Abstract
The huge communities of residential microbes, including bacteria, viruses, Archaea, and Eukaryotes, that colonize humans are increasingly recognized as playing important roles in health and disease. A complex populous ecosystem, the human gastrointestinal (GI) tract harbors up to 1011 bacterial cells per gram of luminal content, whose collective genome, the gut metagenome, contains a vastly greater number of individual genes than the human genome. In health, the function of the microbiome might be considered to be in dynamic equilibrium with the host, exerting both local and distant effects. However, ‘disequilibrium’ may contribute to the emergence of disease, including malignancy. In this review, we discuss how the intestinal bacterial microbiome and in particular how an ‘estrobolome,’ the aggregate of enteric bacterial genes capable of metabolizing estrogens, might affect women’s risk of developing postmenopausal estrogen receptor–positive breast cancer. Estrobolome composition is impacted by factors that modulate its functional activity. Exploring variations in the composition and activities of the estrobolome in healthy individuals and in women with estrogen-driven breast cancer may lead to development of microbiome-based biomarkers and future targeted interventions to attenuate cancer risk.
Humans become colonized at birth by microbiota, primarily bacteria (which are the focus of this review) (1,2), and over 90% reside within the gastrointestinal (GI) tract. The GI tract harbors more than 500 different bacterial species, and estimates of the number of bacteria we carry reach 1011 per gram of luminal content (3–5). Bacterial load, along with species diversity, increases from the stomach to the colon, creating a complex microbial ecosystem (6). The composition of the GI tract microbiota reflects host variables, such as delivery mode, genetics, diet, alcohol intake, environmental exposures, and medications, in particular antibiotics. Investigation of bacterial microbiome composition, function, and assessment of the aggregate of its genes (the metagenome) is now possible via advances in 16S ribosomal RNA (rRNA) sequencing and informatics (7–10). Humans and microbes have co-evolved a complex intricate relationship to benefit the host while allowing the intestinal microbiota to thrive in a mutually advantageous equilibrium (11–13).
Microbiome perturbation can, however, be associated with risk of developing inflammatory, autoimmune, and malignant disease (14–17). Microbial community dysbiosis, a pathologic disequilibrium, could potentially favor oncogenesis and tumor progression and affect responses to cancer therapy and toxicity profiles of chemotherapeutics (18–20). The human gut microbiome is functional and exerts both local and long-distance effects involving hormonal intermediates, metabolites, and immunologic messengers (21,22). Host-microbe interactions thus have the potential to influence carcinogenesis through mechanisms such as chronic inflammation, induction of genotoxic responses, alteration of the microenvironment, and metabolism (23,24). This could be mediated by the microbial ecosystem as a whole or via specific microbes such as the bacterium Helicobacter pylori, which is associated with increased risk of adenocarcinoma of the stomach in humans (25–27).
The Gut Bacterial Microbiome Includes an Estrobolome
Plottel and Blaser define ‘estrobolome’ as “the aggregate of enteric bacterial genes whose products are capable of metabolizing estrogens” (28). Estrogens are C-18 steroid hormones derived from the stepwise reduction of C-27 cholesterol. The main forms of endogenous estrogens are estradiol (E2, predominant in nonpregnant women prior to menopause), estrone (E1, predominant after menopause), and estriol (E3, highest during pregnancy) (29). Estrogens circulate in the blood in free or protein-bound form and exert diverse biological effects. Parent estrogens (E2, E1) undergo first-pass hepatic metabolism; irreversible hydroxylation at the C-2, C-4, or C-16 positions of the steroid ring result in estrogen metabolites that vary in hormone potency and half-life (30). In the liver, estrogens and their metabolites (including cathechol estrogens via hydroxylation and subsequent methylation) are conjugated through glucuronidation or also through sulfonation to allow for biliary excretion (31). Conjugated estrogens are excreted in bile, urine, and feces (32,33). Studies of injected radioactively labeled estradiol, estrone, and estriol in women indicate that approximately 65% of injected estradiol, 48% of injected estrone, and 23% of injected estriol are recovered in bile (34). As approximately 10% to 15% of injected radiolabeled estradiol, estrone, and estriol are found in conjugated form in feces (35,36), a biologically significant proportion of estrogens are reabsorbed in the circulation. Hepatically conjugated estrogens excreted in the bile can be deconjugated by bacterial species with ß-glucuronidase activity in the gut, leading to their reabsorption into the circulation. Especially relevant are gut bacteria possessing ß-glucuronidases and ß-glucosidases, hydrolytic enzymes involved in the deconjugation of estrogens (Figure 1) (37–41).
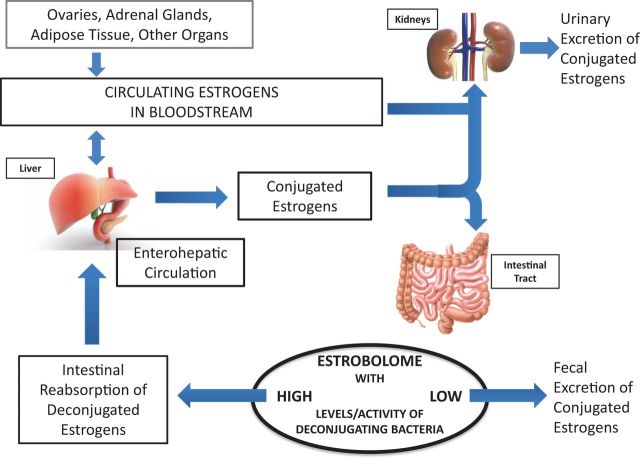
Figure 1.
The estrobolome and enterohepatic circulation of estrogens. Estrogens are primarily produced in the ovaries, adrenal glands, and adipose tissue and circulate in the bloodstream in free or protein-bound form and first undergo metabolism in the liver, where estrogens and their metabolites are conjugated. Conjugated estrogens are eliminated from the body by metabolic conversion to water-soluble molecules, which are excreted in urine or in bile into the feces. The conjugated estrogens excreted in the bile can be deconjugated by bacterial species in the gut with beta-glucuronidase activity (constituents of the ‘estrobolome’), subsequently leading to estrogen reabsorption into the circulation. Circulating estrogens exert effects on target tissues including breast, which stimulate cellular growth and proliferation. By modulating the enterohepatic circulation of estrogens, the estrobolome affects both the excretion and circulation of estrogens. In turn, the composition of the estrobolome can be shaped by factors such as antibiotics, other drugs, and diet that modulate its functional activity. Adapted from Plottel SC, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10(4):324-335.
Bacterial ß-glucuronidases catalyze the hydrolysis of endogenous ß-glucuronides produced in the liver and exogenous ß-glucuronides found in the diet, such as complex carbohydrates (42–44). Many metabolites, steroid hormones, and xenobiotics are excreted into the intestinal tract by bile after hepatic glucuronidation. The removal of glucuronic acid from conjugated substrates (deconjugation) by intestinal bacterial ß-glucuronidases promotes reabsorption of their respective aglycones into the enterohepatic circulation. Distinct bacterial ß-glucuronidase genes from the human gut microbiota have been described (39,40). The well-characterized gus gene is commonly found in gut bacteria (40,45) whereas the BG gene has more recently been described by metagenomic analysis (39). In the human GI tract, the BG gene is well represented in the bacterial phyla Bacteroidetes and Firmicutes whereas gus is more common in Firmicutes (41).
ß-glucuronidase activity can be modulated by diet and by bacterial context. Increased fecal ß-glucuronidase activity has been reported in healthy humans consuming diets high in fat or protein whereas fiber consumption decreases activity (46–48). In Escherichia coli grown in culture, ß-glucuronidase activity was controlled by bacterial population density, suggesting that quorum sensing in vivo affects enzyme levels (49).
