) for fluorescence microscopy. This strain exhibited a high rate of sgRNA-mediated gene disruption, comparable to the strain used in the screen (Figure S3). As controls, we selected several genes known to be either indispensable (MYOA & CDPK1) or dispensable (SAG1, PLP1, & MYOC), and five uncharacterized genes predicted to be dispensable by the screen (controls 1–5, Figure 5A). Examining populations of parasites three days after transfection with guides against selected controls showed high rates of on-target mutations (Figure 5B) and significant loss of the target proteins (Figure 5C). Therefore, we can study the effect of a given sgRNA without isolating clonal populations, allowing us to analyze a large set of candidate genes in arrayed, secondary screens.
Figure 5. Functional Characterization of Indispensable Conserved Apicomplexan Proteins.
(A) Position of analyzed genes within the phenotypic ranking of all T. gondii genes. Known indispensable (red) or dispensable (yellow) genes used as controls are indicated. The ICAPs (blue) and uncharacterized genes predicted to be dispensable (green) are numbered in ascending order according to their rank.
(B–C) Gene disruption observed at a population level three days after transfection with various control constructs. Disruption of the target locus is observed by Surveyor assay comparing, for each locus, the specific sgRNA to an irrelevant sgRNA against the dispensable gene MYOC (B). Loss of the target proteins (red) is observed in samples treated with the targeting sgRNA but not the control, while the loading controls (green) remain unchanged (C).
(D–E) Plaque assays performed immediately following transfection with sgRNAs targeting ICAPs or control genes (D). The number of plaques observed for disruption of each gene relative to the sgRNA against SAG1 (E). Mean ± SEM for n = 2 independent experiments, * FDR-adjusted p < 0.1 relative to the control.
(F) Secondary screen for genes involved in invasion. The period of intracellular growth prior to phenotypic changes was extended by forced release and passaging the day after transfection. Invasion was assayed after the subsequent lysis, and calculated relative to PLP1 disruption. Mean ± SEM for n = 2 independent experiments, * FDR-adjusted p < 0.1 relative to the control. Representative immunofluorescence images are shown.
As an initial measure of gene function, we analyzed plaque formation immediately after transfection with each specific sgRNA construct. Plaques are formed as infection originating from single parasites spreads to adjacent cells clearing a portion of the monolayer, thereby reflecting parasite viability and competency over several lytic cycles. This resulted in reproducible plaque numbers for all sgRNAs against genes known or predicted to be dispensable (Figure 5D–E). In contrast, sgRNAs against known essential proteins and most of the ICAPs led to formation of small or significantly fewer plaques (Figure 5D–E). These experiments confirm our screen’s results and identify several previously uncharacterized genes predicted to be essential for the T. gondii lytic cycle.
Invasion of host cells is a central feature of the apicomplexan life cycle. To identify unknown components of the invasion machinery, we investigated the role of ICAPs in this process. The effect of each sgRNA was measured three days post transfection relative to the disruption of PLP1, which is known to be dispensable for invasion (Kafsack et al., 2009) (Figure 5F). MYOA and CDPK1 served as positive controls based on their documented phenotypes (Lourido et al., 2010; Meissner, 2002). A quarter of the genes tested appeared to impact parasite invasion (Figure 5F). Although this assay could be affected by defects in extracellular survival, slow protein turnover, or sgRNA efficiency, it provides a rapid means to identify candidate invasion factors. ICAP12 had the strongest effect on invasion, which, in light of its micronemal localization, motivated a more detailed characterization.
ICAP12 Is an Invasion Factor Conserved Throughout the Apicomplexa
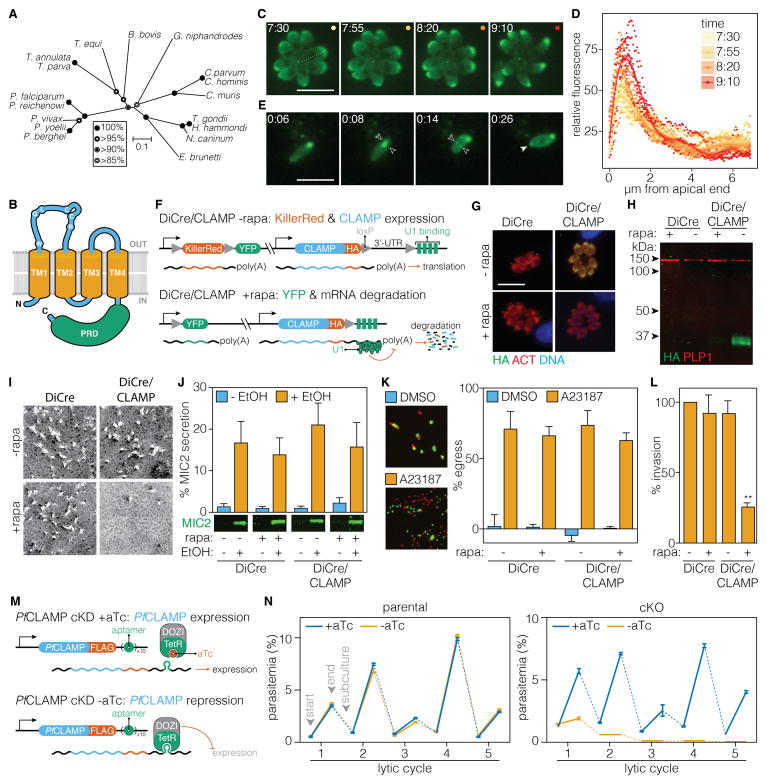
ICAP12 orthologs were present in all available apicomplexan genomes and their alignment recapitulated the known relationship between the species (Figure 6A). Topology prediction supports a model with four transmembrane domains and cytoplasmic N and C termini (Figure 6B). The transmembrane domains and extracellular loops are more conserved than the C-terminal proline-rich domains (Figure S4). No related sequences could be identified outside the Apicomplexa. However, hidden Markov model-based searches suggest structural similarity between ICAP12 and the mammalian tight-junction proteins Claudin-15 and Claudin-19. Based on these features and its localization we named ICAP12 “claudin-like apicomplexan microneme protein” (CLAMP).
Figure 6. CLAMP Mediates T. gondii Invasion and is Essential for the P. falciparum Asexual Cycle.
(A) Neighbor-joining tree showing the phylogenetic relationships of CLAMP homologs in diverse apicomplexans. Bootstrap values for 10,000 trials are displayed. (B) Inferred topology of CLAMP highlighting transmembrane domains (orange) and the proline-rich domain (green). See also Figure S4.
(C–E) CLAMP-mNeonGreen localization during egress and invasion. Intracellular parasites expressing CLAMP-mNeonGreen were stimulated to egress with A23187 (C). Relative fluorescence across the length of each parasite (dotted line) is plotted for the four time-points shown. Lines are polynomial regressions ± 95% CI (D). The localization was also monitored during invasion (E). The position of the moving junction is indicated with paired open arrows. Solid arrowhead indicates a punctum of mNeonGreen at the posterior of the parasite appearing immediately after invasion. Time is expressed in min:sec following addition of the compound (C–D) or initiation of invasion (E). Scale bar = 10 μm.
(F) Diagram of the DiCre/CLAMP strain showing how after rapamycin (rapa) treatment the reporter locus switches from expressing KillerRed to expressing YFP, and CLAMP mRNA degradation is induced.
(G–H) A 2 hour treatment with rapa is sufficient to induce CLAMP degradation as demonstrated by immunofluorescence microscopy 24 hours later (G) or immunoblotting two days later (H). The parental strain (DiCre) is included as a control.
(I–L) The DiCre/CLAMP strain or its parental stain (DiCre) were treated as above. Parasites were harvested and phenotypically assayed for plaque formation (I), microneme secretion (J), egress (K) or invasion (L). Secretion was measured as the percent of total MIC2 present in the parasites (J). Egress was induced with A23187 and compared to a vehicle control (DMSO) over the same period (K). All results are means ± SEM for n = 3 independent experiments, ** p < 0.005 relative to the untreated DiCre strain.
(M) Diagram of the PfCLAMP cKD showing how removing aTc allows the TetR-DOZI regulator to bind and suppress expression.
(N) Growth curves of the parental strain (left) or the cKD (right) ± aTc. Means ± SD for n = 3 technical replicates. See also Figure S5D for an independent replicate.
We tagged the endogenous CLAMP locus with mNeonGreen to study its localization in vivo. The fusion protein concentrated at the apical end of parasites, consistent with micronemal localization (Figure 6C). Micronemes are secreted in response to increased cytosolic Ca2+, which mediates parasite motility, egress and invasion of host cells (Carruthers et al., 1999). We monitored CLAMP localization following stimulation with the Ca2+ ionophore A23187, and observed increased apical fluorescence prior to egress (Figure 6C–D & Movie S1). Following incubation with host cells, formation of CLAMP foci could be detected at the posterior of most parasites (Figure S5A). We also observed active formation of such foci during invasion (arrowheads, Figure 6E, & Movie S2). This relocalization of CLAMP is similar to what has been observed for other membrane-tethered micronemal proteins (Carruthers and Sibley, 1999; Garcia-Reguet et al., 2000).
To directly examine CLAMP function, we used a conditional gene-silencing method (Pieperhoff et al., 2015). In a strain expressing a rapamycin-dimerizable version of the Cre recombinase (DiCre), we modified the endogenous CLAMP locus to include a C-terminal HA-tag and a floxed 3′ UTR followed by four U1-binding sequences (DiCre/CLAMP, Figure 6F). Rapamycin treatment triggers excision of the 3′ UTR, and U1-mediated mRNA degradation. The strain also carries a reporter that switches expression of KillerRed for YFP upon Cre activation. A two-hour rapamycin treatment during initial infection efficiently down-regulated CLAMP expression (green), as measured by immunofluorescence 24 hours later (Figure 6G) or by immunoblot two days later (Figure 6H). To determine the impact of CLAMP on the lytic cycle, we examined plaque formation following treatment with rapamycin. The treatment did not affect the parental strain (DiCre, Figure 6I). However, CLAMP silencing blocked plaque formation, and the few plaques remaining likely represent the 5–10% of parasites that do not undergo recombination (DiCre/CLAMP, Figure 6I).
To determine the precise defect associated with CLAMP loss, we examined several stages of the lytic cycle. We tested whether CLAMP down-regulation might affect microneme secretion, which can be experimentally triggered by ethanol treatment (Carruthers et al., 1999). Rapid shedding of micronemal adhesins from the parasite surface allows quantification of secretion by measuring protein accumulation in the supernatant. Comparing the relative abundance of secreted MIC2—a micronemal adhesin—demonstrates that CLAMP silencing has no effect on microneme secretion (Figure 6J). We also observed normal motility and egress when intracellular parasites were treated with A23187 (Figure 6K) or the phosphodiesterase inhibitor zaprinast (Movie S3). Following stimulated egress, parasites frequently reinvade adjacent host cells. However, CLAMP silencing rendered reinvasion attempts unsuccessful (Movie S4). These events were characterized by repetitive parasite thrusting motion and deformation. To directly measure this defect, CLAMP was silenced during the growth cycle prior to the assaying invasion. Knockdown of CLAMP had a profound effect on invasion, reducing the number of intracellular parasites by 80% (Figure 6L). These results implicate CLAMP in the cellular events immediately preceding invasion of host cells.
CLAMP is Essential During the Asexual Cycle of Malaria
To test whether the essentiality of CLAMP extends to other apicomplexans, we constructed a conditional knockdown (cKD) of its ortholog in P. falciparum. We tagged the endogenous locus of PfCLAMP with a FLAG epitope tag and ten tandem aptamer sequences, which bind the Tet Repressor protein (TetR) when transcribed (Ganesan et al., 2016). TetR is expressed as a fusion with the translational repressor (DOZI) in the same strain, which suppresses expression of the aptamer-tagged transcript unless anhydrotetracycline (aTc) is added to the media (Figure 6M). We confirmed correct integration of the construct into the PfCLAMP locus by sequencing (Figure S5B–C). To test the effect of CLAMP repression on the asexual cycle of P. falciparum, we passaged the parasites into cultures that either contained or lacked aTc. The different conditions had no effect on the growth of the parental strain (Figure 6N, left). In contrast, withdrawal of aTc from the PfCLAMP cKD led to a rapid and complete block in the asexual cycle (Figure 6N, right). These results demonstrate the essentiality of CLAMP in a second apicomplexan species.
DISCUSSION
We present the first genome-wide functional analysis of an apicomplexan. Using CRISPR/Cas9, we targeted all annotated protein-coding genes in the T. gondii genome to generate mutant populations for screens based on positive or negative selection. This method enabled rapid identification of genes that mediate infection of human fibroblasts, or confer drug sensitivity. Our results agree with published observations, follow expected genomic trends, and provide new robust predictions that identify several essential proteins conserved throughout the phylum. We also demonstrate that this method can easily identify mutants resistant to the antiparasitic compound FUDR. For such special cases where gene disruption can mediate resistance, our method will facilitate the identification of drug-resistance pathways, and provide a complementary approach to mapping spontaneous resistance mutations (Flannery et al., 2013).
Essential apicomplexan adaptations represent ideal targets for therapeutic or prophylactic interventions. However, genes involved in such pathways are difficult to identify. Based on results from our genome-wide screen, we characterized 17 indispensable conserved apicomplexan proteins (ICAPs). Individually disrupted, most ICAPs could be shown to independently contribute to growth in fibroblasts. None of the ICAPs have defined domains or resemble proteins outside the phylum, yet most of them localized to distinct subcellular structures, assuming no detrimental effect of epitope tagging on localization. Of the eight that localized to the mitochondrion, only ICAP3 and ICAP14 have predicted signal peptides that might have suggested their compartmentalization. Nonetheless, the preponderance of mitochondrial ICAPs and their conservation in several branches of the Apicomplexa, suggests that this organelle might serve as a focal point for functions conserved across the phylum, in addition to the currently appreciated genus-specific modifications (Seeber et al., 2008). Two other ICAPs localized to apical secretory vesicles called micronemes, which are more typically associated with specific apicomplexan adaptations. During the preparation of this manuscript, the first, ICAP1 (APH), was shown to be required for microneme secretion (Bullen et al., 2016). The second one, ICAP12, was critically important for invasion, as demonstrated by our secondary screens and subsequent analysis. These data argue for an in-depth examination of other ICAPs, which will likely reveal essential processes conserved among apicomplexans.
We renamed ICAP12 CLAMP to reflect its subcellular localization and structural similarity to mammalian claudins. Conditional silencing demonstrated that CLAMP is specifically required during invasion of host cells, and did not affect microneme secretion or dependent processes. CLAMP is thereby distinct from factors that participate in both gliding motility and invasion (Bargieri et al., 2014). Instead CLAMP resembles factors involved in the formation of the tight junction—or moving junction—through which parasites enter host cells. Knockdown of the micronemal protein MIC8 resembles CLAMP disruption by blocking secretion of the rhoptry neck proteins that anchor the micronemal adhesin AMA1 to the host cell membrane (Kessler et al., 2008). Video microscopy showed CLAMP-deficient parasites repeatedly pushing against the host-cell membrane while failing to initiate invasion, much like P. falciparum parasites unable to form a moving-junction (Treeck et al., 2009). Apposition of the parasite and host-cell membrane is a hallmark of apicomplexan parasitism, mediating discharge of rhoptry contents into host cells and complete or partial invasion (Bargieri et al., 2014). Despite the similarities, previously identified invasion factors are not completely conserved throughout the phylum: MIC8 is restricted to the coccidia; and AMA1 and RON2, which are central components of the moving junction, are absent from early-branching apicomplexans (EupathDB). In contrast, CLAMP homologs are found in all sequenced apicomplexan genomes including early-branching members like cryptosporidians and gregarines. Consistent with its conservation, the ortholog of CLAMP in P. falciparum could be readily identified, and its knockdown leads to a complete inhibition of the asexual cycle, although further work will be necessary to define its precise function. These results argue for a pivotal role of CLAMP in all members of the phylum. Its structural resemblance to mammalian claudins suggests the physical involvement of CLAMP in tight-junction formation during invasion. Claudins are known to engage in homotypic and heterotypic interactions, as well as the formation of paracellular channels that restrict the flow of ions across the tight junction (Krause et al., 2008). Whether CLAMP participates in similar processes remains speculative, but its study might shed light into one of the most conserved features of apicomplexan parasitism.