Importantly, bacterial ß-glucuronidases are potential drug targets, as recently shown using ß-glucuronidase inhibitors (19,50). The unique structure of bacterial ß-glucuronidases, containing asparagine and lysine (N-K) residues in the active site modulating functional activity, has been elucidated (19,50). This N-K motif is conserved in ß-glucuronidase but not in ß-galactosidase, a homologue differing at the C-4 and C-5 positions; both catalyze the hydrolysis of similar glycoside substrates (51). Small changes in inhibitor structure can alter conformation within the ß-glucuronidase active site, leading to differences in catalytic activity and pharmacologic inhibition; this was shown by the alleviation of GI toxicity in mice from the chemotherapeutic drug irinotecan (CPT-11) (50). In that study, inhibition of ß-glucuronidase did not affect the serum pharmacokinetics of the drug or its metabolites.
Many bacterial genera and species in the human gut contain genes encoding ß-glucuronidase in humans (Table 1; Supplementary Table 1, available online), underscoring the prevalence of the enzyme and extending earlier observations of individual bacteria obtained from human feces that express ex vivo sulfatase (deconjugative) activity (40,41,52). Circulating inactive steroids are also converted to biologically active estrogens by the hepatic sulfatase pathway (32,52,53). Presumably, gut bacteria expressing sulfatase activity would be capable of hydrolyzing estrogen molecules that had undergone hepatic sulfation and biliary excretion into the GI tract. As such presumed bacterial enzymatic activities and their impact on steroid metabolism and the enterohepatic circulation of estrogens have not been investigated, they remain to be determined.
Table 1.
60 bacterial genera colonizing the human intestinal tract that encode ß-glucuronidase and/or ß-galactosidase + (157)*
| Genus | ß-glucuronidase | ß-galactosidase |
|---|---|---|
| Collinsella | + | — |
| Edwardsiella | + | — |
| Alistipes | + | + |
| Bacteroides | + | + |
| Bifidobacterium | + | + |
| Citrobacter | + | + |
| Clostridium | + | + |
| Dermabacter | + | + |
| Escherichia | + | + |
| Faecalibacterium | + | + |
| Lactobacillus | + | + |
| Marvinbryantia | + | + |
| Propionibacterium | + | + |
| Roseburia | + | + |
| Tannerella | + | + |
| Actinomyces | — | + |
| Alistipes | — | + |
| Anaerostipes | — | + |
| Bacteroides | — | + |
| Barnesiella | — | + |
| Bifidobacterium | — | + |
| Blautia | — | + |
| Butyricicoccus | — | + |
| Butyrivibrio | — | + |
| Catenibacterium | — | + |
| Cedecea | — | + |
| Cetobacterium | — | + |
| Citrobacter | — | + |
| Clostridium | — | + |
| Collinsella | — | + |
| Coprobacillus | — | + |
| Coprococcus | — | + |
| Dorea | — | + |
| Dysgonomonas | — | + |
| Enterobacter | — | + |
| Enterococcus | — | + |
| Eubacterium | — | + |
| Fusobacterium | — | + |
| Hafnia | — | + |
| Holdemania | — | + |
| Klebsiella | — | + |
| Lactobacillus | — | + |
| Megamonas | — | + |
| Mitsuokella | — | + |
| Odoribacter | — | + |
| Paenibacillus | — | + |
| Parabacteroides | — | + |
| Paraprevotella | — | + |
| Pediococcus | — | + |
| Porphyromonas | — | + |
| Prevotella | — | + |
| Pseudoflavonifractor | — | + |
| Roseburia | — | + |
| Ruminococcus | — | + |
| Staphylococcus | — | + |
| Streptococcus | — | + |
| Subdoligranulum | — | + |
| Turicibacter | — | + |
| Weissella | — | + |
| Yokenella | — | + |
* + = Human Microbiome Project (HMP) gut-associated microbial genomes (n = 517) were indexed for the presence of ß-glucuronidase (EC 3.2.1.31) or ß-galactosidase (EC 3.2.1.23) using the Integrated Microbial Genomes database (1). Taxa were classified at the genus level.
Studies of fecal samples from healthy volunteers confirm β-glucuronidase and β-glucosidase bioactivity in humans and define optimal collection and processing techniques (54,55). Additional potential enzymatic activities of the estrobolome include dehydrogenation; studies of ex vivo human fecal extracts from the pregenomic era demonstrated both oxidative and reductive reactions affecting several estrogens (56,57). Intestinal bacterial β-glucuronidase and β-glucosidase activity have also been shown to be induced by a high pH, which may also increase colon cancer the risk (58,59). The role of bacterial β-glucuronidase and β-glucosidase activity in breast cancer risk (or risk reduction) is currently unknown.
The series of reductive reactions that allows synthesis of estrogens from cholesterol precursors is catalyzed in part by hydroxysteroid dehydrogenases (HSD), a group of alcohol oxidoreductases (60,61). Bioinformatic annotation of HSDs, present in essentially all complete bacterial genomes, indicate that HSDs are widely found in the normal human gut microbiota (62). An estrobolome enriched in bacterial hydroxysteroid deconjugative activity may contribute to the modulation of the interconversion of conjugated forms of estrogens as well as androgenic molecules, all relevant to the overall estrogenic host milieu (56). Thus, the reach of estrobolome enzymatic functions may extend beyond deconjugation.
Estrobolome Physiology and Perturbation
Experimental evidence for a central role of gut bacteria in estrogen metabolism was noted decades ago. Germ-free rats excreted amounts of free steroid hormones too small to have been characterized by a gas chromatography mass spectrometry technique whereas conventional rats excreted greater, quantifiable amounts of free steroid hormones, reflecting the deconjugative activity of their residential bacteria (63). Introduction of bacteria into the gut of such germ-free mice shown to exhibit impaired reproductive parameters led to normalization of the estrous cycle in females and increased sperm counts in males, all resulting in restoration of overall fertility and reproductive capacity (64).
Goedert and colleagues evaluated the association of fecal microbiome composition and diversity with urinary levels of estrogens and estrogen metabolites in a cross-sectional study of 60 healthy postmenopausal women (65). Urinary estrogens (E2 and E1) and their metabolites were measured using a standardized approach (66,67), and the fecal microbiome was assessed using bacterial 16S rRNA gene sequencing. Microbial diversity in the fecal specimens was statistically significantly associated with the relationship of estrogen metabolites to parent estrogens (E2 and E1); importantly, the ratio of metabolites to parent estrogens was increased with phylogenetic diversity. These observational findings support the hypothesis that differences in estrogen metabolism and levels are associated with variability in gut microbial diversity. Goedert and colleagues also demonstrated a relationship between gut microbial richness, systemic and fecal estrogens, and beta-glucuronidase activity in healthy postmenopausal women and men (68).
Estrogen Exposure and Breast Cancer Risk
In the United States, breast cancer affects one in eight women and is the second leading cause of female cancer death after lung cancer (69). The most common breast cancer subtype is hormone receptor (HR)–positive/human epidermal growth factor (HER) 2–negative, comprising more than 70% of patients (70), with the majority occurring in postmenopausal women (71).
Based on the Surveillance, Epidemiology, and End Results (SEER) database and projections from the United States Census Bureau, the number of new breast tumors, both in situ and invasive, is expected to increase (72). The incidence of HR-positive invasive tumors is projected to increase by 4% annually between 2011 and 2030 in women age 70 to 84 years (from 47 800 to 95 300 patients/year) and 1.6% among women age 50 to 69 years (from 98 000 to 125 700 patients/year). The number of in situ HR-positive tumors is also expected to increase among all age groups combined (from 53 900 to 127 400 patients/year).
Estrogen is recognized as a causal factor in the etiology of HR-positive breast cancer and plays an important role in initiation and promotion of neoplastic growth (73,74) (Figure 2). Decades ago, Trichopoulos advanced the hypothesis that greater concentrations of estrogens in pregnancy increase the probability of breast cancer in the offspring years later, drawing attention to the potential oncogenic significance of estrogens (75). States of relative estrogen excess in pregnancy, such as twin gestation, are associated with increased relative risks of breast cancer in daughters born of those pregnancies (76), as with intra-uterine exposure to the synthetic estrogen diethylstilbestrol (77).
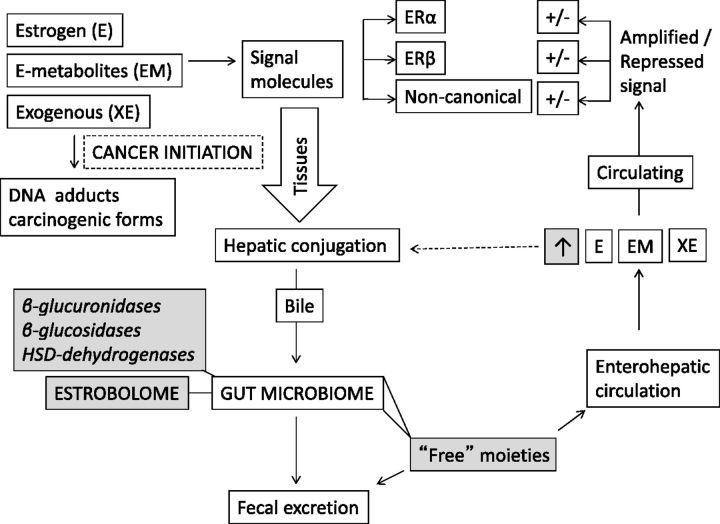
Figure 2.
Potential impact of the estrobolome on estrogens. Forms of endogenous estrogen (E) including E2 (estradiol) and E1 (estrone), estrogen metabolites such as hyroxylated moieties, and exogenous estrogens are relevant to breast tissue carcinogenesis. Estrogens act as signaling molecules via several pathways including canonical paths (alpha and beta receptors) and noncanonical mechanisms. Receptors for estrogen are widely distributed in tissues. Estrogen circulates both freely and protein bound. In the liver, estrogens undergo E2 and E1 interconversion and first pass metabolism. Hepatic conjugation of estrogen allows for biliary excretion of conjugated estrogen and conjugated estrogen metabolites into the gastro-intestinal tract where beta-glucuronidases, glucosidases, and hydroxysteroid dehydrogenases of bacterial origin (the estrobolome fraction of the microbiome) regenerate “free” forms of those molecules. The entero-hepatic circulation therefore contributes to plasma levels of estrogens and their metabolites. An estrobolome enriched in bacteria whose enzymatic activity is higher in deconjugative and hydroxylating function would lead to greater relative levels of circulating free estrogens.
States of relative estrogen excess contribute to hormone-driven breast cancer in postmenopausal women (78), confirmed by a meta-analysis of nine prospective studies (n = 663 women with breast cancer and 1765 women without breast cancer) demonstrating a statistically significant association between endogenous sex hormone levels (including E2 and E1) and breast cancer in postmenopausal women (79). For example, the relative risk of developing breast cancer for women who had estradiol levels in the highest quintile compared with women with levels in the lowest quintile was 2.0 (95% confidence interval [CI] = 1.47 to 2.71).
The ratio of estrogen metabolites to parent estrogens has also been linked to postmenopausal breast cancer risk. The risk increased with higher circulating concentrations of the parent estrogens (E2 and E1). Risk decreased with increasing ratios of 2- and 4-pathway estrogen metabolites to parent estrogens via hydroxylation by hepatic cytochrome P450 (80) and metabolic pathways favoring estrogen 2-hydroxylation over 16α-hydroxylation (81–83). Furthermore, 2-, 4-, and 16α-hydroxyestrogens may be considered carcinogenic (84).
Administration of exogenous estrogens for five to seven years in the placebo-controlled Women’s Health Initiative trials (n = 27 347 postmenopausal women) demonstrated a statistically significant increase in breast cancer during and after the intervention period for combined estrogen and progestin treatment but not for treatment with estrogen alone (85).
How the Estrobolome May Affect Estrogen Levels and Breast Cancer Risk
An important role of the intestinal microbiome is the modulation of systemic estrogens (38–41) as it affects the enterohepatic circulation of estrogens and their reabsorption. We have postulated that, in theory, an estrobolome enriched in enzymes favoring deconjugation would promote reabsorption of free estrogens and thus increase relative total estrogen burden, potentially contributing to the risk of development of hormone-driven malignancies such as breast cancer (Figure 3). The bacterial composition of the estrobolome in turn is likely affected by host factors such as age and ethnicity, as well as lifetime environmental influences including diet, alcohol, and antibiotic use, which may exert selective pressures on bacterial populations (86–88). Some of these factors have also been independently linked to breast cancer risk. Possible overlapping or additive modulators are discussed below.
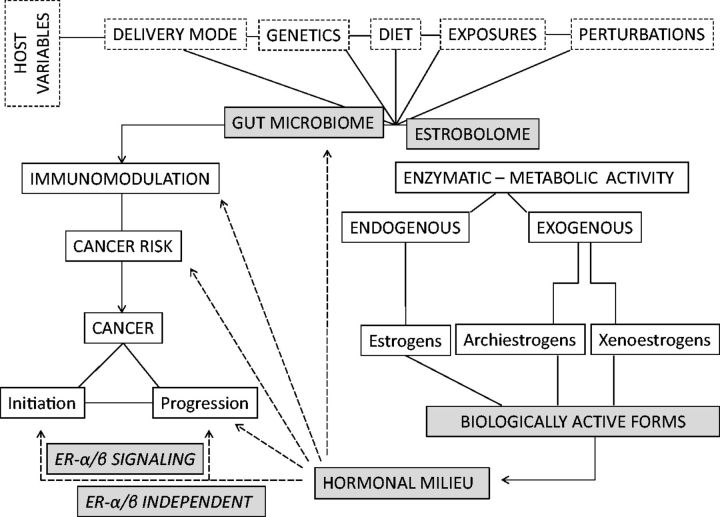
Figure 3.
Potential link of the gut estrobolome and breast cancer risk. The estrobolome/microbiome modulates levels of circulating estrogens and thereby the endogenous hormonal milieu, which can affect the risk of hormone-driven malignancies including breast and endometrial cancer directly or indirectly, for instance via immunomodulation. Higher deconjugating activity in the gut may lead to higher relative plasma levels of estrogen and metabolites, potentially increasing cancer risk, whereas either lower deconjugating activity may have opposite effects. Conversely, the microbiome is modulated by factors such as the environment, host variables, and possibly sex steroid levels.
Antibiotics
A large body of evidence, dating back several decades, indicates that antibiotics perturb bacterial gut populations. Ampicillin administration causes large increases in fecal conjugated estrogens and reduction in urinary estrogen in women (89–91). Healthy men given oxytetracycline had increased fecal conjugated estrogen excretion in parallel with a decrease in urinary estrogen excretion (92). Experimental data point to a decrease in fecal beta-glucuronidase enzyme activities when rats were administered antibiotics (93). These studies suggest that certain antibiotics, by promoting increased excretion of (conjugated) estrogens, reduce the overall deconjugative activity of intestinal bacteria, at least in the short-term and could hypothetically lower breast cancer risk. However, long-term consequences of antibiotics on estrogen excretion have not been studied.