By screening for genes that confer parasite fitness during growth in human fibroblasts, we provide a baseline for gene function under arguably permissive conditions. Future screens will need to define the genes required during other life stages, in different hosts, under varying nutrient conditions, and in response to immune pressures. The potential to obscure or exacerbate deleterious mutations in pooled screens due to competition with wild-type parasites should be mentioned here. However, this screening format has the advantage of simultaneously determining the fate of hundreds of independently generated mutants for a given gene. As such, they are less influenced by rare compensatory mutations that can confound interpretation of outcomes when using clonal strains (Lamarque et al., 2014; Ma et al., 2008). Studies in yeast have revealed that adaptations can overcome the need for ~9% of genes previously considered essential (Liu et al., 2015). These issues argue for a nuanced view of gene essentiality. Although our experiments demonstrate the strong predictive value of this method, careful follow-up experiments are necessary to fully explore the role of individual genes.
Genome-wide functional analyses have transformed the study of many organisms. We demonstrate the power of this approach to identify genes that contribute to T. gondii fitness during infection of human fibroblasts. Although important adaptations distinguish different parasite genera, this method provides a unique tool to model conserved apicomplexan processes in T. gondii, and its success is demonstrated by the identification of a previously uncharacterized protein essential for the malaria parasite P. falciparum. Coupled with the diverse tools available for genetic and chemical manipulation of T. gondii, the genome-wide screens will provide a framework for the systematic examination of genetic interactions. The unconstrained study of apicomplexan genomes will help us understand their unique biology and broaden the scope of interventions to control these widespread parasitic infections.
Methods and Resources
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Sebastian Lourido (lourido@wi.mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
T. gondii tachyzoites from the strain RH and derived strains were maintained at 37°C with 5% CO2 growing in human foreskin fibroblasts (HFFs) cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum and 10 μg/ml gentamicin. When appropriate, chloramphenicol was used at 40 μM and pyrimethamine at 3 μM (1.5 μM for plaque assays).
P. falciparum parasites of the strain NF54attB (kindly provided by David Fidock) and the derived strain were grown in human erythrocytes (Research Blood Components) at 5% haematocrit under 5% O2 and 5% CO2 in RPMI 1640 media supplemented with 5 mg/ml Albumax II (Life Technologies), 2 mg/ml NaHCO3, 25 mM HEPES pH 7.4, 1 mM hypoxanthine and 50 μg/ml gentamicin.
METHOD DETAILS
Plasmid Design and Construction
To construct pCas9/CAT, the chloramphenicol acetyltransferase (CAT) gene, under the TUB1 promoter and SAG1 3′ UTR (Soldati and Boothroyd, 1993), was amplified using primers P1 and P2 (see Table S2 for a complete list of primers and oligonucleotides), and ligated into the PciI and XbaI sites of pU6-Universal (Addgene, #52694). To generate pCas9/decoy, oligonucleotides P3 and P4 were hybridized and cloned into the BsaI sites of pU6-Universal. H2B-YFP (Hu et al., 2004) was excised with NsiI and NotI, and used to replace Cas9 in pCas9/decoy by ligating the fragment into the same restriction sites to generate pH2B-YFP/decoy.
The plasmid for sgRNA expression was constructed by amplifying the pyrimethamine-resistance cassette (Donald and Roos, 1993) with primers P5 and P6, and cloning it into the NsiI and SbfI sites of pU6-Universal. Three BsaI sites in DHFR were eliminated using the QuikChange multi-site directed mutagenesis kit (Agilent Technologies) with primers P7, P8, and P9 resulting in the plasmid pU6-DHFR. The SAG1 protospacer, encoded by primers P10 and P11, was cloned into pU6-DHFR as described above for the decoy protospacer. Guides for gene disruption (P14–P63) and ICAP tagging (P80–P114) were synthesized for Gibson Assembly (New England Biolabs) into pU6-DHFR or pU6-Universal, respectively. Such guides and their reverse complements were hybridized, and Gibson cloned into their respective vectors linearized with BsaI. Guides to knock out RAB4 were similarly generated by cloning sgRNAs against the 5′ (P160 and P161) and 3′ (P162 and P163) ends of the coding sequence into pU6-Universal.
For C-terminal HA-FLAG epitope tagging and U1 mediated knockdown, a 3′ flank of the CLAMP gene, upstream of the stop codon, was amplified by PCR with primers P149 and P150 and inserted into pLIC-HA-FLAG-(3′UTRSAG1-pDHFR-HXGPRT-5′UTRDHFR)flox-4xU1 (Pieperhoff et al., 2015) by ligation-independent cloning (Huynh and Carruthers, 2009) to generate pCLAMP-U1.
The plasmid for conditional knockdown of CLAMP in P. falciparum was generated by amplifying the 5′ and 3′ homology regions from the CLAMP locus using primers P152–P155. P155 included an sgRNA targeting the 3′ end of the CLAMP locus, placed under the regulation of the T7 promoter in the final construct. The fragments were cloned by Gibson assembly into the plasmid containing the aptamers, and the Renilla luciferase-blasticidin deaminase fusion separated from the TetR-DOZI fusion by a T2A self-cleaving peptide, to generate pPfCLAMP-cKD/TetR-DOZI (Ganesan et al., 2016).
Library Design and Construction
Ten guides were designed against each gene in the 14 annotated chromosomes of the T. gondii GT1 genome (release 28, ToxoDB.org), according to published guidelines (Wang et al., 2014). Briefly, selection of sgRNAs was weighted based on targeting of exons, number of potential off-target sequences, and overlap between sgRNAs for a given gene. A ‘G’ was prepended to any sgRNAs that did not start with one to ensure proper RNA polymerase III initiation. Guides against DHFR and HXGPRT were removed from the library to preclude interference with drug-resistance markers. The guide library was synthesized by CustomArray Inc., and includes guides flanked by sequences for Gibson Assembly into pU6-DHFR. The sgRNA library was amplified using primers P12 and P13, and Gibson cloned into pU6-DHFR linearized with BsaI. Assembled constructs were transformed into Mega-X DH10B electrocompetent Escherichia coli (Life Technologies), allowed to recover for 1 h, and grown overnight with 100 μg/ml ampicillin, prior to large-scale plasmid isolation (Macherey Nagel) or frozen storage. Cloning and electroporation efficiencies were monitored to ensure proper library coverage.
T. gondii Strain Generation
All transfections were performed as described previously with a square-wave electroporator (Sidik et al., 2014). RH/Cas9 and RH/Cas9/H2B-YFP were generated by co-transfecting RH with 50 μg each of pCas9/CAT and pCas9/decoy, or pH2B-YFP/decoy and pCas9/CAT, respectively. Stable transgenic strains were selected with 40 μM chloramphenicol. Single clones were isolated by limiting dilution and screened for the presence of Cas9 using immunofluorescence and immunoblotting against the triple-FLAG tag. The decoy protospacer was amplified by PCR from RH/Cas9 genomic DNA using primers P64 and P65, and confirmed by sequencing.
To generate the RAB4 knockout, the pyrimethamine-resistance cassette was amplified from pU6-DHFR with primers P164 and P165 to contain homology regions to replace the entire open reading frame. The amplicon was cotransfected along with two pU6-Universal plasmids carrying guides against the 5′ and 3′ end of the coding sequence into RH/ΔKU80 parasites (Huynh and Carruthers, 2009). Stable transformants were selected for with pyrimethamine and clones were isolated by limiting dilution. Correct integration of the pyrimethamine-resistance cassette into the RAB4 locus was confirmed using P166 and P169 to amplify the 5′ junction, and P167 and P168 to amplify 3′ junction. Deletion of the RAB4 locus was assessed using P170 and P171 to amplify a portion of the open reading frame. Fitness of the RAB4 knockout was determined by plaque assay, as described below.