Several epidemiologic studies, in contrast, have suggested a possible association of antibiotic use and breast cancer risk (94–98). In a North American case-control study involving 2266 women with breast cancer and 7953 healthy controls, increased prior antibiotic use was associated with an increased risk of breast cancer, with an estimated odds ratio of 2.07 (95% CI = 1.48 to 2.88) in women who had received long-term antibiotic treatment (94). In the study, all classes of antibiotics were associated with increased breast cancer risk, and the association remained after adjustment for factors including premenopausal or postmenopausal status, age at menarche, and family history. Other studies showed a possible association (94), but this was not universally observed (96,97). In a nine-year follow-up of 2.1 million women, a slightly increased risk of breast cancer with antibiotic exposure (hazard ratio [HR] = 1.14, 95% CI = 1.10 to 1.18) was observed, but there was little evidence of a dose response, with a hazard ratio of 1.17 (95% CI = 0.97 to 1.42) for long-term use (98). A recent population based study involving 31 131 women with breast cancer found, after multivariable adjustment for body mass index (BMI), smoking, alcohol use, diabetes, and hormone replacement therapy use (99), only a moderate increase in breast cancer risk with the subset that received antibiotics (100). These studies therefore suggest a possible but small increase in breast cancer risk with use of some antibiotics, but it is important to keep in mind that antibiotic use is so ubiquitous (101) that unaccounted exposures could substantially affect epidemiologic findings. As the microbiome may be most prone to modulation by antibiotics during the first years of life (102), additional information and adjustment for antibiotic exposure in childhood may be needed.
Adiposity and Diet
Adiposity has been associated with higher circulating estrogen levels in postmenopausal women, as well as with increased breast cancer risk (103,104). Approximately 35% of the adult world population is either overweight (BMI = 25–30 kg/m2) or obese (BMI ≥ 30 kg/m2), with a rising incidence (105). A meta-analysis of 50 prospective observational studies confirmed a relationship between adult weight gain in women and risk for cancer; each 5 kilogram increase in weight was associated with increases in postmenopausal breast (+11%), ovarian (+13%), and endometrial cancers (+39%) (102). In postmenopausal women, obesity and excess adiposity may lead to increased circulating estrogens through the peripheral aromatization of androgens but also can induce insulin resistance, increase insulin-like growth factor (IGF)–1, and suppress production of hepatic hormone-binding proteins, thereby increasing total and bioavailable estrogens (106).
Changes in diet are known to affect overall gut microbiome composition and function (107). Strict vegetarians have increased fecal excretion of conjugated estrogens compared with nonvegetarians, leading to decreased plasma estrogen concentrations (108,109). Diets rich in fats and red meat promote the production of bile acids necessary for fat digestion and absorption. Microbiota then metabolize these compounds into secondary bile acids (110,111). An American study compared 10 premenopausal women consuming a “Western diet” that included high fat (40% of calories) and low fiber with 10 age-matched vegetarians consuming a high fiber and moderate fat (30%) diet; the vegetarians had triple the estrogens in feces and 15% to 20% lower serum estrogen levels (108). Fecal bacterial ß-glucuronidase activity was also statistically significantly lower in the vegetarians than in the omnivores (P < .05). Immigrants from Asia consuming a low fat diet (20%-25% of calories) had systemic estrogen levels 30% lower than American women eating a high-fat diet (109) . Diet is possibly associated with estrogen metabolism and levels, possibly via the estrobolome, although additional factors such as lifestyle, exercise, and supplements may contribute. In addition, the potential impact of complex dietary fibers that might induce bacterial enzyme selectivity in the estroblome is currently unknown and remains investigational.
Hypercholesteremia is a risk factor for ER-positive breast cancer (112). The cholesterol metabolite 27-hydroxycholesterol (27HC) has been shown to possess estrogenic activities and to promote breast tumor growth in xenograft mouse models by binding to the ER on epithelial cells of mammary glands and stimulating cellular proliferation (113). In humans, 27HC is enriched in ER-positive breast tumors although its role in the pathogenesis of breast cancer remains to be defined (114,115).
Alcohol
Alcohol consumption increases the risk of breast cancer, in particular ER-positive tumors in postmenopausal women (116–120). A recent study showed a positive association between alcohol consumption and endogenous estrogen levels and mammographic density in premenopausal women (121). Similarly, alcohol intake after breast cancer diagnosis is associated with both increased risk of recurrence and death (122). While the pathophysiology is likely multifactorial (120), daily alcohol consumption increases serum estrogen levels, particularly E1 (123,124) but also E2 (125) and other forms of estrogen (126–128). Several biological mechanisms may account for the association of alcohol consumption with the development of breast cancer: 1) alcohol may increase the activity of ER signaling in breast tumors or may increase endogenous steroid hormone levels (123,128–130); 2) ethanol may stimulate the transcriptional activity of ER-α ligand in human breast cancer cell lines in a dose-dependent manner while downregulating expression of BRCA1, an inhibitor of ER-α transcriptional activity (129); and 3) among healthy postmenopausal women who were not on HRT and who consumed 15 to 30 g of alcohol per day, concentrations of serum estrone sulfate were increased by 7.5% and 10.7%, respectively, compared with levels in postmenopausal women who did not consume alcohol (123).
One mechanism contributing to the observed association of alcohol and elevated circulating estrogens could be mediated via gut microbial populations although precise interactions require definition. Alcohol consumption may lead to small intestinal bacterial overgrowth (SIBO) (131). Both gut anaerobic and aerobic bacteria are present at higher levels in subjects with chronic alcohol abuse and alcoholic cirrhosis compared with healthy controls (132–134). SIBO has also been observed in animal models of alcoholic liver disease. Alcohol consumption alters the composition of the colonic microbiome in rats (135). Ethanol may affect the metabolism of intestinal bacteria although it has not been well studied. In a rat model of metabolic alterations of GI tract luminal contents following chronic ethanol consumption (136), pathways that were altered affected fatty, bile, and amino acids, as well as steroids, including 4-hydroxyestrone. In animal models, 4-hydroxyestrone, a catechol estrogen metabolite, has biologically significant estrogenic activity and has been shown to be involved in estrogen-induced tumorigenesis (137). The interactions between alcohol, estrogens, and breast carcinogenesis in humans need greater definition.
Probiotics and fermented foods containing lactic acid bacteria have been explored for anticancer properties, which may involve modulation of the intestinal microbiome and the host immune response (138). Human subjects who consumed oral supplements of Lactobacillus acidophilus demonstrated a reduction in the activity of fecal enzymes, including β-glucuronidase (139,140). In several epidemiologic studies, consumption of fermented milk products was associated with decreased risk of breast cancer (141,142), possibly through changes in the intestinal microbiota altering the enterohepatic cycling of estrogenic compounds.
Microbiome of Human Breast Tissue
Decades ago, a link between the bile acids in the gut and cystic breast tissue in humans was identified (143,144). The bile salt lithocholate, proven to be originating from the intestines, was found in the aspirate of cyst fluid from the breast in women with fibrocystic disease at much higher concentrations than in the serum (143). While the mechanism for maintenance of high bile concentrations in the breast cysts remains to be studied, the farnesoid X receptor (FXR), a bile acid receptor, has been identified in the benign and malignant breast microenvironment (145). Bile acids such as deoxycholic acid (DCA) have been shown to stimulate the growth and metastases of breast cancer cells through FXR, suggesting that bile acids may play a role in breast tumor carcinogenesis.
While beyond the scope of this review of the gut microbiome and breast cancer, microbiota of human breast tissue has also been studied and revealed differences between breast tumor tissues compared with paired normal tissue (146–148). It is unknown if these findings reflect local dysbiosis creating an environment that favors breast tumor formation, ie, oncogenic triggers, or reflect host selection for microbes adapted to the fatty acid-rich environment in the breast tissue. This, however, suggests that, as with other parts of the body, breast tissue has a unique microbiome.
Conclusion and Further Directions
Interest is growing in the dynamic roles of the microbiome in human health and disease. Investigations exploring the potential interactions of the gut microbiome and breast cancer span the translational research spectrum and require collaborations between basic scientists, including immunologists, cell biologists, and microbiologists, clinicians such as oncologists and endocrinologists, animal researchers, epidemiologists, biostatisticians, and bioinformaticians. The ongoing Microbiome Quality Control Project (149) and the International Human Microbiome Standards (IHMS) Project (150) address sample collection, sample storage, standardization, and reproducibility (151–153); microbiome researchers should follow their recommendations in planning studies. In particular, fecal sample biobanking should be incorporated in the study design of longitudinal epidemiologic investigations.
Much of our understanding of the enterohepatic circulation of estrogens is informed by studies performed before 1970. Recent advances in analytical methods, including accurately and sensitively measuring different conjugated and unconjugated estrogens in serum and urine should provide tools for meaningful comparisons between individuals (67,154). Animal models serve to test hypotheses under controlled conditions. Observational large-scale human studies are needed to identify and confirm associations while controlling for other (genetic, epigenetic, dietary, and environmental) variables affecting the microbiome and confounders of cancer risk. A recent study investigating differences in gut microbiome composition among postmenopausal women showed a less diverse fecal microbiome and a statistically significantly altered composition in newly diagnosed breast cancer patients (87% had ER-positive tumors) compared with healthy controls (155), and this finding deserves follow-up.
Further studies are needed to evaluate the estrobolome hypothesis and to define the impact of the microbiome’s metabolic capacity on estrogen metabolism and host physiology. Determining the “functionality” of differing microbiomes vis-à-vis deconjugative activity is essential. We are currently conducting a prospective case-control study in postmenopausal women with recently diagnosed ER-positive breast cancer as well as healthy controls to assess the relationship between the gut bacterial microbiome/β-glucuronidase activity, circulating estrogen levels, and breast cancer. If the estrogen metabolism–gut microbiome axis is functional with underlying individual variations in estrogen levels, it is plausible that the estrobolome could contribute to the risk of hormone-driven malignancies including breast cancer and as such could serve as a potential biomarker (156). Furthermore, interventions that may include use of prebiotics, probiotics, or antimicrobial agents could be designed specifically to target gut bacterial species with β-glucuronidase activity (19,50) to decrease estrogen-related cancer risk or become components of future therapies. In conclusion, links between the microbiome and estrogen-driven breast cancer are growing, and we hope that research will identify specific characteristics of the gut microbiome that can be used to develop novel approaches for breast cancer risk assessment, prevention, and treatment.
Funding
This work was supported in part by the National Cancer Institute: R01 CA161891 (SA), the Shifrin-Myers Breast Cancer Discovery Fund (SA), R01 DK098989 (MB) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and the Diane Belfer Program for Human Microbial Ecology (MB).
Notes
The funders had no role in the writing of the review or the decision to submit it for publication.
We thank Thomas Battaglia for his help in the construction of Table 1 and Supplementary Table 1 (available online).
Supplementary Material
References
- 1.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489(7415):250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. [DOI] [PubMed] [Google Scholar]
- 7.Clarridge JE., 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17(4):840–862, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. [DOI] [PubMed] [Google Scholar]
- 9.Goodrich JK, Di Rienzi SC, Poole AC, et al. Conducting a microbiome study. Cell. 2014;158(2):250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas T, Gilbert J, Meyer F. Metagenomics - a guide from sampling to data analysis. Microb Inform Exp. 2012;2(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449(7164):843–849. [DOI] [PubMed] [Google Scholar]
- 12.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quercia S, Candela M, Giuliani C, et al. From lifetime to evolution: timescales of human gut microbiota adaptation. Front Microbiol. 2014;5:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer J. 2014;20(3):181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yurkovetskiy LA, Pickard JM, Chervonsky AV. Microbiota and autoimmunity: exploring new avenues. Cell Host Microbe. 2015;17(5):548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan AA, Shrivastava A, Khurshid M. Normal to cancer microbiome transformation and its implication in cancer diagnosis. Biochim Biophys Acta. 2012;1826(2):331–337. [DOI] [PubMed] [Google Scholar]
- 17.Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16(10):406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2014;35(2):249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace BD, Wang H, Lane KT, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330(6005):831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreamer BL, Siegel FL, Gourley GR. A novel inhibitor of beta-glucuronidase: L-aspartic acid. Pediatr Res. 2001;50(4):460–466. [DOI] [PubMed] [Google Scholar]
- 21.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapira I, Sultan K, Lee A, Taioli E. Evolving concepts: how diet and the intestinal microbiome act as modulators of breast malignancy. ISRN Oncol. 2013;2013:693920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hullar MA, Fu BC. Diet, the gut microbiome, and epigenetics. Cancer J. 2014;20(3):170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. [DOI] [PubMed] [Google Scholar]
- 26.Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61(5):1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325(16):1132–1136. [DOI] [PubMed] [Google Scholar]
- 28.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10(4):324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and actions of estrogens. N Engl J Med. 2002;346(5):340–352. [DOI] [PubMed] [Google Scholar]
- 30.Zhu BT, Han GZ, Shim JY, Wen Y, Jiang XR. Quantitative structure-activity relationship of various endogenous estrogen metabolites for human estrogen receptor alpha and beta subtypes: Insights into the structural determinants favoring a differential subtype binding. Endocrinology. 2006;147(9):4132–4150. [DOI] [PubMed] [Google Scholar]
- 31.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis. 1998;19(1):1–27. [DOI] [PubMed] [Google Scholar]
- 32.Raftogianis R, Creveling C, Weinshilboum R, Weisz J. Estrogen metabolism by conjugation. J Natl Cancer Inst Monogr. 2000(27):113–124. [DOI] [PubMed] [Google Scholar]
- 33.Bongiovanni AM, Cohn RM. Chapter 9: Clinical aspects of steroid conjugation In. Chemical and Biological Aspects of Steroid Conjugation. Bernstein S, Solomon S, eds. New York: Springer; 1970. [Google Scholar]
- 34.Sandberg AA, Slaunwhite WR., Jr. Studies on phenolic steroids in human subjects. II. The metabolic fate and hepato-biliary-enteric circulation of C14-estrone and C14-estradiol in women. J Clin Invest. 1957;36(8):1266–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adlercreutz H, Martin F. Biliary excretion and intestinal metabolism of progesterone and estrogens in man. J Steroid Biochem. 1980;13(2):231–244. [DOI] [PubMed] [Google Scholar]
- 36.Adlercreutz H, Jarvenpaa P. Assay of estrogens in human feces. J Steroid Biochem. 1982;17(6):639–645. [DOI] [PubMed] [Google Scholar]
- 37.Cole CB, Fuller R, Mallet AK, Rowland IR. The influence of the host on expression of intestinal microbial enzyme activities involved in metabolism of foreign compounds. J Appl Bacteriol. 1985;59(6):549–553. [DOI] [PubMed] [Google Scholar]
- 38.Gadelle D, Raibaud P, Sacquet E. beta-Glucuronidase activities of intestinal bacteria determined both in vitro and in vivo in gnotobiotic rats. Appl Environ Microbiol. 1985;49(3):682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gloux K, Berteau O, El Oumami H, Beguet F, Leclerc M, Dore J. A metagenomic beta-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4539–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. 2008;66(3):487–495. [DOI] [PubMed] [Google Scholar]
- 41.McIntosh FM, Maison N, Holtrop G, et al. Phylogenetic distribution of genes encoding beta-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ Microbiol. 2012;14(8):1876–1887. [DOI] [PubMed] [Google Scholar]
- 42.McBain AJ, Macfarlane GT. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J Med Microbiol. 1998;47(5):407–416. [DOI] [PubMed] [Google Scholar]
- 43.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr. 2007;137(3 Suppl 2):751S–755S. [DOI] [PubMed] [Google Scholar]
- 44.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Polyphenols: food sources and bioavailability. Am J Clin Nutr. 2004;79(5):727–747. [DOI] [PubMed] [Google Scholar]
- 45.Beaud D, Tailliez P, Anba-Mondoloni J. Genetic characterization of the beta-glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology. 2005;151(Pt 7):2323–2330. [DOI] [PubMed] [Google Scholar]
- 46.Reddy BS, Hanson D, Mangat S, et al. Effect of high-fat, high-beef diet and of mode of cooking of beef in the diet on fecal bacterial enzymes and fecal bile acids and neutral sterols. J Nutr. 1980;110(9):1880–1887. [DOI] [PubMed] [Google Scholar]
- 47.Domellof L, Darby L, Hanson D, Mathews L, Simi B, Reddy BS. Fecal sterols and bacterial beta-glucuronidase activity: a preliminary metabolic epidemiology study of healthy volunteers from Umea, Sweden, and metropolitan New York. Nutr Cancer. 1982;4(2):120–127. [DOI] [PubMed] [Google Scholar]
- 48.Reddy BS, Engle A, Simi B, Goldman M. Effect of dietary fiber on colonic bacterial enzymes and bile acids in relation to colon cancer. Gastroenterology. 1992;102(5):1475–1482. [DOI] [PubMed] [Google Scholar]
- 49.Leung J, Liu Y, ed. Population density-dependent regulation of E. coli beta-glucuronidase gene expression. Abstract S1691. Digestive Disease Week; 2003.