To generate the CLAMP conditional knockdown (DiCre/CLAMP), 25 μg of pCLAMP-U1 were linearized with MfeI for efficient homologous recombination and transfected into DiCre/Δku80/KillerRedflox-YFP (DiCre) (Pieperhoff et al., 2015). Following selection with 25 μg/ml mycophenolic acid and 50 μg/ml xanthine, individual clones were obtained by limiting dilution.
Pooled CRISPR Screens
For each biological replicate, 400 μg of the sgRNA library were linearized with AseI, dialyzed against water, and transfected into approximately 4 × 108 RH or RH/Cas9 parasites divided between 8 separate cuvettes. Transfections were used to infect 8 T-175 flasks with confluent HFF monolayers, and pyrimethamine was added 24 hours later. The parasites were allowed to egress naturally from host cells two days after infection, isolated by filtration, and 1.5 × 108 parasites were passaged onto 8 T-175 flasks with fresh monolayers. The remaining parasites (~107) were pelleted and stored at −80°C for analysis. This process was repeated again 5 days and 7 days post-transfection. For the drug-resistance screens, 5 μM 5-fluorodeoxyuridine (FUDR; Sigma) was added to 1.2 × 107 parasites collected on day 7 post transfection, and parasites were cultured until their first lysis. Untreated mutant pools were maintained in parallel for the duration of FUDR selection. Parasite DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) and integrated sgRNA constructs were amplified using a nested PCR with primers P74 and P75 followed by P76 and P77. The resulting libraries were sequenced on a HiSeq 2500 (Illumina) with single-end reads using primers P150 and P151.
RT-PCR
RAB4 expression was assessed in both the parental and ΔRAB4 strains by RT-PCR. Total RNA was prepared from isolated parasites using the RNeasy Plus Kit (Qiagen). The ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs) was used to prepare cDNA, and reverse transcriptase (RT) was excluded from the control reactions. PCR was performed on the cDNA samples with primers specific for RAB4 (P174 and P175) or the ACT1 control (P172 and P173).
Functional Analysis of ICAPs and Controls
1.25 × 107 RH/Cas9 or RH/Cas9/H2B-YFP parasites were transfected with 50–100 μg of pU6-DHFR containing guides against different ICAPs or controls. If aiming for integration, plasmids were linearized with AseI and dialyzed against water prior to transfection. Transfected parasites were seeded on HFFs at a multiplicity of infection of 10, and pyrimethamine was added 24 hours after transfection. Parasites transected with sgSAG1 were allowed to egress from host cells two days after transfection, and used to infect host cells seeded on coverslips. SAG1 loss was quantified by immunofluorescence 24 hours after infecting coverslips, which was equivalent to three days after transfection.
To screen for ICAPs that participate in invasion, parasites were released from host cells 24 hours after transfection by passage through a 27.5-gauge needle, and used to infect fresh monolayers, thus extending the time allowed for protein depletion. To test invasion, freshly lysed parasites were suspended in invasion media (DMEM supplemented with 1% FBS, 20 mM HEPES, pH 7.4). 2 × 105 parasites per well were added to confluent HFF monolayers grown in 96-well plates and centrifuged at 290 × g for 5 minutes. Invasion was allowed to proceed for 10 minutes at 37°C, before the monolayers were fixed in 4% formaldehyde for 20 minutes on ice. Extracellular parasites were stained using mouse-anti-SAG1 (Burg et al., 1988) conjugated to Alexa-Fluor-594 (Life Technologies), and host cell nuclei were stained using Hoechst (Santa Cruz). Images were acquired using a Cytation 3 imager (BioTek), and analyzed using custom FIJI (Schindelin et al., 2012) macros to count the number of parasites and host-cell nuclei.
ICAP Tagging
CRISPR-mediated C-terminal Ty tagging was performed as previously described (Sidik et al., 2014). 30 μg of a repair oligonucleotide containing an in-frame Ty epitope (Bastin et al., 1996) flanked by homology regions to the C terminus of each gene (P115–P148) were co-transfected with 100 μg of pU6-Universal carrying the appropriate sgRNA into TATi/ΔKU80 parasites (Sheiner et al., 2011). Transfected parasites were cultured until their first lysis and used to infect confluent HFF monolayers grown on coverslips. Localization of the Ty-tagged ICAPs was determined 24 post infection by immunofluorescence microscopy.
C-terminal tagging of CLAMP with mNeonGreen, was accomplished by amplifying the fluorescent protein coding sequence with primers P78 and P79 from a template plasmid (kindly provided by Ke Hu). 30 μg of the resulting product were co-transfected into TATi/ΔKU80 parasites along with 100 μg of pU6-Universal carrying an sgRNA against the C-terminal sequence of the endogenous CLAMP locus. Fluorescent parasites were isolated by FACS two days post transfection, and cloned by limiting dilution. Correct integration of mNeonGreen into the CLAMP locus was confirmed by sequencing.
CLAMP Phylogeny and Topology Predictions
CLAMP homologs were readily identified by BLAST searches against all sequenced apicomplexan genomes (EupathDB.org). Sequences were curated for Babesia and Theileria spp. to correct errors in the gene models. Alignment was performed using ClustalW (Larkin et al., 2007) and the phylogenetic tree was generated by neighbor-joining excluding positions with gaps. Bootstrap values were calculated for 10,000 trials. A hidden Markov model-based search was performed for the alignment using HHpred (Meier and Söding, 2015). Significant structural similarity was found between CLAMP and Claudin-19 (p = 4.1 × 10−9), Claudin-15 (p = 7.3 × 10−9), and the voltage-gated calcium channel γ subunit 15 (p = 10−10), which all belong to the same tetraspan family (Simske, 2013). Structural similarity between CLAMP and Claudin-19 (95% confidence) was also found using Phyre2 (Kelley et al., 2015). Topology prediction was performed against several representative orthologs with CCTOP (Dobson et al., 2015) and in all cases arrived at the same prediction of four transmembrane domains with cytoplasmic N and C termini (Figure S4).
CLAMP Conditional Knockdown
DiCre/CLAMP parasites were treated with 50 nM rapamycin or vehicle control for 2 hours, then cultured for 48 hours before phenotypic analysis by immunoblot, plaque formation, MIC2 secretion, or invasion assay.
Immunofluorescence Microscopy and Immunoblotting
Mouse monoclonal antibodies were used to detect SAG1 (clone DG52; Burg et al., 1988), TUB1 (clone 12G10, Developmental Studies Hybridoma Bank at the University of Iowa), MIC2 (clone 6D10; Achbarou et al., 1991), Ty-tagged proteins (clone BB2; Bastin et al., 1996), FLAG-tagged proteins (clone M2; Sigma-Aldrich), and HA-tagged proteins (clone 16B12; BioLegend). CDPK1 was detected using the alpaca-derived nanobody 1B7 (Ingram et al., 2015). Rabbit polyclonal sera were used to detect MyoA (Frénal et al., 2014), PLP1 (Kafsack et al., 2009) and ACT1 (Dobrowolski et al., 1997).
Prior to immunoblotting, DiCre and DiCre/CLAMP parasites were suspended in lysis buffer (137 mM NaCl, 10 mM MgCl2, 1% Triton X-100, Halt protease inhibitors [Thermo Fisher], 20 mM HEPES pH 7.5). An equal volume of 2X Laemmli buffer (4% SDS, 20% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue, 120 mM Tris-HCl pH 6.8) was added, and the samples were heated to 37°C for 10 minutes prior to separation of proteins by SDS-PAGE. After transferring separated proteins to nitrocellulose, the membrane was incubated in stripping buffer (100 mM β-mercaptoethanol, 2% sodium dodecyl sulfate, 62.5 mM Tris-HCl, pH 6.8) for 15 minutes at 50°C, then washed twice in TBS-T (20 mM Tris-HCl, 138 mM NaCl, 0.1% Tween-20, pH 7.5). Samples for all other blots were prepared similarly, but boiled 10 minutes prior to separation by SDS-PAGE, and membranes were not incubated in stripping buffer. β-mercaptoethanol was not included when probing for SAG1.