- 50.Wallace BD, Roberts AB, Pollet RM, et al. Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol. 2015;22(9):1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumura I, Ellington AD. In vitro evolution of beta-glucuronidase into a beta-galactosidase proceeds through non-specific intermediates. J Mol Biol. 2001;305(2):331–339. [DOI] [PubMed] [Google Scholar]
- 52.Van Eldere JR, De Pauw G, Eyssen HJ. Steroid sulfatase activity in a Peptococcus niger strain from the human intestinal microflora. Appl Environ Microbiol. 1987;53(7):1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hobkirk R. Steroid sulfation Current concepts. Trends Endocrinol Metab. 1993;4(2):69–74. [DOI] [PubMed] [Google Scholar]
- 54.Flores R, Shi J, Gail MH, Ravel J, Goedert JJ. Assessment of the human faecal microbiota: I. Measurement and reproducibility of selected enzymatic activities. Eur J Clin Invest. 2012;42(8):848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flores R, Shi J, Gail MH, Gajer P, Ravel J, Goedert JJ. Assessment of the human faecal microbiota: II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42(8):855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombardi P, Goldin B, Boutin E, Gorbach SL. Metabolism of androgens and estrogens by human fecal microorganisms. J Steroid Biochem. 1978;9(8):795–801. [DOI] [PubMed] [Google Scholar]
- 57.Jarvenpaa P, Kosunen T, Fotsis T, Adlercreutz H. In vitro metabolism of estrogens by isolated intestinal micro-organisms and by human faecal microflora. J Steroid Biochem. 1980;13(3):345–349. [DOI] [PubMed] [Google Scholar]
- 58.Kim DH, Kang HJ, Park SH, Kobashi K. Characterization of beta-glucosidase and beta-glucuronidase of alkalotolerant intestinal bacteria. Biol Pharm Bull. 1994;17(3):423–426. [DOI] [PubMed] [Google Scholar]
- 59.Thornton JR. High colonic pH promotes colorectal cancer. Lancet. 1981;1(8229):1081–1083. [DOI] [PubMed] [Google Scholar]
- 60.Holder G, Makin HLJ, Bradlow HL. The measurement of estrogens In. Steroid Analysis, 2nd ed London: Springer; 2010; 605–742. [Google Scholar]
- 61.Berg JM, Tymoczko JL, Stryer L. Chapter 26 (Section 26.4): Important derivatives of cholesterol include bile salts and steroid hormones Biochemistry, 5th ed. New York: WH Freeman; 2002. [Google Scholar]
- 62.Kisiela M, Skarka A, Ebert B, Maser E. Hydroxysteroid dehydrogenases (HSDs) in bacteria: a bioinformatic perspective. J Steroid Biochem Mol Biol. 2012;129(1-2):31–46. [DOI] [PubMed] [Google Scholar]
- 63.Eriksson H, Gustafsson JA, Sjovall J. Steroids in germfree and conventional rats. Free steroids in faeces from conventional rats. Eur J Biochem. 1969;9(2):286–290. [DOI] [PubMed] [Google Scholar]
- 64.Shimizu K, Muranaka Y, Fujimura R, Ishida H, Tazume S, Shimamura T. Normalization of reproductive function in germfree mice following bacterial contamination. Exp Anim. 1998;47(3):151–158. [DOI] [PubMed] [Google Scholar]
- 65.Fuhrman BJ, Feigelson HS, Flores R, et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab. 2014;99(12):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu X, Keefer LK, Ziegler RG, Veenstra TD. A liquid chromatography-mass spectrometry method for the quantitative analysis of urinary endogenous estrogen metabolites. Nat Protoc. 2007;2(6):1350–1355. [DOI] [PubMed] [Google Scholar]
- 67.Ziegler RG, Faupel-Badger JM, Sue LY, et al. A new approach to measuring estrogen exposure and metabolism in epidemiologic studies. J Steroid Biochem Mol Biol. 2010;121(3-5):538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flores R, Shi J, Fuhrman B, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. [DOI] [PubMed] [Google Scholar]
- 70.Kohler BA, Sherman RL, Howlader N, et al. Annual Report to the Nation on the Status of Cancer, 1975-2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J Natl Cancer Inst. 2015;107(6):djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson WF, Katki HA, Rosenberg PS. Incidence of breast cancer in the United States: current and future trends. J Natl Cancer Inst. 2011;103(18):1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenberg PS, Barker KA, Anderson WF. Estrogen Receptor Status and the Future Burden of Invasive and In Situ Breast Cancers in the United States. J Natl Cancer Inst. 2015;107(9):djv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Russo J, Russo IH. The role of estrogen in the initiation of breast cancer. J Steroid Biochem Mol Biol. 2006;102(1-5):89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furth PA, Cabrera MC, Diaz-Cruz ES, Millman S, Nakles RE. Assessing estrogen signaling aberrations in breast cancer risk using genetically engineered mouse models. Ann N Y Acad Sci. 2011;1229:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trichopoulos D. Hypothesis: does breast cancer originate in utero? Lancet. 1990;335(8695):939–940. [DOI] [PubMed] [Google Scholar]
- 76.Cerhan JR, Kushi LH, Olson JE, et al. Twinship and risk of postmenopausal breast cancer. J Natl Cancer Inst. 2000;92(3):261–265. [DOI] [PubMed] [Google Scholar]
- 77.Lagiou P. Intrauterine exposures, pregnancy estrogens and breast cancer risk: where do we currently stand? Breast Cancer Res. 2006;8(6):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hankinson SE, Willett WC, Manson JE, et al. Plasma sex steroid hormone levels and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 1998;90(17):1292–1299. [DOI] [PubMed] [Google Scholar]
- 79.Key T, Appleby P, Barnes I, Reeves G, Endogenous H, Breast Cancer Collaborative G. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94(8):606–616. [DOI] [PubMed] [Google Scholar]
- 80.Falk RT, Brinton LA, Dorgan JF, et al. Relationship of serum estrogens and estrogen metabolites to postmenopausal breast cancer risk: a nested case-control study. Breast Cancer Res. 2013;15(2):R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta M, McDougal A, Safe S. Estrogenic and antiestrogenic activities of 16alpha- and 2-hydroxy metabolites of 17beta-estradiol in MCF-7 and T47D human breast cancer cells. J Steroid Biochem Mol Biol. 1998;67(5-6):413–419. [DOI] [PubMed] [Google Scholar]
- 82.Kabat GC, Chang CJ, Sparano JA, et al. Urinary estrogen metabolites and breast cancer: a case-control study. Cancer Epidemiol Biomarkers Prev. 1997;6(7):505–509. [PubMed] [Google Scholar]
- 83.Muti P, Bradlow HL, Micheli A, et al. Estrogen metabolism and risk of breast cancer: a prospective study of the 2:16alpha-hydroxyestrone ratio in premenopausal and postmenopausal women. Epidemiology. 2000;11(6):635–640. [DOI] [PubMed] [Google Scholar]
- 84.Cavalieri E, Chakravarti D, Guttenplan J, et al. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim Biophys Acta. 2006;1766(1):63–78. [DOI] [PubMed] [Google Scholar]
- 85.Chlebowski RT, Rohan TE, Manson JE, et al. Breast Cancer After Use of Estrogen Plus Progestin and Estrogen Alone: Analyses of Data From 2 Women's Health Initiative Randomized Clinical Trials. JAMA Oncol. 2015;1(3):296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hullar MA, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer Treat Res. 2014;159:377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124(10):4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin F, Peltonen J, Laatikainen T, Pulkkinen M, Adlercreutz H. Excretion of progesterone metabolites and estriol in faeces from pregnant women during ampicillin administration. J Steroid Biochem. 1975;6(9):1339–1346. [DOI] [PubMed] [Google Scholar]
- 90.Adlercreutz H, Martin F, Pulkkinen M, et al. Intestinal metabolism of estrogens. J Clin Endocrinol Metab. 1976;43(3):497–505. [DOI] [PubMed] [Google Scholar]
- 91.Adlercreutz H, Martin F, Tikkanen MJ, Pulkkinen M. Effect of ampicillin administration on the excretion of twelve oestrogens in pregnancy urine. Acta Endocrinol (Copenh) . 1975;80(3):551–557. [DOI] [PubMed] [Google Scholar]
- 92.Hamalainen E, Korpela JT, Adlercreutz H. Effect of oxytetracycline administration on intestinal metabolism of oestrogens and on plasma sex hormones in healthy men. Gut. 1987;28(4):439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goldin BR, Gorbach SL. Alterations of the intestinal microflora by diet, oral antibiotics, and Lactobacillus: decreased production of free amines from aromatic nitro compounds, azo dyes, and glucuronides. J Natl Cancer Inst. 1984;73(3):689–695. [PubMed] [Google Scholar]
- 94.Velicer CM, Heckbert SR, Lampe JW, Potter JD, Robertson CA, Taplin SH. Antibiotic use in relation to the risk of breast cancer. JAMA. 2004;291(7):827–835. [DOI] [PubMed] [Google Scholar]
- 95.Knekt P, Adlercreutz H, Rissanen H, Aromaa A, Teppo L, Heliovaara M. Does antibacterial treatment for urinary tract infection contribute to the risk of breast cancer? Br J Cancer. 2000;82(5):1107–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sorensen HT, Skriver MV, Friis S, McLaughlin JK, Blot WJ, Baron JA. Use of antibiotics and risk of breast cancer: a population-based case-control study. Br J Cancer. 2005;92(3):594–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garcia Rodriguez LA, Gonzalez-Perez A. Use of antibiotics and risk of breast cancer. Am J Epidemiol. 2005;161(7):616–619. [DOI] [PubMed] [Google Scholar]
- 98.Friedman GD, Oestreicher N, Chan J, Quesenberry CP, Jr, Udaltsova N, Habel LA. Antibiotics and risk of breast cancer: up to 9 years of follow-up of 2.1 million women. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2102–2106. [DOI] [PubMed] [Google Scholar]
- 99.Setiadi AF, Ray NC, Kohrt HE, et al. Quantitative, architectural analysis of immune cell subsets in tumor-draining lymph nodes from breast cancer patients and healthy lymph nodes. PLoS One. 2010;5(8):e12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boursi B, Mamtani R, Haynes K, Yang YX. Recurrent antibiotic exposure may promote cancer formation - Another step in understanding the role of the human microbiota? Eur J Cancer. 2015;51(17):2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hicks LA, Taylor TH, Jr, Hunkler RJ. U.S. outpatient antibiotic prescribing, 2010. N Engl J Med. 2013;368(15):1461–1462. [DOI] [PubMed] [Google Scholar]
- 102.Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158(4):705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Keum N, Greenwood DC, Lee DH, et al. Adult weight gain and adiposity-related cancers: a dose-response meta-analysis of prospective observational studies. J Natl Cancer Inst. 2015;107(2):djv088. [DOI] [PubMed] [Google Scholar]
- 104.Arnold M, Pandeya N, Byrnes G, et al. Global burden of cancer attributable to high body-mass index in 2012: a population-based study. Lancet Oncol. 2015;16(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stevens GA, Singh GM, Lu Y, et al. National, regional, and global trends in adult overweight and obesity prevalences. Popul Health Metr. 2012;10(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iyengar NM, Hudis CA, Dannenberg AJ. Obesity and cancer: local and systemic mechanisms. Annu Rev Med. 2015;66:297–309. [DOI] [PubMed] [Google Scholar]
- 107.Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goldin BR, Adlercreutz H, Gorbach SL, et al. Estrogen excretion patterns and plasma levels in vegetarian and omnivorous women. N Engl J Med. 1982;307(25):1542–1547. [DOI] [PubMed] [Google Scholar]
- 109.Gorbach SL, Goldin BR. Diet and the excretion and enterohepatic cycling of estrogens. Prev Med. 1987;16(4):525–531. [DOI] [PubMed] [Google Scholar]
- 110.Reddy BS, Wynder EL. Large-bowel carcinogenesis: fecal constituents of populations with diverse incidence rates of colon cancer. J Natl Cancer Inst. 1973;50(6):1437–1442. [DOI] [PubMed] [Google Scholar]
- 111.Vogt SL, Pena-Diaz J, Finlay BB. Chemical communication in the gut: Effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe. 2015;34:106–115. [DOI] [PubMed] [Google Scholar]
- 112.Boyd NF, McGuire V. Evidence of association between plasma high-density lipoprotein cholesterol and risk factors for breast cancer. J Natl Cancer Inst. 1990;82(6):460–468. [DOI] [PubMed] [Google Scholar]
- 113.Wu Q, Ishikawa T, Sirianni R, et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 2013;5(3):637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nelson ER, Wardell SE, Jasper JS, et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342(6162):1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Warner M, Gustafsson JA. On estrogen, cholesterol metabolism, and breast cancer. N Engl J Med. 2014;370(6):572–573. [DOI] [PubMed] [Google Scholar]
- 116.Willett WC, Stampfer MJ, Colditz GA, Rosner BA, Hennekens CH, Speizer FE. Moderate alcohol consumption and the risk of breast cancer. N Engl J Med. 1987;316(19):1174–1180. [DOI] [PubMed] [Google Scholar]
- 117.Lew JQ, Freedman ND, Leitzmann MF, et al. Alcohol and risk of breast cancer by histologic type and hormone receptor status in postmenopausal women: the NIH-AARP Diet and Health Study. Am J Epidemiol. 2009;170(3):308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li CI, Chlebowski RT, Freiberg M, et al. Alcohol consumption and risk of postmenopausal breast cancer by subtype: the women's health initiative observational study. J Natl Cancer Inst. 2010;102(18):1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deandrea S, Talamini R, Foschi R, et al. Alcohol and breast cancer risk defined by estrogen and progesterone receptor status: a case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17(8):2025–2028. [DOI] [PubMed] [Google Scholar]
- 120.Suzuki R, Orsini N, Mignone L, Saji S, Wolk A. Alcohol intake and risk of breast cancer defined by estrogen and progesterone receptor status–a meta-analysis of epidemiological studies. Int J Cancer. 2008;122(8):1832–1841. [DOI] [PubMed] [Google Scholar]