Intracellular parasites were fixed with either methanol at 4°C for 2 min, or 4% formaldehyde for 10 minutes. Staining was performed with the antibodies described above and detected with Alexa-Fluor-labeled secondary antibodies. Formaldehyde-fixed samples were permeablized with 0.25% Triton X-100 in PBS for 8 minutes. Nuclei were stained with Hoechst (Santa Cruz) or DAPI (Life Technologies) and coverslips were mounted in Prolong Diamond (Thermo Fisher). Images were acquired using an Eclipse Ti epifluorescence microscope (Nikon) using the NIS elements imaging software. FIJI was used for image analysis, and Adobe Photoshop for image processing.
Surveyor Assays
Pools of mutant parasites were suspended in PBS containing 200 μg/ml Proteinase K (Sigma-Aldrich) and 1X Taq PCR Buffer (Sigma-Aldrich) and heated to 37°C for 1 hour, 50°C for 2 hours, and 95°C for 15 min. Mutated regions were then amplified using primers for the SAG1 locus (P66 and P67), the MyoA locus (P68 and P69), the PLP1 locus (P70 and P71) or the CDPK1 locus (P72 and P73). Surveyor reactions were performed using a kit according to the manufacturer’s instructions (Integrated DNA Technologies).
Plaque Formation
4 × 106 RH/Cas9/H2B-YFP parasites were transfected with 50 μg of pU6-DHFR encoding guides against ICAPs or controls. 2,000 transfected parasites were added to HFF monolayers in 6-well plates. 1.5 μM pyrimethamine was added one day post-transfection. Ten days post transfection, the monolayers were rinsed with PBS, fixed in 95% ethanol for 10 minutes and stained with crystal violet (2% crystal violet, 0.8% ammonium oxalate, 20% ethanol) for 5 minutes. 500 parasites per well were used to analyze the effect of CLAMP or RAB4 on plaque formation over the course of 8 days.
Microneme Secretion
Microneme secretion assays were performed as previously described (Lourido et al., 2012). 2 × 107 DiCre or DiCre/CLAMP parasites were suspended in DMEM, then treated with 3% FBS with or without 1% ethanol for 10 minutes at 37°C, 5% CO2. Supernatants were collected by centrifugation 10 minutes at 400 g, 4°C. Proteins were separated by SDS-PAGE, and secreted MIC2 was quantified and normalized to MIC2 levels in total lysates measured by immunoblotting.
Egress Assays
4 × 106 DiCre or DiCre/CLAMP parasites were treated with 50 nM rapamycin or vehicle upon infection of HFF monolayers in 96-well plates. Rapamycin was removed after two hours, and parasites were allowed to grow for 24–30 hours. Egress was induced with 1 μM A23187 (EMD Millipore) for 10 minutes. Knockdown parasites in the rapamycin-treated samples were identified by expression of the YFP reporter. The number of intact vacuoles before or after the induction of egress was quantified. Only YFP-expressing vacuoles were counted for the rapamycin-treated samples.
Invasion Assays
DiCre and DiCre/CLAMP parasites were suspended in invasion media and 5 × 106 parasites were added per well of a 24-well plate containing HFF monolayers seeded on coverslips. Invasion was allowed to proceed at 37°C with 5% CO2 for 20 minutes. HFF cells and parasites were fixed with 4% formaldehyde for 20 minutes on ice. Extracellular parasites were stained with Pacific Blue-conjugated anti-SAG1 prior to permeabilization, and all parasites were stained with Alexa-Fluor-594-conjugated anti-SAG1 after permeabilization with 0.25% Triton. The average number of host cells per field of view was obtained from coverslips prepared and processed in parallel and stained with Hoechst. The number of invaded parasites per fields were manually counted and normalized to the number of host cells in the same area. CLAMP-mNeonGreen parasites were prepared similarly and allowed to invade for 30 minutes before being fixed with 4% formaldehyde for 20 minutes on ice. Extracellular parasites were stained with Alexa-Fluor-594-conjugated anti-SAG1.
Video Microscopy
To capture egress, DiCre or DiCre/CLAMP parasites were prepared as described for the egress assays in glass-bottom 35 mm dishes (MatTek Corp.). Parasites were stimulated to egress with 1 μM A23187 or 500 μM zaprinast, and recorded at 2–5 frames per second for ten minutes, using an Eclipse Ti microscope (Nikon) with an enclosure heated to 37°C. The same setup was used to capture invasion, with the exception that freshly lysed parasites were added directly to monolayers under observation.
P. falciparum Strain Generation and Analysis
The P. falciparum NF54attB strain (kindly provided by David Fidock) was modified to generate a strain NF54Cas9+T7 Polymerase, which contained Cas9 and T7 RNA polymerase expression cassettes integrated into the attB site. Linearized pPfCLAMP-cKD/TetR-DOZI was transfected into NF54Cas9+T7 Polymerase. 50 μg of plasmid were used per 200 μl packed red blood cells (RBCs), adjusted to 50% haematocrit, and electroporated as previously described (Ganesan et al., 2016). Transfected parasites were selected with a combination of 2.5 μg/ml Blasticidin S and 2.5 nM WR99210 beginning 4 days after transfection. The PfCLAMP cKD strain was maintained in 0.5 μM anhydrotetracycline (aTc). Correct integration of the construct was confirmed by PCR and sequencing using primer pairs P156/P157 and P158/P159.
To analyze P. falciparum growth upon CLAMP down regulation, parasites were synchronized to rings using 0.5 M alanine in 10 mM HEPES (pH 7.4), adjusted to 1% parasitemia, and seeded in triplicate wells of a 12-well plate at 2% hematocrit in 5 ml of media with or without aTc. The parental and PfCLAMP cKD strains were treated in parallel. Expansion was measured over six lytic cycles. Samples were collected after each cycle to measure parasitemia by incubating the cells with a 1:5000 dilution of SYBR Green I (Thermo Fisher) for 15 minutes at 37 °C, prior to flow-cytometry on an Accuri C6 instrument (BD Biosciences). Following the analysis, all cultures were subcultured using the same dilution factor, as required to maintain the pre-invasion parasitemia of the parental lines at 1%, and avoid over-expansion of the cultures. After subculturing, the pre-invasion parasitemia was directly measured as indicated above. Parasite growth was expressed as percent parasitemia at the start and end of each lytic cycle.
QUANTIFICATION AND STATISTICAL ANALYSIS
Bioinformatic Analysis of the Screening Results
Sequencing reads were aligned to the sgRNA library. The abundance of each sgRNA was calculated and normalized to the total number of aligned reads. Guides that were not found were assigned a pseudo-count corresponding to 90% of the lowest value in that sample. Only guides whose abundance was above the 5th percentile in the original plasmid preparation of the sgRNA library were taken into account for subsequent analyses. For FUDR selection experiments, the log2 fold-change between treated and untreated samples was calculated for each sgRNA, whereas negative selection experiments used the plasmid preparation for comparison. The “phenotype” score for each gene was calculated as the mean log2 fold change for the top five scoring guides, which minimized the effect of stochastic losses and decreased the error between biological replicates. The mean phenotype score for each gene in four replicates of the screen is reported. Fitness-conferring genes were identified by comparison to 40 control genes known to be dispensable for parasite growth in fibroblast (see Table S1). For a given gene, the four biological replicates were compared to the mean phenotype of the controls using a one-sided t test, and the log2 fold changes for the sgRNAs against that gene were compared to the sgRNAs against the controls using a one-sided Mann-Whitney U test. The p values for each test were corrected using the Benjamini-Hochberg method. Genes were considered fitness-conferring if they met a significance threshold of 0.05 for both tests. Ten-fold cross-validation was performed using the set of previously described essential and dispensable genes (Table S1). In each trial, the test sample was compared using the statistical tests described to the control genes in the training set. The cross-validation was performed 100 times to estimate the error rate.