- 121.Frydenberg H, Flote VG, Larsson IM, et al. Alcohol consumption, endogenous estrogen and mammographic density among premenopausal women. Breast Cancer Res. 2015;17:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kwan ML, Kushi LH, Weltzien E, et al. Alcohol consumption and breast cancer recurrence and survival among women with early-stage breast cancer: the life after cancer epidemiology study. J Clin Oncol. 2010;28(29):4410–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dorgan JF, Baer DJ, Albert PS, et al. Serum hormones and the alcohol-breast cancer association in postmenopausal women. J Natl Cancer Inst. 2001;93(9):710–715. [DOI] [PubMed] [Google Scholar]
- 124.Hankinson SE, Willett WC, Manson JE, et al. Alcohol, height, and adiposity in relation to estrogen and prolactin levels in postmenopausal women. J Natl Cancer Inst. 1995;87(17):1297–1302. [DOI] [PubMed] [Google Scholar]
- 125.Muti P, Trevisan M, Micheli A, et al. Alcohol consumption and total estradiol in premenopausal women. Cancer Epidemiol Biomarkers Prev. 1998;7(3):189–193. [PubMed] [Google Scholar]
- 126.Reichman ME, Judd JT, Longcope C, et al. Effects of alcohol consumption on plasma and urinary hormone concentrations in premenopausal women. J Natl Cancer Inst. 1993;85(9):722–727. [DOI] [PubMed] [Google Scholar]
- 127.Seitz HK, Maurer B. The relationship between alcohol metabolism, estrogen levels, and breast cancer risk. Alcohol Res Health. 2007;30(1):42–43. [PubMed] [Google Scholar]
- 128.Ginsburg ES, Mello NK, Mendelson JH, et al. Effects of alcohol ingestion on estrogens in postmenopausal women. JAMA. 1996;276(21):1747–1751. [DOI] [PubMed] [Google Scholar]
- 129.Fan S, Meng Q, Gao B, et al. Alcohol stimulates estrogen receptor signaling in human breast cancer cell lines. Cancer Res. 2000;60(20):5635–5639. [PubMed] [Google Scholar]
- 130.Singletary KW, Frey RS, Yan W. Effect of ethanol on proliferation and estrogen receptor-alpha expression in human breast cancer cells. Cancer Lett. 2001;165(2):131–137. [DOI] [PubMed] [Google Scholar]
- 131.Chen P, Schnabl B. Host-microbiome interactions in alcoholic liver disease. Gut Liver. 2014;8(3):237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bode JC, Bode C, Heidelbach R, Durr HK, Martini GA. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology. 1984;31(1):30–34. [PubMed] [Google Scholar]
- 133.Casafont Morencos F, de las Heras Castano G, Martin Ramos L, Lopez Arias MJ, Ledesma F, Pons Romero F. Small bowel bacterial overgrowth in patients with alcoholic cirrhosis. Dig Dis Sci. 1996;41(3):552–556. [DOI] [PubMed] [Google Scholar]
- 134.Mutlu EA, Gillevet PM, Rangwala H, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G966–G978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mutlu E, Keshavarzian A, Engen P, Forsyth CB, Sikaroodi M, Gillevet P. Intestinal dysbiosis: a possible mechanism of alcohol-induced endotoxemia and alcoholic steatohepatitis in rats. Alcohol Clin Exp Res. 2009;33(10):1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xie G, Zhong W, Zheng X, et al. Chronic ethanol consumption alters mammalian gastrointestinal content metabolites. J Proteome Res. 2013;12(7):3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu BT, Bui QD, Weisz J, Liehr JG. Conversion of estrone to 2- and 4-hydroxyestrone by hamster kidney and liver microsomes: implications for the mechanism of estrogen-induced carcinogenesis. Endocrinology. 1994;135(5):1772–1779. [DOI] [PubMed] [Google Scholar]
- 138.de Moreno de Leblanc A, Perdigon G. The application of probiotic fermented milks in cancer and intestinal inflammation. Proc Nutr Soc. 2010;69(3):421–428. [DOI] [PubMed] [Google Scholar]
- 139.Goldin BR, Gorbach SL. The effect of milk and lactobacillus feeding on human intestinal bacterial enzyme activity. Am J Clin Nutr. 1984;39(5):756–761. [DOI] [PubMed] [Google Scholar]
- 140.Goldin BR, Swenson L, Dwyer J, Sexton M, Gorbach SL. Effect of diet and Lactobacillus acidophilus supplements on human fecal bacterial enzymes. J Natl Cancer Inst. 1980;64(2):255–261. [DOI] [PubMed] [Google Scholar]
- 141.van't Veer P, Dekker JM, Lamers JW, et al. Consumption of fermented milk products and breast cancer: a case-control study in The Netherlands. Cancer Res. 1989;49(14):4020–4023. [PubMed] [Google Scholar]
- 142.Le MG, Moulton LH, Hill C, Kramar A. Consumption of dairy produce and alcohol in a case-control study of breast cancer. J Natl Cancer Inst. 1986;77(3):633–636. [DOI] [PubMed] [Google Scholar]
- 143.Raju U, Levitz M, Javitt NB. Bile acids in human breast cyst fluid: the identification of lithocholic acid. J Clin Endocrinol Metab. 1990;70(4):1030–1034. [DOI] [PubMed] [Google Scholar]
- 144.Javitt NB, Budai K, Miller DG, Cahan AC, Raju U, Levitz M. Breast-gut connection: origin of chenodeoxycholic acid in breast cyst fluid. Lancet. 1994;343(8898):633–635. [DOI] [PubMed] [Google Scholar]
- 145.Swales KE, Korbonits M, Carpenter R, Walsh DT, Warner TD, Bishop-Bailey D. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer Res. 2006;66(20):10120–10126. [DOI] [PubMed] [Google Scholar]
- 146.Xuan C, Shamonki JM, Chung A, et al. Microbial dysbiosis is associated with human breast cancer. PLoS One. 2014;9(1):e83744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Urbaniak C, Cummins J, Brackstone M, et al. Microbiota of human breast tissue. Appl Environ Microbiol. 2014;80(10):3007–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Banerjee S, Wei Z, Tan F, et al. Distinct microbiological signatures associated with triple negative breast cancer. Sci Rep. 2015;5:15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.International Human Microbiome Standards (IHMS) Project. http://www.microbiome-standards.org. Accessed December 27, 2015.
- 151.Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Flores R, Shi J, Yu G, et al. Collection media and delayed freezing effects on microbial composition of human stool. Microbiome. 2015;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Santiago A, Panda S, Mengels G, et al. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 2014;14:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu X, Roman JM, Issaq HJ, Keefer LK, Veenstra TD, Ziegler RG. Quantitative measurement of endogenous estrogens and estrogen metabolites in human serum by liquid chromatography-tandem mass spectrometry. Anal Chem. 2007;79(20):7813–7821. [DOI] [PubMed] [Google Scholar]
- 155.Goedert JJ, Jones G, Hua X, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst. 2015;107(8):djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shahanavaj K, Gil-Bazo I, Castiglia M, et al. Cancer and the microbiome: potential applications as new tumor biomarker. Expert Rev Anticancer Ther. 2015;15(3):317–330. [DOI] [PubMed] [Google Scholar]
- 157.Markowitz VM, Chen IM, Palaniappan K, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.