Gene-set enrichment analysis was performed as previously described (Subramanian et al., 2005), using gene sets specifically curated for T. gondii (Croken et al., 2014). Gene expression data (kindly provided by David S. Roos and Maria Alejandra Diaz-Miranda, available through ToxoDB) consisted of RNA samples collected at several time points following infection with T. gondii strain GT1 tachyzoites, and analyzed by RNAseq. The maximum log2(RPKM + 1) was used for the analysis and compared to the mean phenotype score for each gene. To compare the phenotype scores with the rate of evolution, syntenic orthologs that did not display copy number variation were obtained from ToxoDB release 26 for T. gondii GT1, Neospora caninum Liverpool, and Hammondia hammondi HH34. dN/dS ratios were determined as previously described (Lorenzi et al., 2016). Briefly, average dN/dS values were calculated according to Goldman Yang (Goldman and Yang, 1994), following alignment of cDNA sequences based on their protein sequence by ClustalW (Larkin et al., 2007). The distribution of phenotype scores in the top and bottom third of the dN/dS distribution were compared by a Komolgorov-Smirnov test. Depth of conservation of T. gondii genes was estimated using ortholog groupings of 79 eukaryotic genomes available through OrthoMCL DB release 5 (Chen et al., 2006). Assignment to the different levels of conservation was performed by asking whether an ortholog was present in at least one of the genomes from a neighboring branch. For simplicity, genes that did not conform to a simple assignment were excluded from this analysis (Other) and specific losses in either haematozoa (piroplasmida and Plasmodium spp.) or cryptosporidia were allowed when assigning a gene to the apicomplexan category.
Statistical Testing
The ratio of plaquing efficiency following mutation of each ICAP relative to SAG1 mutation was compared to a mean of 1 using one-tailed t tests, and the resulting p values were corrected using the Benjamini-Hochberg method. Based on the small sample size, mutants with a corrected p value lower than 0.1 were considered significant. ICAP mutant invasion efficiency was analyzed similarly using PLP1as the control. The effects of inducing CLAMP knockdown on invasion, egress, and MIC2 secretion were analyzed in Prism (Graphpad) using two-tailed t tests with a p value of 0.05 or lower indicating significance. Where appropriate, statistical parameters including the exact value of n, the definition of center, dispersion and precision measures and statistical significance are reported in the figures and corresponding legends.
DATA AND SOFTWARE AVAILABILITY
Software
Guide selection and screen analysis were performed using custom software that will be provided upon request. Statistical analyses were performed in R (www.R-project.org) using built-in packages unless otherwise indicated.
Data Resources
All data from the CRISPR screens is available in Table S3, and will be integrated into an upcoming release of ToxoDB (ToxoDB.org).
Additional Resources
Plasmids used to construct RH/Cas9 as well as pU6-DHFR and pU6[sgSAG1]-DHFR and the genome-wide sgRNA library are available through Addgene and have been assigned the following ID numbers: 80329 (pU6-DHFR), 80324 (pCas9/decoy), 80323 (pCas9/CAT), 80322 (pU6[sgSAG1]-DHFR), and 806363 (genome-wide library).
Supplementary Material
(A) Diagram of the RAB4 locus illustrating the knockout strategy and position of the primers used for validation. Specific primers were used to amplify the 5′ (PCR1) and 3′ (PCR2) junctions, and a segment of the coding sequence (PCR3).
(B) PCR assay for integration of the pyrimethamine-resistance cassette into the RAB4 locus and deletion of the RAB4 coding sequence.
(C) RT-PCR probing for expression of RAB4 or the control ACT1 on cDNA samples prepared with or without reverse transcriptase (RT).
(D) Plaque assay demonstrating normal growth of the RAB4 knockout in human fibroblasts.
Genes previously identified as dispensable or essential for T. gondii growth in culture. Essential genes are further divided into those identified through overexpression screens or other methods. The PubMed ID (PMID) for the citation providing the respective experimental evidence is indicated.
The raw sgRNA counts for the paired library and P3 samples, the calculated fold-change for each sgRNA, and the phenotype score for each gene, are all provided in separate sheets. All calculations were performed as described in the Supplemental Experimental Procedures.
(A) Distribution of published dN/dS values for 16 reference T. gondii genomes (Lorenzi et al., 2016). The lowest and highest third of the dataset is highlighted in orange and blue, respectively.
(B) Genes binned according to dN/dS show higher phenotype scores for genes under positive selection (blue), compared to those under negative selection (orange). Bars indicate the group median. The distributions were compared using a Kolmogorov-Smirnov test.
(A) Diagram of the constructs used to generate the T. gondii strain expressing H2B-YFP and Cas9. The sequence of the decoy sgRNA is highlighted in the top construct, followed by the Cas9-binding sequence (orange).
(B) Expression of H2B-YFP in the nucleus of the clonal parasites detected by live fluorescence video microscopy.
(C) Representative micrographs showing intracellular parasites three days post transfection. Parasites were stained for SAG1 (red), and TgACT1 (ACT; green). Host-cell and parasite nuclei were stained with DAPI (blue). Scale bar = 10 μm. SAG1 retention in wild-type (wt) and Cas9-expressing parasites was measured following the different treatments.
CLAMP orthologs representing the entire breadth of the apicomplexan radiation were aligned using Clustal X. Bars indicate the topology of four representative sequences determined by CCTOP (Dobson et al., 2015). The predicted transmembrane domains (TMs), extracellular loops (ECLs) and proline-rich domain (PRD) are indicated.
(A) Localization of CLAMP After Invasion. CLAMP-mNeonGreen parasites were fixed following invasion. Extracellular parasites were stained for the surface antigen SAG1 (red) prior to permeabilization. Arrowheads indicate puncta of mNeonGreen at the posterior various parasites. Several sections have been magnified to highlight the mNeonGreen localization in invaded (i. and ii.) and non-invaded (iii. and iv.) parasites.
(B–C) Generation and Testing of the PfCLAMP cKD. Diagram of the CLAMP locus for wild-type (wt) and conditional knockdown (cKD) parasites (B). Specific primer pairs (P156/P157 and P158/P159) were used to test for the integration sites (C) and fully sequence them.
(D) Growth curves of the P. falciparum parental strain (left) or the cKD (right) ± aTc. Means ± SD for n = 3 technical replicates. This is an independent replicate of the experiment shown in Figure 6N.
Intracellular parasites expressing CLAMP-mNeonGreen were stimulated with 1 μM A23187. The left panel shows the bright-field and mNeonGreen composite image, while the right panel shows the relative intensity of mNeonGreen fluorescence. Time following the addition of the ionophore is indicated in seconds.
Freshly lysed parasites expressing CLAMP-mNeonGreen were allowed to invade human fibroblasts. The right panel shows the relative mNeonGreen fluorescence, which is present in the composite with the bright-field image in the left panel. Two independent events are shown sequentially. Time is indicated in seconds.
DiCre/CLAMP parasites were treated for 2 hours with rapamycin (+rapa; right) or a vehicle control (−rapa; left) during invasion of human fibroblasts. 30 hours later, egress was induced by treatment with 0.5 mM zaprinast. The change in fluorescence from red to green is indicative of recombination and down-regulation of CLAMP. Time following the addition of zaprinast is indicated in seconds.
DiCre/CLAMP parasites were treated for 2 hours with rapamycin (+rapa) and 30 hours later, treated with 0.5 mM zaprinast to induce egress. Video shows parasites trying to reinvade following egress. The change in fluorescence from red to green is indicative of recombination and down-regulation of CLAMP. Failed invasion attempts by CLAMP-deficient parasites (green arrowheads) and successful invasion by a CLAMP-expressing parasite (red arrowhead) are highlighted. Time is indicated in seconds.
Acknowledgments
We thank Emily Shortt for technical support; George Bell for bioinformatics advice; L. David Sibley, Dominique Soldati-Favre, and Lilach Sheiner for the SAG1, MIC8, MYOA, and MYS antibodies; Ke Hu for the NeonGreen plasmid; David S. Roos and Maria Alejandra Diaz-Miranda for the RNAsequencing data; Markus Meissner for the DiCre strain; and Gail Eskes for naming CLAMP. This work would not have been possible without EupathDB, and we thank all members of the community who have worked to generate this resource. This work was supported by NIGMS Center for Integrative Synthetic Biology Grant P50GM098792, NIH National Research Service Award F31 CA189437 to T.W., NIH Research Project Grant R01AI46675 to V.B.C., and the NIH Director’s New Innovator Award 1DP2OD007124 to J.C.N. and Early Independence Award 1DP5OD017892 to S.L.
Footnotes
AUTHOR CONTRIBUTIONS
S.M.S., D.H. and S.L. conceived this study and performed most of the experiments. S.M.G., A.S.N. and J.C.N. designed and performed the malaria work. M.H. and V.B.C. provided essential reagents and insight. T.W. provided the scripts to design the library, which P.T. adapted and executed. J.P.J.S. advised the experimental design and drafting of the manuscript. S.M.S., D.H. and S.L. wrote the manuscript, which was read and approved by all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achbarou A, Mercereau-Puijalon O, Autheman JM, Fortier B, Camus D, Dubremetz JF. Characterization of microneme proteins of Toxoplasma gondii. Mol Biochem Parasitol. 1991;47:223–233. doi: 10.1016/0166-6851(91)90182-6. [DOI] [PubMed] [Google Scholar]
- Bargieri D, Lagal V, Andenmatten N, Tardieux I, Meissner M, Ménard R. Host cell invasion by apicomplexan parasites: the junction conundrum. PLoS Pathog. 2014;10:e1004273. doi: 10.1371/journal.ppat.1004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Bullen HE, Jia Y, Yamaryo-Botté Y, Bisio H, Zhang O, Jemelin NK, Marq JB, Carruthers V, Botté CY, Soldati-Favre D. Phosphatidic Acid-Mediated Signaling Regulates Microneme Secretion in Toxoplasma. Cell Host Microbe. 2016;19:349–360. doi: 10.1016/j.chom.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Burg JL, Perelman D, Kasper LH, Ware PL, Boothroyd JC. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J Immunol. 1988;141:3584–3591. [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD. Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol Microbiol. 1999;31:421–428. doi: 10.1046/j.1365-2958.1999.01174.x. [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Moreno SN, Sibley LD. Ethanol and acetaldehyde elevate intracellular [Ca2+] and stimulate microneme discharge in Toxoplasma gondii. Biochem J. 1999;342(Pt 2):379–386. [PMC free article] [PubMed] [Google Scholar]
- Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb BS, de Koning-Ward TF, Gilson PR. Toward forward genetic screens in malaria-causing parasites using the piggyBac transposon. BMC Biol. 2011;9:21–21. doi: 10.1186/1741-7007-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croken MM, Qiu W, White MW, Kim K. Gene Set Enrichment Analysis (GSEA) of Toxoplasma gondii expression datasets links cell cycle progression and the bradyzoite developmental program. BMC Genomics. 2014;15:515. doi: 10.1186/1471-2164-15-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski JM, Carruthers VB, Sibley LD. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol Microbiol. 1997;26:163–173. doi: 10.1046/j.1365-2958.1997.5671913.x. [DOI] [PubMed] [Google Scholar]
- Dobson L, Reményi I, Tusnády GE. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408–W412. doi: 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Roos DS. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc Natl Acad Sci USA. 1993;90:11703–11707. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Roos DS. Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proc Natl Acad Sci USA. 1995;92:5749–5753. doi: 10.1073/pnas.92.12.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell A, Coleman BI, Benenati B, Brown KM, Blader IJ, Marth GT, Gubbels MJ. Whole genome profiling of spontaneous and chemically induced mutations in Toxoplasma gondii. BMC Genomics. 2014;15:354. doi: 10.1186/1471-2164-15-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery EL, Fidock DA, Winzeler EA. Using genetic methods to define the targets of compounds with antimalarial activity. J Med Chem. 2013;56:7761–7771. doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frénal K, Marq JB, Jacot D, Polonais V, Soldati-Favre D. Plasticity between MyoC- and MyoA-Glideosomes: An Example of Functional Compensation in Toxoplasma gondii Invasion. PLoS Pathog. 2014;10:e1004504. doi: 10.1371/journal.ppat.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan SM, Falla A, Goldfless SJ, Nasamu AS, Niles JC. Synthetic RNA-protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat Commun. 2016;7:10727–10727. doi: 10.1038/ncomms10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reguet N, Lebrun M, Fourmaux MN, Mercereau-Puijalon O, Mann T, Beckers CJ, Samyn B, Van Beeumen J, Bout D, Dubremetz JF. The microneme protein MIC3 of Toxoplasma gondii is a secretory adhesin that binds to both the surface of the host cells and the surface of the parasite. Cell Microbiol. 2000;2:353–364. doi: 10.1046/j.1462-5822.2000.00064.x. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gomes AR, Bushell E, Schwach F, Girling G, Anar B, Quail MA, Herd C, Pfander C, Modrzynska K, Rayner JC, et al. A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite. Cell Host Microbe. 2015;17:404–413. doi: 10.1016/j.chom.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Roos DS, Angel SO, Murray JM. Variability and heritability of cell division pathways in Toxoplasma gondii. J Cell Sci. 2004;117:5697–5705. doi: 10.1242/jcs.01494. [DOI] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryotic Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram JR, Knockenhauer KE, Markus BM, Mandelbaum J, Ramek A, Shan Y, Shaw DE, Schwartz TU, Ploegh HL, Lourido S. Allosteric activation of apicomplexan calcium-dependent protein kinases. Proceedings of the National Academy of Sciences. 2015;112:201505914–E4984. doi: 10.1073/pnas.1505914112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryotic Cell. 2014;13:1465–1469. doi: 10.1128/EC.00213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack BFC, Pena JDO, Coppens I, Ravindran S, Boothroyd JC, Carruthers VB. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science. 2009;323:530–533. doi: 10.1126/science.1165740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler H, Herm-Götz A, Hegge S, Rauch M, Soldati-Favre D, Frischknecht F, Meissner M. Microneme protein 8--a new essential invasion factor in Toxoplasma gondii. J Cell Sci. 2008;121:947–956. doi: 10.1242/jcs.022350. [DOI] [PubMed] [Google Scholar]
- Kim K, Boothroyd JC. Toxoplasma gondii: stable complementation of sag1 (p30) mutants using SAG1 transfection and fluorescence-activated cell sorting. Exp Parasitol. 1995;80:46–53. doi: 10.1006/expr.1995.1006. [DOI] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Del Castillo Velasco-Herrera M, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2008;1778:631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Kremer K, Kamin D, Rittweger E, Wilkes J, Flammer H, Mahler S, Heng J, Tonkin CJ, Langsley G, Hell SW, et al. An overexpression screen of Toxoplasma gondii Rab-GTPases reveals distinct transport routes to the micronemes. PLoS Pathog. 2013;9:e1003213–e1003216. doi: 10.1371/journal.ppat.1003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarque MH, Roques M, Kong-Hap M, Tonkin ML, Rugarabamu G, Marq JB, Penarete-Vargas DM, Boulanger MJ, Soldati-Favre D, Lebrun M. Plasticity and redundancy among AMA-RON pairs ensure host cell entry of Toxoplasma parasites. Nat Commun. 2014;5:4098. doi: 10.1038/ncomms5098. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Levine ND. The protozoan phylum Apicomplexa. I. II. CRC Press, Inc; 1988. [Google Scholar]
- Liu G, Yong MYJ, Yurieva M, Srinivasan KG, Liu J, Lim JSY, Poidinger M, Wright GD, Zolezzi F, Choi H, et al. Gene Essentiality Is a Quantitative Property Linked to Cellular Evolvability. Cell. 2015;163:1388–1399. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- Lorenzi H, Khan A, Behnke MS, Namasivayam S, Swapna LS, Hadjithomas M, Karamycheva S, Pinney D, Brunk BP, Ajioka JW, et al. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat Commun. 2016;7:10147–10147. doi: 10.1038/ncomms10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, Sibley LD. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010;465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido S, Tang K, Sibley LD. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. Embo J. 2012;31:4524–4534. doi: 10.1038/emboj.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Tran J, Li C, Ganesan L, Wood D, Morrissette N. Secondary mutations correct fitness defects in Toxoplasma gondii with dinitroaniline resistance mutations. Genetics. 2008;180:845–856. doi: 10.1534/genetics.108.092494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae JI, Sheiner L, Nahid A, Tonkin C, Striepen B, McConville MJ. Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe. 2012;12:682–692. doi: 10.1016/j.chom.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AG, Rug M, O’Neill MT, Brown M, Chakravorty S, Szestak T, Chesson J, Wu Y, Hughes K, Coppel RL, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A, Söding J. Automatic Prediction of Protein 3D Structures by Probabilistic Multi-template Homology Modeling. PLoS Comput Biol. 2015;11:e1004343. doi: 10.1371/journal.pcbi.1004343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M. Role of Toxoplasma gondii Myosin A in Powering Parasite Gliding and Host Cell Invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- Pappas G, Roussos N, Falagas ME. Toxoplasmosis snapshots: global status of Toxoplasma gondii seroprevalence and implications for pregnancy and congenital toxoplasmosis. Int J Parasitol. 2009;39:1385–1394. doi: 10.1016/j.ijpara.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL. CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio. 2015;6:e02097–14. doi: 10.1128/mBio.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieperhoff MS, Pall GS, Jiménez-Ruiz E, Das S, Melatti C, Gow M, Wong EH, Heng J, Müller S, Blackman MJ, et al. Conditional U1 Gene Silencing in Toxoplasma gondii. PLoS ONE. 2015;10:e0130356. doi: 10.1371/journal.pone.0130356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JPJ, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F, Soldati-Favre D. Metabolic Pathways in the Apicoplast of Apicomplexa. Elsevier; 2010. pp. 161–228. [DOI] [PubMed] [Google Scholar]
- Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008;24:468–478. doi: 10.1016/j.pt.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, White MW, Striepen B. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7:e1002392. doi: 10.1371/journal.ppat.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Brown KM, Lee TD, Sibley LD. Efficient Gene Disruption in Diverse Strains of Toxoplasma gondii Using CRISPR/CAS9. MBio. 2014;5 doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. Efficient Genome Engineering of Toxoplasma gondii Using CRISPR/Cas9. PLoS ONE. 2014;9:e100450. doi: 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simske JS. Claudins reign: The claudin/EMP/PMP22/γ channel protein family in C. elegans. Tissue Barriers. 2013;1:e25502–e25502. doi: 10.4161/tisb.25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D, Boothroyd JC. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science. 1993;260:349–352. doi: 10.1126/science.8469986. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- Treeck M, Zacherl S, Herrmann S, Cabrera A, Kono M, Struck NS, Engelberg K, Haase S, Frischknecht F, Miura K, et al. Functional Analysis of the Leading Malaria Vaccine Candidate AMA-1 Reveals an Essential Role for the Cytoplasmic Domain in the Invasion Process. PLoS Pathog. 2009;5:e1000322. doi: 10.1371/journal.ppat.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci USA. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- World Health Organization. World Malaria Report. 2014. pp. 1–142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Diagram of the RAB4 locus illustrating the knockout strategy and position of the primers used for validation. Specific primers were used to amplify the 5′ (PCR1) and 3′ (PCR2) junctions, and a segment of the coding sequence (PCR3).
(B) PCR assay for integration of the pyrimethamine-resistance cassette into the RAB4 locus and deletion of the RAB4 coding sequence.
(C) RT-PCR probing for expression of RAB4 or the control ACT1 on cDNA samples prepared with or without reverse transcriptase (RT).
(D) Plaque assay demonstrating normal growth of the RAB4 knockout in human fibroblasts.
Genes previously identified as dispensable or essential for T. gondii growth in culture. Essential genes are further divided into those identified through overexpression screens or other methods. The PubMed ID (PMID) for the citation providing the respective experimental evidence is indicated.
The raw sgRNA counts for the paired library and P3 samples, the calculated fold-change for each sgRNA, and the phenotype score for each gene, are all provided in separate sheets. All calculations were performed as described in the Supplemental Experimental Procedures.
(A) Distribution of published dN/dS values for 16 reference T. gondii genomes (Lorenzi et al., 2016). The lowest and highest third of the dataset is highlighted in orange and blue, respectively.
(B) Genes binned according to dN/dS show higher phenotype scores for genes under positive selection (blue), compared to those under negative selection (orange). Bars indicate the group median. The distributions were compared using a Kolmogorov-Smirnov test.
(A) Diagram of the constructs used to generate the T. gondii strain expressing H2B-YFP and Cas9. The sequence of the decoy sgRNA is highlighted in the top construct, followed by the Cas9-binding sequence (orange).
(B) Expression of H2B-YFP in the nucleus of the clonal parasites detected by live fluorescence video microscopy.
(C) Representative micrographs showing intracellular parasites three days post transfection. Parasites were stained for SAG1 (red), and TgACT1 (ACT; green). Host-cell and parasite nuclei were stained with DAPI (blue). Scale bar = 10 μm. SAG1 retention in wild-type (wt) and Cas9-expressing parasites was measured following the different treatments.
CLAMP orthologs representing the entire breadth of the apicomplexan radiation were aligned using Clustal X. Bars indicate the topology of four representative sequences determined by CCTOP (Dobson et al., 2015). The predicted transmembrane domains (TMs), extracellular loops (ECLs) and proline-rich domain (PRD) are indicated.
(A) Localization of CLAMP After Invasion. CLAMP-mNeonGreen parasites were fixed following invasion. Extracellular parasites were stained for the surface antigen SAG1 (red) prior to permeabilization. Arrowheads indicate puncta of mNeonGreen at the posterior various parasites. Several sections have been magnified to highlight the mNeonGreen localization in invaded (i. and ii.) and non-invaded (iii. and iv.) parasites.
(B–C) Generation and Testing of the PfCLAMP cKD. Diagram of the CLAMP locus for wild-type (wt) and conditional knockdown (cKD) parasites (B). Specific primer pairs (P156/P157 and P158/P159) were used to test for the integration sites (C) and fully sequence them.
(D) Growth curves of the P. falciparum parental strain (left) or the cKD (right) ± aTc. Means ± SD for n = 3 technical replicates. This is an independent replicate of the experiment shown in Figure 6N.
Intracellular parasites expressing CLAMP-mNeonGreen were stimulated with 1 μM A23187. The left panel shows the bright-field and mNeonGreen composite image, while the right panel shows the relative intensity of mNeonGreen fluorescence. Time following the addition of the ionophore is indicated in seconds.
Freshly lysed parasites expressing CLAMP-mNeonGreen were allowed to invade human fibroblasts. The right panel shows the relative mNeonGreen fluorescence, which is present in the composite with the bright-field image in the left panel. Two independent events are shown sequentially. Time is indicated in seconds.
DiCre/CLAMP parasites were treated for 2 hours with rapamycin (+rapa; right) or a vehicle control (−rapa; left) during invasion of human fibroblasts. 30 hours later, egress was induced by treatment with 0.5 mM zaprinast. The change in fluorescence from red to green is indicative of recombination and down-regulation of CLAMP. Time following the addition of zaprinast is indicated in seconds.
DiCre/CLAMP parasites were treated for 2 hours with rapamycin (+rapa) and 30 hours later, treated with 0.5 mM zaprinast to induce egress. Video shows parasites trying to reinvade following egress. The change in fluorescence from red to green is indicative of recombination and down-regulation of CLAMP. Failed invasion attempts by CLAMP-deficient parasites (green arrowheads) and successful invasion by a CLAMP-expressing parasite (red arrowhead) are highlighted. Time is indicated in seconds.